Abstract
It is extremely rare for a single experiment to be so impactful and timely that it shapes and forecasts the experiments of the next decade. Here, we review how two such experiments --the generation of human induced pluripotent stem cells (iPSCs) and the development of CRISPR/Cas9 technology-- have fundamentally reshaped our approach to biomedical research, stem cell biology and human genetics. We will also highlight the previous knowledge that iPSC and CRISPR/Cas9 technologies were built on as this groundwork demonstrated the need for solutions and the benefits that these technologies provided, and have set the stage for their success.
Reprogramming: “The Yamanaka experiment”
Ten years ago Takahashi and Yamanaka reported on the “Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors” (Takahashi and Yamanaka, 2006). The hypothesis of this work was daring and stated that a small set of transcription factors, when ectopically expressed in a somatic cell, can reprogram them back into a pluripotent state. Retrospectively, the simplicity of the experiments that Yamanaka and colleagues used to test this hypothesis were beautiful: take a set of 24 candidate genes, selected mostly for their high and specific expression in pluripotent cells, and simultaneously express them in differentiated cells using integrating retroviruses. Identify cells that induced pluripotency via a selectable marker gene that is not expressed in somatic cells, but is preferentially activated in pluripotent cells. Next, narrow down the cocktail of genes to the minimal set of reprogramming factors (Klf4, Sox2, Oct4 and Myc, a.k.a. KSOM) by process of elimination. Lastly, demonstrate that the resulting induced pluripotent cells have all the key features of their embryonic stem cell counterparts, such as a stem cell-like expression profile, the ability to give rise to differentiated cells in teratoma formation assays and their contribution to tissues in chimeric mice after blastocyst injections (Takahashi and Yamanaka, 2006).
These experiments had an immediate impact. They came at a time when the potential of pluripotent stem cells in research applications and regenerative medicine had widely been appreciated (Rideout et al., 2002) (Figure 1), but technical and ethical limitations presented a challenge that severely impeded major progress towards realizing their full potential. Decades before the study by Yamanaka, John Gurdon (Gurdon, 1962, 1963) had demonstrated that the epigenetic profile of a fully differentiated cell can be reprogrammed to a pluripotent state. From a set of key experiments Gurdon demonstrated that a nucleus taken from a differentiated frog cell and injected into an enucleated oocyte can gives rise to a fully developed frog. This experiment illustrated that during differentiation no essential genetic material is lost and secondly that the epigenetic changes that drive cellular differentiation can be reprogrammed to totipotency. Decades later, the cloning of the sheep “Dolly” also by somatic cell nuclear transfer (SCNT) demonstrated that Gurdon’s finding extended to mammals as well (Campbell et al., 1996). SCNT and cell fusion experiments gave two additional insights that set the stage for the Yamanaka experiment. First, they demonstrated that the cytoplasm of an oocyte or an ESC contained diffusible transacting factors capable of reprogramming a somatic nucleus (reviewed in (Ambrosi and Rasmussen, 2005)). Second, successful derivation of mice by SCNT with nuclei of B-cells as a donor, which had undergone VDJ-recombination, provided genetic evidence that terminally differentiated cells can be reprogrammed (Hochedlinger and Jaenisch, 2002). Though more challenging, SCNT was eventually successful in reprogramming human cells into hESCs in 2014 (Yamada et al., 2014). While these experiments spoke for the possibility of cellular reprogramming, they also suggested highly sophisticated machinery and a complex biological process, making the success of the basic Yamanaka experimental approach even more astounding. Even today, the gradual pace of transcription factor-mediated reprogramming remains one of the most fascinating facets of the Yamanaka experiment: epigenetic changes after fertilization as well as reprogramming by SCNT occur within a few hours, while reprogramming by the Yamanaka experiment requires significantly more time, generally several days and multiple cell divisions. Yet, both processes result in a functionally equivalent cellular pluripotent state in in vitro cultures that is capable of forming an entirely new organism.
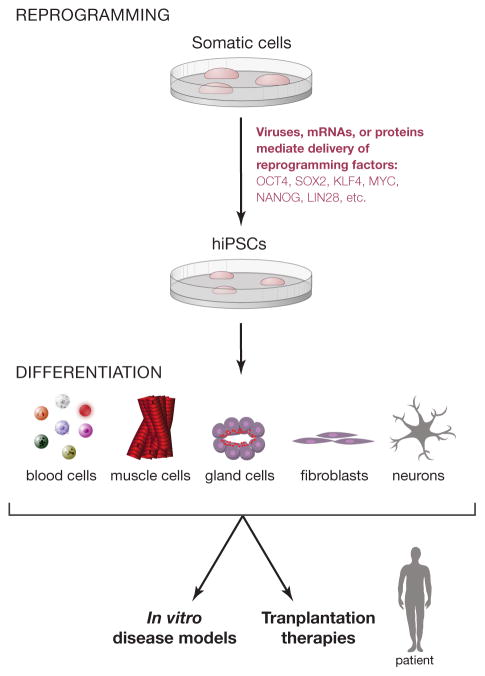
Figure 1. Overview of the iPSC technology.
Patient cells can be reprogrammed into iPSCs using optimized reprogramming protocols that involve small molecules, microRNAs, and combinations of reprogramming factors. iPSCs can be differentiated into somatic cells that could be used either in transplantation therapies or alternatively to model human diseases.
Around the same time as the first mammalian SCNT efforts, James Thomson derived the first human embryonic stem cell lines (Thomson et al., 1998). He used a very similar strategy that had proven successful for Evans and Martin (Evans and Kaufman, 1981; Martin, 1981), culturing the inner cell mass outgrowth of explanted blastocysts. However, it is interesting to note that human and mouse embryonic stem cell maintenance requires distinct signaling networks and culture conditions. LIF/Stat3 is required for maintaining the undifferentiated state in mESCs and BMP4 can inhibit the MEK/ERK differentiation pathway resulting in mESC self-renewal. In contrast hESCs and hiPSCs do not require hLIF, and maintenance of pluripotency seems to rely mostly on FGF and MEK/ERK signaling indicating species-specific requirements for culturing pluripotent cells. It seems likely that this difference can be attributed to a difference in the developmental stage that is captured in vitro from the outgrowth of the inner cell mass, where hPSCs cultured under standard conditions represent a later epiblast-like pluripotent state (Brons et al., 2007; Tesar et al., 2007; Theunissen et al., 2014) and (reviewed in (Nichols and Smith, 2009).
Proof of concept experiments with cells differentiated from hESCs suggested that pluripotent stem cells could be a source for cell replacement transplantation therapies and could provide a model system to understand early human development and cellular differentiation. However, ethical concerns, limited access to embryos, and the possibility of immune rejection were roadblocks that impeded the promise of hESCs.
In 2006 the “Yamanaka experiments” made the ethical debate about pluripotent stem cell research largely obsolete, as they established a robust method to derive human pluripotent cells without the use of human embryos. Furthermore iPSC technology promised to solve complications that were anticipated from immune rejections of heterologous hESC-derived tissues, as it would allow for the generation of patient-specific autologous pluripotent cells and derived tissue. The race to perform the key functional follow-up experiments began immediately. For the mouse system it was essential to establish that iPSCs could pass the most stringent test for pluripotency: germ line transmission (Maherali et al., 2007; Okita et al., 2007; Wernig et al., 2007) and tetraploid complementation (Kang et al., 2009; Zhao et al., 2009).
For the human system the initial question was whether the same set of factors capable of reprogramming mouse cells would also work for human cells (Takahashi et al., 2007). Yamanaka and Takahashi quickly demonstrated that their factors also worked in human cells (Takahashi et al., 2007). However, additional experiments over that last ten years in mouse and human cells also revealed that other sets of transcription factor combinations can be equally potent in reprogramming cells to a pluripotent state, providing valuable insights into the transcriptional pluripotency networks and how cells establish pluripotency (Buganim et al., 2012) (Apostolou and Hochedlinger, 2013; Park et al., 2008; Takahashi and Yamanaka, 2015; Yu et al., 2007).
For the anticipated clinical application of iPSCs it was important to demonstrate that reprogramming could be achieved without stably integrating the KSOM factors into the genome of the somatic cell. Such factor-free iPSCs were generated by independent methods such as the excision of reprogramming factors using the Cre/LoxP (Soldner et al., 2009) or the piggyBack system (Kaji et al., 2009; Woltjen et al., 2009), by avoiding integration of the reprogramming factors all together using non-integrating viruses (Fusaki et al., 2009), episomal vectors (Yu et al., 2009) or direct transfection of the reprogramming factors as either mRNA (Warren et al., 2010) or protein (Kim et al., 2009). Initially, human cell reprogramming was quite inefficient compared to mouse cells, and thus several technical improvements were made to optimize hiPSC reprogramming protocols, culture conditions and iPSC characterization procedures to test for the pluripotency of newly isolated iPSCs. Eventually, these optimizations made iPSC technology increasingly more accessible to laboratories without previous stem cell experience and are now so streamlined that iPSC derivation, maintenance and differentiation are a widely used research tool in all aspects of biomedical research. In addition, efficient and robust reprogramming techniques provided insight into the mechanistic steps of reprogramming and the order of events involved in reverting the epigenome from a differentiated to a pluripotent state. A detailed understanding of the forces at work is necessary to answer the key questions of whether reprogramming of human cells results in a cell state that is equivalent to human embryonic stem cells or whether iPSCs retain to some extent an epigenetic memory (Kim et al., 2010; Polo et al., 2010). For example, do iPSCs derived from liver cells retain some characteristics of liver cells and do they preferentially differentiate into liver tissue relative to other cell types? Tetraploid complementation and germ line transmission experiments gave the clear answers that mouse iPSCs were fully reprogrammed to pluripotency. However these tests are not available for hiPSCs. Moreover, it is not clear to what extent the miPSC’s epigenome is reset during the reprogramming process and how much of the resetting occurs in vivo or when the cells pass through the germ line. It is not surprising that early cellular stages of the reprogramming process will show epigenetic differences, yet all these differences eventually will converge on the same pluripotent cell state as ESCs. Thus, it is interesting to further examine the level and functional relevance of epigenetic memory; yet it seems that such epigenetic differences in cellular state are at best small and overshadowed by differences caused by the reprograming method of choice, cell selection during propagation, culture conditions, and more importantly genetic background of the parental somatic cell (Guenther et al., 2010; (Kyttala et al., 2016); (Rouhani et al., 2014)). For example, it has been demonstrated that the epigenetic memory, i. e. epigenetic characteristics reflecting the state of the donor cells seen initially in the iPSCs, is lost upon prolonged cell passages suggesting that this donor cell-specific memory may be of little functional relevance (Polo et al., 2010).
The most attractive application of the iPSC technology is that it allows the isolation of patient-derived cells that carry all genetic alterations that cause the particular disease. Thus, these cells provide an experimental system to study pathogenesis of the disease in an in vitro system and to possibly devise therapeutic strategies (Robinton and Daley, 2012). Importantly, the iPSC technology allows comparison of the neuroanatomical features, and physiology of the iPSCs to the clinical features of the donor patient.
The power and limitations of iPSCs
In addition to the prospect of future iPSC-based cell replacement therapies, the ability to derive iPSCs from patients’ cells had a striking effect on human disease modeling. Some of the most remarkable advances were made in diseases such as neurodegenerative diseases that are only partially recapitulated in animal models. Here, iPSC technology was particularly transformative, as it made it possible to study the effects of familial monoallelic diseases as well as complex idiopathic diseases in the context of patient-derived neurons and tissue, systems that were previously not readily available for experimental investigation. For example, studying dopaminergic neurons differentiated from patient derived iPSCs yielded insights into the molecular causes of the disease and the identification of cellular stressors that might exacerbate the phenotype (Soldner et al, 2011, 2012);. As a result of such advances, iPSC-based and primary tissue culture systems have largely replaced previous experimental systems that studied human genetic diseases using overexpression studies in cancer cell lines. Indeed, the number of human diseases modeled in culture using patient derived iPS cells (“disease in the dish”) is growing rapidly (summarized in (Avior et al., 2016; Sterneckert et al., 2014)).
While the approach of studying human disease in the disease-relevant cell type resulted in many success stories and insights, several challenges of iPSC disease modeling quickly became apparent. For one, it became evident that many protocols that were developed for the differentiation of hPSCs into functional tissue resulted in embryonic rather than adult human cell types (Bedada et al., 2015; Forster et al., 2014; Hrvatin et al., 2014; Spence et al., 2011; Takebe et al., 2013). This observation might not pose a problem for studies that aim to recapitulate cell-autonomous defects of developmental diseases that likely will become apparent after a few weeks of in vitro differentiation. However, iPSC differentiation experiments that aim to understand human disease and pathologies within the context of the adult or as a function of human aging suffer from a lack of cellular maturity as well as a relatively short timespan limited by culture conditions. One approach to increase the maturity of in vitro cell systems and to mimic cellular aging is to expose these cells to stressors that are associated with aging (Miller et al., 2013; Studer et al., 2015). Significant progress has also been made to current strategies of investigating cell non-autonomous biological problems, including the development of co-culture experiments and protocols to differentiate hPSCs into tissue stem cells and organoid cultures. Organoid cultures are small functional tissue units comprised of several distinct cell types that can be maintained and used to recapitulate features of tissues rather than that of individual cell types in vitro (Lancaster et al., 2013; Sato et al., 2011; Sato et al., 2009) (reviewed in (Lancaster and Knoblich, 2014) (Sato and Clevers, 2013).
An important and often ignored challenge of iPSC technology is the variability between individual iPSC lines in their potential to differentiate into functional cells of a given lineage. This variation between cell lines is unpredictable and mostly caused by genetic background differences as well as the reprogramming history of a given cell line. Thus, in efforts to model a disease, detection of small phenotypic differences between cells differentiated from a patient or control iPSCs may not reveal a disease-relevant phenotypic difference but rather reflect the system’s immanent variation between individual iPSC lines (Soldner and Jaenisch, 2012). The generation of isogenic pairs of disease-specific and control iPSCs that differ exclusively at the disease-causing mutation has been used to control for the variation and have lead to defining subtle disease-relevant differences in monogenic diseases (Soldner et al, 2011). The problem is, however, exacerbated when studying more clinically important sporadic or polygenic diseases where low effect size disease-causing loci are defined by genome wide association studies (GWAS). Since phenotypic differences would be expected to be small, the use of isogenic pairs of disease-specific and control cells would be even more important. Finally, ongoing efforts to learn about human genetic variation by studying dozens or even hundreds of iPSC lines derived from healthy donors may give little interpretable information because of the unpredictable system-inherent phenotypic variability between individual iPSC lines (differing in millions of SNPs within each genome) and experimental variations in their differentiation. Making isogenic iPSC controls by genome editing that differ only in single or few SNPs could reduce variations due to genomic variability.
The challenge associated with the genetic variability of hPSCs is compounded by another remarkable difference between mouse and human pluripotent stem cells: the striking resilience of hPSCs to conventional gene targeting approaches. This dearth of genetic control in hPSCs prevented genetic experiments that were considered standard in mouse embryonic stem cells. Nevertheless conventional gene targeting has been accomplished in hPSCs (Zwaka and Thomson, 2003). Protocols for conventional gene targeting have been optimized to modify human pluripotent stem cells (Costa et al., 2007; Davis et al., 2008a; Irion et al., 2007; Ruby and Zheng, 2009) and have been successfully used to establish hPSC models for human disease such as Lesch-Nyhan syndrome (Urbach et al., 2004). Moreover, this approach has been used to correct the disease-causing mutation with ornithine-d-aminotranferase that is mutated in patients with gyrate atrophy (Howden et al., 2011), or to alter the amount of disease-causing CAG repeat expansions in the huntingtin gene of patient-specific iPSCs (An et al., 2012). Furthermore, these protocols have been used to generate linage reporters for genes such as MIXL and Olig2 to study cell fate decision of differentiating human stem cells (Davis et al., 2008b; Xue et al., 2009). Overall however, these approaches are very time consuming, as they generally require the generation of large targeting constructs and even then are very inefficient and in many cases not successful. It appears that cell-intrinsic features such as low homologous recombination and single-cell survival rates make conventional genome modification as described by Capecchi and Smithies for mESCs (Doetschman et al., 1987; Thomas and Capecchi, 1987) very inefficient in hPSCs.
Both of these challenges have been overcome: the development of the Rho-kinase inhibitor Y-27632 to suppress anoikis during the disaggregation of hPSC colonies dramatically increased single-cell survival of hPSCs (Watanabe et al., 2007). The low frequency of spontaneous homology mediated gene targeting in hPSCs was dramatically increased through the development of site-specific nucleases (SSN) as a tool for their genetic engineering (reviewed in (Carroll, 2014; Hsu et al., 2014; Urnov et al., 2010)).
Genome editing BC (before CRISPR/Cas9)
The development of SSNs as research tools parallels the development of iPSCs: key experiments uncovered the biological principles and highlight how a generalized platform for genome editing would advance basic and biomedical research. Repurposing of the CRISPR/Cas9 system as an engineered SSN removed the impediments that limited the full potential of genome editing by providing this general platform.
Key experiments more than 15 years ago in mammalian cells demonstrated that a double strand break (DSB) generated by a SSN at a defined genomic site can be repaired either by the endogenous homology-mediated repair machinery using an exogenous provided repair template or by the error-prone Non-homologous end joining (NHEJ)-DNA repair pathway (Rouet et al., 1994a, b). The crucial observation made during these experiments was that a DSB increased the rate of homology-mediated genomic changes at the break site by several orders of magnitude compared to conditions in which only an exogenous repair template was provided without the induction of a DSB. Importantly, this principle of employing a DSB to facilitate DNA-repair mediated editing of genomes proved to be almost universal and applies to hPSCs as well as other systems such as C. elegans (Morton et al., 2006; Wood et al., 2011) and Drosophila melanogaster (Beumer et al., 2008; Bibikova et al., 2003; Bibikova et al., 2002), which are similarly resilient to conventional gene-targeting strategies as hPSCs.
Already in 2005, Urnov et al. demonstrated that engineered zinc finger nucleases (ZFN) can serve as a designer SSN to correct X-linked SCID disease-relevant mutations in patient-specific cells (Urnov et al., 2005). It was this study that coined the term “genome editing”. Ten years later the first clinical trials based on this ZFN technological platform are underway to disrupt CCR5 in T cells to treat HIV patients (Tebas et al., 2014).
Based on these pioneering experiments, we and others implemented the use of SSNs such as ZFNs and transcription activator like effector nucleases (TALENs) to engineer hPSCs (DeKelver et al., 2010; Hockemeyer and Jaenisch, 2010; Hockemeyer et al., 2009; Hockemeyer et al., 2011; Lombardo et al., 2007; Sexton et al., 2014; Soldner et al., 2011; Zou et al., 2009). These experiments provided proof of principle for SSN-mediated gene knockouts, for the insertion of transgenes into expressed and non-expressed genes to generate cell type-specific lineage reporters, for the over expression of transgenes from genetically defined loci, and for the insertion or repair of disease-relevant point mutations in hPSCs (Figure 2).
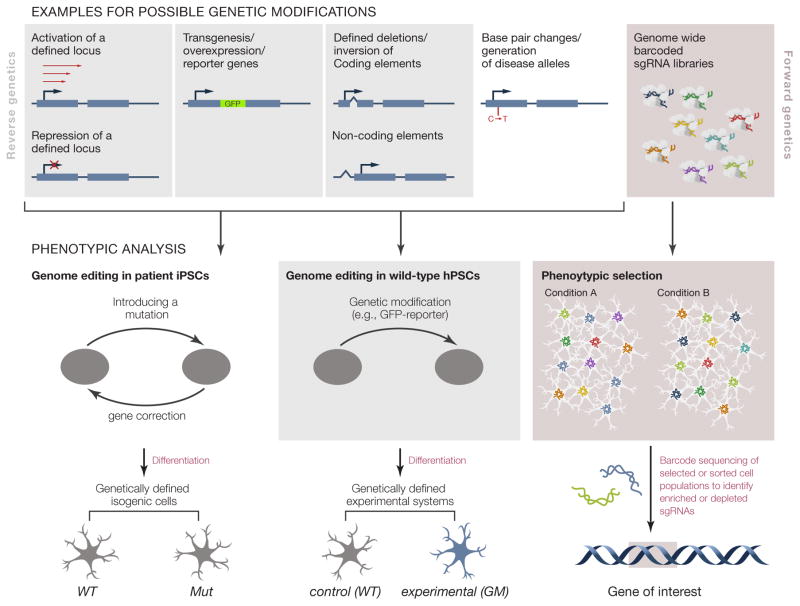
Figure 2. Genome editing applications in hiPSCs.
Genome editing allows for the genetic modification of hiPSCs. The top panel (left side) depicts examples of reverse genetic approaches to study hPSCs using genome editing. Gene expression can be modulated (activated or repressed: CRISPRi and CRISPRa) by reversibly targeting their endogenous promoter. Genes can be inserted to generate reporter genes or to achieve ectopic expression. Genetic information can be deleted or inverted and modifications as small as single base pairs changes can be generated to introduce mutations, polymorphisms or repair disease relevant mutations. The resulting genetically engineered hPSCs differ from wild type cells exclusively at the edited locus and are otherwise isogenic (bottom left). Parallel differentiation of these isogenic cell lines into disease relevant cell-types can provide the basis for the phenotypic analysis of disease specific cellular pathologies. Phenotypes found in these cells can be directly attributed to the genetic manipulation. In addition, forward genetic approaches to study hPSCs (top right panel) became available with the development of genome editing as a screening tool. Bulk transduction of hPSCs with either Cas9 or dCas9 in combination with genome wide barcoded sg RNA libraries --“CRISPR cutting, CRISPRi and CRISPRa”—can be used to identify genes who’s loss- or gain-of-function changes the cellular representation within the infect cell pool. Enrichment or depletion of sgRNAs can be determined by sequencing of the sgRNAs, yielding candidate genes of interest (bottom right panel).
The technical advances that established genetic control in hPSCs proved to be highly synergistic with the development of iPSC technology. Genome editing in hPSCs overcame the issue of enormous genetic background variability inherent to iPSC-based disease models. Independent proof of concept studies demonstrated that SSNs can be used to repair or introduce disease relevant mutations in hPSCs (Soldner et al., 2011; Yusa et al., 2011). The resulting pairs of pluripotent stem cell lines are isogenic, except for the disease-relevant mutation. Parallel differentiation of such isogenic sets of cells into disease-relevant cells and tissues can be used to directly assess the contribution of a mutation to cellular pathology (Chung et al., 2013; Ryan et al., 2013; Wang et al., 2014b; Yusa et al., 2011).
The initial ZFN and TALEN platforms to generate SSNs for genome editing in stem cells were costly and labor-intensive and their implementation as research tools therefore developed comparatively slowly. However, extensive work with ZFNs and TALENs has demonstrated the power of genome editing and highlighted the impact that a universal, cheaper, and simpler platform to make SSNs would have.
CRISPR/Cas9: everyone can edit anything
The need for a simple and unified platform to generate SSNs was met and resolved, similarly to the need for an easy way to make iPSCs, through a single experiment: by repurposing the bacterial Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) adaptive immune systems (reviewed in (Marraffini, 2015) as a SSN. In 2012 the collaborative work of the Jennifer Doudna and Emmanuelle Charpentier laboratories demonstrated that in CRISPR type-2 systems a single protein, Cas9, can function as a designer SSN, by associating with an engineered single guide RNA (sgRNA) that bears homology to a genetic locus of interest (Jinek et al., 2012). In this process, the sgRNA substitutes the natural Cas9-associated bacterial RNAs that normally confer target specificity for the bacterial pathogen DNA, and instead directs Cas9 to introduce a blunt DSB in any target DNA with complementarity to a 20nt long sequence in the sgRNA. Doudna and colleagues predicted that this simple way of engineering SSNs could be exploited to streamline genome editing (Jinek et al., 2012). In less than four years this prediction became reality and Cas9-mediated genome engineering was developed into the platform of choice to generate SSNs and to genetically modify hPSCs (Chen et al., 2015; Cong et al., 2013; Fu et al., 2014; Gonzalez et al., 2014; Gu et al., 2015; Hou et al., 2013; Hsu et al., 2014; Kleinstiver et al., 2016; Liao and Karnik, 2015; Lin et al., 2014; Mali et al., 2013; Ran et al., 2015; Slaymaker et al., 2016; Tsai et al., 2014; Wu et al., 2014b). Some important adaptations and improvements to increase the ease and scope of Cas9-mediated genome engineering in hPSCs was the establishment of CRISPR/CAS-systems from different organisms (Hou et al., 2013; Zetsche et al., 2015) that respond to different PAM sequences or by engineering spCas9 to associate with alternative PAMs by structure-based engineering of Cas9 and thereby extending genomic target range and specificity of spCas9 (Kleinstiver et al., 2015a; Kleinstiver et al., 2015b). Furthermore, several detailed protocols that describe the implementation of genome editing techniques in pluripotent stem cell systems have been optimized and published (Blair et al., 2016; Byrne and Church, 2015; Chiba and Hockemeyer, 2015; Yusa, 2013).
The key advantage of the CRISPR/Cas9 system over previous systems lies in the fact that DNA-binding specificity is encoded solely by the sgRNA and so unlike previous platforms does not require laborious engineering of DNA binding proteins. Thus CRISPR/Cas9-based editing has largely replaced previous SSN technologies. Combining the cellular versatility of iPSC differentiation with the ease of CRISPR/Cas9-mediated genome editing proved to be a very powerful experimental approach and by now genome editing in hPSCs has become a standard tool in stem cell research and human disease modeling (Johnson and Hockemeyer, 2015; Matano et al., 2015; Schwank et al., 2013).
One of the most exciting experiments that became possible since the development of robust and highly efficient editing technologies in hPSCs is to genetically and functionally test the onslaught of empirical data generated by GWAS. Similar to the disease-modeling approach, genome editing allows us to engineer variant alleles observed in these studies found to be associated with a specific disease in an otherwise isogeneic cellular setting. Phenotypic comparison of such cells can reveal how non-coding mutations, enhancer polymorphisms, and balancer mutations, can impact tissue type-specific cellular behaviors that are relevant to the particular condition.
For example, this approach has been used successfully to identify the molecular principles underlying the most frequent non-coding mutations associated with human cancer (Bojesen et al., 2013; Fredriksson et al.; Horn et al., 2013; Huang et al., 2013; Killela et al., 2013). Genetic engineering of these mutations, which occur in the promoter of the catalytic subunit of human telomerase or TERT, revealed that the mutations result in the failure of cells to silence TERT transcription upon cellular differentiation and explains how these mutations function in tumorigenesis (Chiba et al., 2015).
Gene-correction frequencies in hPSCs are generally much lower than in tumor cell lines such as K578 or HCT116 cells that are commonly used for gene editing in cancer cells. A very elegant approach to overcome this challenge and to increase the efficiency of homology-mediated events in iPSCs was used in experiments that employed zinc finger nucleases to correct mutations in iPSCs derived from patients with alpha trypsin deficiency. In these experiments gene targeting efficiencies were increased by the use of a positive selection marker that allowed for the efficient isolation of the edited clones and that could subsequently be removed without leaving residual genetic material using PiggyBac transposition. This editing strategy allowed for the generation of bi-allelic editing events in patient-derived iPSCs to restore alpha trypsin enzymatic function in disease-relevant iPSC-derived hepatocytes in vitro and after xenotransplantation (Yusa et al., 2011).
A similar approach to overcome the challenges associated with the low frequency of gene-correction events in hPSCs was used to correct point mutations in the beta-globin gene of iPSCs derived from patients with sickle cell disease (Zou et al., 2011). In this case a loxP-site flanked selection cassette that was used to increase the genome editing efficiency initially, but was then subsequently removed using Cre-recombinase. This approach results in a single residual loxP site in an in intron of the beta-globin gene. Similarly, two independent studies demonstrated the SSN can be used to directly correct b-thalassemia mutations in patient derived iPSCs and to restore hematopoietic differentiation (Ma et al., 2013; Xie et al., 2014).
Alternative strategies for increasing editing efficiencies include methods to more efficiently detect and subclone cells that have undergone rare editing events (Miyaoka et al., 2014) as well as to enhance delivery methods for the nuclease and donor template (Lin et al., 2014). An orthogonal approach to simplify the generation of isogeneic hPSC lines was the derivation of an inducible Cas9-expressing cell line by editing a Cas9 expression cassette into the AAVS1 locus. In this system Cas9 expression can be induced by doxycycline so that efficient editing afterwards only requires the expression or delivery of the sgRNA (Gonzalez et al., 2014). This system has been used to generate loss of function alleles in EZH2 and to demonstrated the effects of haploinsufficeny for EZH2 in hematopoietic differentiation (Kotini et al., 2015). Further developments that facilitate the derivation of genome engineered iPSC cell lines are protocols that directly combine genome editing with reprogramming. Howden et al demonstrated that human fibroblasts could be simultaneously reprogrammed and edited resulting in edited iPSCs going through only one single-cell cloning event without the need for drug selection (Howden et al., 2015).
Demonstration for how far-reaching the implementation of genome editing in patient-specific iPSCs can be for disease modeling was demonstrated by editing experiments that inserted an inducible Xist lncRNA into chromosome 21 of Down syndrome patient-derived iPSCs. Using this approach Jiang et al. showed that ectopic expression of Xist was sufficient to transcriptionally suppress the targeted third copy of chromosome 21 and to reverse the cellular disease phenotypes in in vitro differentiated cells (Jiang et al., 2013).
Since the implementation of genome editing in hPSCs several diseases have been modeled using isogenic cell lines that have either corrected a disease-relevant mutation in iPSCs or introduced a disease relevant allele in wild-type hPSCs. For example, the genetic correction of mutations in Niemann-Pick Type C patient-specific iPSCs to rescue metabolic defects in cholesterol metabolism and autophagy, which are responsible for the pathology, represents just one demonstration of how this approach has been successfully implemented (Maetzel et al., 2014). Furthermore, genome editing in hPSCs has been used to establish models for Rett-syndrome disrupting MECP2 function in hPSCs (Li et al., 2013), to generate HIV-resistant variants alleles of the CCR5 gene into iPSCs (Ye et al., 2014), to repair MYO15A in iPSCs derived from patients affected by deafness (Chen et al., 2016) and to derive isogeneic cell pairs of COL7A1-corrected iPSCs derived from patients with dystrophic epidermolysis bullosa (Sebastiano et al., 2014).
In a growing number of cases, such approaches have also been used to provide new insight into disease pathology. For example, SSN-mediated correction of disease-causing mutations in LRKK2 that are associated in Parkinson’s disease revealed the transcriptional changes caused by disease-associated alleles in patient cells (Reinhardt et al., 2013). Likewise, genome editing of patient-specific iPSCs followed by in vitro differentiation was also used to generate an isogenic disease model for cystic fibrosis by correcting disease-relevant mutations in CFTR followed by differentiation into airway epithelium (Crane et al., 2015; Firth et al., 2015; Suzuki et al., 2016).
The challenge of studying sporadic (polygenic) diseases
The application of iPSC technology for the study of sporadic diseases poses particular challenges because disease-specific phenotypic changes are expected to be subtle. The genetic basis of sporadic or idiopathic diseases is thought to be a combination of multiple low effect size risk alleles, mostly in regulatory regions such as enhancers, which are identified by GWAS (Gibson, 2011; Merkle and Eggan, 2013). The “common disease-common variant hypothesis” proposes that multiple risk variants with small effect size in combination with additional environmental factors are the drivers of sporadic diseases. Thus, a major challenge of using human-derived cells is that risk variants are not only present in patients but also in unaffected individuals, albeit with lower frequency. Thus, individual risk variants are not sufficient to cause disease-associated phenotypes in carrier individuals or in hiPSCs derived from carriers or patients. While an iPSC isolated from a patient would harbor all risk variants that contribute to the disease, any in vitro study to gain mechanistic insights is complicated by the high system immanent variability in differentiation into the disease-relevant cells (Soldner and Jaenisch, 2012). Another complicating factor is that the likely effect of a GWAS-identified risk regulatory allele on the target gene (or genes) would be predicted to be subtler than would be expected for monogenic diseases as discussed above. Thus, it would be impossible to compare the disease specific cells to a suitable control cell line because any control cells would have a different genetic background which will affect the differentiation potential of the cells and thus would prevent a meaningful comparison.
Thus, a major challenge for using iPSCs for the study of sporadic diseases is how to generate pairs of isogenic cells that differ at one or multiple risk alleles. Figure 3 outlines a possible strategy of how the CRISPR/Cas9 gene editing approach could be used to generate isogenic cells that differ at multiple risk loci and thus would enable the mechanistic study of polygenic diseases. This approach was recently used to decipher the impact of Parkinson disease (PD)-associated risk variants. Genetic engineering of a common PD-associated risk variant in a non-coding distal enhancer resulted in deregulation of SNCA expression, a key gene implicated in the pathogenesis of PD, by as little as 10% (Soldner et al, 2016). In order to detect such subtle gene expression differences, an allele-specific assay was developed that allowed the analysis of cis-acting effects of candidate variants on allele-specific gene expression as a consequence of deletion or exchange of disease-associated regulatory elements. Detailed analysis of isogenic cells with and without the risk allele further demonstrated that a single base pair change causes loss of transcription factor-binding sites for the transcription factors that otherwise function as a suppressor of SNCA-transcription on a non-risk associated allele.
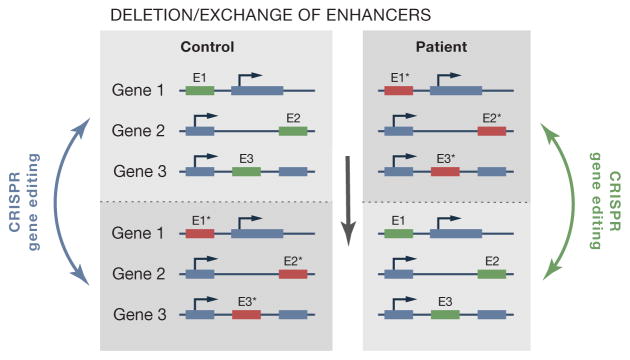
Figure 3.
Strategy to generate isogenic iPSCs that differ at multiple risk alleles. GWAS have identified genomic loci that may slightly increase the risk of developing a sporadic disease. The key challenge of using patient-derived iPSCs to get mechanistic insight into risk alleles is to create meaningful control cells. CRISPR/Cas9 mediated gene editing would allow to exchange risk (red squares) and protective (green squares) alleles and to generate appropriate control cells that differ exclusively at the risk loci under study.
Epidemiology and population genetics suggest that Sporadic Alzheimer Disease (SAD) results from complex interactions between genetic risk variants and environmental factors. In another approach to study risk alleles, patient-derived hiPSCs were used to dissect the effect of a common SAD-associated non-coding genetic variants in the 5′ region of the SORL1 (sortilin-related receptor, L(DLR class) A repeats containing) gene involved in intracellular vesicular trafficking (Young et al., 2015). While initial experiments did not identify a consistent correlation between SORL1 expression and either disease status or risk haplotype, a small but significant correlation between the SAD-associated SORL1 haplotype and the BDNF-dependent response of SORL1 expression was found.
Nuclease specificity and off-target considerations
SSNs are enzymes that are targeted to specific site in the genome, but their specificity can vary and promiscuous binding to so called off-target sites can lead to unwanted cutting and modifications. Strategies to predict, identify, and reduce these off-target events are largely dependent on the SSN design, organism, and cell type and have already been to some extent implemented in hPSCs. Understanding the frequency and impact of off-targets is highly relevant to develop the use of SSN for clinical applications as well as for their reliable use in basic research (Gabriel et al., 2011).
Several studies recently addressed the specificity of Cas9 and its off-target action (reviewed in (Wu et al., 2014a)). Genome-wide binding studies of dCas9 expressed in mouse ESCs demonstrated that Cas9 can associate with a large number of genomic sites, but off-target cutting of the catalytically active Cas9 at a subset of these site was infrequent (Wu et al., 2014b). Similarly, single molecule imaging of Cas9 in living cells have demonstrated that Cas9 searches for target sites by three-dimensional diffusion, and that in contrast to on-target events off-target binding events are, on average, short-lived (<1 second) (Knight et al., 2015).
While these data argue for the high specificity of Cas9, data in cancer cells suggest that off-targets can be frequently detected (Frock et al., 2015; Fu et al., 2014; Tsai et al., 2015; Wang et al., 2015b). For example, when using GUIDE-Seq (Tsai et al., 2015), a protocol optimized in U2OS and HEK293 to detect off-targets more reliable than other methods such as ChIp-seq, found many off target that computational algorithms failed to predicted. Based on these data sets Tsai et al proposed that shorter guide sequences that only have about 17nt homology to the target sequence would improve specificity (Fu et al., 2014). Moreover, the GUIDE-Seq. protocol was also used to engineer CRISPR-Cas9 nucleases with altered PAM specificities (Kleinstiver et al., 2015a; Kleinstiver et al., 2015b) and reduced off-targets (Kleinstiver et al., 2016).
An alternative protocol called BLES-seq that is based on directly labeling the DSBs generated by the nuclease in situ followed by enrichment through streptavidin affinity purification and next-generation sequencing (Crosetto et al., 2013) was originally developed to detect DSBs caused by replicative stress by stalled replication folks in HeLa cells and mouse B lymphocytes. This protocol was further developed to assess Cas9 off-target frequencies of Cas9 and to rationally engineer Cas9 nucleases with improved specificity (Ran et al., 2015; Slaymaker et al., 2016).
Most experiments that have detected significant off-targets have been performed in cancer cells, which may have altered repair pathways that could affect recombination (Fu et al, 2013; Hsu et al, 2013). In contrast, experiments in whole organisms such as mice ((Wang et al., 2013), primates (Niu et al., 2014), Zebrafish (Auer et al., 2014) or C. elegans (Dickinson et al., 2013) reported off-target frequencies that were low or not detectable consistent with high specificity of the CRISPR/Cas9 mediated gene targeting. It is also possible that in non-transformed cells off-target cleavages are efficiently counter-selected by the endogenous DNA-damage response. As hPSCs are primary cells with genetically intact check-points it seems possible that off-targets will accumulate less frequently in hPSCs than it has been observed in cancer cells. To address this it will be important to determine to what extent of-targets are the result of impaired checkpoint control of cancer cells and whether there are specific cell-types and conditions that predispose for the accumulation of off-targets. Data form conventional whole genome sequencing of hPSCs exposed to Cas9 have so far been limited and not yet fully address the issues due to small sample sizes (Park et al., 2015; Smith et al., 2014).
Understanding how to avoid off-target SSN modification is of particular concern for the eventual clinical application of edited cells. For basic research, however, it seems that the necessary experiments are readily available to control for the effects of eventual off-target action of SSNs. Experiments to adequately address off-target concern include: (1) the use several independent guide RNAs to generate a mutant cell line, (2) complementation of loss-of-function phenotypes, (3) secondary editing to the mutant cell line to revert the mutation to wild-type allele followed by confirmation of phenotypic rescue.
Large-scale screens, epigenetic editing and other applications for Cas9 is iPSCs
In addition to allowing for easy, fast, and inexpensive editing of hPSCs the advent of Cas9 as a programmable DNA-binding protein allowed for the development of forward genetics methodologies that were previously not readily available in hESCs. It is trivial to multiplex guide RNA synthesis allowing for the generation of large barcoded libraries of sgRNAs with several fold coverage of every gene in the human genome. These libraries can be employed in loss-of-function screens, for example, to identify gene products that are required for drug resistance or to mediate viral cell death (Gilbert et al., 2014; Hart et al., 2015; Parnas et al., 2015; Shalem et al., 2014; Shi et al., 2015; Wang et al., 2015a; Wang et al., 2014a; Zhou et al., 2014) (Figure 2). Most of the experiments that employ genome-wide screens have been done in cancer cells that can be expanded to accommodate the large numbers of cells that are required to perform these type of genome-wide screens. For the general implementation of these screening approaches in hPSCs or cells differentiated form hPSCs it will be important to develop protocols that allow the expansion of these cells into large homogenous populations that allow robust selection or enrichment for cellular phenotypes.
It is worth mentioning that the combination of iPSCs and genome editing has not only become a game changer for our approaches to human disease modeling but also provides an unprecedented opportunity to study the fundamental principles of cell biology. Previously, cell biologists mostly used aberrant cancer cell lines with often unstable and poorly defined genomes to describe human cellular behavior. This is mainly because human cancer cells presented the only reliable source of human immortal cells that could be expanded sufficiently to facilitate biochemical and genetic experimentation and that could be indefinitely propagated, frozen, shipped, and shared between labs. This monopoly of cancer cells as a model system was broken with the advent of hiPSCs and the general availability of hPSCs. Like cancer cells, hPSCs are immortal, but they do not suffer from the disadvantages of the pathologically altered genomes of cancer cells and yet they still retain the capacity to differentiate into any cell type of interest. The combination of hPSCs with the power of genome editing can now be used to study the specific aspects of human cell biology. Exploiting this potential will be particularly important in areas of research where fundamental biological processes diverge between human and other species such as tumor suppression, and cellular immortality, or neuronal biology. Efforts such as the one launched by the Allen Institute for Cell Science to create an industrial scale library of characterized iPSCs that will be used to create a visual, animated model of the cell, suggest that iPSCs will soon replace cancer cells as a model system for basic cell biology (Callaway, 2014).
In the same way that iPSC technology had broad impacts far beyond regenerative medicine and disease modeling, the impact of the discovery of CRISPR/Cas9 on hPSCs is not only its ability to act as a SSN. Catalytically inactive forms of Cas9 (dCas9) have been successfully derived by fusions with functional protein that bind specific loci, or to activate or repress gene activity at the target site (Chen et al., 2013; Gilbert et al., 2014; Konermann et al., 2015; Mandegar et al., 2016; Tanenbaum et al., 2014) (CRISPRa and CRISPRi, Figure 2). Some of these platforms have been successfully implemented for genome-wide screens and to manipulate hPSCs. As demonstrated for TALE proteins and Zinc finger DNA binding domains, the range of Cas9 could be extended in the future to also methylate or demethylate DNA or histones/chromatin at precise locations in the genome (Maeder et al., 2013; Meister et al., 2010). Moreover, dCas9 fused to fluorescent reporters have been developed to indicate nuclear organization by visualizing individual genomic loci (Chen et al., 2013; Gilbert et al., 2014; Tanenbaum et al., 2014). Excitingly more applications are being developed; recently Cas9 has now also been programmed to target RNA in vitro and in vivo (O’Connell et al., 2014) (Nelles et al., 2016), raising the possibly that it could be used to better understand the transcriptome in addition to the genome.
Arguably the most far-reaching consequence of CRISPR/Cas9 gene targeting is the potential to edit the germ line. Because gene editing by homologous recombination is inefficient, cells carrying the desired targeting event need to be selected in culture. Thus, germ line modification in the past was restricted to mice as chimera-competent ES cells are not available in other species. Because CRISPR/Cas9 gene editing is so efficient, it requires no selection for the desired targeting events rendering ES cells superfluous for the generation of mutant animals. CRISPR/Cas9 enabled gene editing in the zygote and was used to efficiently generate animals carrying defined mutations in multiple species including fish, drosophila, mice and primates (Bassett et al., 2013; Chang et al., 2013; Gratz et al., 2013; Hwang et al., 2013a; Hwang et al., 2013b; Niu et al., 2014; Wang et al., 2013; Yang et al., 2013; Yu et al., 2013).
Challenges and next steps
Despite the obvious advances that have been made as a result of iPSC and editing technologies, several challenges remain. A key limitation remains that human cells prefer to choose the imprecise NHEJ pathway to repair a DSB rather than using the more precise homologous DNA repair pathway using an exogenous repair template (Chapman et al., 2012). Due to this pathway choice, editing events often result in NHEJ-mediated insertions and deletions at the DSB rather than the intended homology mediated modification. NHEJ-mediated gene disruption can be useful when the researcher or clinician intends to generate a loss-of-function event. However, in most clinical treatment settings the generation of a defined allele with high frequency will be essential to devise treatment options that require editing to result in gain-of-function at endogenous genes. Approaches to shift the balance away from NHEJ and towards homology-mediated repair included NHEJ inhibition with small molecules or by controlling the timing of CRISPR/Cas9 delivery with respect to the cell cycle stage (Chu et al., 2015; Maruyama et al., 2015; Robert et al., 2015; Yu et al., 2015). These approaches are promising, yet we are currently far away from testing the efficacy of treatment strategies that rely on gene repair or gain-of-function approaches using high frequency HR repair events of endogenous genes.
Facing this challenge, recent studies used creative ways to take advantage of NHEJ-meditated genome editing and the fact that the simultaneous expression of two nucleases can meditate the excision or inversion of the sequence internal to the two SSN (Chiba et al., 2015; (Chen et al., 2011; Young et al., 2016). In the specific case of Duchenne muscular dystrophy, Cas9 was employed to excise 725 kb of genomic sequences, which removed a premature STOP codon in the disease-causing DMD gene and thereby restored the reading frame and partial protein function (Young et al., 2016).
Similarly, Cas9 mediated genome editing in patient specific iPSCs was used to genetically correct the disease causing chromosomal inversions found in patients with Hemophilia A, demonstrating that NHEJ based approaches can be used to model and correct large scale genomic alterations underlying human disease (Park et al., 2015).
Elegant work that also takes advantage of the fact that genomic sequences between two SSN cuts can reinsert into back into the locus in an inverted manner was recently used demonstrated that CTCF sites interact with each other in an orientation dependent manner (Guo et al., 2015). Using this approach Guo et al elucidate the impact of the directionality of CTCF sites in the mediation of large-scale genome interactions and transcriptional regulation.
Another challenge of genome editing in human cells is that human cells have relatively short conversion tracts (Elliott et al., 1998). This means that even when a DSB is repaired by homology directed repair and not the NHEJ machinery, modifications can only be made with reasonable frequency very close to one side of the double stranded break. This presents a major obstacle towards the introduction of complex genetic changes in hPSCs. The use of Cpf1, a class 2 CRISPR effector that uses the same basic principles as Cas9, but cleaves DNA further away from the PAM sequence and generates a single stranded overhang, may help increase the rate of HDR over NHEJ events (Zetsche et al., 2015). Overcoming this challenge will significantly facilitate the engineering of human stem cells, as it will allow us to refine the human genome more efficiently. Eventually this could result in similar resources that have been used in yeast and mESCs, such as a comprehensive collection of conditional human knockout iPSC libraries, with a homozygous iPSC line for each human gene carrying an exon flanked by loxP sites.
Rethinking the ethical debate
It will be important in the near future to navigate the ethical debate that arises from the confluence of genome editing with stem cell technology. This requires a policy framework that supports scientific progress that is independent of special interest groups that would bias a rational risk benefit assessment of this technology. The rampant progress that has been made over the last few years to improve genome editing technologies and to detect and reduce potential off-targets of SSNs has already lead to the first clinical trials for HIV and are trail blazing through the necessary regulatory hurdles (Tebas et al., 2014). Somatic cell editing and editing in hPSCs in vivo and/or ex vivo coupled with transplantation will progress to become a standard clinical application. These efforts have to be clearly distinguished from editing human germ cells or totipotent cells of the early human embryo. Indeed, the efficiency of altering the genome of mammals by injecting CRISPR/Cas9 RNA or DNA into the fertilized egg (Wang et al, 2013) sparked a debate whether this technology should be used to modify the human germ line (Sheridan, 2015). While technical challenges currently limit the potential application of such modifications, two recent papers describe gene editing of the embryo’s genome following injection of gRNAs, CRSPR/Cas9 RNA and targeting oligos into human zygotes (Kang et al., 2016; Liang et al., 2015). These studies raise a number of scientific issues such as off-target rate, mosaicism and the likely alteration of the non-targeted wild type allele when targeting a mutant allele. More importantly, the technology raises serious ethical issues: do we want to irreversibly alter the human germ line? Thus, the clinical application of this gene editing technology for medical purposes raises important ethical issues that will need to be widely discussed and agreed upon as it would affect future generations.
Acknowledgments
We thank the members of the Hockemeyer lab for helpful discussion. Dirk Hockemeyer is a New Scholar in Aging of the Ellison Medical Foundation and is supported by the Glenn Foundation as well as the The Shurl and Kay Curci Foundations. The work in the Hockemeyer laboratory is supported by National Institutes of Health R01 CA196884-01 and in the Jaenisch lab by NIH grants 1R01NS088538-01 and 2R01MH104610-15. RJ is an advisor to Stemgent and Fate Therapeutics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ambrosi DJ, Rasmussen TP. Reprogramming mediated by stem cell fusion. J Cell Mol Med. 2005;9:320–330. doi: 10.1111/j.1582-4934.2005.tb00358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An MC, Zhang N, Scott G, Montoro D, Wittkop T, Mooney S, Melov S, Ellerby LM. Genetic correction of Huntington’s disease phenotypes in induced pluripotent stem cells. Cell stem cell. 2012;11:253–263. doi: 10.1016/j.stem.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolou E, Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature. 2013;502:462–471. doi: 10.1038/nature12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer TO, Duroure K, De Cian A, Concordet JP, Del Bene F. Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome research. 2014;24:142–153. doi: 10.1101/gr.161638.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avior Y, Sagi I, Benvenisty N. Pluripotent stem cells in disease modelling and drug discovery. Nature reviews Molecular cell biology. 2016;17:170–182. doi: 10.1038/nrm.2015.27. [DOI] [PubMed] [Google Scholar]
- Bassett AR, Tibbit C, Ponting CP, Liu JL. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell reports. 2013;4:220–228. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedada FB, Wheelwright M, Metzger JM. Maturation status of sarcomere structure and function in human iPSC-derived cardiac myocytes. Biochim Biophys Acta. 2015 doi: 10.1016/j.bbamcr.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer KJ, Trautman JK, Bozas A, Liu JL, Rutter J, Gall JG, Carroll D. Efficient gene targeting in Drosophila by direct embryo injection with zinc-finger nucleases. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:19821–19826. doi: 10.1073/pnas.0810475105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science (New York, NY) 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair JD, Bateup HS, Hockemeyer DF. Establishment of Genome-edited Human Pluripotent Stem Cell Lines: From Targeting to Isolation. Journal of visualized experiments : JoVE. 2016 doi: 10.3791/53583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojesen SE, Pooley KA, Johnatty SE, Beesley J, Michailidou K, Tyrer JP, Edwards SL, Pickett HA, Shen HC, Smart CE, et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nature Genetics. 2013;45:371–384. 384e371–372. doi: 10.1038/ng.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A, Jaenisch R. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne SM, Church GM. Crispr-mediated Gene Targeting of Human Induced Pluripotent Stem Cells. Current protocols in stem cell biology. 2015;35:5a.8.1–22. doi: 10.1002/9780470151808.sc05a08s35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E. Microsoft billionaire takes on cell biology. Nature. 2014;516:157. doi: 10.1038/516157a. [DOI] [PubMed] [Google Scholar]
- Campbell K, McWhir J, Ritchie W, Wilmut I. Sheep cloned by nuclear transfer from a cultured cell line. Nature. 1996;380:64–66. doi: 10.1038/380064a0. [DOI] [PubMed] [Google Scholar]
- Carroll D. Genome engineering with targetable nucleases. Annual Review of Biochemistry. 2014;83:409–439. doi: 10.1146/annurev-biochem-060713-035418. [DOI] [PubMed] [Google Scholar]
- Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X, Xiong JW, Xi JJ. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013;23:465–472. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Pruett-Miller SM, Huang Y, Gjoka M, Duda K, Taunton J, Collingwood TN, Frodin M, Davis GD. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nature methods. 2011;8:753–755. doi: 10.1038/nmeth.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JR, Tang ZH, Zheng J, Shi HS, Ding J, Qian XD, Zhang C, Chen JL, Wang CC, Li L, et al. Effects of genetic correction on the differentiation of hair cell-like cells from iPSCs with MYO15A mutation. Cell death and differentiation. 2016 doi: 10.1038/cdd.2016.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Cao J, Xiong M, Petersen AJ, Dong Y, Tao Y, Huang CT, Du Z, Zhang SC. Engineering Human Stem Cell Lines with Inducible Gene Knockout using CRISPR/Cas9. Cell stem cell. 2015;17:233–244. doi: 10.1016/j.stem.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba K, Hockemeyer D. Genome editing in human pluripotent stem cells using site-specific nucleases. Methods Mol Biol. 2015;1239:267–280. doi: 10.1007/978-1-4939-1862-1_15. [DOI] [PubMed] [Google Scholar]
- Chiba K, Johnson JZ, Vogan JM, Wagner T, Boyle JM, Hockemeyer D. Cancer-associated TERT promoter mutations abrogate telomerase silencing. Elife. 2015;4 doi: 10.7554/eLife.07918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, Kuhn R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nature biotechnology. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- Chung CY, Khurana V, Auluck PK, Tardiff DF, Mazzulli JR, Soldner F, Baru V, Lou Y, Freyzon Y, Cho S, et al. Identification and rescue of alpha-synuclein toxicity in Parkinson patient-derived neurons. Science (New York, NY) 2013;342:983–987. doi: 10.1126/science.1245296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science (New York, NY) 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Dottori M, Sourris K, Jamshidi P, Hatzistavrou T, Davis R, Azzola L, Jackson S, Lim SM, Pera M, et al. A method for genetic modification of human embryonic stem cells using electroporation. Nature protocols. 2007;2:792–796. doi: 10.1038/nprot.2007.105. [DOI] [PubMed] [Google Scholar]
- Crane AM, Kramer P, Bui JH, Chung WJ, Li XS, Gonzalez-Garay ML, Hawkins F, Liao W, Mora D, Choi S, et al. Targeted correction and restored function of the CFTR gene in cystic fibrosis induced pluripotent stem cells. Stem cell reports. 2015;4:569–577. doi: 10.1016/j.stemcr.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosetto N, Mitra A, Silva MJ, Bienko M, Dojer N, Wang Q, Karaca E, Chiarle R, Skrzypczak M, Ginalski K, et al. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nature methods. 2013;10:361–365. doi: 10.1038/nmeth.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R, Costa M, Grandela C, Holland A, Hatzistavrou T, Micallef S, Li X, Goulburn A, Azzola L, Elefanty A, et al. A protocol for removal of antibiotic resistance cassettes from human embryonic stem cells genetically modified by homologous recombination or transgenesis. Nature protocols. 2008a;3:1550–1558. doi: 10.1038/nprot.2008.146. [DOI] [PubMed] [Google Scholar]
- Davis R, Ng E, Costa M, Mossman A, Sourris K, Elefanty A, Stanley E. Targeting a GFP reporter gene to the MIXL1 locus of human embryonic stem cells identifies human primitive streak-like cells and enables isolation of primitive hematopoietic precursors. Blood. 2008b;111:1876–1884. doi: 10.1182/blood-2007-06-093609. [DOI] [PubMed] [Google Scholar]
- DeKelver RC, Choi VM, Moehle EA, Paschon DE, Hockemeyer D, Meijsing SH, Sancak Y, Cui X, Steine EJ, Miller JC, et al. Functional genomics, proteomics, and regulatory DNA analysis in isogenic settings using zinc finger nuclease-driven transgenesis into a safe harbor locus in the human genome. Genome research. 2010;20:1133–1142. doi: 10.1101/gr.106773.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DJ, Ward JD, Reiner DJ, Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nature methods. 2013;10:1028–1034. doi: 10.1038/nmeth.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetschman T, Gregg RG, Maeda N, Hooper ML, Melton DW, Thompson S, Smithies O. Targetted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature. 1987;330:576–578. doi: 10.1038/330576a0. [DOI] [PubMed] [Google Scholar]
- Elliott B, Richardson C, Winderbaum J, Nickoloff JA, Jasin M. Gene conversion tracts from double-strand break repair in mammalian cells. Molecular and cellular biology. 1998;18:93–101. doi: 10.1128/mcb.18.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Firth AL, Menon T, Parker GS, Qualls SJ, Lewis BM, Ke E, Dargitz CT, Wright R, Khanna A, Gage FH, et al. Functional Gene Correction for Cystic Fibrosis in Lung Epithelial Cells Generated from Patient iPSCs. Cell reports. 2015;12:1385–1390. doi: 10.1016/j.celrep.2015.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster R, Chiba K, Schaeffer L, Regalado SG, Lai CS, Gao Q, Kiani S, Farin HF, Clevers H, Cost GJ, et al. Human intestinal tissue with adult stem cell properties derived from pluripotent stem cells. Stem cell reports. 2014;2:838–852. doi: 10.1016/j.stemcr.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson NJ, Ny L, Nilsson JA, Larsson E. Systematic analysis of noncoding somatic mutations and gene expression alterations across 14 tumor types. Nat Genet. 2014;46:1258–1263. doi: 10.1038/ng.3141. [DOI] [PubMed] [Google Scholar]
- Frock RL, Hu J, Meyers RM, Ho YJ, Kii E. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. 2015;33:179–186. doi: 10.1038/nbt.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nature biotechnology. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel R, Lombardo A, Arens A, Miller JC, Genovese P, Kaeppel C, Nowrouzi A, Bartholomae CC, Wang J, Friedman G, et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nature biotechnology. 2011;29:816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- Gibson G. Rare and common variants: twenty arguments. Nature reviews Genetics. 2011;13:135–145. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez F, Zhu Z, Shi ZD, Lelli K, Verma N, Li QV, Huangfu D. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell stem cell. 2014;15:215–226. doi: 10.1016/j.stem.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O’Connor-Giles KM. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013;194:1029–1035. doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Ziller MJ, Clement K, Tsankov AM, Akopian V, Gifford CA, Donaghey J, Galonska C, Pop R, Reyon D, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nat Genet. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, Jung I, Wu H, Zhai Y, Tang Y, et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell. 2015;162:900–910. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon J. The developmental capacity of nuclei taken from intestinal epithelium cells of feeding tadpoles. J Embryol exp Morph. 1962;10:622–640. [PubMed] [Google Scholar]
- Gurdon J. Nuclear transplantation in Amphibia and the importance of stable nuclear changes in cellular differentiation. Q Rev Biol. 1963;38:54–78. [Google Scholar]
- Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, Mis M, Zimmermann M, Fradet-Turcotte A, Sun S, et al. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell. 2015;163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Jaenisch R. Monoclonal mice generated by nuclear transfer from mature B and T donor cells. Nature. 2002;415:1035–1038. doi: 10.1038/nature718. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Jaenisch R. Gene targeting in human pluripotent cells. Cold Spring Harbor symposia on quantitative biology. 2010;75:201–209. doi: 10.1101/sqb.2010.75.021. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nature biotechnology. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nature biotechnology. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, Kadel S, Moll I, Nagore E, Hemminki K, et al. TERT promoter mutations in familial and sporadic melanoma. Science (New York, NY) 2013;339:959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, Thomson JA. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden SE, Gore A, Li Z, Fung HL, Nisler BS, Nie J, Chen G, McIntosh BE, Gulbranson DR, Diol NR, et al. Genetic correction and analysis of induced pluripotent stem cells from a patient with gyrate atrophy. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:6537–6542. doi: 10.1073/pnas.1103388108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden SE, Maufort JP, Duffin BM, Elefanty AG, Stanley EG, Thomson JA. Simultaneous Reprogramming and Gene Correction of Patient Fibroblasts. Stem cell reports. 2015;5:1109–1118. doi: 10.1016/j.stemcr.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrvatin S, O’Donnell CW, Deng F, Millman JR, Pagliuca FW, DiIorio P, Rezania A, Gifford DK, Melton DA. Differentiated human stem cells resemble fetal, not adult, beta cells. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:3038–3043. doi: 10.1073/pnas.1400709111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly recurrent TERT promoter mutations in human melanoma. Science (New York, NY) 2013;339:957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Kaini P, Sander JD, Joung JK, Peterson RT, Yeh JR. Heritable and precise zebrafish genome editing using a CRISPR-Cas system. PLoS One. 2013a;8:e68708. doi: 10.1371/journal.pone.0068708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nature biotechnology. 2013b;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irion S, Luche H, Gadue P, Fehling HJ, Kennedy M, Keller G. Identification and targeting of the ROSA26 locus in human embryonic stem cells. Nature biotechnology. 2007;25:1477–1482. doi: 10.1038/nbt1362. [DOI] [PubMed] [Google Scholar]
- Jiang J, Jing Y, Cost GJ, Chiang JC, Kolpa HJ, Cotton AM, Carone DM, Carone BR, Shivak DA, Guschin DY, et al. Translating dosage compensation to trisomy 21. Nature. 2013;500:296–300. doi: 10.1038/nature12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science (New York, NY) 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JZ, Hockemeyer D. Human stem cell-based disease modeling: prospects and challenges. Curr Opin Cell Biol. 2015;37:84–90. doi: 10.1016/j.ceb.2015.10.007. [DOI] [PubMed] [Google Scholar]
- Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang L, Wang J, Zhang Y, Kou Z, Gao S. iPS cells can support full-term development of tetraploid blastocyst-complemented embryos. Cell stem cell. 2009;5:135–138. doi: 10.1016/j.stem.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Kang X, He W, Huang Y, Yu Q, Chen Y, Gao X, Sun X, Fan Y. Introducing precise genetic modifications into human 3PN embryos by CRISPR/Cas-mediated genome editing. Journal of assisted reproduction and genetics. 2016 doi: 10.1007/s10815-016-0710-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA, Friedman AH, Friedman H, Gallia GL, Giovanella BC, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proceedings of the National Academy of Sciences. 2013;110:6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell stem cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LIR, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Prew MS, Tsai SQ, Nguyen NT, Topkar VV, Zheng Z, Joung JK. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nature biotechnology. 2015a;33:1293–1298. doi: 10.1038/nbt.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales AP, Li Z, Peterson RT, Yeh JR, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015b;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SC, Xie L, Deng W, Guglielmi B, Witkowsky LB, Bosanac L, Zhang ET, El Beheiry M, Masson JB, Dahan M, et al. Dynamics of CRISPR-Cas9 genome interrogation in living cells. Science (New York, NY) 2015;350:823–826. doi: 10.1126/science.aac6572. [DOI] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotini AG, Chang CJ, Boussaad I, Delrow JJ, Dolezal EK, Nagulapally AB, Perna F, Fishbein GA, Klimek VM, Hawkins RD, et al. Functional analysis of a chromosomal deletion associated with myelodysplastic syndromes using isogenic human induced pluripotent stem cells. Nature biotechnology. 2015;33:646–655. doi: 10.1038/nbt.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyttala A, Moraghebi R, Valensisi C, Kettunen J, Andrus C, Pasumarthy KK, Nakanishi M, Nishimura K, Ohtaka M, Weltner J, et al. Genetic Variability Overrides the Impact of Parental Cell Type and Determines iPSC Differentiation Potential. Stem cell reports. 2016;6:200–212. doi: 10.1016/j.stemcr.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science (New York, NY) 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang H, Muffat J, Cheng AW, Orlando DA, Loven J, Kwok SM, Feldman DA, Bateup HS, Gao Q, et al. Global transcriptional and translational repression in human-embryonic-stem-cell-derived Rett syndrome neurons. Cell stem cell. 2013;13:446–458. doi: 10.1016/j.stem.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P, Xu Y, Zhang X, Ding C, Huang R, Zhang Z, Lv J, Xie X, Chen Y, Li Y, et al. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein & cell. 2015;6:363–372. doi: 10.1007/s13238-015-0153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J, Karnik R. Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. 2015;47:469–478. doi: 10.1038/ng.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Staahl BT, Alla RK, Doudna JA. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife. 2014;3:e04766. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo A, Genovese P, Beausejour C, Colleoni S, Lee Y, Kim K, Ando D, Urnov F, Galli C, Gregory P, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nature biotechnology. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- Ma N, Liao B, Zhang H, Wang L, Shan Y, Xue Y, Huang K, Chen S, Zhou X, Chen Y, et al. Transcription activator-like effector nuclease (TALEN)-mediated gene correction in integration-free beta-thalassemia induced pluripotent stem cells. The Journal of biological chemistry. 2013;288:34671–34679. doi: 10.1074/jbc.M113.496174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Angstman JF, Richardson ME, Linder SJ, Cascio VM, Tsai SQ, Ho QH, Sander JD, Reyon D, Bernstein BE, et al. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nature biotechnology. 2013;31:1137–1142. doi: 10.1038/nbt.2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maetzel D, Sarkar S, Wang H, Abi-Mosleh L, Xu P, Cheng AW, Gao Q, Mitalipova M, Jaenisch R. Genetic and chemical correction of cholesterol accumulation and impaired autophagy in hepatic and neural cells derived from Niemann-Pick Type C patient-specific iPS cells. Stem cell reports. 2014;2:866–880. doi: 10.1016/j.stemcr.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell stem cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-Guided Human Genome Engineering via Cas9. Science (New York, NY) 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandegar MA, Huebsch N, Frolov EB, Shin E, Truong A, Olvera MP, Chan AH, Miyaoka Y, Holmes K, Spencer CI, et al. CRISPR Interference Efficiently Induces Specific and Reversible Gene Silencing in Human iPSCs. Cell stem cell. 2016;18:541–553. doi: 10.1016/j.stem.2016.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526:55–61. doi: 10.1038/nature15386. [DOI] [PubMed] [Google Scholar]
- Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nature biotechnology. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, Watanabe T, Kanai T, Sato T. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med. 2015;21:256–262. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- Meister GE, Chandrasegaran S, Ostermeier M. Heterodimeric DNA methyltransferases as a platform for creating designer zinc finger methyltransferases for targeted DNA methylation in cells. Nucleic Acids Research. 2010;38:1749–1759. doi: 10.1093/nar/gkp1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle FT, Eggan K. Modeling human disease with pluripotent stem cells: from genome association to function. Cell stem cell. 2013;12:656–668. doi: 10.1016/j.stem.2013.05.016. [DOI] [PubMed] [Google Scholar]
- Miller JD, Ganat YM, Kishinevsky S, Bowman RL, Liu B, Tu EY, Mandal PK, Vera E, Shim JW, Kriks S, et al. Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell stem cell. 2013;13:691–705. doi: 10.1016/j.stem.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaoka Y, Chan AH, Judge LM, Yoo J, Huang M, Nguyen TD, Lizarraga PP, So PL, Conklin BR. Isolation of single-base genome-edited human iPS cells without antibiotic selection. Nature methods. 2014;11:291–293. doi: 10.1038/nmeth.2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton J, Davis MW, Jorgensen EM, Carroll D. Induction and repair of zinc-finger nuclease-targeted double-strand breaks in Caenorhabditis elegans somatic cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:16370–16375. doi: 10.1073/pnas.0605633103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelles DA, Fang MY, O’Connell MR, Xu JL, Markmiller SJ, Doudna JA, Yeo GW. Programmable RNA Tracking in Live Cells with CRISPR/Cas9. Cell. 2016;165:488–496. doi: 10.1016/j.cell.2016.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Smith A. Naive and primed pluripotent states. Cell stem cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, Kang Y, Zhao X, Si W, Li W, et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156:836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- O’Connell MR, Oakes BL, Sternberg SH, East-Seletsky A, Kaplan M, Doudna JA. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature. 2014;516:263–266. doi: 10.1038/nature13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Park CY, Kim DH, Son JS, Sung JJ, Lee J, Bae S, Kim JH, Kim DW, Kim JS. Functional Correction of Large Factor VIII Gene Chromosomal Inversions in Hemophilia A Patient-Derived iPSCs Using CRISPR-Cas9. Cell stem cell. 2015;17:213–220. doi: 10.1016/j.stem.2015.07.001. [DOI] [PubMed] [Google Scholar]
- Park I, Zhao R, West J, Yabuuchi A, Huo H, Ince T, Lerou P, Lensch M, Daley G. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Parnas O, Jovanovic M, Eisenhaure TM, Herbst RH, Dixit A, Ye CJ, Przybylski D, Platt RJ, Tirosh I, Sanjana NE, et al. A Genome-wide CRISPR Screen in Primary Immune Cells to Dissect Regulatory Networks. Cell. 2015;162:675–686. doi: 10.1016/j.cell.2015.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, Shioda T, et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nature biotechnology. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt P, Schmid B, Burbulla LF, Schondorf DC, Wagner L, Glatza M, Hoing S, Hargus G, Heck SA, Dhingra A, et al. Genetic correction of a LRRK2 mutation in human iPSCs links parkinsonian neurodegeneration to ERK-dependent changes in gene expression. Cell stem cell. 2013;12:354–367. doi: 10.1016/j.stem.2013.01.008. [DOI] [PubMed] [Google Scholar]
- Rideout W, Hochedlinger K, Kyba M, Daley G, Jaenisch R. Correction of a genetic defect by nuclear transplantation and combined cell and gene therapy. Cell. 2002;109:17–27. doi: 10.1016/s0092-8674(02)00681-5. [DOI] [PubMed] [Google Scholar]
- Robert F, Barbeau M, Ethier S, Dostie J, Pelletier J. Pharmacological inhibition of DNA-PK stimulates Cas9-mediated genome editing. Genome Med. 2015;7:93. doi: 10.1186/s13073-015-0215-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinton DA, Daley GQ. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481:295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouet P, Smih F, Jasin M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 1994a;91:6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouet P, Smih F, Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Molecular and cellular biology. 1994b;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhani F, Kumasaka N, de Brito MC, Bradley A, Vallier L, Gaffney D. Genetic background drives transcriptional variation in human induced pluripotent stem cells. PLoS Genet. 2014;10:e1004432. doi: 10.1371/journal.pgen.1004432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby K, Zheng B. Gene Targeting in a HUES Line of Human Embryonic Stem Cells Via Electroporation. Stem Cells. 2009;27:1496–1506. doi: 10.1002/stem.73. [DOI] [PubMed] [Google Scholar]
- Ryan SD, Dolatabadi N, Chan SF, Zhang X, Akhtar MW, Parker J, Soldner F, Sunico CR, Nagar S, Talantova M, et al. Isogenic human iPSC Parkinson’s model shows nitrosative stress-induced dysfunction in MEF2-PGC1alpha transcription. Cell. 2013;155:1351–1364. doi: 10.1016/j.cell.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Clevers H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science (New York, NY) 2013;340:1190–1194. doi: 10.1126/science.1234852. [DOI] [PubMed] [Google Scholar]
- Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Schwank G, Koo BK, Sasselli V, Dekkers JF, Heo I, Demircan T, Sasaki N, Boymans S, Cuppen E, van der Ent CK, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell stem cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Sebastiano V, Zhen HH, Haddad B, Bashkirova E, Melo SP, Wang P, Leung TL, Siprashvili Z, Tichy A, Li J, et al. Human COL7A1-corrected induced pluripotent stem cells for the treatment of recessive dystrophic epidermolysis bullosa. Science translational medicine. 2014;6:264ra163. doi: 10.1126/scitranslmed.3009540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton AN, Regalado SG, Lai CS, Cost GJ, O’Neil CM, Urnov FD, Gregory PD, Jaenisch R, Collins K, Hockemeyer D. Genetic and molecular identification of three human TPP1 functions in telomerase action: recruitment, activation, and homeostasis set point regulation. Genes Dev. 2014;28:1885–1899. doi: 10.1101/gad.246819.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science (New York, NY) 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, Shao F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science (New York, NY) 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C, Gore A, Yan W, Abalde-Atristain L, Li Z, He C, Wang Y, Brodsky RA, Zhang K, Cheng L, et al. Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell stem cell. 2014;15:12–13. doi: 10.1016/j.stem.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, Hargus G, Blak A, Cooper O, Mitalipova M, et al. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, Jaenisch R. Medicine. iPSC disease modeling. Science (New York, NY) 2012;338:1155–1156. doi: 10.1126/science.1227682. [DOI] [PubMed] [Google Scholar]
- Soldner F, Laganière J, Cheng AW, Hockemeyer D, Gao Q, Alagappan R, Khurana V, Golbe LI, Myers RH, Lindquist S, et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146:318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, Stelzer Y, Shivalila C, Abraham BJ, Latourelle Barrasa IJ, Goldmann J, Myers R, Young R, Jaenisch R. Parkinson-associated risk variant in enhancer element produces subtle effect on target gene expression. Nature. 2016 doi: 10.1038/nature17939. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, Hoskins EE, Kalinichenko VV, Wells SI, Zorn AM, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterneckert JL, Reinhardt P, Scholer HR. Investigating human disease using stem cell models. Nature reviews Genetics. 2014;15:625–639. doi: 10.1038/nrg3764. [DOI] [PubMed] [Google Scholar]
- Studer L, Vera E, Cornacchia D. Programming and Reprogramming Cellular Age in the Era of Induced Pluripotency. Cell stem cell. 2015;16:591–600. doi: 10.1016/j.stem.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Sargent RG, Illek B, Fischer H, Esmaeili-Shandiz A, Yezzi MJ, Lee A, Yang Y, Kim S, Renz P, et al. TALENs Facilitate Single-step Seamless SDF Correction of F508del CFTR in Airway Epithelial Submucosal Gland Cell-derived CF-iPSCs. Molecular therapy Nucleic acids. 2016;5:e273. doi: 10.1038/mtna.2015.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. A developmental framework for induced pluripotency. Development (Cambridge, England) 2015;142:3274–3285. doi: 10.1242/dev.114249. [DOI] [PubMed] [Google Scholar]
- Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, Zhang RR, Ueno Y, Zheng YW, Koike N, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159:635–646. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, Spratt SK, Surosky RT, Giedlin MA, Nichol G, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med. 2014;370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Theunissen TW, Powell BE, Wang H, Mitalipova M, Faddah DA, Reddy J, Fan ZP, Maetzel D, Ganz K, Shi L, et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell stem cell. 2014;15:471–487. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas KR, Capecchi MR. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science (New York, NY) 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nature biotechnology. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nature biotechnology. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbach A, Schuldiner M, Benvenisty N. Modeling for Lesch-Nyhan disease by gene targeting in human embryonic stem cells. Stem Cells. 2004;22:635–641. doi: 10.1634/stemcells.22-4-635. [DOI] [PubMed] [Google Scholar]
- Urnov F, Miller J, Lee Y, Beausejour C, Rock J, Augustus S, Jamieson A, Porteus M, Gregory P, Holmes M. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nature reviews Genetics. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. Onestep generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, Sabatini DM. Identification and characterization of essential genes in the human genome. Science (New York, NY) 2015a;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science (New York, NY) 2014a;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang Y, Wu X, Wang J, Wang Y, Qiu Z, Chang T, Huang H, Lin RJ, Yee JK. Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nature biotechnology. 2015b;33:175–178. doi: 10.1038/nbt.3127. [DOI] [PubMed] [Google Scholar]
- Wang Y, Liang P, Lan F, Wu H, Lisowski L, Gu M, Hu S, Kay MA, Urnov FD, Shinnawi R, et al. Genome editing of isogenic human induced pluripotent stem cells recapitulates long QT phenotype for drug testing. J Am Coll Cardiol. 2014b;64:451–459. doi: 10.1016/j.jacc.2014.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell stem cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Ueno M, Kamiya D, Nishiyama A, Matsumura M, Wataya T, Takahashi JB, Nishikawa S, Nishikawa S, Muguruma K, et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nature biotechnology. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein B, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Woltjen K, Michael I, Mohseni P, Desai R, Mileikovsky M, Hamalainen R, Cowling R, Wang W, Liu P, Gertsenstein M, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X, et al. Targeted genome editing across species using ZFNs and TALENs. Science (New York, NY) 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Kriz AJ, Sharp PA. Target specificity of the CRISPR-Cas9 system. Quantitative biology. 2014a;2:59–70. doi: 10.1007/s40484-014-0030-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Scott DA, Kriz AJ, Chiu AC, Hsu PD, Dadon DB, Cheng AW, Trevino AE, Konermann S, Chen S, et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nature biotechnology. 2014b;32:670–676. doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F, Ye L, Chang JC, Beyer AI, Wang J, Muench MO, Kan YW. Seamless gene correction of beta-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome research. 2014;24:1526–1533. doi: 10.1101/gr.173427.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue H, Wu S, Papadeas S, Spusta S, Swistowska A, Macarthur C, Mattson M, Maragakis N, Capecchi M, Rao M, et al. A Targeted Neuroglial Reporter Line Generated by Homologous Recombination in Human Embryonic Stem Cells. Stem Cells. 2009 doi: 10.1002/stem.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Johannesson B, Sagi I, Burnett LC, Kort DH, Prosser RW, Paull D, Nestor MW, Freeby M, Greenberg E, et al. Human oocytes reprogram adult somatic nuclei of a type 1 diabetic to diploid pluripotent stem cells. Nature. 2014;510:533–536. doi: 10.1038/nature13287. [DOI] [PubMed] [Google Scholar]
- Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-Step Generation of Mice Carrying Reporter and Conditional Alleles by CRISPR/Cas-Mediated Genome Engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Wang J, Beyer AI, Teque F, Cradick TJ, Qi Z, Chang JC, Bao G, Muench MO, Yu J, et al. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Delta32 mutation confers resistance to HIV infection. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:9591–9596. doi: 10.1073/pnas.1407473111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young CS, Hicks MR, Ermolova NV, Nakano H, Jan M, Younesi S, Karumbayaram S, Kumagai-Cresse C, Wang D, Zack JA, et al. A Single CRISPR-Cas9 Deletion Strategy that Targets the Majority of DMD Patients Restores Dystrophin Function in hiPSC-Derived Muscle Cells. Cell stem cell. 2016 doi: 10.1016/j.stem.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JE, Boulanger-Weill J, Williams DA, Woodruff G, Buen F, Revilla AC, Herrera C, Israel MA, Yuan SH, Edland SD, et al. Elucidating molecular phenotypes caused by the SORL1 Alzheimer’s disease genetic risk factor using human induced pluripotent stem cells. Cell stem cell. 2015;16:373–385. doi: 10.1016/j.stem.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Liu Y, Ma T, Liu K, Xu S, Zhang Y, Liu H, La Russa M, Xie M, Ding S, et al. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell stem cell. 2015;16:142–147. doi: 10.1016/j.stem.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. Human induced pluripotent stem cells free of vector and transgene sequences. Science (New York, NY) 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science (New York, NY) 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Yu Z, Ren M, Wang Z, Zhang B, Rong YS, Jiao R, Gao G. Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila. Genetics. 2013;195:289–291. doi: 10.1534/genetics.113.153825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusa K. Seamless genome editing in human pluripotent stem cells using custom endonuclease-based gene targeting and the piggyBac transposon. Nature protocols. 2013;8:2061–2078. doi: 10.1038/nprot.2013.126. [DOI] [PubMed] [Google Scholar]
- Yusa K, Rashid ST, Strick-Marchand H, Varela I, Liu PQ, Paschon DE, Miranda E, Ordonez A, Hannan NR, Rouhani FJ, et al. Targeted gene correction of alpha1-antitrypsin deficiency in induced pluripotent stem cells. Nature. 2011;478:391–394. doi: 10.1038/nature10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XY, Li W, Lv Z, Liu L, Tong M, Hai T, Hao J, Guo CL, Ma QW, Wang L, et al. iPS cells produce viable mice through tetraploid complementation. Nature. 2009;461:86–90. doi: 10.1038/nature08267. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, Wei W. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
- Zou J, Maeder M, Mali P, Pruett-Miller S, Thibodeau-Beganny S, Chou B, Chen G, Ye Z, Park I, Daley G, et al. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell stem cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Mali P, Huang X, Dowey SN, Cheng L. Site-specific gene correction of a point mutation in human iPS cells derived from an adult patient with sickle cell disease. Blood. 2011;118:4599–4608. doi: 10.1182/blood-2011-02-335554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaka T, Thomson J. Homologous recombination in human embryonic stem cells. Nature biotechnology. 2003;21:319–321. doi: 10.1038/nbt788. [DOI] [PubMed] [Google Scholar]