Abstract
Translesion DNA synthesis (TLS) is a DNA damage tolerance mechanism carried out by low-fidelity DNA polymerases that bypass DNA lesions, which overcomes replication stalling. Despite the miscoding nature of most common DNA lesions, several of them are bypassed in mammalian cells in a relatively accurate manner, which plays a key role maintaining a low mutation load. Whereas it is generally agreed that TLS across the major UV and sunlight induced DNA lesion, the cyclobutane pyrimidine dimer (CPD), is accurate, there were conflicting reports on whether the same is true for the thymine–thymine pyrimidine–pyrimidone(6-4) ultraviolet light photoproduct (TT6-4PP), which represents the second most common class of UV lesions. Using a TLS assay system based on gapped plasmids carrying site-specific TT6-4PP lesions in defined sequence contexts we show that the DNA sequence context markedly affected both the extent and accuracy of TLS. The sequence exhibiting higher TLS exhibited also higher error-frequency, caused primarily by semi-targeted mutations, at the nearest nucleotides flanking the lesion. Our results resolve the discrepancy reported on TLS across TT6-4PP, and suggest that TLS is more accurate in human cells than in mouse cells.
Keywords: DNA repair, Error-prone repair, UV mutagenesis, Lesion bypass, DNA damage
1. Introduction
DNA repair mechanisms are conserved in essentially all living organisms, and function to preserve the accuracy of genetic information [1]. Yet the high abundance of DNA damage, caused by both intracellular (e.g., reactive oxygen species) and external (e.g., sunlight, tobacco smoke) agents, estimated at thousands per day, make the encounter of replication with unrepaired damages inevitable [2,3]. When this happens, cells utilize DNA damage tolerance mechanisms, which function to bypass the lesion without actually removing it from DNA. Two DNA damage tolerance mechanisms are known. Translesion DNA synthesis (TLS) utilizes specialized low-fidelity DNA polymerases to bypass the lesions, whereas homology-dependent repair (HDR, also termed postreplication repair, damage avoidance, or template switch) utilizes the homologous sister chromatid as a DNA sequence source for bypassing the lesion [2–4]. While TLS is inherently inaccurate due to the miscoding of most DNA lesions [5,6], HDR is fundamentally accurate [7–10]. Remarkably, despite the great heterogeneity of DNA lesions, TLS often incorporates the correct nucleotide(s) opposite it, leading to accurate TLS [5]. The classical example is the UV and sunlight-induced TT cyclobutane pyrimidine dimer (CPD), which is bypassed by DNA polymerase η (polη) with an accuracy of 99% [11,12]. Similarly, the bulky benzo[a]pyrene-guanine adduct is bypassed with accuracy of nearly 90% in human cells [13].
Short wavelength UV light produced in DNA two main types of lesions: CPD, mentioned above, which account for 66–75% of the photoproducts, and pyrimidine–pyrimidone (6-4) photoproducts (6-4PP), which account for 25–33% of the photoproducts [1]. The 6-4PP are efficiently repaired by nucleotide excision repair, because they cause a strong distortion in DNA [14], whereas the CPD, which cause a mild distortion in DNA, are poorly repaired, and are dealt with primarily during replication via TLS by polη [15,16]. Yet, replication may encounter also unrepaired 6-4PP, a situation in which TLS functions to enable completion of replication. The TT6-4PP lesion was initially reported to be highly mutagenic [17,18], later confirmed by studies from our group [13,19]. Yet, a publication from the Prakash group reported that this lesion is accurately bypassed in mammalian cells, and suggested that the difference stems from the fact that they assayed replicative TLS in a plasmid, whereas we used gap-filling TLS in a non-replicating plasmid [20]. An alternative explanation for the differences observed in the accuracy of TLS in these studies is the DNA sequence context of the lesion. To explore this possibility we examined TLS across a site-specific single TT6-4PP in the sequence context used in the Prakash study to the one used in our study. We found that the DNA sequence context had a major effect on the accuracy of TLS in mammalian cells, and that overall it was more accurate in human cells than in mouse cells.
2. Materials and methods
2.1. Construction of DNA substrates and plasmids
The construction of the control gapped plasmid GP20 was previously described [21–23]. A gap-lesion plasmid with a site-specific TT6-4PP flanked by sequences used previously in the gap-lesion plasmid system, was prepared using the site-specifically modified 11-mer oligonucleotide 5′-GCAAGTTGGAG-3′ containing a single site-specific TT 6-4 PP (underlined) as previously described [19]. Briefly, the 11-mer was extended to a 53-mer oligonucleotide by ligating to its 5′ end the 21-mer 5′-ACCGCAACGAAGTGATTCCTC-3′, and to its 3′ the 21-mer 5′-GAGGCTACTTGAACCAGACCG-3′, using as a scaffold the 33-mer 5′- AAGTAGCCTCCTCCAACTTGCGAGGAATCAC-3′. The 53-mer was separated from the scaffold and excess 21-mers on a 12% denaturing polyacrylamide gel containing 8 M urea, and then annealed to the two complementary 15-mers 5′-GGAATCACTTCGTTG-3′ and 5′-CTGGTTCAAGTAGCC-3′. The resultant gapped duplex oligonucleotide, with the lesion opposite the gap, was used to construct GP-TT6-4PP-L-seq. A gap-lesion plasmid with a site-specific TT6-4PP flanked by sequences used previously in the gap free-lesion plasmid replicative system, was similarly prepared, except that the 11-mer oligonucleotide was 5′-AGCAATTGTAC-3′ containing a single site-specific TT6-4PP (underlined), and it was extended by ligating to its 5′ end the 21-mer 5′-ACCGCAACGAAGTGATTCCTC-3′, and to its 3′ the 21-mer 5′-CTGGCTACTTGAACCAGACCG-3′, using as a scaffold the 33-mer 5′- AAGTAGCCAGGTACAATTGCTGAGGAATCAC-3′.
The DNA sequences in the vicinity of the gap-lesion structure of the gap-lesion plasmids used in this study were as follows (the lesions are underlined):
| GP-TT6-4PP-L-seq: | ||
| 5′CAACGAAGTGATTCCTCGCAAGTTGGAGGAGGCTACTTGAACCAG3′ | ||
| 3′GTTGCTTCACTAAGG | CCGATGAACTTGGTC5′ | |
| GP-TT6-4PP-P-seq: | ||
| 5′CAACGAAGTGATTCCTCAGCAATTGTACCTGGCTACTTGAACCAG3′ | ||
| 3′GTTGCTTCACTAAGG | CCGATGAACTTGGTC5 | |
2.2. TLS assay
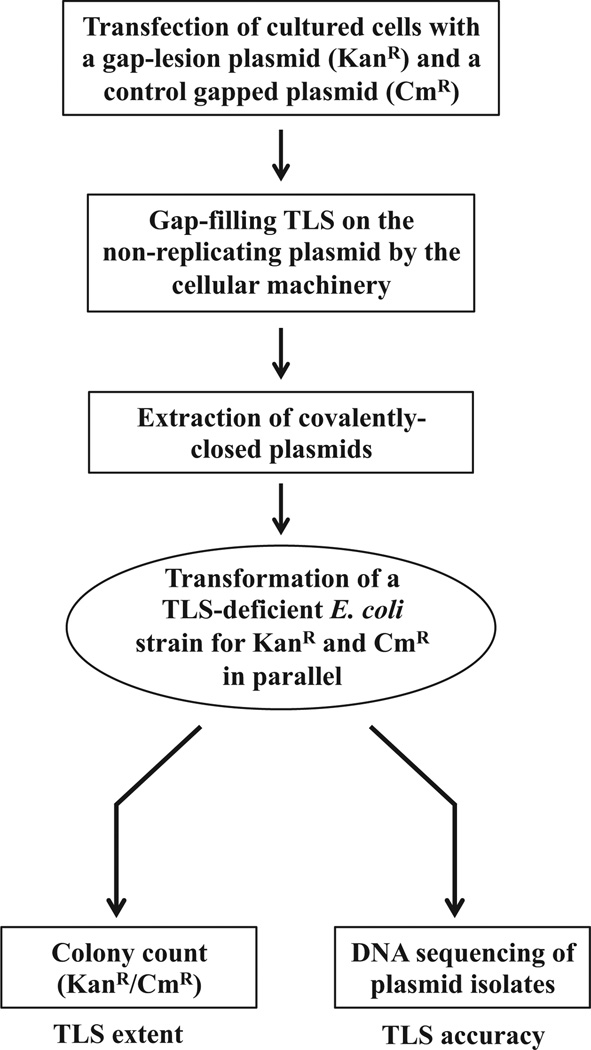
To examine TLS across the TT6-4PP in different sequence contexts we used the gap-lesion plasmid assay, developed in our laboratory more than a decade ago [24], and extensively utilized since to study TLS [22,25–27]. The system is based on a plasmid carrying a site-specific lesion, engineered in a particular sequence context, and opposite a single-stranded gap. When such a plasmid is used to transfect mammalian cells (Fig. 1), the gap is filled in by the cellular TLS machinery, after which product covalently closed plasmids are extracted, and introduced for analysis in an indicator Escherichia coli recA strain, which is TLS-deficient. This enables measurement of the efficiency of TLS by counting the number of E. coli colonies, and its accuracy, by DNA sequence analysis of plasmids extracted from individual colonies (Fig. 1). The system assays TLS of gap filling in the absence of DNA replication, and was proven to depend on the DNA polymerase composition of the cells [19,28], and on regulators of TLS such as ubiquitination of PCNA [22] and on the cell cycle [26].
Fig. 1.
Flow-chart outlining the gap-lesion plasmid-based TLS assay. KanR, kanamycin resistance; CmR, chloramphenicol resistance. See text for details.
The TLS assay was executed as previously described [23,25], using SV40-transformed MRC5sv human fetal lung fibroblasts [27] or mouse embryo fibroblasts [26]. Briefly, transfection was done in 6-well plates (surface area 9.1 cm2) at ~80% confluence using Jet-PEI transfection reagent. After allowing time for TLS (~20 hours), the cells were harvested and plasmids were extracted using a plasmid extraction kit HiYield™ Plasmid Mini Kit/96-well Plasmid Kit/Real Genomics™. Plasmid DNA extraction was done under alkaline lysis conditions followed by renaturation, which leaves only covalently closed plasmids (namely those which underwent complete gap-filling TLS) non denatured. The extracted plasmids were used for transfection of competent E. coli JM109 recA cells. The percentage of plasmid repair, of which most occurs by TLS across TT6-4PP was calculated by dividing the number of colonies obtained from the descendants of gap-lesion plasmids (KanR colonies) by the number of colonies obtained from descendants of the control gapped plasmids (CmR colonies). The DNA sequence opposite the lesion was determined using Sanger sequencing. A small fraction of gap-lesion plasmids was repaired by non-TLS events, which most likely involve formation of a double stranded break as an intermediate. These events were observed as large deletions or insertions in the plasmid isolates. To obtain precise TLS extents, the plasmid repair extents were multiplied by the fraction of TLS events out of all plasmid repair events, as obtained by DNA sequence analysis.
3. Results
3.1. The extent of TLS is sequence context dependent
Using the gapped plasmid assay we assayed TLS across TT6-4PP in the two DNA sequence contexts in human MRC5sv cells. As can be seen in Table 1, TLS across the TT6-4PP in sequence context L-seq amounted to 37%. In contrast, TLS across the same lesion in sequence context P-seq was 12%, namely 3.1-fold lower than in L-seq. This effect was not limited to human cells. Thus, assaying TLS using the same gapped plasmids in mouse embryonic fibroblasts (WT6 cells) yielded similar results, with TLS across TT6-4PP in sequence context L-seq being 2.8-fold higher than in sequence context P-seq (Table 1).
Table 1.
Extent of TLS in mouse embryonic fibroblast WT6 cells and human MRC5sv cell lines.
| Cell line | Gap-lesion plasmid | Transformantsa | Plasmid repairb | TLSc | Relative TLS extent | |
|---|---|---|---|---|---|---|
| KanR | CmR | |||||
| Human cells MRC5sv | TT6-4PP (L-seq) | 95 | 232 | 0.40 ± 0.12 | 0.37 ± 0.11 | 3.1 |
| TT6-4PP (P-seq) | 53 | 358 | 0.13 ± 0.07 | 0.12 ± 0.05 | 1 | |
| Mouse cells WT6 | TT6-4PP (L-seq) | 119 | 392 | 0.28 ± 0.17 | 0.28 ± 0.17 | 2.8 |
| TT6-4PP (P-seq) | 104 | 932 | 0.11 ± 0.1 | 0.10 ± 0.1 | 1 | |
The results of a typical experiment are presented.
Plasmid repair is defined as the number of KanR colonies divided by the number of CmR colonies. The average of 3–4 experiments is presented. See materials and methods section.
TLS values are obtained by dividing plasmid repair values by the fraction of TLS events, as deduced from the DNA sequence analysis. See materials and methods section.
3.2. Sequence context greatly affects the fidelity of TLS acrossTT6-4PP
To examine the accuracy of TLS in the two DNA sequence contexts, we performed DNA sequence analysis of a total of 310 isolates that underwent TLS across TT6-4PP in sequence context L-seq and 268 isolates that underwent TLS in sequence context P-seq. As can be seen in Table 2, in the human cells TLS across TT6-4PP in sequence context L-seq was highly mutagenic, exhibiting 37% mutations during TLS. In contrast, TLS events across TT6-4PP in sequence context P-seq exhibited only 10% mutational event, with 90% of the TLS events leading to the accurate insertion of AA opposite the TT6-4 PP. Thus, TLS across TT6-4PP is 3.7-fold more mutagenic in sequence context L-seq than in sequence context P-seq (chi-squared test P = 1.3 × 10−8).
Table 2.
Mutagenic TLS across TT6-4 PP adducts in two sequence contexts.a
| Cell type | DNA sequence context | TLS across TT6-4PP | Total events | |
|---|---|---|---|---|
| Accurate TLSb | Mutagenic TLSc | |||
| Number of events (%) | ||||
| Human cells MRC5sv | TT6-4PP (L-seq) 5’GCAAGTTGGAGGAGG3’ | 110 (63%) | 64 (37%) | 174 (100%) |
| TT6-4PP (P-seq) 5’AGCAATTGTACCTGG3’ | 138 (90%) | 15 (10%) | 153 (100%) | |
| Chi-squared test P = 1.3E-08 | ||||
| Mouse cells WT6 | TT6-4PP (L-seq) 5’GCAAGTTGGAGGAGG3’ | 58 (43%) | 78 (57%) | 136 (100%) |
| TT6-4PP (P-seq) 5’AGCAATTGTACCTGG3’ | 80 (70%) | 35 (30%) | 115 (100%) | |
| Chi-squared test P = 1.95E-05 | ||||
The plasmid content was isolated from colonies obtained in the experiments described in Table 1, and subjected to DNA sequence analysis.
Accurate TLS is defined as the insertion of AA opposite the TT6-4PP, with no other mutation observed in its vicinity.
The detailed mutation types are provided in Table 3.
The same experiment was performed also in mouse embryonic fibroblasts. As can be seen in Table 2, 57% of the TLS events across TT6-4PP in sequence context L-seq were mutagenic in MEF, compared to 30% in sequence context P-seq. Thus, overall in MEF TLS across TT6-4PP is less accurate than in human MRC5sv cells, yet similarly to the human cells it is still 1.9-fold more mutagenic in sequence context L-seq than in sequence context P-seq (chi-squared test, P = 1.9 × 10−5).
3.3. Sequence context affects mutagenic specificity of TLS across TT6-4PP
The mutations generated during TLS across TT6-4PP are presented in Table 3. If only targeted mutations are compared, namely base changes occurring opposite the TT6-4PP, there is essentially no difference between the two sequence contexts in human cells − 9.8% versus 7.2% of targeted mutations in sequence contexts L-seq and P-seq (P = 0.29), respectively, and a relatively small effect in mouse cells, where P-seq exhibits a 2.3-fold higher frequency than L-seq (21.7% versus 9.6%; P = 0.006). In contrast, big differences were observed in semi-targeted mutations, namely mutations occurring at the 3′ or 5′ nucleotides flanking the TT6-4PP lesion. In human cells pure semi-targeted mutations, namely one or two misinsertions at the nucleotides 3′ or 5′ to the TT6-4PP, but accurate insertion opposite the lesion, accounted for 21.3% of all TLS events sin sequence context L-seq compared to only 2.6% in sequence context P-seq, a 8.2-fold effect (P = 3.74 × 10−7). The effect was observed also in mouse cells, where the pure semi-targeted events accounted for 28.7% and 7%, in L-seq and P-seq, respectively (4.1-fold effect; P = 1.11 × 10−5). Of note, the L-seq mutants contain a significant hotspot located in the base 5′ to the TT6-4PP, leading upon replication the sequence -5′CAAA, which accounted in human cells for 19% of all TLS events, compared to 2% to the total events in P-seq (9.5-fold effect; P = 9.5 × 10−7), and in mouse cells −26.5% compared to 7% in P-seq (3.8-fold effect; P = 5.1 × 10−5).
Table 3.
DNA sequence analysis of descendants of a gap-lesion plasmid carrying a site-specific TT6-4PP flanked by distinct sequences.*
| Nucleotide insertion/site | L-seq context | P-seq context | ||||
|---|---|---|---|---|---|---|
| 5′GCAAGTTGGAGGAGG3′ Sequence opposite TT6-4PP |
Human cells MRC5sv Number (%) |
Mouse cells WT6 Number (%) |
5′AGCAATTGTACCTGG3′ Sequence opposite TT6-4PP |
Human cells MRC5sv Number (%) |
Mouse cells WT6 Number (%) |
|
| Accurate | 5′C-AA-C | 110 (63.2%) | 58 (42.6%) | 5′C-AA-T | 138 (90.2%) | 80 (69.6%) |
| Semi-targeted | C-AA-A | 33 (19%) | 36 (26.5%) | C-AA-A | 3 (2.0%) | 8 (7.0%) |
| A-AA-C | 2 (1.1%) | 3 (2.2%) | G-AA-T | 1 (0.7%) | ||
| C-AA-Δ | 1 (0.6%) | |||||
| Δ-AA-C | 1 (0.6%) | |||||
| Opposite 3′ of 6-4 | C-ΔA-C | 8 (4.6%) | ||||
| C-TA-C | 3 (1.7%) | 1 (0.7%) | C-TA-T | 5 (3.3%) | 16 (13.9%) | |
| C-CA-C | 2 (1.5%) | |||||
| C-GA-C | 1 (0.6%) | C-GA-T | 4 (2.6%) | 7 (6.1%) | ||
| Opposite 3′ of 6-4 + semi-targeted | C-TA-A | 3 (1.7%) | 10 (7.4%) | C-TA-A | 2 (1.7%) | |
| A-TA-C | 2 (1.1%) | |||||
| C-GA-A | 3 (1.7%) | 4 (2.9%) | ||||
| Opposite 5′ of 6-4 | C-AC-C | 2 (1.1%) | 2 (1.5%) | |||
| C-AT-C | 1 (0.6%) | 2 (1.5%) | C-AT-T | 1 (0.7%) | 2 (1.7%) | |
| C-AG-C | 1 (0.6%) | 2 (1.5%) | ||||
| C-AΔ-C | 1 (0.6%) | 2 (1.5%) | ||||
| Opposite 5′ of 6-4 + semi-targeted | C-AT-A | 1 (0.7%) | ||||
| C-AC-Δ | 1 (0.7%) | |||||
| C-AΔ-Δ | 1 (0.6%) | |||||
| Opposite 5′ of 6-4 +2 flanking bases | A-AC-A | 1 (0.6%) | ||||
| Tandem double opposite 6-4 | C-TG-C | 1 (0.7%) | C-GT-T | 1(0.7%) | ||
| C-TT-C | 1 (0.7%) | |||||
| Tandem double opposite 6-4 + semi-targeted | C-GT-A | 1 (0.7%) | ||||
| C-CΔ-A | 7 (5.1%) | |||||
| Tandem double opposite 6-4 +2 flanking bases | T-CΔ-A | 1 (0.7%) | ||||
| A-CT-Δ | 1 (0.7%) | |||||
| Total: | 174 (100%) | 136 (100%) | Total: | 153 (100%) | 115 (100%) | |
Mutations are highlighted in bold type, and are underlined.
Interestingly multiple mutations of 2–4 bases changes, including mixtures of targeted and semi-targeted mutations were more frequent in sequence context L-seq: The effect was striking in mouse cells, where a total of 21% (28/136) multiple mutations were observed: 18 double mutants, 8 triple mutants and 2 quadruplicate mutants, compared to a total of 1.7% (2/115) in sequence context P-seq (12.1-fold, P = 4.5 × 10−6; Table 3). Human cells contained 5.7% (10/174) multiple mutations: 9 double mutants and 1 triple mutant in L-seq compared to only a single tandem double mutant in P-seq (0.7%, 1/153; 8.1-fold effect; P = 0.011).
Another clear difference in the mutational spectra is in the occurrence of minus 1 frameshifts. These mutations were not observed at all in sequence context P-seq. In contrast, in sequence context L-seq they accounted for 6.4% and 8.8% of the mutations caused by TT6-4PP in human and mouse cells, respectively (Table 3).
4. Discussion
The critical role of TLS in coping with the flux of DNA damage that cells face is clearly indicated by the essentiality of the Rev3L gene, encoding the catalytic subunit of DNA polymerase ζ [29], as well as by cancer-predisposition disease xeroderma pigmentosum variant, caused by inactivating mutations in polη [11,12]. This may be one of the reasons that TLS evolved in humans to have a sub-pathway with relatively high accuracy, often termed error-free TLS. Yet, TLS is a major source of mutations, and therefore, under-standing the molecular mechanisms that regulate its accuracy is of importance in understanding how cells maintain their genetic stability.
The data presented shows that DNA sequence context can have a major effect on the fidelity of TLS in mammalian cells. The sequence surrounding a particular DNA lesion is known to affect TLS, as observed in in vitro and in vivo systems [30–34]. The molecular basis for these effects is generally not understood, although local conformations of the DNA and sequence repeats, which may allow misalignment of the template and the proteins involved, as well as their interactions are clearly likely to play an important role.
It was previously suggested that the high mutagenicity of the TT6-4PP in our studies compared to the Prakash studies was due to the assay systems used, namely a gap-filling TLS reaction in a non-replicating plasmid in our study, versus a replicating plasmid in the Prakash study [20]. As we have previously pointed out [9], this was unlikely to be the explanation because the first study reporting the high mutagenicity of TT6-4PP in human cells by Sarasin and colleagues used a replicating plasmid [18]. L-seq was also highly mutagenic in a replicating phage system in E. coli [17]. Moreover, using a chromosomal TLS assay system we have recently shown that the TT6-4PP in sequence context L-seq was mutagenic also during chromosomal TLS [9]. This prompted us to examine the possibility that the difference stems from a sequence context effect. As our results clearly show, the effect does stem primarily from the different sequences flanking the TT6-4PP.
Generally the excessive mutations formed during TLS across the TT6-4PP in sequence context L-seq fall into three categories: (1) semi-targeted mutations in the two nearest nucleotide flanking the TT6-4PP; (2) minus 1 frameshift mutations; (3) multiple (2 to 4) targeted and semi-targeted mutations in the same isolate. We and others have previously shown that polζ is involved in bypass of TT6-4PP [13,35,36], however, it is not yet clear whether additional polymerases are involved, and therefore, it is unknown yet which polymerase is responsible for generating the mutations. Nevertheless, the big differences in the mutational spectra indicate a dramatic loss of fidelity of the inserter polymerase in the L-seq sequence context, or even the participation of different inserter polymerases on the two different templates.
Sequence context effects on TLS may involve both nearest-neighbour and more distant base sequences, as previously reported, for example, for nucleotide excision repair [37]. The most pronounced difference in mutations between the two sequences was a semi-targeted G → T transversion at the base located 5′ to the TT6-4PP, where 19% of the mutations were observed in the L-seq context, and only 2% of the mutations P-seq context (Table 3). This is the site of the first template nucleotide following the lesion, and it is different in the L-seq and P-seq contexts—G and A, respectively. Thus, it can be speculated that this one base change was responsible for at least part of the dramatic change in accuracy, although the molecular basis for this is not clear.
Looking a bit further from the site of the lesion faces the problem of not knowing how many nucleotides should be considered. However, for the sake of discussion, we may consider the length of approximately one helical turn, which means a total of about 10 nucleotides (the first 4 nucleotides from each side of the lesion). In this segment the two sequences, 5′-CAAGTTGGAG-3′ (sequence context L-seq) and 5′-GCAATTGTAC-3′ (sequence context P-seq) differ primarily in the purine/pyrimidine balance flanking the TT6-4 PP: 1 pyrimidine versus 3, respectively, for the first 8 flanking nucleotides (4 on each side). Pyrimidines have usually weaker stacking interactions, which may allow easier adaptation to the polymerase active site, and therefore, higher accuracy. A lower flexibility of sequence context L-seq may be enhanced by the abundance of guanines (4 out of the 8 compared to 2/8 in P-seq), known to be involved in strong stacking interactions. Notice that 3 of the 4 guanines in L-seq are close to the lesion. The occurrence of minus 1 frameshifts in L-seq, but none in P-seq, is consistent with such a possibility, because such mutations are indicative of misalignment, and may be favoured by the 5′GGAGGA3′ direct repeat present 3′ to the TT6-4PP in the L-seq sequence context.
The data presented shows that TLS cross the TT6-4PP lesion was more mutagenic in the mouse cells than in the human cells, for each of the sequence contexts. This is more pronounced for the sequence context P-seq, where TLS is generally more accurate: TLS was 3-fold more mutagenic in mouse cells than in human cells (30% versus 10% mutations; Chi-squared test P = 1.8 × 10−5; Table 2). A more mutagenic effect was observed also for the sequence context L-seq, where TLS was 1.5-fold more mutagenic in mouse cells compared to human cells (57% versus 37% mutations; Chi-squared test, P = 3.1 × 10−4; Table 2). This lower fidelity of TLS in mouse compared to human cells is manifested also in the 3.5-fold higher fraction of multiple mutations observed with the sequence context L-seq in mouse cells (21%) compared to human cells (6%), as described above. A lower fidelity of TLS in mouse cells compared to human cells was observed also for a benzo[a]pyrene-guanine adduct [13,28], and may represent a general higher fidelity of TLS in humans than in mice.
In summary, our results show that DNA sequence context has a major effect on the fidelity of TLS in mammalian cells, and is responsible for the differences in fidelity of TLS across TT6-4PP observed in previous studies. Moreover, our results suggest that TLS is more accurate in human cells than in mouse cells, which may reflect a general requirement for maintaining higher accuracy of the human genome.
Acknowledgments
We thank Alan Lehmann (University of Sussex, Brighton, U.K) and Niels de Wind (Department of Toxicogenetics, Leiden University Medical Center, P.O. Box 9600, 2300 RC Leiden, The Netherlands) for providing cell lines. Z.L. is the incumbent of the Maxwell Ellis Chair for Biomedical Research. This work was supported by grants from the Flight Attendant Medical Research Institute, Florida, USA (to ZL and TPE); the Israel Science Foundation Grant 684/12 to ZL; the Leona M. and Harry B. Helmsley Charitable Trust, NY, USA (to ZL). The preparation of the site-specifically modified oligonucleotides was supported by NIH grant CA168469 to N.E.G.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd. Washington DC: ASM Press; 2006. [Google Scholar]
- 2.Lehmann AR, Fuchs RP. Gaps and forks in DNA replication: rediscovering old models. DNA Repair. 2006;5:1498–1595. doi: 10.1016/j.dnarep.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Friedberg EC. Suffering in silence: the tolerance of DNA damage. Nat. Rev. Mol. Cell Biol. 2005;6:943–953. doi: 10.1038/nrm1781. [DOI] [PubMed] [Google Scholar]
- 4.Livneh Z, Cohen-Fix O, Skaliter R, Elizur T. Replication of damaged DNA and the molecular mechanism of ultraviolet light mutagenesis. CRC Crit. Rev. Biochem. Mol. Biol. 1993;28:465–513. doi: 10.3109/10409239309085136. [DOI] [PubMed] [Google Scholar]
- 5.Livneh Z, Ziv O, Shachar S. Multiple two-polymerase mechanisms in mammalian translesion DNA synthesis. Cell Cycle. 2010;9:729–735. doi: 10.4161/cc.9.4.10727. [DOI] [PubMed] [Google Scholar]
- 6.Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol. 2012;13:141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang H, Lawrence CW. The error-free component of the RAD6/RAD18 DNA damage tolerance pathway of budding yeast employs sister-strand recombination. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15954–15959. doi: 10.1073/pnas.0504586102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berdichevsky A, Izhar L, Livneh Z. Error-free recombinational repair predominates over mutagenic translesion replication in E. coli. Mol. Cell. 2002;10:917–924. doi: 10.1016/s1097-2765(02)00679-2. [DOI] [PubMed] [Google Scholar]
- 9.Izhar L, Ziv O, Cohen IS, Geacintov N, Livneh Z. Genomic assay reveals tolerance of DNA damage by both translesion DNA synthesis and homology-dependent repair in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E1462–E1469. doi: 10.1073/pnas.1216894110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen IS, Bar C, Paz-Elizur T, Ainbinder E, Leopold K, de Wind N, Geacintov N, Livneh Z. DNA lesion identity drives choice of damage tolerance pathway in murine cell chromosomes. Nucl. Acids Res. 2015;43:1637–1645. doi: 10.1093/nar/gku1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 12.Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 13.Shachar S, Ziv O, Avkin S, Adar S, Wittschieben J, Reissner T, Chaney S, Friedberg EC, Wang Z, Carell T, et al. Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J. 2009;28:383–393. doi: 10.1038/emboj.2008.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vreeswijk MP, van Hoffen A, Westland BE, Vrieling H, van Zeeland AA, Mullenders LH. Analysis of repair of cyclobutane pyrimidine dimers and pyrimidine 6-4 pyrimidone photoproducts in transcriptionally active and inactive genes in Chinese hamster cells. J. Biol. Chem. 1994;269:31858–31863. [PubMed] [Google Scholar]
- 15.Tung BS, McGregor WG, Wang YC, Maher VM, McCormick JJ. Comparison of the rate of excision of major UV photoproducts in the human HPRT gene of normal and xeroderma pigmentosum variant cells. Mutat. Res. 1996;362:65–74. doi: 10.1016/0921-8777(95)00034-8. [DOI] [PubMed] [Google Scholar]
- 16.Nakajima S, Lan L, Kanno S, Takao M, Yamamoto K, Eker AP, Yasui A. UV light-induced DNA damage and tolerance for the survival of nucleotide excision repair-deficient human cells. J. Biol. Chem. 2004;279:46674–46677. doi: 10.1074/jbc.M406070200. [DOI] [PubMed] [Google Scholar]
- 17.LeClerc JE, Borden A, Lawrence CW. The thymine–thymine pyrimidine–pyrimidone(6-4) ultraviolet light photoproduct is highly mutagenic and specifically induces 3’ thymine-to-cytosine transitions in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 1991;88:9685–9689. doi: 10.1073/pnas.88.21.9685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gentil A, Le Page F, Margot A, Lawrence CW, Borden A, Sarasin A. Mutagenicity of a unique thymine–thymine dimer or thymine–thymine pyrimidine–pyrimidone (6-4) photoproduct in mammalian cells. Nucl. Acids Res. 1996;24:1837–1840. doi: 10.1093/nar/24.10.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hendel A, Ziv O, Gueranger Q, Geacintov N, Livneh Z. Reduced fidelity and increased efficiency of translesion DNA synthesis across a TT cyclobutane pyrimidine dimer, but not a TT 6-4 photoproduct, in human cells lacking DNA polymerase eta. DNA Repair. 2008;7:1636–1646. doi: 10.1016/j.dnarep.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoon JH, Prakash L, Prakash S. Error-free replicative bypass of (6-4) photoproducts by DNA polymerase zeta in mouse and human cells. Genes Dev. 2010;24:123–128. doi: 10.1101/gad.1872810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reuven NB, Arad G, Stasiak AZ, Stasiak A, Livneh Z. Lesion bypass by the Escherichia coli DNA polymerase V requires assembly of a RecA nucleoprotein filament. J. Biol. Chem. 2001;276:5511–5517. doi: 10.1074/jbc.M006828200. [DOI] [PubMed] [Google Scholar]
- 22.Hendel A, Krijger PH, Diamant N, Goren Z, Langerak P, Kim J, Reissner T, Lee KY, Geacintov NE, Carell T, et al. PCNA ubiquitination is important, but not essential for translesion DNA synthesis in mammalian cells. PLoS Genet. 2011;7:e1002262. doi: 10.1371/journal.pgen.1002262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ziv O, Diamant N, Shachar S, Hendel A, Livneh Z. Quantitative measurement of translesion DNA synthesis in mammalian cells. Methods Mol. Biol. 2012;920:529–542. doi: 10.1007/978-1-61779-998-3_35. [DOI] [PubMed] [Google Scholar]
- 24.Avkin S, Adar S, Blander G, Livneh Z. Quantitative measurement of translesion replication in human cells: evidence for bypass of abasic sites by a replicative DNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 2002;99:3764–3769. doi: 10.1073/pnas.062038699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Avkin S, Sevilya Z, Toube L, Geacintov NE, Chaney SG, Oren M, Livneh Z. p53 and p21 regulate error-prone DNA repair to yield a lower mutation load. Mol. Cell. 2006;22:407–413. doi: 10.1016/j.molcel.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 26.Diamant N, Hendel A, Vered I, Carell T, Reissner T, de Wind N, Geacintov N, Livneh Z. DNA damage bypass operates in the S and G2 phases of the cell cycle and exhibits differential mutagenicity. Nucl. Acids Res. 2012;40:170–180. doi: 10.1093/nar/gkr596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ziv O, Zeisel A, Mirlas-Neisberg N, Swain U, Nevo R, Ben-Chetrit N, Martelli MP, Rossi R, Schiesser S, Canman CE, et al. Identification of novel DNA-damage tolerance genes reveals regulation of translesion DNA synthesis by nucleophosmin. Nat. Commun. 2014;5:5437. doi: 10.1038/ncomms6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Avkin S, Goldsmith M, Velasco-Miguel S, Geacintov N, Friedberg EC, Livneh Z. Quantitative analysis of translesion DNA synthesis across a benzo[a]pyrene-guanine adduct in mammalian cells. The role of DNA polymerase κ. J. Biol. Chem. 2004;279:53298–53305. doi: 10.1074/jbc.M409155200. [DOI] [PubMed] [Google Scholar]
- 29.Gan GN, Wittschieben JP, Wittschieben BO, Wood RD. DNA polymerase zeta (pol zeta) in higher eukaryotes. Cell Res. 2008;18:174–183. doi: 10.1038/cr.2007.117. [DOI] [PubMed] [Google Scholar]
- 30.Paz-Elizur T, Takeshita M, Goodman M, O’Donnell M, Livneh Z. Mechanism of translesion DNA synthesis by DNA polymerase II. comparison to DNA polymerases I and II. J. Biol. Chem. 1996;271:24662–24669. doi: 10.1074/jbc.271.40.24662. [DOI] [PubMed] [Google Scholar]
- 31.Daube SS, Arad G, Livneh Z. Translesion replication by DNA polymerase beta is modulated by sequence context and stimulated by fork-like flap structures in DNA. Biochemistry. 2000;39:397–405. doi: 10.1021/bi991443m. [DOI] [PubMed] [Google Scholar]
- 32.Maccabee M, Evans JS, Glackin MP, Hatahet Z, Wallace SS. Pyrimidine ring fragmentation products. Effects of lesion structure and sequence context on mutagenesis. J. Mol. Biol. 1994;236:514–530. doi: 10.1006/jmbi.1994.1162. [DOI] [PubMed] [Google Scholar]
- 33.Belguise VP, Fuchs RP. N-2-aminofluorene and N-2 acetylaminofluorene adducts: the local sequence context of an adduct and its chemical structure determine its replication properties. J. Mol. Biol. 1995;249:903–913. doi: 10.1006/jmbi.1995.0347. [DOI] [PubMed] [Google Scholar]
- 34.Moriya M, Spiegel S, Fernandes A, Amin S, Liu T, Geacintov N, Grollman AP. Fidelity of translesional synthesis past benzo[a]pyrene diolepoxide-2’-deoxyguanosine DNA adducts: marked effects of host cell, sequence context, and chirality. Biochemistry. 1996;35:16646–16651. doi: 10.1021/bi9608875. [DOI] [PubMed] [Google Scholar]
- 35.Stone JE, Kumar D, Binz SK, Inase A, Iwai S, Chabes A, Burgers PM, Kunkel TA. Lesion bypass by S. cerevisiae Pol zeta alone. DNA Repair (Amst.) 2011;10:826–834. doi: 10.1016/j.dnarep.2011.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jansen JG, Temviriyanukul P, Wit N, Delbos F, Reynaud CA, Jacobs H, de Wind N. Redundancy of mammalian Y family DNA polymerases in cellular responses to genomic DNA lesions induced by ultraviolet light. Nucl. Acids Res. 2014;42:11071–11082. doi: 10.1093/nar/gku779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cai Y, Patel DJ, Broyde S, Geacintov NE. Base sequence context effects on nucleotide excision repair. J. Nucl. Acids. 2010:9. doi: 10.4061/2010/174252. http://dx.doi.org/10.4061/2010/174252, Article ID. [DOI] [PMC free article] [PubMed] [Google Scholar]