Abstract
The detection of protein lysine acylations remains a challenge due to a lack of specific antibodies for acylations with various chain lengths. This problem can be addressed by metabolic labeling techniques using carboxylates with reactive functionalities. Subsequent chemoselective reactions with a complementary moiety connected to a detection tag enable the visualization and quantification of the protein lysine acylome. In this study, we present EDTA-Pd(II) as a novel catalyst for the oxidative Heck reaction on protein-bound alkenes, which allows employment of fully aqueous reaction conditions. We used this reaction to monitor histone lysine acylation in vitro after metabolic incorporation of olefinic carboxylates as chemical reporters.
Introduction
Post-translational modifications (PTMs) are reversible protein alterations strongly linked to a variety of cellular processes, including gene transcription.1 However, the identification and quantification of PTMs faces serious difficulties in cases where the commercially available antibodies are inadequate. Metabolic labeling, in combination with bioorthogonal reactions, offers an attractive alternative that enables the detection of endogenous protein modifications with excellent selectivity.2 A group of PTMs that is suffering from a lack of antibodies is protein lysine acylation. Apart from the well-known protein lysine acetylations, recent evidence indicates the presence of lysine proprionylations, butyrylations and myrystoylations.3,4 The enzymatic conversions of these larger modifications by lysine acyltransferases and lysine deacylases and their functional consequences appear to overlap partially but not completely with lysine acetylations.5 Novel chemistry-based methods can serve as diagnostic tools and enable the elucidation of disease mechanisms connected to these modifications.
The most extensively studied lysine acylation up to date is histone acetylation,6 modulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs). Research has shown that aberrant activity of HATs and HDACs can lead to activation or inhibition of gene transcription in inflammation7,8 and cancer.9,10 In addition, it has been shown that these enzymes also act on non-histone targets such as, for example, the transcription factor p65.11 However, the precise regulatory function of protein lysine acetylations and longer chain acylations of histones and non-histone proteins needs in most cases further investigation.
Radiolabeling using radioactive acetyl-CoA is one of the most traditional techniques that has been employed to obtain the first data on the lysine acetylome.12,13 Afterwards, a combination of immunoprecipitation with acetyl-lysine antibodies and mass spectrometric analysis of histones was developed as a more comprehensive way to map lysine acetylation sites.14 More recently, an antibody-free strategy enabled the investigation of specific acetylation sites, based on the difference in behavior of characteristic 1H/15NHε NMR signals of an acetylated and a non-acetylated amine.15,16 However, the use of NMR spectroscopy falls short on sensitivity especially if applied to cellular proteins.
Currently, a very popular approach to study enzyme activity is chemical labeling of metabolic intermediates using reactive functionalities. A variety of chemical reactions has been developed for the covalent attachment of functionalities to protein-bound functional groups that can be used for enrichment and detection.17 Within this quickly developing field the copper(I)-catalyzed Huisgen cycloaddition, also known as ‘click’ reaction, is the most extensively used strategy.18 More recent developments aim at the replacement of terminal non-conjugated alkynes as chemical reporters by equivalent alkenes due their lower chemical reactivity. Site-specific ligations of protein-bound alkenes have been achieved by alkene-tetrazine ligation.19 Alternatively, our research group described the application of the palladium(II)(Pd(II))-catalyzed oxidative Heck reaction as a novel chemoselective cross-coupling reaction for in vitro detection of alkene-labeled proteins.20
Our current study aims to improve the aqueous oxidative Heck reaction to enable its application to cellular proteins, and to compare its efficiency to the well-known alkyne-azide ‘click’ reaction.18 Previously, we employed the poorly water-soluble bisimine of naphthoquinone (BIAN) ligand, which required solubilization using 20% DMF as a co-solvent. In this work, we describe ethylenediaminetetraacetic acid (EDTA)-Pd(II) as a novel and fully water soluble catalyst that leads to full conversion of protein-bound alkenes via the oxidative Heck reaction. This catalyst enables detection of histone acylation via metabolic labeling with olefinic carboxylates, which demonstrates the applicability of this reaction to detect alkene labeled cellular proteins.
Results and discussion
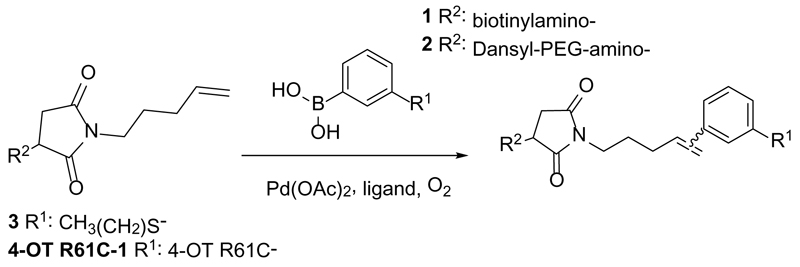
In order to develop water soluble reagents for protein ligation using the oxidative Heck reaction, we employed a cysteine mutant of 4-oxalocrotonate tautomerase, coupled to a terminal alkene at the cysteine residue via a maleimide linker (4-OT R61C-1, SI section 4.1.3), as a model protein.20 In this study, we replaced the previously used poorly water soluble 3-(Dansylamino)phenylboronic acid20 with the newly synthesized water soluble 3-(biotinylamino)phenylboronic acid 1 or 3-(Dansyl-PEG-amino)phenylboronic acid 2 (Scheme 1, SI section 2). Using this starting point, we tested novel water soluble ligands for Pd(II) catalysis.
Scheme 1.
Oxidative Heck reactions between terminal alkenes and arylboronic acids.
In order to investigate the efficiency of alternative ligands, oxidative Heck reactions were performed on protein-bound terminal alkene 4-OT R61C-1 and on its non-protein olefinic equivalent 3 (Scheme 1, SI section 2.6), using three different ligands; the previously used BIAN, 2-amino-4,6-dihydroxypyrimidine (as developed by Davis et al.21) and EDTA (Scheme 1). The reactions were performed with 4-OT R61C-1 using 20 equiv. of the catalyst and 100 equiv. of the phenyl boronic acid 1 20, while 1 equiv. of the catalyst and 10 equiv. excess of phenyl boronic acid were used in the case of the small molecule alkene 3. The reactions were allowed to proceed for 24 h under oxygen atmosphere, at room temperature in aqueous buffer at neutral pH. Firstly, when the BIAN-Pd(II) catalyst (ratio 1.4:1) was employed, the isolated product yield of the reaction between phenyl boronic acid and small molecule alkene 3 was relatively good (Table 1, SI section 3). However, incomplete conversion was observed for the reaction of phenyl boronic acid 1 with 4-OT R61C-1 (Table 1, Fig S3), which is most likely due to limited solubility of the ligand in aqueous buffer at neutral pH. Secondly, for the 2-amino-4,6-dihydroxypyrimidine-Pd(II) catalyst (ratio 2:1), the reaction between phenyl boronic acid 1 and 4-OT R61C-1 was improved but still incomplete (Table 1, Fig S4), whereas the isolated product yield of the reaction between 1 and 3 decreased (Table 1, SI section 3). We note that in case of BIAN and 2-amino-4,6-dihydroxypyrimidine, chelation with EDTA was needed after the completion of the reaction with 4-OT R61C-1 in order to remove the palladium from the protein before mass spectrometric analysis. This indicates that during the reaction Pd(II) also gets bound to proteins, which could hamper the oxidative Heck reaction by limiting the availability of the catalyst.
Table 1.
Yields of oxidative Heck reactions performed on small molecule 3 or protein 4-OT R61C-1.
| Ligand | Reaction 3 with phenylboronic acid [a] | Reaction of 4-OT R61C-1 with 1 [d] |
|---|---|---|
| BIAN | 82% | 58% |
| 2-Amino-4,6-dihydroxypyrimidine | 64%[b] | 84% |
| EDTA | 50%[c] | Full |
Pd(OAc)2 1.0 equiv., BIAN/EDTA/2-amino-4,6-dihydroxypyrimidine 1.4/1/2 equiv., phenylboronic acid 10 equiv., room temperature, 24h, MeOH:H2 O 3:1, pH 7.0. Isolated yield.
pH 8. Isolated yield.
Conversion determined by 1H NMR on the crude reaction mixture.
Pd(OAc)2 20 equiv., BIAN/2-amino-4,6-dihydroxypyrimidine/EDTA 28/40/20 equiv., 100 equiv. of 1, room temperature, 24 h, pH 7.0. The conversion was monitored by LC-MS.
This observation urged us to try EDTA as a ligand to keep Pd(II) in solution and, hence, available for catalysis. Therefore, the EDTA-Pd(II) catalyst (ratio 1:1) was tested in the oxidative Heck reaction on protein-bound alkene 4-OT R61C-1 with phenyl boronic acid 1, which provided full conversion (Table 1, Fig S5). Reaction of 4-OT R61C-1 with phenyl boronic acid 2 under these conditions gave close to full conversion (Fig S6). These findings are interesting because previous studies just show application of EDTA as a ligand for Pd(0) catalyzed reactions and not for Pd(II) catalyzed reactions.21–23 In contrast to the full conversion on protein 4-OT R61C-1, only 50% conversion was found for the reaction between phenyl boronic acid and non-protein bound alkene 3 by EDTA-Pd(II) catalysis as determined by 1H NMR, which indicates that EDTA-Pd(II) is, under these conditions, not the best catalyst for oxidative Heck reactions between phenyl boronic acid and small molecule terminal alkenes. Taking this together, we conclude that EDTA-Pd(II) is a good catalyst for linkage of biotinylated phenyl boronic acid 1 to protein-bound alkenes. We presume that this success originates from the catalyst’s water solubility and superior stability in presence of proteins due to the polar and multivalent nature of the EDTA ligand.
After having established the oxidative Heck reaction as an efficient reaction at neutral pH, we set out to vary the pH of the reaction mixture. Control reactions on the wild type 4-OT demonstrated its stability at pHs ranging from 3 to 11 under the reaction conditions (Fig S17). Subjecting the protein-bound alkene 4-OT R61C-1 to the oxidative Heck reaction using EDTA-Pd(II) catalysis and phenyl boronic acid as a coupling reagent demonstrated limited conversion at pH lower than 7.0, whereas pH 7.0 and higher provides full conversion to the single phenylated product (Fig S11–15). Furthermore, at pH 9.0 a double phenylation of 4-OT R61C-1 was observed, while at pH 11.0 some elimination of the maleimide functionality took place. This indicates that pHs of 7.0 to 8.0 are optimal for the oxidative Heck reaction on protein bound alkenes.
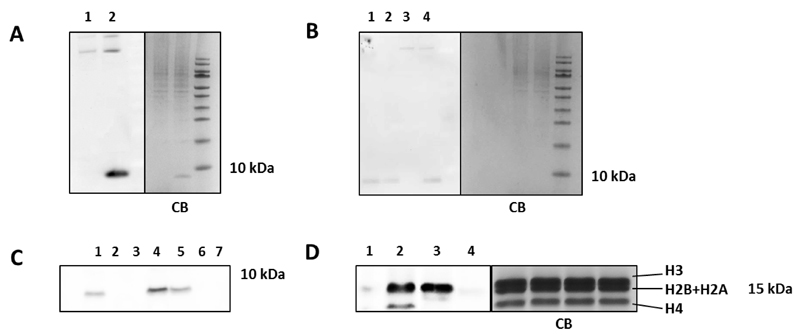
Since the reaction rate decreases with decreasing concentrations, we investigated how well the oxidative Heck reaction performs at low protein concentrations. The on blot detection limit of protein biotinylation, using the Enhanced Chemiluminescence assay (ECL), proved to be 5.0 pmol (0.04 µg), which enables the detection of concentrations down to 250 nM in a reaction mixture (estimated using the fully biotinylated protein 4-OT R61C-2) (SI section 4.5, Fig. S7). Next, the protein-bound terminal alkene 4-OT R61C-1 was reacted with 1 in presence of a cell lysate from RAW 264.7 macrophages mixed in ratios 1:1 and 1:10 based on the protein content (mg/mL) (containing 66 or 50 μM 4-OT R61C-1 respectively). In both cases, when equivalent amounts of protein were loaded on SDS-PAGE, we observed in blot luminescence of the band of biotinylated 4-OT R61C-1 and of some higher molecular weight proteins, that are, presumably, endogenously biotinylated (Fig. 1A and 1B, Fig S8 and S10). Furthermore, LC-MS analysis of the band of reacted 4-OT R61C-1 on SDS-PAGE in ratio 1:1 showed full conversion (Fig. S9). Then, when we performed the reaction on 4-OT R61C-1 at 5 µM concentration, we were pleased to find full conversion of the starting protein, after in blot luminescence detection (Fig. 1B, lane 1 and 4 compared to lane 2 as a reference, Fig. S10).
Fig. 1.
Biotinylation of 4-OT R61C and metabolically labeled histones via the oxidative Heck reaction and the click reaction. A) On blot luminescence imaging and Coomassie Blue staining of 4-OT R61C-1 (0.9 µg) labeled in presence of a cell lysate (protein ratio 1:1). Reaction 1) in absence of 4-OT R61C-1 and 2) in presence of 4-OT R61C-1. B) On blot luminescence imaging and Coomassie Blue staining of 4-OT R61C-1 (0.09 µg) labeled in small scale as well as in presence of a cell lysate (protein ratio 1:10). Reaction 1) 5 µM scale reaction of 4-OT R61C-1, 2) 4-OT R61C-2 (reference), 3) the reaction in absence of 4-OT R61C-1 and 4) in presence of 4-OT R61C-1. C) On blot luminescence imaging of biotinylation of 4-OT R61C-1 and 4-OT R61C-4 in small scale via the oxidative Heck (1, 2, 3) and the click reaction (5, 6, 7). On the SDS-PAGE were loaded 0.1 µg of 1), 5) 3 µM, 2), 6) 1 µM, 3), 7) 0.5 µM scale reaction and 4) 4-OT R61C-2 (reference). D) On blot luminescence imaging and Coomassie Blue staining of biotinylated alkyne-labeled and alkene-labeled histones (7.0 µg) via 2) ‘click’ and 3) oxidative Heck reaction accordingly. Reactions 1) and 4) negative controls for ‘click’ and oxidative Heck reaction accordingly.
Subsequently, we moved on to compare the oxidative Heck reaction with the aqueous ‘click’ reaction at low protein concentrations. For this purpose, the protein-bound terminal alkyne 4-OT R61C-4 was prepared (SI section 6) and subjected to coupling at a protein concentration of 20 μM, using the recently discovered water soluble TABTA ligand,24 Cu(II)sulphate, sodium ascorbate and azide-PEG-biotin, for 2 h at room temperature. Mass spectrometry analysis showed full conversion of protein 4-OT R61C-4 to the desired product (Fig S19). Afterwards, the oxidative Heck reaction and the ‘click’ reaction were performed on 4-OT R61C-1 and 4-OT R61C-4 correspondingly, at protein concentrations of 3, 1 and 0.5 µM. On blot detection showed that both ligation methods resulted in incomplete labeling at 3 µM, while no product was detected at lower concentrations (Fig. 1C, Fig S20). We conclude that both the aqueous oxidative Heck reaction and aqueous ‘click’ reaction give full conversion at higher concentrations and lose their efficacy at protein concentrations in the low micromolar range.
Next we investigated if both methods can be used orthogonal to each other. Exposure of protein-bound alkene 4-OT R61C-1 to the ‘click’ reaction conditions resulted in no change in the protein mass (Fig. S21). On the contrary, application of the oxidative Heck reaction conditions on the protein-bound alkyne 4-OT R61C-4 resulted in a complex mixture with possibly coupling of one or two phenyl groups (Fig. S22), which can be expected due to the Pd(II) catalyzed reactions on alkynes, as reported previously.25,26 This shows that the oxidative Heck reaction on alkenes is not orthogonal to alkynes as chemical reporters.
After having established the aqueous oxidative Heck reaction as a successful and chemoselective cross-coupling method on our model protein 4-OT R61C-1, we moved on to use this reaction for detection of alkene labeled cellular proteins. As a model system we applied histone labeling using 4-pentenoic acid analogously to methods described by Yang et al., who employed alkynic carboxylate 4-pentynoic acid to metabolically label histones in vitro.27 It is expected that these chemical reporters, when supplemented to the medium of cultured cells, are linked to CoA to form acyl-CoA precursors that can subsequently serve as substrates for protein lysine acylation by acyltransferases. In this way, protein acylation can be monitored via bioorthogonal coupling of detection tags to protein-bound alkynes using the ‘click’ reaction or protein bound alkenes using the oxidative Heck reaction (Fig. 2A).
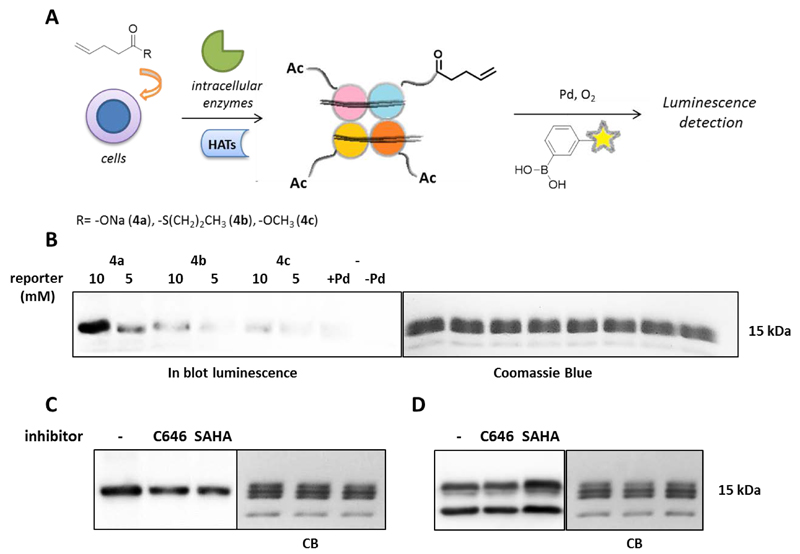
Fig. 2.
A) Schematic procedure of alkene-metabolic labeling and subsequent detection of histones via the oxidative Heck reaction using 3-(biotinylamino)phenylboronic acid 1. B) On blot luminescence imaging and Coomassie Blue staining of histones (5.5 µg) after 6 h incubation with 5 and 10 mM of 4a) sodium pent-4-enoate, 4b) S-propyl pent-4-enethioate and 4c) methyl 4-pentenoate. C) On blot luminescence imaging and Coomassie Blue staining of alkenylated histones (4.0 µg) after 14 h incubation with 30 µM of C646 and 0.41 µM of SAHA followed by treatment with 10 mM of the reporter 4a. D) On blot luminescence detection and Coomassie Blue staining of acetylated histones (3.0 µg) after 20 h incubation with 30 µM of C646 and 0.41 µM of SAHA using the anti-acetyl lysine Antibody.
To investigate the replacement of alkynes with alkenes, we employed sodium 4-pentenoate 4a as an olefinic chemical reporter that was incubated with RAW 264.7 cells at concentrations of 5 and 10 mM for 6 h. The cells were lysed and histones were extracted by acid extraction (Fig. 2B, Fig S23). The oxidative Heck reaction was performed on the alkene labeled histones, using EDTA-Pd(II) as a catalyst and 1 as a coupling partner. Control reactions on untreated histones were also set up, in presence and absence of the catalyst (Fig. 2B, Fig. S23). In the experimental protocol it proved to be important to include 16% of β-mercaptoethanol in the SDS-PAGE loading buffer to minimize non-specific binding of the biotinylated phenyl boronic acid 1 to the proteins.
Additionally, cells were treated with 10 mM of sodium 4-pentynoate for 6 h and the subsequent ‘click’ reaction was performed on the extracted histones, using reagents and conditions mentioned above. In our hands, both methods resulted in a rather equal luminescent signal of histones except for histone H4 that was found to be labeled with the alkynic moiety and not with the olefinic one (Fig. 1D, Fig. S24). We conclude that alkyne or alkene metabolic labeling in combination with the oxidative Heck reaction or the ‘click’ reaction enables the detection of histone acylation with comparable efficacy except for histone H4.
We next investigated a concentration dependency for the metabolic labelling. The labelling efficiency at 5 mM 4-pentenoic acid proved to be much lower compared to 10 mM (Fig 2B, Fig S23). We argued that a poor cellular permeability of this carboxylate could limit the incorporation of 4-pentenoic acid in histones. Therefore, we envisioned that a thioester or a methyl ester derivative could act as a pro-drug type of precursor to facilitate cellular uptake and thus enhance histone pentenoylation. Towards this aim, the thioester S-propyl 4-pentenethioate 4b and the methyl ester methyl 4-penteneoate 4c were employed for metabolic labeling (Fig. 2B, Fig. S23). However, blotting using the ECL assay, demonstrated that 4-pentenoic acid 4a is a more efficient labeling reagent in comparison with 4b and 4c (Fig. 2B, Fig S23). Apparently, pro-drug type of precursors are not more effective in this case.
Finally, the oxidative Heck reaction was employed to detect changes in lysine acylation in presence of HAT and HDAC inhibitors (SI section 8). The HATi C64628 and HDACi SAHA29 were incubated with RAW264.7 cells for 14 h at concentrations of 30 and 0.41 µM respectively, followed by addition of 4-pentenoic acid at 10 mM and subsequent incubation for 6 h. The alkene-labeled histones were detected using the oxidative Heck reaction (Fig. 2C, Fig. S24). For comparison, lysine acetylation was monitored using an Anti-Acetyl Lysine antibody (Fig. 2D, Fig. S26). Pleasingly, metabolic labeling of lysine acylation showed a slight reduction in response to inhibition of HAT activity using C646 (Fig. 2C, Fig. S24). This reduction was however not observed with the Anti-Acetyl Lysine antibody. Interestingly, treatment of the cells with HDACi SAHA resulted also in a reduction of the metabolic labeling, whereas the antibody showed an increase in lysine acetylation (Fig. 2D, Fig. S26). This comparison demonstrates that metabolic labeling of histone lysine acylation using alkene labeled carboxylates responds differently to HATi and HDACi than lysine acetylation as visualized using Anti-Acetyl Lysine antibodies. This could be explained by the idea that incorporation of 4-pentenoic acid needs deacetylation of acetylated lysine residues in order to make the lysine available for acylation with 4-pentenoic acid. An alternative explanation is that histone lysine acylation using 4-pentenoic acid rather mimics larger size lysine acylations such as propionylations and butyrylations and not so much lysine acetylations. Although HATs have been reported to catalyze the transfer of propionyl-groups30 and HDACs the hydrolysis of propionyl- and butyryl-groups,31 it is to be expected that the enzymatic turnover rate and thus the cellular equilibrium between acylation and deacylation depends on the length of the acyl chain, which would explain the differences between the metabolic labeling method and the antibody-based method.
Conclusions
In conclusion, we have developed the Pd(II)-catalyzed oxidative Heck reaction as a powerful tool for protein labeling in a fully aqueous environment. We demonstrated that EDTA is an effective ligand for Pd(II) catalysis on protein-bound terminal alkenes, which provides a fully water soluble catalyst. This catalyst can be employed to link the water soluble 3-(biotinylamino)phenylboronic acid (1) to proteins in quantitative yield at protein concentrations down to 5 µM, whereas the conversion drops at lower concentrations, which is also the case for the aqueous alkyne-azide ‘click’ reaction. The performance of the oxidative Heck reaction on alkene labeled cellular proteins was investigated by metabolic labeling of histone lysine acylation using alkene or alkyne labeled carboxylates. Alkene labeling and detection with the oxidative Heck reaction proved to be equally effective as alkyne labeling and detection with the ‘click’ reaction. Finally, in an attempt to monitor the activity of the responsible enzymes, cells were treated with C646 (HATi) and SAHA (HDACi), which demonstrated that metabolic labeling and detection of lysine acylations deviates from antibody based detection. Taking this together, we conclude that the aqueous oxidative Heck reaction is an effective chemoselective method to biotinylate alkene labeled proteins in biological samples with comparable efficiency as the alkyne to azide ‘click’ reaction.
Supplementary Material
† Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/b000000x/
Acknowledgements
This work was financially supported by a VIDI grant (016.122.302) to F.J.D from the Netherlands Organization for Scientific Research. We thank Prof. Dr. Adriaan J. Minnaard for his helpful advice. We thank Jan-Ytzen van der Meer for technical support with the protein expression.
Notes and references
- 1.Lee DY, Hayes JJ, Pruss D, Wolffe AP. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 2.Sletten EM, Bertozzi CR. Angew Chem Int Edit. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Y, Sprung R, Tang Y, Ball H, Sanfras B, Kim SC, Falck JR, Peng J, Gu W, Zhao Y. Mol Cell Proteomics. 2007;6:812–819. doi: 10.1074/mcp.M700021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stevenson FT, Bursten SL, Locksley RM, Lovett DH. J Exp Med. 1992;176:1053–1062. doi: 10.1084/jem.176.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. Nat Rev Mol Cell Biol. 2014;15:536–550. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- 6.Grunstein M. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 7.Ghizzoni M, Haisma HJ, Maarsingh H, Dekker FJ. Drug Discov Today. 2011;16:505–511. doi: 10.1016/j.drudis.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnes DJ, Adcock IM, Ito K. Eur Respir J. 2005;25:552–563. doi: 10.1183/09031936.05.00117504. [DOI] [PubMed] [Google Scholar]
- 9.Konsoula Z, Barile FA. J Pharmacol Toxicol. 2012;66:215–220. doi: 10.1016/j.vascn.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Egger G, Liang G, Aparicio A, Jones PA. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 11.Dekker FJ, van den Bosch T, Martin NI. Drug Discov Today. 2014;19:654–660. doi: 10.1016/j.drudis.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 12.Brownell JE, Allis CD. Proc Natl Acad Sci USA. 1995;92:6364–6368. doi: 10.1073/pnas.92.14.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu W, roeder RG. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 14.Wolffe AP, Hayes JJ. Nucleic Acid Res. 1999;27:711–720. doi: 10.1093/nar/27.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smet-Nocca C, Wieruszeski J-M, Melnyk O, Benecke A. J Pept Sci. 2010;16:414–423. doi: 10.1002/psc.1257. [DOI] [PubMed] [Google Scholar]
- 16.Liokatis S, Dose A, Schwarzer D, Selenko P. J Am Chem Soc. 2010;132:14704–14705. doi: 10.1021/ja106764y. [DOI] [PubMed] [Google Scholar]
- 17.Lang K, Chin JW. ACS Chem Biol. 2014;9:16–20. doi: 10.1021/cb4009292. [DOI] [PubMed] [Google Scholar]
- 18.Baskin JM, Bertozzi CR. QSAR Comb Sci. 2007;26:1211–1219. [Google Scholar]
- 19.Blackman ML, Royzen M, Fox JM. J Am Chem Soc. 2008;130:13518–13519. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ourailidou ME, van der Meer JY, Baas BJ, Jeronimus-Stratingh M, Gottumukkala AL, Poelarends GJ, Minnaard AJ, Dekker FJ. ChemBioChem. 2014;2:209–212. doi: 10.1002/cbic.201300714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chalker JM, Wood CSC, Davis BG. J Am Chem Soc. 2009;131:16346–16347. doi: 10.1021/ja907150m. [DOI] [PubMed] [Google Scholar]
- 22.Wei J-F, Jiao J, Feng J-J, Lv J, zhang X-R, Shi X-Y, Chen Z-G. J Org Chem. 2009;74:5967–5974. [Google Scholar]
- 23.Korolev DN, Bumagin NA. Tetrahedron Lett. 2005;46:5751–5754. [Google Scholar]
- 24.Rudolf GC, Sieber AS. ChemBioChem. 2013;14:2447–2455. doi: 10.1002/cbic.201300551. [DOI] [PubMed] [Google Scholar]
- 25.Shen K, Han X, Lu X. Org Lett. 2013;15:1732–1735. doi: 10.1021/ol400531a. [DOI] [PubMed] [Google Scholar]
- 26.Yao B, Wang Q, Zhu J. Angew Chem Int Edit. 2012;51:5170–5174. doi: 10.1002/anie.201201640. [DOI] [PubMed] [Google Scholar]
- 27.Yang Y-Y, Ascano JM, Hang HC. J Am Chem Soc. 2010;132:3640–3641. doi: 10.1021/ja908871t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowers EM, Yan G, Mukherjee C, Orry A, Wang L, Holbert MA, Crump NT, Hazzalin CA, Liszczak G, Yuan H, Larocca C, et al. Chem Biol. 2010;17:471–482. doi: 10.1016/j.chembiol.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richon VM, Emiliani S, Verdin E, Webb Y, Breslow R, Rifkind RA, Marks PA. Proc Natl Acad Sci USA. 1998;95:3003–3007. doi: 10.1073/pnas.95.6.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leemhuis H, Packman LC, Nightingale KP, Hollfelder F. ChemBioChem. 2008;9:499–503. doi: 10.1002/cbic.200700556. [DOI] [PubMed] [Google Scholar]
- 31.Smith BC, Denu JM. J Biol Chem. 2007;282:37256–37265. doi: 10.1074/jbc.M707878200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.