Abstract
Although the predominant effect of host restriction APOBEC3 proteins on HIV-1 infection is to block viral replication, they might inadvertently increase retroviral genetic variation by inducing G-to-A hypermutation. Numerous studies have disagreed on the contribution of hypermutation to viral genetic diversity and evolution. Confounding factors contributing to the debate include the extent of lethal (stop codon) and sublethal hypermutation induced by different APOBEC3 proteins, the inability to distinguish between G-to-A mutations induced by APOBEC3 proteins and error-prone viral replication, the potential impact of hypermutation on the frequency of retroviral recombination, and the extent to which viral recombination occurs in vivo, which can reassort mutations in hypermutated genomes. Here, we determined the effects of hypermutation on the HIV-1 recombination rate and its contribution to genetic variation through recombination to generate progeny genomes containing portions of hypermutated genomes without lethal mutations. We found that hypermutation did not significantly affect the rate of recombination, and recombination between hypermutated and wild-type genomes only increased the viral mutation rate by 3.9 × 10−5 mutations/bp/replication cycle in heterozygous virions, which is similar to the HIV-1 mutation rate. Since copackaging of hypermutated and wild-type genomes occurs very rarely in vivo, recombination between hypermutated and wild-type genomes does not significantly contribute to the genetic variation of replicating HIV-1. We also analyzed previously reported hypermutated sequences from infected patients and determined that the frequency of sublethal mutagenesis for A3G and A3F is negligible (4 × 10−21 and1 × 10−11, respectively) and its contribution to viral mutations is far below mutations generated during error-prone reverse transcription. Taken together, we conclude that the contribution of APOBEC3-induced hypermutation to HIV-1 genetic variation is substantially lower than that from mutations during error-prone replication.
Author Summary
HIV-1 populations exhibit high levels of genetic variation, allowing the virus to escape immunological and pharmacological selection pressures, and thwarting efforts to develop an effective vaccine; thus, it is critical to understand all sources of HIV-1 genetic diversity. In addition to error-prone reverse transcription and frequent recombination, APOBEC3 proteins, expressed as part of the host intracellular antiviral defense mechanism, could potentially contribute to viral genetic variation through sublethal hypermutation or rescue of hypermutated portions of the genomes without lethal mutations through recombination. Here, we show that hypermutation by APOBEC3 proteins does not affect the rate of recombination; in addition, even in proviruses generated from heterozygous virions containing wild-type and hypermutated genomes, hypermutation only modestly increased the viral mutation rate by twofold. Since the frequency of copackaged wild-type and hypermutated genomes in patients is extremely low, we conclude that hypermutation does not significantly contribute to HIV-1 genetic variation. Analysis of hypermutated sequences from infected patients indicates that the frequency of sublethal mutagenesis is negligible and its contribution to viral variation is substantially lower than that of error-prone viral replication.
Introduction
Human immunodeficiency virus type 1 (HIV-1) undergoes continuous evolution and adaptation to its host environment resulting in high genetic variation which allows the virus to escape immune responses or to acquire drug resistance [1]. This genetic diversity is generated by three key factors. First, HIV-1, like other retroviruses, has a high mutation rate which has been measured to be between 1.4–3.4 × 10−5 mutations/bp/replication cycle [2–6]. This is mainly attributable to error-prone reverse transcriptase (RT) [7] and a minor contribution by RNA polymerase II [3, 8–11]. Second, HIV-1 copackages two viral RNA genomes per virion, allowing recombination during DNA synthesis that reassorts mutations in the copackaged genomes and leads to a further increase in genetic diversity. For HIV-1, recombination rates of ~9 template switches/genome/single cycle have been measured [12, 13]. Third, HIV-1 produces on average 1011 virions/day/patient, with ~107–108 productively infected CD4+ T cells [1, 14–17], which provides a large population of variants and hence great evolutionary potential.
In 2002, a cellular host protein APOBEC3G (apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G) was identified, which was able to inhibit HIV-1 infection in the absence of the virally-encoded protein Vif (virion infectivity factor) [18]. Some members of the APOBEC3 (A3) family of proteins are potent viral restriction factors and serve as parts of the host’s innate antiviral cellular defense. Among the seven A3 family members, APOBEC3G (A3G), APOBEC3F (A3F), APOBEC3D (A3D), and APOBEC3H (A3H) hapolotypes II, V and VII are packaged into virions in producer cells in the absence of vif. These A3s largely contribute to the inactivation of HIV-1Δvif by causing cytidine deamination (cytosine-to-uracil) during reverse transcription of the newly synthesized minus-strand cDNA in the infected target cells [19, 20]. This process results in extensive guanine-to-adenine (G-to-A) mutations in the viral double-stranded DNA genome, called hypermutation [6], which introduces substitutions and stop codons that often lead to the formation of replication-defective proviruses. Lethal mutagenesis of HIV-1 by A3 proteins can be observed in a single round of viral replication [21–23]. In addition, A3 proteins have also been shown to block the HIV-1 life cycle through non-editing mechanisms, as catalytic site mutants are still able to retain some antiviral activity [24, 25]. A3 proteins can inhibit reverse transcription by blocking RT template binding, reducing tRNA binding and processing, inhibiting strand transfer events, and blocking cDNA synthesis elongation [24–33]. Furthermore, A3G and A3F have been shown to inhibit integration by interfering with tRNA primer removal or blocking the 3’ processing of viral DNA ends by integrase, respectively [32, 34]. HIV-1 has evolved to protect itself against A3 proteins by expressing Vif, which targets the A3 proteins for degradation through the ubiquitin-proteasome pathway [35–41]. This in turn excludes the A3 proteins from being encapsidated into viral particles, leading to a productive viral infection.
Analysis of patient proviral sequences has shown the presence of G-to-A hypermutation, indicating that Vif is not always completely successful at degrading the A3 proteins. Depending upon the study, ~25% (9–43%) of patient-derived proviral sequences are hypermutated [42–46]. A3 proteins are characterized by their preference to introduce cytidine deamination in certain dinucleotide motifs: 5’GG→AG for A3G and 5’GA→AA for A3F, A3D, or A3H [21, 47–54]. Although G-to-A mutations in hypermutated proviral sequences found in patients exhibit specific sequence contexts, these analyses are complicated by mutations introduced by RT. Studies of RT fidelity ex vivo have identified that one of the major substitutions introduced by RT is also G-to-A; however the sequence contexts in which these mutations occur have not been fully defined [2–6, 55–57].
Currently, the extent to which hypermutated genomes in HIV-1 infected patients contribute to viral genetic variation and evolution is being debated and has not been clearly determined. It has been proposed that A3 sublethal mutagenesis has the potential to contribute to viral variation and the emergence of drug resistance mutations [44, 58–67]. However other studies have proposed that A3-induced hypermutation is an “all or nothing” phenomenon [64], failed to find a correlation between hypermutation and emergence of drug resistance mutations [60], and observed purifying selection such that viral RNA incorporated into the virion will contain little to no hypermutation [68].
Therefore, the role of hypermutation on the HIV-1 life cycle and its contribution to genetic diversity are currently unclear. To address these important questions, we determined the effect of hypermutation on recombination and the contribution of hypermutation to the HIV-1 mutation rate per replication cycle. These studies clarify the role of APOBEC3 proteins in HIV genetic variation and significantly contribute to the resolution of a long-debated question in HIV biology.
Results
HIV-1 constructs containing A3G- and A3F-induced hypermutation
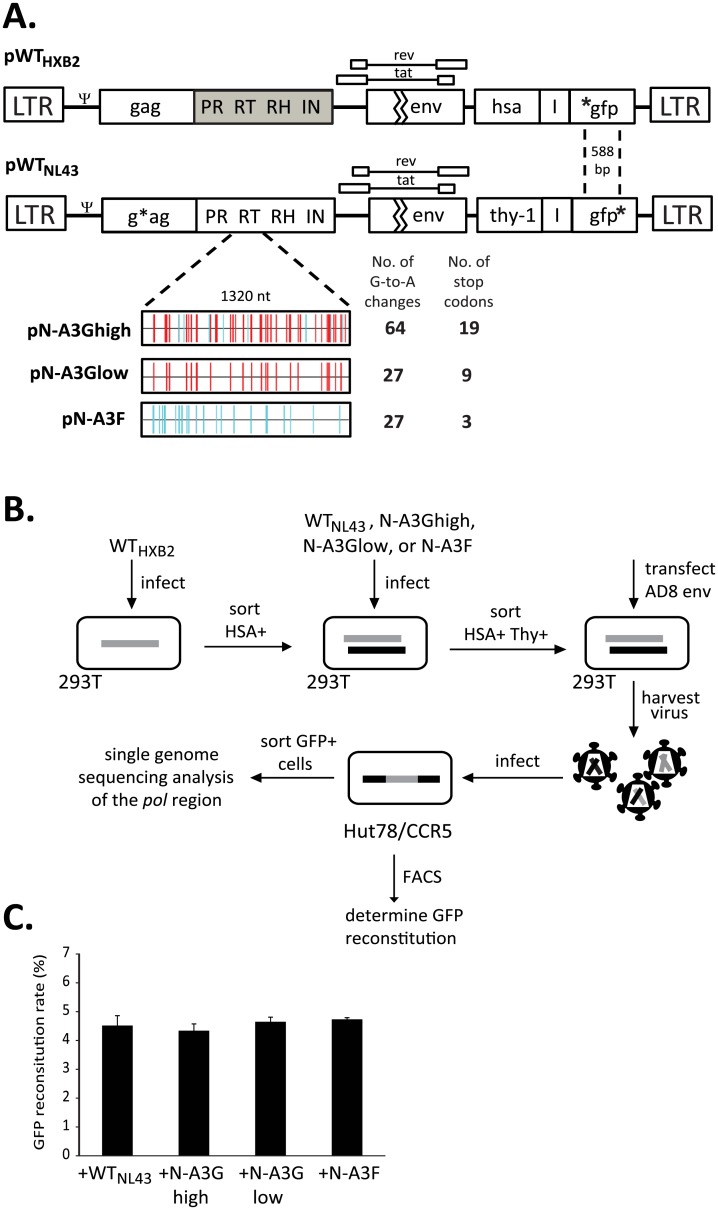
To study the effects of hypermutation on HIV-1 recombination and mutation rates per replication cycle, we generated five HIV-1 constructs (Fig 1A). These constructs are based on HIV-1 NL4-3 (GenBank AF324493.2) and contain the necessary cis-acting elements required for virus expression and replication. They also contain gag and pol genes, and express functional Tat and Rev, but contain inactivating deletions in vif, vpu, vpr, and env. In the construct pWTHXB2, the NL4-3 pol sequence has been replaced with the pol sequence from the HXB2 isolate; in addition, it also expresses from the nef open reading frame, the mouse heat stable antigen (hsa) followed by the internal ribosomal entry site (IRES) from encephalomyocarditis virus and green fluorescent protein gene (gfp) with an inactivating mutation at the 5’ end of the gene.
Fig 1. Experimental system to study the contribution of hypermutation to HIV-1 recombination and genetic variation.
(A) Plasmids pWTHXB2 and pWTNL43 are HIV-1-based constructs expressing hsa-IRES-gfp or Thy-IRES-gfp, respectively, from the nef open reading frame. For pWTHXB2, the pol region is replaced with HXB2 pol sequence (gray shaded region); gfp in the nef open reading frame contains an inactivating mutation at the 5’ end of gfp (*gfp). For pWTNL43, the pol region remains NL4-3 and gfp in the nef open reading frame contains an inactivating mutation at the 3’ end of gfp (gfp*). The inactivating mutations in gfp are 588 bp apart. All constructs contain inactivating mutations in vif, vpu, vpr and env. * denotes inactivating mutations in gfp and gag genes. Constructs derived from pWTNL43 contain a modified RT region hypermutated by A3G (pN-A3Ghigh or pN-A3Glow), or by A3F (pN-A3F). The number of G-to-A changes relative to pWTNL43 and number G-to-A changes leading to a stop codon is shown on the right. Red vertical lines indicate GG→AG mutations, while blue vertical lines indicate GA→AA mutations (Hypermut [102]; www.hiv.lanl.gov). (B) Single cycle recombination system to study the effects of hypermutation. Briefly, 293T cells were infected with WTHXB2-dervied viruses at a low MOI, and HSA+ cells were enriched by sorting. These HSA+ cells were then infected at a low MOI with virus derived from either WTNL43, N-A3Ghigh, N-A3Glow, or N-A3F and sorted for HSA+/Thy+ cells. The respective four cell lines were then transfected with a plasmid expressing HIV-1 Env from the AD8 strain; virus was harvested and used to infect Hut78/CCR5 target cells. The infected cells were used for flow cytometry analysis or sorted for GFP+ cells to be used in single genome sequencing analysis. (C) Rate of GFP reconstitution for each of the four vector pairs after infection into Hut78/CCR5 target cells. Average of 2 independent infections. Paired two-sample t-tests of comparison of each group to WT indicated no significant differences (P > 0.5).
The remaining four constructs contain the NL4-3 pol sequences: either completely wild type (pWTNL43), or a pol sequence in which the RT region contains 64 G-to-A changes (pN-A3Ghigh) or 27 G-to-A changes (pN-A3Glow) at A3G target sites, or contains 27 G-to-A changes (pN-A3F) at A3F target sites. These G-to-A changes are relative to pWTNL43, and the 64 G-to-A changes are high while 27 mutations are low, relative to an average of 42 G-to-A mutations estimated in the same RT region of hypermutated proviral sequences from patients [69]; analysis of hypermutated proviral sequences used to estimate the number of G-to-A mutations in the RT region is discussed later in Results Section. The 64, 27, and 27 G-to-A changes in pN-A3Ghigh, pN-A3Glow, and pN-A3F resulted in 19, 9, and 3 stop codon mutations, respectively. In addition, these four constructs contain in nef a mouse CD90.2 gene (thy) followed by IRES and gfp with an inactivating mutation at the 3’ end of the gene. All thy-containing constructs also contain a frameshift mutation resulting in a premature stop codon in gag to prevent expression of the Gag/Gag-Pol polyproteins; hence, functional Gag and Gag-Pol are only produced from WTHXB2.
Creation of 293T producer cells expressing wild-type and hypermutated proviruses to determine effect of hypermutation on recombination
To study recombination, we generated four producer 293T cell lines, each containing a WTHXB2 provirus and either WTNL43, N-A3Ghigh, N-A3Glow or N-A3F provirus (Fig 1B). These cell lines were generated by sequential infections at low multiplicity of infection (MOI) followed by multiple rounds of cell sorting to ensure that the majority of cells in each cell line contained a single integrated copy of each parental provirus. More than 97% of the cells in the producer cell lines expressed both HSA and Thy markers, with each cell line representing a pool of at least 115,000 independent infection events.
None of the aforementioned constructs express functional HIV-1 Env; to produce infectious viruses, we transfected the four cell lines (WTHXB2/WTNL43, WTHXB2/N-A3Ghigh, WTHXB2/N-A3Glow, and WTHXB2/N-A3F) with a plasmid expressing CCR5-tropic HIV-1 envelope from the AD8 strain [70]. Since 293T cells lack the CD4 receptor, the viruses containing the AD8 Env cannot reinfect the producer cells. We then used the resulting AD8 Env containing viruses to infect Hut78/CCR5 target cells at a low MOI (<0.08) in order to minimize the chance of double infection.
In the producer cells, the full-length RNAs from the two parental proviruses can be assorted randomly prior to being packaged into virions [71–73], resulting in the formation of homozygous virions (both RNAs from same parent) and heterozygous virions (two RNAs from different parents). During DNA synthesis, RT can switch templates between the copackaged RNAs to generate recombinants containing portions of both genomes [74, 75]. Although recombination can occur in all viruses, the two RNAs in the homozygous viruses contain the same inactivating mutation in gfp, and the resulting DNA will have a mutant gfp. In contrast, the two RNAs in the heterozygous viruses have different mutations in gfp, and recombination between the two mutations can reconstitute a functional gfp that confers a GFP+ phenotype to the target cell. Therefore, expression of a functional GFP in target cells can be used to identify proviruses derived from heterozygous virions.
Effect of hypermutation on the recombination rate
The HIV-1 constructs used in this system express either hsa or thy; thus target cells infected with these viruses would be HSA+ or Thy+. However, only recombinant proviruses generated from heterozygous particles can reconstitute a functional gfp and confer the GFP+ phenotype. Therefore, we used the frequency of the gfp reconstitution as a measurement for HIV-1 recombination rate. Flow cytometry analysis of the infected target Hut78/CCR5 population showed that the recombination rate, as measured by the reconstitution of gfp, was not significantly different for all four vector pairs: 4.5%, 4.3%, 4.6% and 4.7% for WTHXB2/WTNL43, WTHXB2/N-A3Ghigh, WTHXB2/N-A3Glow, and WTHXB2/N-A3F, respectively (p > 0.05; one-way ANOVA and paired two-sample t-tests) (Fig 1C). This result indicated that the presence of hypermutation in pol did not affect the frequency of recombination in gfp. Only the proviruses derived from heterozygous viruses can confer GFP+ in target cells; thus, these results also indicated that hypermutation in pol did not affect the copackaging efficiency of RNAs from two parental proviruses into the same viral particle.
Single genome sequencing of retroviral recombinants and determination of recombination junctions
To further analyze recombination events that occurred between wild-type and hypermutated sequences, we harvested viruses from the aforementioned four producer cell lines and infected target Hut/CCR5 cells in two sets of independent experiments. GFP+ cells, which were infected with heterozygous virions, were enriched through multiple rounds of cell sorting until >87% of cells were expressing GFP. Two pools of GFP+ cells were generated for each pair of parental viruses and each target cell pool contained at least 7,700 individual infection events that yielded GFP+ cells. Genomic DNA from GFP+ cell pools was isolated and subjected to single-genome sequencing (SGS) for the pol region, and the results from both GFP+ pools were combined. A total of 152, 140, 141, and 132 single genome sequences were recovered from WTHXB2/WTNL43, WTHXB2/N-A3Ghigh, WTHXB2/N-A3Glow, and WTHXB2/N-A3F samples, respectively. Within the 3096-nt stretch of pol that was sequenced, there are 96 distinct polymorphisms present between WTHXB2 and WTNL43, resulting in 97% nucleotide sequence identity. Within the 1320-nt RT region there are 37 distinct polymorphisms present between WTHXB2 and WTNL43 (97% sequence identity); due to the introduction of hypermutations in RT, there are 95, 62, and 62 polymorphisms present in RT between WTHXB2 and N-A3Ghigh, N-A3Glow and N-A3F, resulting in 92%, 95% and 95% sequence identity, respectively (Fig 2; see polymorphic site distribution).
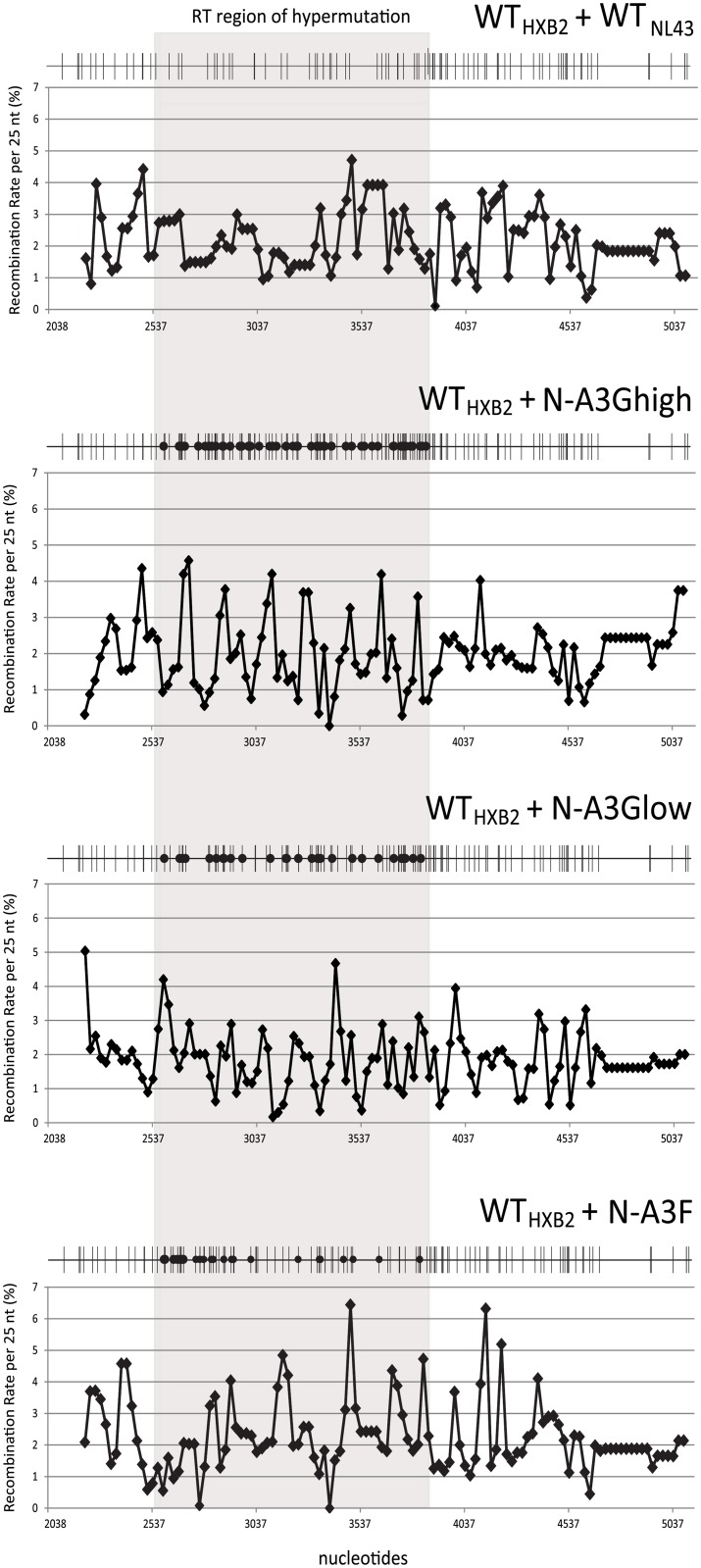
Fig 2. Single-cycle recombination rates across the target pol gene.
The recombination rate per 25-nt region is plotted for each pair of parent viruses. The RT region where hypermutation was introduced is shaded in gray. Nucleotide numbering corresponds to HXB2 (GenBank Accession number K03455). A total of 96, 154, 121, and 121 polymorphic sites are shown as black vertical lines between HXB2 and N-A3Ghigh, N-A3Glow, and N-A3F, respectively, whereas black circles indicate positions of G-to-A hypermutation in the RT region (Highlighter for Nucleotide Sequences v2.2.3 [103]: www.hiv.lanl.gov).
Using polymorphic sites as reference points and the sequences obtained from SGS, we determined the recombination junctions in the entire pol for each progeny recombinant. The average distance between marker sites in pol for WTHXB2/WTNL43, WTHXB2/N-A3Ghigh, WTHXB2/N-A3Glow, and WTHXB2/N-A3F is 31, 19, 24 and 24 nucleotides, respectively. At these marker distances, the probability of unobserved double-crossover events is extremely low. Most of the 565 recombinants recovered had a unique recombination pattern. The average number of crossovers per clone in pol (3 kb) was determined to be 2.4, 2.3, 2.1 and 2.7 for WTHXB2/WTNL43, WTHXB2/N-A3Ghigh, WTHXB2/N-A3Glow, and WTHXB2/N-A3F, respectively (Table 1). In comparison to WTHXB2/WTNL43, there were no significant differences in the numbers of crossovers in pol when one of the parental viruses had hypermutations (all p values > 0.05; Wilcoxon rank sum test). Further analysis of only the RT region where hypermutation was introduced, the average numbers of crossovers per clone was (1.3 kb) were 1.1, 1.0, 1.0 and 1.2 for WTHXB2/WTNL43, WTHXB2/N-A3Ghigh, WTHXB2/N-A3Glow, and WTHXB2/N-A3F, respectively (Table 2). These numbers were not significantly different from WTHXB2/WTNL43 or from each other (all p values > 0.05; Wilcoxon rank sum test), nor were they significantly different from the numbers of crossovers in a region of similar size from RNase H to integrase (1.2 kb) that was not hypermutated in comparison to WTHXB2/WTNL43: 1.0, 1.0, 0.9 and 1.0 for WTHXB2/WTNL43, WTHXB2/N-A3Ghigh, WTHXB2/N-A3Glow, and WTHXB2/N-A3F, respectively (all p values > 0.05; Wilcoxon rank sum test). Additionally, in all four vector pairs the 0.4-kb protease region also contained similar numbers of crossovers per clone in comparison to WTHXB2/WTNL43: 0.4, 0.3, 0.3, and 0.4 for, WTHXB2/WTNL43, WTHXB2/N-A3Ghigh, WTHXB2/N-A3Glow, and WTHXB2/N-A3F, respectively (all p values > 0.05; Wilcoxon rank sum test) (Table 2). Thus, hypermutation did not affect the average number of crossovers in a single replication cycle.
Table 1. Distribution and average number of crossovers in pol observed in recombinants.
| No. of Recombinants | ||||
|---|---|---|---|---|
| WTHXB2 + | WTHXB2 + | WTHXB2 + | WTHXB2 + | |
| No. of crossovers | WTNL43 | N-A3Ghigh | N-A3Glow | N-A3F |
| 0 | 19 | 19 | 24 | 16 |
| 1 | 35 | 29 | 32 | 30 |
| 2 | 36 | 39 | 37 | 25 |
| 3 | 31 | 25 | 23 | 22 |
| 4 | 14 | 9 | 9 | 15 |
| 5 | 5 | 9 | 7 | 12 |
| 6 | 5 | 6 | 7 | 6 |
| 7 | 2 | 3 | 2 | 3 |
| 8 | 3 | 1 | 0 | 1 |
| 9 | 1 | 0 | 0 | 0 |
| 10 | 0 | 0 | 0 | 2 |
| 11 | 1 | 0 | 0 | 0 |
| Total no. clones | 152 | 140 | 141 | 132 |
| Avg. no. of | ||||
| crossovers per clone* | 2.4 | 2.3 | 2.1 | 2.7 |
*The average number of crossovers per clone was calculated as follows: sum of [(number of crossovers) × (number of recombinants with that many crossovers)]/ total number of recombinant clones.
Table 2. Crossover analysis for recombinants throughout pol.
| Vector pair | Avg. no. of crossovers in pol (PR/RT/RNaseH/IN) | Avg. no. of crossovers in PR | Avg. no. of crossovers in RT | Avg. no. of Crossovers in RNaseH + IN |
|---|---|---|---|---|
| WTHXB2 + WTNL43 | 2.4 | 0.35 | 1.07 | 1.01 |
| WTHXB2 + N-A3Ghigh | 2.3 | 0.32 | 0.99 | 1.03 |
| WTHXB2 + N-A3Glow | 2.1 | 0.30 | 0.96 | 0.88 |
| WTHXB2 + N-A3F | 2.7 | 0.39 | 1.23 | 1.04 |
To further characterize the distribution of crossovers for each pair of viruses, we determined the number of recombination events in each region between two neighboring polymorphic sites. We then calculated the recombination rate/nucleotide/genome by dividing the observed events by the number of nucleotides between the two polymorphic sites and then by the number of genomes sequenced. Using the recombination rate of each nucleotide, we summed the rates for 25 nucleotides to generate the recombination rates per 25-nt segment. The results of these analyses are summarized in Fig 2 and the RT regions containing the hypermutations are shaded in grey. These results showed that recombination events can be observed throughout the pol gene; furthermore, crossovers occur throughout the RT regions regardless of the presence of hypermutations in the RT-coding region of one of the parents. Taken together, our results showed that once a genome was packaged into the virion, the presence of hypermutations in the RNA did not affect the frequency or the distribution of the crossovers events.
Contribution of hypermutation to the HIV-1 mutation rate
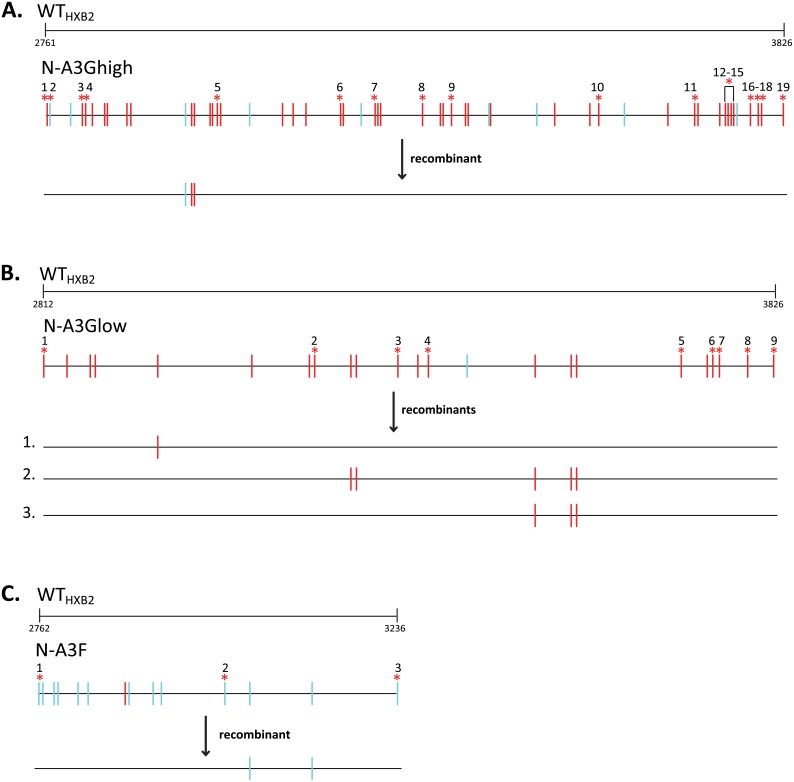
G-to-A substitutions generated by A3 proteins frequently introduce stop codons that lead to loss of expression or expression of aberrant viral proteins resulting in replication defects. Hypermutated sequences in vectors N-A3Ghigh, N-A3Glow and N-A3F contained 19, 9 and 3 stop codons, respectively, due to G-to-A hypermutations (Fig 1A). To determine if any recombinants during template switching acquired G-to-A changes, but not the “lethal” stop codons, we analyzed all recombinants for the presence of G-to-A changes between the first and last stop-codon mutations. Fig 3 depicts the G-to-A changes present between the 19, 9 and 3 stop codons for N-A3Ghigh (Fig 3A), N-A3Glow (Fig 3B) and N-A3F (Fig 3C), respectively. For WTHXB2/N-A3Ghigh, we recovered one out of 140 recombinants that acquired three G-to-A changes, but lacked stop codons (Table 3). For WTHXB2/N-A3Glow, three out of 141 recombinants acquired one, five and three G-to-A changes, respectively, and for WTHXB2/N-A3F one out of 132 recombinants acquired two G-to-A changes without stop codons. Therefore, out of a total of 413 recombinants analyzed (354,642 nts sequenced), 14 G-to-A mutations were rescued from hypermutated genomes without stop codons, and the overall contribution of hypermutation to the HIV-1 mutation rate was 3.9 × 10−5 mutations/bp/replication cycle. The mutation rate was slightly lower for the WTHXB2/N-A3Ghigh population (2.0 × 10−5/bp/replication cycle), and higher for the WTHXB2/N-A3Glow population (6.3 × 10−5/bp/replication cycle). Thus, the contribution of hypermutation to the HIV-1 mutation rate for a population of heterozygous virions containing a hypermutated genome and a nonhypermutated genome was similar to the retroviral mutation rate of 3.4 × 10−5/bp/replication cycle [2].
Fig 3. Recombinants with G-to-A changes after single-cycle recombination assay.
Distribution of G-to-A changes (relative to WTHXB2) for parental N-A3Ghigh (A), N-A3Glow (B), and N-A3F (C) pairs, and for resulting recombinants with non-lethal G-to-A changes. Shown are the G-to-A changes retained by each of the recombinants: one recombinant from WTHXB2/N-A3Ghigh, three recombinants from WTHXB2/N-A3Glow, and one recombinant from WTHXB2/N-A3F. A total of 14 G-to-A changes were observed in these 5 recombinants. G-to-A changes in the GG→AG and GA→AA dinucleotide context are shown as red and blue vertical lines, respectively (Hypermut [102]; www.hiv.lanl.gov), while lethal G-to-A changes that introduce stop codons are marked by * and numbered. Nucleotide numbering corresponds to HXB2 (GenBank Accession number K03455).
Table 3. Contribution of hypermutation to the HIV-1 mutation rate per replication cycle.
| No. of clones | No. of G-to-A mutations rescued | Total no. of nt sequenced | Mutations/bp/ replication cycle | |
|---|---|---|---|---|
| N-A3Ghigh | 140 | 3 | 149100 | 2.0 × 10−5 |
| N-A3Glow | 141 | 9 | 142974 | 6.3 × 10−5 |
| N-A3F | 132 | 2 | 62568 | 3.2 × 10−5 |
| Total | 413 | 14 | 354,642 | - |
| Average | - | - | - | 3.9 × 10−5 |
In silico modeling of the contribution of A3 hypermutation and recombination to HIV-1 mutation rate
We simulated the potential contribution of A3G- and A3F-mediated hypermutation to viral diversity using a custom in-house MATLAB computer program. The NL4-3 genome was used as the baseline sequence, and the input variables were 1) the number of template switches, 2) the number of G-to-A mutations, and 3) the number of heterozygous virions containing a wild-type and a hypermutated genome simulated to undergo one cycle of replication. Our program randomly selected the locations of the template switches and the locations of the G-to-A mutations. For A3G, we simulated 90% of the mutations at GG sites and 10% at GA sites, and for A3F we simulated 86% of the mutations at GA sites and 14% at GG sites; these ratios were based on our previously published [68] observations of the G-to-A mutations induced by A3G and A3F in ex vivo experiments. NL4-3 has 616 GG sites, of which 119 (19.3%) are predicted to generate stop codons upon mutation to AG. NL4-3 also has 756 GA sites, of which 37 (4.8%) are predicted to generate stop codons upon mutation to AA. The program then determined the number of viable recombinants, which was defined by the absence of stop codons, and the mutation rate was then calculated as the number of mutations in nonlethal recombinants divided by the total number of nucleotides.
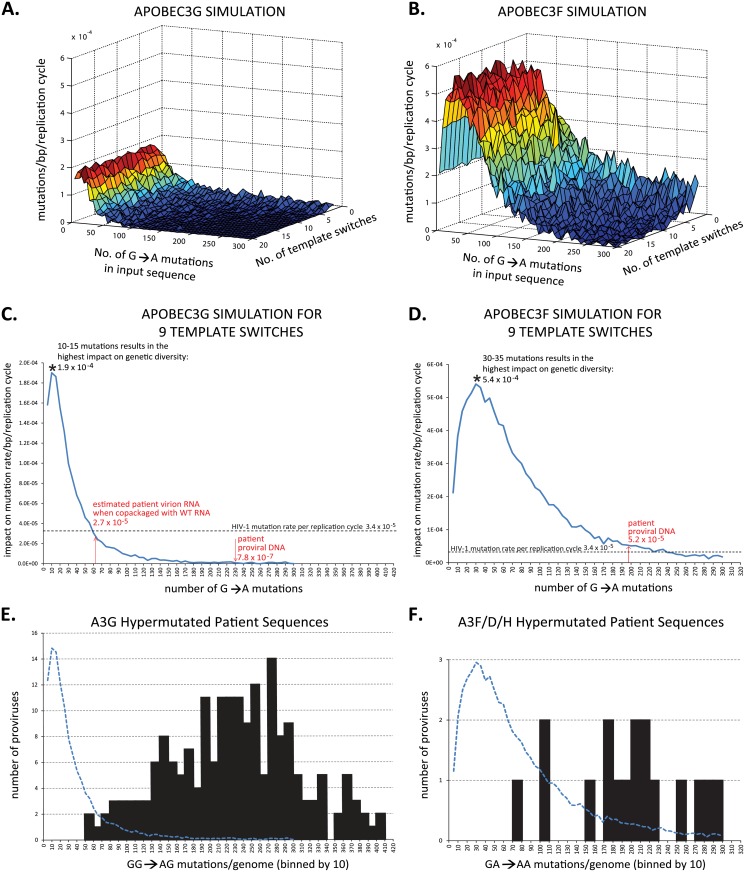
For both A3G and A3F, the contribution of hypermutation to the HIV-1 mutation rate was independent of the number of template switching events in the simulation, regardless of the starting number of G-to-A changes in the input sequence (Fig 4A and 4B). Thus, using 9 template switches as the average number per single replication cycle [12, 13] and with simulations of 10,000 heterozygous virions containing a wild type and a hypermutated genome undergoing one cycle of replication per scenario, our results showed that when the HIV-1 genome contains 10–15 G-to-A mutations, A3G has the most impact on viral genetic diversity with a mutation rate of 1.9 × 10−4 mutations/bp/simulated replication cycle (Fig 4C). However, as the shown in the patient sequence analyses (Fig 4E), none of the 194 hypermutated proviruses that were predominantly mutated at GG sites had the optimal 10–15 GG-to-AG mutations. Instead, hypermutated sequences from patients had an average of 231 GG-to-AG mutations per proviral genome (Table 4 and Tables A and C in S1 File), which is predicted to contribute only 7.8 × 10−7 mutations/bp/cycle, a rate that is >200-fold less than the retroviral mutation rate. One caveat to the estimation above is that purifying selection has been observed previously [68] and HIV-1 RNAs packaged into viral particles contain fewer G-to-A mutations (~27%) than those in hypermutated proviral DNA. Therefore, the virion RNA should have an average of 62 G-to-A mutations/genome (27% of 231). As the distribution of the GG-to-AG hypermutations in proviral genomes shows only three of 194 proviruses (~2%) had <80 mutations, suggesting that very few of the proviral genomes would generate an RNA that will be packaged into virions. For the few mutated RNAs that were able to be packaged into viral particle along with a wild-type RNA, our modeling results predicted that with an average of 62 G-to-A mutations/genome, rescue of GG-to-AG mutations through recombination would result in a mutation rate of 2.7 × 10−5/bp/cycle, which is similar to the HIV-1 mutation rate/bp/cycle (3.4 × 10−5/bp/cycle). Therefore, even in the best case scenario, recombination and rescue of hypermutated portions of proviral genomes without stop codons would only increase the viral mutation rate by twofold. However, since the frequency of copackaging of hypermutated and wild-type genomes is extremely low ([44, 45, 76, 77]; see Discussion), the overall contribution of hypermutation to the viral mutation rate is far less than the mutations generated during reverse transcription.
Fig 4. In silico modeling of the effect of hypermutation on the HIV-1 mutation rate.
(A) and (B) represent the estimated contribution by A3G and A3F to the HIV-1 mutation rate, respectively. The number of G-to-A changes and the number of template switches were varied. One thousand simulations of a heterozygous virion containing a wild-type genome and a hypermutated genome undergoing one cycle of replication were scored per scenario. (C) and (D) show the contribution of GG→AG and GA→AA hypermutations, respectively, to the mutation rate/bp/replication cycle obtained using simulations for 9 template switches/replication cycle. Ten thousand simulations of a heterozygous virion containing a wild-type genome and a hypermutated genome undergoing one cycle of replication were scored per scenario for A3G and A3F. Dotted line indicates the HIV-1 mutation rate of 3.4 × 10−5/bp/replication cycle. Shown on the graph in (C) is the estimated 231 G-to-A changes per patient proviral DNA resulting in a mutation rate of 7.8 × 10−7/bp/replication cycle and the 62 G-to-A changes per virion RNA genome introduced by A3G (with a HIV-1Δvif), resulting in a mutation rate of 2.7 × 10−5/bp/replication cycle. Shown in the graph in (D) is the estimated 197 G-to-A changes per patient proviral DNA introduced by A3F, resulting in a mutation rate of 5.2 × 10−5/bp/replication cycle. (E) and (F) distribution of patient proviruses predominantly mutated at GG sites by A3G or GA sites by A3F, A3D, or A3H, respectively. The numbers of G-to-A mutations for the proviral genomes were estimated as described in Tables A and B in S1 File, and plotted in bins of 10 mutations. The simulated impact of hypermutation and recombination on the retroviral mutation rate plotted in C and D (blue line) is superimposed as a blue dotted line in E and F, respectively, for comparison to the distribution of G-to-A mutations per genome.
Table 4. Summary of analysis to determine sub-lethal mutagenesis probability for patient sequences that were predominantly hypermutated at GG sites 1 .
| Study | No. of Seqs. | Seq. | Total nt | GG→AG Mtns/ genome 2 | GA→AA Mtns/ genome | G→A mtns/ genome | Probability of | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | Stops | Total | Stops | Total | Stops | 0 Stops 3 | ||||
| Eyzaguirre et al. 2013 | 10 | Full-length | 92,440 | 206 | 40 | 38 | 2 | 244 | 42 | 6 × 10−19 |
| Gandhi et al. 2008 | 161 | Env/nef | 73,698 | 238 | 46 | 38 | 2 | 276 | 48 | 1 × 10−21 |
| Ho et al. 2013 | 23 | Gag, Env, nef-LTR | 54,550 | 195 | 38 | 33 | 2 | 228 | 40 | 4 × 10−18 |
| Total | 194 | 220,688 | 44,863 | 8,659 | 7,270 | 349 | 52,076 | 9,068 | ||
| Average 4 | - | - | 231 | 45 | 37 | 2 | 268 | 47 | 4 × 10−21 | |
1 Details of the analysis of sequences are described in Tables A and C in S1 File.
2 Mutations/genome were estimated based on sequence length and average frequency of mutations in the same region of the genome in 11 full-length hypermutated sequences from the Eyzaguirre et al. study [69] to adjust the mutation frequencies for the previously described 5’-to-3’ hypermutation gradient [49, 79–81].
3 The probability of sub-lethal mutagenesis was determined by using the average number of stop codon mutations/genome and Poisson distribution.
4 The average number of mutations per genome was determined by summing up the total number of mutations estimated per genome for all sequences and dividing by the number of sequences.
We performed a similar analysis for patient proviral genomes that were predominantly hypermutated at GA sites, and were likely hypermutated by A3F, A3D or A3H (Table 5). Proviruses that were predominantly hypermutated at GA sites (17 total) had an average of 197 GA-to-AA mutations/genome. GA-to-AA mutations generate stop codons fourfold less frequently (4.8%) than GG-to-AG mutations (19.3%), suggesting that there will be less purifying selection of GA mutations and more GA-to-AA mutations will be present in the virion RNA. When there are an optimal number of GA-to-AA mutations (30-35/genome; Fig 4F, and assuming these RNAs are copackaged with a wild-type RNA, rescue of GA-to-AA mutations without stop codons through recombination would increase the retroviral mutation rate by 16-fold (5.4 × 10−4/bp/cycle). However, the distribution of GA-to-AA mutations/genome in patients (Fig 4F) suggests that none of the hypermutated genomes had an optimal number of mutations (30–35; Fig 4D). The average 197 GA-to-AA mutations/genome is predicted to contribute 5 × 10−5 mutations/bp/replication cycle (Fig 4D), which is similar to the retroviral mutation rate (3.4 × 10−5/bp/cycle). It is worth noting that the simulations above are based on copackaging of the mutated RNA with wild-type RNA. However, since the frequency of copackaging wild-type and hypermutated genomes is extremely low [44, 45, 76, 77] we conclude that A3F-induced hypermutations also contribute very little to the viral variation compared to mutations that are generated during reverse transcription.
Table 5. Summary of analysis to determine sublethal mutagenesis probability for patient sequences predominantly hypermutated at GA sites 1 .
| Study | No. of Seqs. | Seq. | Total nt | GG→AG Mtns/ genome 2 | GA→AA Mtns/ genome | G→A Mtns/ genome | Prob. of | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | Stops | Total | Stops | Total | Stops | 0 Stops 3 | ||||
| Eyzaguirre et al. 2013 | 1 | Full-length | 9,244 | 64 | 12 | 209 | 10 | 273 | 22 | 3 × 10−10 |
| Gandhi et al. 2008 | 5 | Env/nef | 2,282 | 70 | 13 | 167 | 8 | 237 | 22 | 3 × 10−10 |
| Ho et al. 2013 | 11 | Gag, Env, Nef-LTR | 11,460 | 86 | 17 | 209 | 10 | 295 | 27 | 2 × 10−12 |
| Total | 17 | 22,986 | 1,358 | 262 | 3,346 | 161 | 4,703 | 422 | ||
| Average 4 | - | - | 80 | 15 | 197 | 9 | 277 | 25 | 1 × 10−11 | |
1 Details of the sequence analysis are described in Tables B and D in S1 File,
2 Mutations/genome were estimated based on sequence length and average frequency of mutations in the same region of the genome in 11 full-length hypermutated sequences reported by Eyzaguirre et al. [69] to adjust the mutation frequencies for the 5’-to-3’ hypermutation twin gradient [49, 79–81].
3 The probability of sublethal mutagenesis, defined as the probability of generating a viral genome without stop codons, was determined by using the average number of stop codon mutations/genome and Poisson distribution.
4 The average number of mutations per genome was determined by summing up the total number of mutations estimated per genome for all sequences and dividing by the number of sequences.
Estimation of the frequency of sublethal mutagenesis by A3G and A3F
To determine the frequency of sublethal mutagenesis by A3 proteins, we analyzed hypermutated proviral DNA sequences reported in three previous studies. Eyzaguire and colleagues reported 11 near-full-length sequences that were hypermutated throughout the proviral genomes [69]. For 10 of the 11 proviruses, the majority of the G-to-A mutations were in the GG context (A3G type); for one provirus, the majority of the mutations were in the GA dinucleotide context (A3F type) and were likely mutated by A3F, A3D or A3H. Gandhi and colleagues reported 166 hypermutated proviral DNA sequences, of which 161 had a majority of the mutations in the GG context (A3G type) and 5 had mutations primarily in the GA context (A3F type) [43]. Ho and colleagues reported 34 hypermutated proviral DNA sequences, three of which were near-full-length and others were from various regions of the genome [78]. Of these, 23 were predominantly mutated in the GG context (A3G type) and 11 were predominantly mutated in the GA context (A3F type).
We estimated the number of G-to-A changes that arose in each proviral genome, taking into account the lengths of the sequences analyzed as well as their locations in the genome. Furthermore, the sequences analyzed were from patients from whom a consensus sequence for the patient could be derived to further verify that the sequence was indeed hypermutated. For our analysis, proviruses with less than 18 G-to-A mutations would not be identified as hypermutants (Table A in S1 File). It is well known that there is a twin-gradient of hypermutation in the viral genome, which reflects the amount of time the minus-strand DNA is available as a substrate for cytidine deamination by A3 proteins [49, 79–81]. We used the 11 near-full-length hypermutated genomes reported by Eyzaguire et al. [69] to estimate the relative frequency of hypermutation for each region of the genome to adjust for the hypermutation gradient. The average number of GG-to-AG mutations estimated per proviral genome was 231 (Table 4 and Tables A and C in S1 File); the sequences that were predominantly hypermutated at GG sites also had an average of 37 mutations at GA sites/genome.
Since 19.3% of the GG sites generated stop codons in NL4-3, we estimated that mutations at GG sites generated an average of 45 stop codons/genome; since 4.8% of the GA sites in NL4-3 generated stop codons, we estimated that an average of 2 stop codons/genome were generated by GA-to-AA mutations, resulting in an average of 47 stop codons/genome. Assuming a Poisson distribution, we estimated that the probability of generating a provirus without stop codons is 4 × 10−21. Thus, we conclude that the contribution of A3G-induced sublethally mutated proviruses to viral genetic variation is negligible.
Similarly, proviruses that were predominantly mutated at GA sites had an average of 197 GA-to-AA mutations/genome, and 80 GG-to-AG mutations/genome with an average of 25 stop codons/genome (Table 5 and Tables B and D in S1 File); assuming a Poisson distribution, the probability of generating a sublethally mutated provirus is 1 × 10−11. Thus, we conclude that while sublethal mutagenesis can occur, there is a simultaneous overwhelming reduction in the size of the replicating viral population. In the absence of A3F-induced hypermutation, RT and RNA polymerase II would generate 3 × 1010 mutations in 1 × 1011 proviruses, whereas A3F hypermutation would generate 277 mutations in a sublethally mutated provirus.
Discussion
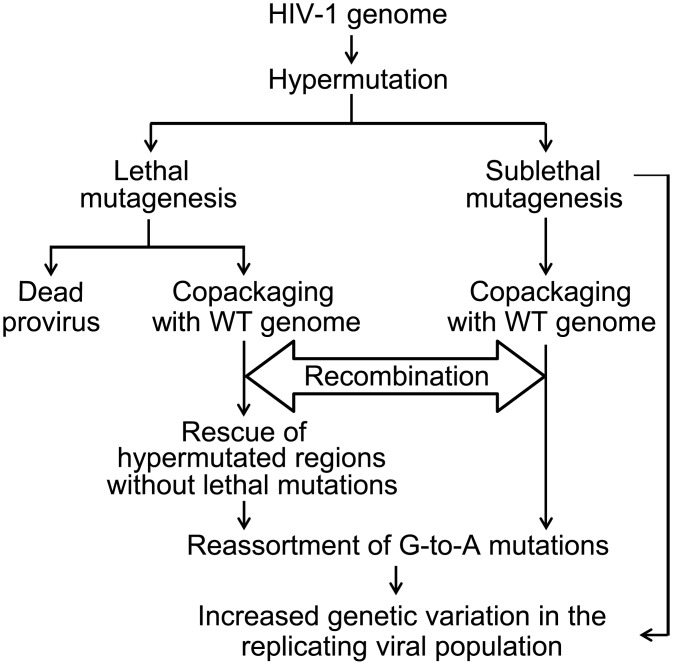
In addition to error-prone reverse transcription and high rates of recombination, G-to-A hypermutation by A3 proteins could increase genetic variation in HIV-1 populations by two mechanisms (Fig 5). First, lethal mutagenesis could generate dead proviruses that cannot increase genetic variation of the replicating viral population; however, parts of such genomes may be rescued when a replication-competent virus infects the same cell through copackaging and recombination. If the resulting recombinants contain portions of hypermutated genomes without lethal mutations, these G-to-A hypermutations can enter the replicating viral population. Second, sublethally mutated viruses can on their own increase genetic variation of the replicating viral population, and through recombination with wild-type genomes, further increase genetic variation. However, hypermutation could also decrease genetic variation by reducing the rate of recombination due to decreased homology between the co-packaged RNAs.
Fig 5. Flow chart of potential contribution of hypermutation and recombination to HIV-1 genetic diversity.
Effect of hypermutation on recombination
To our knowledge, this is the first study to directly examine the effects of hypermutation on the frequency of retroviral recombination and quantify the extent to which recombination can rescue portions of hypermutated genomes without lethal mutations. In our system, hypermutation by A3G or A3F did not affect the efficiency of RNA copackaging or the frequency of recombination as determined by the rate of gfp reconstitution. We also did not observe a decrease in the frequency of recombination in the A3G or A3F hypermutated regions of the pol gene (Table 2 and Fig 2). Our previously proposed dynamic copy-choice mechanism of recombination and results from other studies predict that a decrease in homology between co-packaged RNAs would reduce the frequency of recombination [82–86]. The recombination rate was similar in the absence or presence of hypermutation, which decreased the sequence identity in RT to 92–97%. It is possible that more extensive hypermutation (> 64 mutations/1320 nt of RT) would lead to a reduction in the recombination rate. However, hypermutated full-length patient sequences had on average 42 G-to-A mutations in RT [69], compared to 64 for N-A3Ghigh, suggesting that the majority of hypermutated genomes will not affect the rate of recombination.
Effect of hypermutation on increasing genetic variation through recombination
Our results show that within the population of viruses containing a wild-type and a A3G- or A3F-hypermutated genome, recombination contributed to the retroviral mutation rate to the same extent as mutations during error-prone viral replication (3.9 × 10−5 versus 3.4 × 10−5 mutations/bp/replication cycle, respectively). However, we expect that because of purifying selection on hypermutated genomes, only a small proportion of hypermutated genomes will be packaged into virions [68]. Additionally, it was recently found that, contrary to a previous report [87], >90% of HIV-1 infected CD4+ T cells in lymph nodes of patients contained only one proviral DNA [88], implying that the frequency of copackaging of RNAs from two different proviruses, including RNAs from a wild-type and a hypermutated genome, is likely to be very low [89]. Consistent with purifying selection, Kieffer et al. observed that >9% of the proviral DNAs were hypermutated by A3G or A3F but none of the 2024 viral RNAs isolated from plasma were hypermutated [45]. Since the frequency of copackaging and recombination between hypermutated and wild-type genomes in patients appears to be extremely low (<1/100 –<1/2024; [44, 45, 76, 77]), we conclude that the contribution of hypermutation to viral genetic variation through recombination is far less than mutations that occur during retroviral replication.
Mulder et al. reported that recombination between hypermutated and wild-type genomes resulted in increased resistance to antiviral drug 3TC through acquisition of M184I mutation in RT [65]. In this study, the proviral DNAs underwent one round of DNA transfection (plasmids containing hypermutated genomes and wild-type genomes were co-transfected into cells). DNA recombination is known to occur frequently during co-transfection and has been shown to be sufficient to reconstitute replication-competent retroviruses from defective genomes even in the absence of retroviral recombination [89, 90]. Kim et al. also reported that hypermutation in a T cell line can contribute to selection of M184I mutations and 3TC resistance [66]. It is not clear whether the observed frequencies of M184I mutations (0 of 4 for A3G- cells vs. 3 of 4 for A3G+ cells) are significantly different from each other. It is also possible that, in an experimental system when a limited number of cells and high titers of infectious virus are used, coinfection and recombination occurs at a much higher frequency than in patients, resulting in an increase in 3TC resistance in the presence of A3G. In our studies, we ruled out potential contribution of DNA recombination, and observed the effect of A3G and A3F hypermutation on the viral mutation rate in the absence of selection, which could explain the modest two-fold contribution of hypermutation to the retroviral mutation rate in the progeny from heterozygous viruses.
A recent study determined the HIV-1 mutation rate in vivo by determining the frequency of stop-codon mutations in proviral DNAs in patients [91]. As expected, their analysis included all G-to-A mutations induced by A3 proteins, and concluded that the HIV-1 mutation rate in proviral DNA is extremely high (4.1 x 10−3/bp/cell), and most of the mutations are due to cytidine deaminase activity of A3 proteins. As our analysis points out, most of the hypermutated proviruses are lethally mutated and cannot contribute to the genetic variation of the replicating viral population. Therefore, we have focused our analysis on the potential contribution of hypermutation to genetic variation on which selective forces can act to shape viral evolution.
Our in silico modeling indicated that the number of template-switching events did not affect the contribution of A3G or A3F hypermutation to the retroviral mutation rate. The modeling results indicated that for A3G and A3F, the optimal number of G-to-A mutations (10–15 and 30–35, respectively) would increase the retroviral mutation rate by 3- or 16-fold, respectively, in the population of heterozygous virions. The average numbers of mutations at GG and GA sites in patients (231 and 197, respectively) are much higher than the optimal number of mutations, and their contribution to the retroviral mutation rates are 200-fold lower (7.8 × 10−7 mutations/bp/cycle) or about the same (5 × 10−5/bp/cycle) as the retroviral mutation rate (3.4 × 10−5/bp/cycle), respectively. Even with the optimal number of mutations, given the low frequency of copackaging and recombination (<1/100–<1/2000; [44, 45, 76, 77]), the contribution of hypermutation to the retroviral mutation rate is likely to be far less than the mutation rate during error-prone replication.
Sublethal hypermutation and its effect on HIV-1 genetic variation
Our analysis of hypermutated proviruses reported in three independent studies [43, 69, 78] predicted that proviruses hypermutated predominantly at GG sites or predominantly at GA sites have on average 47 and 25 stop codons, respectively. Based on a Poisson distribution, the frequency of sublethally mutated proviruses predominantly mutated at GG sites (A3G type) and GA sites (A3F type) is predicted to be 4 × 10−21 and 1 × 10−11, respectively. Thus, the vast majority of hypermutation events result in lethal mutagenesis, and very few result in sublethal mutagenesis that can potentially increase genetic variation in the replicating viral population. It is important to point out that these are conservative estimates of sublethal mutagenesis, since nonsynonymous G-to-A mutations, as well as some mutations in the cis-acting viral sequences, also likely result in loss of fitness. Even in the absence of stop codons, the hypermutated viruses with many non-synonymous G-to-A mutations are likely to be highly attenuated in their replication potential, further reducing their capacity to contribute to genetic variation. Therefore, the recombinant viruses containing sublethal mutations are also unlikely to outgrow the nonhypermutated parental viruses, and their contribution to genetic variation will likely diminish with each successive replication cycle.
Simon et al. found Vif alleles in patients that were defective in inducing degradation of A3G or A3F proteins, suggesting that incomplete degradation of A3 proteins could lead to sublethal mutagenesis [92]. Sadler et al. observed that expression of lower amounts of A3G resulted in sublethal mutagenesis in a cell culture system, indicating that sublethal mutagenesis can occur in an ex vivo assay [58]. On the other hand, Armitage et al. found that packaging of single active A3G protein in virions results in substantial levels of hypermutation, and concluded that hypermutation by A3G is typically an all-or-nothing phenomenon [64].
Our analysis does not exclude the possibility of sublethal mutagenesis, but implies that such low levels of hypermutation are likely to be rare. One caveat to our studies is that proviral genomes that are identified as hypermutated genomes requires that the regions sequenced need to have at least two G-to-A mutations in order to be defined as hypermutants; sequences with one G-to-A mutation/~450 nt (the length of env sequence analyzed in [43]) will not be defined as hypermutants, suggesting a lower limit for detection of hypermutation as ~18 G-to-A mutations/proviral genome (Table A in S1 File).
Potential contribution of hypermutation to viral genetic variation and evolution
Many previous studies have concluded that A3 proteins contribute to viral genetic variation and evolution [58, 65, 66, 92–96], while others have concluded that hypermutation by A3 proteins does not increase genetic diversity or contribute to viral evolution [60, 62, 64, 97].
A few studies have analyzed the context in which A3-proteins induce mutations and sought to determine whether mutations in these contexts may have provided a selective advantage to the virus, and thereby contributed to viral evolution [60, 66, 94, 98]. One confounding factor in these analyses is that the contexts in which RT and RNA polymerase II induce mutations are not well defined, and the extent to which error-prone viral replication can induce errors in A3-favored contexts is unknown. HIV-1 RT has a strong bias for inducing G-to-A mutations during replication, with nearly 40% of the substitutions occurring in GA context and GG contexts [2, 4]. Additional studies to define the nucleotide contexts of mutations induced by RT and RNA polymerase II are needed to facilitate these analyses.
In summary, we found that A3G or A3F hypermutation did not affect the rate of recombination and the contribution of A3G and A3F hypermutation to the genetic variation of HIV-1 was significantly less than the rate of mutations induced during error-prone viral replication. Thus, while hypermutation can alter sequences in some proviruses, its contribution to viral variation and evolution is small compared to mutations induced by RT and/or RNA polymerase II.
Material and Methods
Construction of plasmids containing wild-type and hypermutated RT sequences
The names of all plasmids in this study start with ‘p” while the names of viruses and proviruses generated from these plasmids do not. pHCMV-G that expresses the G glycoprotein of vesticular stomatitis virus (VSV-G) [99], pSYNGP that expresses a codon-optimized HIV-1 Gag/Gag-Pol [100], and pIIINL(AD8)env that expresses the HIV-1 CCR5-tropic envelope [70] have been described previously.
To create plasmid pWTHXB2 containing a wild-type pol sequence from the subtype B HXB2 isolate, the region between BamHI and XhoI restriction sites in plasmid pHG(BHXB) [86] was replaced with a corresponding region from plasmid pON-H0 [13]. The resulting construct pWTHXB2 contains all cis-acting elements necessary for virus expression and production, functional gag-pol, as well as hsa and inactivated gfp in the nef gene [13].
The four constructs containing NL4-3-based RT sequences were created as follows. First, SphI and MscI was used to digest NL4-3-based plasmid pON-T6 [13], which in nef contains thy1.2 and IRES followed by an inactivated gfp gene. The distance between the inactivating mutations in the two gfp genes is 588 bp. To create construct pN-A3Ghigh, the SphI-MscI digested pON-T6 backbone was ligated to a synthesized SphI to MscI fragment (GENEWIZ) that contains the following modifications to NL4-3 sequence: 1) a frameshift and a stop codon was introduced to destroy SpeI site in gag; 2) the natural MscI in gag was destroyed by a silent mutation; 3) unique enzyme sites SgrAI, SnaBI, and XbaI flanking the start of RT, RNase H and IN, respectively, were introduced via silent mutations; and 4) RT region contained 64 G-to-A hypermutations by A3G that were previously isolated from an infected cell clone. To create pWTNL43, pN-A3Glow, and pN-A3F, plasmid pN-A3Ghigh was digested with SgrAI and SnaBI, and ligated with a synthesized insert (GENEWIZ) containing either a wild-type RT sequence from pNL4-3 isolate, an NL4-3 RT sequence with 27 G-to-A changes introduced by A3G, or an NL4-3 RT sequence with 27 G-to-A changes introduced by A3F, respectively. All plasmids were verified by sequencing (Macrogen).
Generation, maintenance, and flow cytometry analyses of cell lines
Human embryonic kidney 293T cells and derivatives (American Type Culture Collection) were maintained in Dulbecco’s modified Eagle’s medium (CellGro) supplemented with 10% fetal calf serum (HyClone), and 1% penicillin-streptomycin stock (penicillin 50 U/ml and streptomycin 50 μg/ml, final concentration; Gibco). Hut/CCR5 cells, a human T cell line derived from Hut78 cells to express CCR5 chemokine receptor [101], were maintained in RPMI medium (CellGro) supplemented with 10% fetal calf serum (HyClone), 1% penicillin-streptomycin stock (penicillin 50 U/ml and streptomycin 50 μg/ml final concentration; Gibco), 1 μg/ml of puromycin (Gibco) and 500 μg/ml G418 (ThermoFisher Scientific). All cultured cells were maintained in humidified 37°C incubators with 5% CO2. All transfections were performed using LT1 reagent (Mirus) according to manufacturer’s instructions.
To detect marker gene expression, cells were stained with phycoerythrin-conjugated α-HSA antibody (Becton Dickinson Biosciences) and allophycocyanin-conjugated α-Thy1.2 antibody (eBioscience) at 0.4 μg/ml and 2.0 μg/ml, respectively. Flow cytometry analyses were performed on a FACSCalibur system (BD Biosciences) whereas cell sorting was performed on an ARIA II system (BD Biosciences). Flow cytometry data was analyzed using FlowJo software (Tree Star).
Producer cell lines containing two different proviruses were generated as follows. To generate stock viruses for infection, 293T cells were transfected with viral construct along with plasmids pSYNGP and pHCMV-G that express codon-optimized Gag/GagPol and VSV-G envelope, respectively. Viruses were harvested 48 hours later, filtered with 0.45- μM filters, and used immediately or stored at –80°C. To make producer cell lines, stock virus WTHXB2 was used to infect fresh 293T cells at a multiplicity of infection (MOI) of 0.1. Cells were stained 72 hours post-infection and infected cells expressing HSA surface marker were enriched by multiple rounds of cell sorting until more than 80% of the cells were HSA+. These cells were then infected at an MOI of <0.1 with a second virus, and underwent multiple rounds of cell sorting until >97% of cells were HSA+ and Thy+. Four cell lines were created containing the following pairs of proviruses: WTHXB2/WTNL43, WTHXB2/N-A3Ghigh, WTHXB2/N-A3Glow, and WTHXB2/N-A3F.
Recombination experiments and sorting of recombinants
Producer cell lines WTHXB2/WTNL43, WTHXB2/N-A3Ghigh, WTHXB2/N-A3Glow, and WTHXB2/N-A3F were transfected with pIIINL(AD8)env [70]; 48 hours later viruses were harvested, filtered through 0.45-μM filters, and used to infect 12 × 106 Hut/CCR5 target cells at a low MOI (<0.08) to minimize dual infection. Target cells were stained 72 hours post-infection for marker expression and flow cytometry analysis was used to determine the percentage of HSA, Thy and GFP expressing cells. Target cells expressing GFP were enriched by sorting until 87% of the cells were GFP+.
Viral DNA isolation, single genome sequencing and sequence analysis
Genomic DNA was isolated from the sorted GFP+ cell pools using QIAamp DNA blood kit (Qiagen). Single genome amplification was achieved by serially diluting genomic DNA in 96-well plates to identify a dilution in which PCR-positive wells constituted less than 30% of the total number of reactions. At this dilution, most wells contain amplicons derived from a single DNA molecule. PCR amplification was performed in a 20-μl reaction containing 1× High Fidelity Platinum PCR buffer, 2 mM MgSO4, 0.2 mM of each deoxynucleoside triphosphate, 0.2 μM of each primer, and 0.025 U/μl Platinum Taq High Fidelity polymerase (Invitrogen). For the first round of PCR, sense primer HIV-A GagF1 5′- GTG GCA AAG AAG GAC ACC TAG-3′ and antisense primer HIV-A VifR1 5′-GTC GAC ACC CAA TTC TGA AAT G-3′ were used. PCR was performed with the following parameters: 1 cycle of 94°C for 2 min, 35 cycles of a denaturing step of 94°C for 15 s, an annealing step of 55°C for 30 s, and an extension step of 68°C for 4 min, followed by a final extension of 68°C for 10 min. For the second round of PCR, we used 1 μl of first-round PCR product along with sense primer HIV-A GagF2 5′- GGC TGT TGG AAA TGT GGA AAGG-3′ and antisense primer HIV-A VifR2 5′- ATG GCT TCC AAT CCC ATA TGA TG-3′. The second-round PCR reaction was performed under the same conditions used for first-round PCR, but with a total of 45 cycles. All PCR procedures were performed under PCR clean room conditions with additional procedural safeguards against sample contamination, such as prealiquoting of all reagents, use of dedicated equipment, and physical separation of sample processing from pre- and post-PCR amplification steps. Correctly sized amplicons from the second round of PCR were sequenced directly by cycle-sequencing using BigDye terminator chemistry according to the manufacturer’s recommendations (Applied Biosystems). Individual sequence fragments for each amplicon were assembled and edited using Sequencher (Gene Codes). Individual chromatograms were inspected for the absence of mixed bases at each nucleotide position throughout the entire amplicon; this quality control measure confirmed that the amplicons analyzed were derived from SGS amplification of a single viral template and allowed us to exclude from the analysis amplicons that resulted from PCR-generated in vitro recombination events or Taq polymerase errors. Therefore, the collection of individual sequences obtained via SGS proportionately represents those found in the infected cells.
In order to identify crossover events, we aligned the nucleotide sequences of each genome in a recombinant pair (WTHXB2/WTNL43, WTHXB2/A3Ghigh, WTHXB2/A3Glow, or WTHXB2/A3F). Using the polymorphic differences between two parental sequences, we identified the locations of crossover events for each recombinant sequence.
Computer modeling and simulation
A custom in-house MATLAB program was used to estimate the contribution of hypermutation and recombination to the HIV-1 mutation rate in a population of heterozygous virions containing a wild-type genome and a hypermutated genome. The NL4-3 genome was used as the reference (WT) sequence, and the input variables were 1) the number of template switches, 2) the number of G-to-A mutations in the mutated genome, and 3) the number of heterozygous virions containing a wild-type and a mutated genome simulated to undergo one cycle of replication. All GG and GA sites in NL4-3 (GenBank AF324493.2) were identified and the GG-to-AG and GA-to-AA mutations that would generate stop codons in the appropriate open reading frames were determined. NL4-3 has 616 GG sites, of which 119 (19.3%) are predicted to generate stop codons upon mutation to AG. NL4-3 also has 756 GA sites, of which 37 (4.8%) are predicted to generate stop codons upon mutation to AA. The locations of the template switches and G-to-A mutations were randomly selected. For an A3G-mutated genome, we simulated 90% of the mutations at GG sites and 10% at GA sites, and for an A3F-mutated genome we simulated 86% of the mutations at GA sites and 14% at GG sites; these ratios were based on our previously published [68] observations of the G-to-A mutations induced by A3G and A3F in ex vivo experiments. The viable recombinants, defined by the absence of mutation-induced stop codons, were identified. The mutation rate was calculated as the total number of mutations in viable recombinants divided by the total number of nucleotides from the viable and nonviable recombinants.
Supporting Information
(Table A) Analysis of patient sequences that were predominantly hypermutated at GG sites. (Table B) Analysis of patient sequences that were predominantly hypermutated at GA sites. (Table C) Calculations used to prepare Table 4. (Table D) Calculations used to prepare Table 5.
(XLSX)
Acknowledgments
We thank Drs. John Coffin and Frank Maldarelli for helpful discussions. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and by Intramural AIDS Targeted Antiviral Program grant funding (to VKP and to WSH). This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Coffin J, Swanstrom R. HIV pathogenesis: dynamics and genetics of viral populations and infected cells. Cold Spring Harbor perspectives in medicine. 2013; 3(1):a012526 10.1101/cshperspect.a012526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mansky LM, Temin HM. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. Journal of virology. 1995; 69(8):5087–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim T, Mudry RA Jr., Rexrode CA 2nd, Pathak VK. Retroviral mutation rates and A-to-G hypermutations during different stages of retroviral replication. Journal of virology. 1996; 70(11):7594–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Abram ME, Ferris AL, Shao W, Alvord WG, Hughes SH. Nature, position, and frequency of mutations made in a single cycle of HIV-1 replication. Journal of virology. 2010; 84(19):9864–78. 10.1128/JVI.00915-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mansky LM. Forward mutation rate of human immunodeficiency virus type 1 in a T lymphoid cell line. AIDS research and human retroviruses. 1996; 12(4):307–14. [DOI] [PubMed] [Google Scholar]
- 6. Pathak VK, Temin HM. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: substitutions, frameshifts, and hypermutations. Proceedings of the National Academy of Sciences of the United States of America. 1990; 87(16):6019–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ji JP, Loeb LA. Fidelity of HIV-1 reverse transcriptase copying RNA in vitro. Biochemistry. 1992; 31(4):954–8. [DOI] [PubMed] [Google Scholar]
- 8. O'Neil PK, Sun G, Yu H, Ron Y, Dougherty JP, Preston BD. Mutational analysis of HIV-1 long terminal repeats to explore the relative contribution of reverse transcriptase and RNA polymerase II to viral mutagenesis. The Journal of biological chemistry. 2002; 277(41):38053–61. [DOI] [PubMed] [Google Scholar]
- 9. Thomas MJ, Platas AA, Hawley DK. Transcriptional fidelity and proofreading by RNA polymerase II. Cell. 1998; 93(4):627–37. [DOI] [PubMed] [Google Scholar]
- 10. Wang D, Bushnell DA, Huang X, Westover KD, Levitt M, Kornberg RD. Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution. Science. 2009; 324(5931):1203–6. 10.1126/science.1168729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de Mercoyrol L, Corda Y, Job C, Job D. Accuracy of wheat-germ RNA polymerase II. General enzymatic properties and effect of template conformational transition from right-handed B-DNA to left-handed Z-DNA. European journal of biochemistry / FEBS. 1992; 206(1):49–58. [DOI] [PubMed] [Google Scholar]
- 12. Rhodes T, Wargo H, Hu WS. High rates of human immunodeficiency virus type 1 recombination: near-random segregation of markers one kilobase apart in one round of viral replication. Journal of virology. 2003; 77(20):11193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rhodes TD, Nikolaitchik O, Chen J, Powell D, Hu WS. Genetic recombination of human immunodeficiency virus type 1 in one round of viral replication: effects of genetic distance, target cells, accessory genes, and lack of high negative interference in crossover events. Journal of virology. 2005; 79(3):1666–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996; 271(5255):1582–6. [DOI] [PubMed] [Google Scholar]
- 15. Ramratnam B, Bonhoeffer S, Binley J, Hurley A, Zhang L, Mittler JE, et al. Rapid production and clearance of HIV-1 and hepatitis C virus assessed by large volume plasma apheresis. Lancet. 1999; 354(9192):1782–5. [DOI] [PubMed] [Google Scholar]
- 16. Hazenberg MD, Hamann D, Schuitemaker H, Miedema F. T cell depletion in HIV-1 infection: how CD4+ T cells go out of stock. Nature immunology. 2000; 1(4):285–9. [DOI] [PubMed] [Google Scholar]
- 17. Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997; 387(6629):183–8. [DOI] [PubMed] [Google Scholar]
- 18. Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002; 418(6898):646–50. [DOI] [PubMed] [Google Scholar]
- 19. Lecossier D, Bouchonnet F, Clavel F, Hance AJ. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science. 2003; 300(5622):1112 [DOI] [PubMed] [Google Scholar]
- 20. Harris RS, Sheehy AM, Craig HM, Malim MH, Neuberger MS. DNA deamination: not just a trigger for antibody diversification but also a mechanism for defense against retroviruses. Nature immunology. 2003; 4(7):641–3. [DOI] [PubMed] [Google Scholar]
- 21. Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, et al. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003; 113(6):803–9. [DOI] [PubMed] [Google Scholar]
- 22. Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003; 424(6944):99–103. [DOI] [PubMed] [Google Scholar]
- 23. Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003; 424(6944):94–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Iwatani Y, Chan DS, Wang F, Maynard KS, Sugiura W, Gronenborn AM, et al. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic acids research. 2007; 35(21):7096–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Newman EN, Holmes RK, Craig HM, Klein KC, Lingappa JR, Malim MH, et al. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Current biology: CB. 2005; 15(2):166–70. [DOI] [PubMed] [Google Scholar]
- 26. Adolph MB, Webb J, Chelico L. Retroviral restriction factor APOBEC3G delays the initiation of DNA synthesis by HIV-1 reverse transcriptase. PloS one. 2013; 8(5):e64196 10.1371/journal.pone.0064196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guo F, Cen S, Niu M, Yang Y, Gorelick RJ, Kleiman L. The interaction of APOBEC3G with human immunodeficiency virus type 1 nucleocapsid inhibits tRNA3Lys annealing to viral RNA. Journal of virology. 2007; 81(20):11322–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bishop KN, Verma M, Kim EY, Wolinsky SM, Malim MH. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS pathogens. 2008; 4(12):e1000231 10.1371/journal.ppat.1000231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Anderson JL, Hope TJ. APOBEC3G restricts early HIV-1 replication in the cytoplasm of target cells. Virology. 2008; 375(1):1–12. 10.1016/j.virol.2008.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li XY, Guo F, Zhang L, Kleiman L, Cen S. APOBEC3G inhibits DNA strand transfer during HIV-1 reverse transcription. The Journal of biological chemistry. 2007; 282(44):32065–74. [DOI] [PubMed] [Google Scholar]
- 31. Holmes RK, Koning FA, Bishop KN, Malim MH. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. The Journal of biological chemistry. 2007; 282(4):2587–95. [DOI] [PubMed] [Google Scholar]
- 32. Mbisa JL, Barr R, Thomas JA, Vandegraaff N, Dorweiler IJ, Svarovskaia ES, et al. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. Journal of virology. 2007; 81(13):7099–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mitra M, Singer D, Mano Y, Hritz J, Nam G, Gorelick RJ, et al. Sequence and structural determinants of human APOBEC3H deaminase and anti-HIV-1 activities. Retrovirology. 2015; 12:3 10.1186/s12977-014-0130-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mbisa JL, Bu W, Pathak VK. APOBEC3F and APOBEC3G inhibit HIV-1 DNA integration by different mechanisms. Journal of virology. 2010; 84(10):5250–9. 10.1128/JVI.02358-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Conticello SG, Harris RS, Neuberger MS. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Current biology: CB. 2003; 13(22):2009–13. [DOI] [PubMed] [Google Scholar]
- 36. Kobayashi M, Takaori-Kondo A, Miyauchi Y, Iwai K, Uchiyama T. Ubiquitination of APOBEC3G by an HIV-1 Vif-Cullin5-Elongin B-Elongin C complex is essential for Vif function. The Journal of biological chemistry. 2005; 280(19):18573–8. [DOI] [PubMed] [Google Scholar]
- 37. Marin M, Rose KM, Kozak SL, Kabat D. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nature medicine. 2003; 9(11):1398–403. [DOI] [PubMed] [Google Scholar]
- 38. Mehle A, Strack B, Ancuta P, Zhang C, McPike M, Gabuzda D. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. The Journal of biological chemistry. 2004; 279(9):7792–8. [DOI] [PubMed] [Google Scholar]
- 39. Stopak K, de Noronha C, Yonemoto W, Greene WC. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Molecular cell. 2003; 12(3):591–601. [DOI] [PubMed] [Google Scholar]
- 40. Sheehy AM, Gaddis NC, Malim MH. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nature medicine. 2003; 9(11):1404–7. [DOI] [PubMed] [Google Scholar]
- 41. Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, et al. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003; 302(5647):1056–60. [DOI] [PubMed] [Google Scholar]
- 42. Janini M, Rogers M, Birx DR, McCutchan FE. Human immunodeficiency virus type 1 DNA sequences genetically damaged by hypermutation are often abundant in patient peripheral blood mononuclear cells and may be generated during near-simultaneous infection and activation of CD4(+) T cells. Journal of virology. 2001; 75(17):7973–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gandhi SK, Siliciano JD, Bailey JR, Siliciano RF, Blankson JN. Role of APOBEC3G/F-mediated hypermutation in the control of human immunodeficiency virus type 1 in elite suppressors. Journal of virology. 2008; 82(6):3125–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Neogi U, Shet A, Sahoo PN, Bontell I, Ekstrand ML, Banerjea AC, et al. Human APOBEC3G-mediated hypermutation is associated with antiretroviral therapy failure in HIV-1 subtype C-infected individuals. Journal of the International AIDS Society. 2013; 16:18472 10.7448/IAS.16.1.18472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kieffer TL, Kwon P, Nettles RE, Han Y, Ray SC, Siliciano RF. G—>A hypermutation in protease and reverse transcriptase regions of human immunodeficiency virus type 1 residing in resting CD4+ T cells in vivo. Journal of virology. 2005; 79(3):1975–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Koulinska IN, Chaplin B, Mwakagile D, Essex M, Renjifo B. Hypermutation of HIV type 1 genomes isolated from infants soon after vertical infection. AIDS research and human retroviruses. 2003; 19(12):1115–23. [DOI] [PubMed] [Google Scholar]
- 47. Chelico L, Pham P, Calabrese P, Goodman MF. APOBEC3G DNA deaminase acts processively 3' —> 5' on single-stranded DNA. Nature structural & molecular biology. 2006; 13(5):392–9. [DOI] [PubMed] [Google Scholar]
- 48. Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Current biology: CB. 2004; 14(15):1392–6. [DOI] [PubMed] [Google Scholar]
- 49. Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, et al. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nature structural & molecular biology. 2004; 11(5):435–42. [DOI] [PubMed] [Google Scholar]
- 50. Hache G, Liddament MT, Harris RS. The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. The Journal of biological chemistry. 2005; 280(12):10920–4. [DOI] [PubMed] [Google Scholar]
- 51. Holtz CM, Sadler HA, Mansky LM. APOBEC3G cytosine deamination hotspots are defined by both sequence context and single-stranded DNA secondary structure. Nucleic acids research. 2013; 41(12):6139–48. 10.1093/nar/gkt246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Beale RC, Petersen-Mahrt SK, Watt IN, Harris RS, Rada C, Neuberger MS. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. Journal of molecular biology. 2004; 337(3):585–96. [DOI] [PubMed] [Google Scholar]
- 53. Liddament MT, Brown WL, Schumacher AJ, Harris RS. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Current biology: CB. 2004; 14(15):1385–91. [DOI] [PubMed] [Google Scholar]
- 54. Wiegand HL, Doehle BP, Bogerd HP, Cullen BR. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. The EMBO journal. 2004; 23(12):2451–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pathak VK, Temin HM. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: deletions and deletions with insertions. Proceedings of the National Academy of Sciences of the United States of America. 1990; 87(16):6024–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Parthasarathi S, Varela-Echavarria A, Ron Y, Preston BD, Dougherty JP. Genetic rearrangements occurring during a single cycle of murine leukemia virus vector replication: characterization and implications. Journal of virology. 1995; 69(12):7991–8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Julias JG, Kim T, Arnold G, Pathak VK. The antiretrovirus drug 3'-azido-3'-deoxythymidine increases the retrovirus mutation rate. Journal of virology. 1997; 71(6):4254–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sadler HA, Stenglein MD, Harris RS, Mansky LM. APOBEC3G contributes to HIV-1 variation through sublethal mutagenesis. Journal of virology. 2010; 84(14):7396–404. 10.1128/JVI.00056-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hache G, Mansky LM, Harris RS. Human APOBEC3 proteins, retrovirus restriction, and HIV drug resistance. AIDS reviews. 2006; 8(3):148–57. [PubMed] [Google Scholar]
- 60. Jern P, Russell RA, Pathak VK, Coffin JM. Likely role of APOBEC3G-mediated G-to-A mutations in HIV-1 evolution and drug resistance. PLoS pathogens. 2009; 5(4):e1000367 10.1371/journal.ppat.1000367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Deforche K, Camacho R, Laethem KV, Shapiro B, Moreau Y, Rambaut A, et al. Estimating the relative contribution of dNTP pool imbalance and APOBEC3G/3F editing to HIV evolution in vivo. Journal of computational biology: a journal of computational molecular cell biology. 2007; 14(8):1105–14. [DOI] [PubMed] [Google Scholar]
- 62. Ebrahimi D, Anwar F, Davenport MP. APOBEC3 has not left an evolutionary footprint on the HIV-1 genome. Journal of virology. 2011; 85(17):9139–46. 10.1128/JVI.00658-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wood N, Bhattacharya T, Keele BF, Giorgi E, Liu M, Gaschen B, et al. HIV evolution in early infection: selection pressures, patterns of insertion and deletion, and the impact of APOBEC. PLoS pathogens. 2009; 5(5):e1000414 10.1371/journal.ppat.1000414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Armitage AE, Deforche K, Chang CH, Wee E, Kramer B, Welch JJ, et al. APOBEC3G-induced hypermutation of human immunodeficiency virus type-1 is typically a discrete "all or nothing" phenomenon. PLoS genetics. 2012; 8(3):e1002550 10.1371/journal.pgen.1002550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mulder LC, Harari A, Simon V. Cytidine deamination induced HIV-1 drug resistance. Proceedings of the National Academy of Sciences of the United States of America. 2008; 105(14):5501–6. 10.1073/pnas.0710190105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kim EY, Bhattacharya T, Kunstman K, Swantek P, Koning FA, Malim MH, et al. Human APOBEC3G-mediated editing can promote HIV-1 sequence diversification and accelerate adaptation to selective pressure. Journal of virology. 2010; 84(19):10402–5. 10.1128/JVI.01223-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. McCallum M, Oliveira M, Ibanescu RI, Kramer VG, Moisi D, Asahchop EL, et al. Basis for early and preferential selection of the E138K mutation in HIV-1 reverse transcriptase. Antimicrobial agents and chemotherapy. 2013; 57(10):4681–8. 10.1128/AAC.01029-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Russell RA, Moore MD, Hu WS, Pathak VK. APOBEC3G induces a hypermutation gradient: purifying selection at multiple steps during HIV-1 replication results in levels of G-to-A mutations that are high in DNA, intermediate in cellular viral RNA, and low in virion RNA. Retrovirology. 2009; 6:16 10.1186/1742-4690-6-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Eyzaguirre LM, Charurat M, Redfield RR, Blattner WA, Carr JK, Sajadi MM. Elevated hypermutation levels in HIV-1 natural viral suppressors. Virology. 2013; 443(2):306–12. 10.1016/j.virol.2013.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Freed EO, Englund G, Martin MA. Role of the basic domain of human immunodeficiency virus type 1 matrix in macrophage infection. Journal of virology. 1995; 69(6):3949–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chen J, Nikolaitchik O, Singh J, Wright A, Bencsics CE, Coffin JM, et al. High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proceedings of the National Academy of Sciences of the United States of America. 2009; 106(32):13535–40. 10.1073/pnas.0906822106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Nikolaitchik OA, Dilley KA, Fu W, Gorelick RJ, Tai SH, Soheilian F, et al. Dimeric RNA recognition regulates HIV-1 genome packaging. PLoS pathogens. 2013; 9(3):e1003249 10.1371/journal.ppat.1003249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Moore MD, Fu W, Nikolaitchik O, Chen J, Ptak RG, Hu WS. Dimer initiation signal of human immunodeficiency virus type 1: its role in partner selection during RNA copackaging and its effects on recombination. Journal of virology. 2007; 81(8):4002–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Coffin JM. Structure, replication, and recombination of retrovirus genomes: some unifying hypotheses. The Journal of general virology. 1979; 42(1):1–26. [DOI] [PubMed] [Google Scholar]
- 75. Hu WS, Temin HM. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proceedings of the National Academy of Sciences of the United States of America. 1990; 87(4):1556–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Land AM, Ball TB, Luo M, Pilon R, Sandstrom P, Embree JE, et al. Human immunodeficiency virus (HIV) type 1 proviral hypermutation correlates with CD4 count in HIV-infected women from Kenya. Journal of virology. 2008; 82(16):8172–82. 10.1128/JVI.01115-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kourteva Y, De Pasquale M, Allos T, McMunn C, D'Aquila RT. APOBEC3G expression and hypermutation are inversely associated with human immunodeficiency virus type 1 (HIV-1) burden in vivo. Virology. 2012; 430(1):1–9. 10.1016/j.virol.2012.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DI, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013; 155(3):540–51. 10.1016/j.cell.2013.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Suspene R, Rusniok C, Vartanian JP, Wain-Hobson S. Twin gradients in APOBEC3 edited HIV-1 DNA reflect the dynamics of lentiviral replication. Nucleic acids research. 2006; 34(17):4677–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wurtzer S, Goubard A, Mammano F, Saragosti S, Lecossier D, Hance AJ, et al. Functional central polypurine tract provides downstream protection of the human immunodeficiency virus type 1 genome from editing by APOBEC3G and APOBEC3B. Journal of virology. 2006; 80(7):3679–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kijak GH, Janini LM, Tovanabutra S, Sanders-Buell E, Arroyo MA, Robb ML, et al. Variable contexts and levels of hypermutation in HIV-1 proviral genomes recovered from primary peripheral blood mononuclear cells. Virology. 2008; 376(1):101–11. 10.1016/j.virol.2008.03.017 [DOI] [PubMed] [Google Scholar]
- 82. Nikolaitchik OA, Galli A, Moore MD, Pathak VK, Hu WS. Multiple barriers to recombination between divergent HIV-1 variants revealed by a dual-marker recombination assay. Journal of molecular biology. 2011; 407(4):521–31. 10.1016/j.jmb.2011.01.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Motomura K, Chen J, Hu WS. Genetic recombination between human immunodeficiency virus type 1 (HIV-1) and HIV-2, two distinct human lentiviruses. Journal of virology. 2008; 82(4):1923–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Chin MP, Chen J, Nikolaitchik OA, Hu WS. Molecular determinants of HIV-1 intersubtype recombination potential. Virology. 2007; 363(2):437–46. [DOI] [PubMed] [Google Scholar]
- 85. Chin MP, Lee SK, Chen J, Nikolaitchik OA, Powell DA, Fivash MJ Jr., et al. Long-range recombination gradient between HIV-1 subtypes B and C variants caused by sequence differences in the dimerization initiation signal region. Journal of molecular biology. 2008; 377(5):1324–33. 10.1016/j.jmb.2008.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Galli A, Kearney M, Nikolaitchik OA, Yu S, Chin MP, Maldarelli F, et al. Patterns of Human Immunodeficiency Virus type 1 recombination ex vivo provide evidence for coadaptation of distant sites, resulting in purifying selection for intersubtype recombinants during replication. Journal of virology. 2010; 84(15):7651–61. 10.1128/JVI.00276-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Jung A, Maier R, Vartanian JP, Bocharov G, Jung V, Fischer U, et al. Recombination: Multiply infected spleen cells in HIV patients. Nature. 2002; 418(6894):144 [DOI] [PubMed] [Google Scholar]
- 88. Josefsson L, Palmer S, Faria NR, Lemey P, Casazza J, Ambrozak D, et al. Single cell analysis of lymph node tissue from HIV-1 infected patients reveals that the majority of CD4+ T-cells contain one HIV-1 DNA molecule. PLoS pathogens. 2013; 9(6):e1003432 10.1371/journal.ppat.1003432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Bandyopadhyay PK, Watanabe S, Temin HM. Recombination of transfected DNAs in vertebrate cells in culture. Proceedings of the National Academy of Sciences of the United States of America. 1984; 81(11):3476–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Miller CK, Temin HM. High-efficiency ligation and recombination of DNA fragments by vertebrate cells. Science. 1983; 220(4597):606–9. [DOI] [PubMed] [Google Scholar]
- 91. Cuevas JM, Geller R, Garijo R, Lopez-Aldeguer J, Sanjuan R. Extremely High Mutation Rate of HIV-1 In Vivo. PLoS biology. 2015; 13(9):e1002251 10.1371/journal.pbio.1002251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Simon V, Zennou V, Murray D, Huang Y, Ho DD, Bieniasz PD. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS pathogens. 2005; 1(1):e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kim EY, Lorenzo-Redondo R, Little SJ, Chung YS, Phalora PK, Maljkovic Berry I, et al. Human APOBEC3 induced mutation of human immunodeficiency virus type-1 contributes to adaptation and evolution in natural infection. PLoS pathogens. 2014; 10(7):e1004281 10.1371/journal.ppat.1004281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Armitage AE, Deforche K, Welch JJ, Van Laethem K, Camacho R, Rambaut A, et al. Possible footprints of APOBEC3F and/or other APOBEC3 deaminases, but not APOBEC3G, on HIV-1 from patients with acute/early and chronic infections. Journal of virology. 2014; 88(21):12882–94. 10.1128/JVI.01460-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Fourati S, Malet I, Binka M, Boukobza S, Wirden M, Sayon S, et al. Partially active HIV-1 Vif alleles facilitate viral escape from specific antiretrovirals. Aids. 2010; 24(15):2313–21. 10.1097/QAD.0b013e32833e515a [DOI] [PubMed] [Google Scholar]
- 96. Fourati S, Lambert-Niclot S, Soulie C, Wirden M, Malet I, Valantin MA, et al. Differential impact of APOBEC3-driven mutagenesis on HIV evolution in diverse anatomical compartments. Aids. 2014; 28(4):487–91. 10.1097/QAD.0000000000000182 [DOI] [PubMed] [Google Scholar]
- 97. Noguera-Julian M, Cozzi-Lepri A, Di Giallonardo F, Schuurman R, Daumer M, Aitken S, et al. Contribution of APOBEC3G/F activity to the development of low-abundance drug-resistant human immunodeficiency virus type 1 variants. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2015. [DOI] [PubMed] [Google Scholar]
- 98. Armitage AE, Katzourakis A, de Oliveira T, Welch JJ, Belshaw R, Bishop KN, et al. Conserved footprints of APOBEC3G on Hypermutated human immunodeficiency virus type 1 and human endogenous retrovirus HERV-K(HML2) sequences. Journal of virology. 2008; 82(17):8743–61. 10.1128/JVI.00584-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Yee JK, Miyanohara A, LaPorte P, Bouic K, Burns JC, Friedmann T. A general method for the generation of high-titer, pantropic retroviral vectors: highly efficient infection of primary hepatocytes. Proceedings of the National Academy of Sciences of the United States of America. 1994; 91(20):9564–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Kotsopoulou E, Kim VN, Kingsman AJ, Kingsman SM, Mitrophanous KA. A Rev-independent human immunodeficiency virus type 1 (HIV-1)-based vector that exploits a codon-optimized HIV-1 gag-pol gene. Journal of virology. 2000; 74(10):4839–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wu L, Martin TD, Vazeux R, Unutmaz D, KewalRamani VN. Functional evaluation of DC-SIGN monoclonal antibodies reveals DC-SIGN interactions with ICAM-3 do not promote human immunodeficiency virus type 1 transmission. Journal of virology. 2002; 76(12):5905–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Rose PP, Korber BT. Detecting hypermutations in viral sequences with an emphasis on G—> A hypermutation. Bioinformatics. 2000; 16(4):400–1. [DOI] [PubMed] [Google Scholar]
- 103. Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proceedings of the National Academy of Sciences of the United States of America. 2008; 105(21):7552–7. 10.1073/pnas.0802203105 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(Table A) Analysis of patient sequences that were predominantly hypermutated at GG sites. (Table B) Analysis of patient sequences that were predominantly hypermutated at GA sites. (Table C) Calculations used to prepare Table 4. (Table D) Calculations used to prepare Table 5.
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.