Abstract
In this work we demonstrate a label-free optical imaging technique to assess metabolic status and oxidative stress in human induced pluripotent stem cell-derived cardiomyocytes by two-photon fluorescence lifetime imaging of endogenous fluorophores. Our results show the sensitivity of this method to detect shifts in metabolism and oxidative stress in the cardiomyocytes upon pathological stimuli of hypoxia and cardiotoxic drugs. This non-invasive imaging technique could prove beneficial for drug development and screening, especially for in vitro cardiac models created from stem cell-derived cardiomyocytes and to study the pathogenesis of cardiac diseases and therapy.
OCIS codes: (180.2520) Fluorescence microscopy, (170.3880) Medical and biological imaging
1. Introduction
Many drugs showing promise in preclinical trials fail during clinical development due to the emergence of cardiac side effects [1–3]. The development of in vitro platforms that accurately mimic the biology of human cardiac cells provide a plausible model for high-throughput drug screening to detect potential cardiotoxicity before wide spread human use. The emergence of human induced pluripotent stem cell technology has expanded the possibilities for sourcing human cardiomyocytes [4,5]. However there is a need for non-destructive techniques to visualize the biology and drug response of such in vitro human induced pluripotent stem cell-derived cardiomyocyte (hiPS-CM) models for drug screening. Novel microscopy and analysis methods can serve as powerful tools to study and monitor the physiology of such systems. Indices of metabolism and oxidative stress are particularly relevant in cardiac pathologies and response to drugs [6,7]. In this work, we employ two-photon fluorescence lifetime imaging microscopy (FLIM) to assess the metabolic state and oxidative stress in hiPS-CMs.
FLIM of endogenous fluorophores such as reduced Nicotinamide adenine dinucleotide (NADH), collagen, retinol, melanin, and flavins have been employed for convenient, intrinsic, label-free assessment of metabolism and other physiological functions of biological samples [8–10]. The advantage of this technique is its non-invasiveness. Use of intrinsic fluorophores circumvents administration of external fluorescent dyes, thus avoiding associated nonspecific binding, toxicity, and interference with the biochemical and physiological functions of the biological system being imaged.
FLIM of autofluorescent NADH can serve as a tool to study metabolism due to the extreme sensitivity of fluorescence lifetime to molecular conformations and the fluorophore’s surrounding environment. Also, unlike intensity based imaging, lifetime measurements are independent of concentration. Hence, NADH FLIM is extensively employed as an endogenous biomarker for metabolic imaging [8–15]. The reduced form of NADH is produced during glycolysis and Kreb’s cycle. During oxidative phosphorylation, it is oxidized to NAD+ by donating electrons to the electron transport chain, which are ultimately accepted by oxygen [8,9]. In the case of anaerobic glycolysis, NAD+ is converted to NADH and oxidative phosphorylation is diminished. This decreases the oxidation of NADH, resulting in an overall increase of free (not bound to protein) NADH. Thus, the reduction-oxidation pair NADH:NAD+ serves as an indicator of balance between oxidative phosphorylation and glycolysis. Bird, et al. demonstrated a correlation between the redox ratio NADH:NAD+ and the ratio of free to protein bound NADH, which is advantageous in fluorescent based technique because unlike NADH, NAD+ is not fluorescent [16].
Due to self-quenching, the fluorescence lifetime of NADH in the free state is significanly lower (~0.4ns) compared to protein bound NADH. For example, the lifetime of NADH bound to lactate dehydrogenase (LDH) is 3.4ns [17]. Hence, FLIM can easily differentiate between free and protein bound forms of NADH. In this work, we exploit these properties to assess metabolic activity of hiPS-CMs by FLIM of NADH. NADH was excited at two-photon excitation (TPE) of 740nm and fluorescence signal was collected between 420 – 500 nm. We used the phasor analysis of FLIM to create a map of free/bound NADH [18]. The phasor approach to FLIM for metabolic imaging has been previously applied for various studies, including cancer metabolism and metabolic shifts associated with stem cell differentiation [11,12]. This method simplifies the analysis of FLIM data, eliminating the requirement for fitting the fluorescence decay at each pixel. The technique has been described previously [11,18]. Briefly, data from each pixel are Fourier transformed to obtain the corresponding phasor and the 2-D histogram of the phasor is plotted on the phasor plot. This allows analysis of different lifetime phasor clusters which can be can be mapped back onto the image to create the FLIM map.
Here we employed NADH phasor-FLIM to detect the metabolic response of hiPS-CM to hypoxia. An uninterrupted oxygen supply is essential for cardiac tissue to meet the high metabolic demands and a lack of oxygen can have deleterious effects, leading to numerous pathological conditions. This has led to a growing interest in the effect of hypoxia [19–21]. Using the phasor approach to FLIM, along with metabolic shifts, we also identified an increase in a long lifetime species (LLS) with characteristic clustering in the lifetime phasor plot. Recently, we showed correlation of LLS to products of lipid oxidation formed by reactive oxygen species (ROS) using techniques like third harmonic generation (THG) imaging microscopy, coherent anti-Stokes Raman scattering (CARS) microscopy, and Raman spectroscopy [17]. To correlate the observed LLS to lipid droplets in hiPS-CMs, we performed stimulated Raman scattering (SRS) imaging microscopy. CH2 stretching vibrations, which are abundant in fatty acid chains, are easily excited and detected by SRS microscopy making it a useful technique to identify lipid bodies [22]. The LLS species were also detected by FLIM in hiPS-CMs treated with the anti-cancer drug cis-Diammineplatinum(II) dichloride (cisplatin) and the antiviral compound 3′-Azido-3′-deoxythymidine (AZT), both of which are known to produce oxidative stress and cardiotoxicity [23,24].
This study describes the use of a potentially powerful, non-invasive, label-free optical imaging technique for assessing the metabolic status and oxidative stress in cardiomyocytes. We demonstrate the sensitivity of this imaging technique to detect shifts in hiPS-CM metabolism and the generation of LLS in response to stimuli known to induce such effects like hypoxia and cardiotoxic drugs.
2. Results
2.1 Detection of metabolic shift in response to cyanide
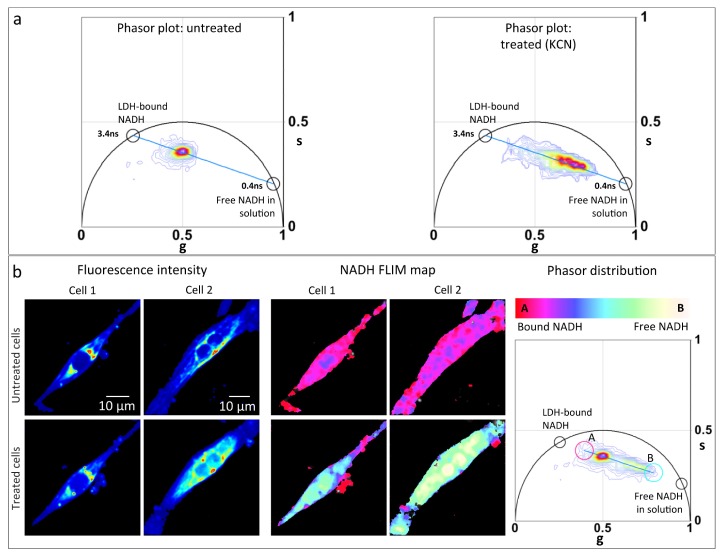
Cyanide is known to block the electron transport chain by inhibiting cytochrome oxidase function [25]. This reduces NADH oxidation causing an increase in the redox ratio, NADH/NAD+ . An increase in NADH fluorescence has been reported in cardiomyocytes treated with cyanide [26]. To test the sensitivity of the FLIM-phasor technique to detect metabolic shifts in hiPS-CMs, we performed NADH FLIM of hiPS-CM treated with 4mM potassium cyanide (KCN). Figure 1(a) shows the phasor distribution of hiPS-CMs before and after treatment with KCN. The phasor distributions lie along the metabolic trajectory (blue line) extending between the phasor fingerprint of free NADH (0.4ns) and protein (LDH) bound NADH (3.4ns). For all the phasor plots in Fig. 1, the experimental points lie along the line from the point at 3.4ns to the point at 0.4ns, on the universal circle. According to the law of linear combination of phasors, the two points on the universal circle are the extrapolated single lifetime components for the bound and free NADH forms, respectively. The phasor distribution of the treated cells is shifted towards the position of free NADH, indicating a higher free/bound NADH ratio. Figure 1(b) shows the total phasor distributions of two hiPS-CMs before and after treatment. The linear decrease in free/bound ratio of NADH has been pseudocolored from white/yellow to red/pink and mapped on the corresponding fluorescence intensity images to create the NADH FLIM map as shown in Fig. 1(b). The color-scale extends between positions ‘A’ and ‘B’ [Fig. 1(b)] which represents two extreme positions of the NADH phasor distribution, i.e., phasors with lowest free/bound NADH ratio and highest free/bound NADH ratio from these particular samples. The cells after KCN treatment display yellow-cyan colors in comparison to the pink-red color map before, consistent with an increase in free/bound NADH ratio after treatment. The NADH phasor shift towards position of free NADH fingerprint establishes the sensitivity of the system to detect shifts in the free/bound NADH ratio and thus metabolic changes in cardiomyocytes.
Fig. 1.
FLIM detects shift of metabolism to glycolysis when subjected to potassium cyanide (KCN). (a) Lifetime phasor distribution of untreated (left panel) and 4mM KCN treated (right panel) hiPS-CMs. Black circles shows phasor fingerprint of pure free NADH in solution (0.4ns) and protein lactate dehydrogenase bound NADH, ‘LDH-bound NADH’ (3.4ns) . Blue line indicates the metabolic trajectory on phasor plot. (b) Fluorescence intensity image (left panel) of two hiPS-CMs (cell 1 and cell 2) before (top row) and after (bottom row) treatment with 4mM KCN excited at 740nm. Middle panel shows the NADH FLIM map of cell 1 and cell 2 before (top row) and after (bottom row). Right panel shows the total phasor distribution of the treated and untreated cells. The color scale white/yellow to red/pink represents linear increase of free to protein bound NADH ratio. To create the NADH FLIM map, this color scale was applied from point A to B of the phasor distribution, dividing it into 32 levels.
2.2 FLIM detects metabolic response to hypoxia
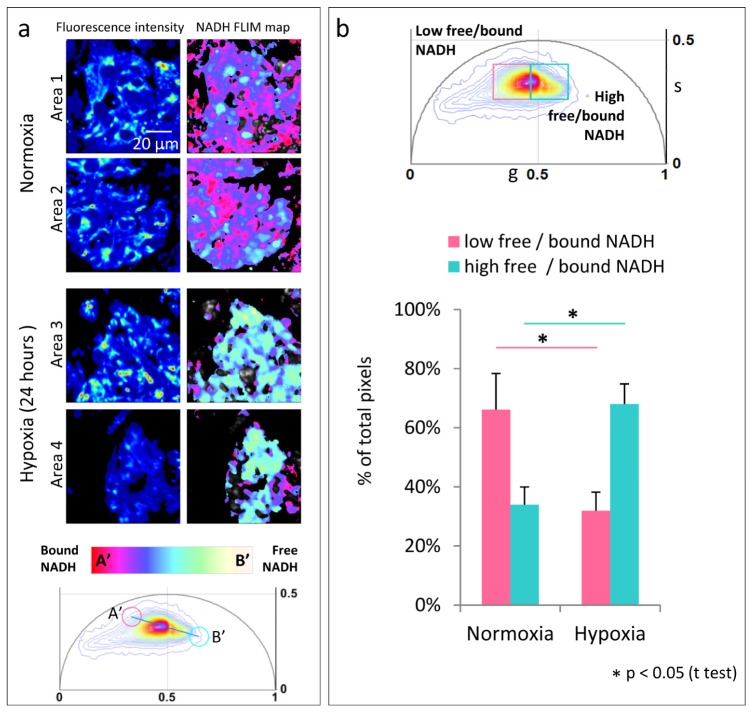
In hypoxic conditions, cells switch to anaerobic glycolysis for energy production. Using the FLIM-phasor technique, we imaged this alteration of cardiac metabolism by subjecting clusters of hiPS-CMs to hypoxia. Figure 2 shows NADH FLIM map of hiPS-CM clusters exposed to normoxia (20% oxygen, normal room oxygen concentration) and hypoxia (1% oxygen) for 24 hours. These oxygen conditions were maintained during FLIM imaging. Figure 2(a) shows the fluorescence intensity and pseudocolored NADH FLIM map of hiPS-CM clusters maintained for 24 hours in normoxia (Area 1 and Area 2) and hypoxia (Area 3 and Area 4). The NADH FLIM map shows distinct difference between hiPS-CM clusters in normoxia and hypoxia with higher free/bound NADH ratio in the latter condition, clearly indicating the glycolytic switch. Figure 2(a) bottom panel shows the corresponding phasor distribution and the positions (A’ and B’) between which the color scale was applied to divide the phasor distribution into 32 levels. The spread of NADH phasor distribution in Fig. 2(a) is smaller compared to Fig. 1(b), hence the positions of A’ and B’ differ from A and B in Fig. 1(b). Figure 2 (b) top panel shows phasor distribution of three different regions of hiPS-CM clusters of about 10 cells each for normoxia and hypoxia. For quantitative analysis, the NADH lifetime phasor distribution was divided into two windows: ‘high free/bound NADH’ (cyan square, center = (0.54, 0.33), side = 0.14) representing higher free/bound NADH ratio while ‘low free/bound NADH’ (pink square, center = (0.398, 0.33), side = 0.14) representing lower free/bound NADH ratio. The windows are of equal size and divide the NADH phasor distribution at the center of mass of the NADH distribution of the control (normoxia) condition. The fraction of pixels in all the acquired images whose corresponding phasor lies within each window was calculated and plotted [Fig. 2(b)]. Comparing the two oxygen conditions, we found the increase of pixel percentage in the ‘high free/bound NADH’ window and decrease in the ‘low free/bound NADH’ window in hypoxia to be statistically significant.
Fig. 2.
Shift in metabolism for 24 hours of hypoxia (a) Fluorescence intensity image (left panel) of clusters of hiPS-CMs under normal 20% oxygen condition (Area 1 and Area 2) and hiPS-CM under hypoxia, 1% oxygen (Area 3 and Area 4) . Right panel shows the corresponding NADH FLIM map. The color scale white/yellow to red/pink represents linear increase of free to protein bound NADH ratio between position A’ and B’ shown on the phasor plot (bottom panel) . (b) Top panel shows phasor distribution obtained from 3 three different regions of hiPS-CM clusters of about 10 cells each for the two oxygen conditions. Bottom panel is quantitative representation of the fraction of pixels with phasors within the ‘low free/bound NADH’ (pink) window and the ‘high free/bound NADH’ (cyan) window (top panel). The windows divide the NADH phasor distribution equally at the center of mass of the NADH phasor distribution of the control cells (normoxia). The error bars show the standard deviation calculated over average value obtained from each image.
2.3 LLS produced in hypoxia indicating oxidative stress
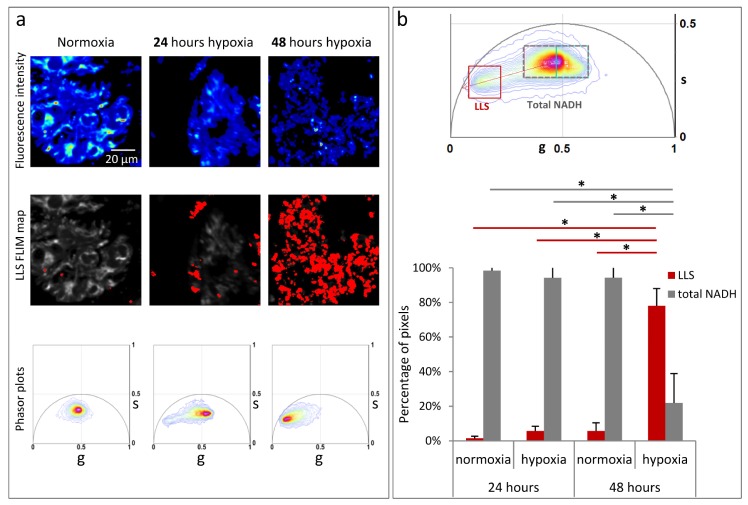
Interestingly, the phasor analysis of the hiPS-CMs in hypoxia when compared to cells subjected to equal duration of normoxia shows presence of long lifetime species (LLS) with characteristic lifetime phasor fingerprint. LLS is indicative of oxidative stress, as previously described [17]. Figure 3(a) shows hiPS-CMs in normoxia, 24 hours hypoxia, and 48 hours hypoxia. LLS FLIM map (in red) has been created by the LLS window on the phasor plot [Fig. 3(b)], which selects phasors with the characteristic LLS lifetime. These areas have granular appearance like lipid droplets similar to the previous observation [17]. Figure 3(a) bottom row shows the corresponding individual phasor distribution. In comparison to normoxia, the long lifetime tail of the phasor distribution moves along the oxidative stress axis shown as red dotted line in Fig. 2(b), towards pure LLS fingerprint on the universal curve (7.8ns) in the hypoxic condition [17].
Fig. 3.
Significant increase in the long lifetime species (LLS) with hypoxia. (a) Top row panels shows fluorescent intensity image of hiPS-CM clusters under normoxia, 24 hours hypoxia, 48 hours hypoxia. Middle row panels show LLS FLIM map (in red) created by the LLS window in (b) top panel. Bottom row shows the corresponding individual phasor plots. (b) Top panel is the phasor distribution of 3 three different regions of hiPS-CM clusters, about 10 cells each for normoxia, 24 hours hypoxia, and 48 hours hypoxia conditions. Gray dotted window (‘Total NADH’) selects the total NADH distribution which covers the total area within pink and cyan windows of Fig. 2(b). ‘LLS’ window (red square) selects the LLS phasor distribution. Red dotted arrow indicated the oxidative stress axis. Bottom panel shows quantitative representation of the percentage pixels with lifetime phasors within the ‘Total NADH’ window (gray dotted rectangle) and ‘LLS’ phasor window (red square) of hiPS-CMs under normoxia, 24 hours hypoxia, and 48 hours hypoxia. The error bars shows the standard deviation calculated over average value obtained from each image. Statistical significance was computed by ANOVA with post-hoc Bonferroni correction.
Figure 3(b) top panel is phasor distribution of three different regions of hiPS-CM clusters of about 10 cells each for the four conditions: 24 hours normoxia, 24 hours hypoxia, 48 hours normoxia and 48 hours hypoxia. For further quantitative analysis, the phasor distribution was divided into ‘LLS’ window: red square, center (0.151, 0.243), size = 0.14 and ‘total NADH’ window: gray dotted rectangle, center (0.468, 0.33), size (∆g = 0.28, ∆s = 0.14). The position of the ‘LLS’ window was determined by the center of mass of the LLS phasor distribution. The ‘total NADH’ window corresponds to the position of pink and cyan windows in Fig. 2(b). Figure 3(b) shows the normalized percentage of pixels in each phasor window for 24 and 48 hours hypoxia and the 24 and 48 hours normoxia controls. An ANOVA with post-hoc Bonferroni correction indicates a significant increase in LLS for cells exposed to 48 hours hypoxia, in comparison to normoxia controls as well as 24 hours hypoxia [Fig. 3(b)].
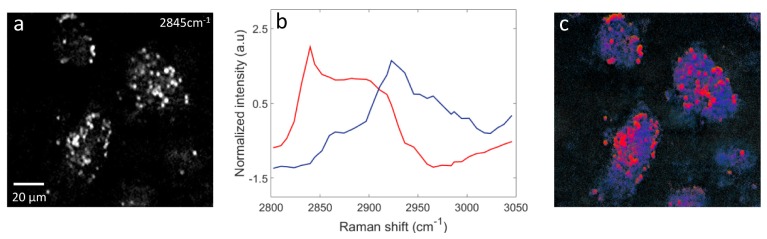
Though the granular areas had the characteristic long lifetime fingerprint on the phasor plot, we confirmed the association with lipids by performing hyperspectral SRS imaging that covers the CH stretching Raman band (2800 - 3050 cm-1) on the hiPS-CM clusters exposed to 48 hour hypoxia. In the SRS hyperspectral images, the strongest signal was observed at 2845 cm−1 (CH2 symmetric stretching Raman mode, characteristic of aliphatic components). Figure 4(a) is the SRS scan at 2845 cm−1 showing intense signal from lipid droplet-like granular structures. Hyperspectral SRS spectra were analyzed using vertex component analysis (VCA) with 3 end members as has been previously described [27]. Figure 4(b) shows the two major spectra retrieved from the VCA analysis (shown in red and blue). The third end member spectrum corresponds to background and has not been shown here. Each spectrum shown in Fig. 4(b) has been normalized to its maximum. Figure 4(c) is the pseudo-colored VCA image where the colors describe the chemical content of each pixel. The red areas in Fig. 4(c) associate with the spectrum in red in Fig. 4(b), which is characteristic of lipids [27]. These areas match very well with the granular areas observed Fig. 4(a), thus consistent with lipid droplets. The blue areas in Fig. 4(c) exhibited spectra typical of cellular protein, represented by the blue curve in Fig. 4(b).
Fig. 4.
Hyperspectral stimulated Raman scattering (SRS) scan of hiPS- CM exposed to 48 hours hypoxia, covering the CH stretching Raman band (2800 - 3050 cm−1) confirms association to lipid droplets. (a) SRS image at 2845cm−1 (CH2 symmetric stretching) characteristic of lipids. (b) Result of 3 endmember vertex component analysis (VCA). The red and blue show the normalized spectra of the 2 major endmembers retrieved. (c) Pseudo-colored VCA image of the SRS hyperspectral scan. The red spectrum is characteristic of lipids. It co-localizes with the lipid droplets observed in (a). The blue spectra have the features of the protein matrix that fills the cellular cytoplasm.
2.4 Cardiotoxic drugs produce LLS
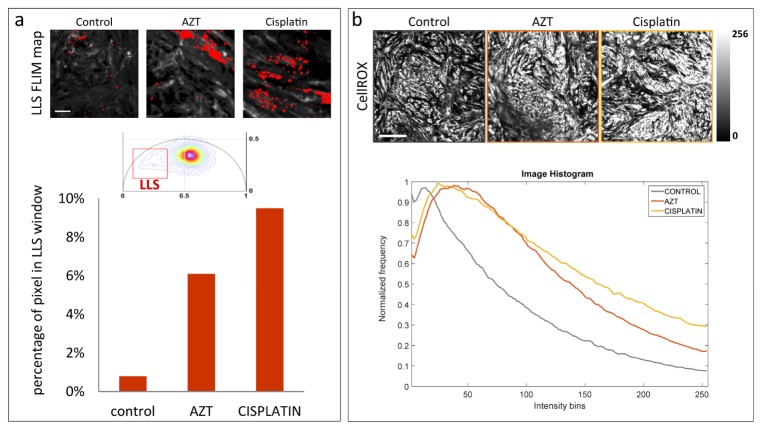
To further substantiate the sensitivity of the FLIM-phasor technique to detect oxidative stress in the cardiomyocyte, we treated hiPS-CMs with two known cardiotoxic drugs, AZT and cisplatin. FLIM was performed along with simultaneous ROS imaging using the ROS sensitive fluorescent dye, CellROX. Figure 5(a) shows the LLS FLIM map (in red) created by the LLS window (red square, center (0.217, 0.236), side = 0.28) on the phasor plot (middle panel). Again, the position of the LLS window was determined by the center of mass of the LLS phasor distribution. Larger numbers of lipid droplets with LLS signal were observed in the FLIM maps of the treated cells [Fig. 5(a)]. For quantitative analysis, the normalized percentage of pixels in each image with phasor in the LLS window was calculated [Fig. 5(a)] bottom panel. Figure 5(b) shows confocal ROS images of control, AZT treated and cisplatin treated cells. Corresponding intensity histograms of the confocal images show more intense CellROX signal in the cells treated with AZT and cisplatin compared to the controls which did not receive any treatment.
Fig. 5.
LLS produced by cardiotoxic drugs 3′-Azido-3′-deoxythymidine (AZT) and cis-Diamineplatinum(II) dichloride (cisplatin). (a) Top panel shows LLS FLIM map (in red) of hiPS-CM clusters (control, AZT treated and cisplatin treated). The FLIM map is generated by the LLS window (red square) selecting the characteristic long lifetime as shown on phasor plot (middle panel). Bottom panel shows the percentage pixels for control, AZT treated and cisplatin treated cells with lifetime phasors within the LLS phasor window. The scale bar is 20µm. (b) Top panel shows confocal image (control, AZT treated and cisplatin treated cells) of ROS indicator CellROX, excited at 633nm and signal collected within 640-740 nm. The scale bar is 200µm. Bottom panel shows the corresponding image intensity histogram.
3. Discussion
In this work, we demonstrate how FLIM imaging can be used to characterize the metabolic state and oxidative stress in hiPS-CMs using phasor analysis of endogenous biomarkers. Our results demonstrate the potential of this technique to detect drug-induced stress and pathological conditions in hiPS-CMs . We performed NADH FLIM of hiPS-CM treated with KCN, a known poison of mitochondria. Comparing the NADH FLIM phasor of the same cells before and after the treatment, we observe a large shift towards the free NADH fingerprint on the phasor plot, indicating a rise in the free/bound NADH ratio. This correlates with the decrease in oxidative phosphorylation which occurs in the mitochondria. Similar results were observed in breast cancer cells, cervical carcinoma HeLa, fibroblast and leukemia cells upon perturbation of the electron transport chain by cyanide [14,15,28]. The result shows the capability of the FLIM-phasor technique to detect metabolic changes in hiPS-CMs.
Lack of sufficient oxygen supply to cardiac tissue leads to pathophysiological conditions which might cause cardiac damage [29]. In the electron transport chain, oxygen is the terminal electron acceptor and its deprivation inhibits oxidative phosphorylation. Hence, hypoxia induces alteration of metabolic functions. Cells tend to depend on anaerobic glycolysis for production of ATP. This metabolic switch can be observed in our results. NADH FLIM-phasor analysis of hiPS-CMs subjected to 24 hours hypoxia (1% oxygen) had higher free/bound NADH ratio indicative of glycolysis as compared to the control cells in normoxia. The direction of this shift follows the same trend as observed in response to KCN treatment, although the shift is greater in the latter condition due to the cyanide poisoning by high concentration of KCN. Such increase in free fraction of NADH compared to its bound form upon hypoxia was reported in NADH fluorescence anisotropy studies in brain slices [30].
Another significant observation of FLIM-phasor analysis of hypoxic hiPS-CMs was the presence of LLS in the form of lipid droplets, which we had previously identified as a biomarker of oxidative stress [17]. This indicates the presence of lipid oxidation by ROS in hypoxic conditions. Interestingly, the amount of LLS increased significantly with 48 hours of hypoxia exposure when compared to 24 and 48 hours normoxia and 24 hours hypoxia. Furthermore, oxidative stress could be injurious to cells ultimately leading to cell death by apoptosis or necrosis [31]. Hence in the 48 hours hypoxia condition, there could be certain degree of cell death causing a loss in metabolism. SRS imaging corroborates the presence of lipid droplets in the cells exposed to 48 hours hypoxia. According to a study by Duranteau et. al. [19] on cardiomyocytes exposed to hypoxia, ROS is generated by the mitochondria which plays the role of oxygen sensor. They hypothesized that in cardiomyocytes, with decrease in oxygen concentration, this ROS signaling elicits adaptive responses like reduced contractile activity and suppressed oxygen consumption. They used an exogenous ROS sensitive dye to show an increase in ROS with a decrease in oxygen concentration which correlates with our observation. Several studies have also attributed generation of ROS during hypoxia or brief periods of ischemia as trigger for cardioprotective preconditioning [20,32]. The stress response initiated by ROS would lead to better adaptation to subsequent prolonged episodes of ischemia and increased chance of survival.
Oxidative stress by ROS was further detected by LLS generation in hiPS-CM treated with known cardiotoxic drugs. LLS were observed in FLIM-phasor analysis of hiPS-CM treated with antiviral drug AZT. Anti-cancer drug cisplatin is often used in ovarian, testicular, and gastrointestinal cancer [24]. However, it has been shown to produce oxidative stress and cardiotoxicity. Our results show an increase in LLS in hiPS-CM with cisplatin treatment. In the imaged areas, quantitative analysis shows a 5-fold and 8-fold increase of LLS signal in AZT treated and cisplatin treated cells respectively. Simultaneous confocal ROS imaging employing CellROX dye confirmed the increase in ROS in the treated cells.
Our results demonstrate that FLIM-phasor is a robust technique for assessing and indexing features of cardiac metabolism and ROS-induced oxidative stress. This method could prove beneficial for drug development and screening studies, especially for in vitro cardiac models derived from stem cell-derived cardiomyocytes. Furthermore, these indices play a major role in the pathogenesis of a myriad of cardiovascular diseases including atherosclerosis, ischemic heart disease, and heart failure. Use of endogenous fluorophores as intrinsic biomarkers of metabolism and oxidative stress makes this technique label-free, creating the possibility of in vivo measurements. Additionally, cardiac systems can be monitored over time. Thus, this non-destructive technique will find potential application in in vitro cardiac models in drug discovery and safety screening, especially unanticipated cardiotoxic effects of anti-cancer drugs as well as understanding the pathogenesis of cardiac diseases and therapy.
4. Materials and methods
4.1 Instrumentation
FLIM was performed on a Zeiss LSM 710 microscope (Carl Zeiss, Jena, Germany) using a 40x, 1.2 N.A. water immersion objective, (Carl Zeiss, Oberkochen, Germany) coupled to an 80MHz multiphoton excitation laser source, Titanium:Sapphire MaiTai laser (Spectra-Physics, Mountain View, CA). 2PE (2 photon excitation) excitation of NADH and LLS was carried out at 740nm. Image scan speed was 25.21 µs/pixel with an image size of 256 × 256 pixels. A dichroic at 690 nm was employed to separate excitation from emission signal. A bandpass emission filter 460/80nm (Semrock, Rochester, NY) coupled to a photomultiplier tube (H7422P-40, Hamamatsu, Japan) was used as the microscope external detector port photo-sensor unit. FLIM data was acquired using A320 FastFLIM FLIMbox (ISS, Champaign, IL). For each image, 60 - 70 frames were collected and integrated for FLIM analysis. SimFCS software (LFD, Irvine) was used for frequency domain FLIM data acquisition. For calibrating the FLIM system, Rhodamine 110 with known lifetime of 4ns was measured for every experiment. All the (g, s) coordinate system used to mention phasor cursor coordinates in this article used the first harmonic phasor plots at 80Mhz (repetition rate of the laser).
Confocal imaging of ROS indicator CellROX was performed on the same Zeiss LSM 710 microscope (Carl Zeiss, Jena, Germany) and with an excitation of 633nm. Fluorescence signal was collected in the range 640-740 nm. The laser excitation power and detector gain was kept constant for all measurements.
Stimulated Raman scattering (SRS) signals were obtained as previously described here [27]. Briefly, a Stokes beam fixed at ∼9400 cm−1 and a pump beam tuned to the wavelength of interest (2800 – 3050 cm−1) were overlapped both temporally and spatially and sent into a laser scanner (Fluoview 300, Olympus, Center Valley, Pennsylvania), attached to an inverted microscope (IX71, Olympus). The combined beams were then focused through a 20 × , 0.75 NA objective lens (UplanS Apo, Olympus) onto the sample. SRS images were obtained by detecting the stimulated Raman loss of the pump beam with a photodiode (FDS1010; Thorlabs, Newton, New Jersey). The average combined power of Stokes and pump beams at the specimen was kept under 50 mW throughout this study to minimize sample photodamage.
4.2 hiPS-CM differentiation and culture
hiPS-CMs were differentiated from the wtc11 line of human induced pluripotent stem (hiPS) cells generously donated by Dr. Bruce Conklin [33]. hiPS cells were differentiated into cardiomyocyte like cells following previously established protocol which entails culture in Roswell Park Memorial Institute (RPMI) medium (Life Technologies, 22400–071) supplemented with B-27 without insulin (Life Technologies, A1895601) [34]. The protocol includes supplementing media with 12 μM CHIR99021 (Selleckchem, S2924) for 24 hours after which it is removed on day 0. Media is supplemented with 5 μM IWP2 (Tocris, 3533) on day 3 which is then removed on day 5. On day 7, the media is supplemented with insulin, and after that day the cells were fed RPMI/B-27 ( + ) insulin (Life Technologies, 17504–044) every 2–3 days for the duration of the experiment. Approximately between days 12–15, the cells started beating spontaneously. hiPS-CM drug exposure and measurements were performed on days 22-30. Cells were qualitatively confirmed to be beating throughout differentiation and culture (including post-hypoxia exposure and post-drug exposure)
4.3 Cyanide treatment
KCN (Sigma, St. Louis, MO) in phosphate buffered saline (PBS) (Sigma-Aldrich, St. Louis, MO) was added to the cell culture media to reach a final concentration of 4mM. FLIM was performed on the cells before the treatment and the same cells were imaged 2-3 mins after addition. During FLIM measurement temperature of 37° C and 5% CO2 was maintained.
4.4 Hypoxia exposure
For hypoxia, the cells were exposed to a gas mixture (1% O2, 5% CO2, 94% N2) in a hypoxic microchamber (Stem Cell Technologies, Vancouver, BC, Canada) and were incubated at 37° C for either 24 or 48 hours. During FLIM imaging, the cells where quickly transferred to the microscope stage incubation chamber which was flooded with the same gas mixture. A temperature of 37° C was maintained throughout the imaging.
4.4 Drug treatments
AZT (Sigma-Aldrich, St. Louis, MO) in nanopore filtered water was added to the hiPS-CMs culture media to reach a final concentration of 50µM. Cisplatin (Sigma-Aldrich, St. Louis, MO) was added to media, also to a final concentration of 50µM. Temperature of 37° C and 5% CO2 was maintained during imaging.
4.5 In vivo staining
To assess the concentration of reactive oxygen species (ROS), hiPS-CMs were stained with ROS indicator, CellROX Deep Red (ThermoFisher Scientific, Waltham, MA), at a final concentration of 5uM after treatment (cisplatin, AZT, or control). Cultures were incubated with CellROX for 30 min at 37° C, then washed 3X with PBS, and imaged immediately.
4.6 Data analysis
All the FLIM data was analyzed using SimFCS software developed at the Laboratory for Fluorescence Dynamics (LFD, UC Irvine). For statistical analysis in section 2.2, the Student’s t-test was used to determine the significance and considered positive for p < 0.05. In section 2.3, statistical significance was determined by an ANOVA with post-hoc Bonferroni correction using MATLAB software.
Acknowledgments
This work was supported by the National Institutes of Health grants P41-GM103540, P50-GM076516, UH2-TR000481 and UH3-TR000481. Also, we want to thank Alba Alfonso-García for help with SRS measurement, analysis and interpretation, Dr. David Tran for preparing hiPS-CM culture for KCN experiment, Yosuke Kurokawa for discussions and Dr. Jenu Chacko for reading the manuscript.
References and links
- 1.Colombo A., Cipolla C., Beggiato M., Cardinale D., “Cardiac toxicity of anticancer agents,” Curr. Cardiol. Rep. 15(5), 362 (2013). 10.1007/s11886-013-0362-6 [DOI] [PubMed] [Google Scholar]
- 2.Heylman C., Sobrino A., Shirure V. S., Hughes C. C., George S. C., “A strategy for integrating essential three-dimensional microphysiological systems of human organs for realistic anticancer drug screening,” Exp. Biol. Med. (Maywood) 239(9), 1240–1254 (2014). 10.1177/1535370214525295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albini A., Pennesi G., Donatelli F., Cammarota R., De Flora S., Noonan D. M., “Cardiotoxicity of anticancer drugs: the need for cardio-oncology and cardio-oncological prevention,” J. Natl. Cancer Inst. 102(1), 14–25 (2010). 10.1093/jnci/djp440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mordwinkin N. M., Burridge P. W., Wu J. C., “A review of human pluripotent stem cell-derived cardiomyocytes for high-throughput drug discovery, cardiotoxicity screening, and publication standards,” J. Cardiovasc. Transl. Res. 6(1), 22–30 (2013). 10.1007/s12265-012-9423-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeevi-Levin N., Itskovitz-Eldor J., Binah O., “Cardiomyocytes derived from human pluripotent stem cells for drug screening,” Pharmacol. Ther. 134(2), 180–188 (2012). 10.1016/j.pharmthera.2012.01.005 [DOI] [PubMed] [Google Scholar]
- 6.Hafstad A. D., Nabeebaccus A. A., Shah A. M., “Novel aspects of ROS signalling in heart failure,” Basic Res. Cardiol. 108(4), 359 (2013). 10.1007/s00395-013-0359-8 [DOI] [PubMed] [Google Scholar]
- 7.Burgoyne J. R., Mongue-Din H., Eaton P., Shah A. M., “Redox signaling in cardiac physiology and pathology,” Circ. Res. 111(8), 1091–1106 (2012). 10.1161/CIRCRESAHA.111.255216 [DOI] [PubMed] [Google Scholar]
- 8.Heikal A. A., “Intracellular coenzymes as natural biomarkers for metabolic activities and mitochondrial anomalies,” Biomarkers Med. 4(2), 241–263 (2010). 10.2217/bmm.10.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Georgakoudi I., Quinn K. P., “Optical imaging using endogenous contrast to assess metabolic state,” Annu. Rev. Biomed. Eng. 14(1), 351–367 (2012). 10.1146/annurev-bioeng-071811-150108 [DOI] [PubMed] [Google Scholar]
- 10.Skala M. C., Riching K. M., Bird D. K., Gendron-Fitzpatrick A., Eickhoff J., Eliceiri K. W., Keely P. J., Ramanujam N., “In vivo multiphoton fluorescence lifetime imaging of protein-bound and free nicotinamide adenine dinucleotide in normal and precancerous epithelia,” J. Biomed. Opt. 12(2), 024014 (2007). 10.1117/1.2717503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stringari C., Sierra R., Donovan P. J., Gratton E., “Label-free separation of human embryonic stem cells and their differentiating progenies by phasor fluorescence lifetime microscopy,” J. Biomed. Opt. 17(4), 046012 (2012). 10.1117/1.JBO.17.4.046012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pate K. T., Stringari C., Sprowl-Tanio S., Wang K., TeSlaa T., Hoverter N. P., McQuade M. M., Garner C., Digman M. A., Teitell M. A., Edwards R. A., Gratton E., Waterman M. L., “Wnt signaling directs a metabolic program of glycolysis and angiogenesis in colon cancer,” EMBO J. 33(13), 1454–1473 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vergen J., Hecht C., Zholudeva L. V., Marquardt M. M., Hallworth R., Nichols M. G., “Metabolic imaging using two-photon excited NADH intensity and fluorescence lifetime imaging,” Microsc. Microanal. 18(4), 761–770 (2012). 10.1017/S1431927612000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghukasyan V. V., Kao F.-J., “Monitoring Cellular Metabolism with Fluorescence Lifetime of Reduced Nicotinamide Adenine Dinucleotide †,” J. Phys. Chem. C 113(27), 11532–11540 (2009). 10.1021/jp810931u [DOI] [Google Scholar]
- 15.Walsh A. J., Cook R. S., Manning H. C., Hicks D. J., Lafontant A., Arteaga C. L., Skala M. C., “Optical metabolic imaging identifies glycolytic levels, subtypes, and early-treatment response in breast cancer,” Cancer Res. 73(20), 6164–6174 (2013). 10.1158/0008-5472.CAN-13-0527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bird D. K., Yan L., Vrotsos K. M., Eliceiri K. W., Vaughan E. M., Keely P. J., White J. G., Ramanujam N., “Metabolic mapping of MCF10A human breast cells via multiphoton fluorescence lifetime imaging of the coenzyme NADH,” Cancer Res. 65(19), 8766–8773 (2005). 10.1158/0008-5472.CAN-04-3922 [DOI] [PubMed] [Google Scholar]
- 17.Datta R., Alfonso-García A., Cinco R., Gratton E., “Fluorescence lifetime imaging of endogenous biomarker of oxidative stress,” Sci. Rep. 5, 9848 (2015). 10.1038/srep09848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Digman M. A., Caiolfa V. R., Zamai M., Gratton E., “The phasor approach to fluorescence lifetime imaging analysis,” Biophys. J. 94(2), L14–L16 (2008). 10.1529/biophysj.107.120154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duranteau J., Chandel N. S., Kulisz A., Shao Z., Schumacker P. T., “Intracellular Signaling by Reactive Oxygen Species during Hypoxia in Cardiomyocytes,” J. Biol. Chem. 273(19), 11619–11624 (1998). 10.1074/jbc.273.19.11619 [DOI] [PubMed] [Google Scholar]
- 20.Vanden Hoek T. L., Becker L. B., Shao Z., Li C., Schumacker P. T., “Reactive Oxygen Species Released from Mitochondria during Brief Hypoxia Induce Preconditioning in Cardiomyocytes,” J. Biol. Chem. 273(29), 18092–18098 (1998). 10.1074/jbc.273.29.18092 [DOI] [PubMed] [Google Scholar]
- 21.Semenza G. L., “Hypoxia-inducible factor 1 and cardiovascular disease,” Annu. Rev. Physiol. 76(1), 39–56 (2014). 10.1146/annurev-physiol-021113-170322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang M. C., Min W., Freudiger C. W., Ruvkun G., Xie X. S., “RNAi screening for fat regulatory genes with SRS microscopy,” Nat. Methods 8(2), 135–138 (2011). 10.1038/nmeth.1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varga Z. V, Ferdinandy P., Liaudet L., Pacher P., “Drug-induced mitochondrial dysfunction and cardiotoxicity,” Am. J. Physiol. 309, 00554 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deavall D. G., Martin E. A., Horner J. M., Roberts R., “Drug-Induced Oxidative Stress and Toxicity,” J. Toxicol. 2012, 645460 (2012). 10.1155/2012/645460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Rourke B., Cortassa S., Aon M. A., “Mitochondrial ion channels: gatekeepers of life and death,” Physiology (Bethesda) 20(5), 303–315 (2005). 10.1152/physiol.00020.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffiths E. J., Lin H., Suleiman M. S., “NADH Fluorescence in Isolated Guinea-Pig and Rat Cardiomyocytes Exposed to Low or High Stimulation Rates and Effect of Metabolic Inhibition with Cyanide,” Biochem. Pharmacol. 56(2), 173–179 (1998). 10.1016/S0006-2952(98)00016-1 [DOI] [PubMed] [Google Scholar]
- 27.Alfonso-García A., Pfisterer S. G., Riezman H., Ikonen E., Potma E. O., “D38-cholesterol as a Raman active probe for imaging intracellular cholesterol storage,” J. Biomed. Opt. 21(6), 061003 (2015). 10.1117/1.JBO.21.6.061003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vergen J., Hecht C., Zholudeva L. V., Marquardt M. M., Hallworth R., Nichols M. G., “Metabolic imaging using two-photon excited NADH intensity and fluorescence lifetime imaging,” Microsc. Microanal. 18(4), 761–770 (2012). 10.1017/S1431927612000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giordano F. J., “Oxygen, oxidative stress, hypoxia, and heart failure,” J. Clin. Invest. 115(3), 500–508 (2005). 10.1172/JCI200524408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vishwasrao H. D., Heikal A. A., Kasischke K. A., Webb W. W., “Conformational dependence of intracellular NADH on metabolic state revealed by associated fluorescence anisotropy,” J. Biol. Chem. 280(26), 25119–25126 (2005). 10.1074/jbc.M502475200 [DOI] [PubMed] [Google Scholar]
- 31.Ryter S. W., Kim H. P., Hoetzel A., Park J. W., Nakahira K., Wang X., Choi A. M. K., “Mechanisms of cell death in oxidative stress,” Antioxid. Redox Signal. 9(1), 49–89 (2007). 10.1089/ars.2007.9.49 [DOI] [PubMed] [Google Scholar]
- 32.Zhou X., Zhai X., Ashraf M., “Direct evidence that initial oxidative stress triggered by preconditioning contributes to second window of protection by endogenous antioxidant enzyme in myocytes,” Circulation 93(6), 1177–1184 (1996). 10.1161/01.CIR.93.6.1177 [DOI] [PubMed] [Google Scholar]
- 33.Okita K., Matsumura Y., Sato Y., Okada A., Morizane A., Okamoto S., Hong H., Nakagawa M., Tanabe K., Tezuka K., Shibata T., Kunisada T., Takahashi M., Takahashi J., Saji H., Yamanaka S., “A more efficient method to generate integration-free human iPS cells,” Nat. Methods 8(5), 409–412 (2011). 10.1038/nmeth.1591 [DOI] [PubMed] [Google Scholar]
- 34.Lian X., Zhang J., Azarin S. M., Zhu K., Hazeltine L. B., Bao X., Hsiao C., Kamp T. J., Palecek S. P., “Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions,” Nat. Protoc. 8(1), 162–175 (2012). 10.1038/nprot.2012.150 [DOI] [PMC free article] [PubMed] [Google Scholar]