Abstract
Lysine acetylations are reversible posttranslational modifications of histone and non-histone proteins that play important regulatory roles in signal transduction cascades and gene expression. Lysine acetylations are regulated by histone acetyltransferases as writers and histone deacetylases as erasers. Because of their role in signal transduction cascades, these enzymes are important players in inflammation. Therefore, applications of histone acetyltransferase inhibitors to reduce inflammatory responses are interesting. Among the few histone acetyltransferase inhibitors described, C646 is one of the most potent (Ki of 0.4 μM for histone acetyltransferase p300). C646 was described to regulate the NF-κB pathway; an important pathway in inflammatory responses, which is regulated by acetylation. Interestingly, this pathway has been implicated in asthma and COPD. Therefore we hypothesized that via regulation of the NF-κB signaling pathway, C646 can inhibit pro-inflammatory gene expression, and have potential for the treatment of inflammatory lung diseases. In line with this, here we demonstrate that C646 reduces pro-inflammatory gene expression in RAW264.7 murine macrophages and murine precision-cut lung slices. To unravel its effects on cellular substrates we applied mass spectrometry and found, counterintuitively, a slight increase in acetylation of histone H3. Based on this finding, and structural features of C646, we presumed inhibitory activity of C646 on histone deacetylases, and indeed found inhibition of histone deacetylases from 7 μM and higher concentrations. This indicates that C646 has potential for further development towards applications in the treatment of inflammation, however, its newly discovered lack of selectivity at higher concentrations needs to be taken into account.
Keywords: C646, inflammation, NF-κB, macrophages, acetylation, histones
1. Introduction
Lysine acetylations are reversible posttranslational modifications that frequently occur on histones and other cellular proteins. These modifications have been recognized to play important regulatory roles in signal transduction cascades and gene expression [1][2]. Protein lysine acetylations are regulated by histone acetyltransferases (HATs) as writers and histone deacetylases (HDACs) as erasers. In the past decades many different isoforms of HATs and HDACs have been discovered [3][4], and aberrations in their activity have been associated with diseases such as inflammatory disorders or cancer [5][6].
For this reason, the development of small molecule inhibitors of these enzymes and their applications in therapeutic areas such as the treatment of inflammatory diseases have received considerable attention. This has resulted in the development of small molecule HAT inhibitors (HATi) [7]. Virtual screening enabled the identification of the small molecule inhibitor C646 as a potent and cell-permeable p300 HATi (Ki 0.4 μM) which is selective for p300 among HAT isoenzymes such as PCAF, GCN5 and MOZ [8].
C646 has shown interesting effects in disease models. For example, C646 has successfully been applied in an animal model of neuropathic pain. In this study, C646 was administered in rats via a lumbar intrathecal catheter, demonstrating the feasibility of local administration of C646 in animals. It was found that C646 treatment diminished both the p300 promoter binding and the expression of COX-2 [9]. In another study on prostate cancer cell lines it was found that C646 mediated inhibition of p300 increased apoptosis, which was, among others, caused by inhibition of the androgen receptor and the NF-κB pathway [10]. These studies indicate that C646 influences signaling cascades such as the NF-κB pathway.
The effects of C646 on the NF-κB pathway could be explained by the role for p300 that has been described in the regulation of the NF-κB pathway [5]. For instance, acetylations of the p65 NF-κB subunit on lysines 218, 221 and 310 are mediated by the HATs p300 and PCAF [11] and increase transcriptional activity. In contrast, acetylations on lysines 122 and 123 decrease transcriptional activity and are also mediated by p300 [12]. Furthermore, upon DNA binding, the transcriptional activation domain of p65 interacts with the HATs p300 and CBP as co-activators of gene transcription [13]. Thus, p300 plays a crucial role in the regulation of acetylation of specific lysine residues of NF-κB, which then determine NF-κB transcriptional capacity, DNA binding and duration of action.
This suggests that inhibition of the HAT p300 using small molecule inhibitors such as C646 may allow for regulation of gene expression via the NF-κB transcription factor [7]. Since the NF-κB pathway is a key factor in inflammatory responses, this calls for investigation of the potential of C646 to suppress these responses. Of particular interest may be applications in inflammatory lung diseases such as asthma and chronic obstructive pulmonary disease (COPD), which still pose a major health problem. Interestingly, a role for the NF-κB pathway has been described in these diseases, as reviewed [14]. Increased NF-κB activity has been observed in the bronchial epithelium and peripheral blood mononuclear cells of asthmatic patients [15][16]. Increased nuclear localization of p65 was observed in sputum macrophages during exacerbations of COPD [17] and also in bronchial biopsies of stable COPD patients [18]. Several lines of research have focused on modulating NF-κB activity as a novel therapeutic strategy for the treatment of asthma and COPD [14]. We hypothesize that pharmacological inhibition of the HAT p300 by small molecule inhibitor C646 will result in inhibition of pro-inflammatory gene transcription via inhibition of the NF-κB signaling pathway.
For these reasons, we set out to explore the p300 HATi C646 in a model system for inflammatory lung diseases. We observed decreased NF-κB reporter gene activity in RAW Blue murine macrophages, and decreased pro-inflammatory gene expression in RAW264.7 murine macrophages as well as in murine lung tissue slices. Next to this we set out to quantify changes in lysine acetylation in the cellular substrates of p300. Acetylation of histone H3 and histone H4 was quantified using mass spectrometry in RAW264.7 macrophages. Counterintuitively, C646 increased acetylation on histone H3 residues 18-26, containing H3 K18 and H3 K23. Based on these findings and the structural properties of C646 we investigated its inhibitory potential on recombinant HDACs and found inhibition from 7 μM and higher concentrations for type I and II HDACs. Importantly, these findings call for further optimization of the selectivity profile of the p300 HATi C646 and its derivatives.
2. Materials and Methods
2.1. Chemicals and Reagents
All chemicals and reagents were purchased from Sigma Aldrich (St. Louis, Zwijndrecht, the Netherlands) unless otherwise stated. C646 was purchased from Axon Medchem (Groningen, the Netherlands) and SAHA from Selleckchem (Munich, Germany). The purity of C646 was assessed by HPLC, MS, and NMR by Axon Medchem, and the same was done for SAHA by Selleckchem.
2.2. Cell culture and viability assay
RAW264.7 macrophages were purchased from American Type Culture Collection (Manassas, Virginia, USA), and cell culture reagents were purchased from Life Technologies. RAW-Blue macrophages were purchased from InvivoGen (Toulouse, France). RAW264.7 macrophages were cultured as described previously [19]. Raw Blue macrophages were cultured in the same manner, with the addition of Zeocin (200 μg/mL) to the culture medium according to the manufacturer's instructions. For the experiments, cells were used until passage 18. Viability of C646 or HDAC inhibitor (HDACi) suberanilohydroxamic acid (SAHA) treated RAW264.7 was assessed by CellTiter 96 Aqueous One Solution Cell Proliferation Assay (Promega, Leiden, the Netherlands). For the assay, cells were seeded at 7500 cells per well in 96 wells plates. On the following day, medium was replaced with medium containing C646 or SAHA at the appropriate concentrations. After 20 hrs of incubation, the CellTiter reagent was added to the wells. After 1 hr of incubation with this reagent, the absorbance at 490 nm was measured using a Synergy H1 Hybrid Multi-Mode Microplate Reader (BioTek, Vermont, USA). The amount of absorbance at this wavelength is indicative of mitochondrial conversion of the reagent, which is linked to cell viability.
2.3. Precision-cut lung slices
Precision-cut lung slices (PCLS) were prepared as described previously [19]. Viability of C646 treated PCLS was assessed by the amount of lactate dehydrogenase (LDH) released by the tissue slices into the culture medium. The measurements were performed as described previously [19]. Viability of SAHA treated PCLS was not determined.
2.4. Treatment of RAW264.7, Raw Blue and PCLS with inhibitors and LPS IFNγ
Cells or PCLS were pre-treated with C646 at 1, 5, 10, 15, 20, 25 or 30 μM (for PCLS the 30 μM concentration was not included due to decreased viability), or with the HDACi SAHA at 0.41 μM for 16 hrs. Inhibitor stocks were prepared in dimethylformamide (DMF) and were further diluted in DMEM culture medium. Vehicle treatment constituted of pre-treatment with 0.3% DMF for the cells (corresponding to the same final DMF% as for 30 μM C646), or with 0.2% DMF for PCLS (corresponding to the same final DMF% as for 20 μM C646), for 16 hrs. Subsequently, cells were stimulated with 10 ng/mL lipopolysaccharide (LPS, Escherichia coli, serotype 0111:B4; Sigma-Aldrich) and 10 ng/mL interferon gamma (IFNγ, cat.#315-05; PeproTech, Hamburg, Germany) for 4 hrs, in continued presence of the inhibitors.
2.5. RAW-Blue NF-κB reporter gene assay
RAW-Blue macrophages are derived from RAW264.7 macrophages by chromosomal integration of a secreted embryonic alkaline phosphatase (SEAP) reporter construct inducible by NF-κB, and were used to monitor NF-κB promoter activity. After treatment with the inhibitors are described above, cell supernatants were used to perform the NF-κB reporter gene assay according to the manufacturer's instructions.
2.6. Gene expression analysis in RAW264.7 or PCLS by RT-q-PCR
Gene expression analysis by RT-q-PCR was performed as described previously [19]. The primers for TNFα (Mm00443258_m1), IL-1β (Mm00434228_m1), IL-6 (Mm00446190_m1), IL-8 (Mm04208136_m1), iNOS (Mm00440502_m1) IL-12b (Mm00434174_m1) and GAPDH (Mm99999915_g1) were purchased as Assay-on-Demand (Applied Biosystems).
2.7. Enzyme inhibition assays and Lineweaver-Burk plots
All human recombinant HDAC inhibition assays were performed as described previously [20]. The Lineweaver-Burk plots of C646 on HDAC2 and HDAC6 were also performed as described previously [20]. Activity of the HAT lysine acetyltransferase 8 (KAT8) was measured using chemical detection of coenzyme A (CoASH) after fluorescent labelling as described previously [21]. To test inhibition of the HAT p300 by C646, the following protocol was used. The substrate (5 μM, Histone H3) was prepared in freshly prepared reaction buffer (50 mM Tris-HCl pH 8.0, 50 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 0.1 mM PMSF, 1% DMSO). Subsequently, the enzyme at 80 nM final concentration (Recombinant Catalytic domain (aa 1284-1673) of human p300, MW= 45.1 kDa, expressed in E. coli, obtained from Enzo Life Sciences Inc. Rome, Italy) was delivered to this substrate solution and mixed gently. The inhibitor in DMSO was then delivered to the enzyme/substrate reaction mixture by using acoustic dispenser from Acoustic Technology (Echo 550, LabCyte Inc. Sunnyvale, CA) in nanoliter range. 2.3 μM [Acetyl-3H]-Acetyl Coenzyme A (PerkinElmer, Waltham, Massachusetts, USA) was then added to this reaction mixture, and incubated for 1 hour at 30°C. The reaction mixture was then delivered to filter-paper (P-81, Upstate, Vimodrone, Italy). The filter-paper was washed with PBS to remove free 3H-CoA, dried and placed in vials containing scintillation fluid for detecting 3H signals transferred to the histone substrate captured on the filter-paper. Radioactive counts were recorded on Tri-Carb 2800TR Liquid Scintillation Analyzer (PerkinElmer, Waltham, Massachusetts, USA). C646 was tested in a 10-dose IC50 mode with 2-fold serial dilution starting at 25 μM. The data were analyzed using Excel and GraphPad Prism 4.0 software (GraphPad Software Inc., San Diego CA, USA) for IC50 curve fit.
2.8. Histone extraction and Micro BCA™ Protein Assay
For the histone extractions, cells were treated with the HAT inhibitor C646 (15, 20, 25 or 30 μM) or the HDACi SAHA (0.41 μM) on the day after seeding. Cells were incubated with C646 for 6 or 20 hrs or SAHA for 20 hrs. Inhibitor stocks were prepared in DMF and were further diluted in DMEM culture medium. Vehicle treatment constituted 0.3% DMF (corresponding to the same final DMF% as for 30 μM C646). Histone extractions were then performed as previously described [22] with minor modifications. After histone extractions the samples were diluted with PBS to determine the total protein concentration using the micro BCA protein assay according to the manufacturer's instructions (Pierce, Rockford, USA, # 23235). Absorbance was measured with a Synergy H1 Hybrid Multi-Mode Microplate Reader (BioTek, Vermont, USA) at 562 nm. A bovine serum albumin standard (2 mg/mL, Pierce, Rockford, USA, # 23209) was used to calibrate the assay.
2.9. Acetylation of histones with acetic anhydride-d6 (in-gel)
For the in-gel reactions of histones with acetic anhydride-d6, 7 μg of histones were loaded on a 15% polyacrylamide gel and resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). After staining with InstantBlue (product #ISB1L from Expedeon, Cambridge, UK) bands for histones H3 and H4 were excised from the gel. Then 50 μL of acetonitrile and 50 μL of ammonium bicarbonate buffer (100 mM) were added to destain the gel bands. Gel bands were dried in acetonitrile. Subsequently, 5 μL of acetic anhydride-d6 was added, after which 100 μL of ammonium bicarbonate (1M) was added immediately. Subsequently, samples were incubated at 37oC for 15 min. Gel bands were washed 3 times with H2O, and the acetic anhydride-d6 reaction was repeated. After the second reaction, gel bands were dried using acetonitrile and trypsin (Promega, Leiden, the Netherlands, # V511A) was added at a 1:20 ratio in ammonium bicarbonate (50 mM). Histones were digested at 37 °C for 16 hrs. Supernatants containing histone peptides were subjected to LC MS/MS analysis as described below.
2.10. NanoChip LC-MS/MS QTOF
For the analysis of acetylation status of the histone peptides by LC-MS/MS, a quadruple time-of-flight mass spectrometer (QTOF, Agilent 6510) with a liquid chromatography-chip cube (# G4240) electrospray ionization interface was coupled to a nanoLC system (Agilent 1200) composed of a nanopump (# G2226A), a capillary loading pump (#G1376A) and a solvent degasser (# G1379B). Injections were performed with an autosampler (# G1389A) equipped with an injection loop of 40 μL and a thermostated cooler maintaining the samples in the autosampler at 4°C during the analysis (#G1377A Micro WPS). The instrument was operated under the MassHunter Data Acquisition software (Agilent Technologies, Santa Clara, USA, version B.04.00, B4033.3). A chip (ProtID-Chip-150 II 300A, #G4240-62006) with a 40 nL trap column and a 75 µm × 150 mm analytical column filled with Zorbax 300SB-C18, 5 µm (Agilent Technologies, Santa Clara, USA) was used for peptide separation.
The identification of peptides was based on data collected in auto MS/MS mode (2 GHz) using the following settings; fragmentor: 175 V, skimmer: 65 V, OCT 1 RF Vpp: 750 V, precursor ion selection: medium (4 m/z), mass range: 200-2500 m/z, acquisition rate for MS: 2 spectra/sec, for MS/MS 3 spectra/sec; MS/MS range: 50-3000 m/z; ramped collision energy: slope 3.8, offset: 0, precursor setting: maximum 3 precursors/cycle; absolute threshold for peak selection was 1000; relative threshold was 0.01 % of the most intense peak, active exclusion enabled after 1 selection, release of active exclusion after 0.6 min, precursors were sorted by abundance only. The MS/MS files were stored in centroid and profile mode. MS1 absolute threshold 50 and MS2 absolute threshold 35 were applied to account for detector noise. A static exclusion range of 200-350 m/z for precursor selection was applied. Gas temperature (nitrogen) was 325 °C and gas flow was 5 L/min. The quantification of peptides was based on data collected in MS mode using the same settings except of the mass range 200-3000 m/z; acquisition rate 1 spectra/sec. In both cases lock masses 1221.990 m/z (HP-1221; Agilent article number G1982-85001) and 299.294 m/z (Methyl Stearate; Agilent article number G1982-85003) were used to recalibrate spectra during the acquisition. The area of the manually extracted ion chromatograms (0.1 m/z tolerance) of the selected peptides was used for label-free quantification.
2.11. Database search
For the database search of the samples subjected to LC-MS/MS, tandem mass spectra were extracted, charge state deconvoluted and deisotoped by the MassHunter Qualitative Analysis software version B.05.00 (Agilent) and saved as .mgf files. All MS/MS data were analyzed using Phenyx (GeneBio, Geneva, Switzerland); version CYCLONE (2010.12.01.1)). The fragment ion mass tolerance of 0.30 Da and a parent ion tolerance of 400 ppm were selected for a database search. The oxidation of methionine (+15.99), light (+42.01) and heavy (+45.03) acetylation of lysine (K), heavy acetylation with light methylation (+59.045) of K, methylation (+14.01), dimethylation (+28.03), trimethylation (+42.04) of K, methylation of arginine (R) (+14.01), dimethylation of arginine (R) (+28.03) N-terminal acetylation (+42.01), deamidation (+0.98) of asparagine (N) and glutamine (Q) were specified in Phenyx as variable modifications.
2.12. Immunoblot protocol
To study α-tubulin acetylation, 20 μg of cell lysate were loaded on a 10% polyacrylamide gel, resolved by SDS-PAGE electrophoresis and electroblotted to PVDF membranes. The membranes were incubated with anti-acetyl α-tubulin antibody (Cell Signalling 5335) or with anti-GAPDH (Cell Signalling 5174), followed by a swine anti-rabbit HRP conjugated antibody (DakoCytomation, P0217). Bands were visualized using a 1B1581 visiglo prime HRP chemiluminescence substrate kit from Amresco (Solon, Ohio, USA). Bands were scanned using a G:BOX iChemi system from Syngene (Cambridge, UK) and quantified using ImageJ quantification software. The GAPDH signal was used as loading control. The experiments were performed at least in triplicate.
2.13. Statistical analysis
GraphPad Prism 5.0 software (GraphPad Software Inc., San Diego CA, USA) was used to perform data analysis in all experiments. Data are presented as mean ± SD from at least 3 independent experiments, and were analyzed by 1-way analysis of variance, followed by Bonferroni post hoc tests in all cases. Significance was assigned at a p value ≤ 0.05.
3. Results
3.1. C646 inhibits NF-κB activity
First, the viability of RAW264.7 cells treated with C646 at different concentrations was assessed (data not shown). As a reference, the well-studied HDACi SAHA was also included at a single concentration and tested in a viability assay (data not shown). Subsequently, to explore the potential of C646 in suppression of (NF-κB mediated) inflammation, an NF-κB reporter gene assay was employed, using RAW-Blue (modified RAW264.7) macrophages. For NF-κB activation, LPS and IFNγ were employed as inflammatory stimulus. An LPS and IFNγ stimulus at 10 ng/mL of each for 4 hours induced NF-κB activity (Fig. 1). We then proceeded to pre-treatment with C646 at concentrations of 1, 5, 10, 15, 20, 25 and 30 μM for 16 hrs before receiving the stimulus, rendering a total incubation time with the inhibitor of 20 hrs. These concentrations were chosen because they are above the Ki value for p300 inhibition of 0.4 μM [8] at different folds. Concentrations above 30 μM were not studied due to decreased viability of the macrophages. A total incubation time of 20 hrs was chosen based on a previous study with a similar incubation time (24 hrs), where C646 affected the NF-κB pathway (decreased expression levels of p65, and decreased binding of p65 at the IkBa promoter were observed) in a disease model of prostate cancer [10], suggesting that effects of C646 on this pathway in cells take place at these relatively long incubation times.
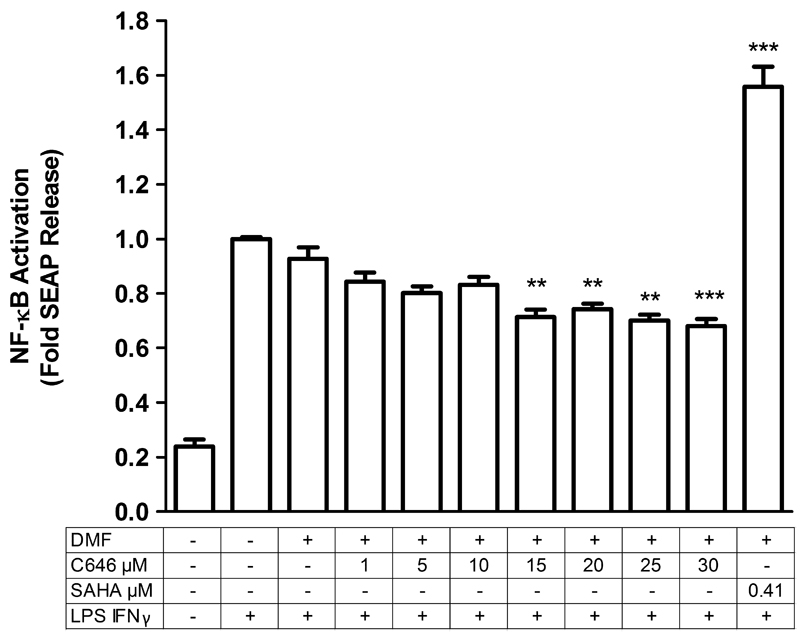
Fig. 1.
C646 pre-treatment reduces NF-κB activity, whereas SAHA pre-treatment increases NF-κB activity. RAW-Blue macrophages were pre-treated with inhibitors C646 (at 1, 5, 10, 15, 20, 25, or 30 μM) or SAHA (at 0.41 μM) for 16 hours, after which an inflammatory LPS and IFNγ stimulus (10 ng/mL of each) was given for 4 hours in continued presence of the inhibitors (making the total incubation time with the inhibitors 20 hrs). Supernatant was then used to determine NF-κB promoter activity. For vehicle treatment, cells were pre-treated with a proportional dilution of the inhibitor solvent DMF. The data shown represent means ± SD of 3-7 independent experiments. *** p < 0.001 and ** p < 0.01 compared to vehicle treated cells.
Interestingly, at 15 μM or higher concentrations of C646, this resulted in significant inhibition of LPS and IFNγ induced NF-κB promoter activity (Fig. 1). This indicates influence of C646 on the NF-κB pathway. In contrast, pre-treatment with the reference compound SAHA at a concentration of 0.41 μM further enhanced LPS and IFNγ induced NF-κB activity (Fig. 1).
3.2. C646 inhibits pro-inflammatory gene expression in RAW264.7 macrophages and precision-cut lung slices
Next, we monitored expression of the NF-κB mediated pro-inflammatory genes TNFα, iNOS, IL-1β, IL-12b IL-6, and IL-8 in RAW264.7 murine macrophages upon receiving the same LPS and IFNγ stimulus, using RT-q-PCR. The stimulus was found to increase expression of all genes (shown for TNFα and IL-12b in Fig. 2; data not shown for the remaining genes), with the exception of IL-8, which was not expressed in RAW264.7 macrophages.
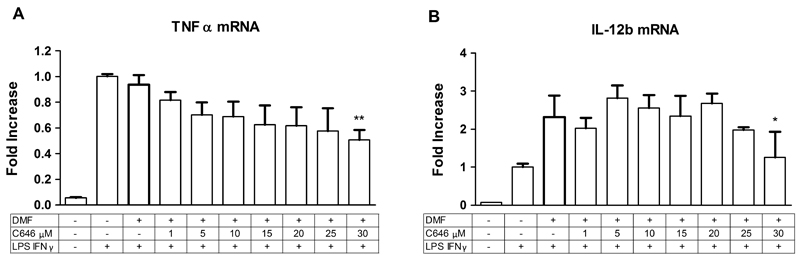
Fig. 2.
C646 pre-treatment reduces TNFα (A) and IL-12b (B) gene expression in RAW264.7 macrophages. RAW264.7 cells were pre-treated with C646 at 1, 5, 10, 15, 20, 25 or 30 μM for 16 hours, after which an inflammatory LPS and IFNγ stimulus (10 ng/mL of each) was given for 4 hours in continued presence of the inhibitors (making the total incubation time with the inhibitors 20 hrs). Subsequently, gene expression was analyzed by RT-q-PCR. For vehicle treatment, cells were pre-treated with a proportional dilution of the inhibitor solvent DMF. Data represent the target gene expression normalized to the reference gene. The values shown are means ± SD of 3-10. * p < 0.05 compared to vehicle treated cells.
Pre-treatment with C646 resulted in a dose dependent decrease in the LPS and IFNγ induced expression of TNFα which became significant at 30 μM (Fig. 2, Table 1). Furthermore, for IL-12b, even though a dose-dependent decrease was less obvious, pre-treatment with C646 at 30 μM significantly inhibited the LPS and IFNγ induced expression of this gene compared to vehicle (DMF dilution) treatment (Fig. 2, Table 1). The expression of other studied genes remained unchanged (Table 1). In contrast, pre-treatment with the reference compound SAHA at 0.41 μM further upregulated LPS and IFNγ induced gene expression of IL-1β (Table 1), but did not affect the expression of the other studied genes (Table 1). This indicates an anti-inflammatory effect of C646 on gene expression in RAW264.7 macrophages, as opposed to a pro-inflammatory effect of SAHA.
Table 1.
Overview of changes in gene expression in RAW264.7 cells upon C646 (at 30 μM) or SAHA (at 0.41 μM) pre-treatment, as well as in PCLS upon C646 (at 25 μM) or SAHA (at 0.41 μM) pre-treatment followed by an inflammatory LPS and IFNγ stimulus. The percentage of change compared to vehicle treatment is indicated. Upreguation is indicated by ↑, downregulation by ↓, and no effect by –. N.E. = gene is not expressed, and N.D. = gene expression was not determined.
| C646 | TNFα | iNOS | IL-1β | IL-6 | IL-8 | IL-12b |
|---|---|---|---|---|---|---|
| RAW264.7 | ↓ (54.1 %) | - | - | - | N.E. | ↓ (54.2 %) |
| PCLS | ↓ (79.1 %) | ↓ (58.4 %) | ↓ (72.3 %) | - | - | ↓ (85.0 %) |
| SAHA | TNFα | iNOS | IL-1β | IL-6 | IL-8 | IL-12b |
| RAW264.7 | - | - | ↑ (317 %) | N.D. | N.E. | - |
| PCLS | - | ↓ (39.9 %) | ↑ (41.5 %) | ↑ (58.8 %) | N.D. | - |
In order to move towards a more physiological environment, and towards a better model system for lung inflammation, we employed murine precision-cut lung slices (PCLS); an ex-vivo model in which structures of lung tissue, including cell-cell and cell-matrix relationships, are maintained [23]. The viability of PCLS treated with C646 was assessed by lactate dehydrogenase (LDH) measurement (data not shown). The viability of PCLS treated with SAHA was not assessed. The LPS and IFNγ stimulus induced expression of the same NF-κB mediated pro-inflammatory genes in PCLS (shown for TNFα, iNOS, IL-1β and IL-12b in Fig. 3; data not shown for the remaining genes).
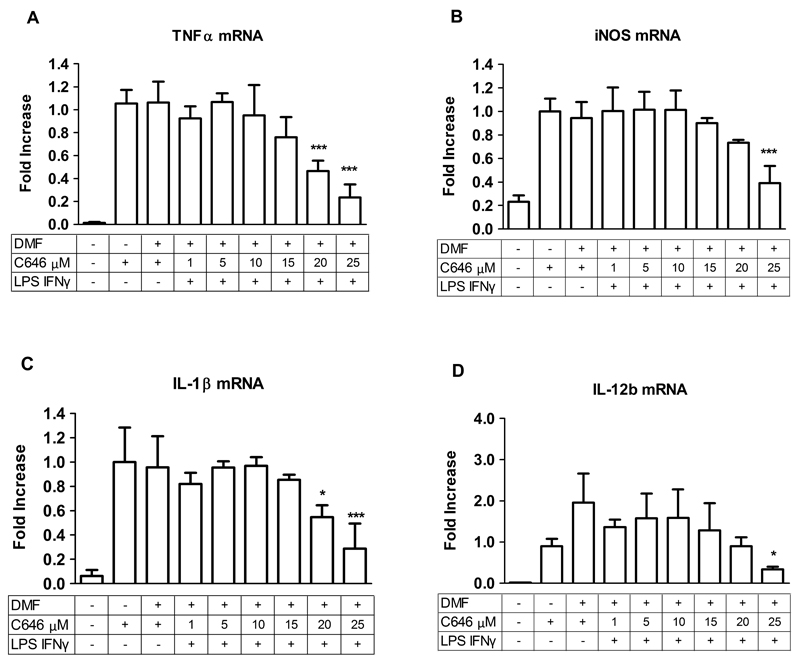
Fig. 3.
C646 pre-treatment reduces pro-inflammatory gene expression of TNFα (A), iNOS (B), IL-1β (C), and IL-12b (D) in precision-cut lung slices (PCLS). PCLS were pre-treated with C646 at 1, 5, 10, 15, 20, 25 or 30 μM for 16 hours, after which an inflammatory LPS and IFNγ stimulus (10 ng/mL of each) was given for 4 hours in continued presence of the inhibitors (making the total incubation time with the inhibitors 20 hrs). Subsequently, gene expression was analyzed by RT-q-PCR. For vehicle treatment, PCLS were pre-treated with a proportional dilution of the inhibitor solvent DMF. Data represent the target gene expression normalized to the reference gene. The values shown are means ± SD of 3-6 independent experiments. *** p < 0.001, ** p < 0.01 and * p < 0.05 compared to vehicle.
For C646 concentration dependency was studied in PCLS until 25 μM. Concentrations above 25 μM were not included due to decreased viability of PCLS. Major decreases in the LPS and IFNγ induced expression of TNFα, iNOS, IL-1β, and IL-12b were observed for pre-treatments at higher concentrations of C646, until 25 μM compared to vehicle treatment (DMF) (Fig. 3, Table 1). Importantly, this indicates an anti-inflammatory effect of C646 on gene expression in PCLS. For IL-6 and IL-8, there were no changes in gene expression at the studied C646 concentrations (Table 1).
Upon pre-treatment of PCLS with SAHA as a reference compound at 0.41 μM, downregulation of the expression of iNOS induced by LPS and IFNγ was observed, whereas the expression of IL-1β and IL-6 were further enhanced (Table 1). No changes in expression of the other genes were found (Table 1).
3.3. C646 increases histone acetylation on H3 residues 18-26 in a time dependent manner
To assist the further development of C646 and potential derivatives towards applications for the treatment of inflammatory diseases, we proceeded to study changes in lysine acetylation in the cellular substrates of p300 upon C646 treatment. To this end, histone acetylation was quantified using a previously described mass spectrometry-based approach in which histones are acetylated on non-acetylated lysine residues using deuterated acetic acid anhydride ((CD3CO)2O) [24][25][26][27]. A deuterated acetyl group is installed only on naturally non-acetylated lysine residues, whereas naturally acetylated lysine residues remain untouched. Subsequent tryptic digestion results in C-terminal cleavage only after arginine residues, generating peptide fragments of suitable length for LC-MS/MS analysis. Within one peptide, the deuterated acetyl groups have comparable physical properties to the natural acetyl groups. Due to their mass difference, the level of lysine acetylation within one peptide can thus be determined.
Treating RAW264.7 cells with SAHA as a reference compound (0.41 µM for 20 hrs) resulted in an increased acetylation for two histone-derived peptides (H4 4-17 and H3 18-26) but not the others (data not shown). Next to this, we found that the LPS and IFNγ stimulus (10 ng/mL each for 4 hours) did not change global histone acetylation (data not shown).
RAW264.7 cells were treated with C646 at 15, 20, 25 or 30 μM for 20 hrs, without subsequent inflammatory stimulus. No changes in histone acetylation were observed at this time point for which changes in gene expression were observed (Fig. 4). We reasoned that effects on histone acetylation upon C646 treatment could take place at earlier time points as this has been reported previously [28], and proceeded to treatment with C646 for 6 hrs at 15, 20, 25 or 30 μM. No significant changes in the acetylation status of histone H4 res. 4-17 were observed (Fig. 5), which is not in line with literature describing that p300 acetylates H4 at K5, K8, and K12 [29]. Furthermore, no changes in acetylation status of other histone H4 peptides were detected.
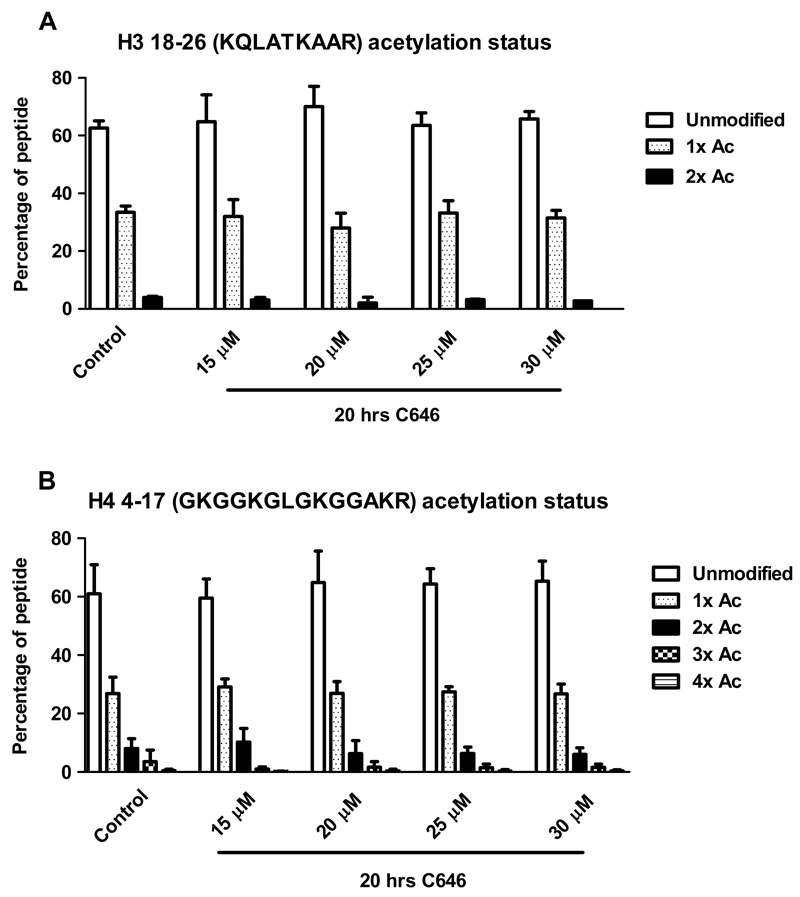
Fig. 4.
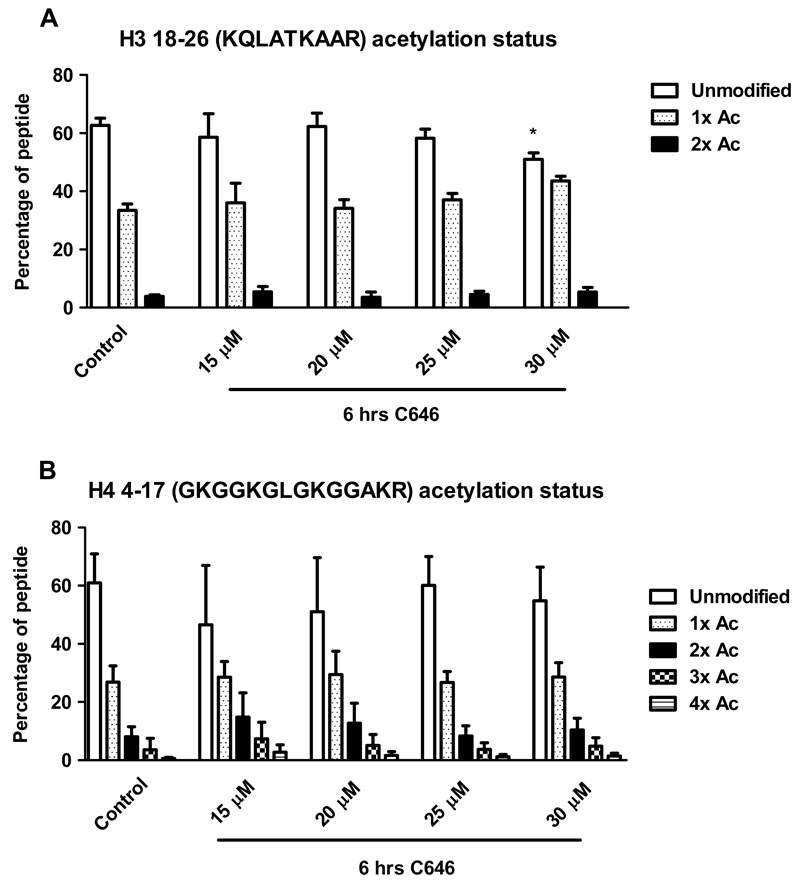
C646 does not affect histone acetylation after 20 hrs of incubation of histone H3 res. 18-26 (A) or H4 res. 4-17 (B). RAW264.7 cells were incubated with C646 at the indicated concentrations for 20 hrs, after which histones were extracted. Histones were resolved by SDS PAGE, and histones H3 and H4 were excised from the gels. Gel pieces were treated with acetic anhydride d6, followed by trypsin digestion. Resulting peptides were subjected to LC-MS/MS analysis. Data are presented as mean values ± SD of 2-12 independent experiments. ** p < 0.01 and * p < 0.05 compared to control (untreated cells). No significant differences were observed between control and vehicle treated cells (data not shown).
Fig. 5.
In RAW264.7 cells, after 6 hrs of incubation C646 increases acetylation status of histone H3 res. 18-26 (A) and does not affect acetylation status of H4 res. 4-17 (B). RAW264.7 cells were incubated with C646 at the indicated concentrations for 6 hrs, after which histones were extracted. Histones were resolved by SDS PAGE, and histones H3 and H4 were excised from the gels. Gel pieces were treated with acetic anhydride d6, followed by trypsin digestion. Resulting peptides were subjected to LC-MS/MS analysis. Data are presented as mean values ± SD of 3-12 independent experiments. ** p < 0.01 and * p < 0.05 compared to control (untreated cells). No significant differences were observed between control and vehicle treated cells (data not shown).
However, while analyzing histone H3, a difference in acetylation status for the peptide containing residues 18-26 was observed after 6 hrs of incubation with C646. Interestingly, a small yet significant increase in acetylation status was detected for this peptide containing H3 K18 and H3 K23 (Fig. 4) upon treatment with 30 μM C646, while this increase was not significant at lower concentrations of C646. Since the increase was not observed after 20 hrs of incubation with C646, this indicates a time dependency for the effect of this inhibitor on acetylation status of this histone peptide. The HAT p300 has indeed been described to acetylate H3 K18 and K23 [29], however, upon HAT inhibition a decrease in acetylation is to be expected instead of the observed increase. Thus, in this case, the observed effects on global histone acetylation are not in line with HAT inhibition. Using the mass spectrometry method, a wide range of acetylation sites was addressed in a single analysis giving an overview of the effect of C646 on histone acetylation (Fig. 6).
Fig. 6.
Coverage of the histone H3 and H4 sequence by LC-MS/MS and the histone acetylation profile after treatment with C646. Detected histone peptides with their residue numbers are indicated in black and bold. Undetected histone peptides are indicated in grey. Compared to the control (untreated cells), increased acetylation was observed after C646 treatment at 30 μM for 6 hrs on one peptide from histone H3 (res. 18-26: KQLATKAAR) containing K18 and K23 (peptide indicated in italics and underlined). Other studied histone peptides (indicated in bold) did not show changes in acetylation status.
3.4. C646 inhibits recombinant HDACs
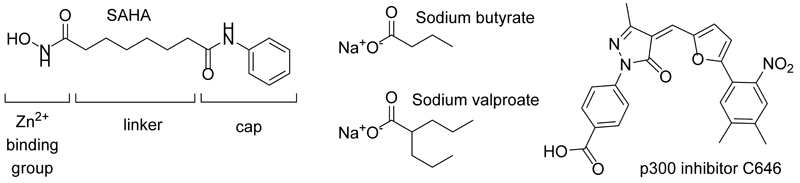
We set out to study the counterintuitive increase in histone acetylation upon C646 treatment in more detail. Based on the structural features of C646 (Fig. 6) we hypothesized that its carboxylate might enable inhibition of HDACs at relatively high concentrations due to coordination of Zn2+ in the active sites of HDACs, similar to HDACi such as sodium butyrate and sodium valproate [30].
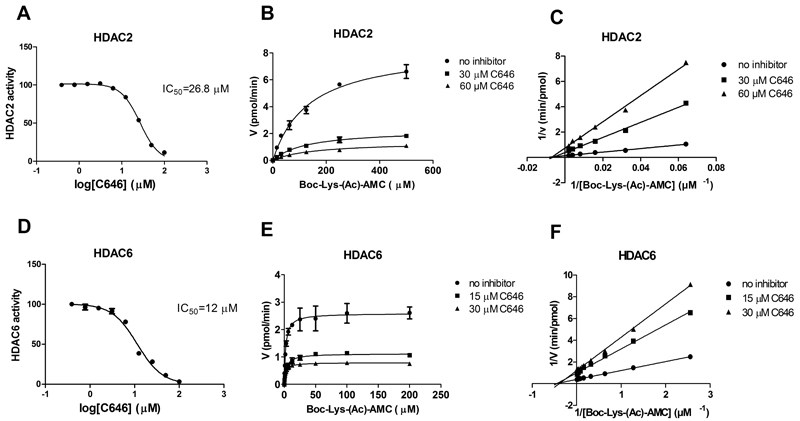
In order to test the hypothesis that the p300 HATi C646 also inhibits HDACs, its inhibitory potency on recombinant HDACs 1, 2, 3, 6 and 8 was measured. In line with our hypothesis C646 indeed inhibited these HDACs (Table 2), except for HDAC1, with potencies in the micromolar range. The observed selectivity profile for the investigated HDACs is interesting since the Ki values for HDAC6 show the highest potency, whereas inhibition of HDAC1 is absent. To confirm that C646 is binding Zn2+, we looked closer into the kinetics of inhibition for the class I member HDAC2 and the class II member HDAC6. Unexpectedly, non-competitive inhibition was observed, with Boc-Lys-(Ac)-AMC as a substrate (Fig. 8). This is not in line with our hypothesis, and indicates either covalent or allosteric binding of C646 to these HDACs. Next to this, we also verified the HAT p300 inhibitory potency of C646 as reported by Bowers et al., and found comparable results with an IC50 value of 0.32 μM. In addition, for the HAT KAT8, we found no inhibition by C646, which supports the previous findings that C646 is a p300 selective HATi (Table 2) [8].
Table 2.
Inhibitory effect of C646 on selected HATs or HDACs. Inhibition was determined by p300, KAT8, or HDAC inhibition assays as described in materials and methods. N.I. = no inhibition.
| Ki value µM | |||||||
|---|---|---|---|---|---|---|---|
| p300 | KAT8 | HDAC1 | HDAC2 | HDAC3 | HDAC6 | HDAC8 | |
| C646 | 0.32 ± 0.02b | N.I. | N.I. | 15 ± 1 | 25 ± 5 | 7.0 ± 1.0 | 11 ± 1 |
| SAHAa | - | - | 0.037 ± 0.005 | 0.09 ± 0.009 | 0.031 ± 0.009 | 0.034 ± 0.007 | 0.001 ± 0.0002 |
Ki values for SAHA were determined at our lab previously [20].
This is an IC50 value.
Fig. 8.
IC50 curves, as well as Michaelis-Menten and Lineweaver-Burk plots after incubation of HDAC2 with C646 (A) (B) and (C), and of HDAC6 with C646 (D), (E) and (F), respectively. For both HDACs, in the Lineweaver-Burk plots, the x-intercepts of the uninhibited and inhibited enzyme conditions appear to be the same, indicating non-competitive inhibition. The data shown represent 2-3 independent experiments (and are displayed ± SD in the Michaelis-Menten plots).
3.5. C646 decreases tubulin acetylation status in a time independent manner
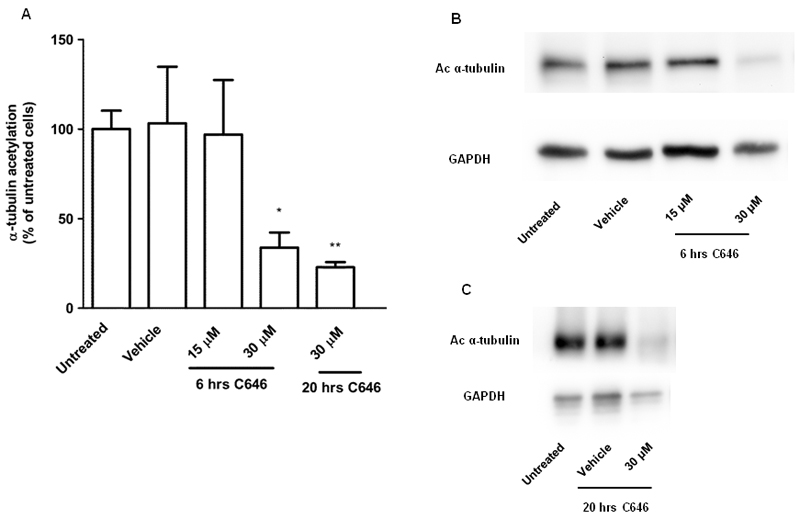
The highest potency for HDAC inhibition by C646 was observed for HDAC6. Therefore, we set out to investigate if HDAC6 inhibition observed for the recombinant enzyme is also reflected in inhibition of deacetylation of α-tubulin, which is a well-known cellular target of HDAC6 [31], in RAW264.7 cells. The α-tubulin acetylation status was monitored by immunoblotting using an anti-acetyl-α-tubulin antibody. After incubation with C646 for 6 hrs at 30 μM, but not at 15 μM, a pronounced inhibition of α-tubulin acetylation was observed (Fig. 9A and B), which is not in line with inhibition of HDAC6. The inhibition of α-tubulin acetylation was also observed after 20 hrs of incubation with C646 at 30 μM (Fig. 9A and C). Treatment with the reference compound SAHA upregulated α-tubulin acetylation (data not shown).
Fig. 9.
C646 reduces α-tubulin acetylation in RAW264.7 cells. Cells were incubated with C646 at 15 or 30 μM for 6 hrs, or at 30 μM for 20 hrs, lysed, and immunoblotted for acetylated α-tubulin or GAPDH (loading control) For vehicle treatment, cells were incubated with the inhibitor solvent DMF. (A) Densitometry results from 3-5 independent experiments. The values shown represent means ± SD. * p < 0.05 compared to vehicle treated cells. (B) and (C) Representative images of 3-5 independent experiments after 6 or 20 hrs of incubation with C646 at the indicated concentrations.
4. Discussion
The inhibitory effects of C646 on pro-inflammatory gene expression RAW264.7 macrophages and in murine precision-cut lung tissue slices upon receiving an LPS and IFNγ stimulus are promising for further development of this inhibitor in model systems of inflammatory lung diseases. The results suggest inhibition of NF-κB signaling, in line with the observed decrease in NF-κB promoter activity in a reporter gene assay upon C646 pre-treatment. This would support previous lines of research focusing on reducing NF-κB activity in inflammatory lung diseases [14] and indicate that this is a promising strategy. This is also in line with previous studies indicating inhibition of the NF-κB pathway upon C646 treatment in prostate cancer cells [10], and decreased expression of the NF-κB mediated pro-inflammatory gene COX-2 upon C646 treatment in rats [9].
Further investigation of the effects of this inhibitor on histone acetylation revealed no HAT inhibitory effects at the level of global histone acetylation after 20 hrs of incubation with C646. After 6 hrs of incubation with C646, however, a slight increase in histone H3 acetylation was observed at lysine residues 18 and/or 23. This indicates a time dependent effect of C646 on histone acetylation, which has not been reported before. Evaluation of the ratios between HAT and HDAC activities under the studied conditions may clarify this.
It should be noted that previous studies indicated that the effects of C646 on histone acetylation are cell type dependent. Previously, effects of C646 which were in line with HAT inhibition have been demonstrated in cells using immunoblot or ChIP assays [32][33]. In other studies, inhibiting effects of this compound on histone acetylation were only shown after pre-treatment with an HDACi. HDACi pre-treatments are done more often in studies with HATi to increase the detection window for inhibition of the acetyl transfer reaction. For example, upon pre-treatment with HDACi trichostatin A (TSA) C646 demonstrated a pronounced inhibition of histone acetylation [34][8].
In contrast, observations similar to ours that C646 increases histone acetylation have been made in a model system employing pancreatic cancer cells [28], also after a short incubation time with C646 (4 hrs). The authors attributed the observed increase in histone H3 acetylation on K18 and K23 to increased cooperative binding of p300 to its substrate acetyl CoA, facilitated by C646 when present at low concentrations. The authors suggest that C646 can be outcompeted by acetyl CoA under these conditions. Although this model may explain the increased histone acetylation in cells at low concentrations of C646, it would be less suited to explain the increase observed in our system in which we employed relatively high concentrations of C646.
The observation that the p300 HATi C646 increases histone acetylation may also be explained through another mechanism. It has recently been reported that the overall histone acetylation levels remain remarkably constant upon depletion of HATs [35]. In Drosophila cells, it was observed that histone acetylation levels were unaffected or in some cases even higher upon siRNA mediated knockdown of specific acetyltransferases. The authors suggest that compensatory modifications occur in the case of a knockdown of specific acetyltransferases in order to compensate structural perturbations, which may be brought about in different ways, including feedback responses to modulate gene expression or enzyme activity of other acetyltransferases or deacetylases. Similar processes could take place when p300 is inhibited by a small molecule inhibitor such as C646.
However, the slight increase in histone acetylation prompted us to pose an alternative hypothesis; direct inhibition of HDACs by C646. Upon investigation, we indeed found inhibition of HDAC2, 3, 6 and 8 by C646 at micromolar concentrations. These observations of HDAC inhibitory activity by C646 place biochemical studies in which C646 has been applied as a HATi in a new perspective. Although the inhibitory potency for HDACs reported here is about 10-fold lower compared to the HAT p300 inhibition, this observation might have important implications. In biochemical experiments, the applied C646 concentrations sometimes exceed the Ki for p300 by a factor of 10. In addition, local concentrations in cells may be even higher. At these concentrations, HDAC inhibition can start playing a role in the observed effects, thus mixing HAT and HDAC inhibitory effects on cellular substrates. Thus, the interpretation of effects connected to presumed inhibition of p300 in the investigated model systems can be hampered by inhibition of HDACs. This clearly calls for a further optimization of C646 with respect to its HDAC inhibitory potency. This is also important in the light of recent studies focusing on applications of this HATi in the treatment of cancer, where HDAC inhibition upon C646 treatment is not presumed [36][37]. Furthermore, the HDAC inhibitory potency of C646 could explain its ambiguous effects on histone acetylation in cells reported in literature.
Based on the inhibitory potency of C646 for HDAC6, we monitored α-tubulin acetylation; an HDAC6 substrate. C646 treatment provided pronounced inhibition of α-tubulin acetylation after both 6 and 20 hrs of incubation, which argues against HDAC6 inhibition by C646 in RAW264.7 cells. The observed effect of C646 on α-tubulin acetylation may originate from inhibition of the HAT p300, which could directly reduce α-tubulin acetylation. However, p300 has not been described to acetylate α-tubulin directly. Alternatively, the results can be explained by the following. p300 has been described to mediate acetylation of HDAC6, which renders this HDAC less active [38][39]. Therefore, inhibition of p300 by C646 could decrease HDAC6 acetylation, thereby increasing HDAC6 activity, and thereby resulting in less α-tubulin acetylation.
In conclusion this study indicates that C646 has potential for further development towards suppression of pro-inflammatory gene expression in inflammatory diseases such as COPD. However, instead of the expected inhibition of histone acetylation a slight increase was observed upon C646 treatment in RAW264.7 macrophages, which is corroborated by inhibition of HDAC activity in enzyme activity assays. It remains unclear if the suppression of pro-inflammatory gene expression is due to its HAT inhibitory activity, due to its HDAC inhibitory activity, or inhibition of other enzymes (or a combination of these factors). We anticipate that the balance between HAT and HDAC inhibition depends on the precise target lysine(s) for acetylation or deacetylation, the enzymes that regulate them, and the extent of inhibition of these enzymes by C646. Despite its potential to suppress pro-inflammatory gene transcription this study indicates that further optimization of this inhibitor with respect to its HDAC inhibitory potency, and to its HAT inhibitory selectivity over HDACs, is needed for further development.
Supplementary Material
Fig. 7.
HDACi contain distinct structural features as described for the marketed HDACi SAHA. They generally consist of a zinc binding group a linker that occupies a relatively narrow tunnel and a cap that binds to the solvent exposed surface of the enzyme. The carboxylate-based HDACi sodium butyrate and sodium valproate and the p300 HATi C646 contain carboxylate groups that could also coordinate to Zinc in the HDAC active site.
Acknowledgements
We acknowledge the EU-COST action TD0905 ‘epigenetics from bench to bedside’ for funding a short term scientific mission of TvdB to the group of AI. We acknowledge Prof. Dr. Reinoud Gosens (Department of Molecular Pharmacology, University of Groningen) for his support with ex vivo experiments regarding precision-cut lung slices. We acknowledge the European Union for funding this project by an ERC starting grant to FJD (309782). Further support was obtained from the Netherlands Organisation of Scientific Research (NWO) by a VIDI grant to FJD (723.012.005), by RF-2010-2318330 grant to AM, by IIT-Sapienza Project to AM, by FP7 Projects BLUEPRINT/282510 and A-PARADDISE/602080 to AM, by Sapienza Ateneo Award Project 2014 (D.R.) and PRIN 2012 (prot.2012CTAYSY) to DR.
Abbreviations
- HAT
histone acetyltransferase
- HATi
HAT inhibitor
- HDAC
histone deacetylase
- PCAF
P300/CBP-associated factor
- MOZ
monocytic leukemic zinc finger
- COX-2
cyclooxygenase-2
- NF-κB
nuclear factor kappa B
- COPD
chronic obstructive pulmonary disease
- FBS
fetal bovine serum
- DMEM
Dulbecco’s modified Eagle’s medium
- SEAP
secreted embryonic alkaline phosphatase
- HDACi
HDAC inhibitor
- SAHA
suberanilohydroxamic acid
- LDH
lactate dehydrogenase
- PCLS
precision-cut lung slices
- LPS
lipopolysaccharide
- IFNγ
interferon gamma
- TNF-α
tumor necrosis factor alpha
- iNOS
inducible nitric oxide synthase
- IL-1β
interleukin 1 beta
- IL-12b
interleukin 12 subunit beta
- IL-6
interleukin 6
- IL-8
interleukin 8
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- AMC
7-Amino-4-methylcoumarin
- KAT8
lysine acetyltransferase 8
- DTT
dithiothreitol
- PMSF
phenylmethylsulfonyl fluoride
- DMF
dimethylformamide
- SDS PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- LC
liquid chromatography
- MS/MS
tandem mass spectrometry
- TSA
Trichostatin A
- siRNA
small interfering RNA
References
- 1.Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol. 2014;15:536–50. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- 2.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–18. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 3.Marmorstein R. Structure and function of histone acetyltransferases. Cell Mol Life Sci. 2001;58:693–703. doi: 10.1007/PL00000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Dekker FJ, Haisma HJ. Histone acetyl transferases as emerging drug targets. Drug Discov Today. 2009;14:942–8. doi: 10.1016/j.drudis.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 6.Chun P. Histone deacetylase inhibitors in hematological malignancies and solid tumors. Arch Pharm Res. 2015;38:933–49. doi: 10.1007/s12272-015-0571-1. [DOI] [PubMed] [Google Scholar]
- 7.Dekker FJ, van den Bosch T, Martin NI. Small molecule inhibitors of histone acetyltransferases and deacetylases are potential drugs for inflammatory diseases. Drug Discov Today. 2014;19:654–60. doi: 10.1016/j.drudis.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 8.Bowers EM, Yan G, Mukherjee C, Orry A, Wang L, Holbert MA, et al. Virtual ligand screening of the p300/CBP histone acetyltransferase: identification of a selective small molecule inhibitor. Chem Biol. 2010;17:471–82. doi: 10.1016/j.chembiol.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu XY, Huang CS, Li Q, Chang RM, Song ZB, Zou WY, et al. p300 exerts an epigenetic role in chronic neuropathic pain through its acetyltransferase activity in rats following chronic constriction injury (CCI) Mol Pain. 2012;8:84. doi: 10.1186/1744-8069-8-84. 8069-8-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santer FR, Hoschele PP, Oh SJ, Erb HH, Bouchal J, Cavarretta IT, et al. Inhibition of the acetyltransferases p300 and CBP reveals a targetable function for p300 in the survival and invasion pathways of prostate cancer cell lines. Mol Cancer Ther. 2011;10:1644–55. doi: 10.1158/1535-7163.MCT-11-0182. [DOI] [PubMed] [Google Scholar]
- 11.Kiernan R, Bres V, Ng RW, Coudart MP, El Messaoudi S, Sardet C, et al. Post-activation turn-off of NF-kappa B-dependent transcription is regulated by acetylation of p65. J Biol Chem. 2003;278:2758–66. doi: 10.1074/jbc.M209572200. [DOI] [PubMed] [Google Scholar]
- 12.Buerki C, Rothgiesser KM, Valovka T, Owen HR, Rehrauer H, Fey M, et al. Functional relevance of novel p300-mediated lysine 314 and 315 acetylation of RelA/p65. Nucleic Acids Res. 2008;36:1665–80. doi: 10.1093/nar/gkn003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerritsen ME, Williams AJ, Neish AS, Moore S, Shi Y, Collins T. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc Natl Acad Sci U S A. 1997;94:2927–32. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwards MR, Bartlett NW, Clarke D, Birrell M, Belvisi M, Johnston SL. Targeting the NF-kappaB pathway in asthma and chronic obstructive pulmonary disease. Pharmacol Ther. 2009;121:1–13. doi: 10.1016/j.pharmthera.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vignola AM, Chiappara G, Siena L, Bruno A, Gagliardo R, Merendino AM, et al. Proliferation and activation of bronchial epithelial cells in corticosteroid-dependent asthma. J Allergy Clin Immunol. 2001;108:738–46. doi: 10.1067/mai.2001.119160. [DOI] [PubMed] [Google Scholar]
- 16.Gagliardo R, Chanez P, Mathieu M, Bruno A, Costanzo G, Gougat C, et al. Persistent activation of nuclear factor-kappaB signaling pathway in severe uncontrolled asthma. Am J Respir Crit Care Med. 2003;168:1190–8. doi: 10.1164/rccm.200205-479OC. [DOI] [PubMed] [Google Scholar]
- 17.Caramori G, Romagnoli M, Casolari P, Bellettato C, Casoni G, Boschetto P, et al. Nuclear localisation of p65 in sputum macrophages but not in sputum neutrophils during COPD exacerbations. Thorax. 2003;58:348–51. doi: 10.1136/thorax.58.4.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Di Stefano A, Caramori G, Oates T, Capelli A, Lusuardi M, Gnemmi I, et al. Increased expression of nuclear factor-kappaB in bronchial biopsies from smokers and patients with COPD. Eur Respir J. 2002;20:556–63. doi: 10.1183/09031936.02.00272002. [DOI] [PubMed] [Google Scholar]
- 19.Eleftheriadis N, Neochoritis CG, Leus NG, van der Wouden PE, Domling A, Dekker FJ. Rational Development of a Potent 15-Lipoxygenase-1 Inhibitor with in Vitro and ex Vivo Anti-inflammatory Properties. J Med Chem. 2015;58:7850–62. doi: 10.1021/acs.jmedchem.5b01121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szymanski W, Ourailidou ME, Velema WA, Dekker FJ, Feringa BL. Light-Controlled Histone Deacetylase (HDAC) Inhibitors: Towards Photopharmacological Chemotherapy. Chemistry. 2015;21:16517–24. doi: 10.1002/chem.201502809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wapenaar H, van der Wouden PE, Groves MR, Rotili D, Mai A, Dekker FJ. Enzyme kinetics and inhibition of histone acetyltransferase KAT8. Eur J Med Chem. 2015;105:289–96. doi: 10.1016/j.ejmech.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duan Q, Chen H, Costa M, Dai W. Phosphorylation of H3S10 blocks the access of H3K9 by specific antibodies and histone methyltransferase. Implication in regulating chromatin dynamics and epigenetic inheritance during mitosis. J Biol Chem. 2008;283:33585–90. doi: 10.1074/jbc.M803312200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morin JP, Baste JM, Gay A, Crochemore C, Corbiere C, Monteil C. Precision cut lung slices as an efficient tool for in vitro lung physio-pharmacotoxicology studies. Xenobiotica. 2013;43:63–72. doi: 10.3109/00498254.2012.727043. [DOI] [PubMed] [Google Scholar]
- 24.Villar-Garea A, Israel L, Imhof A. Analysis of Histone modifications by Mass spectrometry. Curr Protoc Protein Sci. 2008 doi: 10.1002/0471140864.ps1410s51. 14.10.1,14.10.14. [DOI] [PubMed] [Google Scholar]
- 25.Lin S, Garcia BA. Examining histone posttranslational modification patterns by high-resolution mass spectrometry. Methods Enzymol. 2012;512:3–28. doi: 10.1016/B978-0-12-391940-3.00001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hersman E, Nelson DM, Griffith WP, Jelinek C, Cotter RJ. Analysis of Histone Modifications from Tryptic Peptides of Deuteroacetylated Isoforms. Int J Mass Spectrom. 2012;312:5–16. doi: 10.1016/j.ijms.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drogaris P, Villeneuve V, Pomies C, Lee EH, Bourdeau V, Bonneil E, et al. Histone deacetylase inhibitors globally enhance h3/h4 tail acetylation without affecting h3 lysine 56 acetylation. Sci Rep. 2012;2:220. doi: 10.1038/srep00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henry RA, Kuo YM, Bhattacharjee V, Yen TJ, Andrews AJ. Changing the selectivity of p300 by acetyl-CoA modulation of histone acetylation. ACS Chem Biol. 2015;10:146–56. doi: 10.1021/cb500726b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henry RA, Kuo YM, Andrews AJ. Differences in specificity and selectivity between CBP and p300 acetylation of histone H3 and H3/H4. Biochemistry. 2013;52:5746–59. doi: 10.1021/bi400684q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zwergel C, Valente S, Jacob C, Mai A. Emerging approaches for histone deacetylase inhibitor drug discovery. Expert Opin Drug Discov. 2015;10:599–613. doi: 10.1517/17460441.2015.1038236. [DOI] [PubMed] [Google Scholar]
- 31.Perdiz D, Mackeh R, Pous C, Baillet A. The ins and outs of tubulin acetylation: more than just a post-translational modification? Cell Signal. 2011;23:763–71. doi: 10.1016/j.cellsig.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 32.Gao XN, Lin J, Ning QY, Gao L, Yao YS, Zhou JH, et al. A histone acetyltransferase p300 inhibitor C646 induces cell cycle arrest and apoptosis selectively in AML1-ETO-positive AML cells. PLoS One. 2013;8:e55481. doi: 10.1371/journal.pone.0055481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y, Liu K, Liang Y, Chen Y, Chen Y, Gong Y. Histone acetyltransferase inhibitor C646 reverses epithelial to mesenchymal transition of human peritoneal mesothelial cells via blocking TGF-beta1/Smad3 signaling pathway in vitro. Int J Clin Exp Pathol. 2015;8:2746–54. [PMC free article] [PubMed] [Google Scholar]
- 34.Tomioka T, Maruoka H, Kawa H, Yamazoe R, Fujiki D, Shimoke K, et al. The histone deacetylase inhibitor trichostatin A induces neurite outgrowth in PC12 cells via the epigenetically regulated expression of the nur77 gene. Neurosci Res. 2014;88:39–48. doi: 10.1016/j.neures.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 35.Feller C, Forne I, Imhof A, Becker PB. Global and specific responses of the histone acetylome to systematic perturbation. Mol Cell. 2015;57:559–71. doi: 10.1016/j.molcel.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 36.Gaddis M, Gerrard D, Frietze S, Farnham PJ. Altering cancer transcriptomes using epigenomic inhibitors. Epigenetics Chromatin. 2015;8:9. doi: 10.1186/1756-8935-8-9. 8935-8-9. eCollection 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu Y, Chen H, Lu J, Zhang M, Zhang R, Duan T, et al. Acetylation-dependent function of human single-stranded DNA binding protein 1. Nucleic Acids Res. 2015;43:7878–87. doi: 10.1093/nar/gkv707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han Y, Jeong HM, Jin YH, Kim YJ, Jeong HG, Yeo CY, et al. Acetylation of histone deacetylase 6 by p300 attenuates its deacetylase activity. Biochem Biophys Res Commun. 2009;383:88–92. doi: 10.1016/j.bbrc.2009.03.147. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y, Peng L, Seto E, Huang S, Qiu Y. Modulation of histone deacetylase 6 (HDAC6) nuclear import and tubulin deacetylase activity through acetylation. J Biol Chem. 2012;287:29168–74. doi: 10.1074/jbc.M112.371120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.