Abstract
Purpose
As a powerful technology for genome engineering, the CRISPR/Cas system has been successfully applied to modify the genomes of various species. The purpose of this study was to evaluate the technology and establish principles for the introduction of precise genetic modifications in early human embryos.
Methods
3PN zygotes were injected with Cas9 messenger RNA (mRNA) (100 ng/μl) and guide RNA (gRNA) (50 ng/μl). For oligo-injections, donor oligo-1 (99 bp) or oligo-2 (99 bp) (100 ng/μl) or dsDonor (1 kb) was mixed with Cas9 mRNA (100 ng/μl) and gRNA (50 ng/μl) and injected into the embryos.
Results
By co-injecting Cas9 mRNA, gRNAs, and donor DNA, we successfully introduced the naturally occurring CCR5Δ32 allele into early human 3PN embryos. In the embryos containing the engineered CCR5Δ32 allele, however, the other alleles at the same locus could not be fully controlled because they either remained wild type or contained indel mutations.
Conclusions
This work has implications for the development of therapeutic treatments of genetic disorders, and it demonstrates that significant technical issues remain to be addressed. We advocate preventing any application of genome editing on the human germline until after a rigorous and thorough evaluation and discussion are undertaken by the global research and ethics communities.
Electronic supplementary material
The online version of this article (doi:10.1007/s10815-016-0710-8) contains supplementary material, which is available to authorized users.
Keywords: CRISPR/Cas9, Genetic modification, CCR5, Human 3PN embryos
Introduction
Sequencing the genomes of numerous species, including humans, has uncovered the genetic blueprints encoded in DNA that direct the development and progression of organisms on Earth, which are constantly challenged by natural selection [1, 2]. The ability to make random or targeted modifications to genomic sequences allows us to study the functions of genetic components and engineer organisms for research and practical applications. The recent development of site-specific DNA endonuclease technologies, such as zinc finger nuclease (ZFN) [3], transcription activator-like effector nuclease (TALEN) [4], and the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated (Cas) nuclease system (CRISPR/Cas) [5], has greatly accelerated genome engineering applications and broadened the spectrum of species that is applicable to such methods. When these systems bind to a target DNA sequence in the genome, they create a DNA double strand break (DSB), the repair of which leads to specific DNA sequence modifications.
By introducing the CRISPR/Cas system into a zygote, methods have been established for the one-step generation of mice carrying mutations in single and multiple genes and of mice carrying reporter and conditional alleles [6, 7]. Using a similar approach, two groups have demonstrated that precise genetic modifications could be achieved in early mouse embryos to correct disease-causing mutations, leading to the production of healthy adult animals [8, 9]. These studies highlighted that the correction of genetic mutations in germline cells or early embryos could serve as a permanent solution for genetic diseases. Moreover, Niu et al. and Liu et al. recently showed that the injection of zygotes with CRISPR/Cas and TALEN systems could generate targeted genetic mutations in non-human primates [10, 11]. More recently, Liang et al. reported that the CRISPR/Cas9 system can mediate genetic modifications in human tripronuclear zygotes [12]. In the present study, we successfully introduced the naturally occurring CCR5Δ32 allele into early human tripronuclear (3PN) embryos by CRISPR/Cas-mediated genome editing. However, the efficiency of homology directed repair (HDR) of the CCR5Δ32 allele was low, and the edited embryos were mosaic. Our study serves two purposes: first, to evaluate technologies and establish principles for the introduction of precise genetic modifications in early human embryos and to identify any significant technical hurdles; and, second, to raise awareness of the potential legal and ethical challenges that our society will face from the increasing accessibility of genome engineering technologies.
Materials and methods
Source of tripronuclear human zygotes
From April 2014 to September 2014, a total of 213 3PN human zygotes from 87 patients with a visible second polar body were obtained 18–20 h post-insemination from the Third Affiliated Hospital of Guangzhou Medical University.
Tripronuclear human zygote injection
The 3PN zygotes were transferred into HTF medium (Quinn’s Advantage™ Medium with HEPES). They were injected with Cas9 messenger RNA (mRNA) (100 ng/μl) and guide RNA (gRNA) (50 ng/μl). For oligo injections, donor oligo-1 (90 bp) or oligo-2 (90 bp) (100 ng/μl) or dsDonor (1 kb) (100 ng/μl) was mixed with Cas9 mRNA (100 ng/μl) and gRNA (50 ng/μl) and injected into the embryos. The injected embryos were cultured in G1.5 media until day three. All embryos were maintained in a humidified incubator at 37 °C, 5 % CO2, and 20 % oxygen.
Production of Cas9 mRNA, gRNAs, and DNA donors
In vitro transcription (IVT) of Cas9 mRNA was performed as previously described [13]. Briefly, T7 promoter was added to Cas9 coding region by PCR amplification, using px330 vector [14] as template. The PCR product was gel-purified and used as a template for IVT using mMESSAGE mMACHINE T7 ULTRA kit (Life Technologies). For gRNA synthesis, the IVT templates were generated by PCR amplification using primers listed in Table S1. The T7-gRNA PCR product was purified and used as the template for IVT using a MEGAshortscript T7 kit (Life Technologies). Both the Cas9 mRNA and the gRNAs were purified using a MEGAclear kit (Life Technologies). All single-stranded oligos were ordered from Invitrogen. The 1 kb dsDNA donor was ordered as gBlock from Integrated DNA Technologies.
Whole-genome amplification
Whole-genome amplification (WGA) was performed using the REPLI-g Single Cell Kit (no. 150345, Qiagen) based on MDA technologies according to the manufacturers’ recommended protocols. Following completion of the WGA reaction, the products were purified using the HP PCR Product Purification Kit (no. 11732668001, Roche). Purified DNA (1 μl) was run on agarose gels to confirm successful amplification, and the remaining material was kept at −20 °C until processing for PCR.
DNA sequencing analysis
For embryos injected with Cas9 mRNA and gRNAs, with or without single-stranded oligodeoxynucleotides (ssODNs), the 324 bp region flanking the CCR5 target site was amplified by PCR with specific primers (CCR5 geno F and R) from the WGA product of each embryo. For embryos injected with Cas9 mRNA, gRNAs, and 1 kb dsDNA donor, the target region was amplified using primer outside the 1 kb donor sequence. First, the PCR products were sequenced to identify the presence of mutation peaks in the sequencing chromatography. Subsequently, the PCR products from each embryo were cloned into a pMD18T TA cloning vector (Takara) and transformed into competent Escherichia coli cells. The DNA plasmids from 10 to 50 randomly picked colonies were sequenced using M13 universal primers. The sequences of all the primers used are listed in Table S2.
Off-target analysis
Potential off-targets were predicted by searching the human genome (hg19) for matches to the 20 bp CCR5 gRNA1 and gRNA2 sequences, allowing for up to four mismatches followed by NGG PAM sequence. Matches were ranked first by the ascending number of mismatches and then by the ascending distance from the PAM sequence. According to the output of the scan, potential off-targets with less than four mismatches in the seed sequence were selected for PCR amplification using diluted WGA DNA as templates. The PCR products of the candidates were sequenced, and the primers for amplifying the candidate sites are listed in Table S1.
Results
Study design
To address the ethical and public concerns regarding how the CRISPR/Cas9 system could be used to engineer viable human embryos with the intention to produce a living human, we limited our study to using 3PN embryos. The 3PN embryos used in this experiment do not live beyond 3 days and are incapable of in vivo development. As one major type of polyspermy, 3PN zygotes with three pronuclei (often two sperm nuclei and one oocyte nucleus) are commonly found during the in vitro fertilization process. Although polyspermy is considered invariably pathological and the early embryo either fails to develop or develops abnormally [15, 16], 3PN zygotes are capable of substantial in vitro development [17] and therefore serve as a good system for investigating the general principles of genome engineering in early human embryos.
We chose the C–C chemokine receptor type 5 (CCR5) gene, which encodes the major co-receptor used by HIV-1 to infect human cells, as an exemplary target for our studies. The null CCR5Δ32 allele is present at high frequencies in Scandinavian and Northern European populations and is absent in non-European populations [18]. Individuals who are heterozygous or homozygous for this allele are healthy and have a slower progression or resistance to HIV infections [19, 20]. Multiple studies have demonstrated that site-specific nuclease-mediated disruption of CCR5 function occurred in CD34+ hematopoietic stem/progenitor cells (HSPCs) and CD4+ T cells [21–23] and in both human embryonic stem cells and iPS cells [24–26]. We wanted to test whether we could introduce this naturally occurring beneficial CCR5Δ32 allele into early human 3PN embryos.
Generation of CCR5 mutant alleles with high efficiency
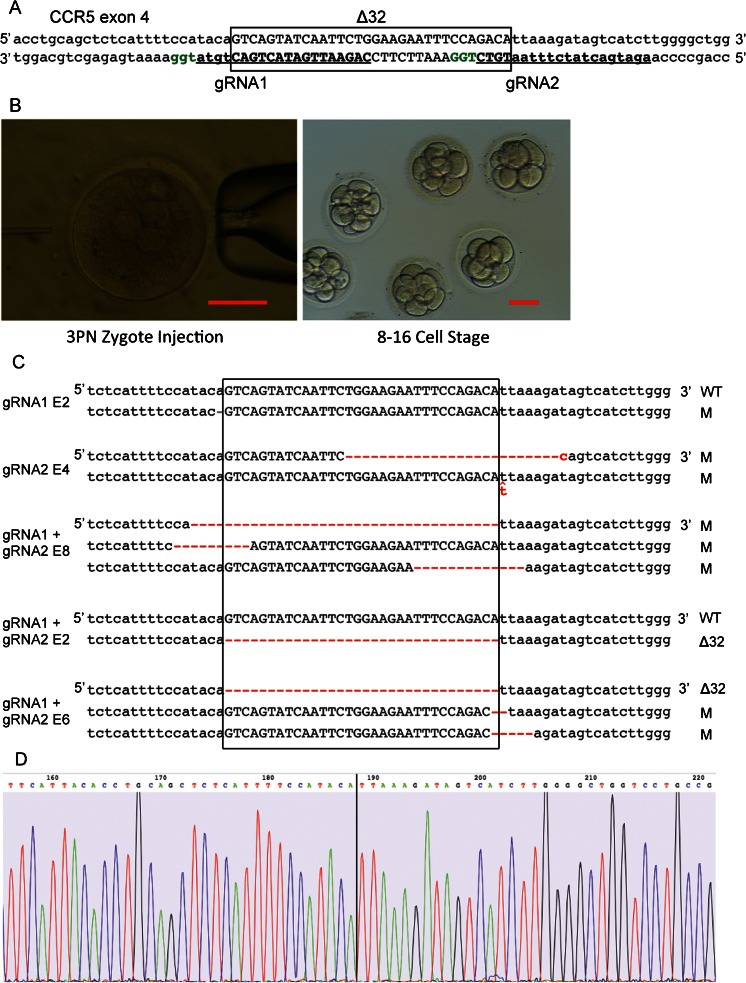
We designed two gRNAs, each targeting at one end of the boundary of the desired 32 bp deletion within CCR5 exon 4 (Fig. 1a). When the CRISPR/Cas-mediated DSBs are repaired through the non-homologous end-joining (NHEJ) pathway, small insertions or deletions (indels) can occur and lead to CCR5 mutant alleles.
Fig. 1.
Gene editing at CCR5 locus in human 3PN embryos. a Schematic of the targeted CCR5 locus. The 32 bp sequence deleted in natural occurring CCR5Δ32 allele is capitalized and marked by black box. The two gRNAs are bold and underlined, and their PAM regions are labeled in green. b Pictures showing the injection of a human 3PN zygote (left panel) and the development to 8–16-cell stage in vitro (right panel). Scale bar = 100 μm. c Schematic of all the alleles identified in each mutant embryos. The gRNAs injected in each embryo are labeled at the left side, and the mutations are labeled in red. d Sequence chromatography of the CCR5Δ32 allele from the targeted human 3PN embryo. The black line indicates the position of 32 bp deletion
First, we sought to evaluate the in vitro developmental potential of 3PN zygotes injected with the CRISPR/Cas system. Cas9 mRNA and gRNA1 or gRNA2 were delivered into 3PN zygotes by cytoplasmic injection. Injected zygotes were cultured in G1.5 media (Vitrolife Sweden AB) for 3 days to reach an 8–16-cell stage (Fig. 1b). Approximately 72 % of the 3PN embryos injected with water (control group) developed into an 8–16-cell stage. In comparison, 64 and 62 % of the zygotes injected with Cas9 mRNA, in combination with gRNA1 and gRNA2, respectively, developed to an 8–16-cell stage, suggesting a minimally detrimental effect.
Each injected embryo that developed to 8-cell stage and beyond was collected independently, and the genomic DNA was amplified using a REPLI-g Single Cell Kit (Qiagen). The targeted region was amplified by PCR and characterized by Sanger sequencing. The PCR products from identified mutant samples were subcloned into plasmids, and each allele was identified by sequencing (Fig. 1c). The efficiency of genetic modification was above 50 % using either gRNA1 or gRNA2, indicating that potentially beneficial genetic modifications could be introduced into early human embryos with high efficiency (Table 1). Because all of the 3PN embryo donors were from non-European populations, we observed a complete absence of the naturally occurring CCR5Δ32 allele in both the control embryos and the injected embryos, as expected.
Table 1.
CRISPR/Cas9-mediated gene editing in human 3PN embryos
| Groupsa | Injected 3PN zygotes | Development (%) | Genetic modification | ||
|---|---|---|---|---|---|
| Cleavage | 8–16 cell | NHEJ mutations (percent) | Δ32 allele (%) | ||
| Control water | 18 | 15 (83) | 13 (72) | 0 | 0 |
| Cas9 + gRNA1 | 11 | 9 (82) | 7 (64) | 4 (57) | 0 |
| Cas9 + gRNA2 | 13 | 10 (77) | 8 (62) | 5 (63) | 0 |
| Cas9 + gRNA1 + ssODN1 (PN injection) | 23 | 14 (61) | 11 (48) | 4 (36) | 0 |
| Cas9 + gRNA1 + ssODN1 | 32 | 28 (88) | 20 (63) | 10 (50) | 1 (5) |
| Cas9 + gRNA2 + ssODN2 | 46 | 39 (85) | 27 (59) | 13 (48) | 0 |
| Cas9 + gRNA1 + 1 kb dsDonor | 25 | 21 (84) | 15 (60) | 7 (47) | 1 (7) |
| Cas9 + gRNA1 + gRNA2 | 45 | 37 (82) | 26 (58) | 13 (50) | 4 (15) |
aCRISPR/Cas system was delivered to 3PN zygotes by cytoplasmic injection in all groups, except the group labeled with PN (pronuclear) injection
Introduction of CCR5Δ32 allele
Next, we tested whether we could introduce the Δ32 allele precisely at the CCR5 locus through the HDR pathway by using ssODNs as donors. Two 90-nt-long ssODNs were designed from the sense strand (ssODN1) and the anti-sense strand (ssODN2) of the CCR5 locus, each containing 45 nt sequences homologous to the sequence from each side of the 32 nt intended deletion (Table S2). We delivered the Cas9 mRNA, gRNA1, and ssODN1 using both cytoplasmic and pronuclear injection methods. Cytoplasmic injection performed better both for embryo development and targeted mutation efficiency (Table 1), although the difference was not statistically significant due to the small sample size. We were able to produce the CCR5Δ32 allele in one of the 20 total embryos by cytoplasmic injection (Fig. 1d, Table 1). We sought to improve this efficiency by providing a double stranded donor with 1 kb total homology but did not see significant improvement (Table 1).
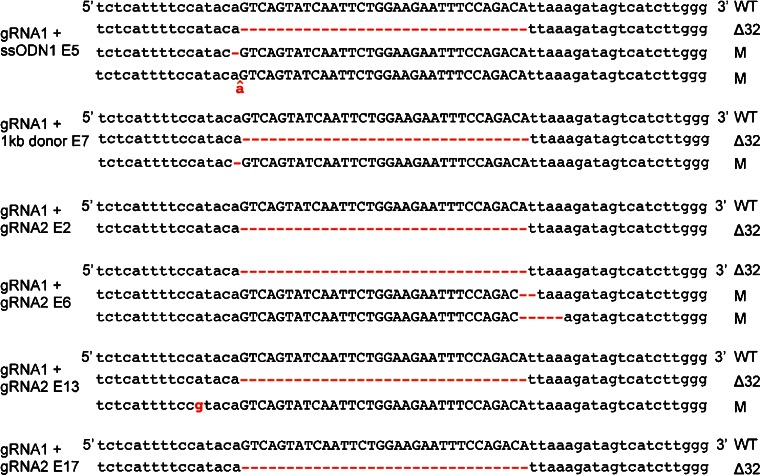
Because NHEJ repair is usually more efficient and because two DSBs can be precisely joined through the NHEJ pathway, we co-injected Cas9 mRNA along with both gRNAs targeting the boundaries of the 32 bp region without providing any donor DNA template. Using this strategy, we were able to generate embryos containing the CCR5Δ32 allele in four of the 26 total samples (Tables 1 and 2). All of the alleles identified from the embryos containing the CCR5Δ32 allele are shown in Fig. 2, and the PCR subcloning results are summarized in Tables 2 and Fig. S1. In the embryos containing the CCR5Δ32 allele, the other alleles at the same locus either remained wild type or contained different indel mutations that were not predefined.
Table 2.
Subcloning of the PCR products from human 3PN zygotes containing CCR5Δ32 allele
| Experiment | WT allele/total (%) | Other mutation/total (%) | Δ32 allele/total (%) |
|---|---|---|---|
| gRNA1 + ssODN1 E5 | 8/21 (38) | 8/21 (38) | 5/21 (24) |
| gRNA1 + 1 kb dsDonor E7 | 11/18 (61) | 4/18 (22) | 3/18 (17) |
| gRNA1 + gRNA2 E2 | 8/19 (42) | 0 | 11/19 (58) |
| gRNA1 + gRNA2 E6 | 0 | 6/25 (24) | 19/25 (76) |
| gRNA1 + gRNA2 E13 | 23/39 (59) | 1/39 (3) | 15/39 (38) |
| gRNA1 + gRNA2 E17 | 30/41 (73) | 0 | 11/41 (27) |
PCR products of the target region were amplified from injected embryos and subcloned into pMD18T TA cloning vector. Plasmid DNA from 18 to 41 E. coli colonies was prepared and sequenced
Fig. 2.
All alleles identified in human 3PN embryos containing CCR5Δ32 allele. Schematic of all the alleles identified in each mutant embryo. The 32 bp sequence deleted in natural occurring CCR5Δ32 allele is capitalized. The gRNAs and donor DNA injected in each embryo are labeled at the left side, and the mutations are labeled in red
Because the majority of 3PN embryos have triploid chromosome complements or mosaic arrangements [16], it is very difficult to assess the mosaicism arising from genome editing. However, the presence of three to four unique alleles in embryos suggests that the mosaicism of genetic modification could occur (Fig. 2) because mosaicism is well documented in similar studies in animal models [7, 27]. In addition, the number of alleles identified from each embryo might be an underestimation due to limitations in the PCR genotyping approach.
To address the potential off-target effects, we predicted the top 11 and 17 potential off-targets for gRNA1 and gRNA2, respectively, based on published findings [7, 28], and we genotyped all 28 loci in three embryos containing CCR5Δ32 allele (Table 2). None of these loci contained indel mutations at the predicted off-targeted regions (Table S2).
Discussion
In this study, we demonstrated that a naturally occurring beneficial allele could be introduced into early human 3PN embryos through zygote injection of the CRISPR/Cas system. By testing different strategies, we successfully introduced the naturally occurring CCR5Δ32 allele into early human 3PN embryos. Similar to the results obtained in other species, NHEJ-mediated indel mutations could be obtained with high efficiency, whereas the HDR-mediated precise modifications had a lower efficiency. Because of the scarcity of human embryos, we had a relatively small number of samples for each group. Therefore, the differences in development and mutation rates among groups are not statistically significant, and we cannot make any absolute conclusions. We would like to highlight the fact that even in the embryos in which we successfully introduced the CCR5Δ32 allele, the other alleles at the same locus either were wild type or contained indel mutations. Liang et al. showed that the efficiency of HDR of the β-globin gene was 4.7 % (per injected zygote) and that the modified embryos displayed mosaicism in which wild-type cells and genetically modified cells coexisted [12]. Our result is consistent with the study by Liang et al. Although mosaicism may be avoided by a more careful consideration of the timing and method of microinjection, Long et al. demonstrated that the degree of muscle phenotypic rescue in mosaic mice exceeded the efficiency of gene correction [9]. Therefore, significant technical hurdles will need to be overcome to gain control of the genotype of both alleles in a viable embryo.
Off-target effects are a major concern for the CRISPR/Cas system. Several studies have reported that the CRISPR/Cas system induced heritable off-target mutations in both mice and humans [13, 29]. In this study, we did not detect off-site targeting as predicted for a total of 28 potential off-target sites. Recent studies used various strategies to experimentally map off-target mutations of the CRISPR/Cas system and found that computational prediction based on sequence similarity to target sites failed to predict many off-target mutations [30–32]. Therefore, the embryos likely carried off-target mutations undetected by our genotyping, and the off-target effects thus merit further inquiry.
Liang et al. reported that off-target mutations were detected in modified 3PN embryos by the T7E1 assay and whole-exome sequencing [12]. However, considering that the off-target effect is site dependent and that more specific strategies using more sophisticated enzymes and meticulous design of the guiding molecule have already been established [33–35], off-target mutagenesis may be minimized by optimizing the procedure [36]. Furthermore, small molecules that enhance CRISPR-mediated HDR have also been reported [37], suggesting that the CRISPR/Cas system could be a reliable genome editing tool for humans. We intend to investigate the effects of these molecules on CRISPR-mediated HDR in human 3PN embryos in our future studies.
Because we used 3PN embryos from donors with no known genetic disorders, we chose to introduce this naturally occurring allele as a test case for generating a precise genetic modification. Compared with 2PN embryos, 3PN embryos only have one more sperm nucleus and are capable of substantial in vitro development. Thus, our results suggest that similar techniques could be applied to correct disease-causing mutations in human early embryos. It is likely that other programmable endonucleases can also be applied in a similar manner, although with potentially different efficiencies and specificities.
These results call for immediate attention being paid to the regulation of the application of genome engineering technologies in humans for genetic modification in early embryos and related work, such as the genetic modification of human germline cells. Because human in vitro fertilization methods are well established and site-specific nuclease technologies are readily available, it is foreseeable that a genetically modified human could be generated. We believe that any attempt to generate genetically modified humans through the modification of early embryos needs to be strictly prohibited until we can resolve both ethical and scientific issues. Several major scientific issues remain. First, for any germline genetic modification in humans, we believe that the resulting allele needs to be precisely predefined. This requires the further improvement of precise modification efficiency as well as assays to select the embryos containing the desired allele. Second, genetic mosaicism, likely due to nuclease action after the one-cell stage, needs to be resolved. Genome engineering of germline stem cells might be one potential solution [38]. Third, the specificity of the technologies needs to be further investigated and improved to ensure that no off-target mutations will be introduced. Fourth, for any introduced allele, even a naturally occurring allele in the human population, the effect of its introduction into a different genetic background needs to be carefully evaluated. Without a much better understanding of the genetic network at a systematic level, we cannot predict the genome-wide effects of changing even a single allele.
Genome engineering technologies for germline editing need to be significantly improved to solve scientific issues that include, but are not limited to, the ones presented above before they may be considered for application in humans. Preimplantation genetic diagnosis (PGD) has already been used clinically in some countries to screen out human embryos with mutations responsible for genetic conditions such as thalassemia and spinal muscular atrophy. The clinical use of PGD appears to justify germline genome editing research because only embryos that contain no suspected mutations, but have undergone the physical intervention of embryonic cell biopsy for genetic testing, are used for embryo transfer. Even with a perfected technology, we believe that genome engineering in the human germline should only be considered if no less invasive alternative approach is available.
A thorough discussion regarding the related ethical issues is beyond the scope of our study, and the topic needs to be addressed comprehensively by the broad international community. However, to start the discussion, we would like to raise several important questions. One of the most important questions is, “how do we define the norm of human and human genome?” With the rapid development of human genomics, a large number of human alleles associated with diseases and human physiology are being discovered. Once we understand the underlying biology, would it be acceptable to introduce alleles that improve human health and life span or to eliminate those imposing serious disease risks (e.g., Huntington’s disease)?
Despite the significant scientific and ethical issues involved, however, we believe that it is necessary to keep developing and improving the technologies for precise genetic modifications in humans by using animal models and in vitro systems such as human 3PN embryos and germline stem cells. The availability and improvement of genome engineering technologies in human early embryos, the proof-of-concept that we and Liang et al. [12] provide, may provide solutions for genetic diseases, improve human health, or play an essential role in surviving future disruptive selection pressures. Recently, a prominent ad hoc group of scientists called for a moratorium on clinical applications of germline gene editing [39]. We also advocate for preventing any application of genome editing in the human germline until after a rigorous and thorough evaluation and discussion are undertaken by the global research and ethics communities.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Oligonucleotides used for making in vitro transcription template, CCR5 genotyping and as HDR-mediated repair template. (PDF 177 kb)
Off-target analysis for CRISPR-Cas9–mediated targeting in human 3PN zygotes. (PDF 27 kb)
The sequences of the CCR5 gene in human 3PN embryos carrying CRISPR/Cas9-induced gene modifications (GIF 192 kb)
Acknowledgments
We thank Dr. Haoyi Wang from Institute of Zoology in Chinese Academy of Sciences for helpful advice. This work was supported in part by the National Natural Science Foundation of China (81370766, 81570101, 81370670), Guangdong Province Higher Education Funding (2013KJCX0149, Yq2013135), and Guangdong Province Science and Technology Project (2014A02011029, 2015B020227002). Y. F. was supported by the State 863 project 2015AA020307.
Authors’ contributions
Y.F. designed the study. Y.F., X.K., and Y.H. recruited the patients and signed the informed consent. Y.F. performed embryo injection. X.K. and Y.H. collected embryos. W.H. and Q.Y. performed the WGA and PCR genotyping and off-target prediction. Y.C., X.G., and X.S. contributed to off-target sites genotyping. Y.F. analyzed the data and wrote the paper.
Compliance with ethical standards
This study was approved by the ethics committee of the Third Affiliated Hospital of Guangzhou Medical University (LLSC2014018). The methods used in the present study closely followed the guidelines legislated and posted by the Ministry of Health of the People’s Republic of China. The patients involved in this study knew about and understood the usage of polyspermic zygotes and voluntarily donated them after providing informed consent.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Capsule As a powerful technology for genome engineering, the CRISPR/Cas system has been successfully applied to modify the genomes of various species.
Xiangjin Kang, Wenyin He and Yuling Huang contributed equally to this work.
Electronic supplementary material
The online version of this article (doi:10.1007/s10815-016-0710-8) contains supplementary material, which is available to authorized users.
An erratum to this article is available at http://dx.doi.org/10.1007/s10815-017-0946-y.
References
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291(5507):1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11(9):636–46. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 4.Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333(6051):1843–6. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- 5.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–78. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–8. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154(6):1370–9. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Y, Liang D, Wang Y, Bai M, Tang W, Bao S, et al. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell. 2013;13(6):659–62. doi: 10.1016/j.stem.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 9.Long C, McAnally JR, Shelton JM, Mireault AA, Bassel-Duby R, Olson EN. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science. 2014;345(6201):1184–8. doi: 10.1126/science.1254445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156(4):836–43. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 11.Liu H, Chen Y, Niu Y, Zhang K, Kang Y, Ge W, et al. TALEN-mediated gene mutagenesis in rhesus and cynomolgus monkeys. Cell Stem Cell. 2014;14:323–8. doi: 10.1016/j.stem.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang P, Xu Y, Zhang X, Ding C, Huang R, Zhang Z, et al. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell. 2015;6(5):363–72. doi: 10.1007/s13238-015-0153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu P, Chen S, Li X, Qin L, Huang K, Wang L et al. Low immunogenicity of neural progenitor cells differentiated from induced pluripotent stem cells derived from less immunogenic somatic cells. PLoS One. 8(7):e69617. [DOI] [PMC free article] [PubMed]
- 14.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 339(6121):819–23. [DOI] [PMC free article] [PubMed]
- 15.Kola I, Trounson A, Dawson G, Rogers P. Tripronuclear human oocytes: altered cleavage patterns and subsequent karyotypic analysis of embryos. Biol Reprod. 1987;37(2):395–401. doi: 10.1095/biolreprod37.2.395. [DOI] [PubMed] [Google Scholar]
- 16.Feenan K, Herbert M. Can “abnormally” fertilized zygotes give rise to viable embryos? Hum Fertil. 2006;9(3):157–69. doi: 10.1080/14647270600636269. [DOI] [PubMed] [Google Scholar]
- 17.Balakier H. Tripronuclear human zygotes: the first cell cycle and subsequent development. Hum Reprod. 1993;8(11):1892–7. doi: 10.1093/oxfordjournals.humrep.a137955. [DOI] [PubMed] [Google Scholar]
- 18.Martinson JJ, Chapman NH, Rees DC, Liu YT, Clegg JB. Global distribution of the CCR5 gene 32-basepair deletion. Nat Genet. 1997;16(1):100–3. doi: 10.1038/ng0597-100. [DOI] [PubMed] [Google Scholar]
- 19.Marmor M, Sheppard HW, Donnell D, Bozeman S, Celum C, Buchbinder S, et al. Homozygous and heterozygous CCR5-Delta32 genotypes are associated with resistance to HIV infection. J Acquir Immune Defic Syndr. 2001;27(5):472–81. doi: 10.1097/00126334-200108150-00009. [DOI] [PubMed] [Google Scholar]
- 20.Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM, et al. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382(6593):722–5. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 21.Holt N, Wang J, Kim K, Friedman G, Wang X, Taupin V, et al. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol. 2010;28(8):839–47. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L, Krymskaya L, Wang J, Henley J, Rao A, Cao LF, et al. Genomic editing of the HIV-1 coreceptor CCR5 in adult hematopoietic stem and progenitor cells using zinc finger nucleases. Mol Ther : J Am Soc Gene Ther. 2013;21(6):1259–69. doi: 10.1038/mt.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Didigu CA, Wilen CB, Wang J, Duong J, Secreto AJ, Danet-Desnoyers GA, et al. Simultaneous zinc-finger nuclease editing of the HIV coreceptors ccr5 and cxcr4 protects CD4+ T cells from HIV-1 infection. Blood. 2014;123(1):61–9. doi: 10.1182/blood-2013-08-521229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yao Y, Nashun B, Zhou T, Qin L, Qin L, Zhao S, et al. Generation of CD34+ cells from CCR5-disrupted human embryonic and induced pluripotent stem cells. Hum Gene Ther. 2012;23(2):238–42. doi: 10.1089/hum.2011.126. [DOI] [PubMed] [Google Scholar]
- 25.Ye L, Wang J, Beyer AI, Teque F, Cradick TJ, Qi Z, et al. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Delta32 mutation confers resistance to HIV infection. Proc Natl Acad Sci U S A. 2014;111(26):9591–6. doi: 10.1073/pnas.1407473111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramalingam S, London V, Kandavelou K, Cebotaru L, Guggino W, Civin C, et al. Generation and genetic engineering of human induced pluripotent stem cells using designed zinc finger nucleases. Stem Cells Dev. 2013;22(4):595–610. doi: 10.1089/scd.2012.0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tesson L, Usal C, Menoret S, Leung E, Niles BJ, Remy S, et al. Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol. 2011;29(8):695–6. doi: 10.1038/nbt.1940. [DOI] [PubMed] [Google Scholar]
- 28.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–32. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hong SG, Winkler T, Wu C, Guo V, Pittaluga S, Nicolae A et al. Path to the clinic: assessment of iPSC-based cell therapies in vivo in a nonhuman primate model. Cell Rep. 7(4):1298–309. [DOI] [PMC free article] [PubMed]
- 30.Frock RL, Hu J, Meyers RM, Ho YJ, Kii E, Alt FW. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol. 2015;33(2):179–86. doi: 10.1038/nbt.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33(2):187–97. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim D, Bae S, Park J, Kim E, Kim S, Yu HR, et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. 2015;12(3):237–43. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
- 33.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–9. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32(6):569–76. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32(3):279–84. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34(2):184–91. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu C, Liu Y, Ma T, Liu K, Xu S, Zhang Y, et al. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell. 2015;16(2):142–7. doi: 10.1016/j.stem.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu Y, Zhou H, Fan X, Zhang Y, Zhang M, Wang Y, et al. Correction of a genetic disease by CRISPR-Cas9-mediated gene editing in mouse spermatogonial stem cells. Cell Res. 2015;25(1):67–79. doi: 10.1038/cr.2014.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baltimore D, Berg P, Botchan M, Carroll D, Charo RA, Church G, et al. Biotechnology: a prudent path forward for genomic engineering and germline gene modification. Science. 2015;348(6230):36–8. doi: 10.1126/science.aab1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Oligonucleotides used for making in vitro transcription template, CCR5 genotyping and as HDR-mediated repair template. (PDF 177 kb)
Off-target analysis for CRISPR-Cas9–mediated targeting in human 3PN zygotes. (PDF 27 kb)
The sequences of the CCR5 gene in human 3PN embryos carrying CRISPR/Cas9-induced gene modifications (GIF 192 kb)