Abstract
Development of functional nanoparticles can be encumbered by unanticipated material properties and biological events, which can negatively impact nanoparticle effectiveness in complex, physiologically relevant systems1–3. Despite the advances in bottom-up nanoengineering and surface chemistry, reductionist functionalization approaches remain inadequate in replicating the complex interfaces present in nature and cannot avoid exposure of foreign materials. Here we report on the preparation of polymeric nanoparticles enclosed in the plasma membrane of human platelets, which are a unique population of cellular fragments that adhere to a variety of disease-relevant substrates4–7. The resulting nanoparticles possess a right-side-out unilamellar membrane coating functionalized with immunomodulatory and adhesion antigens associated with platelets. As compared to uncoated particles, the platelet membrane-cloaked nanoparticles have reduced cellular uptake by macrophage-like cells and are absent of particle-induced complement activation in autologous human plasma. The cloaked nanoparticles also display platelet-mimicking properties such as selective adhesion to damaged human and rodent vasculatures as well as enhanced binding to platelet-adhering pathogens. In an experimental rat model of coronary restenosis and a mouse model of systemic bacterial infection, docetaxel and vancomycin, respectively, show enhanced therapeutic efficacy when delivered by the platelet-mimetic nanoparticles. The multifaceted biointerfacing enabled by the platelet membrane cloaking method provides a new approach in developing functional nanoparticles for disease-targeted delivery.
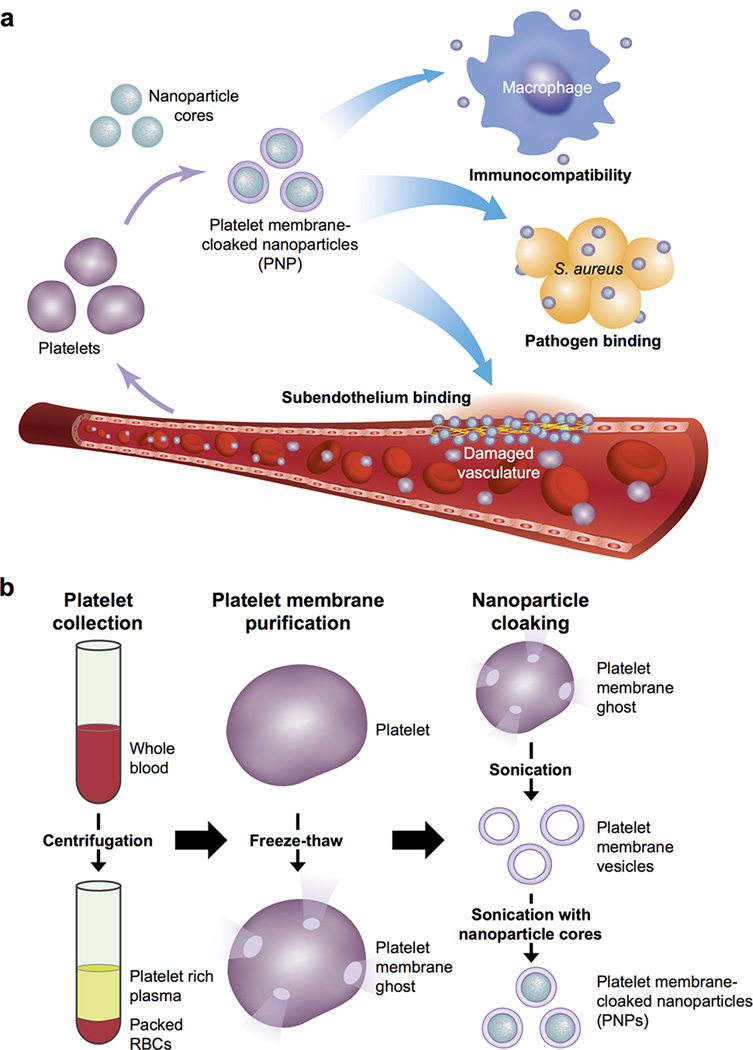
Owing to their role as circulating sentinels for vascular damage and for invasive microorganisms, platelets have inspired the design of many functional nanocarriers8–13. The multitude of platelet functions stem from a unique set of surface moieties responsible for immune evasion14,15, subendothelium adhesion5,16, and pathogen interactions6,7. By adopting a cell membrane cloaking technique17–19, we demonstrate the preparation of platelet membrane-cloaked nanoparticles (PNPs) consisting of a biodegradable polymeric nanoparticle core shielded entirely in the plasma membrane of human platelets. Several inherent platelet properties, including immunocompatibility, binding to injured vasculature, and pathogen adhesion, as well as their therapeutic implications, were studied (Extended Data Fig. 1a).
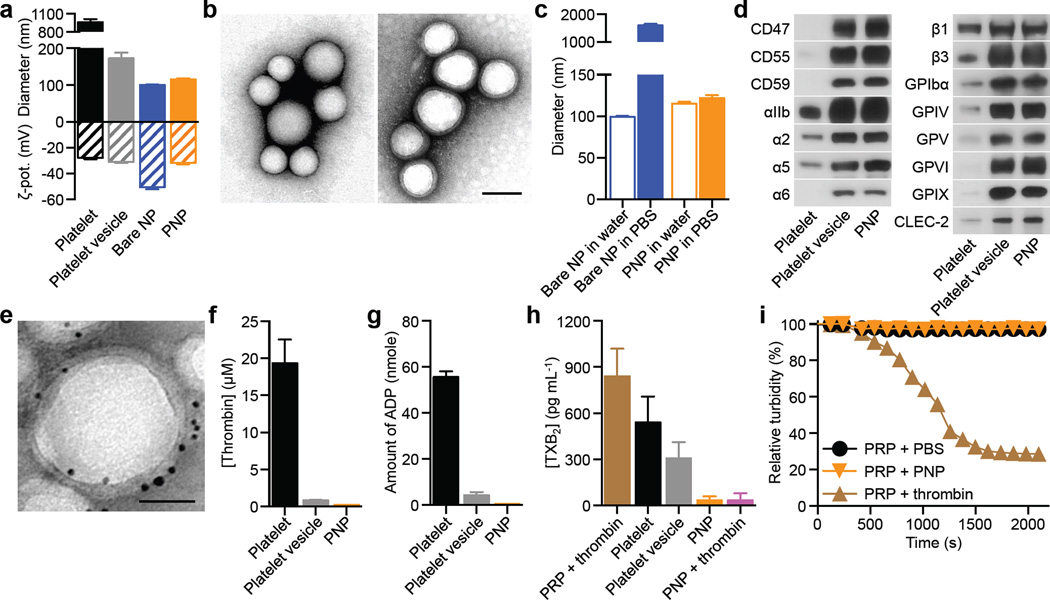
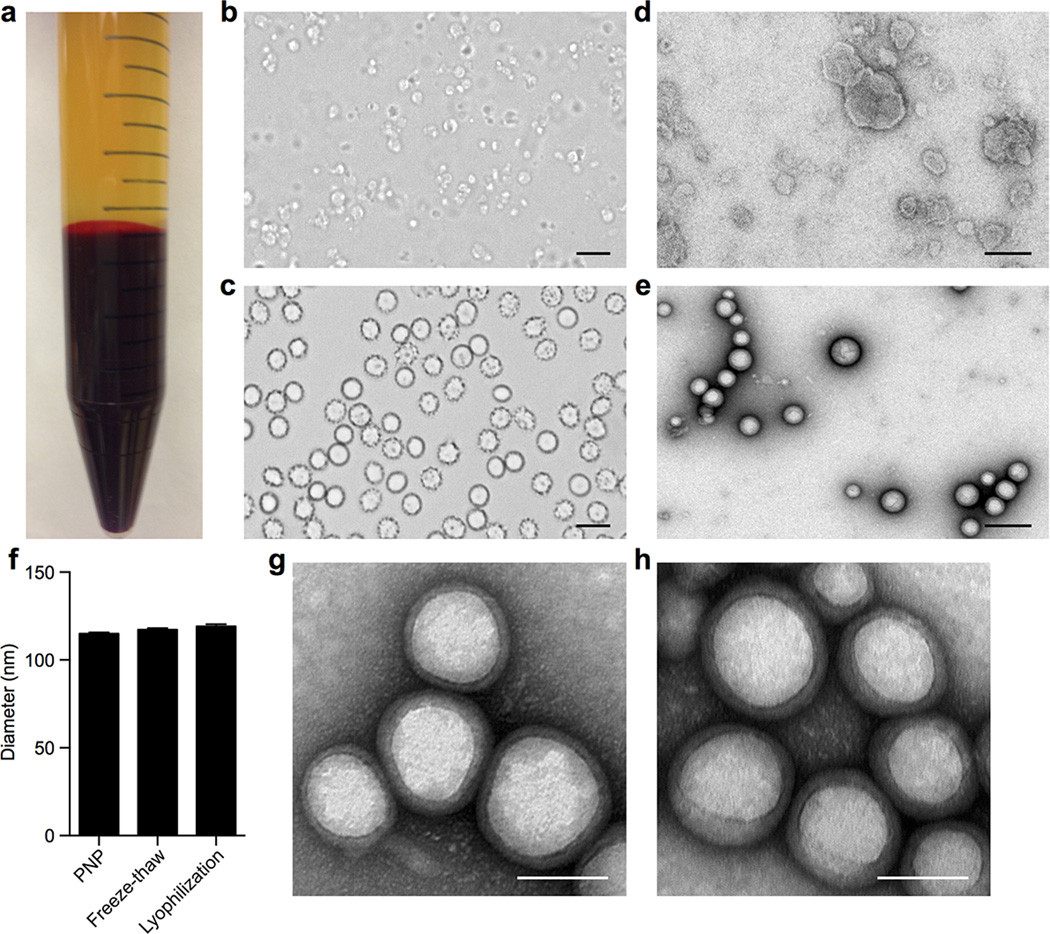
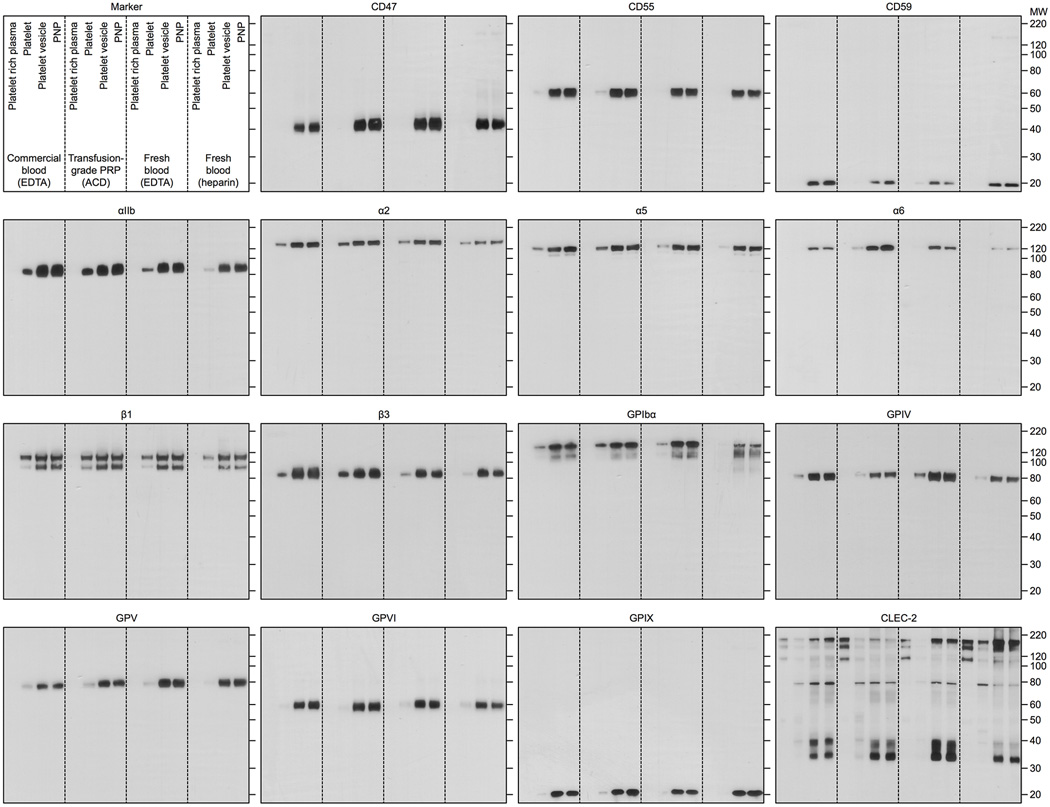
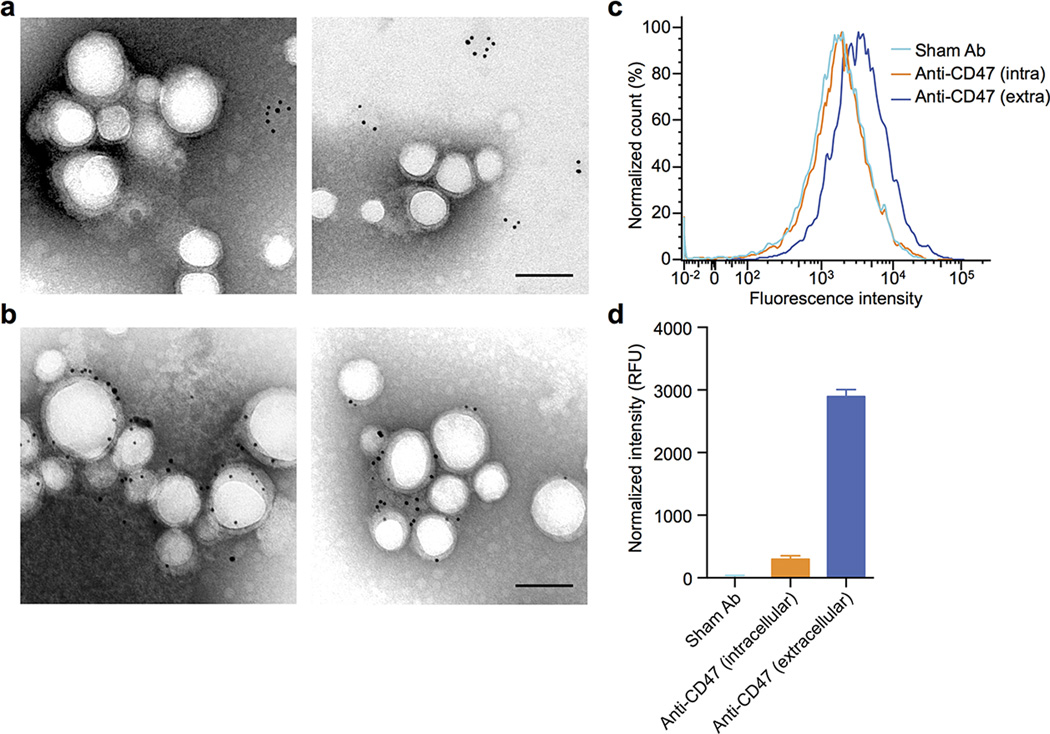
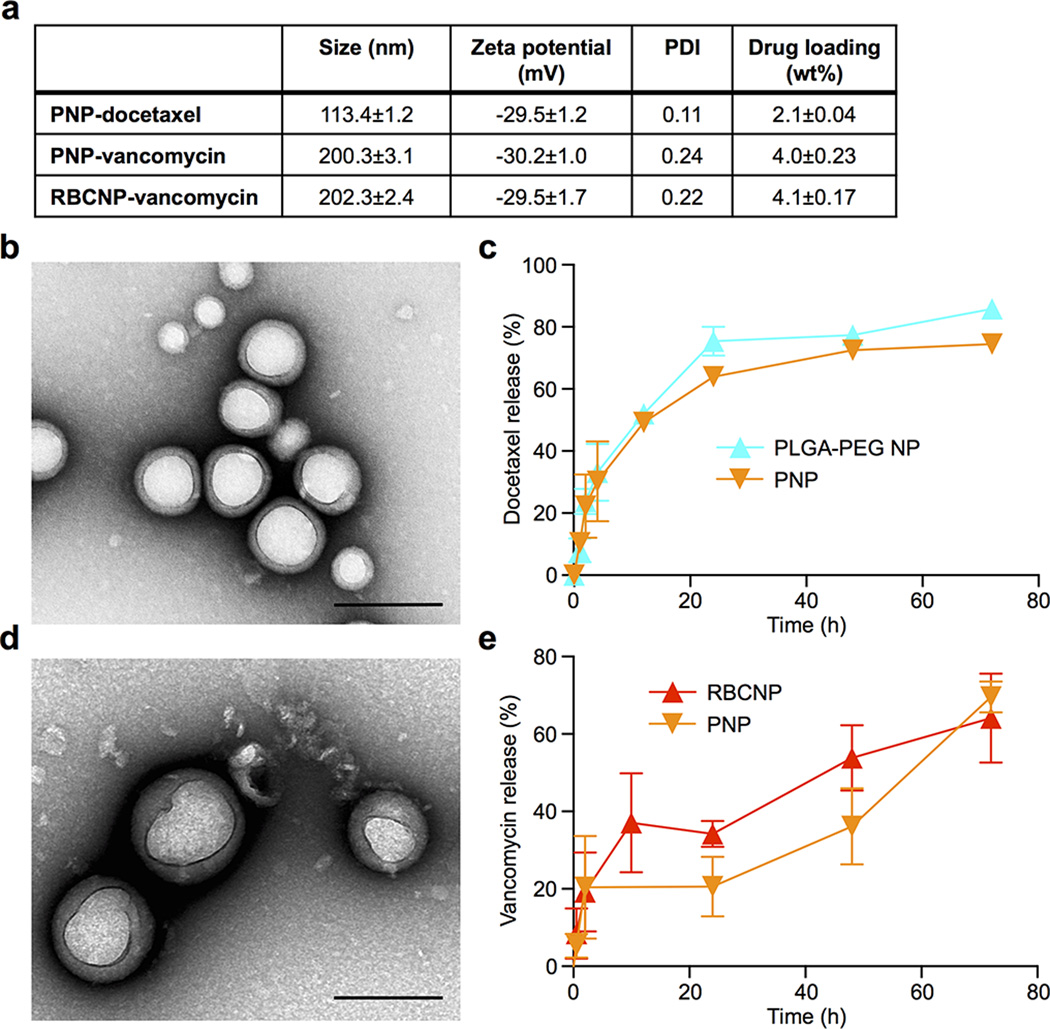
PNPs were prepared by fusing human platelet membrane with 100 nm poly(lactic-co-glycolic acid) (PLGA) nanoparticles. Prior to platelet collection, blood and plasma samples were mixed with EDTA, which prevents platelet aggregation by deactivating fibrinogen-binding integrin αIIbβ320. Platelets were then processed for nanoparticle membrane cloaking (Extended Data Fig. 1b). Physicochemical characterizations revealed that the final PNPs were approximately 15 nm larger than the uncoated PLGA nanoparticles (bare NPs) and possessed an equivalent surface charge to that of platelet and platelet membrane-derived vesicles (platelet vesicles) (Fig. 1a). Transmission electron microscopy (TEM) visualization showed the formation of distinctive nanoparticulates and consistent unilamellar membrane coatings over the polymeric cores (Fig. 1b and Extended Data Fig. 2). Improved colloidal stability was observed with the PNPs compared to bare NPs (Fig. 1c), which is attributable to the stabilizing effect by the plasma membrane’s hydrophilic surface glycans21. Translocation of platelet membrane protein content, including immunomodulatory proteins, CD47, CD55, and CD5914,15, integrin components, αIIb, α2, α5, α6, β1, and β3, and other transmembrane proteins, GPIbα, GPIV, GPV, GPVI, GPIX, and CLEC-25,16, onto the nanoparticles was examined via western blotting (Fig. 1d and Extended Data Fig. 3). Platelets derived from multiple protocols were prepared in parallel for comparison, and it was observed that the PNP preparation resulted in membrane protein retention and enrichment very similar across the different platelet sources (Extended Data Fig. 3). Notably, platelets derived from blood treated with heparin, an anticoagulant that inactivates thrombin rather than platelets, showed evidence of higher platelet activation including increased GPIbα cleavage and CLEC-2 oligomerization22. Using blood anticoagulated with EDTA as the platelet source, a right-side-out membrane orientation on the PNPs was verified via both immunogold staining and flow cytometric analysis with antibodies targeting either the intracellular or extracellular domain of CD47 (Fig. 1e and Extended Data Fig. 4). Pro-thrombotic, platelet-activating molecules such as thrombin, ADP, and thromboxane were removed in the PNP formulation (Fig. 1f–h), thereby permitting PNP administration with little risk of a thrombotic response (Fig. 1i).
Figure 1. Preparation and characterization of PNPs.
(a) Physicochemical characterization of platelets, platelet vesicles, bare NPs, and PNPs (n=3). (b) TEM images of bare NPs (left) and PNPs (right) negatively stained with uranyl acetate. Scale bar = 100 nm. (c) Particle diameter of bare NPs and PNPs in water and in 1X PBS (n=3). (d) Representative protein bands resolved using western blotting. (e) TEM image of PNPs primary stained with extracellular-domain-specific anti-CD47, and secondary stained by an immunogold conjugate. Scale bar = 40 nm. (f–h) Platelet-activating contents including (f) thrombin, (g) ADP, and (h) thromboxane in platelets, platelet vesicles, and PNPs were quantified (n=3). (i) Platelet aggregation assay in which citrate-stabilized platelet rich plasma (PRP) was mixed with PBS, PNPs, or thrombin followed by spectroscopic examination of solution turbidity. All bars represent means ± SD.
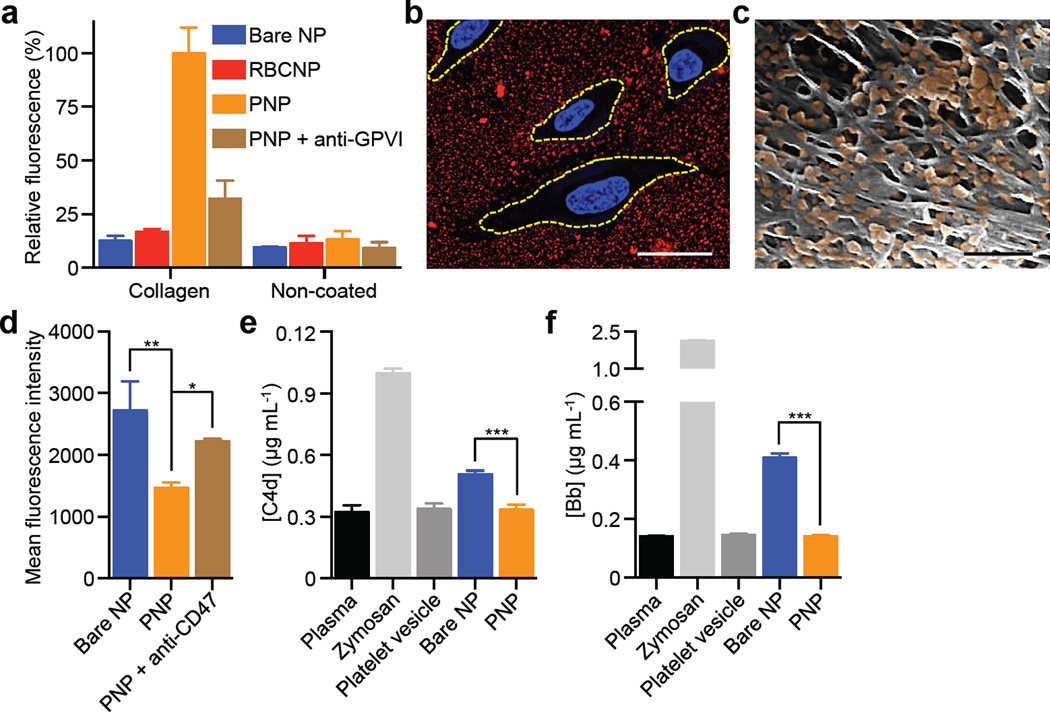
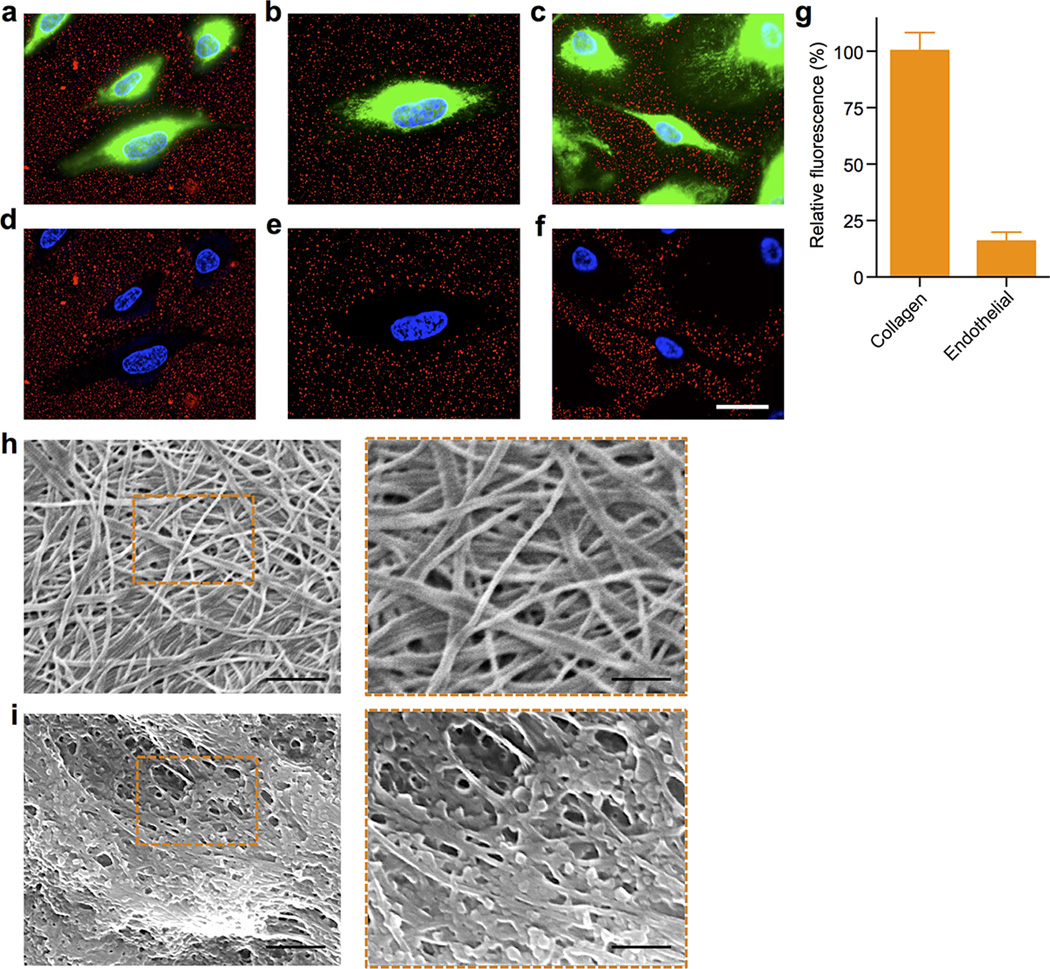
PNPs’ platelet-mimicking functionalities were first studied via the particles’ binding to human type IV collagen, a primary subendothelial component23. Fluorescently labeled PNPs, along with bare NPs and red blood cell membrane-cloaked nanoparticles (RBCNPs), were incubated on collagen-coated plates. The PNPs showed significantly enhanced retention as compared to bare NPs and RBCNPs (Fig. 2a), indicating that the collagen adhesion was membrane type specific. Reduced PNP retention on non-collagen coated plates and in the presence of anti-GPVI antibodies supports a specific collagen/platelet membrane interaction attributable to the presence of membrane glycoprotein receptors for collagen16 (Extended Data Fig. 3). Further examination of PNP’s differential binding to endothelial and collagen surfaces was performed using collagen-coated tissue culture slides seeded with human umbilical vein endothelial cells (HUVECs). PNPs adhered primarily outside of areas encompassed by the cells (Fig. 2b and Extended Data Fig. 5a–g). In addition, the PNPs were incubated with the extracellular matrix derived from decellularized human umbilical cord arteries. Following PBS washes, scanning electron microscopy (SEM) revealed a significant number of PNPs remaining on the fibrous structures on the luminal side of the artery (Fig. 2c and Extended Data Fig. 5h,i).
Figure 2. Collagen binding and immunocompatibility.
(a) Fluorescence quantification of nanoparticle retention on collagen-coated and non-coated plates (n=6). (b) Localization of PNPs (stained in red) on collagen-coated tissue culture slides seeded with HUVECs (nuclei stained in blue). Cellular periphery is outlined based on cytosolic staining. Scale bar = 10 µm. (c) A pseudocolored SEM image of the extracellular matrix of a decellularized human umbilical cord artery following PNP incubation (PNPs colored in orange). Scale bar = 500 nm. (d) Flow cytometric analysis of nanoparticle uptake by human THP-1 macrophage-like cells (n=3). (e) Classical complement activation measured by C4d split products and (f) alternative complement activation measured by Bb split products for bare NPs, platelet vesicles, and PNPs in autologous human plasma (n=4). Zymosan and untreated plasma are used as positive and negative controls respectively. All bars represent means ± SD. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Examination of PNPs’ immunocompatibility was conducted using differentiated human THP-1 macrophage-like cells. The platelet membrane cloaking reduced particle internalization in a CD47-specific manner24, as blocking by anti-CD47 antibodies increased the cellular uptake (Fig. 2d). The PNPs were further investigated for their interactions with the complement system based on quantifications of C4d and Bb split products. Following incubation in human plasma, complement activation was observed with bare NPs, reflecting their susceptibility to opsonization as well as the spontaneous reaction between C3 thioesters and the hydroxyl groups on the PLGA particles25. In contrast, an equal amount of PNPs mixed with autologous plasma showed no observable complement activation (Fig. 2e,f). This suppression of the complement system can be attributed to membrane-bound complement regulator proteins such as CD55 and CD5926 (Extended Data Fig. 3). This result also attests to the completeness of the membrane cloaking, which shields the polymeric cores from plasma exposure and minimizes the risk of anaphylatoxin generation frequently associated with injectable nanocarriers27.
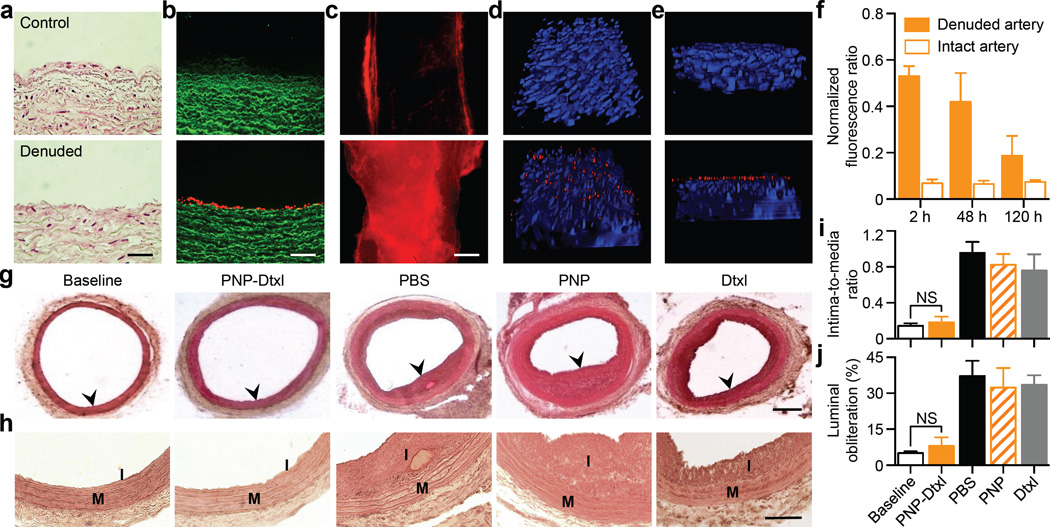
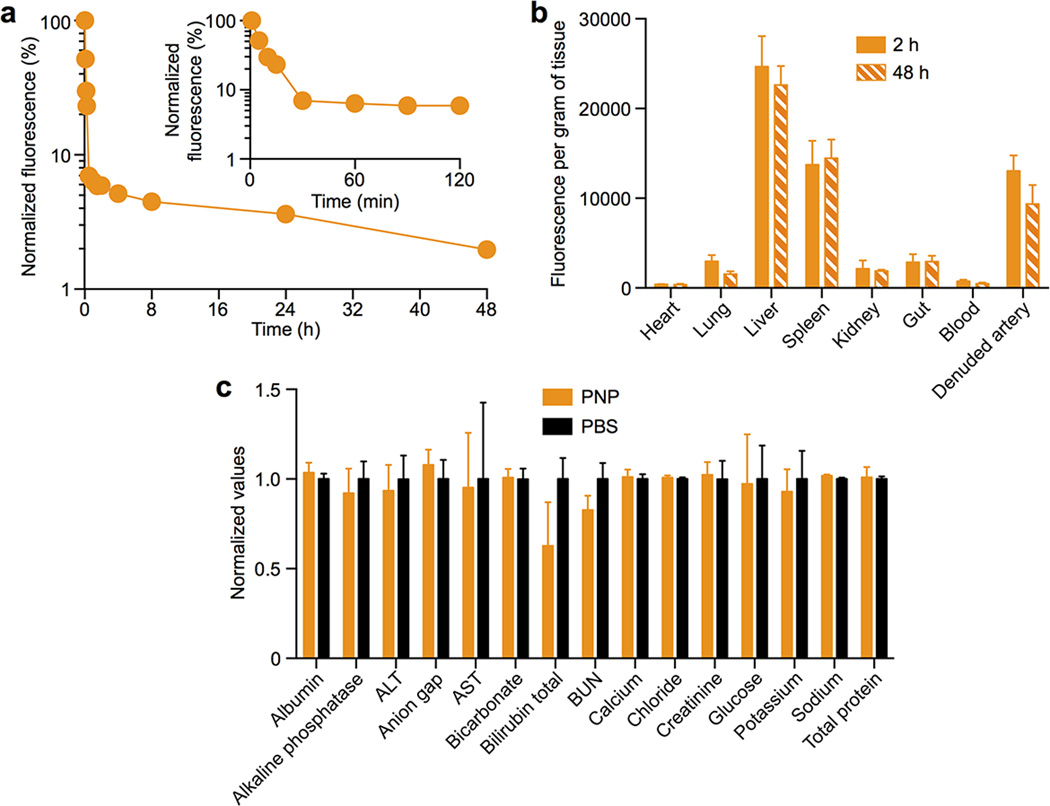
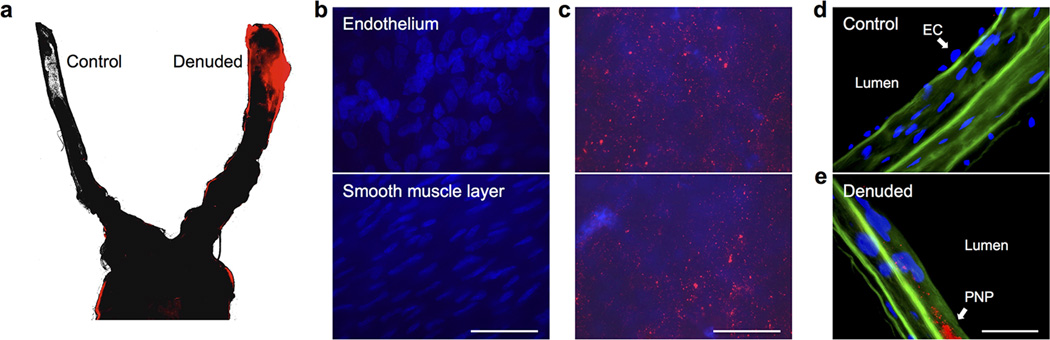
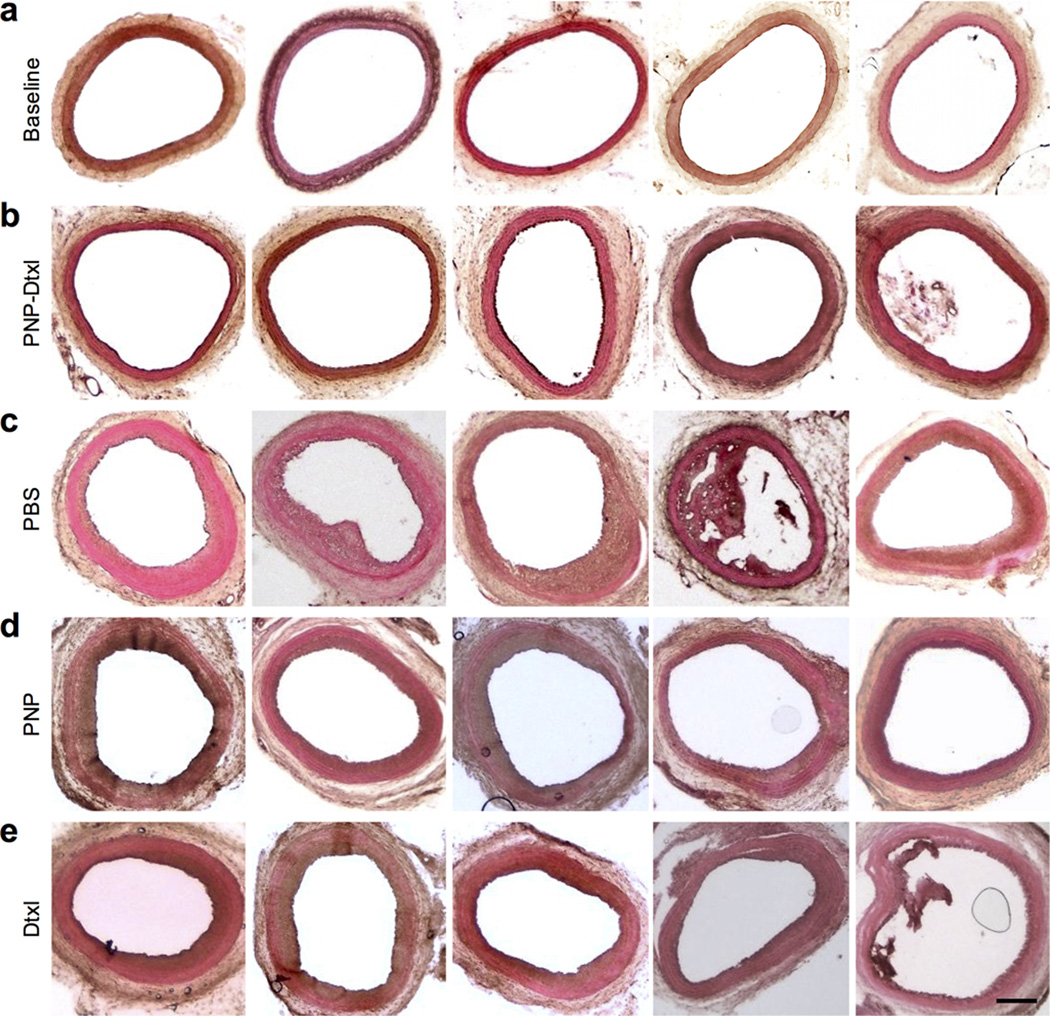
The PNP’s therapeutic potential was first examined by assessing their selective adherence to damaged vasculatures. A segment of the human carotid artery was surgically scraped to expose the subendothelial matrix (Fig. 3a). The intact and damaged artery samples were subsequently incubated with fluorescently labeled PNPs for 30 sec followed by repeated PBS washes. The resulting arterial cross-sections and en face visualizations revealed that the denuded artery was more prone to PNP adhesion than the intact artery (Fig. 3b,c). In Fig. 3c, it can also be observed that PNPs preferentially bind to the edges of the intact artery, where subendothelium was exposed upon tissue incision. This selective PNP adhesion was further validated in a rat model of angioplasty-induced arterial injury. Pharmacokinetic analyses and biodistribution studies showed that >90% of the PNPs were distributed to tissues 30 min after intravenous injection, with liver and spleen being the primary distribution organs (Extended Data Fig. 6a–b). A comprehensive blood chemistry panel analysis revealed that the PNPs did not inflict observable adverse effects in the rats (Extended Data Fig. 6c). Selective particle binding to the denuded artery was observed upon examination of the aortic branch 2 h following PNP administration (Fig. 3d and Extended Data Fig. 7). The PNPs were localized on the luminal side above the smooth muscle layer (Fig. 3e), and retention at the injury site lasted for at least 5 days (Fig. 3f). In a rat model of coronary restenosis, therapeutic relevance of platelet-mimicking delivery was examined using docetaxel-loaded PNPs (PNP-Dtxl) (Extended Data Fig. 8). PNP-Dtxl treatment on day 0 and 5 at 0.3 mg kg−1 of docetaxel dosing potently inhibited neointima growth in balloon-denuded rats as evidenced by the arterial cross-sections collected on day 14 (Fig 3g,h and Extended Data Fig. 9). To quantitatively evaluate the vascular remodeling, intima-to-media ratio (I/M) and luminal obliteration were calculated. As compared to free docetaxel, which resulted in an I/M of 0.76±0.18 and a luminal obliteration of 33.6%, PNP-Dtxl yielded significantly lower values of 0.18±0.06 and 8.0%, respectively (P ≤ 0.0001) (Fig. 3i–j). These results demonstrate the benefit of PNP-directed delivery in improving drug localization to diseased vasculatures.
Figure 3. Adherence to damaged human and rodent vasculatures.
(a) H&E-stained cross-sections of undamaged (top) and damaged (bottom) human carotid arteries. Scale bar = 200 µm. (b) Fluorescence images of the cross-section (scale bar = 200 µm) and (c) the luminal side (scale bar = 500 µm) of undamaged (top) and damaged (bottom) arteries following PNP incubation (tissue in green and PNPs in red). (d,e) 3D reconstructed images of intact (top) and balloon-denuded (bottom) arterial walls from multisectional images following intravenous administration of PNPs in rats (cell nuclei in blue and PNPs in red). Dimensions: 152.5 µm × 116 µm × 41 µm. (f) Retention of PNPs at the denuded and the intact arteries over 120 h following PNP administration (n=6). (g) Representative H&E-stained arterial cross-sections from different treatment groups in a rat model of coronary restenosis. Scale bar = 200 µm (h) Zoomed-in H&E-stained arterial cross-sections highlight the different vascular remodeling from the different treatment groups. I, intima; M, media. Scale bar = 100 µm. (i,j) Quantitative analysis of intima-to-media area ratio and luminal obliteration from the different treatment groups (n=6). All bars represent means ± SD. NS = no statistical significance.
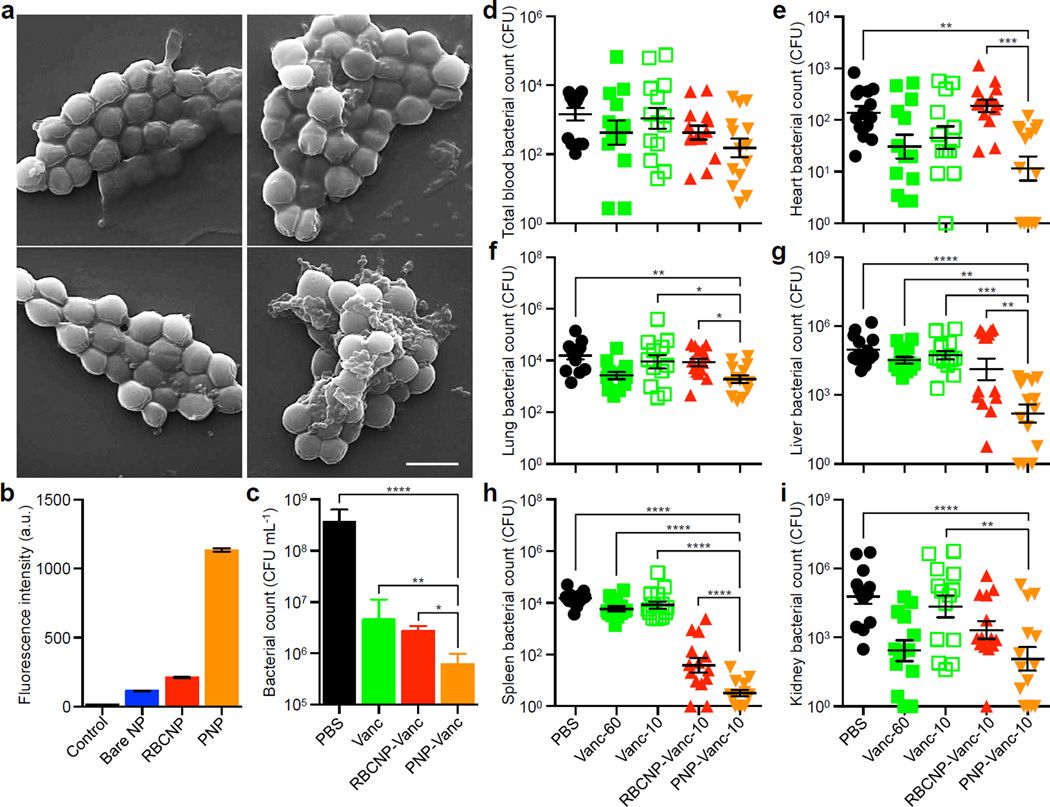
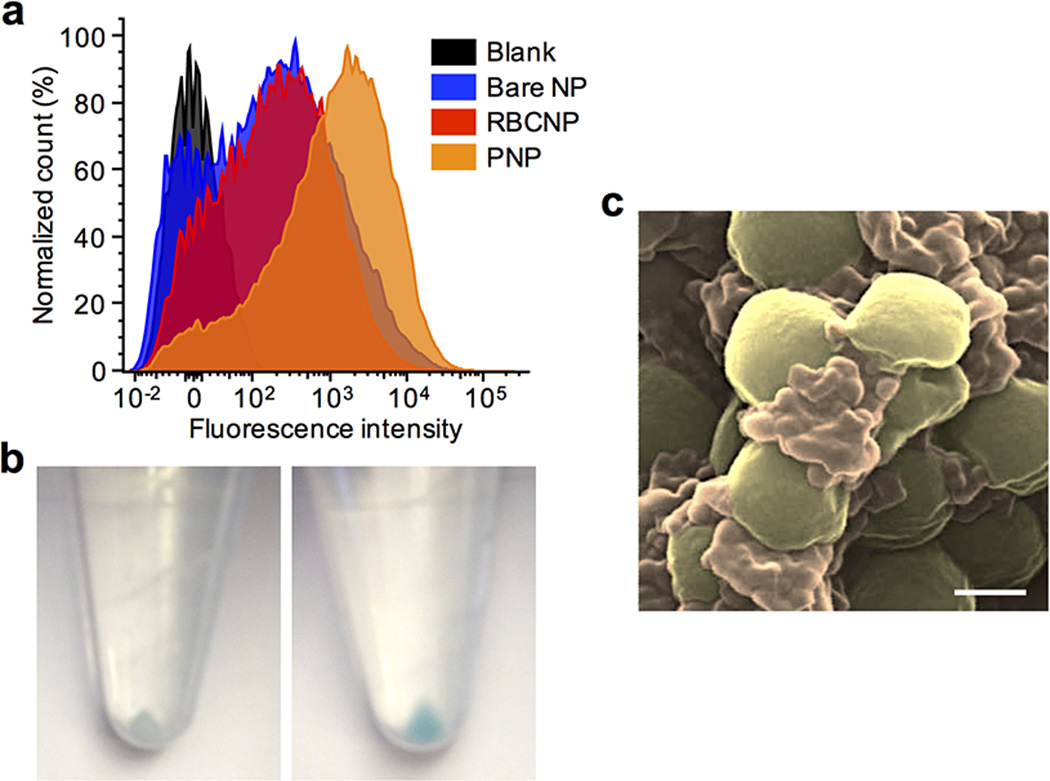
We further examined the therapeutic potential of PNPs against platelet-adhering pathogens. Opportunistic bacteria including several strains of staphylococci and streptococci exploit platelets via both direct and indirect adherence mechanisms for tissue localization and immune evasion6. To demonstrate that PNPs can exploit the inherent bacterial adherence mechanism for targeted antibiotics delivery, MRSA252, a strain of methicillin-resistant Staphylococcus aureus expressing a serine-rich adhesin for platelets (SraP)28, was used as a model pathogen for particle adhesion study. Following 10 min of incubation between formalin-fixed MRSA252 and different nanoformulations, the collected bacteria showed preferential binding by PNPs (Fig. 4a), exhibiting a 12-fold increase in PNP retention as compared to bare NPs (Fig. 4b and Extended Data Fig. 10). This adherence was membrane-specific as RBCNPs showed lower retention than PNPs. The therapeutic potential of PNPs was further evaluated using vancomycin-loaded formulations. In an in vitro antimicrobial study, live MRSA252 bacteria were briefly incubated with free vancomycin, vancomycin-loaded RBCNPs (RBCNP-Vanc), or vancomycin-loaded PNPs (PNP-Vanc) followed by a wash and culturing in fresh media. The PNP-Vanc formulation showed statistically significant improvement in MRSA252 reduction that corroborates the targeting effect of the particles (Fig. 4c). An in vivo antimicrobial efficacy study was further conducted using a mouse model of systemic MRSA252 infection. Mice systemically challenged with 6 × 106 CFU MRSA252 received once daily intravenous treatment of free vancomycin, RBCNP-Vanc, or PNP-Vanc for 3 days at 10 mg kg−1 of vancomycin. A control group of high-dose vancomycin treatment in which infected mice received free vancomycin at 30 mg kg−1 twice daily was conducted in parallel. 24 h following the last treatment, bacterial enumeration at the primary infection organs showed that the PNP-Vanc resulted in the lowest mean bacterial counts across all organs (Fig. 4d–i). Statistical analyses revealed significance between PNP-Vanc and free vancomycin at equivalent dosage in the lung, liver, spleen, and kidney. In comparison to free vancomycin at 6-fold the dosage, PNP-Vanc showed significantly better antimicrobial efficacy in the liver and spleen while being at least equally effective in the blood, heart, lung, and kidney. Notably, as compared to RBCNP-Vanc, PNP-Vanc showed significantly higher potency in the heart, lung, liver, and spleen, reflecting membrane-specific modulation of nanoparticle performance. The study validates the feasibility of harnessing biomembrane interfaces to improve infectious disease treatment.
Figure 4. Binding to platelet-adhering pathogens.
(a) SEM images of MRSA252 bacteria following incubation with PBS (top left), bare NPs (top right), RBCNPs (bottom left), and PNPs (bottom right). Scale bar = 1 µm. (b) Normalized fluorescence intensity of DiD-loaded nanoformulations retained on MRSA252 based on flow cytometric analysis. Bars represent means ± SD (n=3). (c) In vitro antimicrobial efficacy of free vancomycin, vancomycin-loaded RBCNPs (RBCNP-Vanc), and vancomycin-loaded PNPs (PNP-Vanc). Bars represent means ± SD (n=3). (d–i) In vivo antimicrobial efficacy of free vancomycin at 10 mg kg−1 (Vanc-10), RBCNP-Vanc-10, and PNP-Vanc-10, and free vancomycin at 6 times the dosing (Vanc-60, 60 mg kg−1) was examined in a mouse model of systemic infection with MRSA252. Following 3 days of treatments, bacterial loads in different organs including (d) blood, (e) heart, (f) lung, (g) liver, (h) spleen, and (i) kidney were quantified. Bars represent means ± SEM (n=14). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
The vast medical relevance of platelets has inspired many platelet-mimicking systems that target dysfunctional vasculature in cardiovascular diseases8,9, traumas10,11,13, cancers12, and acute inflammations29. The present PNP platform exploits platelet membrane in its entirety to enable biomimetic interactions with proteins, cells, tissues, and microorganisms. Toward translation, the platform would benefit from existing infrastructures and logistics for transfusion medicine, polymeric nanotherapeutics, and cell-derived pharmaceutics. Prior works on the cell membrane cloaking approach demonstrated high cloaking efficiency30 and viable storage18 upon platform optimization (Extended Data Fig. 2f–h). By employing large-scale purification and dispersion techniques commonly applied to biologics, reliable platelet membrane derivation and PNP production can be envisioned.
Methods
Human platelet isolation and platelet membrane derivation
Human type O− blood anti-coagulated with 1.5 mg mL−1 EDTA was purchased from BioreclamationIVT and processed for platelet collection approximately 16 h after blood collection. Unless otherwise stated, platelets derived from this commercial blood source were used in this study. Fresh human type O− blood was also collected with dipotassium EDTA-treated or lithium heparin-treated blood collection tubes (Becton, Dickinson and Company) under the approval of the Institutional Review Board (IRB) at the University of California, San Diego. Patients consented to use of their blood samples for this study prior to collection. The freshly drawn blood was processed for platelet collection approximately 30 min following blood draw. In addition, in-dated human type O− platelet rich plasma (PRP) in acid-citrate-dextrose (ACD) was purchased from the San Diego Blood Bank. Samples not originally drawn in EDTA were adjusted to a concentration of 5 mM EDTA prior to platelet collection. To isolate platelets, the blood and plasma samples were centrifuged at 100 × g for 20 min at room temperature to separate red blood cells and white blood cells. The resulting PRP was then centrifuged at 100 × g for 20 min to remove remaining blood cells. PBS buffer containing 1 mM of EDTA and 2 µM of prostaglandin E1 (PGE1, Sigma Aldrich) was added to the purified PRP to prevent platelet activation. Platelets were then pelleted by centrifugation at 800 × g for 20 min at room temperature, following which the supernatant was discarded and the platelets were resuspended in PBS containing 1 mM of EDTA and mixed with protease inhibitor tablets (Pierce). 1.5 mL aliquots of platelet solution containing ~ 3 × 109 platelets were prepared and used to cloak 1 mg of PLGA nanoparticles.
Platelet membrane was derived by a repeated freeze-thaw process. Aliquots of platelet suspensions were first frozen at −80 °C, thawed at room temperature, and pelleted by centrifugation at 4000 × g for 3 min. Following three repeated washes with PBS solution mixed with protease inhibitor tablets, the pelleted platelet membranes were suspended in water and sonicated in a capped glass vial for 5 min using a Fisher Scientific FS30D bath sonicator at a frequency of 42 kHz and a power of 100 W. The presence of platelet membrane vesicles was verified by size measurement using dynamic light scattering (DLS) and morphological examination by transmission electron microscopy (TEM).
Platelet membrane-cloaked nanoparticle (PNP) preparation and characterization
100 nm polymeric cores were prepared using 0.67 dL g−1 carboxyl-terminated 50:50 poly(lactic-co-glycolic) acid (PLGA) (LACTEL Absorbable Polymers) in a nanoprecipitation process. 1 mL of 10 mg mL−1 PLGA solution in acetone was added dropwise to 3 mL of water. For fluorescently labeled nanoformulations, 1,1’-dioctadecyl-3,3,3’,3’-tetramethylindodicarbocyanine perchlorate (DiD, ex = 644 nm/em = 665 nm, Life Technologies) was loaded into the polymeric cores at 0.1 wt%. The mixture was then stirred in open air for 1 h and placed in vacuum for another 3 h. The resulting nanoparticle solution was filtered with 10 kDa MWCO Amicon Ultra-4 Centrifugal Filters (Millipore). Platelet membrane cloaking was then accomplished by dispersing and fusing platelet membrane vesicles with PLGA particles via sonication using an FS30D bath sonicator at a frequency of 42 kHz and a power of 100 W for 2 min. The size and the surface zeta potential of the replicate PNP samples (n=3) were obtained by DLS measurements using a Malvern ZEN 3600 Zetasizer. PBS stability was examined by mixing 1 mg mL−1 of PNPs in water with 2X PBS at a 1:1 volume ratio. Storability of PNPs was examined by suspending PNPs in 10% sucrose. The nanoparticle solutions were subject to either a freeze-thaw cycle or lyophilization followed by resuspension. The resulting particle solution was then monitored for particle size using DLS. The structure of PNPs was examined with TEM following negative staining with 1 wt% uranyl acetate using an FEI 200 kV Sphera microscope. RBCNPs were prepared using the same polymeric cores and RBC membranes of equivalent total surface area to the platelet membranes using a previously described protocol16. The RBCNPs were characterized using DLS and had similar size and zeta potential as the PNPs.
Docetaxel-loaded PLGA nanoparticle cores were prepared via a nanoprecipitation process. 10 wt% docetaxel was added to 5 mg PLGA in acetone and precipitated dropwise into 3 mL water. The solvent was evaporated as described above and free docetaxel was removed via repeated wash steps. Vancomycin-loaded nanoparticles were synthesized using a double emulsion process. The inner aqueous phase consisted of 25 µL of vancomycin (Sigma Aldrich) dissolved in 1 M NaOH at 200 mg mL−1. The outer phase consisted of 500 µL of PLGA polymer dissolved in dichloromethane at 50 mg mL−1. The first emulsion was formed via sonication at 70% power pulsed (2 sec on/1 sec off) for 2 min on a Fisher Scientific 150E Sonic Dismembrator. The resulting emulsion was then emulsified in aqueous solution under the same dispersion setting. The final w/o/w emulsion was added to 10 mL of water and the solvent was evaporated in a fume food under gentle stirring for 3 h. The particles were collected via centrifugation at 80,000 × g in a Beckman Coulter Optima L-90K Ultracentrifuge. The particles were washed and resuspended in water. Upon preparation of drug-loaded PLGA cores, cell membrane coating was performed by adding the appropriate surface area equivalent of either platelet or RBC membrane followed by 3 min of sonication in a Fisher Scientific FS30D Bath Sonicator. Particle size, polydispersity (PDI), and surface zeta potential were characterized using DLS. Drug loading yield and release rate of replicate samples (n=3) were quantified by high performance liquid chromatography (HPLC). Drug release was determined by dialyzing 500 µL of particle solution at a concentration of 2.67 mg mL−1 in PBS using 3.5K MWCO Slide-A-Lyzers (Thermo Scientific).
Examination of platelet membrane proteins
PNPs were purified from unbound proteins or membrane fragments via centrifugation at 16,000 × g in 10% sucrose. Platelet-rich plasma, platelets, platelet membrane vesicles, and PNPs were then normalized to equivalent overall protein concentration using a Pierce BCA Protein Assay Kit (Life Technologies). To examine the effect of different platelet derivation protocols on the membrane protein expression, platelets collected from commercial blood anti-coagulated in EDTA, freshly drawn blood anti-coagulated in EDTA or heparin, and transfusion-grade PRP in ACD were prepared in parallel. All platelets were processed using the aforementioned platelet membrane derivation protocol for PNP preparation. The samples containing equivalent total proteins were then lyophilized, prepared in lithium dodecyl sulfate (LDS) sample loading buffer (Invitrogen), and separated on a 4–12% Bis-Tris 17-well minigel in MOPS running buffer using a Novex® Xcell SureLock Electrophoresis System (Life Technologies). For membrane protein visualization, the gel was stained using SimplyBlue™ SafeStain solution (Life Technologies) following the manufacturer’s instructions and imaged using a gel imager. Identification of key membrane proteins via western blotting was performed using primary antibodies including mouse anti-human CD47 (eBioscience, B6H12), mouse anti-human CD55 (Biolegend, JS11), mouse anti-human CD59 (Biolegend, p282 (H19)), mouse anti-human αIIb subunit (Biolegend, HIP8), rat anti-human integrin α2 subunit (R&D Systems, 430907), rabbit anti-human integrin α5 subunit (Abgent, AP12204c), mouse anti-human integrin α6 subunit (Abgent, AM1828a), mouse anti-human integrin β1 subunit (R&D Systems, 4B7R), mouse anti-human β3 subunit (Biolegend, VI-PL2), mouse anti-human GPIbα (R&D Systems, 486805), mouse anti-human GPIV (R&D Systems, 877346), mouse anti-human GPV (Santa Cruz Biotech, G-11), rat anti-human GPVI (EMD Millipore, 8E9), rabbit anti-human GPIX (Santa Cruz Biotech, A-9), and mouse anti-human CLEC-2 (Genetex, 8J24). A goat anti-mouse IgG-HRP conjugate (Biolegend, Poly4053), a goat anti-rat IgG-HRP conjugate (Biolegend, Poly4054), or a donkey anti-rabbit IgG-HRP conjugate (Biolegend, Poly4064) was used for secondary staining based on the isotype of the primary antibody. MagicMark XP western protein standard (Invitrogen) was used as a molecular weight ladder. The nitrocellulose membrane was then incubated with ECL western blotting substrate (Pierce) and developed with the Mini-Medical/90 Developer (ImageWorks).
Examination of protein sidedness on PNPs
For immunogold staining, a drop of the PNP solution (1 mg mL−1) was deposited onto a glow-discharged carbon-coated grid. The grid was then washed 3 times with PBS, blocked with 1% BSA for 15 min, and stained with 0.5 mg mL−1 of anti-CD47 targeted to either the intracellular or extracellular domain of the protein. Following 1 h of incubation, the samples were rinsed with PBS containing 1% BSA for 6 times and stained with anti-rabbit IgG gold conjugate (5 nm) solution (Sigma Aldrich) for another hour. Following 6 times of PBS washing, the samples were fixed with 1% glutaraldehyde in PBS for 5 min and washed with water for 6 times. The sample grids were subsequently stained with 2% vanadium solution (Abcam) and visualized using an FEI 200 kV Sphera microscope.
For flow cytometric analysis, 2.0 µm carboxyl-functionalized polystyrene beads at a concentration of 4 wt% (Life Technologies) were functionalized with rabbit N-terminus-targeted (extracellular) anti-human CD47 (Aviva Biosystems, ARP63284), rabbit intracellular-domain-targeted anti-CD47 (Genetex, EPR4150(2)), or rabbit anti-ovalbumin (Abcam, ab1221) as a sham antibody via EDC/NHS chemistry. The resulting antibody-modified beads were re-suspended in 100 µL of DI water. The bead solution was first incubated with 1 mg of bovine serum albumin (BSA, Sigma Aldrich) to block non-specific interactions and then mixed with 1 mL of fluorescently labeled PNPs (200 µg mL−1). The mixture solution was incubated at room temperature for 2 h and then centrifuged to remove the unbound PNPs. The collected polystyrene beads were then subjected to flow cytometric analysis.
Platelet aggregation assay
Platelets, platelet membrane vesicles, and PNPs of equivalent membrane content were prepared and examined for platelet-activating molecules, including thrombin, ADP, and thromboxane, using a SensoLyte 520 Thrombin Activity Assay Kit (AnaSpec), ADP Colorimetric/Fluorometric Assay Kit (Sigma Aldrich), and Thromboxane B2 (TXB2) ELISA Kit (Enzo Life Sciences), respectively, based on the manufacturers’ instructions. Each sample was assayed in replicate (n=3).
Aggregation of platelets in the presence of PNPs was assessed using a spectrophotometric method. 1 mL aliquot of platelet rich plasma (PRP) was first prepared from human whole blood with sodium citrate as the anti-coagulant. The plasma was then loaded into a cuvette followed by addition of 500 µL of 2 mg mL−1 PNPs in PBS solution. As negative and positive controls, the PRP was mixed with 500 µL of PBS or 500 µL of PBS containing 0.5 IU mL−1 of human thrombin (Sigma Aldrich), respectively. The cuvettes were immediately placed in a TeCan Infinite M200 reader and monitored for change in absorbance at 650 nm over time, and platelet aggregation was observed based on the reduction of turbidity.
Collagen binding study
Collagen type IV derived from human placenta (Sigma Aldrich) was reconstituted to a concentration of 2.0 mg mL−1 in 0.25% acetic acid. 200 µL of the collagen solution was then added to a 96-well assay plate and incubated overnight at 4 °C. Prior to the collagen binding study, the plate was blocked with 2% BSA and washed three times with PBS. For the collagen binding study, 100 µL of 1 mg mL−1 DiD-loaded nanoformulations in water were added into replicate wells (n=6) of collagen-coated or non-collagen-coated plates. Following 30 sec of incubation, the plates were washed three times. Retained nanoparticles were then dissolved with 100 µL of DMSO for fluorescence quantification using a TeCan Infinite M200 reader.
Differential adhesion to endothelial and collagen surfaces
Collagen type IV was coated on 8-well Lab-Tek II chamber slides (Nunclon) as described above. The collagen-coated chamber slides were used to seed primary HUVECs obtained from the American Type Culture Collection and cultured in HUVEC Culture Medium (Sigma Aldrich) supplemented with 10% fetal bovine serum for 24 h. The cells were then incubated with 1 mg mL−1 DiD-loaded PNPs in PBS at 4°C for 30 sec. Next the cells were washed with PBS three times and fixed with tissue fixative (Millipore) for 30 min at room temperature. Fluorescence staining was done with 4’6-diamidino-2-phenylinodle (DAPI, Life Technologies) for the nuclei and 22-(n-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-23,24-bisnor-5-cholen-3β-ol (NBD cholesterol, Life Technologies) for the cytosol before mounting the cells in ProLong Gold antifade reagent (Life Technologies) and imaged using a DeltaVision deconvolution scanning fluorescence microscope. Z-stacks were collected at 0.25 µm intervals over 10 µm. The images were deconvolved and superimposed. DiD fluorescence signal over collagen and endothelial surfaces as defined by the boundaries of NBD fluorescence were analyzed using ImageJ. PNP retention over collagen and endothelial surfaces was quantified based on distinct images (n=10) in which the average fluorescence per unit area was analyzed.
Cellular uptake study with macrophage-like cells
THP-1 cells were obtained directly from the American Type Culture Collection and used without further testing. They were maintained in RPMI 1640 media (Life Technologies) supplemented with 10% FBS (Sigma Aldrich). THP-1 cells were differentiated in 100 ng mL−1 phorbol myristate acetate (PMA, Sigma Aldrich) for 48 h and differentiation was visually confirmed by cellular attachment to petri dishes. For the cellular uptake study, the differentiated macrophage-like cells were incubated in replicate wells (n=3) with DiD-loaded PNPs, anti-CD47 blocked PNPs, and bare NPs at 100 µg mL−1 in culture media. Following 30 min of incubation at 37 °C, the macrophage-like cells were scraped off the petri dish and washed three times in PBS to remove non-internalized particles. Flow cytometry was performed to examine nanoparticle uptake by the macrophage-like cells. All flow cytometry studies were conducted on a FACSCanto II flow cytometer (BD Biosciences) and the data was analyzed using FlowJo software from Tree Star. The mean fluorescence was plotted in a bar chart with error bars representing the standard error. Statistical analysis was performed based on a two-tailed, unpaired t-test.
Complement activation study
To assess complement system activation, two complement split products (C4d and Bb) were analyzed using enzyme-linked immunosorbent assay kits (Quidel Corporation). The nanoparticles were incubated in replicate aliquots (n=4) of human serum at a volume ratio of 1:5 in a shaking incubator (80 rpm) at 37 °C for 1 h. The reaction was then stopped by adding 60 volumes of PBS containing 0.05% Tween-20 and 0.035% ProClin 300. Complement system activation of the nanoparticles was assayed following the manufacturer’s instructions, and zymosan was used as a positive control.
PNP adherence to human carotid artery
Human umbilical cord was collected under the approval of the Institutional Review Board (IRB) at the University of California, San Diego, and human carotid arteries were collected under the approval of the IRB at the University of Southern California. Patients consented to use of their samples for this study prior to collection. To derive decellularized arterial extracellular matrix (ECM), human arteries were carefully dissected from the umbilical cord and removed from the surrounding Wharton’s jelly, and subsequently incubated in 2% sodium dodecyl sulfate (SDS, Sigma Aldrich) for 72 h. The decellularized tissue was then rinsed with PBS and incubated in PBS solution containing 200 µg mL−1 PNPs for 30 sec. The sample was then transferred to PBS solution and rinsed extensively prior to examination by scanning electron microscopy (SEM). A control decellularized arterial tissue sample without PNP incubation was prepared and visualized for comparison.
To examine PNP binding on denuded vascular walls, approximately 2 mm thick fresh human carotid artery sections were dissected and placed in normal saline on ice and transported immediately to the laboratory for a PNP binding study. To create the vascular characteristics of damaged arteries, an excised artery sample was surgically scraped on its luminal side with forceps to remove the endothelial layer. Successful denudation was confirmed via microscopy visualization. Prior to the nanoparticle binding experiment, both damaged and non-damaged artery samples were rinsed with PBS solution. The PNP binding experiment was performed by incubating the arterial samples in PBS solution containing 200 µg mL−1 of DiD-loaded PNPs for 30 sec. The samples were then transferred to PBS solution and rinsed extensively prior to visualization by fluorescence microscopy. Endogenous tissue components such as collagen and elastin were identified based on their autofluorescence, which excites and emits maximally at 300~500 nm and was captured using a FITC filter. DiD fluorescence was captured using a Cy5 filter to examine the deposition of PNPs. The arterial samples were imaged via a cross-sectional view of a histological section and a top-down view on the luminal side. The images were normalized to a reference illumination image for proper comparison.
Pharmacokinetics, biodistribution and safety of PNPs in a rat model of angioplasty-induced arterial denudation
All animal experiments were performed in accordance with NIH guidelines and approved by the Animal Care Committee of the University of California, San Diego. For the pharmacokinetics study, adult male Sprague-Dawley rats weighing 300–350 g (Harlan Laboratories) were administered with DiD-labeled PNPs and their blood was collected at specific time points via tail-vein blood sampling for fluorescence quantification. For the safety study, rats were injected with 1 mL of 5 mg mL−1 of PNPs on day 0 and day 5 followed by blood collection on day 10 for comprehensive metabolic panel analysis. Rats receiving equivalent PBS injections were prepared as a control.
For the biodistribution and vasculature-targeting studies, adult male Sprague-Dawley rats weighing 300–350 g (Harlan Laboratories) were subjected to carotid balloon injury. In brief, the animals were anesthetized with intraperitoneal ketamine (Pfizer) at 100 mg kg−1 and xylazine (Lloyd Laboratories) at 10 mg kg−1. A ventral mid-line incision (~2 cm) was made in the neck, and the left common carotid artery and carotid bifurcation was exposed by blunt dissection. Proximal of left carotid artery, inner carotid artery and external carotid artery were temporarily clamped to avoid excessive blood loss during the induction of the 2F Fogarty arterial embolectomy catheter (Edwards Lifesciences). The catheter was introduced into left carotid artery through an arteriotomy on the external carotid artery. The catheter was slowly inflated to a determined volume (0.02 mL) and withdrawn with rotation for 3 times to denude endothelium. The wound was later closed with 4-0 sutures.
Following the wound closure, rats were injected intravenously with 1 mL of 5 mg mL−1 DiD-loaded PNPs in 10% sucrose. At specified time points following the injection, animals were euthanized by CO2 inhalation. After perfusion with PBS, organs including heart, lung, liver, spleen, kidney, gut, blood, and aortic branch including both the left and right carotid arteries were carefully collected and homogenized for biodistribution analysis. The overall PNP distribution at the aortic branch was visualized using Keyence BZ-X700 fluorescence microscope. To examine the local distribution of PNP, damaged and undamaged arteries were cut longitudinally and stained with DAPI solution. En face examination was done on the luminal surfaces of denuded and intact area for the binding of PNPs. Sequences of images along z-axis (0.5 µm per section) from the intima to media layers of the carotid arteries were acquired with and Olympus ix81 fluorescence microscope. The 3D reconstruction of arterial wall from multisectional images was performed using Image J. To analyze the PNP retention, damaged and undamaged arteries collected at specified time points were homogenized, and their respective fluorescence was normalized to the liver fluorescence for comparison. All replicates represent different rats subjected to the same treatment (n=6).
Treatment of experimental coronary restenosis
Sprague-Dawley rats following angioplasty-induced arterial denudation were randomly placed into groups. The nanoparticle treatment group was injected intravenously with 1 mL of 5 mg mL−1 docetaxel-loaded PNPs in 10% sucrose at a docetaxel dose of 0.3 mg kg−1 on day 0 and day 5. As controls, animals receiving PBS, free docetaxel, and empty PNPs were prepared. On day 14, animals were euthanized with an overdose of ketamine-xylazine cocktail and perfused with PBS and 4% paraformaldehyde (PFA) at a pressure of 120–140 mm Hg. Segments of left and right carotid arteries were carefully dissected out, and the PFA-fixed carotid arteries were embedded with Tissue-Tek OCT compound (VWR International) in a tissue base mold and slowly submerged into pre-chilled 2-methyl butane until frozen completely. The frozen tissue block was then immediately stored at −80°C until sectioning. Serial sectioning (15 µm per section) was performed with a Cyrotome cryostat machine (Leica), and the tissue sections were placed on polylysine-treated glass slides. Tissue sections on slides were dried at room temperature for 30 min before staining. For immunohistochemistry, frozen sections on slides were first washed with PBS to remove residual OCT medium and then subjected to standard hematoxylin and eosin (H&E) staining. Areas of intima and media were analyzed using Image J. Luminal obliteration is defined as the intima area / the area within the internal elastic lamina. Statistical analysis was performed using one-way ANOVA. Sample sizes were estimated prior to the study to give statistically interpretable data. Studies were done in a non-blinded fashion. All replicates represent different rats subjected to the same treatment (n=6).
Staphylococcus aureus (MRSA252) bacteria adherence study
MRSA252 obtained from the American Type Culture Collection was cultured on tryptic soy broth (TSB) agar (Becton, Dickinson and Company) overnight at 37 °C. A single colony was inoculated in TSB medium at 37 °C in a rotary shaker. Overnight culture was refreshed in TSB medium at 1:100 dilution at 37 °C under shaking for another 3 h until OD600 of the culture medium reached approximately 1.0 (logarithmic growth phase). The bacteria were harvested by centrifugation at 5,000 × g for 10 min and then washed with sterile PBS twice and then fixed with 10% formalin for 1 h. The fixed bacteria were washed with sterile PBS and suspended in 10% sucrose to a concentration of 1 × 108 CFU mL−1. For the nanoparticle adhesion study, aliquots of 0.8 mL of 1 × 108 CFU mL−1 MRSA252 were mixed with 1.2 mL of 200 µg mL−1 DiD-loaded PNPs, RBCNPs, or bare NPs in 10% sucrose for 10 min at room temperature. The bacteria were then isolated from unbound nanoparticles by repeated centrifugal washes in sucrose solution at 5,000 × g. The purified bacteria were then suspended in 10% sucrose for replicate measurements (n=3) by flow cytometric analysis and SEM imaging.
Antimicrobial efficacy study
For in vitro antimicrobial efficacy study, 5 × 106 CFU of MRSA252 was mixed with 500 µL of 20 mg mL−1 nanoparticles (4 wt% vancomycin loading) in saline. As controls, equivalent amount of bacteria was incubated in either PBS or free vancomycin (0.8 mg mL−1). Following 10 min of incubation, bacteria were isolated from the solution via centrifugation at 2500 × g for 5 min. The collected bacteria pellet was resuspended with 500 µL of TSB culture medium and incubated for 5 h. The resulting samples were serially diluted in PBS and spotted on TSB agar plates. Following 24 h of culturing, the colonies were counted to determine the bacteria count in each sample. Replicates represent separate bacterial aliquots incubated with the same formulation (n=3).
For in vivo antimicrobial efficacy study, vancomycin-loaded PNPs (PNP-Vanc) and vancomycin-loaded RBCNPs (RBCNP-Vanc) were suspended in 10% sucrose solution at 31.25 mg mL−1 (4 wt% vancomycin loading). An equivalent concentration of free vancomycin (1.25 mg mL−1) was also suspended in 10% sucrose. Male CD-1 mice (Harlan Laboratories) weighing ~25 g were challenged intravenously with 6 × 106 CFU of MRSA252 suspended in 100 µL of PBS. 30 min following the bacteria injection, mice were randomly placed into separate groups and injected with 200 µL of PNP-Vanc, RBCNP-Vanc, free vancomycin (daily dosage: 10 mg kg−1 vancomycin), or PBS. To compare to the clinical dosing of vancomycin, a control group treated with twice daily dosing of 30 mg kg−1 free vancomycin was prepared (total daily dosage: 60 mg kg−1 vancomycin). The mice received their corresponding treatments from day 0 to 2. On day 3, blood was collected from the submandibular vein. The mice were then sacrificed, perfused with PBS, and their organs were collected. The organs were homogenized using a Biospec Mini Beadbeater in 1 mL of PBS for 1 min, serially diluted in PBS by 10-fold, and plated onto agar plates with spotting volume of 50 µL. Following 48 h of culture, bacterial colonies were counted to determine the bacterial load in each organ. Under the given experimental conditions, the detection limit was determined to be approximately 20 CFU per organ. Data points on the X-axis represent samples with no detectable bacterial colony. It was confirmed that samples prepared from unchallenged mice had no detectable colonies. The data was tested for normal distribution via the Shapiro-Wilk test. For blood and heart, which contained non-normal distributions, statistical analysis was performed using Kruskal-Wallis test. For the other organs, in which all groups were normally distributed and variance criteria were met, statistical analysis was performed using one-way ANOVA. Grubbs’ test was used to detect and remove statistical outliers. Sample sizes were estimated prior to the study to give statistically interpretable data. Studies were done in a non-blinded fashion. Replicates represent different mice subjected to the same treatment (n=14).
Extended Data
Extended Data Fig. 1. Schematic preparation of PNPs.
(a) Poly(lactic-co-glycolic acid) (PLGA) nanoparticles are enclosed entirely in plasma membrane derived from human platelets. The resulting particles possess platelet-mimicking properties for immunocompatibility, subendothelium binding, and pathogen adhesion. (b) Schematic depicting the process of preparing PNPs.
Extended Data Fig. 2. PNP preparation and storage.
(a) Isolation of platelet rich plasma (PRP) was achieved via centrifugation at 100 × g. PRP was collected from the top layer (yellow) separated from the red blood cells (red, bottom layer). (b) Collected human platelets under light microscopy, which possess a distinctive morphology from (c) red blood cells. Scale bars = 10 µm. (d) Transmission electron micrographs of platelet membrane vesicles and (e) PNPs, both of which were negatively stained with 1% uranyl acetate. Scale bars = 200 nm. (f) Dynamic light scattering measurements of PNPs in 10% sucrose show that the particles retain their size and stability following a freeze-thaw cycle and re-suspension upon lyophilization (n=3). Bars represent means ± SD. (g) Transmission electron micrograph shows retentions of PNPs’ core-shell structure following a freeze-thaw cycle in 10% sucrose. Scale bar = 100 nm. (h) Transmission electron micrograph shows retentions of PNPs’ core-shell structure upon resuspension following lyophilization in 10% sucrose. Scale bar = 100 nm.
Extended Data Fig. 3. Overall protein content on PNPs resolved by western blotting.
Primary platelet membrane protein/protein subunits including CD47, CD55, CD59, αIIb, α2, α5, α6, β1, β3, GPIbα, GPIV, GPV, GPVI, GPIX, and CLEC-2 were monitored in platelet rich plasma, platelets, platelet vesicles, and PNPs. Platelets derived from four different protocols, including commercial blood anti-coagulated in EDTA, freshly drawn blood anti-coagulated in EDTA, freshly drawn blood anti-coagulated in heparin, and transfusion-grade platelet rich plasma anti-coagulated in acid-citrate-dextrose (ACD), were examined to compare the membrane protein expression. Each sample was normalized to equivalent overall protein content prior to western blotting. It was observed that the PNP preparation resulted in membrane protein retention and enrichment very similar across the different platelet sources.
Extended Data Fig. 4. Platelet membrane sidedness on PNPs.
(a) Transmission electron micrograph of PNPs primary stained with anti-CD47 (intracellular), secondary stained with immunogold, and negatively stained with 2% vanadium. The immunogold staining revealed presence of intracellular CD47 domains on collapsed platelet membrane vesicles but not on PNPs. (b) Transmission electron micrograph of PNPs primary stained with anti-CD47 (extracellular), secondary stained with immunogold, and negatively stained with 2% vanadium. PNPs are shown to display extracellular CD47 domains. All scale bars = 100 nm. (c) 2 µm polystyrene beads were functionalized with anti-CD47 against the protein’s extracellular domain, anti-CD47 against the protein’s intracellular domain, or a sham antibody. Flow cytometric analysis of the different beads following DiD-loaded PNP incubation shows the highest particle retention to beads functionalized with anti-CD47 against the protein’s extracellular domain. (d) Normalized fluorescence intensity of PNP retention to the different antibody-functionalized beads. Bars represent means ± SEM.
Extended Data Fig. 5. PNP binding to collagen and extracellular matrix.
(a–f) Collagen-coated tissue culture slides seeded with human umbilical vein endothelial cells (HUVECs) were incubated with PNP solution for 30 sec. Fluorescence microscopy samples demonstrate differential PNP adherence to exposed collagen versus covered endothelial surfaces. (a–c) Representative fluorescence images visualizing DiD-loaded PNPs (red), cellular cytosol (green), and cellular nuclei (blue). (d–f) Images showing only the red and blue channels in order to highlight the differential localization of PNPs. Scale bar = 10 µm. (g) Fluorescence quantification of PNP per unit area on collagen and endothelial surfaces. Bars represent means ± SD (n=10). (h,i) PNP adherence to arterial extracellular matrix (ECM) as visualized by SEM. (h) SEM images of the ECM of a decellularized human umbilical cord artery. Left: Scale bar = 1 µm; Right: Scale bar = 500 nm. (i) SEM images of the ECM of a decellularized human umbilical cord artery following PNP incubation. Left: Scale bar = 1 µm; Right: Scale bar = 500 nm.
Extended Data Fig. 6. Pharmacokinetics, biodistribution, and safety of PNPs.
(a) DiD-loaded PNPs were injected intravenously through the tail vein of Sprague-Dawley rats. At various time points blood was withdrawn via tail vein blood sampling for fluorescence quantification to evaluate the systemic circulation lifetime of the nanoparticles (n=6). (b) Biodistribution of the PNP nanoparticles in balloon-denuded Sprague-Dawley rats at 2 h and 48 h following intravenous nanoparticle administration through the tail vein (n=6). (c) Comprehensive metabolic panel of rats following injections with human-derived PNPs and PBS (n=6). The rats received intravenous injections of PNPs and PBS on day 0 and day 5, and the blood test conducted on day 10 did not reveal significant changes between the two groups, indicating normal liver and kidney functions following the PNP administration. All bars and markers represent means ± SD.
Extended Data Fig. 7. PNP targeting of damaged vasculatures upon intravenous injection to rats with angioplasty-induced arterial denudation.
(a) Fluorescence microscopy of the aortic branch revealed selective PNP binding to the denuded artery (right) as opposed to the undamaged artery (left) (PNP fluorescence in red). (b) Fluorescence images acquired from the control artery, which did not reveal PNP fluorescence upon focusing on either the endothelium (top) or the smooth muscle layer (bottom) (nuclei in blue). (c) Fluorescence images acquired from the denuded artery, which revealed significant PNP retention as fluorescent punctates (PNP fluorescence in red) above the smooth muscle layer. (d) Fluorescence image of arterial cross-section acquired from the control artery, which showed nuclei of endothelial cells above the collagen layer (autofluorescence in green) and an absence of PNP fluorescence. (e) Fluorescence image of arterial cross-section acquired from the denuded artery, which showed PNP retention as fluorescent punctates on the collagen layer (PNP fluorescence in red; collagen autofluorescence in green) and an absence of endothelial cell nuclei. All scale bars = 100 µm.
Extended Data Fig. 8. Characterizations of drug-loaded cell membrane cloaked nanoparticles.
(a) Physicochemical properties of drug-loaded cell membrane cloaked nanoparticles. (b) TEM visualization of docetaxel-loaded PNPs (PNP-Dtxl). Scale bar = 200 nm. (c) Drug release profile of PNP-Dtxl as compared to PEG-PLGA diblock nanoparticles of equivalent size and docetaxel loading (n=3). (d) TEM visualization of vancomycin-loaded PNPs (PNP-Vanc). Scale bar = 200 nm. (e) Drug release profiles of PNP-Vanc and RBCNP-Vanc (n=3). Bars represent means ± SD.
Extended Data Fig. 9. Treatment of an experimental rat model of coronary restenosis.
H&E-stained arterial cross-sections reveal the vascular structure of non-damaged arteries (serving as baseline) and denuded arteries following treatment with PNP-Dtxl, PBS, PNP with no docetaxel content, or free docetaxel. Scale bar = 200 µm.
Extended Data Fig. 10. PNP adherence to MRSA252 bacteria.
(a) Flow cytometric analysis of MRSA252 bacteria following incubation with different DiD-loaded nanoformulations. (b) Pellets of MRSA252 following incubation with DiD-loaded RBCNPs (left) and DiD-loaded PNPs (right) show differential retention of nanoformulation with MRSA252 upon pelleting of the bacteria. (c) A pseudocolored SEM image of PNPs binding to MRSA252 under high magnification (MRSA colored in gold, PNP colored in orange). Scale bar = 400 nm.
Acknowledgments
This work is supported by the National Institutes of Health under Award Numbers R01DK095168 (L.Z), R01HL108735 (S.C) and R01EY25090 (K.Z), and partially by the Defense Threat Reduction Agency Joint Science and Technology Office for Chemical and Biological Defense under Grant Number HDTRA1-14-1-0064 (L.Z). R.F. is supported by a National Institutes of Health R25CA153915 training grant from the National Cancer Institute.
Footnotes
Author contributions
C-M.H., R.F., K-C.W., B.L, K.Z., S.C. and L.Z. conceived and designed the experiments; C-M.H., R.F., K-C.W., B.L, S.T., D.D., P.N., P.A., C.W., A.K., C.C., V.Q., M.R., S.P., J.Z., W.S. F.H., T.C., and W.G. performed all the experiments. The manuscript was written by C-M.H., R.F., B.L, W.G., and L.Z. All authors discussed the results and reviewed the manuscript.
Author information
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper. Correspondence and requests for materials should be addressed to L.Z. (zhang@ucsd.edu), S.C. (shuchien@ucsd.edu) or K.Z. (kang.zhang@gmail.com).
References
- 1.Pelaz B, et al. Interfacing engineered nanoparticles with biological systems: anticipating adverse nanobio interactions. Small. 2013;9:1573–1584. doi: 10.1002/smll.201201229. [DOI] [PubMed] [Google Scholar]
- 2.Salvati A, et al. Transferrin-functionalized nanoparticles lose their targeting capabilities when a biomolecule corona adsorbs on the surface. Nature Nanotech. 2013;8:137–143. doi: 10.1038/nnano.2012.237. [DOI] [PubMed] [Google Scholar]
- 3.Tenzer S, et al. Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nature Nanotech. 2013;8:772–781. doi: 10.1038/nnano.2013.181. [DOI] [PubMed] [Google Scholar]
- 4.Born GV, Cross MJ. The aggregation of blood platelets. J. Physiol. 1963;168:178–195. doi: 10.1113/jphysiol.1963.sp007185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kieffer N, Phillips DR. Platelet membrane-glycoproteins - functions in cellular interactions. Annu. Rev. Cell Biol. 1990;6:329–357. doi: 10.1146/annurev.cb.06.110190.001553. [DOI] [PubMed] [Google Scholar]
- 6.Fitzgerald JR, Foster TJ, Cox D. The interaction of bacterial pathogens with platelets. Nat. Rev. Microbiol. 2006;4:445–457. doi: 10.1038/nrmicro1425. [DOI] [PubMed] [Google Scholar]
- 7.Yeaman MR. Platelets in defense against bacterial pathogens. Cell. Mol. Life. Sci. 2010;67:525–544. doi: 10.1007/s00018-009-0210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peters D, et al. Targeting atherosclerosis by using modular, multifunctional micelles. Proc. Natl. Acad. Sci. U.S.A. 2009;106:9815–9819. doi: 10.1073/pnas.0903369106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan JM, et al. Spatiotemporal controlled delivery of nanoparticles to injured vasculature. Proc. Natl. Acad. Sci. U.S.A. 2010;107:2213–2218. doi: 10.1073/pnas.0914585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bertram JP, et al. Intravenous hemostat: nanotechnology to halt bleeding. Sci. Trans. Med. 2009;1:11ra22. doi: 10.1126/scitranslmed.3000397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Modery-Pawlowski CL, et al. Approaches to synthetic platelet analogs. Biomaterials. 2013;34:526–541. doi: 10.1016/j.biomaterials.2012.09.074. [DOI] [PubMed] [Google Scholar]
- 12.Simberg D, et al. Biomimetic amplification of nanoparticle homing to tumors. Proc. Natl. Acad. Sci. U.S.A. 2007;104:932–936. doi: 10.1073/pnas.0610298104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anselmo AC, et al. Platelet-like nanoparticles: mimicking shape, flexibility, and surface biology of platelets to target vascular injuries. ACS Nano. 2014 doi: 10.1021/nn503732m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olsson M, Bruhns P, Frazier WA, Ravetch JV, Oldenborg PA. Platelet homeostasis is regulated by platelet expression of CD47 under normal conditions and in passive immune thrombocytopenia. Blood. 2005;105:3577–3582. doi: 10.1182/blood-2004-08-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sims PJ, Rollins SA, Wiedmer T. Regulatory control of complement on blood-platelets - modulation of platelet procoagulant responses by a membrane inhibitor of the C5b-9 Complex. J. Biol. Chem. 1989;264:19228–19235. [PubMed] [Google Scholar]
- 16.Nieswandt B, Watson SP. Platelet-collagen interaction: is GPVI the central receptor? Blood. 2003;102:449–461. doi: 10.1182/blood-2002-12-3882. [DOI] [PubMed] [Google Scholar]
- 17.Hu CM, et al. Erythrocyte membrane-camouflaged polymeric nanoparticles as a biomimetic delivery platform. Proc. Natl. Acad. Sci. U.S.A. 2011;108:10980–10985. doi: 10.1073/pnas.1106634108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu CM, Fang RH, Copp J, Luk BT, Zhang L. A biomimetic nanosponge that absorbs pore-forming toxins. Nature Nanotech. 2013;8:336–340. doi: 10.1038/nnano.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu CM, Fang RH, Luk BT, Zhang L. Nanoparticle-detained toxins for safe and effective vaccination. Nature Nanotech. 2013;8:933–938. doi: 10.1038/nnano.2013.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gachet C, et al. Alpha IIb beta 3 integrin dissociation induced by EDTA results in morphological changes of the platelet surface-connected canalicular system with differential location of the two separate subunits. J. Cell. Biol. 1993;120:1021–1030. doi: 10.1083/jcb.120.4.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luk B, et al. Interfacial interactions between natural RBC membranes and synthetic polymeric nanoparticles. Nanoscale. 2013;6:2730–2737. doi: 10.1039/c3nr06371b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hughes CE, et al. CLEC-2 activates Syk through dimerization. Blood. 2010;115:2947–2955. doi: 10.1182/blood-2009-08-237834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalluri R. Basement membranes: structure, assembly and role in tumour angiogenesis. Nature Rev. Cancer. 2003;3:422–433. doi: 10.1038/nrc1094. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez PL, et al. Minimal “self” peptides that inhibit phagocytic clearance and enhance delivery of nanoparticles. Science. 2013;339:971–975. doi: 10.1126/science.1229568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Law SKA, Dodds AW. The internal thioester and the covalent binding properties of the complement proteins C3 and C4. Protein Sci. 1997;6:263–274. doi: 10.1002/pro.5560060201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Terstappen LWMM, Nguyen M, Lazarus HM, Medof ME. Expression of the DAF (CD55) and CD59 antigens during normal hematopoietic-cell differentiation. J. Leukocyte Biol. 1992;52:652–660. doi: 10.1002/jlb.52.6.652. [DOI] [PubMed] [Google Scholar]
- 27.Andersen AJ, Hashemi SH, Andresen TL, Hunter AC, Moghimi SM. Complement: alive and kicking nanomedicines. J. Biomed. Nanotech. 2009;5:364–372. doi: 10.1166/jbn.2009.1045. [DOI] [PubMed] [Google Scholar]
- 28.Siboo IR, Chambers HF, Sullam PM. Role of SraP, a serine-rich surface protein of Staphylococcus aureus, in binding to human platelets. Infect. Immun. 2005;73:2273–2280. doi: 10.1128/IAI.73.4.2273-2280.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamaly N, et al. Development and in vivo efficacy of targeted polymeric inflammation-resolving nanoparticles. Proc. Natl. Acad. Sci. U.S.A. 2013;110:6506–6511. doi: 10.1073/pnas.1303377110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu CM, et al. ‘Marker-of-self’ functionalization of nanoscale particles through a top-down cellular membrane coating approach. Nanoscale. 2013;5:2664–2668. doi: 10.1039/c3nr00015j. [DOI] [PMC free article] [PubMed] [Google Scholar]