Abstract
The fission yeast Schizosaccharomyces pombe has been successfully used as a model to gain fundamental knowledge in understanding how eukaryotic cells acquire copper during vegetative growth. These studies have revealed the existence of a heteromeric Ctr4–Ctr5 plasma membrane complex that mediates uptake of copper within the cells. Furthermore, additional studies have led to the identification of one of the first vacuolar copper transporters, Ctr6, as well as the copper-responsive Cuf1 transcription factor. Recent investigations have extended the use of S. pombe to elucidate new roles for copper metabolism in meiotic differentiation. For example, these studies have led to the discovery of Mfc1, which turned out to be the first example of a meiosis-specific copper transporter. Whereas copper-dependent transcriptional regulation of the Ctr family members is under the control of Cuf1 during mitosis or meiosis, meiosis-specific copper transporter Mfc1 is regulated by the recently discovered transactivator Mca1. It is foreseeable that identification of novel meiotic copper-related proteins will serve as stepping stones to unravel fundamental aspects of copper homoeostasis.
Keywords: copper homoeostasis, copper transporter, fission yeast, meiosis
Introduction
Copper is a redox-active transition metal that serves as a cofactor for a number of important enzymes, including Cco (cytochrome c oxidase), Sod1 (superoxide dismutase 1), multicopper ferroxidase and Cao (copper amine oxidase). These enzymes are respectively involved in fundamental cellular processes such as respiration, superoxide anion detoxification, iron transport and xenobiotic amine metabolism [1]. On the other hand, copper can engage in detrimental chemical reactions or compete with other metal ions at enzyme-active sites, due to its ability to lose or gain a single electron [2]. It has also been reported that excess copper may cause cell-cycle misregulation [3]. It is therefore critical that cells use tightly regulated homoeostatic mechanisms to acquire sufficient amounts of copper, while at the same time preventing its accumulation to cytotoxic levels. The present review focuses on recent progress that has been made in understanding new roles for proteins involved in copper metabolism in the yeast model Schizosaccharomyces pombe.
Copper transport in vegetative cells
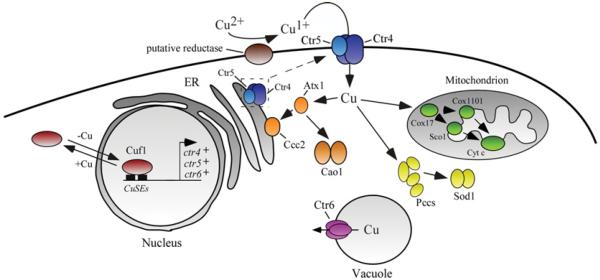
The copper transport machinery has traditionally been studied in dividing cells that grow mitotically (vegetative growth). In these cells, it is thought that copper is reduced from Cu2+ to Cu+ by a cell-surface reductase before being transported by a dual partner complex composed of two proteins encoded by the ctr4+ and ctr5+ genes [4–6] (Figure 1). Co-expression of Ctr4 and Ctr5 proteins is required for proper function and localization of the heteroprotein complex at the cellular surface. The absence of either Ctr4 or Ctr5 results in reciprocal mislocalization of the respective Ctr5 or Ctr4 partner within the secretory pathway [5,6]. As expected, cells with deletion of the ctr4+ or ctr5+ gene exhibit copper starvation phenotypes characterized by the inability to take up 64Cu, poor cell growth on low-copper medium and alterations in copper-dependent enzymes activity (e.g. Sod1 and Cao1). Furthermore, ctr4Δ, ctr5Δ or ctr4Δ ctr5Δ mutants fail to grow on respiratory carbon sources owing to a defect of copper incorporation in mitochondrial Cco. In addition, these mutants are greatly affected when grown in media with limited iron availability because of alteration in copper-dependent ferroxidase Fio1, which is essential for high-affinity iron uptake in fission yeast.
Figure 1. Copper homoeostasis during proliferation in mitosis in S. pombe.
Copper is reduced from Cu2+ to Cu+ by a putative cell-surface reductase. It is then transported across the plasma membrane by a heteroprotein complex formed by the Ctr4 and Ctr5 proteins. There is a mutual dependence between Ctr4 and Ctr5 because trafficking of either protein to the cell surface requires concomitant trafficking of its partner. Results obtained in yeasts S. pombe and S. cerevisiae, in addition to comparison of sequence homology with S. cerevisiae chaperones, suggest that copper chaperones in fission yeast include Atx1, Pccs and Cox17. Atx1 carries copper to Ccc2, a P-type ATPase in the Golgi membrane that pumps copper into the secretory pathway where it can be incorporated into newly synthesized copper proteins. Atx1 is also involved in the delivery of copper to Cao1. In the mitochondrion, Cox17 delivers copper to Cco via Sco1 and presumably Cox1101. Pccs shuttles copper to Sod1 in the cytosol. When the pool of cytoplasmic copper is depleted, Ctr6 participates in copper efflux from the vacuole, providing copper to cytosolic copper-dependent enzymes. In response to copper deficiency, Cuf1 is localized in the nucleus where it binds to CuSEs and activates transport of copper by inducing expression of ctr4+, ctr5+ and ctr6+ genes. In contrast, in cells undergoing a shift from low to sufficient copper concentrations, Cuf1 translocates from the nucleus to the cytoplasm. Brown, putative reductase; blue, Ctr4/Ctr5; orange, Atx1, Ccc2 and Cao1; green, Cox17, Cox1101, Sco1 and cytochrome c oxidase; yellow, Pccs and Sod1; purple, Ctr6; red, Cuf1.
As is the case for most members of the Ctr family, Ctr4 and Ctr5 harbour extracellular N-terminal hydrophilic regions that contain methionine residues organized as Met-Xaa2-Met and/or Met-Xaa-Met motifs (denoted Mets motifs). The extracellularly located N-terminal regions of Ctr4 and Ctr5 can function independently in copper transport, but efficiency is maximal when both N-termini are present [6]. The primary structure of Ctr4 and Ctr5 contains three TMDs (transmembrane domains), a Met-Xaa3-Met motif within their second TMD and a Gly-Xaa3-Gly motif within their third TMD that is essential for trimeric assembly of the Ctr complex [7,8]. Structure–function analysis of Ctr4 and Ctr5 has revealed that the Met-Xaa3-Met motif in TMD2 of Ctr5 is not required for a functional Ctr4–Ctr5 copper-transporting complex [9]. In contrast and, as is the case for human and Saccharomyces cerevisiae Ctr1 proteins, the TDM2 Met-Xaa3-Met motif in Ctr4 is necessary for function and localization of the Ctr4–Ctr5 complex at the plasma membrane [9]. Further functional dissection of the Ctr4 and Ctr5 domains using Ctr4/Ctr5 chimaeric proteins has revealed that the C-terminal region of Ctr4, which is composed of TMD3 and the cytosolic tail, blocks the delivery of Ctr4/Ctr5 chimaeras to the cell surface. In contrast, replacing the Ctr4 C-terminal region with the equivalent C-terminal region of Ctr5 results in migration of the Ctr4/Ctr5 chimaeric proteins to the cell surface. These observations suggest a role for the Ctr5 C-terminal region in the regulation of exit of the heteromeric Ctr4–Ctr5 complex from the secretory pathway [9]. In agreement with these findings, bimolecular fluorescence complementation experiments have shown that the assembly of a functional heteromeric Ctr4–Ctr5 complex on the cell surface requires a combination of two Ctr4 molecules with one Ctr5 molecule [10]. In addition, these studies have indicated that the Ctr4–Ctr5 copper transport complex undergoes internalization in cells in response to high-copper conditions, and can recycle back to the cell surface when cells undergo a transition from copper-sufficient to copper-limiting conditions. After crossing the cell surface, copper chaperones traffic copper to specific targets [11]. In the case of S. pombe, Pccs, Cox17 and Atx1 serve as copper chaperones to deliver copper to cytosolic Sod1, mitochondrial Cco and cytosolic Cao1 respectively. Furthermore, Atx1 carries copper to the secretory compartment by docking specifically with the Ccc2 copper-transporting P-type ATPase [12–14] (Figure 1).
A third member of the Ctr family, denoted Ctr6, localizes on the membrane of vacuoles [15]. The predicted topology of Ctr6 indicates that the protein possesses three TMDs (TMD1–3) and that its N-terminal region is located inside the vacuole, whereas its C-terminus is located in the cytoplasm. The N-terminus of Ctr6 contains a Met-Xaa-His-Cys-Xaa-Met-Xaa-Met motif that is involved in the process of copper transport [15]. This motif is identical with the Mets motifs identified in the Ctr family of copper transporters [16], except for the presence of a cysteine residue in the fourth position of the motif instead of a methionine residue. TMD2 of Ctr6 contains a highly conserved Met-Xaa3-Met motif, whereas TMD3 possesses a trimerization Gly-Xaa3-Gly motif that is predicted to be responsible for the assembly of Ctr6 as a homotrimer [15]. Genetic studies have revealed that the disruption of ctr6+ (ctr6Δ) within the context of a ctr4Δ strain results in decreased activity of Sod1, suggesting a role for Ctr6 in delivering copper to one or more cytosolic copper-dependent enzymes [15]. Cells overexpressing ctr6+ are unable to grow under high-copper conditions. Surprisingly, this phenotype is not due to an increase in copper uptake. Instead, the cell-surface copper transport activity is reduced and this is associated with decreased steady-state levels of ctr4+ mRNA [15]. On the basis of these observations, a model of intercommunication between vacuole and plasma membrane has been proposed. According to the model, as Ctr6 mobilizes stored copper from vacuole to the cytoplasm, Cuf1 senses the increased size of the pool of bioavailable copper which, in turn, represses the Cuf1-dependent transcription of Ctr4. The combination of these effects results in reduced copper uptake [15].
Copper regulator Cuf1
The transcription factor Cuf1 activates the expression of genes encoding components of the copper transport pathway (e.g. Ctr4, Ctr5 and Ctr6) in S. pombe, under conditions of low intracellular levels of copper [4,5,15,17,18]. In contrast, when cells are grown in the presence of high levels of copper, Cuf1 is unable to bind chromatin, which results in repression of the ctr4+, ctr5+ and ctr6+ genes [14]. cuf1Δ mutant cells exhibit phenotypes typical of copper insufficiency, including inability to transport copper, failure to grow on respiratory carbon sources owing to the lack of copper incorporation into Cco, and impaired Sod1 and Cao activities. Although Cuf1 plays an essential role in transactivating copper transport gene expression under copper-limiting conditions, the sequence of its first 61 amino acid residues displays strong homology with S. cerevisiae Ace1 and Candida glabrata Amt1 classes of copper-detoxifying transcription factors [17]. It has been shown that Cuf1 binds specifically to DNA sequences similar to those of Ace1 and Amt1. The Cuf1 DNA-binding sequence 5′-D(T/A)DDHGCTGD-3′ (D = A, G or T; H = A, C or T) denoted CuSE (copper-signalling element) is closely related to the MRE (metal-regulatory element; 5′-HTHNNGCTGD-3′) that is bound by Ace1 and Amt1. In vitro binding studies have revealed that a recombinant protein containing residues 1–174 of Cuf1 binds CuSEs [19]. Further analysis, using chromatin immunoprecipitation assays in S. pombe, have revealed that Cuf1 binds to the CuSEs-containing ctr4+ promoter in vivo under copper-limiting conditions [14]. In contrast, Cuf1 is released from the ctr4+ promoter under conditions of high copper concentrations, resulting in inactivation of ctr4+ gene expression.
N-terminal residues 11–53 of Cuf1 contain a non-canonical NLS (nuclear localization signal) [20]. Deletion of this region or individual mutation of the nine positively charged amino acids (Lys13, Arg16, Arg19, Lys24, Arg28, Lys45, Arg47, Arg50 and Arg53) to alanine in full-length Cuf1 abrogates its translocation to the nucleus. Furthermore, when the same N-terminal sequence is fused with GFP2× (two copies of GFP), the resulting fusion protein is able to accumulate in the nucleus, indicating that Cuf1's NLS property is transferable to a heterologous reporter GFP2× protein [20].
The C-terminal region of Cuf1 contains a cluster of cysteine residues (Cys328-Xaa-Cys330-Xaa3-Cys334-Xaa-Cys336-Xaa2-Cys339-Xaa2-His342), known as the C-rich domain, which exhibits a strong identity with a similar domain found in two copies at the C-terminus of Mac1, its functional orthologue in S. cerevisiae [19,21–24]. Mutations of all five cysteine residues and one histidine residue within the C-rich domain result in cells that exhibit sustained expression of the ctr4+ gene, even in the presence of a high concentration (100 μM) of copper, along with concomitant cellular hypersensitivity to copper ions [19]. This observation, along with other phenotypes observed for this gain-of-function allele, suggests that the C-rich domain of Cuf1 is responsible for sensing the presence of excess copper. Additional structure–function analysis has revealed the presence of a leucine-rich NES (nuclear export signal) located between residues 349 and 358, downstream of the copper-sensing C-rich domain of Cuf1 [25]. Consistent with the amino acid composition of Cuf1 NES (L349AALNHISAL358), nuclear export of Cuf1 requires Crm1, an exportin known to bind proteins containing a leucine-rich NES [26,27]. Protein–protein interaction assays have revealed that Crm1 associates with Cuf1. It has been shown that nuclear retention of Cuf1 is not sufficient to cause a constitutive activation of copper transport genes in a crm1 temperature-sensitive mutant strain [25]. When the Cuf1 on N-terminus (residues 1–61; including the NLS) is co-expressed as a separate molecule with the C-terminal portion of Cuf1 (residues 62–410) in S. pombe, the presence of copper induces cytoplasmic retention of Cuf1-(1–61). The sequestration of Cuf1-(1–61) in the cytosol requires cysteine and histidine residues (Cys328, Cys330, Cys334, Cys336, Cys339 and His342) within the C-rich motif of Cuf1-(62–410). Similarly, a two-hybrid approach has shown that Cuf1-(1–61) interacts with Cuf1-(62–410) in a copper- and C-rich motif-dependent manner [25]. On the basis of these results and other observations, a model for modulation of Cuf1 localization has been proposed. According to the model, Cuf1 associates with an unidentified importin via its accessible NLS (located within its N-terminus) under copper-limiting conditions. Cuf1 is then delivered to the nucleus, activating expression of target genes. In contrast, under conditions of excess copper, Cuf1 binds copper ions through specific cysteine residues found in both N- and C-termini (particularly the C-rich motif), and adopts a conformation that masks the N-terminal NLS. Inaccessibility of the NLS allows Cuf1 to accumulate in the cytosol because the association between Cuf1 and an importin is not possible. In cells undergoing a shift from low to elevated copper concentrations, metallation of the pre-existing nuclear pool of Cuf1 induces intramolecular conformational changes that block Cuf1 binding to CuSE, preventing its transactivation function. To further ensure that expression of copper transport genes will not occur in the presence of copper, the Crm1 exportin interacts with Cuf1 through its accessible NES (within its C-terminus), resulting in export of Cuf1 from the nucleus to the cytoplasm.
Copper transport during meiosis
A number of studies have shown that trace elements such as copper and zinc are necessary for normal progression of meiosis. Insufficient concentrations of these metal ions result in meiotic blocks as well as errors during meiosis [28–30]. Despite the known essential role for copper during meiotic differentiation, copper-requiring and copper transport proteins involved in this developmental process remain poorly understood. Meiosis is a specialized programme of the cell cycle whereby diploid germline cells produce haploid gametes which are required for fertilization or germination [31,32]. The early stage of meiosis involves a round of DNA synthesis during which chromosomal material is duplicated, giving rise to pairs of homologous chromosomes. The subsequent stage consists of meiotic recombination between homologous chromosomes that favours the evolution of new phenotypic traits. This step is followed by two successive meiotic divisions in which homologous chromosomes and then sister chromatids are separated to produce four haploid sets of chromosomes that represent the genetic material included into each of the four gametes.
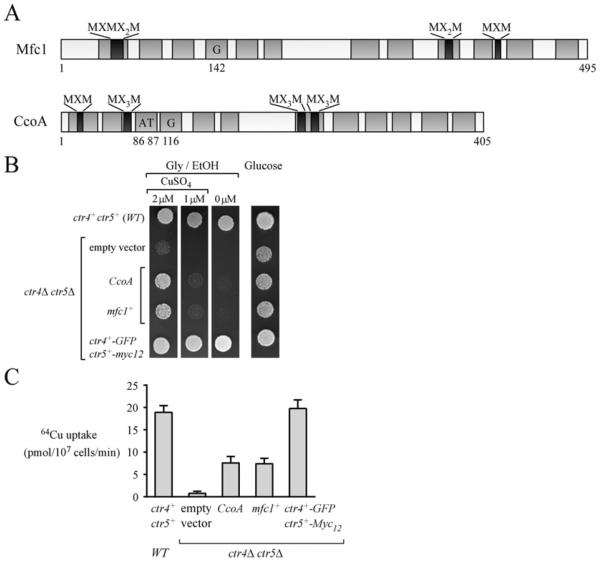
In S. pombe, copper is required for meiosis on the basis of the observation that zygotes undergo a meiotic block at metaphase I when copper levels are insufficient [28]. Copper uptake is critical to provide copper to copper-dependent proteins to maintain their activities. Therefore expression profiles of copper transporters during meiotic differentiation under copper-limiting conditions have been investigated. Expression profiles of ctr4+ and ctr5+, which encode the high-affinity copper-transporting complex at the cell surface, have revealed that their loci are highly expressed within the first 1 h of meiosis, followed by their repression 3 h after induction of meiosis. Consistent with ctr4+ gene expression profile, Ctr4 is observed at the cell surface of zygotic cells within the first 1 h of meiosis and remains there until the 3 h meiotic time point is reached [28]. A third gene, mfc1+, exhibits a distinct, but complementary temporal expression profile when compared with ctr4+. mfc1+ expression is primarily detected at the 3 h meiotic time point and mfc1+ mRNA levels remain sustained throughout the meiotic programme [28]. An important difference between mfc1+ and ctr4+ resides in the fact that the mfc1+ gene is exclusively expressed during meiosis, when it is under the control of its own promoter. On the basis of amino acid composition, mfc1+ is predicted to encode a transmembrane protein related to the MFS (major facilitator superfamily) of transporters [33] (Figure 2). The predicted topology of Mfc1 highlights 12 TMDs that are connected by hydrophilic loops, with both N- and C-termini located in the interior of the cell. The Mfc1 sequence contains 19 methionine residues, four pairs of which are organized as putative copper-binding Mets motifs such as those typically found in members of the Ctr family of copper transporters [16,34]. When Mfc1 is artificially expressed in ctr4Δ ctr5Δ cells proliferating in mitosis in the presence of low concentrations of exogenous copper (e.g. 2 μM), it functionally complements their inability to grow on respiratory carbon sources. This observation is consistent with the interpretation that Mfc1 restores the ability to provide copper to the copper-requiring Cco enzyme. Furthermore, ctr4Δ ctr5Δ cells expressing mfc1+ under the control of a heterologous mitotic nmt1+ promoter are able to mediate uptake of copper, although efficacy is reduced 61–65% when compared with heteromeric Ctr4–Ctr5 complex [28]. Localization of functional Mfc1–GFP shows that it is found at the plasma membrane in cells proliferating in mitosis [28].
Figure 2. Mfc1 and CcoA expressed in ctr4Δ ctr5Δ cells proliferating in mitosis partially restores respiratory defect and copper transport activity.
(A) Primary structures of Mfc1 and CcoA are shown. Both proteins contain 12 TMDs (grey) and putative copper-binding Mets motifs (Met-Xaa-Met, Met-Xaa2-Met and/or Met-Xaa3-Met) (black). CcoA Gly116 is required for CcoA function [37]. This amino acid residue is highly conserved in Mfc1 (position 142). In-frame insertion of a threonine–alanine dipeptide between amino acid residues 86 and 87 of CcoA results in its inactivation [37]. (B) A ctr4Δ ctr5Δ double mutant strain was transformed with an empty vector, CcoA, mfc1+, or ctr4+ –GFP and ctr5+ –Myc12. Cultures were spotted into media containing glucose or glycerol/ethanol (Gly/EtOH) and CuSO4 (0, 1 and 2 μM). WT, isogenic parental strain FY435 (ctr4+ and ctr5+) used as a control. (C) Indicated transformed ctr4Δ ctr5Δ cells were incubated with 64CuCl2 (2 μM) for 10 min. Uptake of 64CuCl2 was performed at 30°C and 4°C, and values were corrected with respect to the number of cells and temperature (data obtained at 30°C were subtracted from those at 4°C). Results are the means ± S.D. of triplicate samples from three independent cultures.
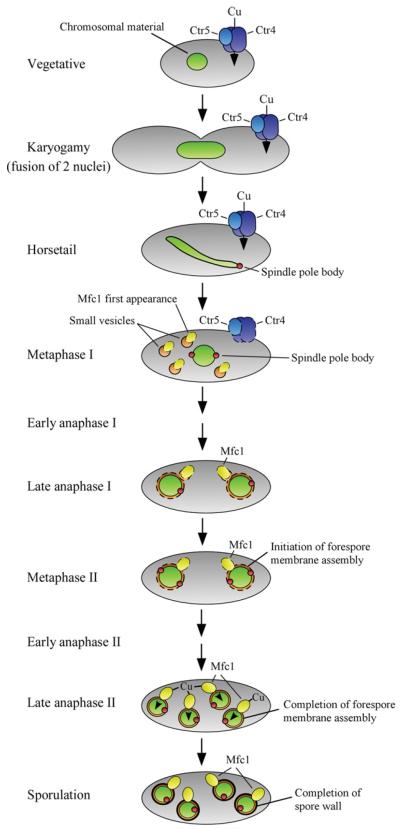
Cellular localization of Mfc1 during meiosis has revealed that the protein is found at the forespore membrane, suggesting a role for Mfc1 in mediating copper transport during spore formation (Figure 3). This suggested function of Mfc1 is supported by the observation that deletion of the mfc1+ gene (mfc1Δ/Δ) results in a strong reduction in Cao1 activity, a copper-dependent enzyme present in the forespore during sporulation [28]. Furthermore, as shown by experiments using the fluorescent copper-binding tracker CS1 (coppersensor-1) [35,36], labile and exchangeable copper pools are primarily detected in the forespores in a mfc1+ /mfc1+ strain, whereas an mfc1Δ/mfc1Δ mutant exhibits an intracellular dispersed punctate pattern of fluorescence that is distributed throughout the ascospores, without any marked preference for the forespores [28].
Figure 3. Model for copper acquisition in meiotic S. pombe cells.
At early stages of meiosis (karyogamy, horsetail and metaphase I), copper uptake is mediated by the heteromeric Ctr4–Ctr5 complex. During metaphase I, whereas the Ctr4–Ctr5 complex disappears from the cell surface, Mfc1 is first observed in small vesicle structures within the cytoplasm. During late anaphase I and metaphase II, Mfc1 is observed in the surroundings of the forespore membrane, which undergoes assembly. Following forespore membrane completion (late anaphase II), Mfc1 is located at the forespore membrane where it remains until matured spores are formed. It has been suggested that Mfc1 mediates copper uptake into the forespore when it is located at the forespore membrane. Blue, Ctr4/Ctr5; green, chromosomal material; red, spindle pole body; orange, small vesicles and forespore membrane; yellow, Mfc1; black, spore wall.
A MFS-type transporter required for copper acquisition has been identified recently in the bacterium Rhodobacter capsulatus [37]. This transmembrane protein, named CcoA, is involved in cellular copper transport for proper assembly of cbb3-type Cco. Mutant cells lacking CcoA (ccoAΔ) exhibit decreased intracellular copper levels. Consequently, ccoAΔ cells are unable to synthesize sufficient amounts of active cbb3-Cco because the steady-state amounts of the copper-dependent subunits of the enzyme are decreased [37]. As is the case for the vast majority of MFS-type transporters, CcoA is predicted to possess 12 TMDs. Although comparison of CcoA with Mfc1 has only revealed approximately 37% sequence similarity, the proteins share a particular feature which is the presence of putative Mets motifs that are organized in potential copper co-ordination arrangements. We investigated whether CcoA could complement the inability of a S. pombe ctr4Δ ctr5Δ mutant strain to grow on non-fermentable carbon sources due to copper limitation. When expressed under the control of a heterologous mitotic S. pombe promoter, CcoA restored growth on a glycerol/ethanol medium containing CuSO4 (2 μM) that was comparable with cells artificially expressing Mfc1 (Figure 2). To further investigate the relative contribution of CcoA compared with Mfc1 for copper acquisition, we measured 64Cu uptake in ctr4Δ ctr5Δ cells expressing CcoA or Mfc1 and compared the results with the parental strain. Although ctr4Δ ctr5Δ cells expressing CcoA showed a 62% reduction rate of 64Cu uptake compared with cells expressing functional Ctr4 and Ctr5 (ctr4+ –GFP and ctr5+ –Myc12), results showed that CcoA could mediate transport of copper when heterologously expressed in fission yeast (Figure 2). These observations open the way for further investigations of the role of Mfc1 and its putative orthologues such as CcoA in copper homoeostasis.
Transactivator Mca1
mfc1+ is induced at the level of gene transcription in response to copper deprivation during the meiotic programme. Similarly to mfc1+, ctr4+ and ctr5+ genes are transcriptionally induced under low-copper conditions. However, there are two important differences between expression profiles of ctr4+ and mfc1+. First, ctr4+ and mfc1+ transcript levels are induced at different times after initiation of meiosis. Secondly, whereas the deletion of cuf1+ (cuf1Δ) impairs the induction of ctr4+, activation of mfc1+ is unaffected, suggesting the existence of a distinct transcriptional regulator for the induction of mfc1+ in response to copper starvation [28]. Analysis of promoter regions in mfc1+ has revealed that two TCGGCG regulatory elements containing CGG triplets are responsible for transcriptional activation of mfc1+ under low-copper conditions [38]. Consistent with the fact that cis-acting elements containing CGG triplets are known to serve as binding sites for members of the Zn2Cys6 binuclear cluster class of regulators [39], Mca1 (a Zn2Cys6 cluster-type transcription factor) was found to be necessary for maximum induction of mfc1+ gene expression [38]. Both mfc1Δ and mca1Δ mutant strains display phenotypes linked to copper starvation. However, the severity of copper-deficient phenotypes owing to the absence of Mca1 is greater than the absence of Mfc1. For example, under copper-limiting conditions, mfc1Δ/mfc1Δ mutant cells exhibit a meiotic progression that is delayed and prolonged (by ~2–3 h) compared with the wild-type strain. Under the same copper-limiting conditions, cells lacking Mca1 (mca1Δ/mca1Δ mutants) undergo a meiotic arrest at metaphase I that blocks cellular differentiation [38]. These observations suggest that Mca1 may play an important role in activation of other meiotic genes aside from mfc1+, especially those expressed in early meiosis that precedes metaphase I. Future experiments are clearly required to identify additional meiotic genes that are induced by low-copper concentrations in relationship with Mca1.
Mca1 has primary features consistent with a role in the induction of mfc1+ gene expression. Its N-terminal region contains one zinc-finger unit (residues 24–51) that includes six cysteine residues, which are predicted to coordinate two zinc ions. The zinc-finger motif of Mca1 is followed by a large linker region (residues 51–117) and a heptad repeat of leucine residues (117–138), which is likely to be responsible for its dimerization. The unusual wide linker region of Mca1 may explain its ability to bind the two CCG triplets that are found in the two copies of the TCGGCGN13TCGGCG sequence of the mfc1+ promoter. Within this TCGGCGN13TCGGCG sequence, the spacing between the two CGG triplets is 16 bp, which represents a long spacer compared with other known similar binding elements containing CGG trinucleotides. On the basis of previous studies on Zn2Cys6 cluster-type transcription factors, it can be suggested that the zinc-finger motif, the linker and the dimerization region form the putative N-terminal DNA-binding domain of Mca1 (residues 1–138). Binding studies have revealed that the N-terminal 150-residue segment of Mca1 (including residues 1–138) specifically recognizes the TCGGCGN13TCGGCG sequence of the mfc1+ promoter region [38]. Given the fact that two zinc-finger units are required to bind a pair of CGG triplets, this restriction implies that Mca1 may bind the TCGGCGN13TCGGCG sequence as a homodimer. Alternatively, the possibility exists that Mca1 forms a heterodimer complex with a second unidentified zinc-binuclear cluster protein or with members of other transcription factor families. Besides its DNA-binding domain and dimerization region, Mca1 contains a homology regulatory region in the middle of its sequence and a transactivation domain. Although the involvement of these two regions has not been investigated in detail in Mca1, the middle homology region may play a role in preventing Mca1 binding to TCGGCGNxTCGGCG sequences with incorrect spacing between the two CGG triplets. The middle homology region may also have a regulatory role on the activation domain by modulating its transcriptional activity. Although Mca1 contains few potential metal-binding motifs (e.g. Met-Xaa-Met, Met-Xaa3-Met, Cys-Xaa-Cys-Xaa-His and Cys-Xaa3-Met-His-Xaa3-His), it is currently unknown whether they are part of a copper-sensing region, allowing Mca1 to adopt an active form to transactivate target gene expression as a function of copper availability. Alternatively, a number of mechanisms, for example specific post-translational modifications, binding of small inducer molecules and interaction with effectors or partner proteins for Mca1 function in regulation of the mfc1+ gene, have to be considered. Answers to these questions await further genetic, biochemical and molecular studies.
Acknowledgement
We are grateful to Dr Gilles Dupuis for critically reading the review and for his valuable comments.
Funding The work in our laboratory on copper homoeostasis is supported by the Canadian Institutes of Health Research (CIHR) [grant number MOP-114986 (to S.L.)].
Abbreviations used
- Cao
copper amine oxidase
- Cco
cytochrome c oxidase
- CuSE
copper-signalling element
- MFS
major facilitator superfamily
- NES
nuclear export signal
- NLS
nuclear localization signal
- Sod1
superoxide dismutase 1
- TMD
transmembrane domain.
References
- 1.Nevitt T, Ohrvik H, Thiele DJ. Charting the travels of copper in eukaryotes from yeast to mammals. Biochim. Biophys. Acta. 2012;1823:1580–1593. doi: 10.1016/j.bbamcr.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Macomber L, Imlay JA. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. U.S.A. 2009;106:8344–8349. doi: 10.1073/pnas.0812808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tarhan C, Sarikaya AT. Does copper stress lead to spindle misposition-dependent cell cycle arrest? Genet. Mol. Res. 2012;11:3824–3834. doi: 10.4238/2012.October.25.1. [DOI] [PubMed] [Google Scholar]
- 4.Labb é S, Peña MMO, Fernandes AR, Thiele DJ. A copper-sensing transcription factor regulates iron uptake genes in Schizosaccharomyces pombe. J. Biol. Chem. 1999;274:36252–36260. doi: 10.1074/jbc.274.51.36252. [DOI] [PubMed] [Google Scholar]
- 5.Zhou H, Thiele DJ. Identification of a novel high affinity copper transport complex in the fission yeast Schizosaccharomyces pombe. J. Biol. Chem. 2001;276:20529–20535. doi: 10.1074/jbc.M102004200. [DOI] [PubMed] [Google Scholar]
- 6.Beaudoin J, Laliberté J, Labbé S. Functional dissection of Ctr4 and Ctr5 amino-terminal regions reveals motifs with redundant roles in copper transport. Microbiology. 2006;152:209–222. doi: 10.1099/mic.0.28392-0. [DOI] [PubMed] [Google Scholar]
- 7.Puig S, Lee J, Lau M, Thiele DJ. Biochemical and genetic analyses of yeast and human high affinity copper transporters suggest a conserved mechanism for copper uptake. J. Biol. Chem. 2002;277:26021–26030. doi: 10.1074/jbc.M202547200. [DOI] [PubMed] [Google Scholar]
- 8.Aller SG, Eng ET, De Feo CJ, Unger VM. Eukaryotic CTR copper uptake transporters require two faces of the third transmembrane domain for helix packing, oligomerization, and function. J. Biol. Chem. 2004;279:53435–53441. doi: 10.1074/jbc.M409421200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beaudoin J, Thiele DJ, Labbé S, Puig S. Dissection of the relative contribution of the Schizosaccharomyces pombe Ctr4 and Ctr5 proteins to the copper transport and cell surface delivery functions. Microbiology. 2011;157:1021–1031. doi: 10.1099/mic.0.046854-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ioannoni R, Beaudoin J, Mercier A, Labbé S. Copper-dependent trafficking of the Ctr4-Ctr5 copper transporting complex. PLoS ONE. 2010;5:e11964. doi: 10.1371/journal.pone.0011964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Halloran TV, Culotta VC. Metallochaperones, an intracellular shuttle service for metal ions. J. Biol. Chem. 2000;275:25057–25060. doi: 10.1074/jbc.R000006200. [DOI] [PubMed] [Google Scholar]
- 12.Laliberté J, Whitson LJ, Beaudoin J, Holloway SP, Hart PJ, Labbé S. The Schizosaccharomyces pombe Pccs protein functions in both copper trafficking and metal detoxification pathways. J. Biol. Chem. 2004;279:28744–28755. doi: 10.1074/jbc.M403426200. [DOI] [PubMed] [Google Scholar]
- 13.Peter C, Laliberté J, Beaudoin J, Labbé S. Copper distributed by Atx1 is available to copper amine oxidase 1 in Schizosaccharomyces pombe. Eukaryotic Cell. 2008;7:1781–1794. doi: 10.1128/EC.00230-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Labbé S, Beaudoin J, Ioannoni R. Copper transport in fungi. In: Culotta VC, Scott RA, editors. Metals in Cells. Encyclopedia of Inorganic and Bioinorganic Chemistry. Wiley-Blackwell; Chichester: 2013. pp. 163–174. [Google Scholar]
- 15.Bellemare DR, Shaner L, Morano KA, Beaudoin J, Langlois R, Labbé S. Ctr6, a vacuolar membrane copper transporter in Schizosaccharomyces pombe. J. Biol. Chem. 2002;277:46676–46686. doi: 10.1074/jbc.M206444200. [DOI] [PubMed] [Google Scholar]
- 16.Puig S, Thiele DJ. Molecular mechanisms of copper uptake and distribution. Curr. Opin. Chem. Biol. 2002;6:171–180. doi: 10.1016/s1367-5931(02)00298-3. [DOI] [PubMed] [Google Scholar]
- 17.Beaudoin J, Labbé S. The fission yeast copper-sensing transcription factor Cuf1 regulates the copper transporter gene expression through an Ace1/Amt1-like recognition sequence. J. Biol. Chem. 2001;276:15472–15480. doi: 10.1074/jbc.M011256200. [DOI] [PubMed] [Google Scholar]
- 18.Rustici G, van Bakel H, Lackner DH, Holstege FC, Wijmenga C, Bahler J, Brazma A. Global transcriptional responses of fission and budding yeast to changes in copper and iron levels: a comparative study. Genome Biol. 2007;8:R73. doi: 10.1186/gb-2007-8-5-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beaudoin J, Mercier A, Langlois R, Labbé S. The Schizosaccharomyces pombe Cuf1 is composed of functional modules from two distinct classes of copper metalloregulatory transcription factors. J. Biol. Chem. 2003;278:14565–14577. doi: 10.1074/jbc.M300861200. [DOI] [PubMed] [Google Scholar]
- 20.Beaudoin J, Labbé S. Copper induces cytoplasmic retention of fission yeast transcription factor Cuf1. Eukaryotic Cell. 2006;5:277–292. doi: 10.1128/EC.5.2.277-292.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jungmann J, Reins HA, Lee J, Romeo A, Hassett R, Kosman DJ, Jentsch S. Mac1, a nuclear regulatory protein related to Cu-dependent transcription factors is involved in Cu/Fe utilization and stress resistance in yeast. EMBO J. 1993;12:5051–5056. doi: 10.1002/j.1460-2075.1993.tb06198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu Z, Labbé S, Pena MMO, Thiele DJ. Copper differentially regulates the activity and degradation of yeast Mac1 transcription factor. J. Biol. Chem. 1998;273:1277–1280. doi: 10.1074/jbc.273.3.1277. [DOI] [PubMed] [Google Scholar]
- 23.Jensen LT, Winge DR. Identification of a copper-induced intramolecular interaction in the transcription factor Mac1 from Saccharomyces cerevisiae. EMBO J. 1998;17:5400–5408. doi: 10.1093/emboj/17.18.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rutherford JC, Bird AJ. Metal-responsive transcription factors that regulate iron, zinc, and copper homeostasis in eukaryotic cells. Eukaryotic Cell. 2004;3:1–13. doi: 10.1128/EC.3.1.1-13.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beaudoin J, Labbé S. Crm1-mediated nuclear export of the Schizosaccharomyces pombe transcription factor Cuf1 during a shift from low to high copper concentrations. Eukaryotic Cell. 2007;6:764–775. doi: 10.1128/EC.00002-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fornerod M, Ohno M, Yoshida M, Mattaj IW. Crm1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 27.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. Crm1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 28.Beaudoin J, Ioannoni R, Lopez-Maury L, Bahler J, Ait-Mohand S, Guerin B, Dodani SC, Chang CJ, Labbé S. Mfc1 is a novel forespore membrane copper transporter in meiotic and sporulating cells. J. Biol. Chem. 2011;286:34356–34372. doi: 10.1074/jbc.M111.280396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim AM, Vogt S, O'Halloran TV, Woodruff TK. Zinc availability regulates exit from meiosis in maturing mammalian oocytes. Nat. Chem. Biol. 2010;6:674–681. doi: 10.1038/nchembio.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernhardt ML, Kong BY, Kim AM, O'Halloran TV, Woodruff TK. A zinc-dependent mechanism regulates meiotic progression in mammalian oocytes. Biol. Reprod. 2012;86:114. doi: 10.1095/biolreprod.111.097253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Handel MA, Schimenti JC. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat. Rev. Genet. 2010;11:124–136. doi: 10.1038/nrg2723. [DOI] [PubMed] [Google Scholar]
- 32.Marston AL, Amon A. Meiosis: cell-cycle controls shuffle and deal. Nat. Rev. Mol. Cell Biol. 2004;5:983–997. doi: 10.1038/nrm1526. [DOI] [PubMed] [Google Scholar]
- 33.Law CJ, Maloney PC, Wang DN. Ins and outs of major facilitator superfamily antiporters. Annu. Rev. Microbiol. 2008;62:289–305. doi: 10.1146/annurev.micro.61.080706.093329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim BE, Nevitt T, Thiele DJ. Mechanisms for copper acquisition, distribution and regulation. Nat. Chem. Biol. 2008;4:176–185. doi: 10.1038/nchembio.72. [DOI] [PubMed] [Google Scholar]
- 35.Miller EW, Zeng L, Domaille DW, Chang CJ. Preparation and use of coppersensor-1, a synthetic fluorophore for live-cell copper imaging. Nat. Protoc. 2006;1:824–827. doi: 10.1038/nprot.2006.140. [DOI] [PubMed] [Google Scholar]
- 36.Zeng L, Miller EW, Pralle A, Isacoff EY, Chang CJ. A selective turn-on fluorescent sensor for imaging copper in living cells. J. Am. Chem. Soc. 2006;128:10–11. doi: 10.1021/ja055064u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ekici S, Yang H, Koch HG, Daldal F. Novel transporter required for biogenesis of cbb3-type cytochrome c oxidase in Rhodobacter capsulatus. mBio. 2012;3:e00293–e00211. doi: 10.1128/mBio.00293-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beaudoin J, Ioannoni R, Mailloux S, Plante S, Labbé S. Transcriptional regulation of the copper transporter Mfc1 in meiotic cells. Eukaryotic Cell. 2013;12:575–590. doi: 10.1128/EC.00019-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacPherson S, Larochelle M, Turcotte B. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol. Mol. Biol. Rev. 2006;70:583–604. doi: 10.1128/MMBR.00015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]