Abstract
Simulations of the folding pathways of two large alpha/beta proteins, the alpha subunit of tryptophan synthase and triose phosphate isomerase, are reported using the knight's walk lattice model of globular proteins and Monte Carlo dynamics. Starting from randomly generated unfolded states and with no assumptions regarding the nature of the folding intermediates, for the tryptophan synthase subunit these simulations predict, in agreement with experiment, the existence and location of a stable equilibrium intermediate comprised of six beta strands on the amino terminus of the molecule. For the case of triose phosphate isomerase, the simulations predict that both amino- and carboxyl-terminal intermediates should be observed. In a significant modification of previous lattice models, this model includes a full heavy atom side chain description and is capable of representing native conformations at the level of 2.5- to 3-A rms deviation for the C alpha positions, as compared to the crystal structure. With a well-balanced compromise between accuracy of the protein description and the computer requirements necessary to perform simulations spanning biologically significant amounts of time, the lattice model described here brings the possibility of studying important biological processes to present-day computers.
Full text
PDF

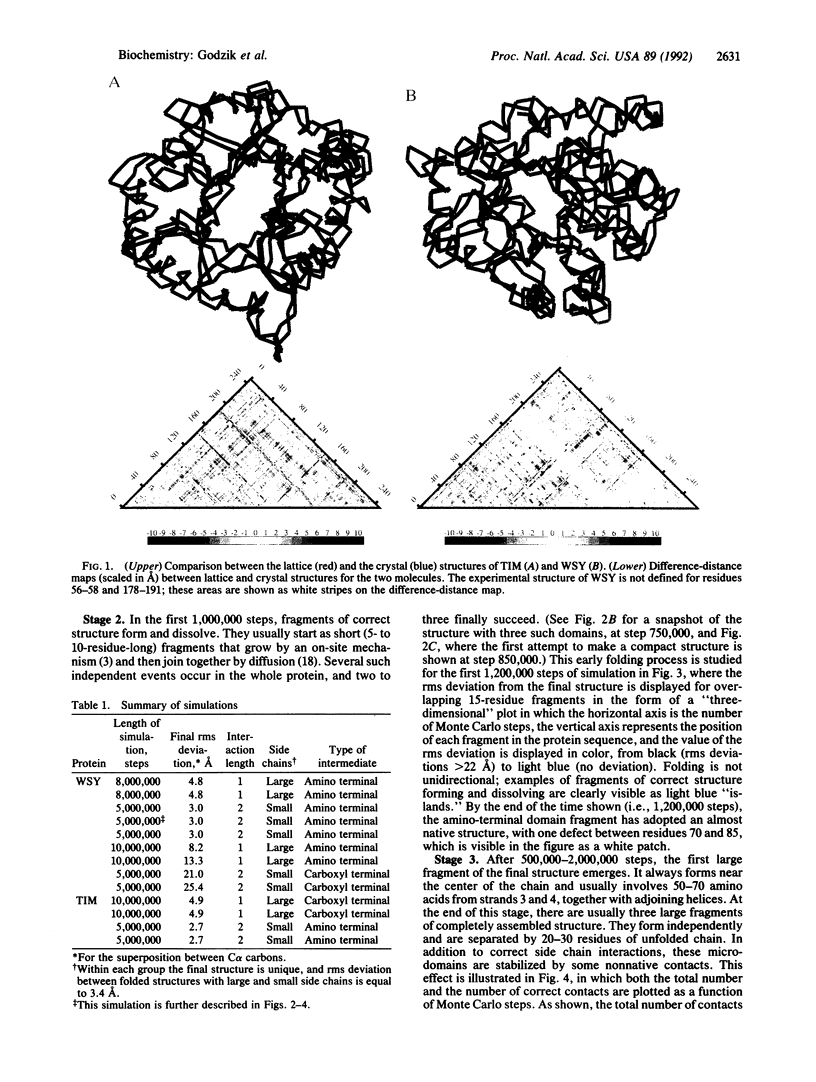
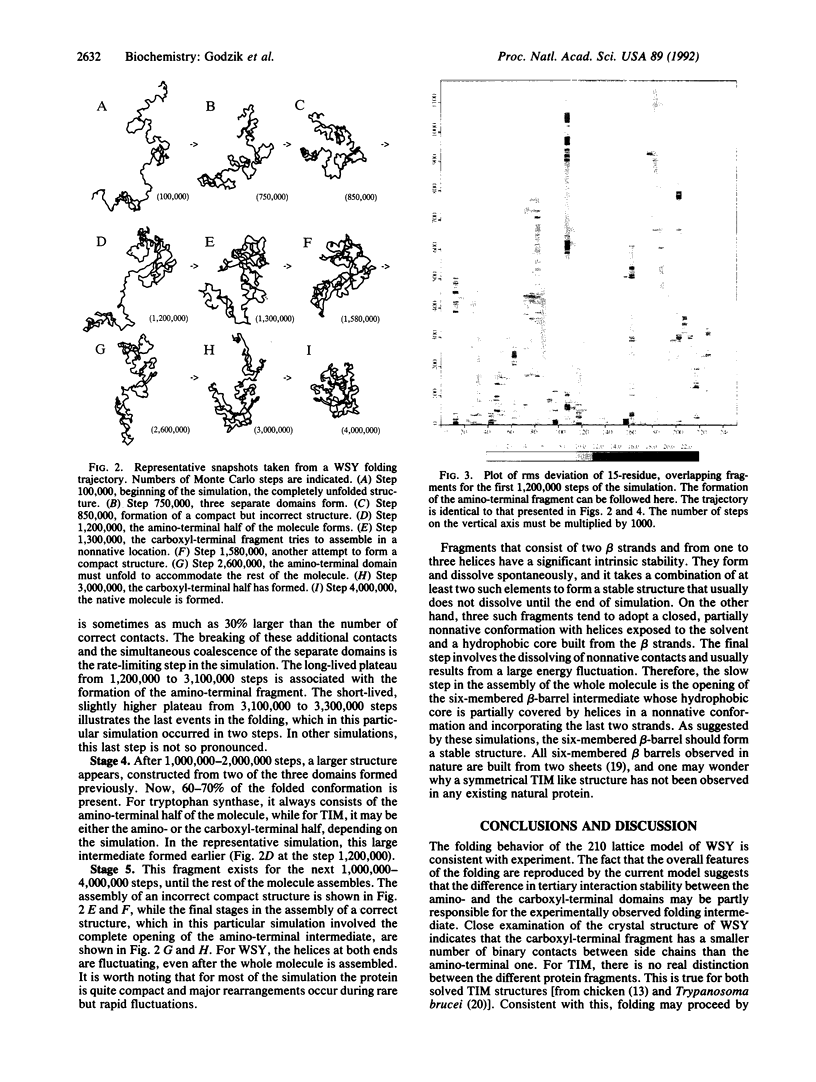
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banner D. W., Bloomer A. c., Petsko G. A., Phillips D. C., Wilson I. A. Atomic coordinates for triose phosphate isomerase from chicken muscle. Biochem Biophys Res Commun. 1976 Sep 7;72(1):146–155. doi: 10.1016/0006-291x(76)90972-4. [DOI] [PubMed] [Google Scholar]
- Behe M. J., Lattman E. E., Rose G. D. The protein-folding problem: the native fold determines packing, but does packing determine the native fold? Proc Natl Acad Sci U S A. 1991 May 15;88(10):4195–4199. doi: 10.1073/pnas.88.10.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton T. E. Protein folding. Biochem J. 1990 Aug 15;270(1):1–16. doi: 10.1042/bj2700001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber G. K., Petsko G. A. The evolution of alpha/beta barrel enzymes. Trends Biochem Sci. 1990 Jun;15(6):228–234. doi: 10.1016/0968-0004(90)90035-a. [DOI] [PubMed] [Google Scholar]
- Hyde C. C., Ahmed S. A., Padlan E. A., Miles E. W., Davies D. R. Three-dimensional structure of the tryptophan synthase alpha 2 beta 2 multienzyme complex from Salmonella typhimurium. J Biol Chem. 1988 Nov 25;263(33):17857–17871. [PubMed] [Google Scholar]
- Karplus M., Petsko G. A. Molecular dynamics simulations in biology. Nature. 1990 Oct 18;347(6294):631–639. doi: 10.1038/347631a0. [DOI] [PubMed] [Google Scholar]
- Lasters I., Wodak S. J., Pio F. The design of idealized alpha/beta-barrels: analysis of beta-sheet closure requirements. Proteins. 1990;7(3):249–256. doi: 10.1002/prot.340070306. [DOI] [PubMed] [Google Scholar]
- Lim W. A., Sauer R. T. The role of internal packing interactions in determining the structure and stability of a protein. J Mol Biol. 1991 May 20;219(2):359–376. doi: 10.1016/0022-2836(91)90570-v. [DOI] [PubMed] [Google Scholar]
- Luger K., Hommel U., Herold M., Hofsteenge J., Kirschner K. Correct folding of circularly permuted variants of a beta alpha barrel enzyme in vivo. Science. 1989 Jan 13;243(4888):206–210. doi: 10.1126/science.2643160. [DOI] [PubMed] [Google Scholar]
- Richardson J. S. The anatomy and taxonomy of protein structure. Adv Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- Sikorski A., Skolnick J. Dynamic Monte Carlo simulations of globular protein folding/unfolding pathways. II. Alpha-helical motifs. J Mol Biol. 1990 Apr 20;212(4):819–836. doi: 10.1016/0022-2836(90)90238-h. [DOI] [PubMed] [Google Scholar]
- Skolnick J., Kolinski A. Dynamic Monte Carlo simulations of a new lattice model of globular protein folding, structure and dynamics. J Mol Biol. 1991 Sep 20;221(2):499–531. doi: 10.1016/0022-2836(91)80070-b. [DOI] [PubMed] [Google Scholar]
- Skolnick J., Kolinski A. Dynamic Monte Carlo simulations of globular protein folding/unfolding pathways. I. Six-member, Greek key beta-barrel proteins. J Mol Biol. 1990 Apr 20;212(4):787–817. doi: 10.1016/0022-2836(90)90237-g. [DOI] [PubMed] [Google Scholar]
- Skolnick J., Kolinski A. Simulations of the folding of a globular protein. Science. 1990 Nov 23;250(4984):1121–1125. doi: 10.1126/science.250.4984.1121. [DOI] [PubMed] [Google Scholar]
- Tweedy N. B., Hurle M. R., Chrunyk B. A., Matthews C. R. Multiple replacements at position 211 in the alpha subunit of tryptophan synthase as a probe of the folding unit association reaction. Biochemistry. 1990 Feb 13;29(6):1539–1545. doi: 10.1021/bi00458a027. [DOI] [PubMed] [Google Scholar]
- Wierenga R. K., Kalk K. H., Hol W. G. Structure determination of the glycosomal triosephosphate isomerase from Trypanosoma brucei brucei at 2.4 A resolution. J Mol Biol. 1987 Nov 5;198(1):109–121. doi: 10.1016/0022-2836(87)90461-x. [DOI] [PubMed] [Google Scholar]
- Wright P. E., Dyson H. J., Lerner R. A. Conformation of peptide fragments of proteins in aqueous solution: implications for initiation of protein folding. Biochemistry. 1988 Sep 20;27(19):7167–7175. doi: 10.1021/bi00419a001. [DOI] [PubMed] [Google Scholar]