Abstract
The possibility that G protein-coupled receptor family C member A (GPRC6A) is the osteocalcin (Ocn)-sensing G protein-coupled receptor that directly regulates pancreatic β-cell functions is controversial. In the current study, we found that Ocn and an Ocn-derived C-terminal hexapeptide directly activate GPRC6A-dependent ERK signaling in vitro. Computational models probe the structural basis of Ocn binding to GPRC6A and predict that the C-terminal hexapeptide docks to the extracellular side of the transmembrane domain of GPRC6A. Consistent with the modeling, mutations in the computationally identified binding pocket of GPRC6A reduced Ocn and C-terminal hexapeptide activation of this receptor. In addition, selective deletion of Gprc6a in β-cells (Gprc6aβ-cell-cko) by crossing Gprc6aflox/flox mice with Ins2-Cre mice resulted in reduced pancreatic weight, islet number, insulin protein content, and insulin message expression. Both islet size and β-cell proliferation were reduced in Gprc6aβ-cell-cko compared with control mice. Gprc6aβ-cell-cko exhibited abnormal glucose tolerance, but normal insulin sensitivity. Islets isolated from Gprc6aβ-cell-cko mice showed reduced insulin simulation index in response to Ocn. These data establish the structural basis for Ocn direct activation of GPRC6A and confirm a role for GPRC6A in regulating β-cell proliferation and insulin secretion.
Osteocalcin (Ocn) released from bone has been proposed to function as a hormone regulating energy metabolism and sex hormone production through the binding to, and activation of G protein-coupled receptor family C member A (GPRC6A), a class-C G protein-coupled receptor in target tissues (1–3). The possibility that Ocn is a ligand for GPRC6A is controversial but supported by many observations. Ocn activates GPRC6A signaling responses in vitro (4, 5). Correlations between expression of GPRC6A and functional responses to Ocn are observed in β-cells and Leydig cells (5–10). Ocn has been shown to stimulate insulin secretion and cell proliferation of β-cells in vitro and in vivo (4, 5, 11–13) and to stimulate testosterone and 25-hydroxy vitamin D biosynthesis in Leydig cells (4). Genetic interactions between GPRC6A and Ocn also support their involvement in common signaling pathways. In this regard, double heterozygous GPRC6A and Ocn-deficient mice exhibit additive effects in impairing glucose homeostasis (11). In addition, Gprc6a−/− mice, β-cell-specific conditionals of Gprc6a mice, and Ocn−/− mice all have glucose intolerance and impaired insulin secretion (5, 6, 11, 14). Effects of GPRC6A on glucose homeostasis may also arise from Ocn activation of GPRC6A in peripheral tissues regulating insulin sensitivity (6).
In spite of these data, whether Ocn is a direct ligand for GPRC6A has recently been questioned (15). First, the structural basis for Ocn binding to GPRC6A has not been established. Indeed, Ocn, whose crystal structure shows 3 α-helices surrounding an hydrophobic core, differs significantly from the known physiological amino acid and cation ligands for GPRC6A (15–17). Second, Ocn failed to stimulate a GPRC6A receptor constructs overexpressed in CHO or FlpIN-TREx-HEK293 cells (15, 18), suggesting that Ocn activity is not directly mediated by GPRC6A. Indeed, orally administered Ocn has the capacity to indirectly regulate insulin secretion through stimulation of intestinal Glucagon-like peptide-1 secretion (19), and/or production of other insulin-regulating hormones, such as testosterone (10). Third, a different Gprc6a−/− mouse model, created by deleting the transmembrane and intracellular domains, does not develop glucose intolerance and insulin resistance (20, 21). Finally, a direct function of GPRC6A in β-cells has also been questioned (15, 18, 22). In this regard, GPRC6A is a pertussis toxin sensitive receptor, a class of G protein-coupled receptors (GPCRs) that typically inhibit, rather than stimulate insulin secretion; and some studies failed to observe an effect of the GPRC6A ligand, L-arginine (L-Arg), to stimulate insulin release from islets (23).
In the current study, we reexamined the question of whether Ocn binds to and activates GPRC6A to regulate β-cell function. To this end, we explored the structural basis and the functional effects of Ocn binding to and activation of GPRC6A and examined the role of this receptor in regulating β-cell function by selectively disrupting GPRC6A in pancreatic β-cells.
Materials and Methods
Animals
We generated both global Gprc6a−/− and Gprc6aflox/flox mice as previously reported (6) and detailed below. We used Ins2-Cre mice (24) obtained from The Jackson Laboratory (B6.Cg-Tg(Ins2-cre)25Mgn/J) to delete Gprc6a in β-cells.
Mice were maintained and used in accordance with recommendations as described (National Research Council 1985; Guide for the Care and Use of Laboratory Animals Department of Health and Human Services Publication NIH 86-23, Institute on Laboratory Animal Resources, Rockville, MD) and following guidelines established by the University of Tennessee Health Science Center Institutional Animal Care and Use Committee. The animal study protocol was approved by the institutional review boards at University of Tennessee Health Science Center Institutional Animal Care and Use Committee.
Reagents and antibodies
Insulin (Mouse) Ultrasensitive ELISA kit and mouse C-peptide ELISA kit were obtained from ALPCO Diagnostics. Marker of proliferation Ki-67 (Ki-67) antibody was purchased from Novus Biologicals. Ocn was purified from bovine tibial bone extracts (25, 26). Decarboxylated Ocn was produced by treating Ocn in vacuo at 110°C (26, 27). Purity and decarboxylation state were confirmed by native gel electrophoresis (25), or by blotting followed by reaction with 4-diazobenzene sulfonic acid staining for γ-carboxyglutamic acid (26, 28). The Ocn-6aa-C, consisting of 6 residues (NH2-Arg-Phe-Tyr-Gly-Pro-Val-COOH) from the C-terminal of human Ocn (hOcn) (NP_954642.1) was synthesized by Molecular Resource Center at University of Tennessee Health Science Center, the molecular weight of Ocn-6aa-C is 738.405 determined by Matrix-assisted laser desorption ionization Time-of-Flight mass spectrometry. The antibodies of phospho-AMP-activated protein kinase (AMPK)-α, phospho-Liver kinase B1 (LKB1), and phospho-Phosphoinositide 3-kinase (PI3K) were purchased from Cell Signaling Technology. Insulin antibody was purchased from Santa Cruz Biotechnology, Inc. An antibody preparation for bovine Ocn (bOcn) is made in rabbit challenged with purified Ocn following methods described previously (29). The estimated titer is 1 μL of antiserum will bind 0.5-ng Ocn (radioligand-binding dose dilution) (for antibodies, please see Table 1).
Table 1.
Antibody Table
| Name | Company |
|---|---|
| Phospho-ERK1/2 MAPK antibody | Cell Signaling Technology |
| pERK1/2 MAPK antibody | Cell Signaling Technology |
| Phospho-AMPK-α (T172) antibody | Cell Signaling Technology |
| Phospho-LKB1 (S428) antibody | Cell Signaling Technology |
| Phospho-PI3K p85 (Tyr458)p55(Tyr199) antibody | Cell Signaling Technology |
| Insulin antibody | Santa Cruz Biotechnology, Inc |
Measurement of total and phospho-ERK by Western blotting
HEK-293 and HEK-293 transfected with a mouse GPRC6A cDNA cells (10, 30) were cultured in DMEM (25mM glucose, catalog number 11995; Invitrogen) supplemented with 10% fetal bovine serum and 1% PBS for 48 hours followed by overnight incubation in DMEM/F12 (17.5mM glucose, catalog number 11330; Invitrogen) containing 0.1% BSA to achieve quiescence. Quiescent cells were treated with various concentrations of GPRC6A ligands, including L-arginine, Ocn, and Ocn-6aa-C in quiescent media for 20 minutes at 37°C. ERK activation will be assessed by immunoblotting using antiphospho-ERK1/2 MAPK antibody corrected for the total amount of ERK using an anti-ERK1/2 MAPK antibody (Cell Signaling Technology).
Measurement of cAMP accumulation
Untransfected HEK-293 and HEK-293 cells transfected with a mouse GPRC6A cDNA (105 cells/well) (10) were incubated in DMEM/F12 containing 0.1% BSA to achieve quiescence and treated with vehicle control, 10mM L-arginine (31), or 60-ng/mL bOcn for 30 minutes at 37°C. cAMP levels were measured by using cAMP EIA kit (Cayman Chemical).
GPRC6A homology modeling
To investigate the molecular mechanism for Ocn binding to GPRC6A, structural models of GPRC6A and Ocn were constructed and used to identify potential Ocn binding poses. In multiple sequence alignments the helix regions are highly conserved among the GPCRs, whereas the loop regions exhibit low sequence similarity. The GPRC6A sequence exhibits a similarity of 43.2% and 44.7% to related family members, the Metabotropic glutamate receptor-1 (mGluR-1) and mGluR-5 receptors, respectively, for which there are existing crystal structures of the transmembrane domain (32). These receptor structures were selected as templates for the homology modeling calculations and the corresponding structural models selected for docking studies. Missing regions in the cytoplasmic loop-2 (sequence 688–691) and C terminus (sequence 844–845) of the mGluR-1 template structure were modeled using mGluR-5, and missing regions in the cytoplasmic loop-2 (sequence 683–688) and the extracellular loop-2 (sequence 721–728) of mGluR-5 template structure were modeled using mGluR-1. Ten mainchain models with ten sidechain conformers per mainchain model were generated for each template using the MOE-2012 (Molecular Operating Environment, 2013.08; Chemical Computing Group, Inc) homology modeling facility with the CHARMM27 force-field (33). The GPRC6A homology models were validated using PolyPhobius (34). The best-scoring homology models; 1 from using mGluR-1 as a template and 1 from using mGluR-5 as a template, were selected for docking studies based on their predicted Generalized Born/volume integral scores (35), which rank the models based on Coulomb and Generalized Born interaction energies.
The highest scoring models are shown in figure 2 below. These models exhibit similar structures, with a root mean square deviation (RMSD) of approximately 4 Å (2) between the backbone atoms. These models were validated against Hidden Markov secondary structure predictions generated by PolyPhobius, and found to be consistent (Supplemental Tables 1 and 2), with only 1 loop region and 1 transmembrane helix in the homology models deviating from the PolyPhobius-predicted structure for 5 or more residues.
bOcn (NP_776674.1), similarity score of 93.9% to hOcn (NP_954542.1), was used as the main template for modeling the hOcn structure. The alignment between hOcn, bOcn, and pig Ocn (NP_001157476.1) shows that Ocn is well conserved among different species, with the hOcn exhibiting a similarity of 93.9% and 87.8% to the bOcn and pig Ocn, respectively. The highest scoring model, based on Generalized Born/volume integral scores, was generated using bOcn as the main template (see figure 2 below). The Ocn-6aa-C, consisting of 6 residues was obtained from the hOcn homology model by deleting the rest of the protein in MOE-2012 (see figure 2 below).
Docking of Ocn C terminus (Ocn-6aa-C)
Docking of Ocn-6aa-C to the GPRC6A homology models was carried using Fast Fourier Transform-based rigid docking program Cluspro (36). The docking was carried out agnostically, ie, without prespecifying the possible binding site residues, to the extracellular side of the transmembrane domain of the receptor. In addition, explicit repulsions were added between the peptide and the residues present on the cytoplasmic side of the receptor, and the receptor residues interacting with the surrounding membrane regions.
Cluspro first ranks the generated models using a pairwise interaction potential scoring function, and then clusters the top 1000 models, ranked by scoring function, using pairwise C-α RMSD between the 2 proteins. The goal of clustering is to isolate highly populated low-energy basins on the energy landscape, large clusters being more likely to include native structures. After clustering, the ranked complexes were subjected to a 300 steps of van der Waals minimization with the backbone fixed, using the CHARMM (33) potential, to remove potential side chain clashes. Cluspro then output the centers of the largest clusters.
Metabolic studies
For glucose tolerance test (GTT) glucose (2 g/kg body weight) was injected ip (37) after a 5-hour fast, and blood glucose was monitored using blood glucose strips and the Accu-Check glucometer (Roche) at indicated times. For insulin tolerance test (ITT) (38), mice were fasted for 5 hours, injected ip with insulin (0.75-U/kg body weight; Sigma), and blood glucose levels were measured at indicated times as described (39). ITT data are presented as percentage of initial blood glucose concentration.
Mouse islets isolation and ligand stimulation
Primary islets were isolated using modified method (40, 41). The insulin stimulation index was calculated as the ratio of stimulation media insulin concentrations in Ocn divided by the insulin concentration in control stimulation media (without Ocn) at low glucose conditions (5, 42).
β-Cell area was calculated as the ratio of the surface positive for insulin immunostaining divided by the total pancreatic surface. β-Cell mass was calculated as the β-cell area multiplied by pancreatic weight. To measure this ratio, slides from wild-type (WT) and Gprc6aβ-cell-cko mice immunostained for insulin (insulin antibody; Santa Cruz Biotechnology, Inc) were examined using ImageScope (University of Tennessee Health Science Center Imaging CORE) and analyzed using the NIH ImageJ system software. At least 5 animals were analyzed per genotype.
We also obtained an INS-1 832/13 dp45 rat β-cell line from Christopher Newgard (Sarah W. Stedman Nutrition and Metabolism Center, Duke University School of Medicine) that was selected for its high expression of GPRC6A (43). INS-1 cells were cultured in RPMI 1640 (A10491, Invitrogen; contains 10mM HEPES, 2mM glutamine, 1mM Na-pyruvate, and 25mM glucose) with 10% fetal bovine serum, 100-U/mL penicillin, 100-μg/mL streptomycin, and 50 μmol/L β-mercaptoethanol. The cells were plated onto 24-well plates at a density of approximately 0.5 × 106 cells/well and were grown to 100% confluence before assay. At 18 hours before secretion experiments, the standard tissue culture medium containing 11.1 mmol/L glucose was switched to fresh medium containing 5 mmol/L glucose. Insulin secretion was assayed in HEPES-balanced salt solution (114 mmol/L NaCl, 4.7 mmol/L KCl, 1.2 mmol/L KH2PO4, 1.16 mmol/L MgSO4, 20 mmol/L HEPES, 2.5 mmol/L CaCl2, 25.5 mmol/L NaHCO3, and 0.2% bovine serum albumin [essentially fatty acid free]; pH 7.2) (43).
Real-time RT-PCR
For quantitative real-time RT-PCR assessment of insulin, glucagon, sterol regulatory element-binding factor-1 (Srebf1), and carbohydrate-responsive element-binding protein (Chrebp) gene expression, we isolated total RNA from the islets or pancreas or other tissues of Control and Gprc6aβ-cell-cko mice by standard TRIzol method (Invitrogen) and reverse transcribed 2.0 μg of total RNAs using cDNA synthesis kit (Bio-Rad). PCR reactions were described in previously publication (5, 6). The primers for mouse insulin (NM_008386) consisted of mIns1.F313: ggggagcgtggcttcttcta and mIns1.R452: acaatgccacgcttctgcc; for mouse glucagon (NM_008100) consisted of mGCG.For145: gaagacaaacgccactcaca and mGCG.Rev472: tggtgctcatctcgtcagag; for Srebf1 gene (NM_011480) consisted of mSrebp1c.F3294: tgttggcatcctgctatctg and mSrebp1c.R3483: agggaaagctttggggtcta; for mouse Chrebp gene (NM_021455) consisted of mChrebp.For1973: agatggagaaccgacgtatca and mChrebp.Rev2076: actgagcgtgctgacaagtc; and for the cyclophilin A (NM_008907) consisted of CycA.For: ctgcactgccaagactgaat and CycA.Rev: ccacaatgttcatgccttct. The RT-PCR primers for rat Gprc6a (NM_001271106) consisted of rGPRC6A.For1460: ctgtcacgaagatggcagaa and rGPRC6A.Rev1898: cagaccactaatcccccaga (primer set 1–2); rGPRC6A.For535: aaaatccgctttccttcgttr and GPRC6A. Rev1400: tgggcatcaaaatgaaatgar (primer set 3–4); GPRC6A.For122: tgtttgccattcacgaaaaa and rGPRC6A.Rev694: cctggattgcaaatgtgttg (primer set 5–6).
Statistics
We evaluated differences between groups by one-way ANOVA, followed by a post hoc Tukey's test. Significance was set at P < .05. All values are expressed as means ± SEM. All computations were performed using the Statgraphic statistical graphics system (STSC, Inc).
Results
Signal transduction pathways stimulated by Ocn activation of GPRC6A
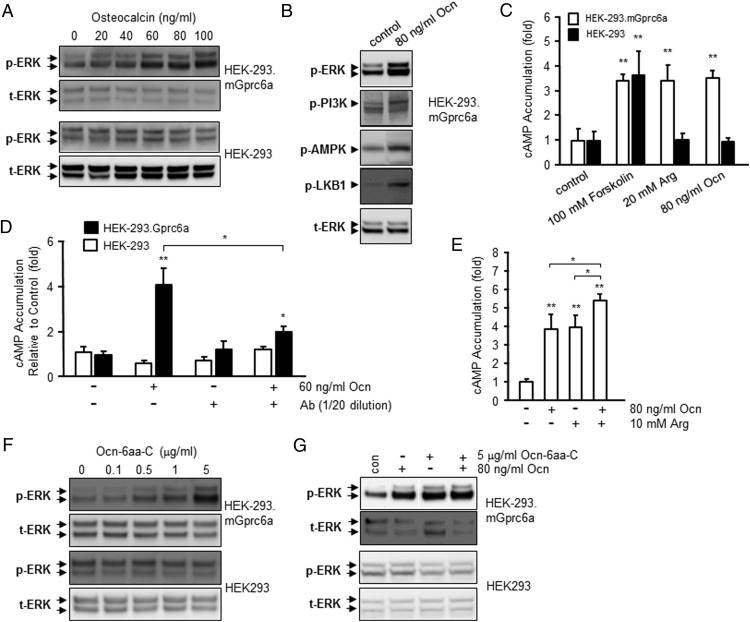
To establish that Ocn activates GPRC6A and that GPRC6A is coupled to signaling pathways known to regulate insulin production and β-cell proliferation, we examined the effects of Ocn in activating ERK phosphorylation, AMPK, and cAMP signaling in HEK-293 cells transfected with GPRC6A or untransfected HEK-293 cells that lack GPRC6A expression (10). We observed a dose-dependent effect of Ocn in stimulating ERK phosphorylation with an estimated EC50 of 49.9-ng/mL (Figure 1A). A similar ERK phosphorylation response is observed for synthetic human uncarboxylated Ocn (H-7534; (Glu17,21,24)-Ocn; BACHEM) at 40–100-ng/mL (data not shown). An effect of Ocn is seen for PI3K phosphorylation, AMPK phosphorylation and LKB1 phosphorylation (Figure 1B) and cAMP accumulation (Figure 1C) in HEK-293 cells transfected with GPRC6A, with a significant response observed from 20-ng/mL (Figure 1A). No response to Ocn was observed in nontransfected HEK-293 cells (Figure 1, A and C) (7). Consistent with previous reports (5, 9), L-Arg (20mM), a ligand for GPRC6A, and Ocn (80-ng/mL) stimulated cAMP only in HEK-293 cells overexpressing GPRC6A (Figure 1C). The magnitude of the cAMP response was similar to forskolin, which stimulated cAMP in HEK-293 cells in a receptor independent manner (ie, with and without GPRC6A overexpression). Interestingly, we found that Ocn-stimulated cAMP accumulation in HEK-293 cells transfected with GPRC6A was significantly blocked by Ocn antibody (Figure 1D).
Figure 1.
Evidence for Ocn activation of GPRC6A. A, Dose-dependent effects of Ocn on GPRC6A-mediated ERK phosphorylation in HEK-293 cells overexpressing GPRC6A. B, Ocn actives GPRC6A-mediated PI3K, AMPK, and LKB1 phosphorylation in HEK-293 cells overexpressing GPRC6A. C, cAMP response to forskolin, Ocn, or L-arginine (31), a known GPRC6A ligand, in HEK-293 cells with and without GPRC6A transfection. D, An Ocn blocking antibody inhibited Ocn-stimulated cAMP accumulation in HEK-293 cells transfected with GPRC6A. E, Ocn and L-Arg show additive cAMP responses. * and **, significant differences from control and stimulated groups at P < .05 and P < .01 (n ≥ 4). F, Ocn-derived 6-aa C-terminal peptide (Ocn-6aa-C) activates GPRC6A-mediated ERK phosphorylation in HEK-293 cells expressing GPRC6A but not in control HEK-293 cells (lower panel). G, Ocn-6aa-C and Ocn show nonadditive effects on GPRC6A-dependent ERK activation.
To look at interactions between Ocn and L-Arg (31), we assessed the effects of Ocn (80-ng/mL) in stimulating cAMP accumulation in the presence and absence of L-Arg (10mM). We found that the addition of L-Arg augmented the effects of Ocn in stimulating cAMP in HEK-293 cells expressing GPRC6A (Figure 1E). These additive effects suggest that L-Arg and Ocn may be binding to different sites on GPRC6A that are complementary. This possibility is supported by structural modeling and GPRC6A receptor mutagenesis (see below). Ocn is cleaved by the circulating serine protease, plasmin, at a single R43-R44 site near its carboxyl end to create an N-terminal 1- to 43-amino acid peptide and C-terminal 44–49 6-amino acid peptide (44). Although it has been suggested that Ocn is too large to activate GPRC6A (15), a 7-amino acid peptide has recently been shown to activate the related receptor calcium-sensing receptor (CASR) (45). Therefore, we examined the ability of the hexapeptide cleavage product of Ocn (Ocn-6aa-C; sequence NH2-Arg-Phe-Tyr-Gly-Pro-Val-COOH) to stimulate GPRC6A transfected into HEK-293 cells. The identical RFYGPV sequence is a highly evolutionarily conserved sequence in land vertebrates from mammals (human) to amphibians (clawed frog) (46). To expand on the number of species from mammals, birds, reptiles, and amphibians: chimpanzees, domestic cow, chicken, dog, American alligator, and Mexican axolotl share the RFYGPV C-terminal sequence determined by uniprot protein sequence alignment (www.uniprot.org). Although mouse Ocn has the RIYGITI C-terminal sequence, the activity of human and bOcn for mouse gprc6a and modeling studies provoked a test of the Ocn-6aa-C as an agonist. As assessed by ERK phosphorylation, we observed a dose-dependent activation of GPRC6A by Ocn-6aa-C, with an estimated EC50 of approximately 1 μg/mL (Figure 1F). This concentration of Ocn-6aa-C required for receptor activation, however, was roughly 20-fold greater than for intact Ocn, which has an estimated EC50 of 49.9 ng/mL. The maximal response to Ocn-6aa-C was achieved at concentrations of 5 μg/mL (6.77μM) (Figure 1F), whereas the maximal effects of intact Ocn were observed at 80 ng/mL (Figure 1A). Ocn and the Ocn-6aa-C peptide, at functional concentrations, did not have additive effects on GPRC6A activation (Figure 1G). Neither intact Ocn nor Ocn-6aa-C stimulated ERK activity in HEK-293 cells not transfected with GPRC6A (Figure 1, A, C, D, F, and G).
Docking of Ocn-6aa-C
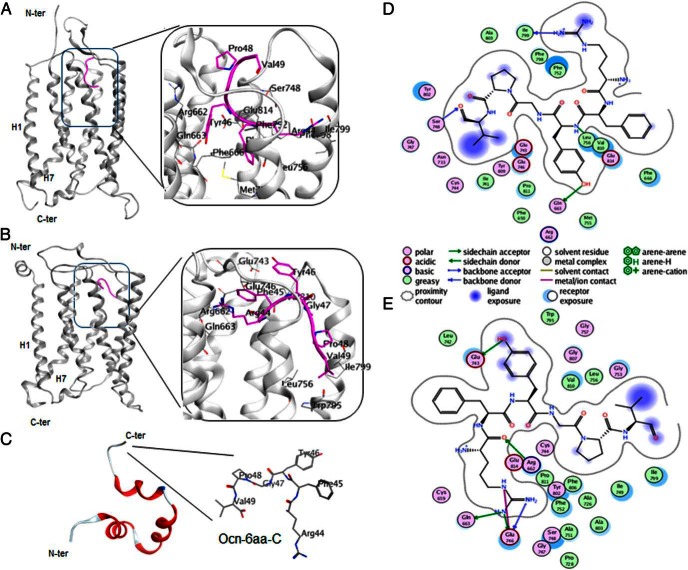
Computational docking of Ocn-6aa-C to the extracellular side of the transmembrane domain of the GPRC6A receptor homology models was carried out as described in Materials and Methods. The binding sites found in the mGluR-1 and mGluR-5 based models of GPRC6A, generated using the rigid-body docking protocol, are shown in Figure 2, A and B, respectively, and the binding pocket residues are shown in Figure 2, D and E (and Supplemental Table 3). The cluster sizes and weighted energy scores for these models are given in Table 2. The top ranked cluster center in mGluR-1 represents more than 90% of the top 1000 predicted models (based on pairwise interaction potential scoring function), and in mGluR-5 represents more than 60% of the top 1000 predicted models.
Figure 2.
Docking of Ocn C-terminal (Ocn-6aa-C) to GPRC6A. A, Binding site in the mGluR-1-based template model, Ocn-6aa-C is shown in pink ribbon-stick representation. B, Binding site in the mGlur-5-based model. C, Ocn and Ocn-6aa-C models based on bOcn structure. Ocn C-terminal consisting of 6 residues: Arg44, Phe45, Tyr46, Gly47, Pro48, and Val49. D and E, Ocn-6aa-C binding sites. Binding pocket residues found in the (D) mGluR-1- and (E) mGluR-5-based models of GPRC6A, generated using the rigid-body docking protocol.
Table 2.
The Cluster Sizes and Weighted Energy Scores of mGluR-1 and mGluR-5 Models Generated Using Cluspro by Clustering the Top 1000 Models
| Cluster | Members | Representative | Weighted Score |
|---|---|---|---|
| mGluR-1 model | |||
| 1 | 641 | Center | −1220.3 |
| 2 | 193 | Center | −1226.4 |
| 3 | 166 | Center | −1227.4 |
| mGluR-5 model | |||
| 1 | 922 | Center | −1207.8 |
| 2 | 54 | Center | −1151.8 |
| 3 | 24 | Center | −1164.8 |
Cluster: number of clusters generated by clustering the top-1000 docking models based on the RMSD between the models. Members: number of structural models (out of 1000) present in each cluster. Representative: the model selected to represent the cluster; this is the centroid/center of the cluster with the maximum number of neighbors (smallest RMSD distances with the other members of the cluster). Weighted score: the predicted binding free energy values generated by the Cluspro scoring function in kcal/mol for the cluster centers. The rigid-docking program Cluspro clusters the top-1000 docking models predicted by their binding-free energies based on a scoring function. The best model is predicted as the center of the largest cluster.
In the 2 alternative predicted binding modes shown in Figure 2, Ocn-6aa-C appears to bind to roughly the same region of the receptor. Fifteen of the predicted binding residues in the 2 sites are common in both models, including Arg662, Gln663, Glu743, Cyt744, Glu746, Gly747, Ser748, Phe752, Leu756, Ile799, Tyr802, Ala803, Val810, Pro811, and Glu814 (Supplemental Table 3 and Figure 2, D and E). Most of the interactions between the Ocn C terminus and GPRC6A involve a mix of hydrophobic and hydrophilic (hydrogen bonds and ionic) interactions. In both predicted models, the hydrophobic residues in the Ocn C-terminal peptides Phe45, Tyr46, Pro48, Gly47, and Val49 are surrounded by aliphatic side-chains and/or aromatic residues in GPRC6A. The peptide is also located in hydrogen bond acceptor-rich regions, with the side chains (Arg44, Tyr46) and the backbone of the peptide forming possible polar interactions with the receptor.
Mutagenesis of GPRC6A predicted binding pocket residues blocks activation by Ocn
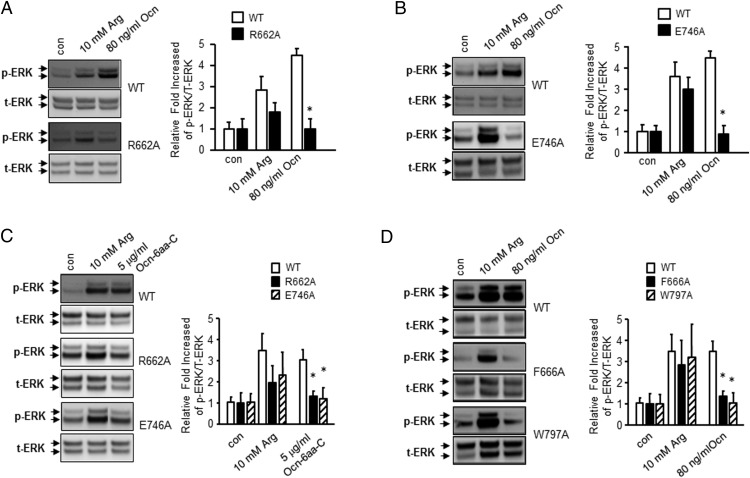
To test the above modeling we mutated Arg662 (R-662), Glu746 (E-746), Phe666 (F-666), and Trp797 (W-797) in mouse GPRC6A into alanine by site-directed mutagenesis (Figure 2, D and E) (47). Arg662 and Glu746 are present in the binding site in both the models and conserved in human GPRC6A. Phe666 is present in the mGluR1 model and conserved in human GPRC6A (Figure 2D), and Trp797 (which corresponds to human W-795) is present in the mGluR5 model (Figure 2E). Mutant and the WT mGPRC6A cDNAs were transiently transfected into HEK-293 cells. We confirmed that R662A and WT mGRPC6A proteins were equally expressed, as assessed by Western blotting using a Myc antibody, which recognized the Myc epitope located at the amino-terminal tail of the WT and mutant receptors (Supplemental Figure 1). Ocn stimulated ERK phosphorylation in the WT receptor but not the R662A mutant receptor-transfected HEK-293 cells (Figure 3A). Interestingly, L-Arg, which is believed to bind to the venous fly trap motif of GPRC6A, activated the R662A mutant, although the response was less than the WT GPRC6A (Figure 3A). Cells transfected with the E746A mutant GPRC6A also showed no response to Ocn stimulation, but remained responsive to L-Arg (Figure 3B). The R662A and E746A mutant receptors also lost their response to Ocn-6aa-C stimulation (Figure 3C). Similarly, the F666A and W797A mutant receptors lost their response to Ocn, but maintained their responsiveness to L-Arg (Figure 3D). These results indicate that the binding sites identified by the computational modeling are probably correct, and are distinct from the L-Arg agonist site.
Figure 3.
Mutagenesis of residues in predicted Ocn-binding pocket of GPRC6A. A, Comparison of Ocn and L-Arg activation of WT (upper panel) and R662A GPRC6A transfected in HEK-293 cells (lower panel). Bar graph depicting fold increase in ERK activation in response to Ocn and L-Arg in WT and mutant GPRC6A. B, Comparison of Ocn and L-Arg activation of WT (upper panel) and E746A GPRC6A transfected in HEK-293 cells (lower panel). Bar graph depicting fold increase in ERK activation in response to Ocn and L-Arg in WT and mutant GPRC6A. C, Comparison of Ocn-6aa-C and L-Arg activation of WT (upper panel) and R662A GPRC6A (middle panel) and E746A GPRC6A (lower panel) transfected in HEK-293 cells. Bar graph depicting fold increase in ERK activation in response to Ocn-6aa-C and L-Arg in WT and mutant GPRC6As. D, Comparison of Ocn and L-Arg activation of WT (upper panel), F666A (middle panel), and W797A (lower panel) GPRC6A transfected in HEK-293 cells. Bar graph depicting fold increase in ERK activation in response to Ocn and L-Arg in WT and mutant GPRC6A.
Gprc6aβ-cell-cko mice have impaired insulin production, glucose intolerance, and resistance to Ocn stimulation of insulin secretion
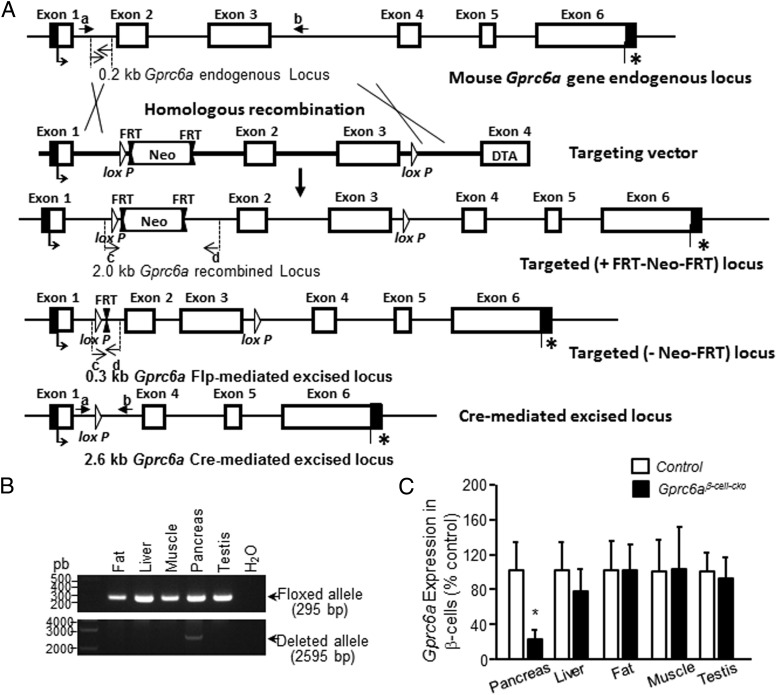
We created Gprc6aflox/flox mice to investigate the tissue-specific effects of loss-of-GPRC6A function. Gprc6aflox/flox mice were produced by using the loxP-FRT system to successfully delete the Gprc6a exons 2 and 3 and schematically shown in Figure 4A. To test the function of GPRC6A in β-cells, we generated Gprc6aβ-cell-cko (Ins2-Cre/+;Gprc6aflox/−) mice by crossing Gprc6aflox/− mice with Ins2-Cre mice. This approach selectively deleted exons 2 and 3 in the pancreas, but not in other organs that were tested, including the liver, fat, muscle, and testis (Figure 4B). Gprc6aβ-cell-cko mice exhibited a 77% reduction in Gprc6a mRNA levels in β-cells (Figure 4C).
Figure 4.
Generation of a conditional allele of Gprc6a mouse model. A, Schematic representation of the targeting strategy. Exon 1–6 open reading frames are represented by open boxes, and thin lines represent untranslated regions of the Gprc6a locus. The neomycin resistance gene (for positive selection) flanked by 2 FRT sites and LoxP (open triangle) are indicated. B, Specificity of Gprc6a deletion was tested by PCR in the indicated tissues. C, Efficiency of Gprc6a deletion by Ins2-Cre in pancreas was tested by real-time PCR using specific Gprc6a primers as described in Materials and Methods. Expression was assessed by real-time PCR using total RNA derived from control group (WT, +/+;Gprc6aflox/+, +/+;Gprc6aflox/−, or Ins2-Cre/+;Gprc6aflox/+) and Gprc6aβ-cell-cko mouse tissues as indicated. Gprc6a expression is relative to the level of the cyclophilin control gene. Values represent the mean ± SEM. *, significant difference between control group and Gprc6aβ-cell-cko mice (P < .05; n ≥ 4).
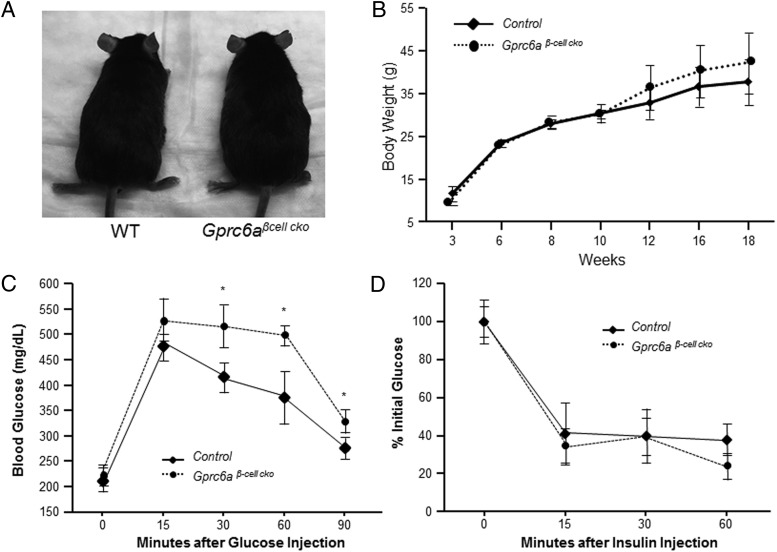
The gross appearance (Figure 5A) and body weight (Figure 5B) of Gprc6aβ-cell-cko mice were not different from the WT male mice. The fasting blood glucose levels were not different between 10-week-old Gprc6aβ-cell-cko both male and female mice (223.0 ± 10.7 mg/dL) and male and female controls (211.7 ± 10.87 mg/dL; P = .0625) (9). However, fasting serum insulin (0.818 ± 0.098 ng/mL) and C-peptide (763.8 ± 5.77pM) concentrations in Gprc6aβ-cell-cko mice were both significantly lower than respective values in controls (1.24 ± 0.084 ng/mL and 1151 ± 72.2pM, P = .02 and 0.02, respectively) in male and female mice.
Figure 5.
Phenotype of Gprc6aβ-cell-cko mice. A, Gross appearance of adult WT and Gprc6aβ-cell-cko male mice. B, Comparison of the body weight in control group and Gprc6aβ-cell-cko male mice at ages ranging from 3 to 18 weeks. Data represent the mean ± SEM from 4 to 6 mice in each group. GTT (C) and ITT (D) (38) in WT and Gprc6aβ-cell-cko mice. Shown is blood glucose (mg/dL) during GTT in 10-week-old control and Gprc6aβ-cell-cko male and female mice. ITT data are presented as percentage of initial blood glucose concentration. Data represent the mean ± SEM from more than 5 male and female mice in each group. *, difference from control group and Gprc6aβ-cell-cko mice at P < .05.
Next, we performed GTT and ITT (38) in Gprc6aβ-cell-cko mice in both male and female mice. After injection of glucose (2 g/kg) Gprc6aβ-cell-cko mice had a significantly higher serum glucose levels than controls (Figure 5C), similar to the impaired glucose tolerance observed in global Gprc6a−/− (6). In contrast, Gprc6aβ-cell-cko and control mice exhibited a similar sensitivity to insulin administration (0.75 U/kg) (Figure 5D). These findings contrast with the hyperglycemia and insulin resistance observed in global Gprc6a−/− mice (6).
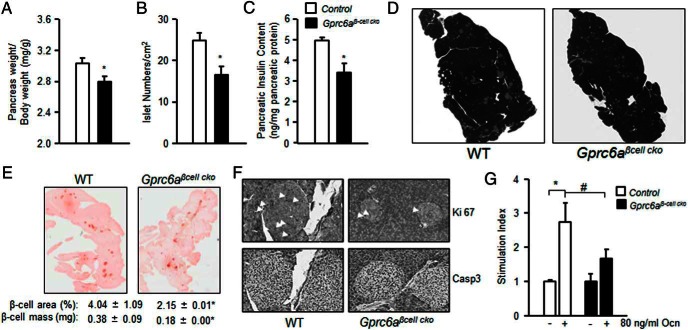
We found that the pancreas weight (Figure 6A) and the number of islets (Figure 6B) were significantly decreased in Gprc6aβ-cell-cko mice compared with the controls. In hematoxylin and eosin-stained slides, the size of islets from Gprc6aβ-cell-cko mice is smaller compared with the controls (Figure 6D, upper panel). Furthermore, pancreatic insulin content was decreased in Gprc6aβ-cell-cko mice compared with the controls (Figure 6C). To determine the functional bases of the phenotype observed in Gprc6aβ-cell-cko mice, pancreas sections were analyzed by histomorphometry. Insulin immunolabeling showed that β-cell area and β-cell mass were significantly reduced in Gprc6aβ-cell-cko compared with control mice (Figure 6E).
Figure 6.
Characterization of the phenotype of Gprc6aβ-cell-cko mice. A, Comparison of pancreas weight in 10-week-old control group and Gprc6aβ-cell-cko mice. The weight of pancreas was normalization by body weight. Comparison of islet number (B) and insulin content (C) in pancreas from in control group and Gprc6aβ-cell-cko mice. Values represent the mean ± SEM. *, significant difference between control group and Gprc6aβ-cell-cko mice (P < .05; n ≥ 4). D, Representative hematoxylin and eosin staining. E, Immunostaining for insulin (red staining), the quantified average stained area for WT and Gprc6aβ-cell-cko mouse pancreas sections is indicated below the image. F, The immunostaining for Ki-67 (upper panel, arrowheads show Ki-67-positive staining) and cysteine-aspartic acid protease 3 (Casp3) in WT and Gprc6aβ-cell-cko mice (bottom panel). G, Islets from Gprc6aβ-cell-cko mice showed impaired insulin stimulation index by Ocn. Stimulation index was attenuated in response to 60-ng/mL Ocn in isolated islets from Gprc6aβ-cell-cko mice. Values represent the mean ± SEM. *, significant different between control group and Gprc6aβ-cell-cko mice (P < .05; n ≥ 3); #, significant different between Ocn-treated control group and Gprc6aβ-cell-cko mice (P < .05; n ≥ 3).
We also analyzed β-cell proliferation in islet β-cells, as assessed by Ki-67 antibody immunostaining. As shown in Figure 6F, upper panel, β-cells proliferated significantly decreased in Gprc6aβ-cell-cko than in control mice. However, we did not detect any difference between Gprc6aβ-cell-cko mice and controls in the expression of the apoptosis marker cysteine-aspartic acid protease 3 (Figure 6F, bottom panel).
Next, we evaluated the effects of agonist stimulation of GPRC6A in islets derived from Gprc6aβ-cell-cko mice. Treatment of isolated islets from control mice with Ocn at concentrations of 80 ng/mL resulted in stimulation indices of 2.8, in the presence of low glucose (5.6mM). In contrast, islets isolated from Gprc6aβ-cell-cko mice had an attenuated response to Ocn, with respective stimulation indices of 1.7, a response significantly lower than observed in control mice (Figure 6G).
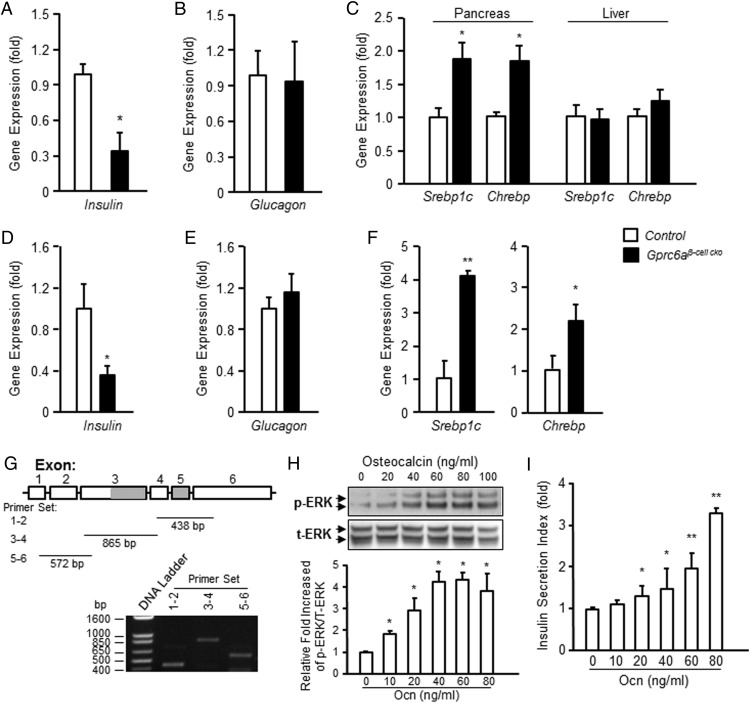
To further investigate the abnormality of islets from Gprc6aβ-cell-cko mice, we assess the gene expression in Gprc6aβ-cell-cko mice compared with WT mice (Figure 7). We found that insulin message expression was also significantly lower in the pancreas obtained from Gprc6aβ-cell-cko mice (ie, decreased by 65%) (Figure 7A). In contrast, glucagon expression was not attenuated (Figure 7B). The expression of both sterol regulatory element-binding protein-1c (Srebp1c) and Chrebp (48) mRNA levels were increased in the pancreas of Gprc6aβ-cell-cko mice compared with controls (Figure 7C). In contrast, we observed no difference in expression of Srebp1c or Chrebp mRNA levels in the liver (Figure 7C). Similar alterations of insulin, glucagon, Srebp1c, and Chrebp message expression were observed in isolated islets derived from Gprc6aβ-cell-cko and control mice. Insulin message expression was reduced by 65% in isolated pancreatic islets derived from Gprc6aβ-cell-cko mice compared with control group mice (Figure 7D). This response was selective for insulin, because glucagon expression was not attenuated in islets from Gprc6aβ-cell-cko mice (Figure 7E). Srebp-1c and Chrebp mRNA expression were also significantly increased in islets from the Gprc6aβ-cell-cko strain (Figure 7F).
Figure 7.
Selective deletion of Gprc6a in pancreatic β-cell attenuated insulin, Srebp1c, and Chrebp expression but not glucagon expression in pancreas. Comparison of insulin (A) and glucagon (B) expression in pancreas from in control group and Gprc6aβ-cell-cko mice. C, Comparison of Srebp1c and Chrebp expression in pancreas and liver from in control group and Gprc6aβ-cell-cko mice. Expression was assessed by real-time PCR using total RNA derived from pancreas or liver from control group and Gprc6aβ-cell-cko mice. Insulin, glucagon, Srebp1c, and Chrebp expression is relative to the level of the cyclophilin control gene. Comparison of insulin (D) and glucagon (E) expression in pancreatic islets. Expression was assessed by real-time PCR using total RNA derived from isolated islets from control group and Gprc6aβ-cell-cko mice. Insulin and glucagon expression is relative to the level of the cyclophilin control gene. F, Comparison of Srebp1c and Chrebp expression in isolated islets from control and Gprc6aβ-cell-cko mice. Expression of insulin, Srebp1c, and Chrebp, but not glucagon, were significantly different in islets from Gprc6aβ-cell-cko mice compared with the islets from control group mice. G, Expression of GPRC6A transcript in INS-1 β-cells. The expected sized fragment was generated from 3 different primer sets for gprc6a, set 1–2 spanning exons 1 and 2 of 438-bp length, set 3–4 spanning exons 3 and 4 of 865 bp, and set 5–6 spanning exons 5–6 of 572 bp. Dose-dependent effects of Ocn on GPRC6A-mediated ERK phosphorylation (H) and insulin secretion (I) in INS-1 β-cells. Values represent the mean ± SEM. *, significant difference between control group and Gprc6aβ-cell-cko mice (P < .05; n ≥ 3).
Finally, because changes in β-cell mass in the in vivo studies rather than direct regulation of insulin secretion by β-cells could account for our findings, we sought to test whether Ocn directly stimulates insulin secretion in a β-cell line that expresses Gprc6a. We selected an INS-1 clone that expressed the full-length Gprc6a message (Figure 7G). Ocn resulted in a dose-dependent stimulation of ERK phosphorylation, achieving activation at concentrations of 20 ng/mL (Figure 7H), similar to the response of Gprc6a expressed in HEK-293 cells (Figure 1A). Ocn resulted in a dose-dependent increase in insulin secretion from these INS-1 cells (Figure 7I).
Discussion
In the current study, we address uncertainties regarding the physiological and pathological roles of the widely expressed G protein-coupled receptor, GPRC6A. Specifically, we address whether Ocn, a bone-derived peptide, is a ligand for GPRC6A and the discrepancies in the phenotype of different Gprc6a knockout mouse models and results of in vitro studies regarding GPRC6A regulation of β-cell functions. We show that GPRC6A is a physiologically relevant receptor for Ocn and demonstrate a direct role for GPRC6A in regulating β-cell functions, including β-cell mass and insulin secretion, using both in vivo and in vitro model systems.
Several lines of evidence support that Ocn is a direct ligand for GPRC6A. First, we show that Ocn dose dependently activates ERK and cAMP second-messenger pathways in HEK-293 cells expressing GPRC6A but not in HEK-293 lacking GPRC6A expression. Ocn also stimulated AMPK phosphorylation, which has important functions in pancreatic β-cells, including the regulation of insulin secretion, cell proliferation, and survival (49). Second, we performed structural modeling, suggesting sites in the heptahelical domain of GPRC6A that bind Ocn, as well as showing where a hexapeptide (Ocn-6aa-C) derived from the Ocn C-terminal docks with GPRC6A. We derived 2 alternative computational models of Ocn binding to GPRC6A and identified residues specifically interacting with the hormone in the models (Table 2). Mutagenesis of Arg662, Phe666, Glu746, and Trp797 in GPRC6A, residues suggested from the modeling to be important for binding Ocn, indeed resulted in reduced Ocn receptor activation in vitro.
Analogous to our findings, previous mutagenesis-based studies of class-C GPCR allosteric sites show that the positions of the peptide binding-pocket residues predicted to bind Ocn are also implicated in the binding of allosteric modulators of the related receptors mGluR-1, mGluR-5, and CASR (50–52). These include Arg662, Phe666, Leu756, Trp795, Phe798, and Glu814. Hence, the modeling and mutagenesis results presented here are consistent with Ocn interacting with residues in the common allosteric site for class-C GPCRs, which may follow a similar mode of activation of GPRC6A.
We found that full-length Ocn activated GPRC6A at lower concentrations than a C-terminal hexapeptide of Ocn (46). We speculate that the C-terminal is able to bind with higher affinity to GPRC6A in the presence of full-length Ocn, because the rest of the Ocn protein may act as a scaffold to keep the peptide in place in the allosteric binding site. We also found that Ocn and L-Arg had additive effects on GPRC6A receptor activation and that the GPRC6A R662A and E746A mutants lost functional responses to Ocn and Ocn-6aa-C but not L-Arg. The molecular basis for the functional interactions between Ocn and L-Arg may be due to Ocn activation of binding sites in the transmembrane domain that are distinct from the amino acid ligand binding sites in the venous fly trap motif (22, 47, 53). This observation makes it unlikely that Ocn actions are indirectly L-Arg released by the breakdown of Ocn or effects on L-Arg metabolism (15).
Most importantly, we confirmed previous studies that GPRC6A regulates β-cell functions (11). Indeed, we found that GPRC6A directly regulates insulin production and secretion by β-cells and that Ocn stimulation of insulin secretion was inhibited in islets derived from mice with conditional deletion of Gprc6a in β-cells (Gprc6aβ-cell-cko mice). Gprc6aβ-cell-cko mice exhibited impaired glucose tolerance due to diminished insulin secretion. Gprc6aβ-cell-cko mice also exhibited reduced circulating insulin and C-peptide concentrations, diminished pancreatic insulin content, and impaired glucose tolerance. Finally, we confirmed that Ocn stimulated insulin secretion in INS-1 clone that expressed GPRC6A transcripts.
The loss of pancreatic β-cell mass represents a critical step in the progression of type 2 diabetes, and it is known that β-cell numbers and insulin secretory capacity can increase to meet metabolic demands. We found a decrease in the size, number of islets and β-cell insulin content and cell proliferation by Ki-67 immunostaining in Gprc6aβ-cell-cko β-cell mice, a pancreatic phenotype similar to what we observed in Gprc6a−/− mice (9). The lack of increased apoptosis and diminished proliferation found in the Gprc6aβ-cell-cko β-cell mice adds additional support to the idea that the reduction in islet size is due to positive effects of GPRC6A in regulating β-cell proliferation. The related observation that Ocn augments insulin content and enhanced human β-cell proliferation of cultured human islets and increased the production of insulin and C-peptide in human islets grafted into NOD-SCID mice, suggests our observations in mice are relevant to human β-cell function (26). We also found that Srebp1c and Chrebp expression were increased in the pancreas of Gprc6aβ-cell-cko mice, suggesting that these transcription factors are downstream of GPRC6A in β-cells, the significance of which will need to be determined (54–57). It is noteworthy that global Gprc6a−/− mice exhibit a fatty liver, and Chrebp up-regulates genes involved in fatty acid synthesis in a glucose-dependent manner. Infusion of Ocn in high-fat diet-fed mice has been shown to attenuate nonalcoholic hepatosteatosis and decrease liver fat content (58). Chrebp is expressed in liver as well as pancreatic β-cells and adipocytes. Overall, our findings, in addition to other reports (5, 13, 26, 39, 48), define a direct role of Ocn in the regulation of serum insulin levels and β-cell proliferation through activation of GPRC6A.
The pancreatic islet and β-cell phenotype in our Gprc6aβ-cell-cko mice is the same as that observed with a different Gprc6aflox/flox mouse model using Pdx1-Cre to delete Gprc6a in β-cells. Thus, 2 independent studies indicate that GPRC6A directly controls β-cell proliferation and insulin production (11).
Both of these studies, that targeted the extracellular domain of GPRC6A (11), differ from another Gprc6a knockout mouse model that deleted the transmembrane (TM) domain. Indeed, mice with deletion of the TM domain of GPRC6A failed to exhibit increased fat mass, alterations in testosterone production, or abnormalities in insulin secretion or glucose homeostasis (20, 22, 23) that were observed in Gprc6a knockout mice created by targeting the extracellular domain.
The reasons for the discrepancies in these in vivo studies are not certain. It is noteworthy, that disruption of the extracellular domain the related CASR also results in a different phenotype than targeting the TM domain of CASR (59). This has been explained by either residual dominant negative actions of the extracellular domain in TM knockouts or residual functions due to alternatively splicing in the extracellular domain knockouts, and could similarly explain the differences in the 2 Gprc6a targeting strategies. Environmental factors may also explain the discrepancies in the different GPRC6A-deficient mice. For example, the GPRC6A-deficient mice, that were previously reported to exhibit no alterations in body composition or glucose metabolism, demonstrated increased basal plasma glucose levels, impaired oral glucose tolerance, and insulin resistance, when placed on a high-fat diet; albeit these changes were attributed to a central effect of GPRC6A in stimulating appetite, rather than a direct effects of this receptor to regulate insulin secretion by β-cells (21)
In addition, there are inconsistent findings regarding activation of GPRC6A in β-cells. Some investigators failed to show that Ocn activates mouse GPRC6A expressed in heterologous cell lines. The GPRC6A construct used in these studies, however, was not a WT cDNA but contained an mGluR signal peptide and c-myc insert in N terminus that may have altered Ocn sensing (15, 18). Interestingly, Ocn was found to stimulate insulin secretion in INS-1 rat β-cell line, but unlike our findings that show INS-1 cells express GPRC6A, GPRC6A expression was not detected in their INS-1 cells (18). Moreover, these investigators found that GPRC6A is expressed in islets but failed to show that L-arginine, which was shown to activate GPRC6A in vitro, stimulates insulin secretion in vivo (23). Differences in the GPRC6A constructs, cell lines used and/or the study conditions may account for these negative in vitro findings.
From previous studies, we know that calcium modulates Ocn activation of GPRC6A (5, 7). Circular dichroism and nuclear magnetic resonance have shown that in the absence of Ca2+ binding Ocn is predominantly a random coil but with Ca2+ is a folded protein (38, 60–62). These observations are consistent with the fact that Ocn does not activate GPRC6A in the absence of calcium in the media but activates GPRC6A in the presence of 1mM calcium (7, 63). High-resolution structures from nuclear magnetic resonance and x-ray crystallography of porcine and fish γ-carboxylated Glu (Gla) Ocn show that the structure is characterized by a protein core formed by 3 α-helical segments stabilized by a disulfide salt-bridge between 2 cysteines, with Ca2+ binding to the 3 Gla residues (64, 65). A recently available crystal structure of bovine Glu-Ocn shows a similar structure with Ca2+ bound, along with highly flexible N- and C-terminal regions (17). Interactions between Ocn and GPRC6A are likely to require an increased local concentration of Ca2+ binding in the orthosteric site of the receptor, which results in Ocn adopting the 3-helical tertiary structure needed for forming proper binding/interactions with the receptor.
The fact that GPRC6A is expressed in liver, adipocytes, muscle, and bone, raises the possibility that GPRC6A regulates insulin sensitivity in peripheral tissues. In contrast to the insulin resistance and hyperglycemia in global Gprc6a−/− mice, Gprc6aβ-cell-cko mice exhibited normal insulin sensitivity (9). The higher fasting glucose levels in Gprc6a−/− mice compared with Gprc6aβ-cell-cko mice could be explained by diminished glucose utilization in peripheral tissues in the global knockout mouse model. Conditional deletion of Gprc6a in these other organs will be necessary to determine the potential role of GPRC6A in regulating gluconeogenesis, fatty acid metabolism and insulin resistance. The fact that Ocn promotes insulin-induced glucose uptake in C2C12 myotubules (66), however, is consistent with a role of GPRC6A in muscle glucose utilization. A dual role for GPRC6A in regulating β -cell function and peripheral tissue insulin sensitivity would make this receptor an attractive therapeutic target for treating type 2 diabetes.
In conclusion, there is a consensus that GPRC6A can sense cations and L-amino acids (16), and recently, data have been presented showing that testosterone can bind to and activate GPRC6A at sites that overlap those identified for Ocn (32). Although a single receptor that can sense at least 4 structurally distinct ligands, including cations, amino acids, testosterone and Ocn, at first seems questionable (16), we demonstrate both functional and structural data supporting that Ocn is a ligand for GPRC6A. As such, GPRC6A is a point of integration for diverse ligands ranging from nutrients to hormones, heretofore thought to be separate and unrelated. More importantly, GPRC6A regulation of insulin production, β-cell proliferation and peripheral tissue sensitivity defines a potential new target for treatment for type 2 diabetes. Future structural modeling and development of small molecules that mimic the action of Ocn in activating GPRC6A may represent an important drug development opportunity.
Acknowledgments
Author contributions: M.P., K.K., R.Y., and Y.W. performed experiments; S.K.N. provided important reagents and reviewed the manuscript; M.P., K.K., J.C.S., J.B., and L.D.Q. designed the study and wrote and reviewed the manuscript; and M.P. and L.D.Q. are guarantors of this work and, as such, had full access to the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
This work was supported by National Institutes of Health Grant R01-AR37308 and Americans Diabetes Association Grant 1-13-BS-149-BR (to L.D.Q.).
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- AMPK
- AMP-activated protein kinase
- L-Arg
- L-arginine
- bOcn
- bovine Ocn
- Casp3
- cysteine-aspartic acid protease 3
- CASR
- calcium sensing receptor
- Chrebp
- carbohydrate-responsive element-binding protein
- GPCR
- G protein-coupled receptor
- GPRC6A
- G protein-coupled receptor family C member A
- GTT
- glucose tolerance test
- hOcn
- human Ocn
- ITT
- insulin tolerance test
- Ki-67
- marker of proliferation Ki-67
- L-Arg
- L-arginine
- LKB1
- Liver kinase B1
- MALDI-TOF
- Matrix-assisted laser desorption ionization Time-of-Flight
- mGluR
- Metabotropic glutamate receptor
- Ocn
- osteocalcin
- PI3K
- Phosphoinositide 3-kinase
- RMSD
- root mean square deviation
- Srebp1c
- sterol regulatory element-binding protein-1c
- TM
- transmembrane
- WT
- wild type.
References
- 1. Karsenty G, Oury F. Regulation of male fertility by the bone-derived hormone osteocalcin. Mol Cell Endocrinol. 2014;382:521–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Oury F, Khrimian L, Denny CA, et al. Maternal and offspring pools of osteocalcin influence brain development and functions. Cell. 2013;155:228–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Oury F, Ferron M, Huizhen W, et al. Osteocalcin regulates murine and human fertility through a pancreas-bone-testis axis. J Clin Invest. 2013;123:2421–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Oury F, Sumara G, Sumara O, et al. Endocrine regulation of male fertility by the skeleton. Cell. 2011;144:796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pi M, Wu Y, Quarles LD. GPRC6A mediates responses to osteocalcin in β-cells in vitro and pancreas in vivo. J Bone Miner Res. 2011;26:1680–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pi M, Chen L, Huang MZ, et al. GPRC6A null mice exhibit osteopenia, feminization and metabolic syndrome. PLoS One. 2008;3:e3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pi M, Parrill AL, Quarles LD. GPRC6A mediates the non-genomic effects of steroids. J Biol Chem. 2010;285:39953–39964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pi M, Quarles LD. Multiligand specificity and wide tissue expression of GPRC6A reveals new endocrine networks. Endocrinology. 2012;153:2062–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pi M, Quarles LD. GPRC6A regulates prostate cancer progression. Prostate. 2012;72:399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pi M, Zhang L, Lei SF, et al. Impaired osteoblast function in GPRC6A null mice. J Bone Miner Res. 2010;25:1092–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wei J, Hanna T, Suda N, Karsenty G, Ducy P. Osteocalcin promotes β-cell proliferation during development and adulthood through Gprc6a. Diabetes. 2014;63:1021–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fulzele K, Riddle RC, DiGirolamo DJ, et al. Insulin receptor signaling in osteoblasts regulates postnatal bone acquisition and body composition. Cell. 2010;142:309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ferron M, Wei J, Yoshizawa T, et al. Insulin signaling in osteoblasts integrates bone remodeling and energy metabolism. Cell. 2010;142:296–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ducy P, Desbois C, Boyce B, et al. Increased bone formation in osteocalcin-deficient mice. Nature. 1996;382:448–452. [DOI] [PubMed] [Google Scholar]
- 15. Jacobsen SE, Nørskov-Lauritsen L, Thomsen AR, et al. Delineation of the GPRC6A receptor signaling pathways using a mammalian cell line stably expressing the receptor. J Pharmacol Exp Ther. 2013;347:298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Clemmensen C, Smajilovic S, Wellendorph P, Bräuner-Osborne H. The GPCR, class C, group 6, subtype A (GPRC6A) receptor: from cloning to physiological function. Br J Pharmacol. 2014;171:1129–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Malashkevich VN, Almo SC, Dowd TL. X-ray crystal structure of bovine 3 Glu-osteocalcin. Biochemistry. 2013;52:8387–8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rueda P, Harley E, Lu Y, et al. Murine GPRC6A mediates cellular responses to L-amino acids, but not osteocalcin variants. PLoS One. 2016;11:e0146846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mizokami A, Yasutake Y, Gao J, et al. Osteocalcin induces release of glucagon-like peptide-1 and thereby stimulates insulin secretion in mice. PLoS One. 2013;8:e57375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Clemmensen C, Pehmøller C, Klein AB, Ratner C, Wojtaszewski JF, Bräuner-Osborne H. Enhanced voluntary wheel running in GPRC6A receptor knockout mice. Physiol Behav. 2013;118:144–151. [DOI] [PubMed] [Google Scholar]
- 21. Clemmensen C, Smajilovic S, Madsen AN, Klein AB, Holst B, Bräuner-Osborne H. Increased susceptibility to diet-induced obesity in GPRC6A receptor knockout mice. J Endocrinol. 2013;217:151–160. [DOI] [PubMed] [Google Scholar]
- 22. Wellendorph P, Bräuner-Osborne H. Molecular basis for amino acid sensing by family C G-protein-coupled receptors. Br J Pharmacol. 2009;156:869–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Smajilovic S, Clemmensen C, Johansen LD, et al. The L-α-amino acid receptor GPRC6A is expressed in the islets of Langerhans but is not involved in L-arginine-induced insulin release. Amino Acids. 2013;44:383–390. [DOI] [PubMed] [Google Scholar]
- 24. Lee JY, Ristow M, Lin X, White MF, Magnuson MA, Hennighausen L. RIP-Cre revisited, evidence for impairments of pancreatic β-cell function. J Biol Chem. 2006;281:2649–2653. [DOI] [PubMed] [Google Scholar]
- 25. Nishimoto SK, Price PA. Proof that the γ-carboxyglutamic acid-containing bone protein is synthesized in calf bone. Comparative synthesis rate and effect of coumadin on synthesis. J Biol Chem. 1979;254:437–441. [PubMed] [Google Scholar]
- 26. Sabek OM, Nishimoto SK, Fraga D, Tejpal N, Ricordi C, Gaber AO. Osteocalcin effect on human β cells mass and function. Endocrinology. 2015;156:3137–3146. [DOI] [PubMed] [Google Scholar]
- 27. Nishimoto SK, Price PA. The vitamin K-dependent bone protein is accumulated within cultured osteosarcoma cells in the presence of the vitamin K antagonist warfarin. J Biol Chem. 1985;260:2832–2836. [PubMed] [Google Scholar]
- 28. Nishimoto SK. A colorimetric assay specific for γ-carboxyglutamic acid-containing proteins: its utility in protein purification procedures. Anal Biochem. 1990;186:273–279. [DOI] [PubMed] [Google Scholar]
- 29. Price PA, Nishimoto SK. Radioimmunoassay for the vitamin K-dependent protein of bone and its discovery in plasma. Proc Natl Acad Sci USA. 1980;77:2234–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kuang D, Yao Y, Lam J, Tsushima RG, Hampson DR. Cloning and characterization of a family C orphan G-protein coupled receptor. J Neurochem. 2005;93:383–391. [DOI] [PubMed] [Google Scholar]
- 31. Bhindi B, Locke J, Alibhai SM, et al. Dissecting the association between metabolic syndrome and prostate cancer risk: analysis of a large clinical cohort. Eur Urol. 2015;67:64–70. [DOI] [PubMed] [Google Scholar]
- 32. Pi M, Kapoor K, Wu Y, et al. Structural and functional evidence for testosterone activation of GPRC6A in peripheral tissues. Mol Endocrinol. 2015;29:1759–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. MacKerell AD, Bashford D, Bellott M, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. [DOI] [PubMed] [Google Scholar]
- 34. Kall L, Krogh A, Sonnhammer EL. An HMM posterior decoder for sequence feature prediction that includes homology information. Bioinformatics. 2005;21(suppl 1):i251–i257. [DOI] [PubMed] [Google Scholar]
- 35. Labute P. The generalized Born/volume integral implicit solvent model: estimation of the free energy of hydration using London dispersion instead of atomic surface area. J Comput Chem. 2008;29:1693–1698. [DOI] [PubMed] [Google Scholar]
- 36. Kozakov D, Brenke R, Comeau SR, Vajda S. PIPER: an FFT-based protein docking program with pairwise potentials. Proteins. 2006;65:392–406. [DOI] [PubMed] [Google Scholar]
- 37. Echeverri CJ, Beachy PA, Baum B, et al. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat Methods. 2006;3:777–779. [DOI] [PubMed] [Google Scholar]
- 38. Atkinson RA, Evans JS, Hauschka PV, et al. Conformational studies of osteocalcin in solution. Eur J Biochem. 1995;232:515–521. [DOI] [PubMed] [Google Scholar]
- 39. Lee NK, Sowa H, Hinoi E, et al. Endocrine regulation of energy metabolism by the skeleton. Cell. 2007;130:456–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu M, Shapiro ME. A new method for isolation of murine islets with markedly improved yields. Transplant Proc. 1995;27:3208–3210. [PubMed] [Google Scholar]
- 41. Gerling IC, Serreze DV, Christianson SW, Leiter EH. Intrathymic islet cell transplantation reduces β-cell autoimmunity and prevents diabetes in NOD/Lt mice. Diabetes. 1992;41:1672–1676. [DOI] [PubMed] [Google Scholar]
- 42. Pi M, Wu Y, Lenchik NI, Gerling I, Quarles LD. GPRC6A mediates the effects of L-arginine on insulin secretion in mouse pancreatic islets. Endocrinology. 2012;153:4608–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hohmeier HE, Mulder H, Chen G, Henkel-Rieger R, Prentki M, Newgard CB. Isolation of INS-1-derived cell lines with robust ATP-sensitive K+ channel-dependent and -independent glucose-stimulated insulin secretion. Diabetes. 2000;49:424–430. [DOI] [PubMed] [Google Scholar]
- 44. Novak JF, Hayes JD, Nishimoto SK. Plasmin-mediated proteolysis of osteocalcin. J Bone Miner Res. 1997;12:1035–1042. [DOI] [PubMed] [Google Scholar]
- 45. Walter S, Baruch A, Dong J, et al. Pharmacology of AMG 416 (Velcalcetide), a novel peptide agonist of the calcium-sensing receptor, for the treatment of secondary hyperparathyroidism in hemodialysis patients. J Pharmacol Exp Ther. 2013;346:229–240. [DOI] [PubMed] [Google Scholar]
- 46. Novak JF, Judkins MB, Chernin MI, et al. A plasmin-derived hexapeptide from the carboxyl end of osteocalcin counteracts oxytocin-mediated growth inhibition [corrected] of osteosarcoma cells. Cancer Res. 2000;60:3470–3476. [PubMed] [Google Scholar]
- 47. Faure H, Gorojankina T, Rice N, et al. Molecular determinants of non-competitive antagonist binding to the mouse GPRC6A receptor. Cell Calcium. 2009;46:323–332. [DOI] [PubMed] [Google Scholar]
- 48. Tiano JP, Mauvais-Jarvis F. Molecular mechanisms of estrogen receptors' suppression of lipogenesis in pancreatic β-cells. Endocrinology. 2012;153:2997–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fu A, Eberhard CE, Screaton RA. Role of AMPK in pancreatic β cell function. Mol Cell Endocrinol. 2013;366:127–134. [DOI] [PubMed] [Google Scholar]
- 50. Wu H, Wang C, Gregory KJ, et al. Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science. 2014;344:58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Doré AS, Okrasa K, Patel JC, et al. Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain. Nature. 2014;511:557–562. [DOI] [PubMed] [Google Scholar]
- 52. Petrel C, Kessler A, Dauban P, Dodd RH, Rognan D, Ruat M. Positive and negative allosteric modulators of the Ca2+-sensing receptor interact within overlapping but not identical binding sites in the transmembrane domain. J Biol Chem. 2004;279:18990–18997. [DOI] [PubMed] [Google Scholar]
- 53. Wellendorph P, Hansen KB, Balsgaard A, Greenwood JR, Egebjerg J, Bräuner-Osborne H. Deorphanization of GPRC6A: a promiscuous L-α-amino acid receptor with preference for basic amino acids. Mol Pharmacol. 2005;67:589–597. [DOI] [PubMed] [Google Scholar]
- 54. Takahashi A, Shimano H, Nakagawa Y, et al. Transgenic mice overexpressing SREBP-1a under the control of the PEPCK promoter exhibit insulin resistance, but not diabetes. Biochim Biophys Acta. 2005;1740:427–433. [DOI] [PubMed] [Google Scholar]
- 55. Diraison F, Parton L, Ferré P, et al. Over-expression of sterol-regulatory-element-binding protein-1c (SREBP1c) in rat pancreatic islets induces lipogenesis and decreases glucose-stimulated insulin release: modulation by 5-aminoimidazole-4-carboxamide ribonucleoside (AICAR). Biochem J. 2004;378:769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Metukuri MR, Zhang P, Basantani MK, et al. ChREBP mediates glucose-stimulated pancreatic β-cell proliferation. Diabetes. 2012;61:2004–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Iizuka K, Bruick RK, Liang G, Horton JD, Uyeda K. Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc Natl Acad Sci USA. 2004;101:7281–7286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gupte AA, Sabek OM, Fraga D, et al. Osteocalcin protects against nonalcoholic steatohepatitis in a mouse model of metabolic syndrome. Endocrinology. 2014;155:4697–4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chang W, Tu C, Chen TH, Bikle D, Shoback D. The extracellular calcium-sensing receptor (CaSR) is a critical modulator of skeletal development. Sci Signal. 2008;1:ra1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hauschka PV, Carr SA. Calcium-dependent α-helical structure in osteocalcin. Biochemistry. 1982;21:2538–2547. [DOI] [PubMed] [Google Scholar]
- 61. Delmas PD, Stenner DD, Romberg RW, Riggs BL, Mann KG. Immunochemical studies of conformational alterations in bone γ-carboxyglutamic acid containing protein. Biochemistry. 1984;23:4720–4725. [DOI] [PubMed] [Google Scholar]
- 62. Dowd TL, Rosen JF, Li L, Gundberg CM. The three-dimensional structure of bovine calcium ion-bound osteocalcin using 1H NMR spectroscopy. Biochemistry. 2003;42:7769–7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pi M, Faber P, Ekema G, et al. Identification of a novel extracellular cation-sensing G-protein-coupled receptor. J Biol Chem. 2005;280:40201–40209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hoang QQ, Sicheri F, Howard AJ, Yang DS. Bone recognition mechanism of porcine osteocalcin from crystal structure. Nature. 2003;425:977–980. [DOI] [PubMed] [Google Scholar]
- 65. Frazão C, Simes DC, Coelho R, et al. Structural evidence of a fourth Gla residue in fish osteocalcin: biological implications. Biochemistry. 2005;44:1234–1242. [DOI] [PubMed] [Google Scholar]
- 66. Tsuka S, Aonuma F, Higashi S, et al. Promotion of insulin-induced glucose uptake in C2C12 myotubes by osteocalcin. Biochem Biophys Res Commun. 2015;459:437–442. [DOI] [PubMed] [Google Scholar]