Fig 1, Fig 4A and 4C, Fig 8C and 8D, S2 Fig and S8 Fig are incorrectly labeled. Please see the complete, correct figure captions here.
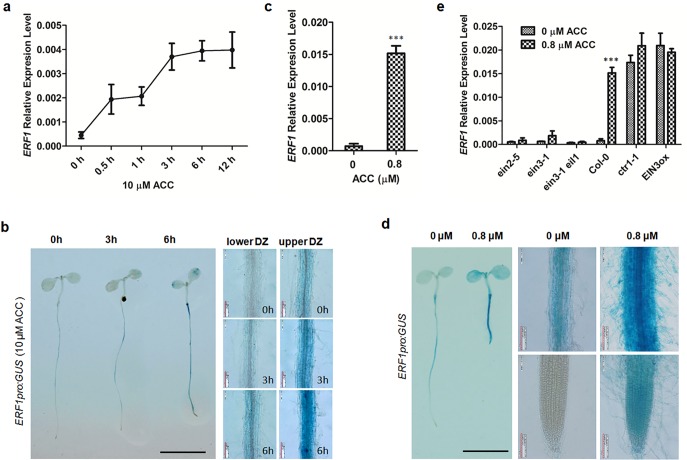
Fig 1. ERF1 expression is responsive to ethylene.
(a) Ethylene-induced ERF1 expression in wildtype. Seeds of Col-0 were germinated on MS medium for 5 d then treated with 10 μM ACC for 0, 0.5, 1, 3, 6, and 12 h. The transcriptional level of ERF1 was detected by quantitative RT-PCR (qRT-PCR). Values are mean ± SD of three replicates. ACC, 1-aminocyclopropane-1-carboxylic acid (precursor of ethylene biosynthesis). (b) Ethylene-induced expression ofERF1pro:GUS. Five-day-old seedlings of transgenic lines were treated with 10 μM ACC for 0, 3, and 6 h before GUS staining. Upper DZ and lower DZ represent different primary root regions. Scale bar, 0.5 cm. (c) Ethylene-induced expression of ERF1 in wildtype. Seeds of Col-0 were germinated on MS medium with 0 or 0.8 μM ACC for 5 d, and relative ERF1 transcription levels were measured by qRT-PCR. Values are mean ± SD of three replicates (***P<0.001). Asterisks indicate Student’s t-test significant differences. (d) Ethylene-activated expression in ERF1pro:GUS lines. Transgenic plants were grown on MS medium with either 0 or 0.8 μM ACC for 5 d before GUS staining assay. Scale bar, 0.5 cm. (e) The relative ERF1 expression level was determined in ethylene signaling related mutants ein2-5, ein3-1, ein3-1eil1and compared to wildtype (Col-0) seedlings. Seedlings of constitutive (ctr1-1) and β-estradiol inducible EIN3-FLAG (iE/qm) (EIN3ox) expression were also examined. Seeds (ein2-5, ein3-1, ein3-1eil1,ctr1-1and Col-0) were geminated on MS medium with either 0 or 0.8 μM ACC for 5 d. Seeds of EIN3ox were grown on medium containing 1 μM β-estradiol and 0 or 0.8 μM ACC for 5 d. Roots of seedlings were used for qRT-PCR analysis. Values are mean ± SD of three replicas (***P<0.001). Asterisks indicate Student’s t-test significant differences.
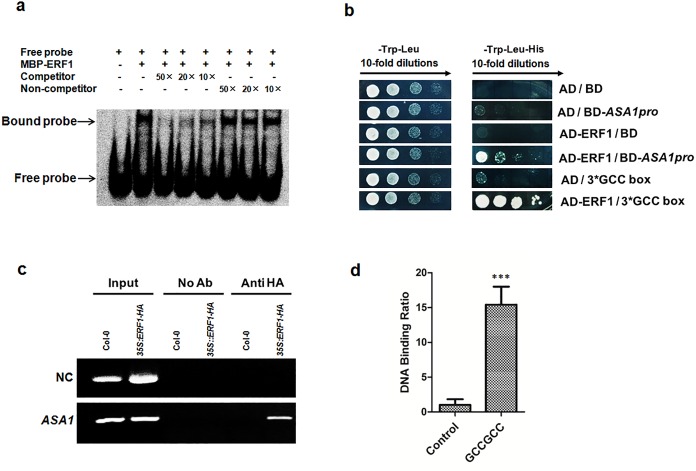
Fig 4. ERF1 directly binds to ASA1 promoter region in vitro and in vivo.
(a) EMSA assay for binding to GCC-box sequence in the promoter of ASA1 byERF1 protein in vitro. Dig-labeled probes were incubated with ERF1-MBP protein. As indicated, unlabelled probes were used as competitors, unlabelled probes with mutated GCC-box sequence were used as non-competitors, and the ERF1-MBP protein bound probes were separated from free probes by an acrylamide gel. (b) Yeast-one-hybrid assay. pGADT7/ERF1 (AD-ERF1) and pHIS2/ASA1pro (BD-ASA1pro) constructs were co-transformed into yeast strain Y187. AD-empty and BD-empty, AD-empty and BD-ASA1pro, AD-ERF1 and BD-empty, AD-empty and BD-3*GCC-box were used as negative controls while AD-ERF1 and BD-3*GCC-box were used as a positive control. (c). Chromatin immunoprecipitation-PCR for ASA1 promoter. Roots of 5-day-old 35S:HA:ERF1 and Col-0 seedlings were used. Anti-HA antibodies were used for the enrichment of the DNA fragments containing GCC-box in the promoter ofASA1. The results were determined by PCR. Tub8 was used as negative control (NC). (d) Quantitative real-time PCR was performed using the same ChIP products and PCR primers flanking GCC-boxes in ASA1 promoter as in c. The region of ASA1 that do not contain GCC-box was used as negative control. Values are mean ± SD of three replicas (***P<0.001). Asterisks indicate Student’s t-test significant differences.
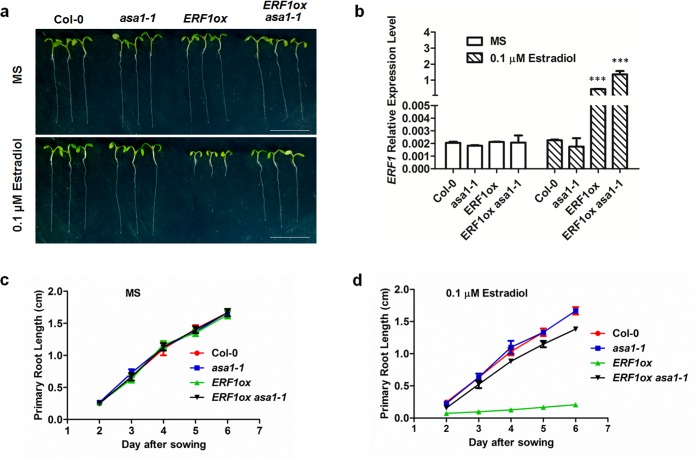
Fig 8. ASA1 acts downstream of ERF1.
(a) The primary root phenotypes of Col-0, asa1-1, ERF1ox and ERF1ox asa1-1 seedlings grown on MS medium with either 0 or 0.1 μM estradiol for 5 d. ERF1ox is the transgenic plants expressing ERF1 protein under control of the estradiol-inducible promoter in Col-0 background. Scale bar, 1 cm. (b) qRT–PCR analysis of transcriptions of ERF1. The roots of 5-day-old Col-0, asa1-1, ERF1ox and ERF1ox asa1-1 seedlings grown on MS medium with either 0 or 0.1 μM estradiol were used. Values are mean ± SD of three replicas (***P<0.001. Asterisks indicate Student’s t-test significant differences). (c-d). Primary root length of Col-0, asa1-1, ERF1ox and ERF1ox asa1-1 seedlings grown on MS medium with either 0 or 0.1μM estradiol were measured. Data shown are average and SD (Values are mean ± SD, n = 20).
Supporting Information
(a) Seeds of the transgenic lines and wildtype were germinated vertically on MS medium for 5 days, and the representative seedlings were photographed. Scale bar, 1 cm. (b) The primary root length of the 5-d-old transgenic lines and wildtype was measured. Data shown are average and SD (n = 20, *P<0.05, **P<0.01, ***P<0.001. Asterisks indicate Student’s t-test significant differences). (c) The expression level of ERF1 in these materials was tested by qRT-PCR. Values are mean ± SD of three replicates (*P<0.05, ***P<0.001. Asterisks indicate Student’s t-test significant differences).
(DOC)
(a) The primary root phenotypes of Col-0, asa1-1, ERF1ox and ERF1ox asa1-1 seedlings grown on MS medium with either 0 or 1 μM ACC for 5 d. Scale bar, 1 cm. (b-c) Primary root length of Col-0, asa1-1, ERF1ox and ERF1ox asa1-1 seedlings grown on MS medium with either 0 or 1 μM ACC were measured. Data shown are average and SD (Values are mean ± SD, n = 20).
(DOC)
Reference
- 1.Mao J-L, Miao Z-Q, Wang Z, Yu L-H, Cai X-T, Xiang C-B (2016) Arabidopsis ERF1 Mediates Cross-Talk between Ethylene and Auxin Biosynthesis during Primary Root Elongation by Regulating ASA1 Expression. PLoS Genet 12(1): e1005760 doi:10.1371/journal.pgen.1005760 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(a) Seeds of the transgenic lines and wildtype were germinated vertically on MS medium for 5 days, and the representative seedlings were photographed. Scale bar, 1 cm. (b) The primary root length of the 5-d-old transgenic lines and wildtype was measured. Data shown are average and SD (n = 20, *P<0.05, **P<0.01, ***P<0.001. Asterisks indicate Student’s t-test significant differences). (c) The expression level of ERF1 in these materials was tested by qRT-PCR. Values are mean ± SD of three replicates (*P<0.05, ***P<0.001. Asterisks indicate Student’s t-test significant differences).
(DOC)
(a) The primary root phenotypes of Col-0, asa1-1, ERF1ox and ERF1ox asa1-1 seedlings grown on MS medium with either 0 or 1 μM ACC for 5 d. Scale bar, 1 cm. (b-c) Primary root length of Col-0, asa1-1, ERF1ox and ERF1ox asa1-1 seedlings grown on MS medium with either 0 or 1 μM ACC were measured. Data shown are average and SD (Values are mean ± SD, n = 20).
(DOC)