Abstract
The current state of screening methods for drug discovery is still riddled with several inefficiencies. Although some widely used high-throughput screening platforms may enhance the drug screening process, their cost and oversimplification of cell–drug interactions pose a translational difficulty. Microfluidic cell-chips resolve many issues found in conventional HTS technology, providing benefits such as reduced sample quantity and integration of 3D cell culture physically more representative of the physiological/pathological microenvironment. In this review, we introduce the advantages of microfluidic devices in drug screening, and outline the critical factors which influence device design, highlighting recent innovations and advances in the field including a summary of commercialization efforts on microfluidic cell chips. Future perspectives of microfluidic cell devices are also provided based on considerations of present technological limitations and translational barriers.
Keywords: : high-throughput/content screening, microfluidic cell chip, physiological microenvironment
The progressive advent of affordable genetic and proteomic sequencing technology has brought to the forefront an increasingly large number of therapeutic targets for various diseases. However, conventional drug screening approaches are time-consuming and costly, with more than 90% of screened drug candidates failing after entering clinical trials [1]. Efficient methods of screening drugs against the desired target still remain fairly intractable. High-throughput screening (HTS) is a promising but exhaustive force approach widely adopted both in academia and pharmaceutical industry to address this issue. At the most basic level, it comprises of an automated workstation that handles solutions, drugs and microplates, permitting multiple drugs and their efficacy on reporter cells to be verified simultaneously. Although HTS improves the screening throughput and seems to be a standard platform for drug discovery, it leaves several unresolved demands, with monetary cost being the first. Current pharmaceutical industries employ HTS primarily for chemical optimization at the early stage of the drug development. HTS products only contribute 19–33%(or less) on the clinical development of drugs from major pharmaceutical companies [2]. HTS in its current commercial state also limits its translatibility from screened drugs to clinical drugs for patients. These limitations are due to the use of conventional culture techniques which are too simple and lacking the ability to mimic the complex cell–cell and cell–microenvironmental interactions found in real tissues.
In contrast, drug screening through a microfluidic cell-culture system may be able to tackle the above-mentioned issues. Microfluidic systems, recognized as ‘lab-on-a-chip’ systems, can integrate many analytical components into a single chip, which reduces the required sample volume significantly, typically anywhere from 10 to 1000-fold less than the conventional counterpart. This significant savings in cell and drug sample size facilitate an increasing number of tests could be prohibitively expensive otherwise. A rough cost estimation and comparison between a microfluidic device [3] and a bioreactor system [4] shows that number of cells and culture medium/drug needed in a well of the bioreactor are at least 100 times more than that in a microchamber on the chip. Most importantly, the dynamic 3D microenvironment for cell-culture and transport mechanism between cells/tissues and microvessels are more faithfully replicated on the microfluidic system, therefore alleviating the translational barrier to in vivo expectations. This review discusses the advantages of microfluidic platforms as well as aspects of manufacturing and translation with emphasis on advances in microfluidic (cell-based) ‘lab-on-a-chip’ systems designed for HTS applications.
Advantages of using a microfluidic cell-culture platform in HTS
The process from drug discovery to commercialization is complicated and time-consuming since cytotoxicity, efficacy and (adverse) side effects of new drugs need to be repetitively validated along each step to ensure consistency and reliability of results. In order to approximate expected therapeutic effects on humans, ideally, drug screening tests should be performed within vivo models. However, employing animal models for drug screening is expensive and time-consuming. As such, in vitro drug screening prior to animal studies help to eliminate and select from thousands of candidates, but their efficacy and physiological effects cannot be guaranteed from these screening tests – we can only infer at the initial possible extent of cytotoxicity. Recent trends show an increasing number of pharmaceutical companies enlisting cell-based assays in drug screening and optimizations instead of using commercially available in vitro biochemical assays. Several drugs have been successfully discovered through cell-based assays and the efficacies and safety of those selected drugs have been further acknowledged by the US FDA [2]. Although 3D culture techniques were introduced to conventional multiple-well plate-based assays to construct a more natural extracellular microenvironment, most of these conventional screening platforms are still limited in applicability due to their reliance on static culture maintenance. In contrast, microfluidic devices can provide 3D microenvironment with microvascular perfusion and diffusion between mimicked microvessels and 3D cell culture, which is closer to what cells encounter in real tissues or organs [5–7].
The microscale construction of channels and chambers vastly reduces the required volume of reagents that their conventional counterpart needs. This downscaling of reagent requirements accompanied with the resulting ease and increase of parallelized testing compliments HTS practices well, especially in the scope of prohibitively expensive drug discovery or personalized medicine. For example, on a conventional 96-well plate the volume needed for cell culture is around 100–200 µl per well. In contrast, taking a 3D microfluidic cell array that we published previously as an example [6], the required volume is only 50 nl per microchamber as the dimension of a chamber is 780 µm in diameter and 110 µm in height.
The on-board construction of microscale fluidic channels also permit the versatility to replicate the dimensional facets of the natural cellular environment. A typically common technique utilizes microfabrication methods borrowed from the electronics industry to create intricately patterned features on substrates (commonly silicon wafers) to provide the mold on which a biocompatible elastomer such as polydimethylsiloxane (PDMS) is cast, which is then cured to construct a microfluidic component or device. The ability to generate patterns of any variety through this method provides a flexible modularity and compatibility to the devices thus constructed.
Some concerns regarding cell-based microfluidic device need to be taken into consideration in their use. Conventional cell culture is done in an incubator that provides ideal and consistent support of oxygen and carbon dioxide. Ideally, if cultured inside an incubator, microfluidic systems also allow sufficient gas exchange as PDMS is a gas permeable material. However, the gas permeability is affected by the thickness of the PDMS layer, surface coating for cell attachment and plasma oxidation [8]. On the other hand, the high surface area-to-volume ratio in a microfluidic device can be seen as an advantage for the purposes of biochemical reactions, but it is also the cause of fast nutrient depletion in cell culture medium and liquid evaporation from the device leading directly to air bubbles [9]. As a result, these characteristics of cell culture in microfluidic device need to be addressed and considered before conducting drug screening.
Critical factors on designing a microfluidic cell-culture platform
Biocompatible materials & surface modifications
In constructing a microfluidic device for cell culture, the selection of biocompatible materials becomes a primary consideration. Well-established technologies of microelectromechanical system (MEMS) provide silicon-based materials with highly favorable advantages. While crystal silicon is completely opaque, biocompatible glass, owing to its transparency, becomes important for integration with optical observation and detection approaches on cell-based chips. Although the price of glass with better optical clarity is high, it is still a promising candidate for its well-understood surface chemistry, fabrication technologies and tolerance to organic solvents [10,11]. The majority of current cell-based microfluidic devices are made out of PDMS with glass as supporting substrates. The predominance of PDMS on cell-based chipis based strongly on its gas permeability which is suitable for cell proliferation, as well as its optical clarity and biocompatibility. The current scope of soft lithography techniques allow the versatility to fabricate devices on the micron scale using PDMS, and that limit can be pushed to the nanoscale through newer approaches researched over the past decade [12,13]. In addition, since PDMS is an elastic material, it enables nonleaky fluidic interconnections which facilitates implementation of mechanical microvalves and micropumps [14]. Furthermore, a wide range of adjustable elastic moduli of PDMS can be achieved by the ratio of base to the curing agent to match most soft tissues, relevant in cell mechanobiology studies [15].
The inherent surface characteristic of PDMS is strongly hydrophobic and must therefore be rendered hydrophilic to facilitate cell attachment and growth. The majority of mammalian cells in vitro grow as monolayers on a flat substrate. It has been demonstrated that the absence of proper cell adhesion may suppress cellular functions or even cause cell death. Many different approaches for reducing hydrophobicity or surface modification have been extensively developed to overcome this major drawback [16]. Plasma oxidation of PDMS is commonly used to convert methyl groups into hydroxyl groups on the surface. Unfortunately, this effect is severely short-lived, and the plasma treated surface returns to hydrophobicity due to the diffusion of uncrosslinked PDMS oligomers from the bulk to the surface. This phenomenon of hydrophobic recovery can be attenuated to some degree. Vickers et al. used solvent extraction to remove the unreacted oligomers prior to plasma oxidation to extend hydrophilic state from 3 h for native PDMS to 7 days [17]. Recently, Park et al. presented a simple and cheap method to create hydroxyl group on the surface by immersing common Pt-cured PDMS in boiling deionized water [18]. Besides surface chemistry analysis, this research also demonstrated the improved cell attachment and proliferation on boiling water treated PDMS.
Another approach to increase cell attachment is coating with extracellular matrix proteins such as fibronectin, collagen, laminin or charged molecules. Wang et al. systematically investigated Caco-2 cell adhesion on different substrates and different surface modifications on PDMS including ECM proteins coating (laminin, fibronectin, collagen and Matrigel) and charged molecules deposition (poly-D-lysine,L-α-phosphatidycholine and layer-by-layer) [19]. The results showed adsorption of fibronectin on PDMS could sufficiently increase cell adhesion to the same level as cell culture treated polystyrene (PS). Furthermore, the amount of attached cells on PDMS with oxygen plasma treatment followed by fibronectin coating was twice as that on the PS surface by the third day. Another concern of the hydrophobic property of PDMS is nonspecific adsorption of molecules. Since cell-based microfluidic devices involve medium flow, proteins and hydrophobic components in medium or even some hydrophobic drugs may attach to the hydrophobic sites of the PDMS channels. The nonspecific absorption may consume nutrients or relay false results from drug screening tests. The nonspecific absorption at undesired locations may also cause fluidic congestion between channels and malfunction of microvalves. More detailed discussions on nonfouling surface chemistries can be found in the literature presented by Zhang et al. [20].
While PDMS has been used widely in construction of biomicrofluidic devices, the manually operative nature of PDMS fabrication becomes a critical limitation of using PDMS in mass production. Alternative materials have been utilized in building biomicrofluidic devices, such as thermoplastic materials (e.g., PMMA: poly methyl-methacrylate, PC: polycarbonate, PTFE: polytetrafluoroethylene, PVDF: polyvinyledene fluoride, and PGS: polyglycerol sebacate) [21–28], paper (e.g., nitrocellulose, off-stoichimetry-thoil-ene(OSTE)) [29–31], and gelatin with a high melting temperature [32]. In addition, hybrid microfluidic devices using PMMA and PDMS would allow devices to be manufactured easily due to PMMA and have some complexity from PDMS features. One such example is the use of PMMA devices with PDMS valves [33]. Though many standard industry manufacture techniques – such as injection molding, lithography, laser ablation, 2 photon polymerization and 3D printing – can be used to fabricate biomicrofluidic devices once these alternative materials are introduced, usually the feature resolution lower than a couple of hundred microns becomes problematic. The currently most promising technique in terms of speed and reliability seems to be stereolithography [23,26,34–35] which can print biocompatible polymers at applicable resolutions (100 µm). Stereolithography, similar to 3D printing, generates layer-by-layer patterns to form a 3D structure [23,35–36]. Stereolithography holds promise as a rapid prototyping and manufacture method for future microfluidic devices.
Microvalves & pumping mechanisms
For cell-based microfluidic HTS platforms, across-linked array is extensively utilized to investigate cellular responses to multiple drugs on different cell-culture chambers at the same time. This makes microvalves that control the route and timing of fluids within a microfluidic device a crucial component in separating different analytes. The microvalves can be actuated by electrostatic, electromagnetic, pneumatic and piezoelectric forces [37]. Among these methods, pneumatic microvalves show greatly promising characteristics and have been widely used in cell-based microfluidic devices because they can be integrated into the soft lithography fabrication process. Pneumatic microvalves exploit an air or liquid channel to control the pressure applied on elastic PDMS membrane. The deformation of the membrane caused by the pressure change will interfere with or permit the fluid flow in the microchannel directly adjacent to it. By integrating microvalves, cell-based microfluidic device has capability to achieve high-throughput analysis. For instance, King et al. used 144 pneumatic microvalves to separate a 16 × 16 cell array into 64 subarrays for monitoring dynamic gene expression in a high-throughput platform [38]. The normally-closed microvalves in rows/columns were actuated by applying negative pressure on the control channel to allow cell seeding/stimulation. In this way, total 64 conditions (8 cell types and 8 soluble stimulus) can be evaluated in quadruplicate at one time. Generally, the choice of using normally-open or normally-closed microvalves is decided by their individual roles in amicrofluidic device.
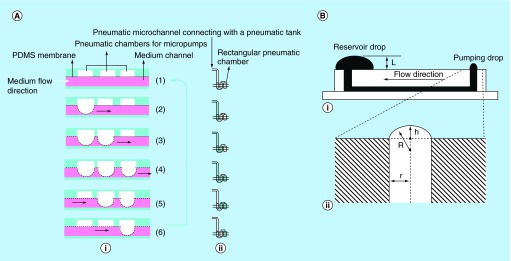
As alluded to previously, providing a continuous medium in a cell-based microfluidic device is necessary due to rapid nutrient consumption. Although medium delivery can be performed by commercial syringe or peristaltic pumps- commonly used in academic labs or small scale studies – they are too costly and bulky for multiplex delivery especially when the screening is scaled up to high throughput. In order to reduce the amount of external hardware required, micropumpsare introduced onto the chip to generate temporal and volumetric fluid movement. Generally, micropumps can be classified into two categories: active and passive micropumps. Pneumatic microvalves working as micropumps show promising properties on controlling multiplex liquid delivery. The working principle of pneumatic pumps is integrating several pneumatic microvalves and actuating them sequentially to generate peristaltic fluid movement in the channels. As a variation to using parallel air channel to actuate a series of microvalves, Wu et al. proposed an S-shaped pneumatic microchannel intersecting a straight liquid flow microchannel underneath to overcome the limitation of throughput of medium delivery due to the inevitable time delay caused by fluidic resistance [39]. As shown in Figure 1A, the fluidic resistance of air (i.e., directional transmission of the pressure change) in the S-shaped channel could generate a sequential rise of pressure to actuate the microvalves at intersections sequentially in an automated fashion. In this study,30 pneumatic micropumps were integrated with one pneumatic tank to simultaneously activate all channels on the cell-based microfluidic device with a uniform pumping rate. Oral cancer cells cultured in 3D agarose gel has been demonstrated to retain 95–98% viability during 48 h of culture in this perfusion-based cell culture platform. Because of the increased demand for use of a great number of microvalves on one chip, recently, Lau et al. integrated 100 microvalves to investigate the response of individual microvalves in different operation configurations such as membrane thickness and driving pressures through experiments, modeling and simulation [40]. This study provides a general guideline for the designs of microvalves integrated high-throughput microfluidic device.
Figure 1. . Working principles of pneumatic micropumps.
(A) The peristaltic sequence of three microvalves in an S-shape pneumatic micropump to pump the fluid forward. (i) Schematic representation of the sequence; (ii) a top view of the pumping motion corresponding to (i). (B) Side view of a passive pump composed of two reservoirs and bridge. (i) Schematic representation of the pump; (ii) the critical parameters involved in the working mechanism of this pump.
(A) Reproduced with permission from [39] © Springer (2007).
(B) Reproduced with permission from [41] © The Royal Society of Chemistry (2002).
Passive micropumps are also promising for HTS applications due to their straightforward implementation and the possibility of tubeless microfluidics design [42]. For instance, Walker et al. proposed a pumping method that using the differential internal pressure of droplets at inlets and outlets to drive fluid flow [41]. Since the internal pressure of a smaller drop at the inlet is higher than a larger drop at the reservoir (outlet), the different pressure gradient in a liquid-filled microchannel causes the fluid to flow toward the reservoir (Figure 1B). Although this strategy reveals the possible application for high-throughput testing based on its adaptability for robotic multipipettes, this design with the open air–liquid interface is sensitive to evaporation which may cause a nonconstant temporal flow rate. In addition, an open device brings more challenges to maintain a sterile cell culture environment on chip.
Detection methods
The detection schemes adaptable to the microfluidic platforms also guides their manufacture. The most common detection schemes are optical, electrochemical, and possibly coupled to a mass spectrometric readout. For example, cell viability determination using MTT assay is commonly used in conventional drug screening. This absorbance-based colorimetric detection can be applied to on-chip detection, and the absorbance change can be measured either by coupling the chip with a plate reader [43] or by a CCD camera [44]. Fluorescence measurement is another common optical mode of detection that is the most commonly used with the microfluidic drug-screening platform. Live-dead dye staining allow cell viability to be visualized directly under microscope [45–50]. Microscope detection is the simplest approach for the platform design, especially for those microscopes that come with a motorized heating stage (e.g., Zeiss AxioOberver Z1), permitting on stage culture of cells and imaging without manual intervention. However, for such detection the sensitivity, resolution and throughput is limited by the microscope's hardware (as well as by limitations posed by the visible wavelength). On the other hand, coupling with a microarray scanner, which is widely used for high-throughput DNA-DNA [51] and DNA-protein [52] screening, may improve the sensitivity and throughput of the on-chip detection. Lee et al., demonstrated a 3D cell array with 560 features for HTS application, and the results correlated with the results from conventional MTT assay [53]. However, it is still challenging to get a high resolution when the microfluidic cell chip becomes more complex to mimic physiologically relevant functionality.
Electrochemistry-based approaches are also common for on-chip detection. Impedimetric detection can be used for cytotoxicity evaluation on chip. The general principle relies on a microelectrode system embedded in each bioreactor region measuring the electrochemical impedance. For instance, adherent cells detaching from the substrate surface due to causes changes in the impedance. Based on this principle, a label-free and real-time measurement could be performed during drug screening and toxicity testing [54–56]. Although this detection approach shows potential for real-time, nonoptical, high-throughput detection that circumvents the need for an automated microscope stage, its requirement of cell type (only suitable for adherent cells) and highly customized circuit design may limit its translation. In addition to cytotoxicity evaluation, drug kinetics and relevant pathway could be detected on the microfluidic platform as well. For example, amperometric measurement can be carried out on the chip for ion channel drug studies [57]. Furthermore, the microfluidic platform can be potentially coupled with mass spectroscopy so that the downstream product of the drug could be detected [58]. In addition, cancer cell secreted molecules can be detected by surface plasmon resonance imaging technology on chip [59]. All these qualities allow the detection on the microfluidic platform to be versatile and comparable with that on conventional HTS platforms.
Advanced microfluidic cell-culture systems on drug screening
Precise concentration gradient generator
Drug screening and therapeutic optimization usually requires various drug concentrations to be investigated for dose-dependent cellular response. Additionally, the combination chemotherapy is widely accepted to have better efficacy and less side effects at a lower dose than single chemotherapy with a high dose. Therefore, the gradient generator becomes a powerful tool in this field. Compared to conventional methods such as Transwell assays and Dunn chambers, microfluidic devices can not only increase throughput and reduce experimental cost but also possess a higher gradient resolution. Moreover, it can be further combined with real-time observation system. In 2004, Hung et al. first developed a microfluidic cell culture array which integrated a concentration gradient generator and optical analysis for long-term cellular monitoring [46]. The established culture environment was believed to be suitable for continuous operation. Not only was cell viability in this device demonstrated, the cultures were also successfully passaged and regrown within the device by introducing trypsin. Although drug testing was not conducted in this study, the platform's ability to provide long-term cell culture potentiates its possible utility in drug screening applications.
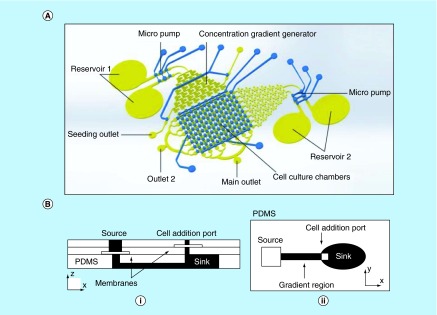
In general, microfluidic-based gradient generators can be classified into two major categories, steady-state and time-evolving. In the steady-state type of microfluidic systems, laminar flow is utilized to mix chemical species at the interfacial regions between parallel fluid streams to generate a gradient. The typical design composes a Y-junction configuration where the chemical solution flow and buffer flow converge together to produce gradients which are perpendicular to flow direction. A more controllable gradient module can be created by integrating multiple cascaded-mixing stages in a stepwise manner. Based on this method, arbitrary gradients such as linear, logarithmic and Gaussian gradients have been developed by a mathematical model which was established as an analog of the equivalent electrical model and verified by fluorescence measurements [60]. A Christmas-tree design has also been broadly applied to studies of cellular response to drug concentration gradients [45,48,50,57]. Recently, An et al. developed a fully automated microfluidic system for high-throughput drug screening in the combinational chemotherapy [49]. In order to evaluate the effect of combinational treatment of TRAIL and curcumin as a sensitizer in PC3 human prostate cancer cell line, an eight-by-eight cell chamber was connected to two orthogonal concentration gradient generators, giving sixty-four pair-wise concentration combinations of drug and sensitizer that could be tested at the same time (Figure 2A). The LC50 value of curcumin for chemosensitizer treated with TRAIL in microfluidic device showed that the drug treatment performed in themicrofluidic device were more sensitive than in a 96-well plate due to frequent replenishment of fresh media and drug solution. Unlike the conventional utilization of gradient generator by connecting to cell array directly, Wang et al. connected the outputs of the Christmas-tree generator as vertical injection ports to the flow of buffer solution in the main channel [61]. This convection-driven microfluidic device created a linear gradient profile which could be tuned just by changing the flow rate.
Figure 2. . Two types of microfluidic-based gradient generators.
(A) An automated microfluidic array integrating two ‘Christmas-tree’ concentration gradient generators. (B) (i) Side and (ii) top representation of a time-evolving static gradient generator.
(A) Reproduced with permission from [49] © The Korean Society of Applied Pharmacology (2014).
(B) Reproduced with permission from [62] © The Royal Society of Chemistry (2006).
The second type of gradient generator is usually classified as a time-evolving static gradient generator. As shown in Figure 2B, the general configuration of this type of microfluidic system includes two reservoirs with different concentrations as source and sink. The gradient is passively developed in the connecting channel between these two compartments by diffusion under quasi-static conditions. Because of the absence of fluid flow, incorporation of cell array and this type of gradient generator has less impact on shear stress. In addition, more signaling factors secreted by cells could be retained [62]. However, the concentration resolution of this type of gradient generator is limited by the volume of chambers.
Mimicked physiological environment
2D cell microarrayswere first developed for high-throughput protein identification by transfected cells without fluidic components [63]. Several versions of 2D microfluidic cell arrays were developed to meet the need of screening multiple compound screening on a single chip [38,64]. However, more studies have decisively demonstrated that cells cultured in monolayer conditions have different behaviors than in vivo phenotypes. Indeed, cells are subject to the properties and dynamics of surrounding extracellular matrix, including biochemical and mechanical cues from their native environment through soluble factors, cell–cell, and cell–stromal interactions [65]. Recently, some 3D culture technologies such as cell encapsulation in gel or spheroid culture were used to construct a mimicked microenvironment. 3D cell microarrays without fluidics have been reported by encapsulating cells in matrix followed by 3D cell droplets using a robotic spotter [47].
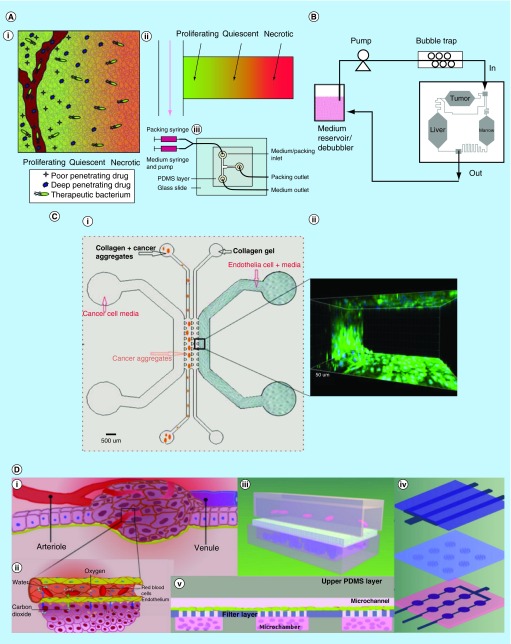
Combining microfluidics with 3D cell matrix, a more physiologically realistic microenvironment can be established. For instance, Walsh et al. have proposed a simple concept to mimic the microenvironment gradients present in tumors [66]. A relatively long flow channel was connected along one side of cell chamber where spheroidal human LS174T colon carcinoma cells were inserted (Figure 3A). This established a controllable linear gradient along the distance apart from flow channel to imitate the conditions found in blood vessels surrounding tumors. With continuously flowing media, viable cells (proximal to the channel), apoptotic and necrotic cells (distal into the chamber) were found which corresponded to theoretically surmised regions of proliferating, quiescent and necrotic cells, respectively. In addition, the pH decreased with positions away from the channel. The diffusion coefficient of doxorubicin was accurately measured in the device and the value perfectly matched with previously clinical studies and mathematical models. Compared to conventional 2D uniform culture condition, the device provides an inherent gradient microenvironment for determining the efficacy of an anticancer drug limited by its penetration depth.
Figure 3. . Microfluidic devices with physiologically mimicked microenvironment.
(A) A microfluidic device mimic the microenvironment surrounding blood vessels in tumors. (i) Nutrient and waste gradients away from vessels; (ii) the corresponding microenvironment gradients which can be created in the microfluidic device; (iii) top view of the device. (B) A micro cell culture analog reproduces a mimicked multiorgan environment. Three cell chambers are connected by channels mimicking blood flow with the specific retained time for each chamber. (C) A microfluidic system co-cultured endothelial cells and cancer cell spheroids in a mimicked tumor microenvironment. (i) The schematic illustration of the microfluidic system; (ii) the photograph of 3D projection represents an endothelial monolayer at the interface. (D) A microfluidic array reconstructed a 3D tumor microenvironment with cancer cells and microvascular endothelial cells along vertical axis. (i) Schematic representation of tumor microenvironment; (ii) nutrient and gas transport between microvessels and tumor cells; (iii) schematics of the microfluidic system; (iv) schematics of each layer of the device; (v) cross-section view of the device.
(A) Adapted with permission from [66] © The Royal Society of Chemistry (2009).
(B) Reproduced with permission from [5] © The Royal Society of Chemistry (2009).
(C) Reproduced with permission from [7] © American Chemical Society (2014).
(D) Reproduced from [6] http://pubs.acs.org/doi/full/10.1021/ac403899j © American Chemical Society (2014).
In order to analyze the pharmacological effect and complex process of drug behavior from absorption, distribution, metabolism and elimination, Sung et al. developed a microfluidic device with 3D hydrogel cell cultures to reproduce multiorgan interactions [5]. The device contained three distinct compartments for liver, colon cancer, and bone marrow cells. Each chamberwas interconnected with channels mimicking the blood flow pattern in human – the various chamber sizes and channel lengths were designed to match the physiological blood residence time of each organ type as shown in Figure 3B. Colon cancer cells and hepatoma cells were embedded in Matrigel. Myeloblasts were encapsulated in alginate due to their greater mobility. Tegafur, an oral prodrug of the cancer drug 5-fluorourail, revealed a cytotoxic effect as tested on the device. The results were consistent with previously published animal and human clinical studies. In contrast, the viability of colon cancer cell line was not affected when the testing was performed in a 96-well microtiter plate. It demonstrates that this kind of microfluidic devices can provide a more realistic environment than conventional static assay to emulate the physiologically expected behavior.
At the cellular scale, the local microenvironment can also be recapitulated in microfluidic devices. Recently, many researchers continue to combine the 3D cell culture microenvironment and co-culture technique as a biomimetic strategy. Niuet al. developed a microfluidic device supporting co-culture of endothelial and cancer cells to validate the antimetastatic effect of 12 native compounds [7]. In this device, four parallel channels were separated by trapezoidal posts with 100–125 μm spacing in the reaction region (Figure 3C). Rat tail collagen was formed in the central channel as ECM which also supported the attachment of endothelial cells on a channel running adjacent to the collagen channel. On the other side of the collagen channel, spheroids of human lung carcinoma cell line A549 were mixed with collagen solution. Cancer cell medium was supplied through the channel next to the channel of cancer aggregates. Drug molecules and secreted stimulants from endothelial cells could diffuse into collagen matrix and then affect cancer cells. The dispersion of cancer cell spheroids was quantified to evaluate drug efficacy. The authors picked three compounds to evaluate their on-chip and off-chip antimetastatic effect. The on-chip antimetastatic or antiangiogenic effect of those drugs resonated with the results of respective biological assays performed in the conventional 3D environment while contrasting to dissimilar behaviors found in 2D culture environment. It demonstrates that the physiologically relevant microenvironment created in this kind of microfluidic device is necessary to reproduce in vivo behavior.
The interorgan interaction device [5] discussed above can provide basic physiologically relevant results from the activity of cells representing different organs, it becomes a challenge to scale it up to HTS applications. Similarly, the device used for metastatic studies [7], while providing insightful results on the antimetastic efficiencies of drugs, does not provide the most amenable configuration to be adopted to a HTS platform. Meanwhile, our group also developed a 3D microfluidic cell array (3D μFCA)to reconstruct the in vivo spatial relationship between microvessels and cancer cells embedded in extracellular matrix by a three-layered PDMS assembly [6]. The cell array architecture in this device is amenable to high-throughput screening as well. As shown in Figure 3D, the three-layer design enables 3D hydrogel-encapsulated cell culture in an array of microchambers running next to membrane-separated microchannels seeded with endothelial cells (HMVEC) to serve as bioartificial blood vessels. The microfluidic cell array realizes the diffusion process for the transportation of nutrients, metabolic waste products, and other molecules by way of its design. In this study, real-time visualization and quantitative analysis of the apoptotic response to four anticancer drugs via caspase-3 activities in PC9 cells co-cultured with HMVECs were recorded in single experiment on single chips. The results highlightthe system's potential for high-throughput ex vivo drug screening in a mimicked vascular microenvironment.
Organs-on-chips
Devices categorized as ‘organs-on-chips’ focus on the development of physiologically organ-level functions on a microfluidic device [67–71]. A human lung-on-a-chip was developed to mimic the cyclic mechanical strain on cells during breathing motion [68]. The alveolar-capillary interface was recapitulated through two channels separated by a thin, porous and flexible ECM-coated membrane. The upper channel was lined with human lung alveolar epithelial cells and the lower channel was lined with lung microvascular endothelial cells. Two full-height channels located at the both sides of cell channels were applied with cyclic suction to provide the rhythmical deflection of PDMS side wall and the porous membrane with cells. The inflammatory responses of lung to airborne nanoparticles was imitated through the introduction of silica nanoparticle. The similar effects of on-chip mechanical strain on nanoparticle absorption was also observed in whole mouse lung. The same group further developed a disease model of drug toxicity-induced pulmonary edema on this chip [72]. A similar device structure could also be used for mimicking physiological peristaltic motions of the human intestine [69]. The cyclic mechanical strain on the chip showed the ability to promote the formation of 3D villi-like structure and a high-integrity barrier. Moreover, it demonstrated the ability of long-term co-culture with microbial flora that normally lives in human intestine without decreasing epithelial cell viability.
Recent studies also show the potential amplification of drug efficacy assessment [73]. Table 1 listed recent studies about organs-on-chip which have potential ability for drug testing. A human kidney proximal tubule-on-a-chip was developed by constructing luminal and interstitial flow channels separated by porous membrane [74]. The toxicity of cisplatin, a known proximal tubule nephrotoxin and chemotherapeutic drug, was tested on the chip and the results were similar to the effects seen in most patients. Although an organ or a tissue-level functionality and features can be mimicked on these organs-on-chips, most the chips are only designed for single drug testing. Therefore, approaches to improve the sample size per chip to gear towards high-throughput screening should be considered for the future generations of these devices. Recently, Agarwal et al. developed a heart-on-chip with a potential high-throughput design [75]. The chip contains 50 thin film cantilevers lined with anisotropic rat cardiac microtissues to recapitulate the laminar architecture of heart ventricle. During muscle contraction, diastolic and systolic stresses could be calculated from the deflection of these cantilevers. The detected inotropic effect of isoproterenol was consistent with previous studies in rat heart. A more detail discussion about organs-on-chips studies can be found in a recently published review paper [76].
Table 1. . Organs-on-chip for drug testing.
| Organ | Device description | Tested drug/molecules | 2D vs 3D culture | Co-culture | Signal detection method | Scalable to HTS | Ref. |
|---|---|---|---|---|---|---|---|
| Liver |
Artificial liver sinusoid was created with a microfluidic endothelial-like barrier which has mass transport properties similar to the liver acinus |
Diclofenac |
2D |
No |
Fluorescent microscope |
No |
[67] |
| Heart (rat) |
50 muscular thin films (MTFs): sub millimeter sized thin film cantilevers of soft elastomers. Anisotropic cardiac microtissues which recapitulate the laminar architecture of the heart ventricle are engineered on these cantilevers |
Isoproterenol |
2D |
No |
Stereo-microscope |
Yes |
[75] |
| Lung |
1. Two closely apposed microchannels separated by a thin, porous, flexible membrane. (upper: epithelial cells; lower: endothelial cells)2. Two side channels for vacuum-mimic breathing |
No |
2D |
Human alveolar epithelial cells and microvascular endothelial cells |
Fluorescent microscope |
No |
[68,72] |
| Bone marrow |
Culture of living marrow with a functional hematopoietic niche in vitro by first engineering new bone in vivo, removing it and perfusing it with culture medium in a microfluidic device |
Radiation countermeasure drugs |
3D (engineered bone marrow) |
No |
Off-chip flow cytometric analysis |
No |
[70] |
|
in vitro model of BBB |
The device has two perpendicularly-crossing channels to introduce dynamic flows, a porous membrane at the intersection of the flow channels for cell culture, and multiple embedded electrodes to monitor TEER across the barrier |
Histamine, fluorescent tracer for permeability |
2D |
Endothelial and astrocytic cells |
TEER electrodes, fluorescent microscope |
No |
[77] |
| Gut |
Similar as the structure of lung-on-a-chip (same group) |
Microbial studies |
2D |
No for device but can co-culture normal intestinal microbe (Lactobacillus rhamnosus GG) |
Fluorescent microscope (transport), off-chip OD measurement for microbial studies |
No |
[69] |
| Renal (rat) |
Kidney epithelial cells were cultured on the upper surface of an extracellular matrix-coated, porous membrane that splits the main channel of the device into two adjacent channels, thereby creating an apical ‘luminal’ channel and a basal ‘interstitial’ space |
Water and Na uptake after hormonal stimulations of vasopressin and aldosterone |
2D |
No |
Fluorescent microscope |
No |
[73] |
| Kidney | Same as above | Cisplatin toxicity and PGP efflux transporter activity | 2D | No | Fluorescent microscope | No | [74] |
BBB: Blood–brain barrier; OD: Optical density; TEER: Transendothelial electrical resistance.
Devices with increased throughput & mimicked physiological environment
As aforementioned, the most common strategy to construct a biomimetic microenvironment is to introduce compatible 3D matrices. Most of microfluidic-based gradient generators are only applied to 2D cell culture for drug testing. Recent research has begun to acclimate integration of microfluidic gradient generators to a 3D cell culture platform to increase the throughput of drug screening within a more realistic environment instead of traditional 2D culture [49,78]. Unlike the direct combination of microfluidic gradient generators to 2D cell culture array, Occhetta et al. introduced a branched channel from the connection between the output of gradient generator and culture area for the generation and culture of 3D micromasses of adult hBM-MSCs under continuous and controlled laminar flow perfusion [79]. The secondary channels were used as waste lines to avoid cell clogging in the upstream channels, as well as to reduce shear stress experienced by cells.
On the other hand, in order to increase the throughput of drug screening, the capability of automated operation is another goals. Recently, some microfluidic devices were invented as commercial microplate-reader compatible format or allowing the incorporation into automated liquid handlers with 3D culture platform [80–82]. For instance, Huang et al. developed a three-layer microfluidic cell culture chip in which the waste medium collector module could be removed for directly subsequent bioassay using a commercial microplate reader.
To build up a more realistic physiological environment usually there is a need to utilize the complex arrangement, compartmentalization, and design networks and chambers as mentioned in above sections. However, the spatial limitation of replicates is a big challenge for scaling up the throughput. One of the practical designs for high-throughput drug testing is arranging the layout of compartments along vertical axis and positioning each unit with mimicked physiological environment in a horizontal array format. For instance, as in our current design of the 3D μFCA, the in vivo spatial relationship between microvessels and cancer cells is imitated by setting up the compartments of cancer cells below the arrayed channels of endothelial cells. However, the throughput of the current design is still limited by the parallel configuration of the artificial vascular channels and the channels connecting the chambers for cancer cells. By simply changing the bonding orientation between the top microchannel layer and the bottom microchamber layer from its current parallel configuration to an orthogonal alignment, and integrating the techniques of microvalve and tubeless cell seeding, the second generation of 3D μFCA will be amendable for high-throughput drug screening with closely mimicked 3D microenvironment.
Commercialized microfabricated cell chips
With rapid growing of an emphasis on translational research and more matured bio-microfabrication technologies, currently in the USA alone there are over 20 companies developing microfabricated chips for DNA/RNA/protein biochemical assay instruments, at least 20 BioMEM outsourcing/prototyping companies, over ten microfluidic chip companies for infectious disease diagnostics, three companies using microchips for in vivo drug delivery, at least five microchip-based cancer cell/biomarker diagnostic companies and about 10 companies working on microfabricated cell chips for drug screening and discovery [83], from which eight are listed in Table 2 with brief descriptions of the device features and functions. As shown in Table 2, the cell responses to drugs on these microfabricated cell chips are through image detection of fluorescence signals. Quantitative fluorescence image analysis software is an essential and critical part to obtain biologically accurate and quantitative data for a drug testing report. Due to the complexity of 3D culture, quantitative 3D fluorescence image analysis especially its automation over long dynamic drug testing periods remains a challenge in the field.
Table 2. . Commercialized microfabricated cell chips.
| Companyname | Brief device description | 2D vs 3D culture | Structured co-culture | Signal detection method | Device materials | Scalable for mid† to high‡ throughput screening | Static or Perfusive | One example of highlighted functions | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| ACEA Biosciences |
Multiple-well format with embedded gold electrodes using impedance measurement to detect cell proliferation and migration |
2D culture |
No |
Impedance |
Mutiple well plastic culture plates |
Yes |
Static |
Label-free cell analysis |
[84] |
| AIM Biotech |
3 microchannels side-by-side separated by micropilars with the center channel for 3D hydrogel culture and two side channels for medium feeding; three units on a microscope slide (75 mm × 25 mm) |
3D (center); 2D (side channels) |
Yes |
Fluorescence by microscopic imaging |
Gas permeable polymer on the bottom |
Difficult |
Perfusive |
Angiogenesis study in tumor |
[3] |
| Bell Brook Labs |
Multiple microchannels connected with passive pumping wells enabled by surface tension |
2D or 3D |
No |
Fluorescence by microscopic imaging |
192 microchannels from PMMA or PDMS |
Mid throughput |
Static |
iuvoMicroconduit Array Platform |
[85] |
| Cellasic bought by Millipore |
CellASIC® ONIX Microfluidic Plates, 4 independent open wells on a 96-well plate with multiple inlets for each well |
3D or 2D |
No |
Fluorescence by microscopic imaging |
Glass bottom |
Difficult |
Perfusive |
Compatible for mammalian, yeast and bacteria culture |
[86] |
| Cytoo |
3D cell culture matrix created by micropatterning |
3D |
Possible |
Fluorescence by microscopic imaging |
Glass bottom |
Possible |
Static |
3D cell culture on patterned surface |
[87] |
| Celsee Diagnostics |
On-chip circulating tumor cell (CTC) capture, grow and gene-level analysis |
Similar to suspension culture |
No |
Fluorescence by microscopic imaging |
Plastic chip |
Difficult |
Perfusive |
CTC |
[88] |
| Fluxion Biosciences |
6∼24 samples per multiple-well plate format with flow, could handle adhesive cells and liquid samples |
2D culture |
No |
Fluorescence by microscopic imaging |
Glass bottom |
Difficult |
Perfusive |
Flow shear stress study on cells |
[89] |
| Xona microfluidics | 2 channels separated by different width of grooves allowing axons and dendrites to be fluidically isolated from cell bodies | 2D culture | Possible | Fluorescence by microscopic imaging | PDMS chambers with glass bottom | Difficult | Static or short term Perfusive | Neuron cell culture with microgrooves axons and dendrites separated from cell body | [90] |
†Mid throughput: a couple of hundred compounds.
‡High-throughput: a couple of thousand compounds.
Conclusion & future perspective
Currently a robotic liquid handling system coupled with biochemical assays (e.g., live/dead, apoptosis markers, O2 uptake, etc.) usually uses 96, 384 or 1536-well plates for 2D cell culture and 96 or 384-well plates for 3D spheroid culture without 3D matrixin HTS and high content screening (HCS). Expensiveness of this robotic system and lack of mimicked 3D microenvironment are two major disadvantages which actually have been motivating the development of microfluidic 3D cell chips for HTS and HCS. The intrinsic property of microfabrication will enable an easy scaling-up of the current 3D microfluidic cell chips to accommodate 3D cell culture in several hundred microchambers. The challenge following the scaling-up is to reduce the number of tubing for massive input and output channels caused by hundreds of compounds during screening and complicated flow patterns mimicking in vivo drug transport. A tubing-free 3D microfluidic cell chip is ideal. In general, there is a dilemma between the desire of making a 3D microfluidic cell chip simple (e.g., open-device to simplify 3D cell seeding and minimize tubing) and enabling complicated on-chip features to mimic in vivo microenvironment (e.g., 3D cell encapsulation culture with perfusion and diffusion in a mid-to-high-throughput format). A good balance point is the key to successfully push a device into the commercialization direction.
The next generation of 3D microfluidic cell chips should take full advantages of its own on-chip microcirculation property to maximally simulate in vivo microenvironment that not only includes all types of epithelial, endothelial and mesenchymal cells with proper ECM but also blood cells in flow. Thus, a 3D microfluidic cell chip will be able to be applied in drug screening/discovery targeting immune diseases and immunotherapies, such as immunotherapy for cancer. One bioengineering obstacle must be solved here is how to keep white blood cells live and healthy in on-chip circulating flow.
With mimicked close-to-in vivo microenvironment and organ-on-chip designs, there is no doubt that 3D microfluidic cell chips will increase the in vitro drug screening accuracy that in turn would reduce the drug failing rate through clinical trials in the near future. The social benefit of using 3D microfluidic cell chips in drug discovery and screening is to reduce the cost of new drug development by accurate in vitro screening and thus ultimately reduce healthcare expenses. On the other hand, with improved resolutions of advanced 3D printing technologies such as stereolithography using biocompatible materials more and more microchips from research labs will be able to get into production lines and eventually provide tools to advance pharmaceutical and biomedical fields.
Executive summary.
Introduction
Commercial high-throughput drug screening lacks the ability to reproduce in vivo-like cell response to drugs, which limits its translatability to clinical drugs.
The physiologically relevant microenvironment for cell culture can be more faithfully replicated by a microfluidic system.
Advantages of using a microfluidic cell culture platform in HTS
The microscale size of microfluidic device robustly reduces the required volume of reagents.
Mature manufacturing technology enables the flexibility of the platform for a wide spectrum of applications.
Critical factors on designing a microfluidic cell-culture platform
- Biocompatible materials & surface modifications
- – Polydimethylsiloxane (PDMS) is the majority of the materials used for platform design because it is gaspermeable, optically clear and compatible with soft lithography.
- – Due to the hydrophobicity of PDMS, surface modification is necessary for cell-culture applications.
- – The techniques to modify PDMS surface includes plasma treatment, solvent extraction and extracellular matrix (ECM)-coating.
- – Since PDMS has a limitation in mass production, alternative materials such as PMMA were investigated. Hybrid PMMA-PDMS microfluidic devices are also discussed.
- – Stereolithography shows promising properties as a rapid prototyping and manufacture method for future devices.
- Microvalves & pumping mechanisms
- – For HTS applications, crosslinked channels need to be controlled by the microvalves. Pneumatic microvalves are widely used for this purpose.
- – To supply continuous media and nutrients, pump should be included in the design. Passive pump may be advantageous compared with active pump as it provides the possibility to build up a tubing-less system.
- Detection methods
- – Currently available detection methods, such as absorbance, fluorescence, electrochemistry or mass spectroscopy, can be applied to microfluidic cell-culture platforms.
- – Fluorescent detection is the most common method for detection on cell-based microchips.
- – Impedimetric detection provides another method for real-time, high-throughput and label-free detection.
Advanced microfluidic cell-culture systems on drug screening
- Precise concentration gradient generator
- – Creating a concentration gradient of a certain drug on chip is essential for drug screening, and this can be achieved via the channel design.
- – Steady-state and time-evolving generators are the two major types of microfluidic-based gradient generators.
- Mimicked physiological environment
- – A microenvironment close to the physiological condition can be established on chip with 3D culturing techniques.
- – Co-culture with 3D microenvironment can be also achieved on microfluidic platforms, which may provide more accurate outcome for drug screening applications.
Organs-on-chips
- Devices with increased throughput and mimicked physiological environment
- – Spatial implication of replicates is the main challenge for improving the throughput, but this may be resolved via rearrangement on the vertical compartments.
- Commercialized microfabricated cell chips
- – In the US, currently over 60 companies work on the applications of microfluidic devices.
Perspective, future directions & conclusions
How to reduce the numbers of connected tubing is the major issue for scaling up the microfluidic cell culture platforms.
A balance between simplifying the design and enabling on-chip complicated features for mimicking in vivo microenvironment is required for the translation from lab-devices to industry/clinical tools.
Advanced 3D printing technologies with fine resolutions such as stereolithography enable fast paces to transfer a microchip from a research lab to industry.
Footnotes
Financial & competing interests disclosure
This work was partially supported by NIH/NCI 1U54CA137788 (CCNY-MSKCC Partnership), NSF CBET-1055608, and Pershing Square Sohn Cancer Research Alliance. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.FDA Issues Advice to Make Earliest Stages Of Clinical Drug Development More Efficient. www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/2006/ucm108576.htm
- 2.Macarron R, Banks MN, Bojanic D, et al. Impact of high-throughput screening in biomedical research. Nat. Rev. Drug Discov. 2011;10(3):188–195. doi: 10.1038/nrd3368. [DOI] [PubMed] [Google Scholar]; • Gives a broad concept and detailed consideration about high-throughput screening.
- 3.AIM Biotech. www.aimbiotech.com/chips.html
- 4.3D Biotek Perfusion Bioreactor System with Pump. www.sigmaaldrich.com/catalog/product/aldrich/z755354?lang=en®ion=US
- 5.Sung JH, Shuler ML. A micro cell culture analog (mu CCA) with 3-D hydrogel culture of multiple cell lines to assess metabolism-dependent cytotoxicity of anti-cancer drugs. Lab Chip. 2009;9(10):1385–1394. doi: 10.1039/b901377f. [DOI] [PubMed] [Google Scholar]
- 6.Dereli-Korkut Z, Akaydin HD, Ahmed AHR, Jiang XJ, Wang SH. Three dimensional microfluidic cell arrays for ex vivo drug screening with mimicked vascular flow. Anal. Chem. 2014;86(6):2997–3004. doi: 10.1021/ac403899j. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The system has potential for high-throughput ex vivo drug screening in a mimicked vascular microenvironment.
- 7.Niu Y, Bai J, Kamm RD, Wang Y, Wang C. Validating antimetastatic effects of natural products in an engineered microfluidic platform mimicking tumor microenvironment. Mol. Pharm. 2014;11(7):2022–2029. doi: 10.1021/mp500054h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mehta G, Mehta K, Sud D, et al. Quantitative measurement and control of oxygen levels in microfluidic poly(dimethylsiloxane) bioreactors during cell culture. Biomed. Microdevices. 2007;9(2):123–134. doi: 10.1007/s10544-006-9005-7. [DOI] [PubMed] [Google Scholar]
- 9.Zheng B, Roach LS, Ismagilov RF. Screening of protein crystallization conditions on a microfluidic chip using nanoliter-size droplets. J. Am. Chem. Soc. 2003;125(37):11170–11171. doi: 10.1021/ja037166v. [DOI] [PubMed] [Google Scholar]
- 10.Yamashita T, Tanaka Y, Idota N, Sato K, Mawatari K, Kitamori T. Cultivation and recovery of vascular endothelial cells in microchannels of a separable micro-chemical chip. Biomaterials. 2011;32(10):2459–2465. doi: 10.1016/j.biomaterials.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 11.Zheng XT, Yu L, Li PW, et al. On-chip investigation of cell-drug interactions. Adv. Drug Del. Rev. 2013;65(11–12):1556–1574. doi: 10.1016/j.addr.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 12.Gates BD, Xu QB, Stewart M, Ryan D, Willson CG, Whitesides GM. New approaches to nanofabrication: molding, printing, and other techniques. Chem. Rev. 2005;105(4):1171–1196. doi: 10.1021/cr030076o. [DOI] [PubMed] [Google Scholar]
- 13.Byun I, Park J, Kim J, Kim B. Fabrication of PDMS Nano-stamp by replicating Si Nano-moulds fabricated by interference lithography. Proc. Prec. Eng. Nanotechnol. (Aspen2011) 2012;516:25–29. [Google Scholar]
- 14.Lei KF. Microfluidics in Detection Science: Lab-on-a-chip Technologies. The Royal Society of Chemistry; Cambridge, UK: 2015. Materials and fabrication techniques for nano- and microfluidic devices; pp. 1–28. [Google Scholar]
- 15.Palchesko RN, Zhang L, Sun Y, Feinberg AW. Development of polydimethylsiloxane substrates with tunable elastic modulus to study cell mechanobiology in muscle and nerve. PLoS ONE. 2012;7(12):e51499. doi: 10.1371/journal.pone.0051499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou JW, Khodakov DA, Ellis AV, Voelcker NH. Surface modification for PDMS-based microfluidic devices. Electrophoresis. 2012;33(1):89–104. doi: 10.1002/elps.201100482. [DOI] [PubMed] [Google Scholar]
- 17.Vickers JA, Caulum MM, Henry CS. Generation of hydrophilic poly(dimethylsiloxane) for high-performance microchip electrophoresis. Anal. Chem. 2006;78(21):7446–7452. doi: 10.1021/ac0609632. [DOI] [PubMed] [Google Scholar]
- 18.Park JY, Ahn D, Choi YY, et al. Surface chemistry modification of PDMS elastomers with boiling water improves cellular adhesion. Sensors and Actuators B-Chemical. 2012;173:765–771. [Google Scholar]
- 19.Wang L, Sun B, Ziemer KS, Barabino GA, Carrier RL. Chemical and physical modifications to poly(dimethylsiloxane) surfaces affect adhesion of Caco-2 cells. J. Biomed. Mater. Res. A. 2010;93a(4):1260–1271. doi: 10.1002/jbm.a.32621. [DOI] [PubMed] [Google Scholar]
- 20.Zhang HB, Chiao M. Anti-fouling coatings of poly(dimethylsiloxane) devices for biological and biomedical applications. J. Med. Biol. Eng. 2015;35(2):143–155. doi: 10.1007/s40846-015-0029-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, Bettinger CJ, Langer RS, Borenstein JT. Biodegradable microfluidic scaffolds for tissue engineering from amino alcohol-based poly(ester amide) elastomers. Organogenesis. 2010;6(4):212–216. doi: 10.4161/org.6.4.12909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Domachuk P, Tsioris K, Omenetto FG, Kaplan DL. Bio-microfluidics: biomaterials and biomimetic designs. Adv. Mater. 2010;22(2):249–260. doi: 10.1002/adma.200900821. [DOI] [PubMed] [Google Scholar]
- 23.Waldbaur A, Rapp H, Lange K, Rapp BE. Let there be chip-towards rapid prototyping of microfluidic devices: one-step manufacturing processes. Anal. Methods. 2011;3(12):2681–2716. [Google Scholar]
- 24.Ren K, Zhou J, Wu H. Materials for microfluidic chip fabrication. Acc. Chem. Res. 2013;46(11):2396–2406. doi: 10.1021/ar300314s. [DOI] [PubMed] [Google Scholar]
- 25.Cabodi M, Choi NW, Gleghorn JP, Lee CSD, Bonassar LJ, Stroock AD. A microfluidic biomaterial. J. Am. Chem. Soc. 2005;127(40):13788–13789. doi: 10.1021/ja054820t. [DOI] [PubMed] [Google Scholar]
- 26.Tseng P, Murray C, Kim D, Di Carlo D. Research highlights: printing the future of microfabrication. Lab Chip. 2014;14(9):1491–1495. doi: 10.1039/c4lc90023e. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez-Rivera V, Weidner JW, Yost MJ. Three-dimensional biomimetic technology: novel biorubber creates defined micro- and macro-scale architectures in collagen hydrogels. J. Vis. Exp. 2016;12(108):e53578. doi: 10.3791/53578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bettinger CJ, Weinberg EJ, Kulig KM, et al. Three-dimensional microfluidic tissue-engineering scaffolds using a flexible biodegradable polymer. Adv. Mater. (Deerfield Beach, Fla.) 2005;18(2):165–169. doi: 10.1002/adma.200500438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yetisen AK, Akram MS, Lowe CR. Paper-based microfluidic point-of-care diagnostic devices. Lab Chip. 2013;13(12):2210–2251. doi: 10.1039/c3lc50169h. [DOI] [PubMed] [Google Scholar]
- 30.Cate DM, Adkins JA, Mettakoonpitak J, Henry CS. Recent developments in paper-based microfluidic devices. Anal. Chem. 2015;87(1):19–41. doi: 10.1021/ac503968p. [DOI] [PubMed] [Google Scholar]
- 31.Hansson J, Yasuga H, Haraldsson T, Van Der Wijngaart W. Synthetic microfluidic paper: high surface area and high porosity polymer micropillar arrays. Lab Chip. 2016;16(2):298–304. doi: 10.1039/c5lc01318f. [DOI] [PubMed] [Google Scholar]
- 32.Paguirigan A, Beebe DJ. Gelatin based microfluidic devices for cell culture. Lab Chip. 2006;6(3):407–413. doi: 10.1039/b517524k. [DOI] [PubMed] [Google Scholar]
- 33.Zhang W, Lin S, Wang C, et al. PMMA/PDMS valves and pumps for disposable microfluidics. Lab Chip. 2009;9(21):3088–3094. doi: 10.1039/b907254c. [DOI] [PubMed] [Google Scholar]
- 34.Au AK, Lee W, Folch A. Mail-order microfluidics: evaluation of stereolithography for the production of microfluidic devices. Lab Chip. 2014;14(7):1294–1301. doi: 10.1039/c3lc51360b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang HW, Cho DW. Development of an indirect stereolithography technology for scaffold fabrication with a wide range of biomaterial selectivity. Tissue Eng. C Methods. 2012;18(9):719–729. doi: 10.1089/ten.tec.2011.0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khalil S, Sun W. Bioprinting endothelial cells with alginate for 3D tissue constructs. J. Biomech. Eng. 2009;131(11):111002. doi: 10.1115/1.3128729. [DOI] [PubMed] [Google Scholar]
- 37.Au AK, Lai HY, Utela BR, Folch A. Microvalves and micropumps for BioMEMS. Micromachines-Basel. 2011;2(2):179–220. [Google Scholar]
- 38.King KR, Wang SH, Irimia D, Jayaraman A, Toner M, Yarmush ML. A high-throughput microfluidic real-time gene expression living cell array. Lab Chip. 2007;7(1):77–85. doi: 10.1039/b612516f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu MH, Huang SB, Cui ZF, Cui Z, Lee GB. A high throughput perfusion-based microbioreactor platform integrated with pneumatic micropumps for three-dimensional cell culture. Biomed. Microdevices. 2008;10(2):309–319. doi: 10.1007/s10544-007-9138-3. [DOI] [PubMed] [Google Scholar]
- 40.Lau ATH, Yip HM, Ng KCC, Cui X, Lam RHW. Dynamics of microvalve operations in integrated microfluidics. Micromachines-Basel. 2014;5(1):50–65. [Google Scholar]
- 41.Walker GM, Beebe DJ. A passive pumping method for microfluidic devices. Lab Chip. 2002;2(3):131–134. doi: 10.1039/b204381e. [DOI] [PubMed] [Google Scholar]
- 42.Pedro JR, David JB, Justin CW. Microfluidics and Nanotechnology. CRC Press; FL, USA: 2014. A review of tubeless microfluidic devices; pp. 221–264. [Google Scholar]
- 43.Chen YL, Gao D, Liu HX, Lin S, Jiang YY. Drug cytotoxicity and signaling pathway analysis with three-dimensional tumor spheroids in a microwell-based microfluidic chip for drug screening. Anal. Chim. Acta. 2015;898:85–92. doi: 10.1016/j.aca.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 44.Park J, Yoon TH. Microfluidic image cytometry (mu FIC) assessments of silver nanoparticle cytotoxicity. Bull. Korean Chem. Soc. 2012;33(12):4023–4027. [Google Scholar]
- 45.Ye NN, Qin JH, Shi WW, Liu X, Lin BC. Cell-based high content screening using an integrated microfluidic device. Lab Chip. 2007;7(12):1696–1704. doi: 10.1039/b711513j. [DOI] [PubMed] [Google Scholar]
- 46.Hung PJ, Lee PJ, Sabounchi P, Lin R, Lee LP. Continuous perfusion microfluidic cell culture array for high-throughput cell-based assays. Biotechnol. Bioeng. 2005;89(1):1–8. doi: 10.1002/bit.20289. [DOI] [PubMed] [Google Scholar]
- 47.Flaim CJ, Chien S, Bhatia SN. An extracellular matrix microarray for probing cellular differentiation. Nat. Methods. 2005;2(2):119–125. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- 48.Englert DL, Manson MD, Jayaraman A. Flow-based microfluidic device for quantifying bacterial chemotaxis in stable, competing gradients. Appl. Environ. Microbiol. 2009;75(13):4557–4564. doi: 10.1128/AEM.02952-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.An D, Kim K, Kim J. Microfluidic system based high throughput drug screening system for Curcumin/TRAIL combinational chemotherapy in human prostate cancer PC3 cells. Biomol. Ther. (Seoul) 2014;22(4):355–362. doi: 10.4062/biomolther.2014.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu D, Wang L, Zhong R, et al. Parallel microfluidic networks for studying cellular response to chemical modulation. J. Biotechnol. 2007;131(3):286–292. doi: 10.1016/j.jbiotec.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 51.Lau C, Nygard S, Fure H, et al. CD14 and complement crosstalk and largely mediate the transcriptional response to escherichia coli in human whole blood as revealed by DNA microarray. PLoS ONE. 2015;10(2) doi: 10.1371/journal.pone.0117261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lao YH, Chiang HY, Yang DK, Peck K, Chen LC. Selection of aptamers targeting the sialic acid receptor of hemagglutinin by epitope-specific SELEX. Chem. Commun. 2014;50(63):8719–8722. doi: 10.1039/c4cc03116d. [DOI] [PubMed] [Google Scholar]
- 53.Lee MY, Kumar RA, Sukumaran SM, Hogg MG, Clark DS, Dordick JS. Three-dimensional cellular microarray for high-throughput toxicology assays. Proc. Natl Acad. Sci. USA. 2008;105(1):59–63. doi: 10.1073/pnas.0708756105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yeon JH, Park JK. Cytotoxicity test based on electrochemical impedance measurement of HepG2 cultured in microfabricated cell chip. Anal. Biochem. 2005;341(2):308–315. doi: 10.1016/j.ab.2005.03.047. [DOI] [PubMed] [Google Scholar]
- 55.Caviglia C, Zor K, Montini L, et al. Impedimetric toxicity assay in microfluidics using free and liposome-encapsulated anticancer drugs. Anal. Chem. 2015;87(4):2204–2212. doi: 10.1021/ac503621d. [DOI] [PubMed] [Google Scholar]
- 56.Caviglia C, Zor K, Canepa S, et al. Interdependence of initial cell density, drug concentration and exposure time revealed by real-time impedance spectroscopic cytotoxicity assay. Analyst. 2015;140(10):3623–3629. doi: 10.1039/c5an00097a. [DOI] [PubMed] [Google Scholar]
- 57.Pihl J, Sinclair J, Sahlin E, et al. Microfluidic gradient-generating device for pharmacological profiling. Anal. Chem. 2005;77(13):3897–3903. doi: 10.1021/ac050218+. [DOI] [PubMed] [Google Scholar]
- 58.Mao SF, Gao D, Liu W, Wei HB, Lin JM. Imitation of drug metabolism in human liver and cytotoxicity assay using a microfluidic device coupled to mass spectrometric detection. Lab Chip. 2012;12(1):219–226. doi: 10.1039/c1lc20678h. [DOI] [PubMed] [Google Scholar]
- 59.Berthuy OI, Blum LJ, Marquette CA. Cancer-cells on a chip for label-free optic detection of secreted molecules. Biosensors (Basel). 2016;6(1) doi: 10.3390/bios6010002. pii: E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee K, Kim C, Ahn B, et al. Generalized serial dilution module for monotonic and arbitrary microfluidic gradient generators. Lab Chip. 2009;9(5):709–717. doi: 10.1039/b813582g. [DOI] [PubMed] [Google Scholar]
- 61.Wang H, Chen CH, Xiang ZL, Wang M, Lee C. A convection-driven long-range linear gradient generator with dynamic control. Lab Chip. 2015;15(6):1445–1450. doi: 10.1039/c4lc01451k. [DOI] [PubMed] [Google Scholar]
- 62.Abhyankar VV, Lokuta MA, Huttenlocher A, Beebe DJ. Characterization of a membrane-based gradient generator for use in cell-signaling studies. Lab Chip. 2006;6(3):389–393. doi: 10.1039/b514133h. [DOI] [PubMed] [Google Scholar]
- 63.Ziauddin J, Sabatini DM. Microarrays of cells expressing defined cDNAs. Nature. 2001;411(6833):107–110. doi: 10.1038/35075114. [DOI] [PubMed] [Google Scholar]
- 64.Lee PJ, Hung PJ, Rao VM, Lee LP. Nanoliter scale microbioreactor array for quantitative cell biology. Biotechnol. Bioeng. 2006;94(1):5–14. doi: 10.1002/bit.20745. [DOI] [PubMed] [Google Scholar]
- 65.Baker BM, Chen CS. Deconstructing the third dimension - how 3D culture microenvironments alter cellular cues. J. Cell Sci. 2012;125(13):3015–3024. doi: 10.1242/jcs.079509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walsh CL, Babin BM, Kasinskas RW, Foster JA, Mcgarry MJ, Forbes NS. A multipurpose microfluidic device designed to mimic microenvironment gradients and develop targeted cancer therapeutics. Lab Chip. 2009;9(4):545–554. doi: 10.1039/b810571e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee PJ, Hung PJ, Lee LP. An artificial liver sinusoid with a microfluidic endothelial-like barrier for primary hepatocyte culture. Biotechnol. Bioeng. 2007;97(5):1340–1346. doi: 10.1002/bit.21360. [DOI] [PubMed] [Google Scholar]
- 68.Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Reconstituting organ-level lung functions on a chip. Science. 2010;328(5986):1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The chip can preproduce the rhythmic breathing like a living lung.
- 69.Kim HJ, Huh D, Hamilton G, Ingber DE. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip. 2012;12(12):2165–2174. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- 70.Torisawa YS, Spina CS, Mammoto T, et al. Bone marrow-on-a-chip replicates hematopoietic niche physiology in vitro . Nat. Methods. 2014;11(6):663–+. doi: 10.1038/nmeth.2938. [DOI] [PubMed] [Google Scholar]
- 71.Yasotharan S, Pinto S, Sled JG, Bolz SS, Gunther A. Artery-on-a-chip platform for automated, multimodal assessment of cerebral blood vessel structure and function. Lab Chip. 2015;15(12):2660–2669. doi: 10.1039/c5lc00021a. [DOI] [PubMed] [Google Scholar]
- 72.Huh D, Leslie DC, Matthews BD, et al. A human disease model of drug toxicity-induced pulmonary edema in a lung-on-a-chip microdevice. Sci. Transl. Med. 2012;4(159):159ra147. doi: 10.1126/scitranslmed.3004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jang KJ, Suh KY. A multi-layer microfluidic device for efficient culture and analysis of renal tubular cells. Lab Chip. 2010;10(1):36–42. doi: 10.1039/b907515a. [DOI] [PubMed] [Google Scholar]
- 74.Jang KJ, Mehr AP, Hamilton GA, et al. Human kidney proximal tubule-on-a-chip for drug transport and nephrotoxicity assessment. Integrative Biology. 2013;5(9):1119–1129. doi: 10.1039/c3ib40049b. [DOI] [PubMed] [Google Scholar]
- 75.Agarwal A, Goss JA, Cho A, Mccain ML, Parker KK. Microfluidic heart on a chip for higher throughput pharmacological studies. Lab Chip. 2013;13(18):3599–3608. doi: 10.1039/c3lc50350j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bhatia SN, Ingber DE. Microfluidic organs-on-chips. Nat. Biotechnol. 2014;32(8):760–772. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]; • Most of organ-on-chip only test one drug at one time; in contrast, this chip has 50 samples per chip and holds the potential to be scaled up.
- 77.Booth R, Kim H. Characterization of a microfluidic in vitro model of the blood–brain barrier (mu BBB) Lab Chip. 2012;12(10):1784–1792. doi: 10.1039/c2lc40094d. [DOI] [PubMed] [Google Scholar]
- 78.Kim J, Taylor D, Agrawal N, et al. A programmable microfluidic cell array for combinatorial drug screening. Lab Chip. 2012;12(10):1813–1822. doi: 10.1039/c2lc21202a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Occhetta P, Centola M, Tonnarelli B, Redaelli A, Martin I, Rasponi M. High-throughput microfluidic platform for 3D cultures of mesenchymal stem cells, towards engineering developmental processes. Sci. Rep. 2015;5:10288. doi: 10.1038/srep10288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang SB, Wang SS, Hsieh CH, Lin YC, Lai CS, Wu MH. An integrated microfluidic cell culture system for high-throughput perfusion three-dimensional cell culture-based assays: effect of cell culture model on the results of chemosensitivity assays dagger. Lab Chip. 2013;13(6):1133–1143. doi: 10.1039/c2lc41264k. [DOI] [PubMed] [Google Scholar]
- 81.Wen Y, Zhang X, Yang S-T. Microplate-reader compatible perfusion microbioreactor array for modular tissue culture and cytotoxicity assays. Biotechnol. Prog. 2010;26(4):1135–1144. doi: 10.1002/btpr.423. [DOI] [PubMed] [Google Scholar]
- 82.Montanez-Sauri SI, Sung KE, Puccinelli JP, Pehlke C, Beebe DJ. Automation of three-dimensional cell culture in arrayed microfluidic devices. J. Lab. Autom. 2011;16(3):171–185. doi: 10.1016/j.jala.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.List of microfluidics companies. http://fluidicmems.com/list-of-microfluidics-lab-on-a-chip-and-biomems-companies; • Using this map, the idea about the market of microfluidic chips in the world can be had.
- 84.ACEA Biosciences. www.aceabio.com/about/
- 85.Bell Brook Labs. www.bellbrooklabs.com/products-services/iuvo-microconduit-array-platform/microchannel-5250/
- 86.Cellasic bought by Millipore. www.emdmillipore.com/US/en/life-science-research/cell-culture-systems/cellASIC-live-cell-analysis/microfluidic-plates/68eb.qB.wfkAAAFBWmVb3.sJ,nav
- 87.Cytoo. http://cytoo.com/about-us
- 88.Celsee Diagnostics. http://celsee.com/about-us/
- 89.Fluxion Biosciences. http://fluxionbio.com/bioflux/
- 90.Xona mcirofluidics. http://xonamicrofluidics.com/