Abstract
Our current understanding of immunology was largely defined in laboratory mice because of experimental advantages including inbred homogeneity, tools for genetic manipulation, the ability to perform kinetic tissue analyses starting with the onset of disease, and tractable models. Comparably reductionist experiments are neither technically nor ethically possible in humans. Despite revealing many fundamental principals of immunology, there is growing concern that mice fail to capture relevant aspects of the human immune system, which may account for failures to translate disease treatments from bench to bedside1–8. Laboratory mice live in abnormally hygienic “specific pathogen free” (SPF) barrier facilities. Here we show that the standard practice of laboratory mouse husbandry has profound effects on the immune system and that environmental changes result in better recapitulation of features of adult humans. Laboratory mice lack effector-differentiated and mucosally distributed memory T cells, which more closely resembles neonatal than adult humans. These cell populations were present in free-living barn populations of feral mice, pet store mice with diverse microbial experience, and were induced in laboratory mice after co-housing with pet store mice, suggesting a role for environment. Consequences of altering mouse housing profoundly impacted the cellular composition of the innate and adaptive immune system and resulted in global changes in blood cell gene expression patterns that more closely aligned with immune signatures of adult humans rather than neonates, altered the mouse’s resistance to infection, and impacted T cell differentiation to a de novo viral infection. These data highlight the impact of environment on the basal immune state and response to infection and suggest that restoring physiological microbial exposure in laboratory mice could provide a relevant tool for modeling immunological events in free-living organisms, including humans.
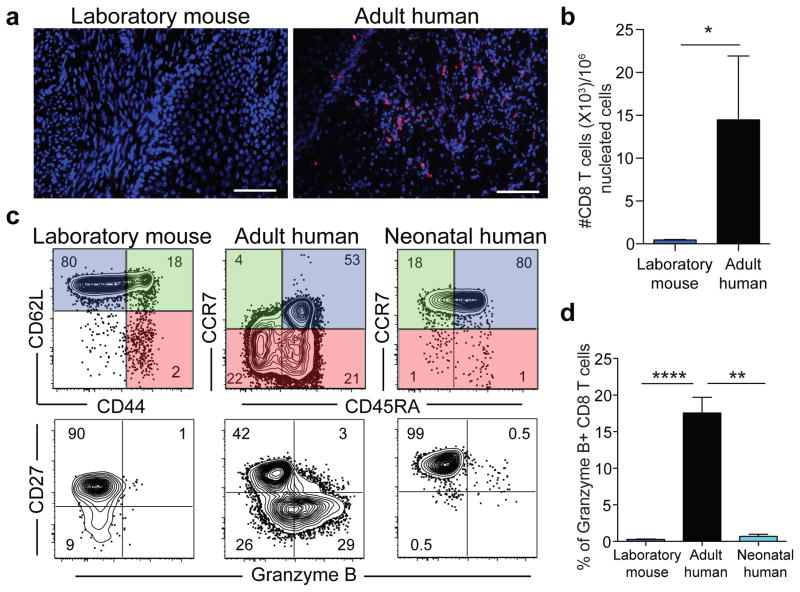
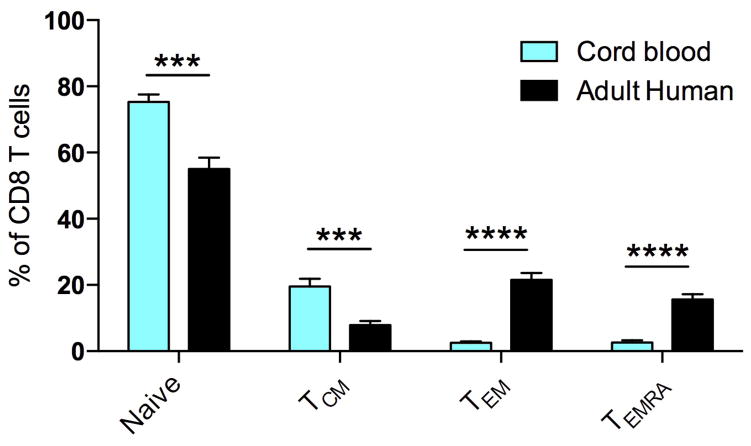
Given reported species-specific differences in immune responses1–8 we wished to compare memory CD8 T cell distribution and differentiation between mice and humans. CD8 T cells are critical for adaptive immune control of intracellular infections and cancer and their distribution and differentiation relate directly to their function. We assessed nonlymphoid distribution by examining available specimens of normal cervical tissue from premenopausal adult women. We found that CD8 T cells were integrated within the mucosa, exhibited a tissue resident memory T cell (TRM) phenotype9, and comprised ~15,000 of every million cells (Fig. 1a&b and data not shown). This agrees with previous reports that adult (unlike neonatal) human nonlymphoid tissues are abundantly populated with TRM10–12. In contrast, CD8 T cells were almost completely absent from cervical sections from adult inbred laboratory mice (C57BL/6 strain) housed under SPF conditions (Fig. 1a&b). We then compared major CD8 T cell lineages in blood, focusing on species-specific markers that define functionally homologous populations of naïve, central memory (TCM), and terminally differentiated effector memory CD8 T cells (TEM or TEMRA) (Fig. 1c). Relative to adult humans, memory CD8 T cells were much scarcer in laboratory mice, and almost entirely comprised of TCM rather than TEM or TEMRA. Also in contrast to humans, laboratory mice lacked CD27lo/Granzyme B+ effector differentiated memory CD8 T cells that are thought to respond most immediately to infection (Fig. 1c)13,14. Thus, memory CD8 T cells in laboratory mice were less frequent and strikingly different from those in adult humans, and in fact, appeared much more similar to neonates (Fig. 1c&d and extended data Fig. 1).
Figure 1. Laboratory mice lack differentiated memory CD8 T cell subsets, which is more similar to neonates than adult humans.
a,b) CD8 T cell density within adult laboratory mouse (n=5) and human cervix (n=3). Representative immunofluorescence staining of frozen sections (scale bars, 50 μm) is shown. CD8β (red), DAPI (nuclei, blue). c) CD8 T cell phenotype was compared between adult human blood (n=13), adult laboratory mouse blood (n=10), and human cord blood (n=8) among 2 independent experiments by fluorescent flow cytometry (representative plots shown). Top panels gated on CD8+/CD3+ cells. Bottom panels gated on antigen experienced CD8+/CD3+ cells, as defined by conventional lineage markers in each species (red and green quadrants). d) Enumeration of granzyme B+ CD8 T cell frequencies amongst antigen experienced subsets. Significance was determined using unpaired two-sided Mann-Whitney U-test (in b) or Kruskal-wallis (ANOVA) test (in d). * p<0.05, ** p<0.01, **** p<0.0001, bars indicate mean ± S.E.M.
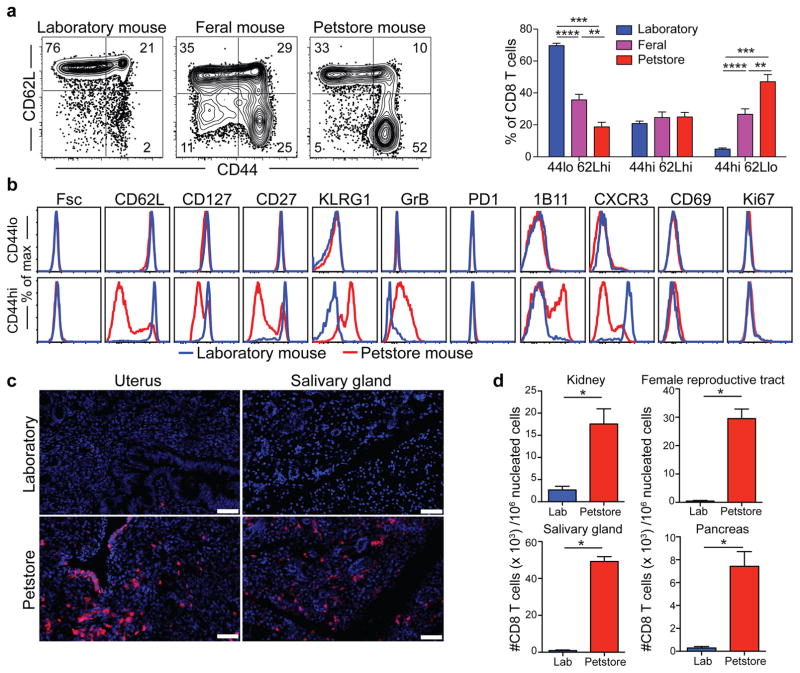
Mice are amongst the closest relatives of primates, but diverged from human ancestors 65–75 million years ago15. We wished to test whether the differences we observed between mice and adult humans were intrinsic to all mice (divergent immune system evolution), or unique to laboratory mice. To this end, we trapped free-living barn populations of feral mice and also procured mice from commercial pet stores. Compared to inbred SPF laboratory mice, feral and petstore mice were enriched in antigen-experienced CD8 T cells, particularly those that displayed a differentiated effector memory phenotype (Fig. 2a). More extensive phenotypic characterization of differentiation markers previously defined among CD8 T cells in laboratory mice revealed that naïve (CD44lo) cells were phenotypically identical between SPF and petstore mice (Fig. 2b). These data validated the consistency of markers used to discriminate between naïve and antigen experienced CD8 T cell populations in the different mouse cohorts. However, examination of antigen-experienced (CD44hi) CD8 T cells indicated enrichment of granzyme B+ and CD27lo cells in petstore mice, as well as other signatures of terminal effector differentiation including CD62Llo, CXCR3lo, CD127lo, and 1B11hi phenotypes. However, cell size (Fsc), proliferation (Ki67), and other measures of recent antigen exposure (CD69 and PD-1) were equivalent between both cohorts of mice, indicating that differences in CD8 T cell differentiation state were not due to an ongoing acute infection in petstore mice. Moreover, the relative density of CD8 T cells in petstore mouse nonlymphoid tissues was up to 50-fold greater than that observed in SPF laboratory mice (Fig. 2c,d).
Figure 2. CD8 T cell subsets vary between feral, petstore, and laboratory mice.
a) CD8 T cell subsets were compared in PBMC between laboratory mice (n=9), feral mice that were trapped in the wild (n=10), and mice obtained from a pet store (n=6), among 2 independent experiments. b) Phenotype of CD44lo/CD62Lhi (naïve) and CD44hi (antigen-experienced) CD8+ PBMC was compared by fluorescent flow cytometry. c,d) CD8 T cell density in non-lymphoid tissues was compared between laboratory and pet store mice by quantitative immunofluorescence microscopy (QIM). Immunofluorescence staining of frozen sections of indicated tissues (n=8 animals per group, scale bars = 50 μm). CD8β (red), DAPI (nuclei, blue). Significance was determined using unpaired two-sided Mann-Whitney U-test. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001, bars indicate mean ± S.E.M.
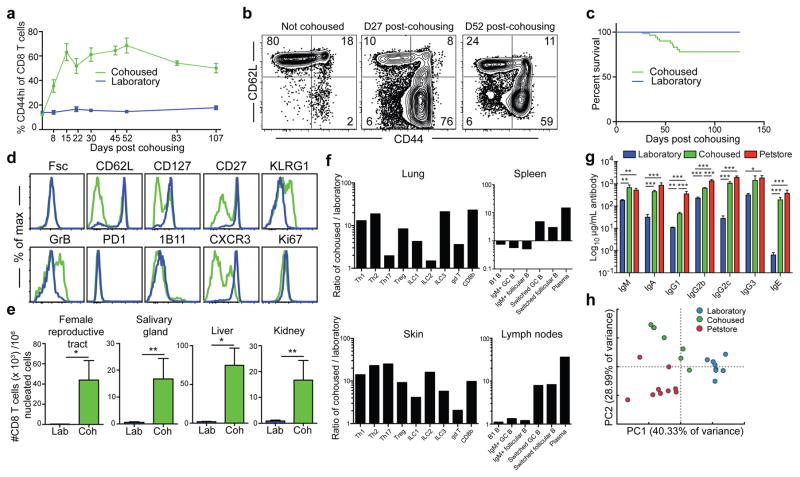
Petstore mice recapitulated aspects of human CD8 T cell differentiation and distribution that were absent in laboratory mice. These differences could be solely due to genetics, or dependent on environment. Inbred mice are genetically homogenous, which permits refined approaches and reductionist comparisons not possible in outbred populations, but may be accompanied by mutations that impair immune system development or function. Laboratory mice live in filtered microisolator housing that provides extreme hygiene16. Conditions are so clean, mice harboring fatal immunodeficiency diseases often thrive in these environments due to the absence of pathogen exposure. Thus, modern husbandry has evolved to the point in which laboratory mice, the standard model for biomedical research, exhibit far less infectious experience than free-living (i.e., dirty) humans or mice naturally acquire17–20. To test whether environmental conditions might impact the differentiation state and distribution of CD8 T cells, we co-housed inbred laboratory mice with petstore mice, which were not raised in ultra hygienic barrier facilities. Adult petstore mice were introduced into cages containing adult laboratory (C57BL/6 strain) inbred mice. Within four weeks of co-housing, the CD44hi cells increased from 15% to ~70% of CD8+ peripheral blood mononuclear cells (PBMC), and plateaued at ~50% after eight weeks for the duration of the study (Fig. 3a,b). 22% of laboratory mice died during the first eight weeks of co-housing, after which no further mortality was observed (Fig. 3c).
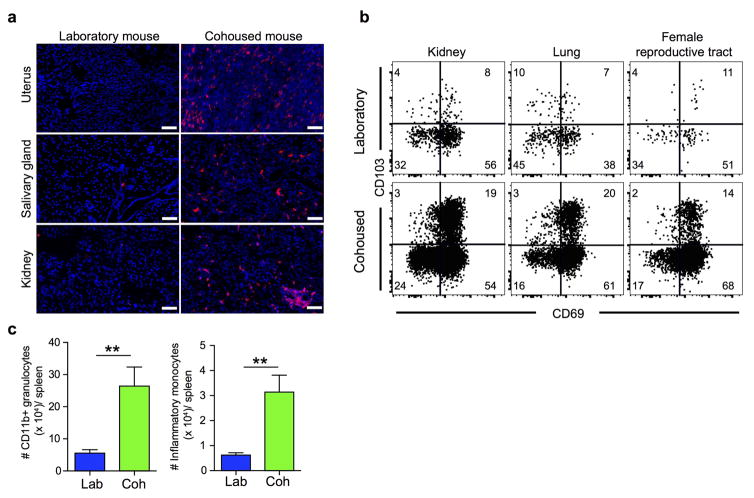
Figure 3. Co-housing with petstore mice changes the immune system of laboratory mice.
a) Proportion of CD44hi (antigen-experienced) CD8+ PBMC in laboratory mice housed in either SPF conditions (blue, n=10 for each time point) or co-housed with pet store mice (green, n=9 for each time point) among 2 independent experiments. b) CD8 T cell phenotype in blood of laboratory mice after co-housing with pet store mice. Representative flow plots (n=15) are shown. c) Survival of laboratory mice after co-housing with pet store mice (n=65 mice sampled). d) Representative phenotypes of CD44hi CD8+ PBMC isolated from laboratory mice co-housed with pet store mice for 100 days (n=15) and non-cohoused age-matched controls (n=10) was compared by fluorescent flow cytometry. e) Enumeration of CD8 T cells in non-lymphoid tissues between laboratory and cohoused mice by QIM (n=8 animals per group). f) Ratio of indicated cell types isolated and enumerated by flow cytometry from cohoused (n=6) and laboratory mice (n=6) among 2 independent experiments. ILC = innate lymphoid cells, Th = CD4 T helper cells, Treg = Regulatory CD4 T cells, GC = Germinal center. Single positive staining for T-bet, Gata3, and Rorγt were used to determine Th1, Th2, and Th17 lineages, respectively. g) Serum antibody concentrations (n=7 per group). h) Principal component analysis of gene expression data from PBMC of laboratory (n=8), cohoused (n=7) and pet store mice (n=8). Significance was determined using unpaired two-sided Mann-Whitney U-test (in e) or one-way ANOVA with Bonferroni post-hoc analysis (in g). *p<0.05, ** p<0.01, ***p<0.001. Bars indicate mean ± S.E.M.
Serology was performed for common mouse pathogens, and revealed exposure to viral, bacterial, and helminth pathogens (although not murine cytomegalovirus, see extended data table 1). Co-housing resulted in a constitutive increase in highly differentiated effector memory cells that matched signatures in outbred mice and humans, including the accrual of granzyme B+ and CD27lo cells (Fig. 3d). Moreover, nonlymphoid tissues became populated by CD8 T cells expressing a TRM phenotype (Fig. 3e and extended data Fig. 2). Expanding the cellular analysis beyond CD8 T cells revealed extensive and profound changes to many innate and adaptive immune cell lineages throughout the host and increased levels of serum antibody (Fig. 3f&g and extended data Fig. 2). Furthermore, Principal Components Analysis of PBMC gene expression data in the space of all detected genes (~18000) revealed a spatial shift of cohoused samples away from laboratory samples and towards pet store along PC1 (Fig. 3h). Together, these data demonstrate that co-housing profoundly impacted the status of the immune system.
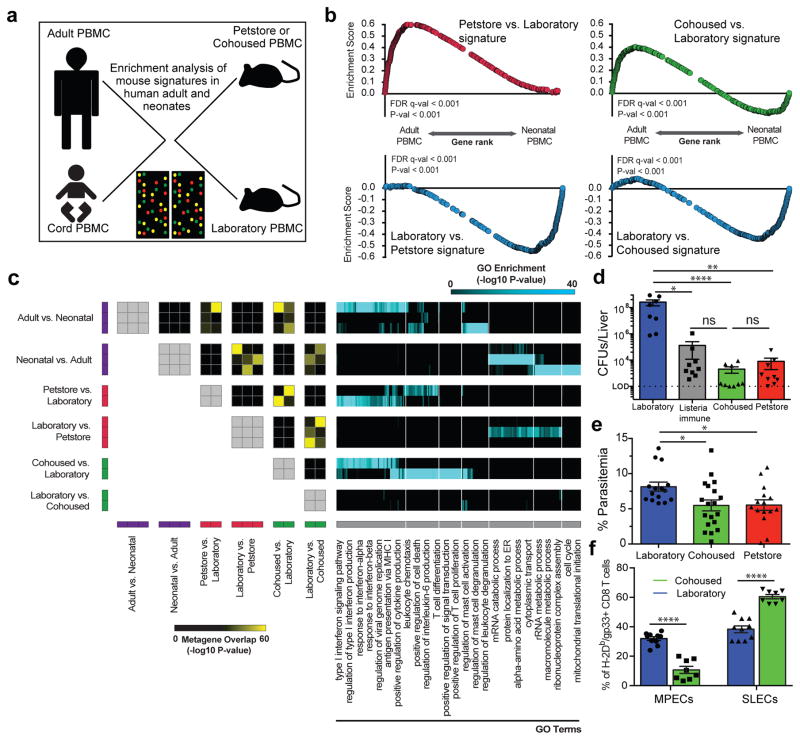
Our data suggested that laboratory mice have features in common with neonatal humans, and that altering the environment reproduced phenotypic signatures of adult humans. To test this hypothesis more broadly, we queried expression-profiling data of maternal and neonatal cord PBMC from an unaffiliated study21 with petstore vs. laboratory and cohoused vs. laboratory signatures via gene set enrichment analysis (GSEA) (Fig. 4a and 4b). The top 400 genes up-regulated in petstore mice showed highly significant enrichment in human adult expression data, while the top 400 down-regulated genes showed enrichment in neonates. Laboratory mice acquired this gene expression program after co-housing.
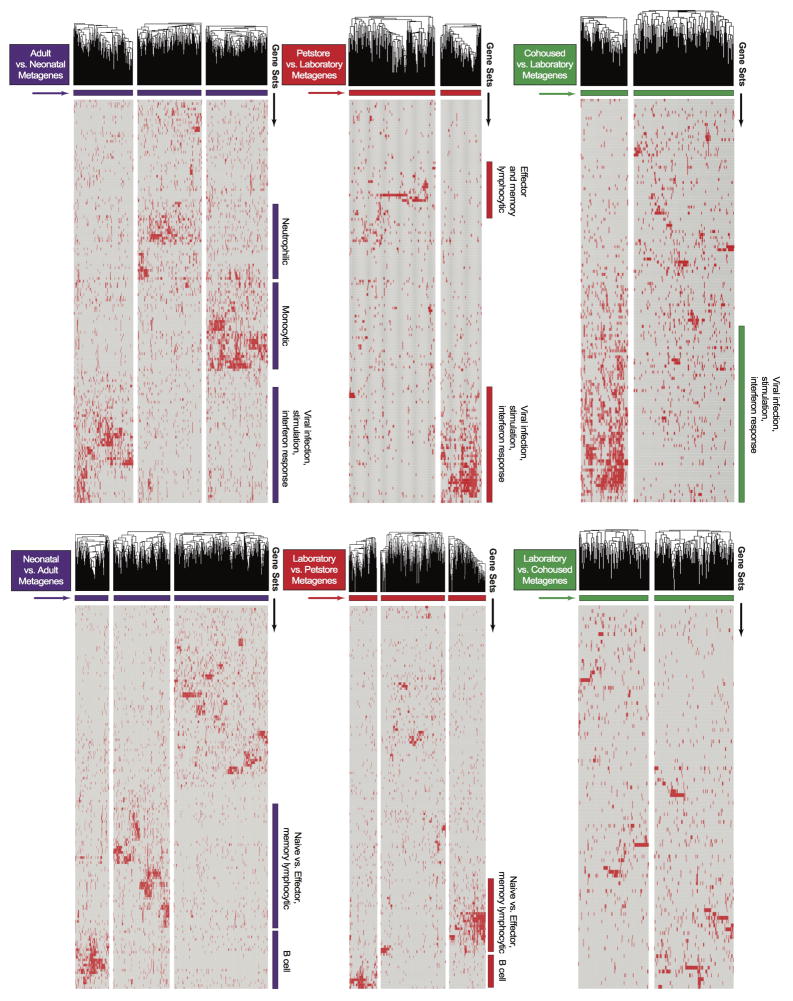
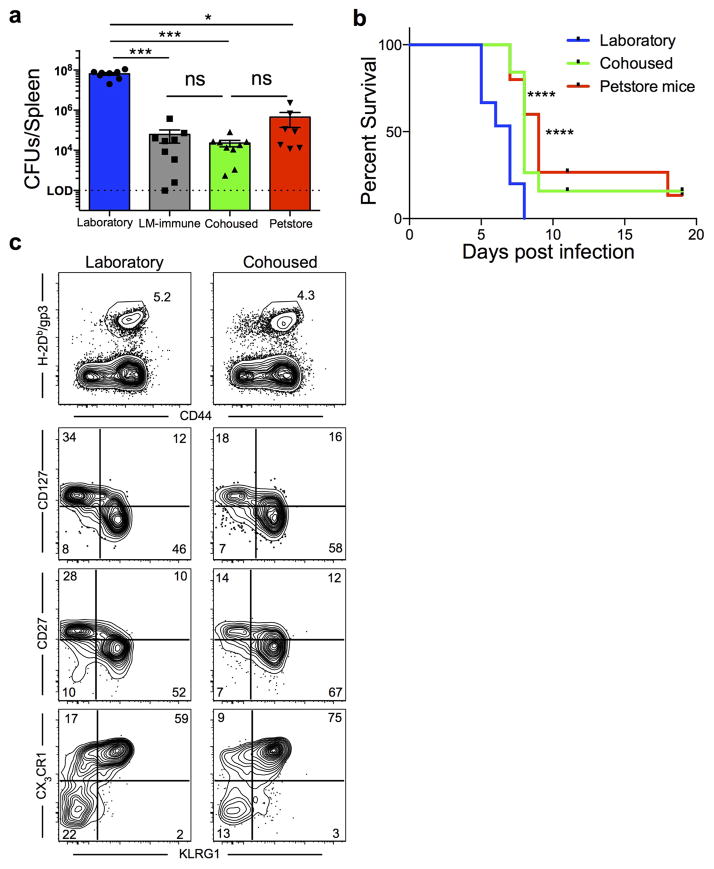
Figure 4. Microbial experience matures mouse immune transcriptome from neonatal to adult human and impacts immune system function.
a) Experimental design and b) GSEA plots showing the enrichment of gene signatures among the indicated mouse group comparisons relative to human adult vs. neonatal comparison. Signatures consist of top 400 significantly differentially expressed genes of laboratory (n=8), cohoused (n=7) and petstore mice (n=8). c) Pairwise overlaps of metagenes identified through leading-edge metagene analysis and corresponding GO terms. d) Bacterial load in the liver 3 days post-challenge with 8.5 × 104 CFU of Listeria monocytogenes (LM) among indicated groups (n=9 for all except laboratory mice where n=8). e) Parasitic load in peripheral blood 5 days post Plasmodium berghei ANKA parasitized RBC challenge among the indicated groups of laboratory (n=15), cohoused (n=19), and petstore mice (n=15). f) 28 days after LCMV Armstrong infection, the proportion of H-2Db/gp33-specific CD8 T cell MPECs (KLRG1-, CD127+) and SLECs (KLRG1+, CD127−) in PBMC of cohoused (n=8) and laboratory (n=9). Data are cumulative of 2 independent experiments. d–f) Data points represent individual mice. Significance was determined using Kruskal-wallis (ANOVA) test (in d), one-way ANOVA test (in e) and unpaired two-sided t-test (in f) * p<0.05, ** p<0.01, **** p<0.0001. Bars indicate mean ± S.E.M.
To more deeply interrogate transcriptional concordances between petstore mice, cohoused mice, and adult humans relative to laboratory mice and neonatal humans, we applied GSEA with the ImmuneSigDB database of immunological signatures22. We then used leading edge metagene analysis of GSEA results to identify modules of coregulated genes upregulated in human adult vs. cord PBMC, and those found in pairwise comparisons of laboratory, petstore, and co-housed mice. Overlap between the resulting metagenes was used to identify global similarities between each dataset. We observed highly significant overlaps between metagenes upregulated in adult vs. cord human blood and metagenes upregulated in petstore or co-housed vs. laboratory mouse blood. These included numerous pathways related to innate and adaptive immune functions (Fig. 4c, extended data Fig. 3 and supplemental data Tables 1&2). Conversely, metagenes upregulated in human cord blood overlapped with those upregulated in laboratory mice. Thus, these functional modules represent a major axis of similarity in immune status between petstore mice and adult humans relative to laboratory mice and neonatal humans, and can be conferred to laboratory mice via cohousing with petstore mice.
We next questioned whether mouse husbandry affected immune responses. We performed challenges with the intracellular pathogen Listeria monocytogenes (LM), which is a bacterial infection often used to gauge immune function in laboratory mice. Compared to laboratory mice, both petstore and co-housed mice exhibited a >10,000 fold reduction in bacterial burden three days after challenge, which actually matched bacterial control in laboratory mice that were previously vaccinated against LM (Fig. 4d and extended data Fig. 4a). Hence, physiological exposure to environmental microbes revealed that C57BL/6 mice exhibit considerably more innate resistance to LM infection than indicated from studies using SPF animals. The impact of mouse husbandry on infection control extended to a cerebral malaria model (Plasmodium berghei ANKA, Fig. 4e, extended data Fig. 4b). Lymphocytic choriomeningitis virus infection is an oft-used model to examine critical questions of adaptive T cell differentiation, including the regulation of memory precursor vs. short lived effector cell (MPEC vs. SLEC) development23,24. In this case, we observed that the proportion of MPECs and SLECs were significantly impacted by mouse husbandry (Fig. 4f and extended data Fig. 4c).
Experiments in mice have informed much of our understanding of immune regulation, and have directly informed the development of life-saving clinical therapies. However, our study reveals an unanticipated impact of SPF husbandry on the immune system. Our study does not advocate for an end to SPF studies. But for studies of the immune system it is ironic that such an immunologically inexperienced organism has become de rigueur, as our data show that this compromises development of an adult-like human immune system. For maximizing opportunities for bench to bedside translational medicine, it may be opportune to add dirty mice to our repertoire of investigative tools. Much like the analysis of truly sterile “germ-free” mice has revealed how influential the commensal flora is on “normal” physiology and immune system function, our study suggests that the immune system in mice may not be fully “normalized” without more complete microbial exposure10. Indeed, just as many autoimmune diseases do not manifest in genetically predisposed mice in the absence of commensal flora, certain infectious experiences have been shown to induce heterologous and innate immune memory, trigger autoimmune disease, and affect transplantation tolerance 25–30. Forward genetic screens are ongoing in mice to reveal the function of immunological genes, and it might be beneficial to conduct these screens in a “dirty” mouse model.
More generally, dirty mice might be valuable for interrogating aspects of the hygiene hypothesis, immune function and treatment from disease in the settings of transplantation, allergy, autoimmunity, and vaccination, and perhaps in disparate diseases that may have an immune system or inflammatory component (such as cardiovascular disease and cancer)4. Such mice could supplement current models to either increase translation potential for human disease or to better inform efficacy of preclinical prophylactic and therapeutic modalities, without sacrificing powerful experimental tools and approaches that are unavailable to human studies.
Material and Methods
Mice, Cohousing and Infections
Petstore mice were purchased from a nation-wide retailer. Feral mice were trapped on a horse farm or rural outdoor petting zoo in Minnesota or Georgia, USA. Male or female petstore mice were introduced into the cages of six to eight week old C57BL/6 mice of the same sex purchased from the National Cancer Institute. Co-housing occurred within a BSL-3 facility. Age-matched C57BL/6 laboratory mice maintained in SPF facilities served as controls. Number of animals was determined based on previous experience in order to reach statistical significance. All animals surviving the experimental treatment were included in the final analysis. No method of randomization was used to allocate animals to experimental groups. Investigators were not blinded to the group allocation during experiments. Listeria monocytogenes (LM) was grown in tryptic soy broth containing streptomycin to log phase growth. The indicated groups of mice were infected i.v. with 8.5 × 104 CFU of wild type LM (provided by John Harty, University of Iowa). Bacterial load in the spleen and liver was determined 3 days post-challenge as previously described31,32. LM immune mice were generated by primary infection with LM-OVA, (provided by Hao Shen, University of Pennsylvania School of Medicine)33 5 months prior to secondary challenge. Plasmodium berghei ANKA (provided by Susan K. Pierce, NIH/NIAID) was propagated by passage in mice and blood collection. A million parasitized RBCs were injected i.p. into the indicated mice. Parasitemia was determined by flow cytometry on peripheral blood34. All mice were used in accordance with the Institutional Animal Care and Use Committees guidelines at the University of Minnesota.
Human specimens
Adult PBMC samples were collected from healthy volunteers at the University of Minnesota35. Fresh cord blood samples were acquired from the Clinical Cell Therapy Laboratory at the University of Minnesota Medical Center. The PBMC isolation has been described in detail elsewhere36. After isolation, cells were frozen in 107cells/ml aliquots in a cryopreservative solution (Sigma-Aldrich) for future phenotyping. Premenopausal, human cervical tissue was obtained from the Tissue Procurement Facility (BioNet, University of Minnesota). Cervical samples were frozen roughly 1–2 hours after surgical resection. Informed consent was obtained from all subjects. The University of Minnesota Institutional Review Board approved all protocols used.
Intravascular staining, leukocyte isolation and phenotyping
An intravascular staining method was used to discriminate between cells present in the vasculature from cells in the tissue parenchyma37. Briefly, animals were injected i.v. with biotin/fluorochrome-conjugated anti-CD45 through the tail vein. Three minutes post-injection, animals were sacrificed, and tissues were harvested as described before38. Isolated mouse cells were surface-stained with antibodies against CD3 (145-2C11), CD45 (30F-11), CD11b (M1/70), CD11c (N418), NKp46 (29A1.4), Ly6G (1A8), MHC II (Ia-Ie) (M5/114.15.2), CD8α (53-6.7), CD45.2 (104), CD4 (RM4-5), CD62L (MEL-14), CD44 (IM7), CD69 (H1.2F3), CD103 (M290), Ly6C (AL21), CD43 (1B11), CD43 (S7), CD27 (LG.3A10), PD-1 (RMP1-30 and J43), KLRG1 (2F1), CXCR3 (CXCR3-173), CD127 (SB/199), α4β7 (DATK32), F4/80 (Cl-A3-1), CXCR5 (2G8), CD38 (90), IgM (RMM-1), IgD (11-26c.2a), GL7 (GL7), CD19 (6D5), B220 (RA3-6B2). Isolated human cells were surface-stained with antibodies against CD8α (3B5), CD45RA (HI100), CCR7 (G043H7), CD27 (O323), and CD3 (SK7). All of the above antibodies were purchased from BD Biosciences, Biolegend or Affymetrix eBiosciences. Cell viability was determined using Ghost Dye 780 (Tonbo Biosciences). Phycoerythrin (PE)-conjugated granzyme B (Invitrogen), Fluorescein isothiocyanate (FITC)-conjugated Ki67 (Invitrogen) and AF488 conjugated goat anti-mouse IgG (H+L) antibody intracellular staining was performed using the Cytofix/Cytoperm kit (BD Pharmingen) following manufacturer’s instructions. Intracellular staining for transcription factors was performed using Transcription factor staining buffer set (Affymetrix eBiosciences) with antibodies against Foxp3 (FJK-16s), T-bet (4B10), Eomes (Dan11mag), Gata3 (L50-823), Rorγt (Q31-378) following manufacturer’s guidelines. Single positive staining for T-bet, Gata3, and Rorγt were used to determine Th1, Th2, and Th17 lineages, respectively. FITC-conjugated mouse lineage cocktail (Tonbo Biosciences) was used in combination with other recommended lineage markers to identify various innate lymphoid cell subsets. The stained samples were acquired using LSRII or LSR Fortessa flow cytometers (BD) and analyzed with FlowJo software (Tree Star, Inc.).
Infectious agent screening
Laboratory mice, cohoused laboratory mice (after at least 30 days of cohousing) and petstore mice were screened using EZ-spot and PCR Rodent Infectious Agent (PRIA) array methods (Charles River Laboratories). Dried whole blood, feces, oral swab and body swabs were collected as per the sample submission guidelines of Charles River Laboratories.
Tissue freezing, immunofluorescence and microscopy
Harvested murine tissues were fixed in 2% Paraformaldehyde for 2hrs before being treated with 30% sucrose overnight for cryoprotection. The sucrose treated tissue is embedded in tissue freezing medium OCT and frozen in an isopentane liquid bath. Human cervix specimens were embedded in tissue freezing medium OCT and snap frozen in an isopentane liquid bath. Frozen blocks were processed, stained, imaged, and enumerated by quantitative immunofluorescence microscopy as described39,40 and included staining with the following antibodies: anti-mouse CD8β (YTS156.7.7), anti-human CD8β (SIDI8BEE, eBioscience), anti-mouse CD4 (RM4-5), anti-mouse CD11b (M1/70), and counterstaining with DAPI (mouse) or Cytox Green (human) to detect nuclei.
Serum antibody quantification
Mouse serum antibody titers were quantified using Ready-Set-Go! ELISA kits (Affymetrix eBioscience) following manufacturer’s instructions.
RNA isolation and Microarray hybridization
For each sample, 1–3×106 PMBCs were used for each RNA extraction. Cells were first homogenized using QIAshredder columns (Qiagen) then RNA was then extracted using RNeasy kit (Qiagen) as per manufacturer’s instructions. Following quality control, total RNA samples were processed using the Illumina TotalPrep-96 RNA Amplification Kit for High-Throughput RNA Amplification for Array Analysis. Samples were loaded onto the MouseRef-8 v2.0 Expression BeadChip (Illumina) and hybridized Beadchips were scanned using the Illumina iScan Beadarray Reader. Basic quality metrics were checked using Illumina Genomestudio.
Bioinformatics analysis
Prior to analysis, mouse microarray data were quantile normalized using preprocessCore (Bioconductor) and batch correction was performed using the ComBat algorithm. Principal components analysis was performed in R. Raw human adult and neonatal cord PBMC microarray data were obtained from a previous unaffiliated study profiling the peripheral blood of 72 smoking or non-smoking women and the cord blood of their neonates (Gene Expression Omnibus, accession code GSE27272)21. Human microarray data were quantile normalized as previously described. To obtain lists of genes upregulated or downregulated between Petstore, Cohoused, and Laboratory cohorts, differential expression analysis was performed using the linear modeling and empirical Bayesian method implemented in limma (Bioconductor). Gene set enrichment analysis (GSEA) was performed as described previously41. LEM (Leading Edge Metagene) analysis was performed downstream of GSEA to yield groups of genes, termed metagenes, which are coordinately upregulated in a given phenotypic comparison and common to multiple enriched gene sets. Briefly, for a given phenotypic comparison, gene set enrichment analysis was performed using ImmuneSigDB, a curated compendium of 4872 gene sets describing a wide range of cell states and experimental perturbations from immunology literature22. The top 150 significantly enriched gene sets, as restricted by an FDR < 0.25 and ranked by P < 0.05, were subsetted for their leading edge genes. These genes were then clustered into metagenes using non-negative matrix factorization. The significance of overlap between pairs of metagenes was determined using a Fisher exact test (P < 1e-05). Metagenes were functionally annotated based on the significance of overlap between member genes and Gene Ontology terms 41, as measured by hypergeometric test using the GOrilla enrichment analysis tool42.
Statistics
Data were subjected to D’Agostino & Pearson omnibus normality test to determine if they are sampled from a Gaussian distribution. If a Gaussian model of sampling was satisfied, parametric tests (unpaired two-tailed Student’s t-test for two groups and one-way analysis of variance (ANOVA) with Bonferroni multiple comparison test for more than two groups) were used. If the samples deviated from Gaussian distribution, non-parametric tests (Mann-Whitney U test for two groups, Kruskal-Wallis with Dunn’s multiple comparison test for more than two groups) were used unless otherwise stated. Variances between groups were compared using F test and found to be equal. All statistical analysis was done in GraphPad Prism (GraphPad Software Inc.). P-values <0.05 are considered significant and are indicated in figure legends.
Extended Data
Extended data figure 1.
Frequency of CD8 T cell subsets in neonates vs. adult humans. CD8 T cell subsets were defined in adult PBMC (n=13) and cord blood PBMC (n=8) by fluorescence flow cytometry based on the following markers: Naïve=CD45RAhi CCR7hi, TCM=CD45RAlo CCR7hi, TEM=CD45RAlo CCR7lo and TEMRA=CD45RAhi CCR7lo. Significance was determined using unpaired two-sided t-test. *** p<0.001, **** p<0.0001. Bars indicate mean ± S.E.M.
Extended data figure 2. Co-housing laboratory mice with petstore mice induces accumulation of TRM-phenotype CD8 T cells and other innate cells in tissues of laboratory mice.
a) CD8 T cell density within the indicated tissues of adult laboratory mice (n=5) and cohoused mice (n=7). Representative immunofluorescence staining, CD8β (red), DAPI (nuclei, blue), scale bars = 50 μm. b) Phenotype of CD8 T cells was compared between laboratory mice (n=9) and age-matched laboratory mice that were co-housed (n=9, representative flow cytometry plots shown). Samples gated on CD44hi cells isolated from the indicated tissue (vasculature populations were excluded, see methods). c) Enumeration of CD11b+ granulocytes and Ly6Chi inflammatory monocytes in spleens of laboratory (n=6) and cohoused (n=6) mouse. Significance was determined using unpaired two-sided Mann-Whitney U-test. ** p<0.01, bars indicate mean ± S.E.M.
Extended data figure 3.
LEM metagene analysis. For each comparison, standard GSEA was performed using the ImmSigDB database of gene-sets. Genes in the top 150 enriched sets (FDR<0.001, ranked by P-value) were filtered to only leading edge genes and subsequently clustered into groups (metagenes) using an NMF algorithm. Hierarchical clustering of genes within individual metagenes was performed to obtain the final heatmap. Metagenes with qualitatively discernible ‘blocks’ of gene-set membership were annotated according to the identity of corresponding enriched gene-sets. Heatmaps for Adult vs. Neonatal, Petstore vs. Laboratory, Cohoused vs. Laboratory, Neonatal vs. Adult, Laboratory vs. Petstore, and Laboratory vs. Cohoused comparisons are shown. Individual genes within each metagene are listed in Supplemental table 1. Pairwise overlaps between metagenes from different comparisons are visualized in Figure 4c.
Extended data figure 4.
Environment altered antimicrobial resistance and CD8 T cell differentiation. Laboratory mice were co-housed with petstore mice as described in figure 3. a) Bacterial load in the spleen 3 days post-challenge with 8.5 × 104 CFU of Listeria monocytogenes (LM) among laboratory (n=8), LM-immune (n=9), cohoused (n=9) and petstore mice (n=9) among 2 independent experiments. b) Survival of laboratory mice (n=15), cohoused mice (n=19) and petstore mice (n=15) after challenge with 106 Plasmodium berghei ANKA parasitized RBCs among 2 independent experiments. c) Laboratory (n=9) and cohoused (n=8) mice were infected with LCMV. Four weeks later, LCMV-specific CD8 T cells (identified with H-2Db/gp33 MHC I tetramers) were evaluated for expression of the indicated markers. Top row, gated on live CD8α+ T cells. Bottom 3 rows, gated on live CD8α+ H-2Db-gp33+ T cells. Significance was determined using Kruskal-wallis (ANOVA) test (in a) and log-rank (Mantel-Cox) test (in b). * p<0.05, *** p<0.001, **** p<0.0001. Bars indicate mean ± S.E.M.
Extended data table 1.
Microbial exposure in laboratory, petstore and cohoused mice.Frequency of each indicated microbial exposure within the population was evaluated by serological analysis and/or PCR among laboratory (n=4), petstore (n=15) and cohoused (n=13) mice. Each cohoused sample was collected from a different cage. Pinworm and mite specific-PCR was performed on pooled petstore and cohoused samples.
| Petstore | Laboratory | Cohoused | |
|---|---|---|---|
|
|
|||
| Viruses | |||
|
|
|||
| Rotavirus (EDIM) | 0 | 0 | 0 |
| Mouse Hepatitis Virus | 93.3 | 0 | 61.5 |
| Murine norovirus | 60 | 0 | 38.5 |
| Mouse parvovirus NS1 | 53.3 | 0 | 0 |
| Mouse parvovirus type 1 | 40 | 0 | 7.7 |
| Mouse parvovirus type 2 | 46.7 | 0 | 0 |
| Minute virus of mice | 46.7 | 0 | 0 |
| Theiler’s murine encephalomyelitis virus | 60 | 0 | 38.5 |
| Sendai virus | 66.7 | 0 | 23.1 |
| Ectromelia virus | 0 | 0 | 0 |
| Lymphocytic Choriomeningitis virus | 6.7 | 0 | 7.7 |
| Mouse adenovirus 1 and 2 | 0 | 0 | 0 |
| Mouse cytomegalo virus | 0 | 0 | 0 |
| Polyoma virus | 6.7 | 0 | 0 |
| Pneumonia virus of mouse | 53.3 | 0 | 0 |
| Reovirus | 0 | 0 | 0 |
|
|
|||
| Bacteria | |||
|
|
|||
| Cillia-Associated Respiratory Bacillus | 0 | 0 | 0 |
| Mycoplasma pulmonis | 73.3 | 0 | 30.8 |
| Clostridium piliforme | 26.7 | 0 | 0 |
|
|
|||
| Parasites/Protozoa/Fungi | |||
|
|
|||
| Enchephalitozoon cuniculi | 40 | 0 | 0 |
| Pinworm | 100 | 0 | 100 |
| Mites | 100 | 0 | 100 |
Supplementary Material
Acknowledgments
This study was supported by National Institutes of Health grants 1R01AI111671, R01AI084913 (to D.M.), R01AI116678, R01AI075168 (to S.C.J.) and a BSL-3 suite rental waiver grant from the University of Minnesota. We thank Rafi Ahmed (Emory University) for providing reagents for pilot studies, David McKenna (University of Minnesota) for cord blood, and all members of the BSL-3 mouse team (University of Minnesota).
Footnotes
Author contributions
L.K.B., S.E.H, J.M.S., O.A.O., K.A.C., E.A.T., K.A.F, P.C.R, V.V., and D.M. performed and analyzed the experiments. K.B. and W.N.H. analyzed the transcriptome data. M.K.J., A.F., and R.P.S. provided input on research design. L.K.B., S.E.H., W.N.H., S.C.J., and D.M. wrote the manuscript.
The authors declare no competing financial interests
References
- 1.Mestas J, Hughes CCW. Of mice and not men: differences between mouse and human immunology. J Immunol Baltim Md 1950. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 2.Seok J, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2013;110:3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shay T, et al. Conservation and divergence in the transcriptional programs of the human and mouse immune systems. Proc Natl Acad Sci U S A. 2013;110:2946–2951. doi: 10.1073/pnas.1222738110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mak IW, Evaniew N, Ghert M. Lost in translation: animal models and clinical trials in cancer treatment. Am J Transl Res. 2014;6:114–118. [PMC free article] [PubMed] [Google Scholar]
- 5.Rivera J, Tessarollo L. Genetic background and the dilemma of translating mouse studies to humans. Immunity. 2008;28:1–4. doi: 10.1016/j.immuni.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 6.Payne KJ, Crooks GM. Immune-cell lineage commitment: translation from mice to humans. Immunity. 2007;26:674–677. doi: 10.1016/j.immuni.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 7.von Herrath MG, Nepom GT. Lost in translation: barriers to implementing clinical immunotherapeutics for autoimmunity. J Exp Med. 2005;202:1159–1162. doi: 10.1084/jem.20051224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takao K, Miyakawa T. Genomic responses in mouse models greatly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2015;112:1167–1172. doi: 10.1073/pnas.1401965111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schenkel JM, Masopust D. Tissue-resident memory T cells. Immunity. 2014;41:886–897. doi: 10.1016/j.immuni.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thome JJC, et al. Early-life compartmentalization of human T cell differentiation and regulatory function in mucosal and lymphoid tissues. Nat Med. 2015 doi: 10.1038/nm.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thome JJC, et al. Spatial map of human T cell compartmentalization and maintenance over decades of life. Cell. 2014;159:814–828. doi: 10.1016/j.cell.2014.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Machado CS, Rodrigues MA, Maffei HV. Gut intraepithelial lymphocyte counts in neonates, infants and children. Acta Paediatr Oslo Nor 1992. 1994;83:1264–1267. doi: 10.1111/j.1651-2227.1994.tb13011.x. [DOI] [PubMed] [Google Scholar]
- 13.Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol. 2004;22:745–763. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- 14.Olson JA, McDonald-Hyman C, Jameson SC, Hamilton SE. Effector-like CD8+ T cells in the memory population mediate potent protective immunity. Immunity. 2013;38:1250–1260. doi: 10.1016/j.immuni.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mouse Genome Sequencing Consortium et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 16.Pritchett-Corning KR, Cosentino J, Clifford CB. Contemporary prevalence of infectious agents in laboratory mice and rats. Lab Anim. 2009;43:165–173. doi: 10.1258/la.2008.008009. [DOI] [PubMed] [Google Scholar]
- 17.Pedersen AB, Babayan SA. Wild immunology. Mol Ecol. 2011;20:872–880. doi: 10.1111/j.1365-294X.2010.04938.x. [DOI] [PubMed] [Google Scholar]
- 18.Maizels RM, Nussey DH. Into the wild: digging at immunology’s evolutionary roots. Nat Immunol. 2013;14:879–883. doi: 10.1038/ni.2643. [DOI] [PubMed] [Google Scholar]
- 19.Cadwell K. The virome in host health and disease. Immunity. 2015;42:805–813. doi: 10.1016/j.immuni.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Virgin HW, Wherry EJ, Ahmed R. Redefining chronic viral infection. Cell. 2009;138:30–50. doi: 10.1016/j.cell.2009.06.036. [DOI] [PubMed] [Google Scholar]
- 21.Votavova H, et al. Transcriptome alterations in maternal and fetal cells induced by tobacco smoke. Placenta. 2011;32:763–770. doi: 10.1016/j.placenta.2011.06.022. [DOI] [PubMed] [Google Scholar]
- 22.Godec J, et al. Compendium of immune signatures identifies conserved and species-specific biology in response to inflammation. Immunity. 2016;44:194–206. doi: 10.1016/j.immuni.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaech SM, et al. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol. 2003;4:1191–1198. doi: 10.1038/ni1009. [DOI] [PubMed] [Google Scholar]
- 24.Joshi NS, et al. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27:281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jordan MB, Hildeman D, Kappler J, Marrack P. An animal model of hemophagocytic lymphohistiocytosis (HLH): CD8+ T cells and interferon gamma are essential for the disorder. Blood. 2004;104:735–743. doi: 10.1182/blood-2003-10-3413. [DOI] [PubMed] [Google Scholar]
- 26.Selin LK, et al. Memory of mice and men: CD8+ T-cell cross-reactivity and heterologous immunity. Immunol Rev. 2006;211:164–181. doi: 10.1111/j.0105-2896.2006.00394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun JC, Ugolini S, Vivier E. Immunological memory within the innate immune system. EMBO J. 2014;33:1295–1303. doi: 10.1002/embj.201387651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adams AB, Pearson TC, Larsen CP. Heterologous immunity: an overlooked barrier to tolerance. Immunol Rev. 2003;196:147–160. doi: 10.1046/j.1600-065x.2003.00082.x. [DOI] [PubMed] [Google Scholar]
- 29.Taurog JD, et al. The germfree state prevents development of gut and joint inflammatory disease in HLA-B27 transgenic rats. J Exp Med. 1994;180:2359–2364. doi: 10.1084/jem.180.6.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pozzilli P, Signore A, Williams AJK, Beales PE. NOD mouse colonies around the world-recent facts and figures. Immunol Today. 1993;14:193–196. doi: 10.1016/0167-5699(93)90160-M. [DOI] [PubMed] [Google Scholar]
- 31.Hamilton SE, Wolkers MC, Schoenberger SP, Jameson SC. The generation of protective memory-like CD8+ T cells during homeostatic proliferation requires CD4+ T cells. Nat Immunol. 2006;7:475–481. doi: 10.1038/ni1326. [DOI] [PubMed] [Google Scholar]
- 32.Hamilton SE, Schenkel JM, Akue AD, Jameson SC. IL-2 complex treatment can protect naive mice from bacterial and viral infection. J Immunol Baltim Md 1950. 2010;185:6584–6590. doi: 10.4049/jimmunol.1001215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pope C, et al. Organ-specific regulation of the CD8 T cell response to Listeria monocytogenes infection. J Immunol Baltim Md 1950. 2001;166:3402–3409. doi: 10.4049/jimmunol.166.5.3402. [DOI] [PubMed] [Google Scholar]
- 34.Gordon EB, et al. Inhibiting the Mammalian target of rapamycin blocks the development of experimental cerebral malaria. mBio. 2015;6:e00725. doi: 10.1128/mBio.00725-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balfour HH, et al. Behavioral, virologic, and immunologic factors associated with acquisition and severity of primary Epstein-Barr virus infection in university students. J Infect Dis. 2013;207:80–88. doi: 10.1093/infdis/jis646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Odumade OA, et al. Primary Epstein-Barr virus infection does not erode preexisting CD8+ T cell memory in humans. J Exp Med. 2012;209:471–478. doi: 10.1084/jem.20112401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson KG, et al. Intravascular staining for discrimination of vascular and tissue leukocytes. Nat Protoc. 2014;9:209–222. doi: 10.1038/nprot.2014.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beura LK, et al. Lymphocytic choriomeningitis virus persistence promotes effector-like memory differentiation and enhances mucosal T cell distribution. J Leukoc Biol. 2015;97:217–225. doi: 10.1189/jlb.1HI0314-154R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schenkel JM, Fraser KA, Vezys V, Masopust D. Sensing and alarm function of resident memory CD8+ T cells. Nat Immunol. 2013;14:509–513. doi: 10.1038/ni.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steinert EM, et al. Quantifying Memory CD8 T Cells Reveals Regionalization of Immunosurveillance. Cell. 2015;161:737–749. doi: 10.1016/j.cell.2015.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.