Abstract
Trib2 is highly expressed in human T cell acute lymphoblastic leukemia (T-ALL) and is a direct transcriptional target of the oncogenic drivers Notch and TAL1. In human TAL1-driven T-ALL cell lines, Trib2 is proposed to function as an important survival factor, but there is limited information about the role of Trib2 in primary T-ALL. In this study, we investigated the role of Trib2 in the initiation and maintenance of Notch-dependent T-ALL. Trib2 had no effect on the growth and survival of murine T-ALL cell lines in vitro when expression was blocked by shRNAs. To test the function of Trib2 on leukemogenesis in vivo, we generated Trib2 knockout mice. Mice were born at the expected Mendelian frequencies without gross developmental anomalies. Adult mice did not develop pathology or shortened survival, and hematopoiesis, including T cell development, was unperturbed. Using a retroviral model of Notch-induced T-ALL, deletion of Trib2 unexpectedly decreased the latency and increased the penetrance of T-ALL development in vivo. Immunoblotting of primary murine T-ALL cells showed that the absence of Trib2 increased C/EBPα expression, a known regulator of cell proliferation, and did not alter AKT or ERK phosphorylation. Although Trib2 was suggested to be highly expressed in T-ALL, transcriptomic analysis of two independent T-ALL cohorts showed that low Trib2 expression correlated with the TLX1-expressing cortical mature T-ALL subtype, whereas high Trib2 expression correlated with the LYL1-expressing early immature T-ALL subtype. These data indicate that Trib2 has a complex role in the pathogenesis of Notch-driven T-ALL, which may vary between different T-ALL subtypes.
Introduction
T cell acute lymphoblastic leukemia (T-ALL) is an aggressive hematologic malignancy resulting from the oncogenic transformation of T cell progenitors and accounting for 25% of adult and 15% of pediatric ALL cases [1]. Although intensified chemotherapy has markedly improved long-term survival, survival following relapse is poor, highlighting the need for novel therapies in recurrent disease. Activating mutations of Notch1 occur in about 50% of T-ALL cases, most of which occur in the heterodimerization domain (HD) and proline-glutamic acid- serine-threonine (PEST) domains [2]. NOTCH1 is a potentially important therapeutic target in T-ALL given the high prevalence of mutations and the significant role of NOTCH signaling in T-ALL. Although inhibiting Notch in mouse models of T-ALL caused marked anti-leukemic effects in vivo [3, 4], success in patients has met with difficulties, in part due to “on target” toxicity and possible drug resistance [5]. Therefore, identifying downstream Notch effectors may lead to the identification of novel therapeutic targets in T-ALL.
Previous studies identified Trib2, a member of the Tribbles protein family, as a direct transcriptional target of Notch1 in T-ALL cell lines [6, 7]. Tribbles are an evolutionarily conserved protein family that is implicated in diverse cellular events that include proliferation, migration, metabolism, and oncogenic transformation (reviewed in [8]). Tribbles was first characterized in Drosophila as an important cell cycle regulator [9]. In mammals, the Trib protein family members Trib1, Trib2 and Trib3 are characterized by a conserved pseudokinase domain [10], and COP1 [11] and MEK1 [12] binding domains. Trib proteins function as scaffolding molecules that facilitate protein degradation via a proteasome-dependent mechanism. In mammals, Trib1 and Trib2 promote C/EBPα degradation [6, 13] by recruiting the E3 ligase, COP1. Similarly, Trib3 promotes COP1-dependent degradation of acetyl CoA carboxylase (ACC), an enzyme involved in fatty acid synthesis [11]. The Trib proteins also modulate signaling pathways such as AKT [14] and MAPK [15]. Trib2 can inhibit AKT phosphorylation [16, 17] and Trib proteins interact with MEK1 and enhance ERK phosphorylation [15] through the MEK1 binding motif [12].
Trib2 is highly expressed in human T-ALL and T cells [18]. In addition to its association with Notch expression in T-ALL, a recent study showed that Trib2 is required for the growth and survival of human T-ALL cell lines driven by TAL1 [19]. In contrast, we found that shRNA-mediated knockdown of Trib2 in murine T-ALL cell lines did not affect cell growth or survival. To investigate the importance of Trib2 in T cell development and in T-ALL pathogenesis in vivo, we generated Trib2 knockout mice. Our results show that global deletion of Trib2 had no obvious defects on mouse development or survival. In particular, T cell development was normal. Strikingly, the absence of Trib2 resulted in a significant decrease in disease latency and increased disease penetrance in a murine model of Notch-driven T-ALL. Tumor cells from moribund mice expressed increased C/EBPα protein, while no change was seen in the amounts of phosphorylated AKT or ERK. Taken together, these data indicate that the influence of Trib2 expression on T cell leukemogenesis may vary in different contexts; however, in murine T-ALL, we identified an important role for Trib2 as a negative regulator of Notch-induced T-ALL in vivo.
Results
Murine T-ALL cells do not depend on persistent Trib2 expression
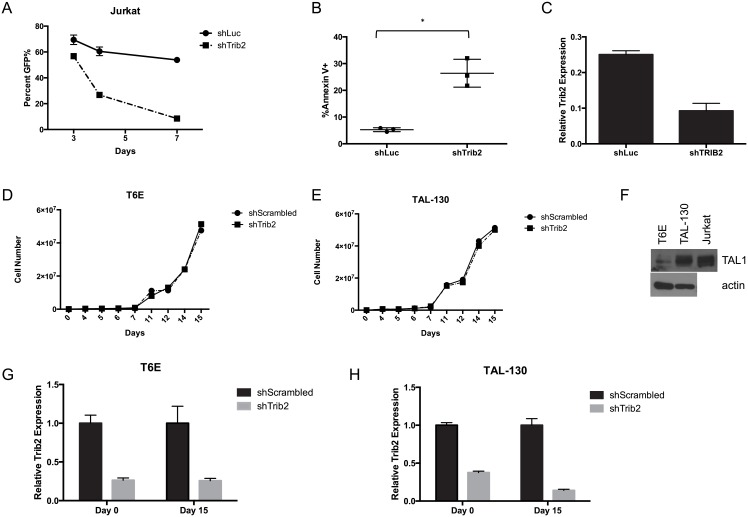
In order to determine the importance of Trib2 in the maintenance of T-ALL, we investigated the effect of Trib2 loss in T-ALL cell lines. A previous study identified Trib2 as an important survival factor in human TAL1-driven T-ALL cell lines, including Jurkat [19]. Consistent with these data, we observed a severe decrease in Jurkat cell growth (Fig 1A) due to apoptosis (Fig 1B) when Trib2 was knocked down (Fig 1C), suggesting an important role for Trib2 in the growth/survival of these cells. Activating mutations in NOTCH1 occur in over 50% of human T-ALLs [2]. Although Notch1 is mutated in Jurkat cells, growth and survival of these cells is Notch-independent [20, 21]. As Trib2 is a direct Notch target in Notch-induced murine T-ALL cells [7], we investigated the requirement for persistent Trib2 expression in these Notch-dependent cells. T6E cells [22], a murine Notch1-dependent T-ALL cell line, were transduced with shRNAs targeting Trib2 or a scrambled control sequence. Unlike Jurkat cells, Trib2 knockdown did not affect cell growth or survival (Fig 1D and data not shown). To determine if Trib2 dependency is specific for Tal1 expressing cell lines, we assayed the effects of Trib2 knockdown in TAL-130 cells, a T-ALL cell line derived from mice expressing a Tal1 transgene [23]. Knockdown of Trib2 by shRNA in TAL-130 cells showed no changes in cell growth (Fig 1E) or survival (data not shown). Immunoblotting confirmed that TAL-130 cells express higher levels of Tal1 than T6E cells (Fig 1F). Furthermore, both T6E and TAL-130 cells maintained Trib2 knockdown for at least 15 days after expressing the shRNA (Fig 1G and 1H), indicating that neither cell line requires Trib2 for proliferation or survival. Together, these data indicate that the requirement for persistent Trib2 expression varies between different T-ALL cell lines.
Fig 1. Trib2 is not required to maintain murine T-ALL cell lines.
Jurkat cells were transduced with normalized viral particles to express shRNAs against Luciferase (shLuc) or Trib2 (shTrib2) along with a GFP reporter. A) The percentage of GFP+ cells within the population was monitored for 7 days post-infection. B) At 3 days post-transduction, apoptosis was measured by Annexin V staining (“*”, p = 0.018) and C) Trib2 expression was measured by qPCR 3 days post-transduction in sorted GFP+ cells. Errors bars represent the standard deviation of biological replicates. D) T6E cells were transduced with shRNAs against a scrambled sequence (shScrambled) or Trib2 (shTrib2) and GFP as a surrogate marker. GFP+ cells were sorted 48 hours after transduction (day 0) and growth was monitored. E) TAL-130 cells were transduced with shRNAs against a scrambled sequence (shScrambled) or Trib2 (shTrib2) and GFP as a surrogate marker. GFP+ cells were sorted 48 hours after transduction (day 0) and growth was monitored. F) TAL1 expression was measured in T6E and TAL-130 cells by immunoblot. Jurkat lysates were used as a positive control. G & H) Trib2 expression was measured at days 0 and 15 in T6E (Panel G) or TAL-130 cells (Panel H). Error bars indicate standard deviation. Data are representative of 3 experiments.
Absence of T cell defects in Trib2 knockout mice
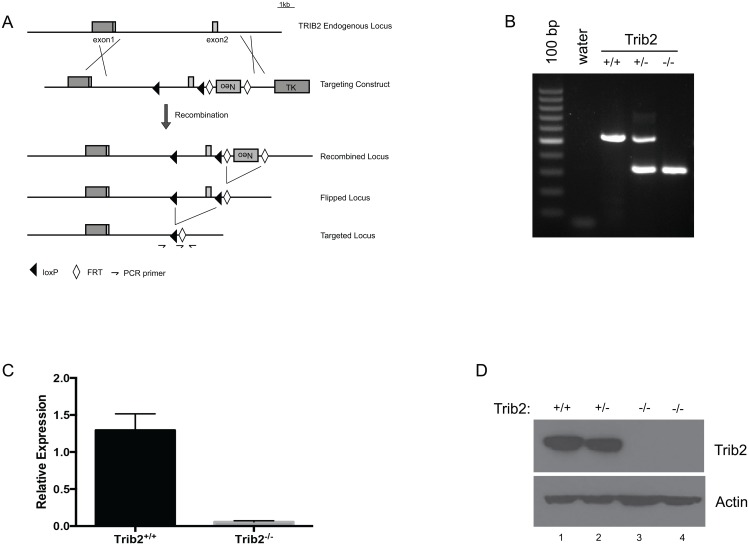
Trib2 is highly expressed in developing murine T cells and murine and human T-ALLs [18]. We therefore generated a genomic Trib2 knockout mouse to determine the importance of Trib2 in T cell development and disease initiation. LoxP sites flanking exon 2 of Trib2 were inserted into a targeting vector and germline transmission was achieved through homologous recombination. The mice were subsequently crossed to FLP1-transgenic mice to remove the neomycin cassette and then backcrossed for greater than 8 generations to the C57BL/6 background (Fig 2A). Mice were born at the expected Mendelian frequencies without gross developmental anomalies (data not shown). Adult mice did not develop pathology or shortened survival (data not shown). Deletion of exon 2 of Trib2 was confirmed by PCR genotyping of genomic DNA from offspring (Fig 2B). Trib2 mRNA and protein expression was not detected in Trib2-/- thymocytes as compared to Trib2+/+ or Trib2+/- littermate controls (Fig 2C and 2D).
Fig 2. Generation and characterization of Trib2 null mice.
A) The targeting strategy to delete Trib2 exon 2 is shown. B) The deletion of the targeted region is shown by PCR performed on tail genomic DNA. C) qPCR; and D) immunoblot showing the deletion of Trib2 mRNA and protein in mouse thymocytes. Error bars indicate standard deviation.
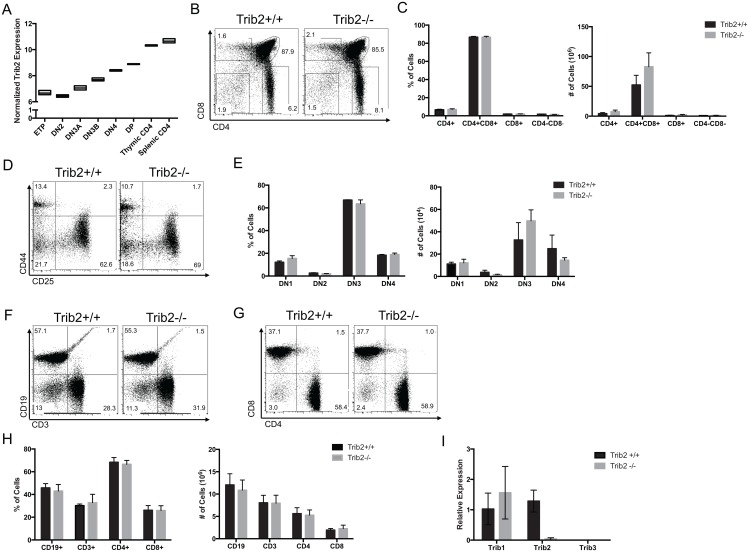
Trib2 mRNA expression increases throughout T cell differentiation from T lineage precursors to mature, resting T cells (Fig 3A). We therefore analyzed the effect of Trib2 loss in both developing T cells and early hematopoietic progenitor populations in the bone marrow. Deletion of Trib2 did not affect the percentage of hematopoietic stem cells or progenitors in the bone marrow (S1A and S1B Fig). Additionally, the survival of Trib2-/- mice did not differ from Trib2+/+ littermates in response to serial 5-fluorouracil treatment, suggesting an equivalent response to stress (S1C Fig). Immunophenotyping by flow cytometry showed no differences in the percentage or absolute numbers of T-lineage committed thymocytes (Fig 3B and 3C). Furthermore, no difference in the percentage or numbers of CD4/CD8 double negative cells at each stage of development was observed (Fig 3D and 3E). Additional data showed no difference in the percentage or numbers of CD19+ B cells or CD3+ T cells in the spleen (Fig 3F and 3H) or peripheral blood (data not shown) of Trib2-/- mice. Finally, no difference in the percentage or numbers of CD4 or CD8-expressing T cells was observed in the spleens of mice lacking Trib2 (Fig 3G and 3H). In addition, there did not appear to be compensatory changes in either Trib1 or Trib3 expression in Trib2-deficient thymocytes (Fig 3I). Thus, Trib2 absence does not impair T cell development.
Fig 3. Deletion of Trib2 does not affect T cell development.
A) Levels of Trib2 expression throughout T cell development as determined in the Immgen microarray data set are shown. B) Thymic subsets were analyzed by flow cytometry for CD4 and CD8 expression. Representative scatter plots (B) and C) graphs depicting the absolute numbers and percentages of cells in the total population are shown (n = 5–6 mice per group). D) CD4−CD8− double negative thymocytes were further assessed for the surface expression of CD44 and CD25. Representative scatter plots and E) graphs depicting the percentages and absolute numbers of cells in the total population are shown (n = 3 mice per group). F) The surface expression of CD3 and CD19, and G) CD4 and CD8 on splenocytes was assessed (n = 6 mice per group). H) Representative scatter plots and graphs depicting the percentages and absolute numbers of cells in the total splenocyte population are shown (n = 6 mice per group). Error bars indicate standard deviation. I) The expression of Trib1 and Trib3 were measured by qPCR (n = 3 per group).
Trib2 absence decreases latency and increases penetrance of Notch-induced T-ALL
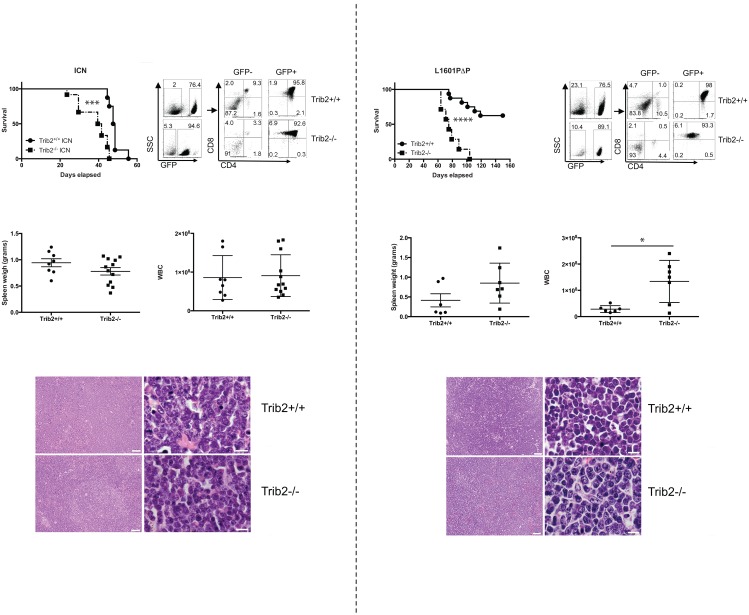
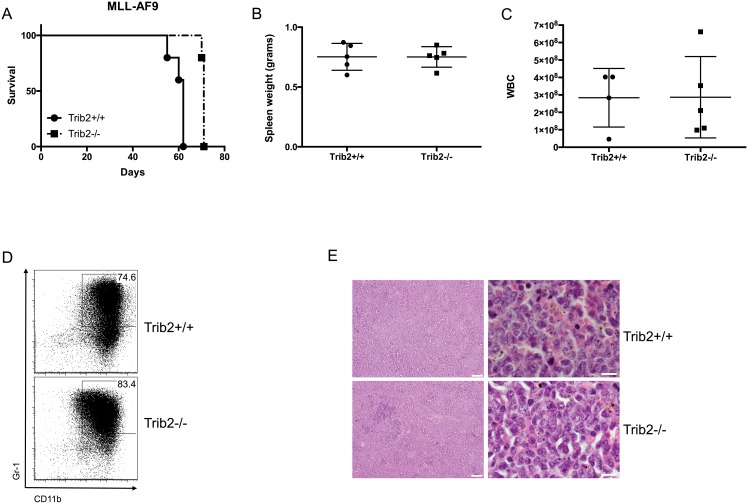
As Trib2 absence did not affect steady-state T cell development, we employed a murine T-ALL model to determine the importance of Trib2 in disease pathogenesis. Hematopoietic stem and progenitor cells from either Trib2-/- or wild-type (Trib2+/+) controls were transduced with retroviral supernatants expressing oncogenic forms of Notch along with GFP as a surrogate marker [24]. The cells were then injected into lethally irradiated syngeneic C57BL/6 recipients. Two different forms of oncogenic Notch were used for these studies; Notch1 L1601PΔP expresses a full length Notch1 allele that contains a destabilizing mutation in the HD domain and lacks the regulatory PEST domain, whereas ICN1 (intracellular Notch1) encodes only the intracellular Notch1 domain [24]. Notch1 L1601PΔP exhibits weaker activity than ICN1 in Notch1 reporter assays and induces T-ALL with longer latency and reduced penetrance [24]. Mice receiving Trib2-/- cells expressing ICN1 succumbed to T-ALL more quickly than their wild-type counterparts, with a median survival of 40 and 47.5 days, respectively (Fig 4A). Despite a decrease in survival time, the absence of Trib2 did not affect the expression of CD4 and CD8 on the surface of leukemic cells (Fig 4B). Additionally, moribund recipient mice showed similar spleen weights (Fig 4C), white blood cell counts (WBC; Fig 4D) and immature blast morphology (Fig 4E). The leukemic cells filled the spleen, obliterating the normal splenic architecture (Fig 4E). Leukemic cells from both Trib2-/- and Trib2+/+ donors also filled the bone marrow and infiltrated non-hematopoietic organs, such as liver and lung (data not shown). Consistent with previous results [24], L1601PΔP induced a partially penetrant phenotype with approximately 40% of the mice receiving Trib2+/+ cells transduced with L1601PΔP succumbing to T-ALL with a median survival of 97 days. In contrast, all of the mice receiving Trib2-/- cells transduced with L1601PΔP developed leukemia with a median survival of 75 days (Fig 4F). When comparing the mice that developed leukemia induced by L1601PΔP, no difference was observed between the Trib2+/+ and the Trib2-/- mice with regards to CD4 and CD8 expression on the leukemic cells (Fig 4G) or spleen weights (Fig 4H). Moribund Trib2-/- recipients had significantly higher WBCs than the control group (Fig 4I). The immature blast histopathology of these T-cell leukemias was similar to that previously reported [24] and between the two groups (Fig 4J and not shown). The leukemic cells were capable of serial transfer; however, mice receiving sorted Trib2-/- cells expressing either ICN or L1601PΔP succumbed to leukemia more quickly than those receiving sorted Trib2+/+ cells (S3 Fig). Thus, the absence of Trib2 increases the penetrance and decreases the latency of Notch1-induced T-ALL.
Fig 4. Loss of Trib2 decreases disease latency of Notch-driven T-ALL.
A) Kaplan-Meier curve of lethally irradiated mice reconstituted with Trib2+/+ or Trib2-/- cells expressing intracellular Notch1 (ICN; “***”, p = 0.0002). Mice with a body condition score of ≤2 and decreased mobility were euthanized (n = 8 Trib2+/+ and n = 12 Trib2-/- recipients. B) Flow cytometry was used to assess the immunophenotype of leukemic cells in the spleen. C) Spleen weights and D) WBC of moribund mice are shown. E) Representative H&E staining of spleens harvested from moribund mice (left panels, 10X magnification, scale bar = 50μM; right panels, 100X magnification, scale bar = 10μM). F) Kaplan-Meier curve of lethally irradiated mice reconstituted with Trib2+/+ or Trib2-/- cells expressing the weakly oncogenic Notch mutant, Notch 1 L1601PΔP (“****”, p<0.0001), (n = 16 Trib2+/+ and n = 7 Trib2-/- recipients). G) Flow cytometry was used to assess the immunophenotype of leukemic cells in the spleen. H) Spleen weights and I) WBC of moribund mice are shown (“*”, p = 0.013). J) H&E staining of spleens from moribund mice (left panels, 10X magnification, scale bar = 50μM; right panels, 100X magnification, scale bar = 10μM).
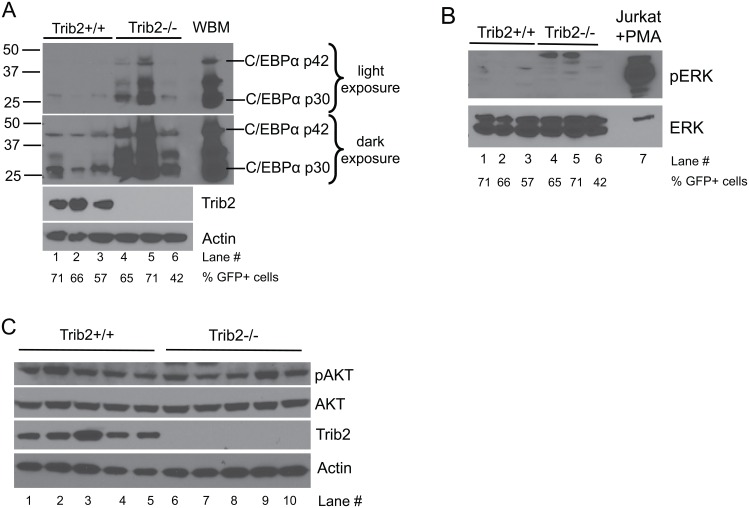
Absence of Trib2 is associated with increased C/EBPα expression in T-ALL cells
Trib2 regulates multiple signaling pathways including AKT [14, 16], MAPK [15] and C/EBPα [6]. There are two C/EBPα isoforms, p42 and p30, that are derived from post-transcriptional processing of a single transcript [25, 26]. C/EBPα p42 is anti-proliferative, whereas p30 is proliferative and a decrease in the p42/p30 ratio is associated with tumorigenesis [27, 28]. In myeloid cells, retroviral Trib2 expression decreased the p42/p30 ratio [6]. In the Trib2-deficient T-ALL cells, C/EBPα was markedly increased. Both the p42 and p30 isoforms were increased; however, p30 remained greater than p42 (Fig 5A). Despite a substantial increase in protein levels, the C/EBPα target genes Mpo [29], Ltf [30], and Lyz2 [31] remained undetectable in sorted tumor cells from Trib2+/+ and Trib2-/- mice, and expression of the myeloid marker Itgam did not change (data not shown). Phosphorylated ERK was not detectable in either group (Fig 5B) and no change in AKT phosphorylation was observed between Trib2-/- and Trib2+/+ tumor cells expressing ICN1 (Fig 5C). Together, these data implicate Trib2 as a suppressor of disease initiation in Notch-driven T-ALL through a process that may involve C/EBPα.
Fig 5. C/EBPα expression increases in the absence of Trib2 in primary tumor cells expressing oncogenic Notch.
Immunoblotting was used to visualize A) the expression of C/EBPα, and B) the phosphorylation status of ERK in Trib2+/+ or Trib2-/- splenocytes of moribund mice. Percentage of GFP+ cells in each sample; Lane 1: 71, Lane 2: 66, Lane 3: 57, Lane 4: 65, Lane 5: 71, Lane 6: 42. Whole bone marrow (WBM) (A) or Jurkat cells stimulated with PMA (B) were used as controls. In Panel A, lighter and darker exposures are provided. C) The phosphorylation status of AKT (pS473 AKT) was determined by immunoblot in GFP+ sorted splenocytes from moribund mice.
Absence of Trib2 does not affect AML induction by MLL-AF9
One potential explanation for the decreased latency of Notch-induced T-ALL in the Trib2-null cells was that the hematopoietic progenitors were more susceptible to retroviral transduction. To address this possibility, we used the MLL-AF9 retroviral model of AML induction [32]. We used identical methods to those used for the Notch bone marrow experiments with the exception that the MigR1 retrovirus expressed MLL-AF9. In contrast to Notch-induced T-ALL, absence of Trib2 did not decrease disease latency in this model. Instead, there was a slight increase in disease latency with a median survival of 62 days or 71 days for recipients of Trib2+/+ or Trib2-/- cells, respectively (Fig 6A). Both groups showed splenomegaly (Fig 6B) and increased WBCs (Fig 6C), while the animals that received empty vector were not affected (S2 Fig). The cells were largely Gr1+CD11b+ immature myeloid blasts that infiltrated multiple organs and filled the bone marrow (Fig 6D and 6E and data not shown). Thus, the decreased latency observed in the Notch T-ALL model is specific to T-ALL and not a general feature of leukemia induction in retroviral models transducing Trib2-null donor cells.
Fig 6. Trib2 does not suppress the initiation of MLL-AF9-driven myeloid leukemia.
A) Kaplan-Meier curve of lethally irradiated mice reconstituted with Trib2+/+ or Trib2-/- cells expressing MLL-AF9. Mice with a body condition score of ≤2 and decreased mobility were euthanized (n = 5 per group). B) Spleen weights and C) WBC of moribund mice are shown. D) Flow cytometry was used to assess the immunophenotype of leukemic cells in the spleen. E) H&E staining of spleens harvested from moribund mice (left panels, 10X magnification, scale bar = 50μM; right panels, 100X magnification, scale bar = 10μM).
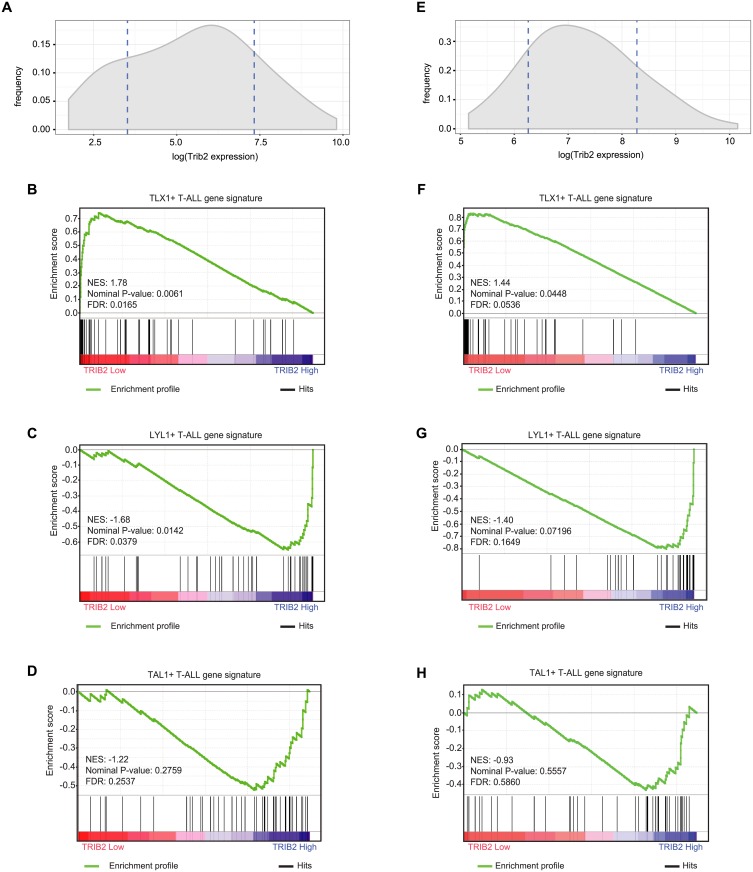
Trib2 expression correlates with different T-ALL subtypes
Our finding that absence of Trib2 promoted Notch-induced T-ALL was surprising as TRIB2 expression correlates with activated NOTCH1 in primary patient tumor samples [18]. T-ALL is not a uniform disease; instead, molecular profiling revealed signatures that specify different subtypes. Primary samples from two separate datasets (GSE13159 and GSE26713 including adult and pediatric T-ALL samples, respectively) were identified based on the expression of the oncogenes TLX1, LYL1 and TAL1, and grouped based on unique expression profiles, which correlate with recognized stages of normal thymocyte development; LYL1+ samples resemble thymocytes in the pre-T double negative stage of development, while TLX1+ and TAL1+ samples resemble thymocytes at the early and late cortical stages, respectively [33]. To determine whether TRIB2 expression varied between subtypes, we correlated the level of TRIB2 expression with the gene signatures of T-ALL molecular subtypes. The Gene Set Enrichment Analysis (GSEA) [34] analysis revealed a marked enrichment of the TLX1+ gene signature (cortical mature T-ALL) in low TRIB2 expressing T-ALL patients (Fig 7A, 7B, 7E and 7F), whereas the gene signature of LYL1+ (early immature T-ALL) significantly associated with samples expressing high levels of TRIB2 (Fig 7A, 7C, 7E and 7G). Interestingly, the gene signature of the TAL1+ T-ALL subtype (late cortical T-ALL) did not significantly correlate with the TRIB2 expression status (Fig 7D and 7H). Of particular significance, these correlations held true across two independent cohorts of T-ALL patients, including 174 adult (Fig 7A–7D) and 117 pediatric samples (Fig 7E–7H), respectively [35, 36]. Thus, these data suggest that Trib2 expression varies across human T-ALL subtypes.
Fig 7. TRIB2 expression correlates with the gene signatures of human early immature and cortical mature T-ALL subtypes.
A) Adult T-ALL samples with low (mean-SD) and high (mean+SD) TRIB2 gene expression (average of 202478_at and 202470_s_at) were identified in the GSE13159 dataset. Dashed lines indicate mean-SD and mean+SD of TRIB2 gene expression. B, C) GSEA analysis shows enrichment of gene signatures of the TLX1+ (B) and LYL1+ (C), but not TAL1+ (D), adult T-ALL molecular subtypes in samples with low and high TRIB2 expression, respectively (FDR < 0.05). E) Pediatric T-ALL samples with low (mean-SD and high (mean+SD) TRIB2 gene expression (average of 202478_at and 202470_s_at) were identified in the GSE26713 dataset. Dashed lines indicate mean-SD and mean+SD of TRIB2 gene expression. F, G) GSEA analysis shows enrichment of gene signatures of the TLX1+ (F) and LYL1+ (G), but not TAL1+ (H), pediatric T-ALL molecular subtypes in samples with low and high TRIB2 expression, respectively (FDR < 0.05). NES, normalized enrichment score.
Discussion
Our data show an important role for Trib2 in the induction of T-ALL by oncogenic Notch. While Trib2 is not required to maintain either a Notch-induced or a Tal1-induced murine T-ALL cell line, disease latency was decreased in mice transplanted with Trib2-/- cells expressing both strong and weak oncogenic Notch1 alleles. The weak oncogenic Notch1 allele L1601PΔP induced T-ALL in ~40% of mice with increased survival time relative to ICN1 when expressed in wild-type BM progenitors. However, when expressed in Trib2-deficient progenitors, penetrance increased to 100% while survival decreased compared to the wild-type controls. Together, these data suggest that Trib2 suppresses T-ALL pathogenesis. One possible explanation for our data is that the hematopoietic progenitors lacking Trib2 are more susceptible to retroviral transduction. If so, we would predict that leukemia survival would be reduced regardless of the oncogenic driver. This does not seem to be the case, however, as transduction of Trib2-deficient cells with the myeloid oncogene, MLL-AF9, did not decrease leukemia latency. This suggests that the decrease in leukemia survival is not a general property of Trib2-deficient hematopoietic progenitors.
In contrast to the effects of Trib2 absence on leukemogenesis, we did not observe effects on thymocyte development. This was somewhat surprising as the amount of active Notch1 in DN3 cells is similar to the amount in T-ALL cells [37], and Trib2 is relatively highly expressed (at least compared to Trib1 and Trib3) during T cell development. Of particular relevance to our study, defects were not observed in the DN3 to DP populations that are suggested to contain the leukemic stem cells in different models of Notch1-induced T-ALL [4, 38–40]. In addition to the thymus, the splenic CD4+ and CD8+ populations were present in the expected ratios; however, we cannot rule out the possibility that T cell function may be perturbed in the absence of Trib2.
Given the relatively high expression of Trib2 in developing T cells and its positive correlation with human T-ALL [18], we were surprised to find that Trib2 functioned as a tumor suppressor in our murine Notch1-induced T-ALL model. This led us to investigate Trib2 expression in the different subtypes of human T-ALL. The results from two independent T-ALL cohorts showed that TRIB2 was negatively correlated with the TLX1+ gene signature (cortical mature T-ALL) and positively correlated with the LYL1+ gene signature (early immature T-ALL). Interestingly, many of the genes associated with the expression of TLX1 are involved in cell growth and proliferation, including MYC [33]. Additional work is required to understand the implications of these associations.
Although weak activating Notch1 mutations including L1601PΔP are frequent in human T-ALL [2], few cooperating events that result in increased disease penetrance in vivo have been identified. Similar to our data showing that the loss of Trib2 results in disease penetrance of 100%, the absence of the tumor suppressor PTEN [41] or expression of oncogenic KRAS [24] both increase penetrance and decrease the latency of T-ALL induced by L1601PΔP. It remains to be determined whether the comparable effects on L1601PΔP-mediated disease are due to similar mechanisms by these different perturbations.
Previous studies suggest that Trib proteins can function as both dominant oncogenes [6, 42, 43] and tumor suppressors [44]. Trib3 was shown to enhance the tumorigenicity of a variety of tumor cell lines and xenografts via a mechanism that involved dysregulation of AKT phosphorylation by the mTORC2 complex and subsequent inactivation of the transcription factor FOXO3 [44]. Trib1 and Trib2 are also implicated as tumor suppressors in some subtypes of AML through inhibition of JNK signaling [45]. In contrast, Trib2 transformation in a murine AML model depends on C/EBPα degradation by a COP1-dependent mechanism. The overexpression of Trib2 in AML does not result in total C/EBPα degradation in the tumor cells; instead, there appears to be preferential degradation of the p42 C/EBPα isoform that results in a relative increase in expression of the p30 C/EBPα isoform [6]. The two C/EBPα isoforms are generated by post-transcriptional processes [25, 26] and differ in their amino termini; the N-terminal region unique to p42 contains transactivation elements that interact with transcriptional machinery. The two isoforms share a common C-terminal region that interacts with the chromatin remodeling complex and other transcription factors [46]. Functionally, C/EBPα p42 is anti-proliferative via its suppression of E2F. Conversely, C/EBPα p30 is unable to interact with E2F resulting in unchecked proliferation [27]. Expression of C/EBPα p30 in the absence of p42 is sufficient to induce AML in mice through a mechanism involving loss of proliferative control in myeloid progenitor populations [28]. In developing T cells, C/EBPα transcription is repressed by the Notch target, Hes1, which is important for T cell specification [47], a mechanism that may be reinforced by Trib2-induced degradation of C/EBPα protein. Unlike the scenario in Trib2-induced AML, the p42:p30 C/EBPα ratio is maintained in the Trib2-deficient leukemic T-ALL cells; however, the major distinction is that there is much more total C/EBPα protein in the absence of Trib2. One potential explanation is that the increased amount of C/EBPα promotes the pro-proliferative effects of the p30 isoform. Nevertheless, how the increased levels of C/EBPα promote the more aggressive disease in our Notch1-induced T-ALL models remains to be determined.
In summary, we show that Trib2 absence does not have gross effects on mouse development, and in particular on T cell development. This suggests that in some contexts, therapeutically targeting Trib2 may be a well-tolerated treatment strategy. However, our studies also describe a new role for Trib2 in the pathogenesis of T-ALL by a mechanism that may involve increased C/EBPα expression. Although our data clearly show that Trib2 absence enhances the severity of Notch1-induced murine T-ALL, additional studies are required to determine the relevance of these findings to human T-ALL.
Materials and Methods
Animals
To generate the Trib2 null mice, a 129S7/SvEv BAC clone, which covers the entire TRIB2 genomic locus, was obtained from the Sanger Institute (http://www.sanger.ac.uk/). The BAC recombineering system was utilized to construct a targeting vector (http://recombineering.ncifcrf.gov/) [48]. Two loxP sites were inserted to sandwich exon2. The FRT-flanked Neomycin cassette was cloned downstream of the 3’ loxP site. TL1 ES cells were electroporated with the linearized targeting vector. Homologous recombinants were screened with a combination of PCR and Southern blotting. Once germline transmission was observed, the mice were mated with FLP1-transgenic mice (B6;SJL-Tg(ACTFLPe)9205Dym/J, The Jackson Laboratory) to remove the Neomycin cassette. The flippase gene was bred out in the subsequent matings. The mice were then backcrossed to the C57BL/6 background for more than 8 generations. All mice were housed in specific pathogen-free facilities at the University of Pennsylvania. Experiments were performed according to the guidelines from the National Institutes of Health with approved protocols from the University of Pennsylvania Animal Care and Use Committee. The Body Condition Score Index (BCSI) [49] was an important criterion for monitoring the health of the mice. Mice were monitored at least three times per week and euthanized by CO2 inhalation followed by cervical dislocation.
Bone marrow transduction and transplantation
Bone marrow cells were collected from 6- to 12-week-old mice 4 days after intravenous administration of fluorouracil (5-FU) (250 mg/kg). The cells were cultured overnight in the presence of IL-3 (6 ng/ml), IL-6 (5 ng/ml), and SCF (100 ng/ml). The cells were then washed, resuspended in media containing cytokines and 4 μg/mL hexadimethidrine bromide (Sigma). Retroviral supernatants, normalized by titer, were added and the cells were centrifuged at 1,290 g for 90 minutes. A second round of spinoculation was performed the following day. After washing with PBS, at least 5 × 105 cells were injected intravenously into lethally irradiated (9 Gy) recipients. For secondary recipients, 2x106 sorted, GFP+ splenocytes were injected into sublethally irradiated (5 Gy) recipients. Mice were maintained on antibiotics in the drinking water for 2 weeks after BMT. Mice were bled every 2 weeks to monitor blood counts and evaluate the presence of circulating immature T cell progenitors by flow cytometry. Mice with a body condition score of ≤2 [49] and decreased mobility were euthanized and tissues were harvested for analysis.
Serial fluorouracil treatment
Mice received an intravenous dose of 5-FU on Day 0 (250mg/kg), Day 7 (150mg/kg) and Day 14 (150mg/kg). Mice with a body condition score of ≤2 and decreased mobility were euthanized.
Constructs and retroviruses
Production of high-titer virus was performed as described previously [50, 51]. Lentiviral pLKO.1 shRNA constructs were co-transfected into 293T/17 cells (ATCC, CRL-11268) with pMDL (gag-pol), pRSV-Rev and pHIT123 (envelope). Retroviral constructs were co-transfected into 293T cells with pCGP (gag-pol) and pHIT123 (envelope). Viral titers were determined using 3T3 fibroblasts. Viral supernatants were stored at -80°C.
Cell culture and transduction
All cell lines were maintained in RPMI 1640 (Cellgro) supplemented with 10% fetal bovine serum (FBS) (Hyclone), 1% penicillin/streptomycin (Gibco), 1% L-glutamine (Gibco) and 100 μM 2-mercaptoethanol (Sigma). Cells were centrifuged with viral supernatant and 8 μg/mL hexadimethidrine bromide (Sigma) at 1,290 g for 90 min at 25°C. The cells were analyzed or sorted at the indicated times after transduction. For growth curves, 5x104 cells were seeded. Cells were counted over time. Cell density was maintained at 1x106 cells/ml.
Flow cytometry and cell sorting
For the analysis of primary mouse cells, bone marrow, blood thymocytes and splenocytes were harvested and resuspended in PBS supplemented with 2% heat-inactivated FBS (Gibco). Antibodies used for cell surface staining (all obtained from eBiosciences except where noted) were CD3 (145-2C11), CD19 (1D3), CD4 (RM4-5), CD8 (53–6.7), CD25 (PC61; Biolegend), CD44 (IM7), Thy1.2 (53–2.1), CD11b (M1/70) and Gr-1 (RB6-8C5). Dead cells were excluded from analyses with 4',6-diamidino-2-phenylindole (DAPI). To detect Annexin V, Jurkat cells were stained following the manufacturer’s protocol. Briefly, an equal number of cells were resuspended in Annexin V binding buffer (10 mM Hepes pH 7.4, 140 mM NaCl, 2.5 mM CaCl2) with Annexin V antibody and incubated at room temperature in the dark for 15 minutes. The cells were then stored on ice and analyzed immediately. Cells were run on an LSR II flow cytometer (BD) and analyzed with FlowJo software v9.7 (TreeStar).
For cell sorting, primary splenocytes or cell lines were resuspended in PBS supplemented with 2% FBS, 1% penicillin/streptomycin and DAPI (for the exclusion of dead cells). GFP+ cells were sorted on a BD FACS Aria II using a 70μm nozzle at 70psi.
Quantitative PCR
RNA was extracted using TRIzol. cDNA was synthesized from RNA with the SuperScript III kit (Invitrogen). Transcripts were amplified with Taqman PCR master mix (Applied Biosystems), and qPCR was performed on the ABI ViiA 7 system (Applied Biosystems). mRNA quantities were normalized to 18S. Primer/probe sets to Trib2 (Mm01270457_m1) and 18S (4319413E) were purchased from Life Technologies.
Immunoblotting
Whole-cell lysates were prepared with RIPA buffer supplemented with protease inhibitors (Roche), NaF (10mM), PMSF (1mM) and Na3VO4 (1mM) or cells were directly lysed in SDS sample buffer. Proteins were separated using SDS-PAGE and transferred to PVDF membranes. Antibodies used for immunoblot from Cell Signaling Technology were pS473 AKT (4051), AKT (4691), pT202/Y204 ERK (9101), ERK (9102), Trib2 (13533), and C/EBPα (8178). The antibody against TAL1 was purchased from Santa Cruz Biotechnology (sc-12984). β-actin (A5316; Sigma) was used as a loading control. Secondary anti-mouse-HRP (NA931V) and anti-rabbit-HRP (NA934V) were obtained from GE Healthcare. Blots were visualized with SuperSignal west pico chemilumenscence (34080; Thermo Scientific) or SuperSignal west femto chemilumenscence substrate (34095; Thermo Scientific).
Histology
Freshly dissected organs were fixed in 10% neutral buffered formalin, paraffin embedded, sectioned, and stained with hematoxylin and eosin. Images were captured using an Olympus BX41 or a Nikon Eclipse Ni microscope.
Microarray data analysis
CEL files from GSE15907, Immunological Genome Project data Phase 1 [52], were normalized using robust multichip average (RMA) with the Bioconductor [53] “affy” package [54]. The normalized expression values of Trib2 in different T cell subsets were plotted in GraphPad Prism. Variation between biological replicates is indicated using a box and whisker plot.
GSEA analysis
The microarray-based gene expression measurements of human T-ALL samples were obtained from the GSE13159 and GSE26713 datasets. The samples from GSE13159 were GCRMA background adjusted, quantile normalized, PM corrected and batch corrected [53]. The samples from GSE26713 were normalized using RMA. Based on the expression level of TRIB2 (average of TRIB2 probes 202478_at and 202470_s_at) the low and high expressing TRIB2 T-ALL groups were identified as the samples with less than mean-SD or higher than mean+SD TRIB2 expression, respectively. Standard GSEA analyses were performed using the gene sets defined in C2:CGP collection of MSigDB V5.1.
Statistical analysis
All statistical analyses were performed using GraphPad Prism.
Supporting Information
A) Flow cytometry was used to assess the presence of lineage−Kit+Sca-1+ (LSK) hematopoietic progenitors and lineage−Kit+Sca-1+CD150+CD48− (LSK SLAM) hematopoietic stem cells in the bone marrow of Trib2+/+ and Trib2-/- mice. Representative plots are shown B) The average percentage of LSK and LSK SLAM cells in the bone marrow of Trib2+/+ and Trib2-/- mice is shown (n = 3 per group). C) Kaplan-Meier curve of mice serially injected with 5-FU. Mice received an intravenous dose on Day 0 (250mg/kg), Day 7 (150mg/kg) and Day 14 (150mg/kg). Mice with a body condition score of ≤2 and decreased mobility were euthanized (n = 5 per group; p = 0.06).
(EPS)
A) Flow cytometry was used to assess the immunophenotype of GFP+ cells in the spleen. B) The percentage of each cell type in the spleen is shown.
(EPS)
(EPS)
A) Jurkat, or B) TAL-130 cells were treated with 1μM of the GSI Compound E or DMSO for 48 hours and then the GSI was “washed out” and replaced with DMSO to permit Notch signaling. After 4 hrs, RNA was isolated and subjected to reverse transcription followed by quantitative PCR to determine the expression levels of Trib2 or Hes1 relative to EF1α.
(EPS)
Acknowledgments
We thank Michelle Kelliher for providing the TAL-130 cell line and A. Thomas Look for providing the shRNAs specific to human Trib2. The following cores at the University of Pennsylvania contributed to this study: mouse husbandry (University Laboratory Animal Resources), the Abramson Cancer Center Flow Cytometry Core (P30-CA016520), the Abramson Cancer Center Histology Core, and the Abramson Family Cancer Research Institute Cores. This work was supported by the National Institute of Health (NIH) T32 CA009140 and the American Cancer Society PF-15-065-01-TBG (to S.J.S.), NIH T32 HL0074 and T32 GM0071 (to E.A.M.), National Science Foundation DGE-1321851 and NIH F31 CA189661 (K.S.R.), the Leukemia and Lymphoma Society Fellow Award 5669–11 and NIH T32 HD007516 (to K.V.P.); the Children Cancer Free Foundation (Stichting Kinderen Kankervrij) KiKa-2010-082 (to J.P.P.M.), NIH P01 CA119070, NIH R01 AI047833, the Samuel Waxman Foundation for Cancer Research, and Alex's Lemonade Stand Foundation (to W.S.P.).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Institute of Health (NIH) T32 CA009140 and the American Cancer Society PF-15-065-01-TBG (to S.J.S.), NIH T32 HL0074 and T32 GM0071 (to E.A.M.), National Science Foundation DGE-1321851 and NIH F31 CA189661 (K.S.R.), the Leukemia and Lymphoma Society Fellow Award 5669-11 and NIH T32 HD007516 (to K.V.P.); the Children Cancer Free Foundation (Stichting Kinderen Kankervrij) KiKa-2010-082 (to J.P.P.M.), NIH P01 CA119070, NIH R01 AI047833, the Samuel Waxman Foundation for Cancer Research, and Alex’s Lemonade Stand Foundation (to W.S.P.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Van Vlierberghe P, Ferrando A. The molecular basis of T cell acute lymphoblastic leukemia. J Clin Invest. 2012;122:3398–406. 10.1172/JCI61269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–71. [DOI] [PubMed] [Google Scholar]
- 3.Cullion K, Draheim KM, Hermance N, Tammam J, Sharma VM, Ware C, et al. Targeting the Notch1 and mTOR pathways in a mouse T-ALL model. Blood. 2009;113:6172–81. 10.1182/blood-2008-02-136762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tatarek J, Cullion K, Ashworth T, Gerstein R, Aster JC, Kelliher MA. Notch1 inhibition targets the leukemia-initiating cells in a Tal1/Lmo2 mouse model of T-ALL. Blood. 2011;118:1579–90. 10.1182/blood-2010-08-300343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tosello V, Ferrando AA. The NOTCH signaling pathway: role in the pathogenesis of T-cell acute lymphoblastic leukemia and implication for therapy. Ther Adv Hematol. 2013;4:199–210. 10.1177/2040620712471368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keeshan K, He Y, Wouters BJ, Shestova O, Xu L, Sai H, et al. Tribbles homolog 2 inactivates C/EBPalpha and causes acute myelogenous leukemia. Cancer Cell. 2006;10:401–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wouters BJ, Jorda MA, Keeshan K, Louwers I, Erpelinck-Verschueren CA, Tielemans D, et al. Distinct gene expression profiles of acute myeloid/T-lymphoid leukemia with silenced CEBPA and mutations in NOTCH1. Blood. 2007;110:3706–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stein SJ, Mack EA, Rome KS, Pear WS. Tribbles in normal and malignant haematopoiesis. Biochem Soc Trans. 2015;43:1112–5. 10.1042/BST20150117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grosshans J, Wieschaus E. A genetic link between morphogenesis and cell division during formation of the ventral furrow in Drosophila. Cell. 2000;101:523–31. [DOI] [PubMed] [Google Scholar]
- 10.Hegedus Z, Czibula A, Kiss-Toth E. Tribbles: A family of kinase-like proteins with potent signalling regulatory function. Cell Signal. 2007;19:238–80. [DOI] [PubMed] [Google Scholar]
- 11.Qi L, Heredia JE, Altarejos JY, Screaton R, Goebel N, Niessen S, et al. TRB3 links the E3 ubiquitin ligase COP1 to lipid metabolism. Science. 2006;312:1763–6. [DOI] [PubMed] [Google Scholar]
- 12.Yokoyama T, Kanno Y, Yamazaki Y, Takahara T, Miyata S, Nakamura T. Trib1 links the MEK1/ERK pathway in myeloid leukemogenesis. Blood. 2010;116:2768–75. 10.1182/blood-2009-10-246264 [DOI] [PubMed] [Google Scholar]
- 13.Dedhia PH, Keeshan K, Uljon S, Xu L, Vega ME, Shestova O, et al. Differential ability of Tribbles family members to promote degradation of C/EBPalpha and induce acute myelogenous leukemia. Blood. 2010;116:1321–8. 10.1182/blood-2009-07-229450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du K, Herzig S, Kulkarni RN, Montminy M. TRB3: a tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science. 2003;300:1574–7. [DOI] [PubMed] [Google Scholar]
- 15.Kiss-Toth E, Bagstaff SM, Sung HY, Jozsa V, Dempsey C, Caunt JC, et al. Human tribbles, a protein family controlling mitogen-activated protein kinase cascades. J Biol Chem. 2004;279:42703–8. [DOI] [PubMed] [Google Scholar]
- 16.Naiki T, Saijou E, Miyaoka Y, Sekine K, Miyajima A. TRB2, a mouse Tribbles ortholog, suppresses adipocyte differentiation by inhibiting AKT and C/EBPbeta. J Biol Chem. 2007;282:24075–82. [DOI] [PubMed] [Google Scholar]
- 17.Keeshan K, Bailis W, Dedhia PH, Vega ME, Shestova O, Xu L, et al. Transformation by Tribbles homolog 2 (Trib2) requires both the Trib2 kinase domain and COP1 binding. Blood. 2010;116:4948–57. 10.1182/blood-2009-10-247361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hannon MM, Lohan F, Erbilgin Y, Sayitoglu M, O'Hagan K, Mills K, et al. Elevated TRIB2 with NOTCH1 activation in paediatric/adult T-ALL. Br J Haematol. 2012;158:626–34. 10.1111/j.1365-2141.2012.09222.x [DOI] [PubMed] [Google Scholar]
- 19.Sanda T, Lawton LN, Barrasa MI, Fan ZP, Kohlhammer H, Gutierrez A, et al. Core transcriptional regulatory circuit controlled by the TAL1 complex in human T cell acute lymphoblastic leukemia. Cancer Cell. 2012;22:209–21. 10.1016/j.ccr.2012.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med. 2007;204:1813–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao SS, O'Neil J, Liberator CD, Hardwick JS, Dai X, Zhang T, et al. Inhibition of NOTCH signaling by gamma secretase inhibitor engages the RB pathway and elicits cell cycle exit in T-cell acute lymphoblastic leukemia cells. Cancer Res. 2009;69:3060–8. 10.1158/0008-5472.CAN-08-4295 [DOI] [PubMed] [Google Scholar]
- 22.Pear WS, Aster JC, Scott ML, Hasserjian RP, Soffer B, Sklar J, et al. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med. 1996;183:2283–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma VM, Calvo JA, Draheim KM, Cunningham LA, Hermance N, Beverly L, et al. Notch1 contributes to mouse T-cell leukemia by directly inducing the expression of c-myc. Mol Cell Biol. 2006;26:8022–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiang MY, Xu L, Shestova O, Histen G, L'Heureux S, Romany C, et al. Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia. J Clin Invest. 2008;118:3181–94. 10.1172/JCI35090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin FT, MacDougald OA, Diehl AM, Lane MD. A 30-kDa alternative translation product of the CCAAT/enhancer binding protein alpha message: transcriptional activator lacking antimitotic activity. Proc Natl Acad Sci U S A. 1993;90:9606–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calkhoven CF, Muller C, Leutz A. Translational control of C/EBPalpha and C/EBPbeta isoform expression. Genes Dev. 2000;14:1920–32. [PMC free article] [PubMed] [Google Scholar]
- 27.Slomiany BA, D'Arigo KL, Kelly MM, Kurtz DT. C/EBPalpha inhibits cell growth via direct repression of E2F-DP-mediated transcription. Mol Cell Biol. 2000;20:5986–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirstetter P, Schuster MB, Bereshchenko O, Moore S, Dvinge H, Kurz E, et al. Modeling of C/EBPalpha mutant acute myeloid leukemia reveals a common expression signature of committed myeloid leukemia-initiating cells. Cancer Cell. 2008;13:299–310. 10.1016/j.ccr.2008.02.008 [DOI] [PubMed] [Google Scholar]
- 29.Ford AM, Bennett CA, Healy LE, Towatari M, Greaves MF, Enver T. Regulation of the myeloperoxidase enhancer binding proteins Pu1, C-EBP alpha, -beta, and -delta during granulocyte-lineage specification. Proc Natl Acad Sci U S A. 1996;93:10838–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Radomska HS, Huettner CS, Zhang P, Cheng T, Scadden DT, Tenen DG. CCAAT/enhancer binding protein alpha is a regulatory switch sufficient for induction of granulocytic development from bipotential myeloid progenitors. Mol Cell Biol. 1998;18:4301–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma O, Hong S, Guo H, Ghiaur G, Friedman AD. Granulopoiesis requires increased C/EBPalpha compared to monopoiesis, correlated with elevated Cebpa in immature G-CSF receptor versus M-CSF receptor expressing cells. PLoS One. 2014;9:e95784 10.1371/journal.pone.0095784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neff T, Sinha AU, Kluk MJ, Zhu N, Khattab MH, Stein L, et al. Polycomb repressive complex 2 is required for MLL-AF9 leukemia. Proc Natl Acad Sci U S A. 2012;109:5028–33. 10.1073/pnas.1202258109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferrando AA, Neuberg DS, Staunton J, Loh ML, Huard C, Raimondi SC, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1:75–87. [DOI] [PubMed] [Google Scholar]
- 34.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haferlach T, Kohlmann A, Wieczorek L, Basso G, Kronnie GT, Bene MC, et al. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: report from the International Microarray Innovations in Leukemia Study Group. J Clin Oncol. 2010;28:2529–37. 10.1200/JCO.2009.23.4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Homminga I, Pieters R, Langerak AW, de Rooi JJ, Stubbs A, Verstegen M, et al. Integrated transcript and genome analyses reveal NKX2-1 and MEF2C as potential oncogenes in T cell acute lymphoblastic leukemia. Cancer Cell. 2011;19:484–97. 10.1016/j.ccr.2011.02.008 [DOI] [PubMed] [Google Scholar]
- 37.Yashiro-Ohtani Y, He Y, Ohtani T, Jones ME, Shestova O, Xu L, et al. Pre-TCR signaling inactivates Notch1 transcription by antagonizing E2A. Genes Dev. 2009;23:1665–76. 10.1101/gad.1793709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiang MY, Shestova O, Xu L, Aster JC, Pear WS. Divergent effects of supraphysiologic Notch signals on leukemia stem cells and hematopoietic stem cells. Blood. 2013;121:905–17. 10.1182/blood-2012-03-416503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tremblay M, Tremblay CS, Herblot S, Aplan PD, Hebert J, Perreault C, et al. Modeling T-cell acute lymphoblastic leukemia induced by the SCL and LMO1 oncogenes. Genes Dev. 2010;24:1093–105. 10.1101/gad.1897910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X, Gounari F, Protopopov A, Khazaie K, von Boehmer H. Oncogenesis of T-ALL and nonmalignant consequences of overexpressing intracellular NOTCH1. J Exp Med. 2008;205:2851–61. 10.1084/jem.20081561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Medyouf H, Gao X, Armstrong F, Gusscott S, Liu Q, Gedman AL, et al. Acute T-cell leukemias remain dependent on Notch signaling despite PTEN and INK4A/ARF loss. Blood. 2010;115:1175–84. 10.1182/blood-2009-04-214718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grandinetti KB, Stevens TA, Ha S, Salamone RJ, Walker JR, Zhang J, et al. Overexpression of TRIB2 in human lung cancers contributes to tumorigenesis through downregulation of C/EBPalpha. Oncogene. 2011;30:3328–35. 10.1038/onc.2011.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jin G, Yamazaki Y, Takuwa M, Takahara T, Kaneko K, Kuwata T, et al. Trib1 and Evi1 cooperate with Hoxa and Meis1 in myeloid leukemogenesis. Blood. 2007;109:3998–4005. [DOI] [PubMed] [Google Scholar]
- 44.Salazar M, Lorente M, Garcia-Taboada E, Perez Gomez E, Davila D, Zuniga-Garcia P, et al. Loss of Tribbles pseudokinase-3 promotes Akt-driven tumorigenesis via FOXO inactivation. Cell Death Differ. 2015;22:131–44. 10.1038/cdd.2014.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gilby DC, Sung HY, Winship PR, Goodeve AC, Reilly JT, Kiss-Toth E. Tribbles-1 and -2 are tumour suppressors, down-regulated in human acute myeloid leukaemia. Immunol Lett. 2010;130:115–24. 10.1016/j.imlet.2009.12.007 [DOI] [PubMed] [Google Scholar]
- 46.Koschmieder S, Halmos B, Levantini E, Tenen DG. Dysregulation of the C/EBPalpha differentiation pathway in human cancer. J Clin Oncol. 2009;27:619–28. 10.1200/JCO.2008.17.9812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Obaldia ME, Bell JJ, Wang X, Harly C, Yashiro-Ohtani Y, DeLong JH, et al. T cell development requires constraint of the myeloid regulator C/EBP-alpha by the Notch target and transcriptional repressor Hes1. Nat Immunol. 2013;14:1277–84. 10.1038/ni.2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ullman-Cullere MH, Foltz CJ. Body condition scoring: a rapid and accurate method for assessing health status in mice. Lab Anim Sci. 1999;49:319–23. [PubMed] [Google Scholar]
- 50.Aster JC, Xu L, Karnell FG, Patriub V, Pui JC, Pear WS. Essential roles for ankyrin repeat and transactivation domains in induction of T-cell leukemia by notch1. Mol Cell Biol. 2000;20:7505–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci U S A. 1993;90:8392–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heng TS, Painter MW, Immunological Genome Project C. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–4. 10.1038/ni1008-1091 [DOI] [PubMed] [Google Scholar]
- 53.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) Flow cytometry was used to assess the presence of lineage−Kit+Sca-1+ (LSK) hematopoietic progenitors and lineage−Kit+Sca-1+CD150+CD48− (LSK SLAM) hematopoietic stem cells in the bone marrow of Trib2+/+ and Trib2-/- mice. Representative plots are shown B) The average percentage of LSK and LSK SLAM cells in the bone marrow of Trib2+/+ and Trib2-/- mice is shown (n = 3 per group). C) Kaplan-Meier curve of mice serially injected with 5-FU. Mice received an intravenous dose on Day 0 (250mg/kg), Day 7 (150mg/kg) and Day 14 (150mg/kg). Mice with a body condition score of ≤2 and decreased mobility were euthanized (n = 5 per group; p = 0.06).
(EPS)
A) Flow cytometry was used to assess the immunophenotype of GFP+ cells in the spleen. B) The percentage of each cell type in the spleen is shown.
(EPS)
(EPS)
A) Jurkat, or B) TAL-130 cells were treated with 1μM of the GSI Compound E or DMSO for 48 hours and then the GSI was “washed out” and replaced with DMSO to permit Notch signaling. After 4 hrs, RNA was isolated and subjected to reverse transcription followed by quantitative PCR to determine the expression levels of Trib2 or Hes1 relative to EF1α.
(EPS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.