Abstract
For the catabolism of D-glucuronate, different pathways are used by different life forms. The pathways in bacteria and animals are established, however, a fungal pathway has not been described. In this communication, we describe an enzyme that is essential for D-glucuronate catabolism in the filamentous fungus Aspergillus niger. The enzyme has an NADH dependent 2-keto-L-gulonate reductase activity forming L-idonate. The deletion of the corresponding gene, the gluC, results in a phenotype of no growth on D-glucuronate. The open reading frame of the A. niger 2-keto-L-gulonate reductase was expressed as an active protein in the yeast Saccharomyces cerevisiae. A histidine tagged protein was purified and it was demonstrated that the enzyme converts 2-keto-L-gulonate to L-idonate and, in the reverse direction, L-idonate to 2-keto-L-gulonate using the NAD(H) as cofactors. Such an L-idonate forming 2-keto-L-gulonate dehydrogenase has not been described previously. In addition, the finding indicates that the catabolic D-glucuronate pathway in A. niger differs fundamentally from the other known D-glucuronate pathways.
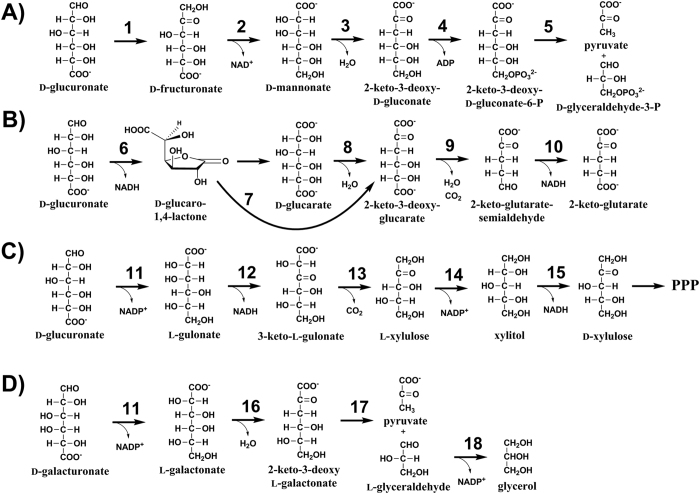
D-Glucuronic acid is a hexuronic acid derived from D-glucose by the oxidation of C6. It has several essential functions in the cellular metabolism including glucuronidation in xenobiotic metabolism1 and being a precursor in L-ascorbic acid biosynthesis2. D-Glucuronate occurs also as component of cell wall polysaccharides such as glucuronoxylan3 in hemicellulose and ulvan4 in algae. D-Glucuronate is catabolised in different life forms through different metabolic pathways. Two bacterial pathways and an animal pathway are known.
In some bacteria, such as Escherichia coli, D-glucuronate is in the first step converted by a D-glucuronate isomerase (EC 5.3.1.12) to D-fructuronate which is then reduced by an NADH utilizing D-fructuronate reductase (EC 1.1.1.57) to D-mannonate5 (Fig. 1A). In the next step, a D-mannonate dehydratase (EC 4.2.1.8) is removing a water molecule from the D-mannonate producing 2-keto-3-deoxy-D-gluconate5. In E. coli, the genes for these enzymes are called uxaC, uxuB and uxuC, respectively6. The resulting 2-keto-3-deoxy-D-gluconate is phosphorylated to 2-keto-3-deoxy-D-gluconate-6-phosphate which is then split by an aldolase to D-glyceraldehyde-3-phosphate and pyruvate. The genes for these enzymes in E. coli are kdgK and kdgA, respectively6. The later part of the pathway is also used in the catabolism of D-galacturonate. The D-galacturonate pathway is very similar; uxaC, kdgK and kdgA are used by both pathways6.
Figure 1.
The first reactions of the D-glucuronate catabolism in (A) bacterial isomerase pathway, (B) bacterial oxidative pathway, (C) animals and (D) D-galacturonate catabolism in fungi. The enzymes are: (1) D-glucuronate isomerase (EC 5.3.1.12), (2) D-fructuronate reductase (EC 1.1.1.57), (3) D-mannonate dehydratase (EC 4.2.1.8), (4) 2-keto-3-deoxy-D-gluconate kinase (EC 2.7.1.45), (5) aldolase (EC 4.1.2.25), (6) uronate dehydrogenase (EC 1.1.1.203), (7) cycloisomerase (EC 5.5.1.-), (8) dehydratase (EC 4.2.1.40), (9) decarboxylating 2-keto-3-deoxy-glucarate dehydratase (EC 4.2.1.41), (10) 2-keto-glutarate semialdehyde dehydrogenase (EC 1.2.1.26), (11) hexuronate reductase (EC 1.1.1.19), (12) L-gulonate 3-dehydrogenase (EC 1.1.1.45), (13) 3-keto-L-gulonate decarboxylase (EC 4.1.1.34), (14) L-xylulose reductase (EC 1.1.1.10), (15) xylitol dehydrogenase (EC 1.1.1.9), (16) L-galactonate dehydratase (EC 4.2.1.146), (17) 2-keto-3-deoxy-L-galactonate aldolase (EC 4.1.2.54) and (18) L-glyceraldehyde reductase (EC 1.1.1.372).
A second bacterial pathway for D-glucuronate catabolism (Fig. 1B) exists in which D-glucuronate is oxidised to D-glucaro-1,4-lactone. The lactone is then hydrolysed to D-glucarate which is then converted by a dehydratase to 2-keto-3-deoxy-glucarate7,8. Alternatively, the lactone is directly converted to the 2-keto-3-deoxy-glucarate by a cycloisomerase9. A combined dehydratase and decarboxylase is then producing 2-keto-glutarate-semialdehyde which is then converted in the last step of the pathway by a dehydrogenase to 2-keto-glutarate8. Also this pathway can be partly used for D-galacturonate catabolism. The first dehydrogenase, the cycloisomerase and the last two enzymes of the pathway are used for D-glucuronate and D-galacturonate catabolism.
In animals, D-glucuronate is catabolised through a pathway that is sometimes called the glucuronate-xylulose-pentose phosphate pathway or uronate cycle10 (Fig. 1C). It was estimated that about 5% of D-glucose is metabolised through this pathway in mammals11. The enzymes of this pathway are: D-glucuronate reductase, (EC 1.1.1.19) L-gulonate-3-dehydrogenase (EC 1.1.1.45) 3-keto-L-gulonate decarboxylase (EC 4.1.1.34), L-xylulose reductase (EC 1.1.1.10), xylitol dehydrogenase (EC 1.1.1.9) and xylulokinase (EC 2.7.1.17). The D-glucuronate reductase is an NADPH requiring that was described in human kidney producing L-gulonate12. L-Gulonate is then reduced by L-gulonate-3-dehydrogenase to 3-keto-L-gulonate. This enzyme activity has been known for about 50 years13 but only recently the corresponding gene was identified14. The 3-keto-L-gulonate decarboxylase is then acting on the 3-keto-L-gulonate to produce L-xylulose. A gene coding for a protein with this enzyme activity has not been described. L-Xylulose is then reduced to xylitol by an L-xylulose reductase. The corresponding gene has been identified in animals15. The first five reactions of this pathway are shown in the Fig. 1. In the subsequent step D-xylulose is phosphorylated to D-xylulose-5-phosphate which is a metabolite in the pentose phosphate pathway.
In fungal microorganisms, a pathway for D-glucuronate catabolism has not been described, although many fungi are able to catabolise D-glucuronate16. However a pathway for D-galacturonate catabolism has been described (Fig. 1D). In this pathway, D-galacturonate is first reduced to L-galactonate by an NADPH requiring reductase17. This enzyme is unspecific and can use also D-glucuronate as a substrate generating L-gulonate. After D-galacturonate is reduced to L-galactonate, a dehydratase is active on the L-galactonate to produce 2-keto-3-deoxy-L-galactonate18. This is followed by an aldolase to form pyruvate and L-glyceraldehyde19. In the last step L-glyceraldehyde is reduced to glycerol20.
In the present manuscript we describe an enzyme that is essential for D-glucuronate catabolism in the filamentous fungus Aspergillus niger. The enzyme has an NADH dependent, L-idonate forming, 2-keto-L-gulonate reductase activity. The finding indicates that the catabolic D-glucuronate pathway in A. niger is different from the bacterial and animal pathway. It is also different from the fungal D-galacturonate pathway, although the first enzyme may be shared between the pathways.
Results
Since the pathways for D-glucuronate and D-galacturonate catabolism are similar in bacteria we tested whether also in fungi similar pathways are used for these two hexuronic acids. In the first step of the fungal D-galacturonate pathway D-galacturonate is reduced to L-galactonate by an NADPH requiring reductase, which is called Gar1 in Trichoderma reesei17 and GaaA in Aspergillus niger21. These enzymes are unspecific and can also reduce D-glucuronate to L-gulonate. The homology in protein sequences between the Gar1 in T.reesei and GaaA in A.niger is low; however A. niger has a gene encoding a close homologue of Gar1 while T. reesei has a gene encoding a homologue for GaaA. In order to test whether the gaaA or the gar1 homologue gene is induced on D-glucuronate and to identify the other genes that are involved in the catabolism of D-glucuronate, we made transcription analysis in A. niger during growth on D-glucuronate. The complete mRNA was sequenced after 0, 4 and 20 h after shifting fresh mycelia to D-glucuronate as a sole carbon source. In addition, a similar transcription analysis was carried out after 5 hour cultivation on D-galacturonate medium. The results are in the Table 1.
Table 1. RNA sequencing of A. niger wild type strain cultivated on D-glucuronate (D-glcUA) and D-galacturonate (D-galUA).
| Transcript level (FPKM) | |||||||
|---|---|---|---|---|---|---|---|
| Gene | JGI protein ID | Genomic location | D-glcUA |
D-galUA | Function/InterPro prediction | ||
| 0 h | 4 h | 20 h | 5 h | ||||
| gaaA | 1158309 | chr_402:1949209–1950762 | 77 | 1618 | 1371 | 2563 | D-galacturonate/D-glucuronate reductase |
| gaaB | 1186088 | chr_502:1524792–1526872 | 104 | 520 | 851 | 4714 | L-galactonate dehydratase |
| gaaC | 1145855 | chr_503:270465–271586 | 98 | 506 | 578 | 827 | 2-dehydro-3-deoxy-L-galactonate aldolase |
| gaaD | 1127092 | chr_701:333409–334937 | 241 | 670 | 1009 | 2887 | L-glyceraldehyde reductase |
| gar1 | 1108731 | chr_502:1395768–1397124 | 89 | 59 | 130 | 134 | Homolog of T. reesei D-galacturonate reductase |
| gatA | 1202517 | chr_102:1065966–1067890 | 6 | 184 | 177 | 391 | D-galacturonate transporter |
| gluC | 1220513 | chr_601:1159357–1160584 | 4 | 1285 | 645 | 9 | IPR006139: D-isomer specific 2-hydroxyacid DH |
| – | 1013899 | chr_304:170864–172922 | 1015 | 916 | 929 | 1213 | IPR004000: Actin/actin-like |
Transcript levels are presented as fragments per kilobase of exon per million fragments mapped (FPKM).
The gar1 homologue is not induced whereas the transcription of the gaaA is induced on both carbon sources. Since the gaaA was upregulated on D-glucuronate we tested a gaaA deletion mutant for the utilization of D-glucuronate (Fig. 2A) and D-galacturonate (Fig. 2B) in submerged cultivations. The ΔgaaA had a slightly reduced utilization rate of D-glucuronate while the phenotype was more pronounced on D-galacturonate. The observation of a reduced D-glucuronate utilization rate suggests that the gaaA may be part of the D-glucuronate pathway. However, the gaaA deletion neither blocked the utilization of D-glucuronate nor D-galacturonate completely. Thus, most likely there is an additional reductase(s) functioning in these pathways. We tested also other mutants of the D-galacturonate pathway: The ΔgaaB (L-galactonate dehydratase) and ΔgaaC (2-keto-3-deoxy-L-galactonate aldolase) strains. In the wild type strain, these genes were induced on D-galacturonate while the induction was lower on D-glucuronate (Table 1). The ΔgaaB and ΔgaaC strains did not utilize D-galacturonate (Fig. 2B). The observed D-galacturonate concentrations were slightly increasing most likely due to the evaporation during the cultivations. In contrast, the utilization of D-glucuronate was not reduced (Fig. 2A) suggesting that gaaB and gaaC are not involved in the D-glucuronate pathway. In a previous study all the genes of the A. niger ATCC 1015 that had some homology to a sugar acid dehydratase, including gaaB, were expressed in yeast and the activities tested with a small library of sugar acids22. Since none of these dehydratases exhibited activity with L-gulonate we concluded that there must be some other enzyme activity to convert L-gulonate.
Figure 2.
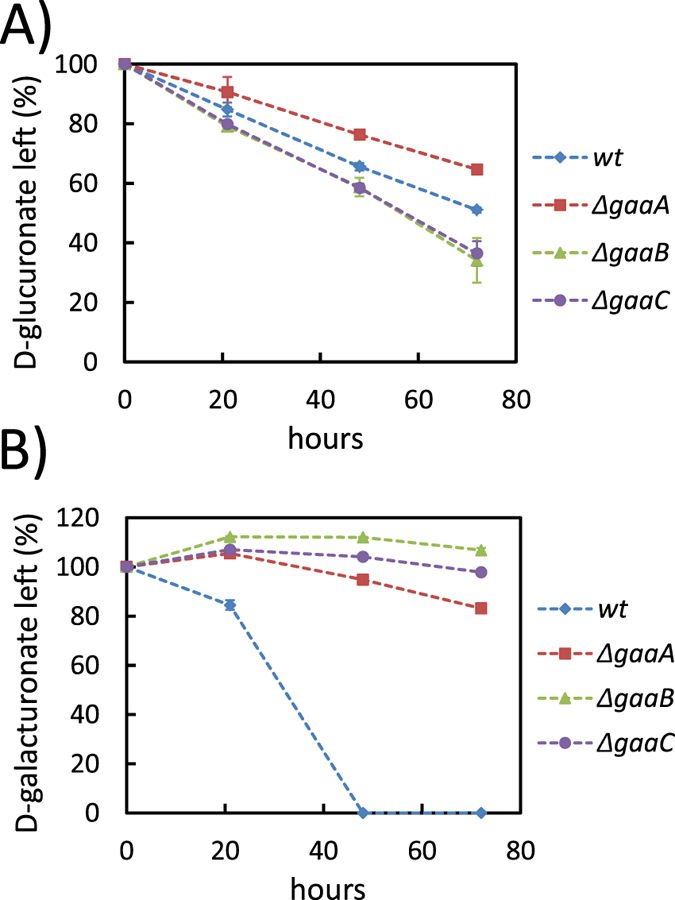
Consumption of (A) D-glucuronic acid and (B) D-galacturonic acid in submerged cultivations by the A. niger strains wt, ∆gaaA, ∆gaaB and ∆gaaC. Data represent means ± standard deviation from three biological repeats. If error bars are not visible they are smaller than the symbol.
In the RNA sequence analysis a gene with the JGI protein ID 122051323 was upregulated in D-glucuronate but not in D-galacturonate (Table 1). The gene putatively codes for an enzyme of the protein family of D-isomer specific 2-hydroxyacid dehydrogenases. The open reading frame of the gene, as predicted in JGI genome database23, was ordered as custom synthesized gene and expressed on a multicopy plasmid in S. cerevisiae. A few sugar acids including L-gulonate were tested for the activity with the resulting yeast crude extract. In the extract, the protein did not show L-gulonate dehydrogenase activity; however we found 2-keto-L-gulonate reductase activity as well as L-idonate dehydrogenase activity. The activity was strictly dependent on the cofactors NAD+/NADH and no activity was observed with NADP+/NADPH. We called the gene encoding this enzyme gluC.
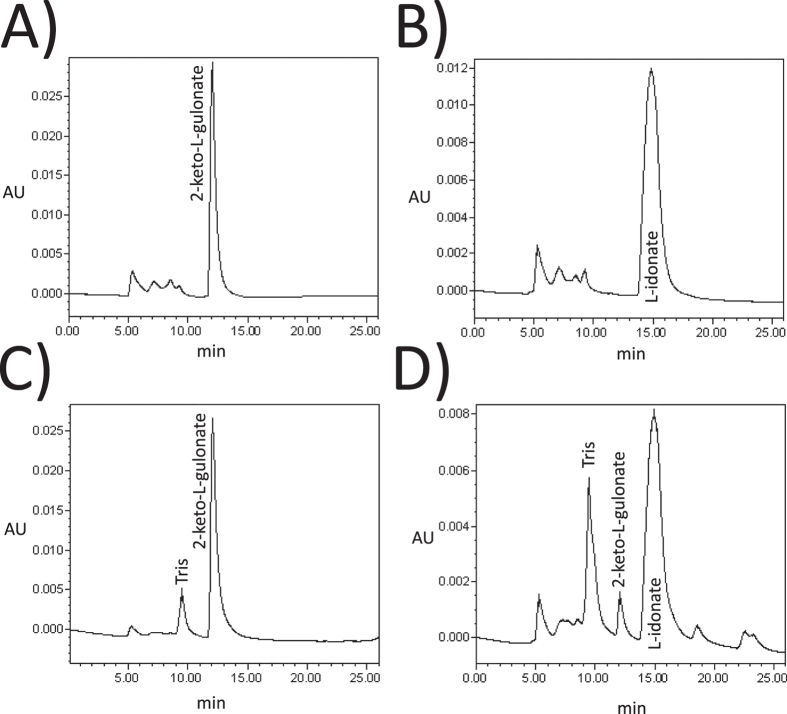
As the next step, the histidine-tagged GluC protein was purified and analysed with more detail. The reaction product of the purified protein was confirmed from in vitro reactions containing 2-keto-L-gulonate as substrate using HPLC (Fig. 3) The reaction product was indeed L-idonate and not L-gulonate (L-gulonate has a retention time at 16.2 min). We concluded that the gluC codes for a NAD+ specific L-idonate forming 2-keto-L-gulonate reductase. Next the kinetic parameters of the purified GluC protein were determined for 2-keto-L-gulonate (Fig. 4A) and L-idonate (Fig. 4B). The Vmax values 120 and 6 μmol min−1 mg−1 and the Km values 30 and 20 mM were determined for 2-keto-L-gulonate and L-idonate, respectively.
Figure 3.
HPLC analysis of (A) 10 mM 2-keto-L-gulonate, (B) 10 mM L-idonate, (C) in vitro reaction mixture without GluC and (D) in vitro reaction mixture with purified GluC.
Figure 4.
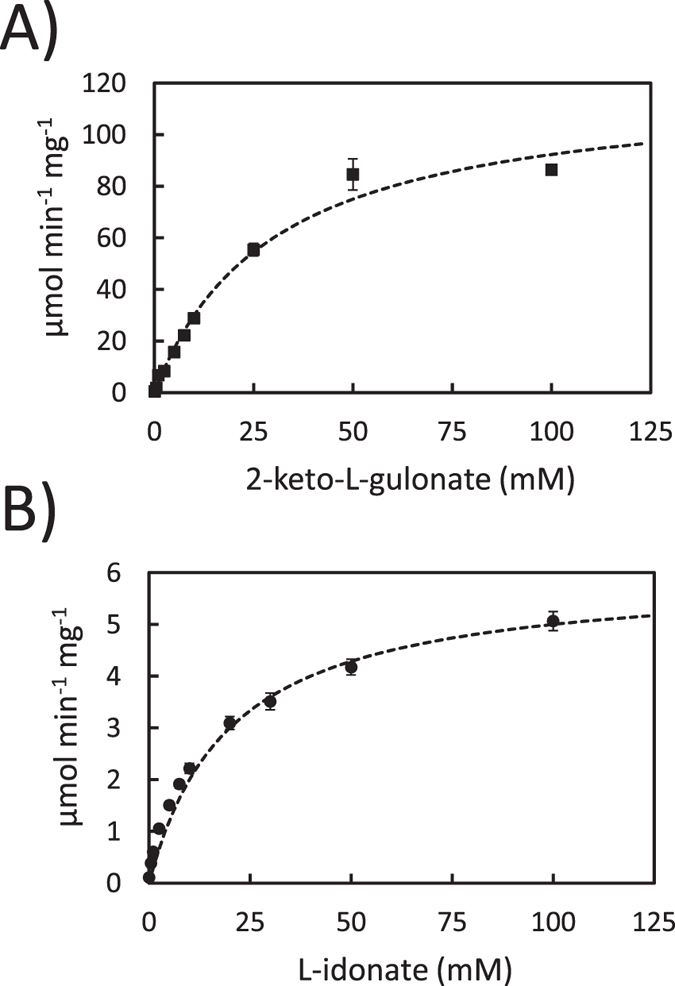
(A) 2-keto-L-gulonate reductase and (B) L-idonate dehydrogenase activity of the purified GluC with NADH/NAD+. Data represent means ± standard deviation from three technical repeats. If error bars are not visible they are smaller than the symbol.
The gene gluC was deleted from A. niger for the phenotypic characterization of the mutant strain. The resulting ∆gluC strain was tested for growth on D-glucuronate containing agar plates (Fig. 5) and for utilization of D-glucuronate in submerged cultivations (Fig. 6). The ∆gluC showed only minor growth on D-glucuronate plate probably due to the amino acid supplementation. In the liquid cultivations, utilization of D-glucuronate was almost completely blocked. A slight consumption of D-glucuronate was observed in the submerged cultivation indicating that there may be an additional enzyme for the reaction. However, these results suggest that GluC is the main enzyme for this reaction in the pathway for D-glucuronate catabolism in A. niger.
Figure 5. Growth of the A. niger strains wt, ∆kusA and ∆gluC on D-galacturonic and D-glucuronic acid on agar plates.
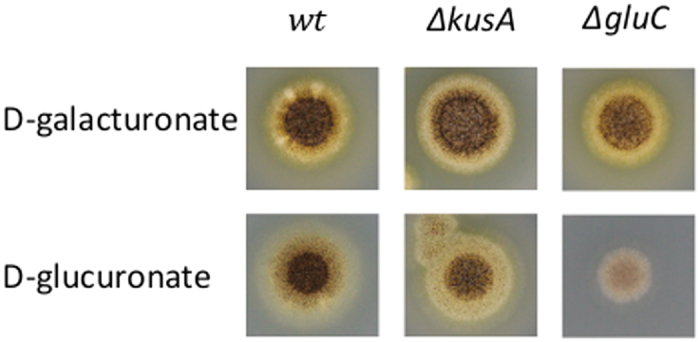
Figure 6. Consumption of D-glucuronic acid in submerged cultivations by wt and ∆gluC.
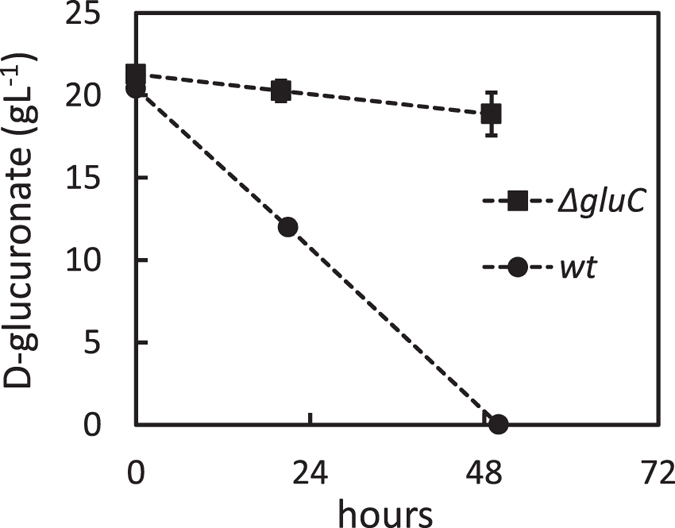
Data represent means ± standard deviation from three biological repeats. If error bars not visible are smaller than the symbol.
Discussion
Several fungal species are able to catabolise D-glucuronate; however they are apparently not using any of the bacterial pathways. Genes encoding homologous enzymes for the bacterial pathways are not found in the fungal genomes. Since there is no other information about a fungal pathway for D-glucuronate catabolism, we investigated whether fungi would use the animal pathway. The first enzyme, the D-glucuronate/D-galacturonate reductase, seemed to be part of the fungal pathway, the corresponding gene gaaA was upregulated on D-glucuronate and the deletion mutant had a reduced D-glucuronate consumption rate. The L-gulonate that is formed in this reaction is in the next step reduced to 3-keto-L-gulonic acid in the animal pathway. We did not find this enzyme activity in the fungal extract. Another possible path would have been an analogous pathway for the fungal D-galacturonate pathway. In the bacterial pathways, D-galacturonate and D-glucuronate are catabolised using partly the same enzymes. However, we could show that the next two enzymes from the fungal D-galacturonate pathway are not part of the catabolic D-glucuronate pathway in A. niger. We also knew from a previous study that A. niger does not have a L-gulonate dehydratase22. In order to identify genes that are involved in the D-glucuronate catabolism, we made an RNA sequencing. Among the most upregulated genes during growth on D-glucuronate was the gluC gene encoding a 2-keto-L-gulonate reductase. This gene was essential for D-glucuronate catabolism in A. niger indicating that the catabolic D-glucuronate pathway differs fundamentally from the fungal catabolic D-galacturonate pathway and from other known D-glucuronate pathways. The 2-keto-L-gulonate reductase that forms L-idonate is also an enzyme activity that has not been reported before. A bacterial enzyme had been described previously that converts 2-keto-D-gluconate to D-gluconate. This enzyme is very unspecific and can catalyse also the reaction from 2-keto-L-gulonate to L-idonate using NADPH as a cofactor24. The reductase identified in this communication is the first specific L-idonate forming 2-keto-L-gulonate reductase and the first L-idonate forming 2-keto-L-gulonate reductase utilizing NADH. An L-idonate forming 2-keto-L-gulonate reductase had been suggested to be in the plant pathway for L-ascorbic acid conversion to L-tartaric acid25,26, however the corresponding gene has not been identified.
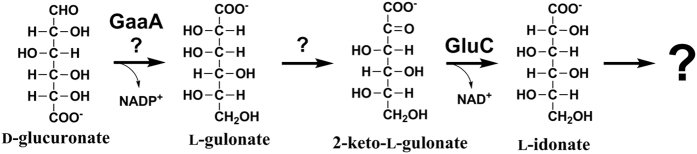
For the fungal pathway for D-glucuronate catabolism, we suggest that the first reaction is a reduction producing L-gulonate. However, deletion of gaaA did not block the catabolism of D-glucuronate nor D-galacturonate completely. Thus, there must be an additional enzyme(s) having this reductase activity. The gaaA is the only gene that is shared by the two pathways. In the D-glucuronate pathway, L-gulonate is most likely then converted to 2-keto-L-gulonate, however there is no information about this step and whether it is a single step. The 2-keto-L-gulonate is then converted to L-idonate by the action of GluC. A metabolic pathway for L-idonate catabolism is described in E. coli27. In this pathway, L-idonate is oxidized to 5-keto-D-gluconate and further reduced to D-gluconate and phosphorylated to 6-phosphogluconate which can be catabolized through Entner-Doudoroff or pentose phosphate pathway. In fungi, no information is available about the reactions after the L-idonate. Nevertheless, the current study is a strong indication that the catabolic D-glucuronate pathway in A. niger starts with the reduction to L-gulonate and rearrangement of its hydroxyl group at C2 via 2-keto-L-gulonate resulting in formation of L-idonate (Fig. 7).
Figure 7. First three suggested reactions in the catabolic D-glucuronate pathway in A. niger.
The enzymes are: the hexuronate reductase (EC 1.1.1.19) GaaA and the 2-keto-L-gulonate reductase GluC.
Methods
Strains
The Aspergillus niger strain ATCC 1015 (CBS 113.46) was used as a wild type. The A. niger gene deletion strains ∆pyrG (deleted orotidine-5′-phosphate decarboxylase), ∆gaaA (deleted D-galacturonate reductase), ∆gaaB (deleted L-galactonate dehydratase) and ∆gaaC (deleted 2-keto-3-deoxy-L-galactonate aldolase) were described earlier28,29,30. All the plasmids were produced in Escherichia coli TOP10 cells. The Saccharomyces cerevisiae strains ATCC 90845 and a modified CEN.PK2 (MATα, leu2-3/112, ura3-52, trp1-289, his3-∆1, MAL2-8c, SUC2) were used in the homologous recombination for the plasmid construction and for the production of the purified GluC enzyme, respectively.
Media and cultural conditions
Luria Broth culture medium supplemented with 100 μg ml−1 of ampicillin and cultural conditions of 37 °C and 250 rpm were used for E. coli cultures. YPD medium (10 g yeast extract l−1, 20 g peptone l−1 and 20 g D-glucose l−1) was used for yeast pre-cultures. After the transformation of an expression plasmid in yeast, SCD-URA (uracil deficient synthetic complete media supplemented with 20 g D-glucose l−1) plates were used for uracil auxotrophic selection. SCD-URA medium was used in protein production. All the yeast cultivations were carried out at 30 °C and the liquid cultivations at 250 rpm. A. niger spores were generated on potato-dextrose plates and ~108 spores were inoculated to 50 ml of YP medium (10 g yeast extract l−1, 20 g peptone l−1) containing 30 g gelatin l−1 for pre-cultures. Mycelia were pre-grown in 250-ml Erlenmeyer flasks by incubating overnight at 28 °C, 200 rpm and harvested by vacuum filtration, rinsed with sterile water and weighted. In A. niger transformations, SCD-URA plates supplemented with 1.2 M D-sorbitol and 20 g agar l−1 (pH 6.5) were used. A. nidulans defined minimal medium31 was used in the A. niger cultivations. The minimal medium used in the phenotypic characterization in liquid cultivations (Fig. 2) contained 20 g D-galacturonate l−1 or 20 g D-glucuronate l−1 and the pH was adjusted to 5. These cultivations were inoculated with 4 g l−1 (wet) of pre-grown mycelia. Similar minimal medium was used in the transcriptional analysis but the pH was adjusted to 6.8 and cultivations were inoculated with 13 g l−1 (wet) of pre-grown mycelia. In the phenotypic characterization of A. niger ∆gluC in liquid cultivations (Fig. 5), similar minimal medium containing D-glucuronate was used but the pH was adjusted to 3. These cultivations were inoculated with 17.5 g l−1 (wet) of pre-grown mycelia. Agar plates used for phenotypic characterization of ∆gluC (Fig. 5) contained SC-medium (synthetic complete), 15 g agar l−1 and 20 g D-glucuronate l−1 or 20 g D-galacturonate l−1. These plates were inoculated with 1.5*106 spores.
Transcriptional analysis
Samples of 2 ml were collected from the cultures and the mycelium was harvested by vacuum filtration. The filtered mycelium was frozen with liquid nitrogen and stored at −80 °C. Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen). RNA library preparation and sequencing was carried out by GATC (Constance, Germany) using the InViewTM Transcriptome Explore package. In brief, a random primed library was prepared for each of the samples and sequenced with Illumina HiSeq 2500 for 50 bp. Read data was trimmed and quality controlled with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were aligned with tophat232 to A. niger ATCC1015 genome version 4.023 retrieved with GFF annotations from http://genome.jgi.doe.gov/Aspni7/Aspni7.home.html and reads counted with R package “GenomicFeatures”33.
Protein production and purification
The yeast codon optimized gluC gene (GenScript, USA) was released with EcoRI and BamHI (both NEB) and was ligated into a modified pYX212 plasmid34 containing TPI1 promoter and URA3 selectable marker. A yeast strain was then transformed with the resulting plasmid using the lithium acetate method35. The yeast strain expressing gluC in the plasmid was cultivated overnight on SCD-URA medium in 1000 ml total volume. Cells were collected by centrifugation and washed. The cell pellet was resuspended to 25 ml of lysis buffer (The QIAexpressionist, Qiagen) containing protease inhibitor (Roche), disrupted five times for 30 seconds using glass beads and BeadBeater (Biospec Products). The extract was centrifuged for 40 min at +4 °C and the supernatant was run through a nickel-nitrilotriacetic acid column (Qiagen). The histidine-tagged GluC protein was purified following the QIAexpressionist protocol (Qiagen). The concentration of purified GluC protein was measured with a protein assay kit (Bio Rad).
Enzymatic assays
The oxidoreductase activity of purified GluC was assayed using Konelab 20XT Clinical Chemistry Analyzer (Thermo Scientific). The reaction mixture contained 50 mM Tris buffer, 400 μM NAD+ or NADH, a substrate in different concentrations and purified GluC in a final concentration of 3.6 mg l−1. The pH 8 was used with NAD+ and L-idonate (Omicron Biochemicals Inc, USA) and pH 7 with NADH and 2-keto-L-gulonate (Omicron Biochemicals Inc, USA). The reaction was started by addition of the purified GluC and the formation/consumption of NADH was followed at 340 nm. For the HPLC analysis, a similar reaction mixture but 10 mM NAD+/NADH and L-idonate/2-keto-L-gulonate were used.
Gene deletions in A. niger
For the deletion of the kusA gene (an ortholog of the Ku70 protein in other eukaryotes), a component of non-homologous end joining pathway, a deletion cassette containing homologous 5′ and 3′ flanks (~1.5 kb) for targeted integration and the selectable marker pyrG (A. niger) was constructed. The 5′ and 3′ flanks were amplified by PCR (KAPA HiFi DNA polymerase, Kapa Biosystems) with the primers P1/P2 and P3/P4, respectively (Table 2). The amplified flanks and pyrG were joined using assembly PCR and the resulting cassette was ligated into pCR®-Blunt II-TOPO (Invitrogen). The cassette was linearized with BcuI (Fermentas) and transformed to A. niger ∆pyrG strain using the protoplast transformation method and selected for growth in the absence of uracil. Correct integration of the transformed cassette into the genome was confirmed with colony PCR using Phire direct PCR kit (Thermo Scientific) and the primers P13/P14. The pyrG gene that was used as selectable marker was deleted again from the ∆kusA strain (pyrG∆, kusA::pyrG) as described by Mojzita et al.28. The resulting strain ∆kusA ∆pyrG was used as parental strain for the gluC deletion. The gluC deletion cassette contained homologous 5′ and 3′ flanks (~1 kb) for targeted integration and the selectable marker pyrG. The 5′ and 3′ flanks were amplified by PCR with the primers P5/P6 and P7/P8, respectively, pRS426 plasmid with P9/P10 and pyrG with P11/P12. The resulting PCR amplified fragments (5′ flank, 3′flank, pyrG and pRS426) contained 40 bp compatible ends for homologous recombination. All the fragments were joined using yeast homologous recombination as described earlier36. The resulting cassette was linearized with NotI (NEB), transformed to A. niger ∆kusA ∆pyrG strain and mutants with successful integration were selected for growth in the absence of uracil. Resulting transformants were screened for the correct integration of the deletion cassette with colony PCR using Phire direct PCR kit (Thermo Scientific) and the primers P15/P16.
Table 2. Oligonucleotides used in the study.
| Name | Sequence |
|---|---|
| P1 | GATAAGCTTGATATCGAATTCCTGCAGCCCGGGGGATCCACTAGTCTTTGCATCACCGCATGCAC |
| P2 | AGCTGGTATAGCCAAACATCGCCAATCACCTCAATCACCCAGTAGAATGTTGTGGAATCGTTTAAAGC |
| P3 | CATGCGGGCTTGGGACGCCATGTCCGTCGCGTGATAACCCCATGGCGGGATTGTTGGATT |
| P4 | CGGTGGCGGCCGCTCTAGAACTAGTATGTTTCGGCGCACTAATAGC |
| P5 | TGATATCGAATTCCTGCAGCGCGGCCGCCAAGGTTCAGGGATCATGGT |
| P6 | CAATCACCTCAATCACCCGGAGAGGATTTGGAAAGTCAAC |
| P7 | CATGTCCGTCGCGTGATAACCGCACATCGTCACCCATTTC |
| P8 | GCTCTAGAACTAGTGGATCCCCCGGGCGGCCGCCTTTCAGAGAGCGACTCGGC |
| P9 | GCTCTCTGAAAGGCGGCCGCCCGGGGGATCCACTAGTTCT |
| P10 | CCCTGAACCTTGGCGGCCGCGCTGCAGGAATTCGATATCAA |
| P11 | GTTGACTTTCCAAATCCTCTCCGGGTGATTGAGGTGATTG |
| P12 | GAAATGGGTGACGATGTGCGTTATCACGCGACGGACATG |
| P13 | ATACTTCCCTCTTT CAATTTCG |
| P14 | AAGACACCACATAACGACATCC |
| P15 | CTACCAATCCTGGAAGGAGA |
| P16 | AGCTGGTATAGCCAAACATC |
Chemical analyses
Samples of 2 ml were removed from liquid cultivations at intervals and mycelium was separated from the supernatant by centrifugation. The concentration of D-glucuronate and D-galacturonate (from cultivations) or L-idonate and 2-keto-L-gulonate (from in vitro reactions) was determined by HPLC using a Fast Acid Analysis Column (100 mm × 7.8 mm, BioRad Laboratories, Hercules, CA) linked to an Aminex HPX-87H organic acid analysis column (300 mm × 7.8 mm, BioRad Laboratories) with 5.0 mM H2SO4 as eluent and a flow rate of 0.5 ml min−1. The column was maintained at 55 °C. Peaks were detected using a Waters 2487 dual wavelength UV (210 nm) detector.
Additional Information
How to cite this article: Kuivanen, J. et al. A novel pathway for fungal D-glucuronate catabolism contains an L-idonate forming 2-keto-L-gulonate reductase. Sci. Rep. 6, 26329; doi: 10.1038/srep26329 (2016).
Acknowledgments
This work was supported by the Academy of Finland through the Sustainable Energy (SusEn) program (grant 271025) and by the CNPq of Brazil (grant 490236/2012-0) in the form of a cooperation project.
Footnotes
Author Contributions J.K. and P.R. designed and J.K. and M.H.S.-G. performed the experimental work. M.A. performed the processing of the raw data from the RNA sequencing. J.K. analysed the data and prepare the figures and tables. J.K. and P.R. wrote the manuscript. All the authors reviewed the manuscript and approved the final version.
References
- Mazerska Z., Mróz A., Pawłowska M. & Augustin E. The role of glucuronidation in drug resistance. Pharmacol. Ther. 10.1016/j.pharmthera.2016.01.009 (2016). [DOI] [PubMed] [Google Scholar]
- Linster C. L. & Van Schaftingen E. Vitamin C biosynthesis, recycling and degradation in mammals. FEBS J. 274, 1–22 (2007). [DOI] [PubMed] [Google Scholar]
- Reis D. et al. Cellulose-glucuronoxylans and plant cell wall structure. Micron 25, 171–187 (1994). [Google Scholar]
- Lahaye M. & Robic A. Structure and function properties of Ulvan, a polysaccharide from green seaweeds. Biomacromolecules 8, 1765–1774 (2007). [DOI] [PubMed] [Google Scholar]
- Ashwell G. Enzymes of glucuronic and galacturonic acid metabolism in bacteria. Methods Enzymol. 5, 190–208 (1962). [Google Scholar]
- Portalier R., Robert-Baudouy J. & Stoeber F. Regulation of Escherichia coli K-12 hexuronate system genes: exu regulon. J. Bacteriol. 143, 1095–1107 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagley S. & Trudgill P. W. The metabolism of galactarate, D-glucarate and various pentoses by species of Pseudomonas. Biochem. J. 95, 48–58 (1965). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. F. & Feingold D. S. D-Glucaric acid and galactaric acid catabolism by Agrobacterium tumefaciens. J. Bacteriol. 102, 85–96 (1970). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andberg M. et al. Characterization of a novel Agrobacterium tumefaciens galactarolactone cycloisomerase enzyme for direct conversion of D-galactarolactone to 3-deoxy-2-keto-L-threo-hexarate. J. Biol. Chem. 287, 17662–17671 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankes L., Politzer W., Touster O. & Anderson L. Myo-inositol catabolism in human pentosurics: the predominant role of the glucuronate-xylulose-pentose phosphate pathway. Ann. N. Y. Acad. Sci. 165, 564–576 (1969). [PubMed] [Google Scholar]
- Kaneko J., Harvey J. & Bruss M. Clinical Chemistry of Domestic Animals 6st edn (Academic Press, 1997). [Google Scholar]
- Sato S. & Kador P. Human kidney aldose and aldehyde reductases. J. Diabetes Complications 7, 179–187 (1993). [DOI] [PubMed] [Google Scholar]
- Smiley J. & Ashwell G. Purification and properties of ß-L-hydroxy acid dehydrogenase II. isolation of ß-keto-L-gulonic acid, an intermediate in L-xylulose biosynthesis. J. Biol. Chem. 236, 357–364 (1961). [Google Scholar]
- Ishikura S., Usami N., Araki M. & Hara A. Structural and functional characterization of rabbit and human L -gulonate 3-dehydrogenase. 314, 303–314 (2005). [DOI] [PubMed]
- Ishikura S. et al. Molecular cloning, expression and tissue distribution of hamster diacetyl reductase. Identity with L-xylulose reductase. Chem. Biol. Interact. 130–132, 879–889 (2001). [DOI] [PubMed] [Google Scholar]
- Fungal Growth Database. (2016). Available at http://www.fung-growth.org (Accessed: 21st March 2016).
- Kuorelahti S., Kalkkinen N., Penttilä M., Londesborough J. & Richard P. Identification in the mold Hypocrea jecorina of the first fungal D-galacturonic acid reductase. Biochemistry 44, 11234–11240 (2005). [DOI] [PubMed] [Google Scholar]
- Kuorelahti S., Jouhten P., Maaheimo H., Penttilä M. & Richard P. L-Galactonate dehydratase is part of the fungal path for D-galacturonic acid catabolism. Mol. Microbiol. 61, 1060–1068 (2006). [DOI] [PubMed] [Google Scholar]
- Hilditch S., Berghäll S., Kalkkinen N., Penttilä M. & Richard P. The missing link in the fungal D-galacturonate pathway: Identification of the L-threo-3-deoxy-hexulosonate aldolase. J. Biol. Chem. 282, 26195–26201 (2007). [DOI] [PubMed] [Google Scholar]
- Liepins J., Kuorelahti S., Penttilä M. & Richard P. Enzymes for the NADPH-dependent reduction of dihydroxyacetone and D-glyceraldehyde and L-glyceraldehyde in the mould Hypocrea jecorina. FEBS J. 273, 4229–4235 (2006). [DOI] [PubMed] [Google Scholar]
- Martens-Uzunova E. S. & Schaap P. J. An evolutionary conserved D-galacturonic acid metabolic pathway operates across filamentous fungi capable of pectin degradation. Fungal Genet. Biol. 45, 1449–1457 (2008). [DOI] [PubMed] [Google Scholar]
- Motter F. A. et al. Categorisation of sugar acid dehydratases In Aspergillus niger. Fungal Genet. Biol. 64, 67–72 (2014). [DOI] [PubMed] [Google Scholar]
- Andersen M. R. et al. Comparative genomics of citric-acid-producing Aspergillus niger ATCC 1015 versus enzyme-producing CBS 513.88. Genome Res. 21, 885–897 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yum D., Lee B., Hahm D. & Pan J. The yiaE gene, located at 80.1 minutes on the Escherichia coli chromosome, encodes a 2-ketoaldonate reductase. J. Bacteriol. 180, 1–6 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malipiero U., Ruffner H. & Rast D. Ascorbic to tartaric acid conversion in grapevines. J. Plant Physiol. 129, 33–40 (1987). [Google Scholar]
- DeBolt S., Cook D. R. & Ford C. M. L-Tartaric acid synthesis from vitamin C in higher plants. Proc. Natl. Acad. Sci. USA. 103, 5608–13 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bausch C. et al. Sequence analysis of the GntII (Subsidiary) system for gluconate metabolism reveals a novel pathway for L-idonic acid catabolism in Escherichia coli. J. Bacteriol. 180, 3704–10 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojzita D. et al. Metabolic engineering of fungal strains for conversion of D-galacturonate to meso-galactarate. Appl. Environ Microbiol. 76, 169–175 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuivanen J. et al. Engineering filamentous fungi for conversion of D-galacturonic acid to L-galactonic acid. Appl. Environ. Microbiol. 78, 8676–8683 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebe M. G., Mojzita D., Hilditch S., Ruohonen L. & Penttilä M. Bioconversion of D-galacturonate to keto-deoxy-L-galactonate (3-deoxy-L-threo-hex-2-ulosonate) using filamentous fungi. BMC Biotechnol. 10, 10.1186/1472-6750-10-63 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt R., Johnson G. & Ogata W. Wild-type and mutant stocks of Aspergillus nidulans. Genetics 52, 233–246 (1965). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M. et al. Software for computing and annotating genomic ranges. Plos Comput. Biol. 9, 1–10 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verho R., Putkonen M., Londesborough J., Penttilä M. & Richard P. A Novel NADH-linked L-xylulose reductase in the L-arabinose catabolic pathway of yeast. J. Biol. Chem. 279, 14746–14751 (2004). [DOI] [PubMed] [Google Scholar]
- Gietz R. & Schiestl R. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG methode. Nat. Protoc. 2, 31–34 (2007). [DOI] [PubMed] [Google Scholar]
- Kuivanen J., Penttilä M. & Richard P. Metabolic engineering of the fungal D-galacturonate pathway for L-ascorbic acid production. Microb. Cell Fact. 14, 1–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]