Abstract
Three species of Myodes voles known to harbor hantaviruses include the bank vole (Myodes glareolus), which serves as the reservoir host of Puumala virus (PUUV), the prototype arvicolid rodent-borne hantavirus causing hemorrhagic fever with renal syndrome (HFRS) in Europe, and the grey red-backed vole (M. rufocanus) and royal vole (M. regulus) which carry two PUUV-like hantaviruses, designated Hokkaido virus (HOKV) and Muju virus (MUJV), respectively. To ascertain the hantavirus harbored by the northern red-backed vole (M. rutilus), we initially screened sera from 233 M. rutilus, as well as from 90 M. rufocanus and 110 M. glareolus, captured in Western and Eastern Siberia during June 2007 to October 2009, for anti-hantaviral antibodies. Thereafter, lung tissues from 44 seropositive voles were analyzed for hantavirus RNA by reverse transcription-polymerase chain reaction. Partial L-, M- and S-segment sequences, detected in M. rutilus and M. rufocanus, were closely related to HOKV, differing from previously published L-, M- and S-segment sequences of HOKV by 17.8–20.2%, 15.9–23.4% and 15.0–17.0% at the nucleotide level and 2.6–7.9%, 1.3–6.3% and 1.2–4.0% at the amino acid level, respectively. Alignment and comparison of hantavirus sequences from M. glareolus trapped in Tiumen Oblast showed very high sequence similarity to the Omsk lineage of PUUV. Phylogenetic analysis, using neighbor-joining, maximal likelihood and Bayesian methods, showed that HOKV strains shared a common ancestry with PUUV and exhibited geographic-specific clustering. This report provides the first molecular evidence that both M. rutilus and M. rufocanus harbor HOKV, which might represent a genetic variant of PUUV.
Keywords: hantavirus, Puumala virus, Myodes voles, Siberia
1. Introduction
Hantaviruses (family Bunyaviridae, genus Hantavirus) possess a negative-sense, single-stranded, tripartite RNA genome, comprising L, M and S segments, which encode an RNA-dependent RNA polymerase (RdRp), envelope glycoproteins (Gn and Gc) and nucleocapsid (N protein), respectively (Plyusnin et al., 1996). Several hantaviruses, harbored by rodents (order Rodentia) belonging to the Muridae and Cricetidae families, cause hemorrhagic fever with renal syndrome (HFRS) (Jonsson et al., 2010). Hantaan virus (HTNV), Dobrava-Belgrade virus (DOBV) and Seoul virus (SEOV) are the principal pathogenic murid rodent-borne hantaviruses, while Puumala virus (PUUV), carried by the bank vole (Myodes glareolus), is the prototype HFRS-causing arvicolid rodent-borne hantavirus.
M. glareolus is one of the most abundant and widely distributed small mammal species in Europe, occurring in most areas except near the coast of the Mediterranean Basin and in the far northern territories (Olsson et al., 2010). In the Asian part of the continent, its range extends to the Sayan Mountains and northern Kazakhstan (Shenbrot and Krasnov, 2005). Following the latest glacial period, M. glareolus recolonized the continent and formed at least eight distinct lineages of co-evolved PUUV (Sironen et al., 2001). Phylogenetic studies suggest that the Omsk lineage of PUUV in western Siberia has a common evolutionary origin with that of the Finnish lineage (Dekonenko et al., 2003).
Three other species of Myodes voles serve as reservoir hosts of hantaviruses: the grey red-backed vole (M. rufocanus), northern red-backed vole (M. rutilus) and royal vole (M. regulus). The former two species have overlapping geographic ranges with M. glareolus in Europe and Asia (Shenbrot and Krasnov, 2005). In particular, M. rufocanus is distributed in the northern Paleoarctic, extending from northern Fennoscandia through northern Russia to Kamchatka, northeastern and northern Korea, Mongolia, China, Sakhalin (Russia) and Hokkaido (Japan), whereas M. rutilus is a Holarctic species found throughout northern Europe, Asia, Alaska and Canada. By contrast, M. regulus is confined to mountainous regions at elevations above 500 m in the Korean Peninsula (Kaneko, 1990).
Hantaviral antigens were initially detected in M. rufocanus in Far East and Siberian Russia (Kosoy et al., 1997; Tkachenko et al., 1983, 1987). Subsequently, genetic evidence of a PUUV-like hantavirus, designated Hokkaido virus (HOKV), was demonstrated in M. rufocanus trapped in Hokkaido, Japan (Kariwa et al., 1995, 1999; Sanada et al., 2012), the Far East region and Buryatia Republic of Russia (Plyusnina et al., 2008; Yashina et al., 2004, 2013) and Jilin province, China (Zhang et al., 2007). Another PUUV-like virus, named Muju virus (MUJV), was detected in M. regulus, trapped at multiple sites in the Republic of Korea (Lee et al., 2014; Song et al., 2007). Both HOKV and MUJV share a common ancestry with PUUV.
Although hantaviral antigens or antibodies have been reported in M. rutilus, captured in Alaska (Lee et al., 1982) and in European, Siberian and Far East Russia (Kosoy et al., 1997; Tkachenko et al., 1987; Yashina et al., 2012), the hantavirus harbored by M. rutilus has not been characterized. In the present study, genetic analysis of hantaviruses in M. rutilus, M. rufocanus and M. glareolus, sharing the same ecological habitats in the Western Siberia, indicates that both M. rutilus and M. rufocanus serve as reservoir hosts of HOKV. Based on whole genome sequence analysis of MUJV (Lee et al., 2014) and findings from this investigation, HOKV and MUJV appear to represent genetic variants, or genotypes, of PUUV, rather than distinct hantavirus species. We, therefore, tentatively propose that HOKV and MUJV be called the Hokkaido and Muju genotypes of PUUV.
2. Materials and methods
2.1. Rodent trapping and screening
During June 2007 to October 2009, rodents were captured in 12 forest localities of seven administrative regions of Western and Eastern Siberia (Altai Republic, Altai and Krasnoyarsk Krais, and Tiumen, Omsk, Kemerovo and Tomsk Oblasts), where two or three Myodes species were sympatric (Table 1 and Fig. 1). Captured Myodes voles were identified according to a complex of morphologic criteria, including configuration of the prisms and triangles of the occlusal surface of the third upper (M3) and first lower (M1) molars, body size, length and hairiness of tail, and fur coloration (Gromov and Erbaeva, 1995; Gromov and Polyakov, 1977). Sera and lung tissues were stored in liquid nitrogen for subsequent analysis for anti-hantavirus antibodies by the indirect immunofluorescent antibody test (IFA) using Vero E6 cells infected with HTNV (76–118), PUUV (CG1820) and SEOV (SR-11) as antigens (Dzagurova et al., 1995), and for hantavirus RNA by reverse transcription-polymerase chain reaction (RT-PCR) (Yashina et al., 2004, 2013). Although hantaviral antigens have been detected before antibody responses in M. glareolus experimentally infected with PUUV (Apekina et al., 2014), we were unable to test all tissues from wild-trapped Myodes voles by RT-PCR, due to budgetary constraints, and instead were able to test tissues only from IFA-seropositive voles.
Table 1.
Prevalence of hantavirus infection, as determined by serology and RT-PCR, in Myodes voles captured in Western and Eastern Siberia, 2007–2009
| Administrative Region | Trap Locality | Trap Year | Myodes Species | Antibody Prevalence | Hantavirus RNA |
|---|---|---|---|---|---|
| Altai Krai | Khmelevka | 2008 | M. rutilus | 0/13 | NT |
| M. rufocanus | 0/1 | NT | |||
| M. glareolus | 1/7 | 0/1 | |||
| Kolyvan | 2008 | M. rutilus | 1/11 | 0/1 | |
| M. rufocanus | 0/1 | NT | |||
| Pokrovka | 2008 | M. rutilus | 0/16 | NT | |
| M. rufocanus | 8/37 | 1/8 | |||
| Solton | 2007 | M. rutilus | 2/14 | 1/2 | |
| M. rufocanus | 1/2 | 0/1 | |||
| M. glareolus | 0/10 | NT | |||
| Altai Republic | Teletskoye Lake | 2007–2009 | M. rutilus | 6/80 | 1/6 |
| M. rufocanus | 1/3 | 1/1 | |||
| M. glareolus | 1/10 | 0/1 | |||
| Kemerovo Oblast | Balakhnino | 2009 | M. rutilus | 0/5 | NT |
| M. rufocanus | 0/6 | NT | |||
| M. glareolus | 0/1 | NT | |||
| Kuzedeyevo | 2007 | M. rutilus | 1/9 | 0/1 | |
| M. rufocanus | 0/6 | NT | |||
| M. glareolus | 0/21 | NT | |||
| Krasnoyarsk Krai | Parnaya | 2008 | M. rutilus | 1/11 | 0/1 |
| M. glareolus | 0/5 | NT | |||
| Srednyaya Shush | 2008 | M. rutilus | 4/26 | 0/4 | |
| M. rufocanus | 9/31 | 0/9 | |||
| Omsk Oblast | Ust-Ishim | 2007 | M. rutilus | 0/11 | NT |
| M. glareolus | 2/11 | 0/2 | |||
| Tiumen Oblast | Kuchuk | 2007 | M. rutilus | 1/29 | 0/1 |
| M. glareolus | 4/42 | 3/4 | |||
| Tomsk Oblast | Sukhorechie | 2009 | M. rutilus | 1/8 | 0/1 |
| M. rufocanus | 0/3 | NT | |||
| M. glareolus | 0/3 | NT |
Number of antibody positive Myodes voles/Number captured
Hantavirus RNA was detected by RT-PCR, using L-segment specific primers, in lungs of anti-hantavirus antibody-positive voles
NT, samples were not analyzed
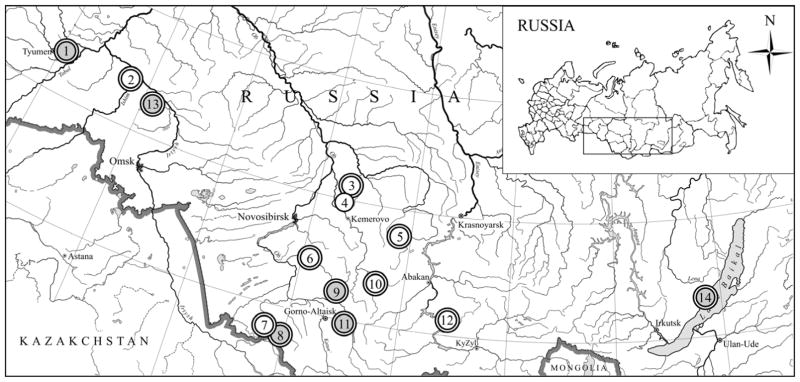
Fig. 1.
Map of Russia, showing the locations of the rodent trapping sites: (1) Kuchuk, (2) Ust-Ishim, (3) Sukhorechie, (4) Balakhnino, (5) Parnaya, (6) Khmelevka, (7) Kolyvan, (8) Pokrovka, (9) Solton, (10) Kuzedeyevo, (11) Teletskoye Lake, (12) and Srednyaya Shush, as well as (13) Omsk region (Dekonenko et. al., 2003) and (14) Olhon district (Yashina et al., 2013). Sites where anti-hantavirus antibody-positive and hantavirus RNA-positive voles were captured are labeled with double and filled circles, respectively
2.2. Ethics statement
All trapping and handling of rodents and processing of their tissues were performed according to well-established protocols (Mills et al., 1995), approved by the Institutional Animal Care and Use Committee of the State Research Center of Virology and Biotechnology “Vector”.
2.3. RT-PCR, mtDNA and DNA sequencing
Total RNA was extracted from lung tissue of anti-hantaviral antibody-positive rodents with the RNeasy MiniKit (Qiagen, Hilden, Germany), and cDNA was synthesized using Expand reverse transcriptase (Roche) and universal primer 5′-TAGTAGTAGACTCC-3′. Four sets of nested primers were used to amplify selected regions of the large (L: positions 180–522), medium (M: 2764–3004) and small (S: 43–1251) segments (nucleotide position numbers are those for PUUV strain CG1820: GenBank M63194, M29979, M32750). Sequences of primers are shown in Table 2. To confirm the taxonomic identity of hantavirus RNA-positive voles, genomic DNA was extracted from frozen lung tissue using the QIAamp DNA Mini Kit (Qiagen), and the partial 426-nucleotide region of the cytochrome b gene of mitochondrial DNA (mtDNA) was amplified by PCR using previously described universal primers: +14115 (5′-CGAAGCTTGATATGAAAAACCATCGTTG-3′); and −14532 (5′-GCAGCCCCTCAGAATGATATTTGTCCAC-3′) (Smith & Patton, 1991). Amplicons were gel-purified with QIAquick Gel Extraction kit (Qiagen) and directly sequenced using ABI Prizm BigDye Terminator kit (PE Applied Biosystem) and an automatic ABI Prism 310 genetic analyzer.
Table 2.
Oligonucleotide primers for nested PCR used in this study
| Gene | Primer sequence (5′-3′) | Positiona, orientation | Reference |
|---|---|---|---|
| L | AGAGAA(G/A)T(T/C)TACTA(C/A)AAATGGAGAAATACA | 20 (+) | Yashina et al., 2013 |
| TTTAAATT(G/A)AACAT(T/G)GC(T/C)TC(A/T)AGTGC | 677 (−) | ||
| GA(C/T)CAGATGATAAA(G/A)CATGA(T/C)TGGTC | 154 (+) | ||
| CTGTGTTGA(G/T)AT(A/G)TTTGA(T/G)CCATCAGT | 549 (−) | ||
| M | ATTTAA(G/A)CAATGGTG(C/T)ACTAC(T/A)AC | 2644 (+) | Yashina et al., 2000 |
| CC(G/A)TAACACATTGC(A/G)GC | 3117 (−) | ||
| TAGAAAGAAATGTGCATTTGC | 2743 (+) | ||
| CCTGA(G/A)CCCCATGC(A/T/C)CCATC | 3024 (−) | ||
| S | TTCTGCAGTAGTAGTAGACTCCTTGAAAAG | 1 (+ −) | Yashina et al., 2010 This study |
| C(A/T)GGTGCACA(G/T)GCAAANACCC | 991 (−) | ||
| G(A/T)GG(A/C/T)CA(G/A)AC(A/T)GCAGA(C/T)TGG | 380 (+) | ||
| AGCTCAGGATCCATGTCATC | 1271 (−) | ||
| CCCAGATCTATGAGTGACTTGACAGACATCCAAG | 42 (+) | ||
| CC(A/T)GGTGT(A/G)AG(T/C)TCTTC(A/T)GC | 629 (−) |
2.4. Genetic and phylogenetic analyses
Pair-wise alignment and comparison of newfound hantavirus nucleotide sequences were performed using ClustalW (Thompson et al., 1994). The distance-based neighbor-joining method, supported by MEGA 5.1 (Tamura et al., 2011), was used to construct phylogenetic trees, which were evaluated by bootstrap analysis of 1,000 neighbor-joining iterations. In addition, unrooted phylogenetic trees were generated by maximum likelihood and Bayesian methods, implemented in PAUP* (Phylogenetic Analysis Using Parsimony, 4.0b10) (Swofford, 2003), RAxML Blackbox webserver (Stamatakis et al., 2008) and MrBayes 3.1 (Ronquist and Huelsenbeck, 2003), under the best-fit GTR+I+Γ model of evolution selected by hierarchical likelihood-ratio test in MrModeltest v2.3 (Posada and Crandall, 1998) and jModelTest version 0.1 (Posada, 2008). Two replicate Bayesian Metropolis–Hastings Markov Chain Monte Carlo runs, each comprising six chains of 10 million generations sampled every 100 generations with a burn-in of 25,000 (25%), resulted in 150,000 trees overall. Each genomic segment (S, M and L) was treated separately in phylogenetic analyses. The posterior node probabilities were based on 2 million generations and estimated sample sizes over 100 (implemented in MrBayes).
3. Results
M. rutilus was the dominant Myodes species captured at six sites, M. rufocanus at three and M. glareolus at two, with equal numbers of M. rutilus and M. glareolus at one site (Omsk Oblast) (Table 1). Sera, diluted 1:10, from 433 voles (233 M. rutilus, 90 M. rufocanus and 110 M. glareolus) were initially screened by IFA for the presence of anti-hantaviral antibodies. Individuals of one, two or three Myodes species in 11 of the 12 trap sites were anti-hantavirus antibody positive (Table 1).
Of the 44 (10.2%) seropositive voles, hantavirus RNA was detected in two of 17 M. rutilus, two of 19 M. rufocanus and three of eight M. glareolus, which were taxonomically verified by mitochondrial DNA analysis. Sequence analysis, based on a 426-nucleotide cytochrome b region of mtDNA, showed that intraspecies differences in Myodes voles (M. glareolus, M. rufocanus and M. rutilus) varied from 0 to 2.3%, while interspecies differences were from 6.8–9.6%. In all studied cases, cytochrome b typing matched morphological specification of hantavirus RNA-positive Myodes voles. The GenBank accession numbers for the cytochrome b sequences are provided in Table 3.
Table 3.
Hokkaido virus (HOKV) sequences from M. rutilus and M. rufocanus and Puumala virus (PUUV) sequences from M. glareolus captured in Western Siberia
| Trap Locality | Myodes species | Virus strain | GenBank accession no.
|
|||
|---|---|---|---|---|---|---|
| S segment | M segment | L segment | Host cyt b | |||
| Pokrovka | M. rufocanus | Pokrovka674/Mrf/2008 | KM245964 | KM245954 | KM245959 | KP859518 |
| Solton | M. rutilus | Solton35/Mrt/2007 | KM245962 | KM245952 | KM245957 | KP859515 |
| Teletskoye | M. rutilus | Teletskoye854/Mrt/2008 | KM245963 | KM245953 | KM245958 | KP859516 |
| M. rufocanus | Teletskoye937/Mrf/2008 | KM245965 | KM245955 | KM245960 | KP859519 | |
| Kuchuk | M. glareolus | Kuchuk170/Mg/2007 | KP292966 | KP292970 | KP292967 | KP859521 |
| Kuchuk197/Mg/2007 | – | KP292972 | KP292968 | KP859522 | ||
| Kuchuk246/Mg/2007 | – | KP292973 | KP292969 | KP859523 | ||
– The short 210-nucleotide sequences of the S segment (position 400–609) of PUUV strains Kuchuk197/Mg/2007 and Kuchuk246/Mg/2007 were not deposited in GenBank and were not included in the phylogenetic analysis.
Hantavirus RNA-positive voles were captured in three localities of the Altai region: near Teletskoye Lake (Artybash village, Altai Republic), Pokrovka and Solton villages (Altai Krai) and in one locality of Tiumen Oblast near Kuchuk Lake (Table 1 and Fig. 1). In one of these sites (Teletskoye Lake), hantavirus RNA was detected in two of three hantavirus-infected Myodes species (M. rutilus and M. rufocanus), and in each of three other sites hantavirus sequences from a single host species were identified.
Hantavirus RNA was detected in lung tissues from seven of 44 anti-hantaviral antibody-positive Myodes voles. Of these, six voles had anti-hantavirus antibody titers ≥1:80 (Table 4). Six of 17 voles with antibody titers ≥1:80 were hantavirus RNA positive, compared to one of 27 voles with anti-hantaviral antibody titers ≤1:40. At titers of ≥1:320, four of five voles were hantavirus RNA positive (Table 4).
Table 4.
Myodes voles with anti-hantavirus antibodies and hantavirus RNA
| Trap locality | Animal no./species | Anti-hantavirus IFA titer | Hantavirus RNAa |
|---|---|---|---|
| Khmelevka | Khmelevka161/Mg | 20 | − |
| Kolyvan | Kolyvan721/Mrt | 40 | − |
| Pokrovka | Pokrovka613/Mrf | 20 | − |
| Pokrovka622/Mrf | 40 | − | |
| Pokrovka630/Mrf | 640 | − | |
| Pokrovka632/Mrf | 10 | − | |
| Pokrovka670/Mrf | 80 | − | |
| Pokrovka674/Mrf | 320 | + | |
| Pokrovka678/Mrf | 160 | − | |
| Pokrovka695/Mrf | 160 | − | |
| Solton | Solton35/Mrt | 640 | + |
| Solton39/Mrt | 40 | − | |
| Solton38Mrf | 40 | − | |
| Teletskoye Lake | Teletskoye338/Mrt | 40 | −b |
| Teletskoye361/Mrt | 40 | − | |
| Teletskoye854/Mrt | 80 | +c | |
| Teletskoye860/Mrt | 20 | − | |
| Teletskoye864/Mrt | 80 | − | |
| Teletskoye2321/Mrt | 80 | − | |
| Teletskoye937/Mrf | 160 | + | |
| Teletskoye894/Mg | 40 | − | |
| Kuzedeyevo | Kuzedeyevo91/Mrt | 20 | − |
| Parnaya | Parnaya1241/Mrt | 80 | − |
| Srednyaya Shush | Shush1004/Mrt | 10 | − |
| Shush1007/Mrt | 40 | − | |
| Shush1010/Mrt | 40 | − | |
| Shush1018/Mrt | 20 | − | |
| Shush1002/Mrf | 40 | − | |
| Shush1008/Mrf | 40 | − | |
| Shush1013/Mrf | 80 | − | |
| Shush1050/Mrf | 80 | − | |
| Shush1056/Mrf | 40 | − | |
| Shush1059/Mrf | 40 | − | |
| Shush1060/Mrf | 10 | − | |
| Shush1066/Mrf | 20 | − | |
| Shush1102/Mrf | 80 | − | |
| Ust-Ishim | Ust-Ishim261/Mg | 20 | − |
| Ust-Ishim277/Mg | 80 | − | |
| Kuchuk | Kuchuk170/Mg | 40 | + |
| Kuchuk197/Mg | 640 | + | |
| Kuchuk207/Mg | 40 | − | |
| Kuchuk246/Mg | 640 | + | |
| Kuchuk232/Mrt | 10 | − | |
| Sukhorechie | Sukhorechie1394/Mrt | 10 | − |
Hantavirus L-segment RNA was detected in rodent lungs by RT-PCR
negative
positive
Abbreviations: Mrt, M. rutilus; Mrf, M. rufocanus; Mg, M. glareolus.
Analysis of partial L- (position 180–522), M- (position 2764–3004) and S- (position 43–1251) segment sequences from M. rutilus and M. rufocanus showed that both species harbored a hantavirus which was most closely related to HOKV. HOKV was detected in M. rutilus in Solton, in M. rufocanus in Pokrovka and in both M. rutilus and M. rufocanus in Teletskoye Lake (Table 1). In both Solton and Teletskoye Lake, M. rutilus was the dominant species, accounting for 53.8% and 86.0% of all captured Myodes voles, respectively.
The nucleotide sequence similarity among the newly identified HOKV strains Teletskoye854/Mrt, Teletskoye937/Mrf, Solton35/Mrt and Pokrovka674/Mrf were 83.6–97.7%, 80.5–98.8% and 84.5–97.6% for the L-, M- and S-segments, respectively. And the deduced amino acid sequence similarity was 99.1–100% for the RdRp, 98.8–100% for the Gc glycoprotein and 97.3–99.3% for the N protein. Minimal nucleotide sequence divergence (<3%) was shown for HOKV strains, Teletskoye854/Mrt and Teletskoye937/Mrf detected in M. rutilus and M. rufocanus, respectively, from one locality. Compared with previously published HOKV L-, M- and S-segment sequences from M. rufocanus, 79.8–82.2%, 76.6–84.1% and 83.0–85.0% similarity was found at the nucleotide level and 92.1–97.4%, 93.7–98.7% and 96.0–98.8% at the amino acid level. The amino acid sequences of the RdRp, Gc and N proteins of the Siberian HOKV strains differed from that of PUUV strains by 6.1–9.7%, 5.0–6.3% and 3.5–4.5%, and from that of MUJV strains by 9.7–10.5%, 5.0–6.3% and 4.2–6.0%.
Hantavirus RNA, detected in three of four seropositive M. glareolus in Tiumen Oblast (Kuchuk170/Mg, Kuchuk197/Mg, Kuchuk246/Mg), showed the highest degree of sequence similarity to the Omsk lineage of PUUV, previously described in M. glareolus captured in a neighboring administrative region of Siberia (Omsk Oblast): 93.3–96.7% and 97.5% for M and 96.6–96.9% and 98.5–99.2% for S segment at the nucleotide and amino acid levels, respectively. Compared with other PUUV strains, Kuchuk170/Mg, Kuchuk197/Mg and Kuchuk246/Mg differed by 14.5–19.6% at the nucleotide level and 1.8–6.1% at the amino acid level for the L segment, and by 14.1–17.4% and 3.7–8.7% for the M segment, and by 11.2–17.4% and 1.7–4.7% for the S segment, respectively. The partial amino acid sequences of the Siberian HOKV strains differed from that of the Siberian PUUV strains by 6.1–7.9% for RdRp, 6.2–7.5% for Gc and 3.7–5.0% for N protein.
Phylogenetic trees based on partial S-, M- and L-segment sequences, using neighbor-joining, maximum-likelihood and Bayesian methods, indicated that HOKV strains were placed within the well-supported clade formed by three hantaviruses associated with Myodes voles: PUUV, HOKV and MUJV (Fig. 2). HOKV strains formed five genetic lineages, represented respectively by previously described strains from Japan (Kamiiso-8Cr-95, Tobetsu60Cr, Kiritappu126S, Nakagawa49S, Ishikari9S) and Russia (Sakhalin99S), Republic Buryatia (Mukhorshibir767, Tunka227), Russia-China (Fusong and Khekhtsir3S) and two new lineages from Russia: Siberia 1 (Solton35/Mrt, Teletskoye854/Mrt, Olhon109/Mrf) and Siberia 2 (Pokrovka674/Mrf). The N protein sequence of the newfound HOKV strains contained specific amino acid signatures of HOKV (Val/Ile68, Val/Ala79 and Ile262) and residues common for all HOKV strains and those also shared by MUJV, Khabarovsk virus (KBRV) and Topografov virus (TOPV) (Arg26, Pro283). Specific amino acid signatures for the Siberia 1 lineage were Met127, Asp302 and Asp307, and for the Siberia 2 lineage Gln237.
Fig. 2.
Phylogenetic trees, generated by the maximum-likelihood and Bayesian methods, using the GTR+I+Γ model of evolution as estimated from the data, were based on the alignment of the coding regions of the (S) partial S segment (position 43–1251), (M) partial M segment (position 2764–3004) and (L) partial L segment (position 180–522) of Myodes vole-borne hantaviruses and other representative hantaviruses. Because the unrooted phylogenetic trees using these methods were very similar, the trees generated by MrBayes were displayed. The phylogenetic positions of the newfound Myodes vole-borne hantaviruses from Siberia are shown in relationship to PUUV CG215 (AF367066); PUUV CRF161 (AF367069, AF367061); PUUV CRF308 (AF367070); PUUV Virrat/25Cg (Z69985, Z70201); PUUV Pallasjarvi/63Cg/98 (AJ314597); PUUV Sotkamo (NC_005224, NC_005223, NC_005225); PUUV Pieksamaki/Mg4/2008 (JN831946); PUUV Pieksamaki/Mg7/2008 (JN831945); PUUV Kazan (Z84204, Z84205, EF405801); PUUV Udmurtia/338Cg (Z30708); PUUV CG1820 (M32750, M29979, M63194); PUUV Samara/147Cg (AB433855); PUUV Samara/49Cg/2005 (AB433850, AB574183); PUUV Samara/94Cg/2005 (AB574184); PUUV DTK/Ufa-97 (AB297667); PUUV Couvin/59Cg/97 (AJ277034, AJ277040); PUUV Cg-Erft (AJ238779, AJ238778); PUUV Omsk222 (AF442616); PUUV Opina916 (AF294652); PUUV Cg13891 (U22418); PUUV Balkan-2 (AJ314601); PUUV Klippitztoerl (AJ888751); PUUV Vranica (U14137); PUUV Vindeln/L20 (Z48586); PUUV Mellansel/Cg47 (AJ223374); PUUV Umea (AY526219, AY526218, AY526217); PUUV Fyn (AJ238791); PUUV Fyn131 (AJ278092); PUUV Eidsvoll/Cg1138 (AJ223369); PUUV Solleftea/Cg6 (AJ223377); HOKV Kitahiyama128 (AB675463, AB676848, AB712372); HOKV Kamiiso-8Cr-95 (AB010730); HOKV Kamiiso-Crf (AB011631); HOKV Tobetsu27S/2004 (AB675465); HOKV Tobetsu35/2010 (AB675451, AB675452); HOKV Ishikari9S/2009 (AB675469); HOKV Kiritappu126S/2000 (AB675474); HOKV Sakhalin99/1998 (AB675453, AB675454, AB675455); HOKV Tunka227 (KP325675, KR072694); HOKV Muhorshibir767 (AM930972, AM930975); HOKV Muhorshibir791 (AM930976); HOKV Olhon109 (KP325674, KM245956, KM245961); HOKV Fusong 200-05 (EF211820); HOKV Fusong-247 (EF442087, EF442094); HOKV Fusong-302 (EF442095); HOKV Fusong8405 (EF422372); HOKV Khekhtsir3S/1998 (AB677476); HOKV Khekhtsir37/2002 (AB677484, AB677488); HOKV CRF74372 (AY491383); HOKV CRF74333 (AY491381); MUJV 11-1 (JX028273, JX028272, JX028271); MUJV 11-4 (JX046484, JX046483, JX046482); MUJV 11-5 (JX046487, JX046486, JX046485); TOPV Ls136v (AJ011646, AJ011647, AJ011649); KBRV MF-43 (U35255, AJ011648, AJ011650); TULV M5302v (NC_005227, NC_005228, NC_005226); PHV PH-1 (Z49098, X55129, EF646763); and LUXV LX309 (HM756286, HM756287, HQ404253). Other rodent-borne hantaviruses included ANDV Chile9717869 (NC_003466, NC_003467, NC_003468); SNV NMH10 (NC_005216, NC_005215, NC_005217); HTNV 76–118 (NC_005218, Y00386, NC_005222); SOOV SOO-1 (AY675349, AY675353, DQ562292); DOBV Greece (NC_005233, NC_005234, NC_005235); and SEOV 80-39 (NC_005236, NC_005237, NC_005238). Shrew- and mole-borne hantaviruses included MJNV Cl05–11 (EF641804, EF641798, EF641806); TPMV VRC66412 (AY526097, EU001329, EU001330); and NVAV MSB95703 (FJ539168, HQ840957, FJ593498). The corresponding geographic areas are indicated as BAL (Balkan), BEL (Belgium), BUR (Buryatia), CHN/RUS (China/Russia), DAN (Denmark), FIN (Finland), JPN/RUS (Japan/Russia), KOR (Korea), NSCA (Northern Scandinavia), OMSK (Omsk), RUS (Russia), SIB-1 (Siberia), SIB-2 (Siberia), and SSCA (Southern Scandinavia). The numbers at each node are posterior node probabilities based on 150,000 trees. The scale bar indicates nucleotide substitutions per site. The GenBank accession numbers for the newfound hantaviral sequences from Siberia are listed in Table 3.
The Siberia 1 lineage of HOKV included three new strains from the Altai region (Solton35, Teletskoye854, Teletskoye937) and the previously described strain Olhon109 from M. rufocanus captured in Olhon district of Irkutsk Oblast (Yashina et al., 2013), located more then 1,750 km to the east, near Baikal Lake. The Siberia 2 lineage was found in M. rufocanus captured at one site near Pokrovka village (Altai Krai), located approximately 260 km southwest from the two other sites, Teletskoye Lake and Solton. The newly identified PUUV strain Kuchuk170 from M. glareolus was placed within the Omsk lineage of PUUV, according to geography.
4. Discussion
The principal objectives of this study were to determine the molecular phylogeny of hantaviruses harbored by three species of Myodes voles inhabiting the same natural foci in Western and Eastern Siberia and to fill a long-standing gap in knowledge about the hantavirus harbored by M. rutilus. This report provides the first genetic evidence that HOKV is harbored by both M. rufocanus and M. rutilus in Siberian Russia. That HOKV was found irrespective of whether the dominant Myodes species was M. rutilus (Teletskoye Lake and Solton) or M. rufocanus (Pokrovka) suggests that both species are natural reservoirs of HOKV.
Host-specificity studies of experimental PUUV infection in wild-trapped and colonized cricetid rodents (Klingström et al., 2002) are compatible with our finding of HOKV RNA in lung tissues of two species of anti-hantaviral antibody-positive, wild-trapped voles, M. rufocanus and M. rutilus. Because M. rutilus was the dominant Myodes species over M. rufocanus in Solton and Teletskoye Lake, spillover infection of HOKV from the latter to the former is unlikely.
Overall, hantavirus RNA was detected in only 15.9% (7/44) of the anti-hantaviral antibody-positive Myodes voles. The discordance between the prevalence of anti-hantaviral antibody and hantavirus RNA was most striking in M. rufocanus (2/19, or 10.5%) and M. rutilus (2/17 or 11.8%), compared to M. glareolus (3/8, or 37.5%) (Tables 1 and 4). However, this discrepancy may have been artificially accentuated by including 12 voles (6 M. rutilus, 4 M. rufocanus and 2 M. glareolus) with anti-hantaviral antibody titers of ≤1:20. That is, hantavirus RNA was more commonly detected in voles with antibody titers ≥1:80 (6/17, or 35.3%) or ≥160 (5/8, or 62.5%), which is consistent with experimental PUUV infection studies, which showed that 48% of voles with high levels of antibodies correlated with PUUV antigen in lung tissues (Apekina et al., 2014).
A possible reason for the low success rate of detecting hantavirus RNA in seropositive voles include the insufficiently sensitive RT-PCR assay; that is, although the L-segment oligonucleotide primers were broadly cross reactive and have been successfully employed for gene amplification of KBRV in Microtus maximowiczii, Vladivostok virus (VLAV) in Microtus fortis and Tula virus (TULV) in Microtus arvalis, the most reasonable explanation for the low RT-PCR positivity in lung tissues among antibody-positive M. rufocanus and M. rutilus was the lower viral load, which was beyond the limits of detection. Few studies have compared two of these hantaviral markers in M. rufocanus. For example, only 5 of 8 seropositive wild Microtus and Myodes voles were RT-PCR positive (Plyusnina et al., 2008).
Another possible reason for the discrepancy is that HOKV infection may be less robust and persistent in M. rutilus and M. rufocanus, resulting in lower viral copy numbers, compared to PUUV infection in M. glareolus. Thus, while 75% (3/4) of seropositive M. glareolus from Kuchuk was PUUV RNA positive, only 12.5% (1/8) of seropositive M. rufocanus and 16.7% (1/6) M. rutilus were HOKV RNA positive in Pokrovka and Teletskoye Lake, respectively.
By contrast, passively acquired maternal antibodies cannot explain the anti-hantaviral antibody prevalence, because all of the captured voles were subadults and adults. Equally unlikely is the occurrence of spillover and the failure to capture another arvicolid rodent species that represents the reservoir host of HOKV. Only two or three species of Myodes voles, which are potential reservoirs of PUUV and PUU-like virus, inhabit the studied territory and all of these species were trapped and analyzed. No other Myodes species are described in Siberia.
We were unable to detect PUUV in one seropositive M. glareolus captured in Teletskoye Lake, where HOKV was found in seropositive M. rutilus and M. rufocanus. Similarly, in the trapping site of Kuchuk in Tiumen Oblast, where PUUV was detected in the dominant M. glareolus, we failed to find HOKV in one seropositive M. rutilus. These results are consistent with that of previous studies, indicating hantavirus antigen in M. glareolus and M. rutilus within the same natural focus, where M. glareolus was the dominant species (Myasnikov et al., 1992).
Geographic-specific clustering within distinct genetic lineages has been recognized for arvicolid rodent-borne hantaviruses, such as PUUV (Plyusnin et al., 1995; Sironen et al., 2001), MUJV (Lee et al., 2014; Song et al., 2007) and TULV (Song et al., 2004; Tkachenko et al., 2015). Available data on PUUV support the hypothesis that the phylogeography of different lineages was formed by separate post-glacial migrations of M. glareolus (Asikainen et al., 2000). We suggest a similar basis for the distribution pattern of distinct lineages of HOKV. Previously, it was shown that the vast territory of European and Asian Russia, including the southern regions of Siberia, was colonized by the same haplogroup of M. rufocanus (Abramson et al., 2012). In accordance with these data, we found genetically closely related HOKV strains of Siberia 1 lineage in the geographically distant Altai region and Baikal Lake area (Teletskoye854/Mrt, Teletskoye937/Mrf, Solton35/Mrt and Olhon109). Geographically close but genetically distant Siberia 2 and Siberia 1 lineages were found in the mountainous area of the Altai region, suggesting that the two lineages are associated with different colonization events of M. rufocanus, which had long periods of isolated evolution.
Discovery of multiple newfound hantaviruses raises critical questions about their taxonomic relationships (Lee et al., 2014; Maes et al., 2009) and evolutionary origins (Yanagihara et al., 2014). Our data and results from other studies on the genetic diversity of PUUV, MUJV and HOKV (Lee et al., 2014; Plyusnina et al., 2008; Sironen and Plyusnin, 2011; Zhang et al., 2007) indicate that the current criterion of at least a 7% amino acid sequence difference in both the complete N protein and GnGc, promulgated by the International Committee on the Taxonomy of Viruses (ICTV) (Plyusnin et al., 2012), is not fulfilled to warrant separate hantavirus species designation for PUUV, HOKV and MUJV. In this study, differences between the N protein sequences (based on nearly the complete N protein, 402 of 433 amino acids) of Siberian HOKV and PUUV strains were 4.8–6.0%. Recently, based on a thorough genetic analysis of DOBV strains, a consortium of European hantavirus experts proposed four genotypes: namely Dobrava, Saaremaa, Kurkino and Sochi (Klempa et al., 2013). Similarly, analysis of publicly available sequences of hantaviruses harbored by Myodes voles supports the concept of three PUUV genotypes: Puumala, Hokkaido and Muju.
Also, the ICTV criterion requiring a unique ecological niche of the reservoir host is invalid for HOKV. M. rufocanus and M. rutilus, which belong to the same family (Cricetidae), subfamily (Arvicolinae) and genus (Myodes) and which inhabit the same localities in coniferous and broad-leaved forests of Siberian and Far East Russia, harbor HOKV. Thus, this criterion might need to be revised to simply requiring that reservoir hosts belong to the same genus.
Although approximately 7% of the human population in the Omsk region and 9% in the Tiumen region have anti-hantaviral antibodies (Myasnikov et al., 1987), only low numbers of HFRS cases have been registered in Western Siberia. Serological studies in the Altai Republic and Altai Krai have also revealed anti-hantaviral antibody prevalence of approximately 1–2% (Malkin et al., 1996). It is unclear if the rarity of HFRS in Siberia is due to under-reporting or to the lower pathogenicity of HOKV. Future in-depth studies are warranted to clarify if HOKV and MUJV, like PUUV, cause HFRS.
Highlights.
Three species of Myodes voles occupy the same ecological niche in Siberia
Myodes glareolus harbors Puumala virus in Siberia
Myodes rufocanus and Myodes rutilus serve as the reservoir hosts of the Hokkaido genotype of Puumala virus in Siberia
Acknowledgments
This work was supported in part by grants from the Biotechnology Engagement Program, through the International Science and Technology Center (#0805.2), and the National Institutes of Health (R01AI075057, P20GM103516). We thank our colleagues, Dr. Alexander Pozdnyakov, Dr. Dmitry Petrovski, Dr. Anton Krivopalov and Dr. Pavel Zadubrovskiy for help during the field expeditions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abramson NI, Petrova TV, Dokuchaev NE, Obolenskaya EV, Lissovsky AA. Phylogeography of the gray red-backed vole Craseomys rufocanus (Rodentia: Cricetidae) across the distribution range inferred from nonrecombining molecular markers. Russian J Theriol. 2012;11:137–156. [Google Scholar]
- Apekina NS, Bernshtein AD, Demina VT, Gavrilovskaya IN. Variants of the immunoreactivity and infectious process in bank vole (Myodes glareolus) experimentally infected with the hantavirus Puumala (PUUV) Vopr Virusol. 2014;59:42–46. in Russian. [PubMed] [Google Scholar]
- Asikainen K, Hanninen T, Henttonen H, Niemimaa J, Laakkonen J, Andersen HK, Bille N, Leirs H, Vaheri A, Plyusnin A. Molecular evolution of Puumala hantavirus in Fennoscandia: phylogenetic analysis of strains from two recolonization routes, Karelia and Denmark. J Gen Virol. 2000;81:2833–2841. doi: 10.1099/0022-1317-81-12-2833. [DOI] [PubMed] [Google Scholar]
- Dekonenko A, Yakimenko V, Ivanov A, Morozov V, Nikitin P, Khasanova S, Dzagurova T, Tkachenko E, Schmaljohn C. Genetic similarity of Puumala viruses found in Finland and western Siberia and of the mitochondrial DNA of their rodent hosts suggests a common evolutionary origin. Infect Genet Evol. 2003;3:245–257. doi: 10.1016/s1567-1348(03)00088-1. [DOI] [PubMed] [Google Scholar]
- Dzagurova T, Tkachenko E, Slonova R, Ivanov L, Ivanidze E, Markeshin S, Dekonenko A, Niklasson B, Lundkvist A. Antigenic relationships of hantavirus strains analysed by monoclonal antibodies. Arch Virol. 1995;140:1763–1773. doi: 10.1007/BF01384340. [DOI] [PubMed] [Google Scholar]
- Gromov IM, Erbaeva MA. Mammals of Russia and Adjacent Territories: Lagomorphs and Rodents. St. Petersburg: Zoological Institute of the Russian Academy of Sciences; 1995. in Russian. [Google Scholar]
- Gromov IM, Polyakov IY. Voles (Microtinae) 8. Vol. 3. Smithsonian Institution Libraries and the National Science Foundation; Washington, DC: 1977. Fauna of the USSR: Mammals. [Google Scholar]
- Jonsson CB, Figueiredo LTM, Vapalahti O. A global perspective on hantavirus ecology, epidemiology, and disease. Clin Microbiol Rev. 2010;23:412–441. doi: 10.1128/CMR.00062-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko Y. Identification and some morphological characters of Clethrionomys rufocanus and Eothenomys regulus from USSR, northeast China, and Korea in comparison with C. rufocanus from Finland. J. Mammal. Soc. Japan. 1990;14:129–148. [Google Scholar]
- Kariwa H, Yoshimatsu K, Sawabe J, Yokota E, Arikawa J, Takashima I, Fukushima H, Lundkvist A, Shubin FN, Isachkova LM, Slonova RA, Leonova GN, Hashimoto N. Genetic diversities of hantaviruses among rodents in Hokkaido, Japan and Far East Russia. Virus Res. 1999;59:219–228. doi: 10.1016/s0168-1702(98)00141-5. [DOI] [PubMed] [Google Scholar]
- Kariwa H, Yoshizumi S, Arikawa J, Yoshimatsu K, Takahashi K, Takashima I, Hashimoto N. Evidence for the existence of Puumula-related virus among Clethrionomys rufocanus in Hokkaido, Japan. Am J Trop Med Hyg. 1995;53:222–227. doi: 10.4269/ajtmh.1995.53.222. [DOI] [PubMed] [Google Scholar]
- Klempa B, Avsic-Zupanc T, Clement J, Dzagurova TK, Henttonen H, Heyman P, Jakab F, Krüger DH, Maes P, Papa A, Tkachenko EA, Ulrich RG, Vapalahti O, Vaheri A. Complex evolution and epidemiology of Dobrava-Belgrade hantavirus: definition of genotypes and their characteristics. Arch Virol. 2013;158:521–529. doi: 10.1007/s00705-012-1514-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingström J, Heyman P, Escutenaire S, Sjolander KB, Jaegere FD, Henttonen H, Lundkvist A. Rodent host specificity of European hantaviruses: evidence of Puumala virus interspecific spillover. J Med Virol. 2002;68:581–588. doi: 10.1002/jmv.10232. [DOI] [PubMed] [Google Scholar]
- Kosoy M, Slonova R, Mills J, Mandel E, Childs J. Community structure and prevalence of hantavirus infection in rodents: a geographic division of the enzootic area in far eastern Russia. J Vect Ecol. 1997;22:52–63. [PubMed] [Google Scholar]
- Lee JG, Gu SH, Baek LJ, Shin OS, Park KS, Kim HC, Klein TA, Yanagihara R, Song JW. Muju virus, harbored by Myodes regulus in Korea, might represent a genetic variant of Puumala virus, the prototype arvicolid rodent-born hantavirus. Viruses. 2014;6:1701–1714. doi: 10.3390/v6041701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P-W, Yanagihara R, Franko MC, Amyx HL, Gibbs CJ, Jr, Gajdusek DC, Traub R. Preliminary evidence that Hantaan or a closely related virus is enzootic in domestic rodents. N Engl J Med. 1982;307:624–625. doi: 10.1056/NEJM198209023071013. [DOI] [PubMed] [Google Scholar]
- Maes P, Klempa B, Clement J, Matthijnssens J, Gajdusek DC, Krüger DH, van Ranst M. A proposal for new criteria for the classification of hantaviruses, based on S and M segment protein sequences. Infect Genet Evol. 2009;9:813–820. doi: 10.1016/j.meegid.2009.04.012. [DOI] [PubMed] [Google Scholar]
- Malkin AE, Myasnikov YA, Ryltseva EV, Tkachenko EA. Landscape distribution of the natural HFRS foci in Russia. Med Parazitol. 1996;2:27–32. in Russian. [PubMed] [Google Scholar]
- Mills JN, Childs JE, Ksiazek TG, Peters CJ. Methods for trapping and sampling small mammals for virologic testing. Atlanta: US Department of Health and Human Services, Center for Disease Control and Prevention; 1995. [Google Scholar]
- Myasnikov YA, Tkachenko EA, Rezapkin GV, Petrov VA, Gasanova TA, Bagan RN, Spiridonova AG, Loginov AI, Vereschagin NN, Zebreeva GA, Malyshev VA, Azeeva KY, Kalashnikova ON, Khalitova SD, Zelenkina FN, Stepanenko AG, Nurgaleeva RG, Fadeev ES, Safonova NM, Grebenschikov VE, Zuevsky AP. Specification of the eastern border of the European area endemic for hemorrhagic fever with renal syndrome. Vopr Virusol. 1987;32:610–615. in Russian. [PubMed] [Google Scholar]
- Myasnikov YA, Apekina NS, Zuevskii AP, Khitrin AV, Bernshtein AD. The distribution of natural foci of hemorrhagic fever with renal syndrome in different landscape zones of Tyumen region. Vopr Virusol. 1992;37:161–164. in Russian. [PubMed] [Google Scholar]
- Olsson GE, Leirs H, Henttonen H. Hantaviruses and their hosts in Europe: reservoirs here and there, but not everywhere. Vector Borne Zoonotic Dis. 2010;10:549–561. doi: 10.1089/vbz.2009.0138. [DOI] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Plyusnin A, Beatty BJ, Elliott RM, Goldbach R, Kormelink R, Lundkvist A, Schmaljohn CS, Tesh RB. Bunyaviridae. In: King AMQ, Lefkowitz EJ, Adams MJ, Carstens EB, editors. Virus Taxonomy: Classification and Nomenclature of Viruses. Elsevier Academic Press; San Diego, CA, USA: 2012. pp. 725–741. [Google Scholar]
- Plyusnin A, Vapalahti O, Lehvaslaiho H, Apekina N, Mikhailova T, Gavrilovskaya I, Laakkonen J, Niemimaa J, Henttonen H, Brummer-Korvenkontio M, Vaheri A. Genetic variation of wild Puumala viruses within the serotype, local rodent populations and individual animal. Virus Res. 1995;38:25–41. doi: 10.1016/0168-1702(95)00038-r. [DOI] [PubMed] [Google Scholar]
- Plyusnin A, Vapalahti O, Vaheri A. Hantaviruses: Genome structure, expression and evolution. J Gen Virol. 1996;77:2677–2687. doi: 10.1099/0022-1317-77-11-2677. [DOI] [PubMed] [Google Scholar]
- Plyusnina A, Laakkonen J, Niemimaa J, Nemirov K, Muruyeva G, Pohodiev B, Lundkvist A, Vaheri A, Henttonen H, Vapalahti O, Plyusnin A. Genetic analysis of hantaviruses carried by Myodes and Microtus rodents in Buryatia. Virol J. 2008;5:4. doi: 10.1186/1743-422X-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Sanada T, Seto T, Ozaki Y, Saasa N, Yoshimatsu K, Arikawa J, Yoshii K, Kariwa H. Isolation of Hokkaido virus, genus Hantavirus, using a newly established cell line derived from the kidney of the grey red-backed vole (Myodes rufocanus bedfordiae) J Gen Virol. 2012;93:2237–2246. doi: 10.1099/vir.0.045377-0. [DOI] [PubMed] [Google Scholar]
- Shenbrot I, Krasnov B. An Atlas of the Geographic Distribution of the Arvicoline Rodents of the World (Rodentia, Muridae: Arvicolinae) Sofia: Pensoft Publication; 2005. [Google Scholar]
- Sironen T, Plyusnin A. Genetics and evolution of hantaviruses. In: Plyusnin A, Elliott RM, editors. Bunyaviridae: Molecular and Cellular Biology. Caister Academic Press; Norfolk, U.K: 2011. pp. 61–94. [Google Scholar]
- Sironen T, Vaheri A, Plyusnin A. Molecular evolution of Puumala hantavirus. J Virol. 2001;75:11803–11810. doi: 10.1128/JVI.75.23.11803-11810.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MF, Patton JL. The diversification of South American murid rodents: evidence from mitochondrial DNA sequence data for the akodontine tribe. Biol J Linnean Soc. 1993;50:149–177. [Google Scholar]
- Song JW, Baek LJ, Song KJ, Skrok A, Markowski J, Bratosiewicz-Wasik J, Kordek R, Liberski PP, Yanagihara R. Characterization of Tula virus from common voles (Microtus arvalis) in Poland: evidence for geographic-specific phylogenetic clustering. Virus Genes. 2004;29:239–247. doi: 10.1023/B:VIRU.0000036384.50102.cf. [DOI] [PubMed] [Google Scholar]
- Song KJ, Baek LJ, Moon S, Ha SJ, Kim SH, Park KS, Klein TA, Sames W, Kim HC, Lee JS, Yanagihara R, Song J-W. Muju virus, a novel hantavirus harboured by the arvicolid rodent Myodes regulus in Korea. J Gen Virol. 2007;88:3121–3129. doi: 10.1099/vir.0.83139-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web-Servers. Syst Biol. 2008;75:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods) Sinauer Associates; Sunderland, Massachusetts: 2003. Version 4. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucl Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkachenko EA, Ivanov AP, Donets MA, Miasnikov YA, Ryltseva EV, Gaponova LK, Bashkirtsev VN, Okulova NM, Drozdov SG, Slonova RA, Somov GP, Astakhova TI. Potential reservoir and vectors of haemorrhagic fever with renal syndrome (HFRS) in the U.S.S.R. Ann Soc Belg Med Trop. 1983;63:267–269. [PubMed] [Google Scholar]
- Tkachenko EA, Ryltseva EV, Myasnikov YA, Ivanov AP, Rezapkin GV, Tatiyanchenko LA, Pashkov AY. Study of circulation of hemorrhagic fever with renal syndrome virus among small mammals in the USSR. Vopr Virusol. 1987;32:709–715. in Russian. [PubMed] [Google Scholar]
- Tkachenko EA, Witkowski PT, Radosa L, Dzagurova TK, Okulova NM, Yunicheva YV, Vasilenko L, Morozov VG, Malkin GA, Krüger DH, Klempa B. Adler hantavirus, a new genetic variant of Tula virus identified in Major’s pine voles (Microtus majori) sampled in southern European Russia. Infect Genet Evol. 2015;29:156–163. doi: 10.1016/j.meegid.2014.11.018. [DOI] [PubMed] [Google Scholar]
- Yanagihara R, Gu SH, Arai S, Kang HJ, Song J-W. Hantaviruses: rediscovery and new beginning. Virus Res. 2014;187:6–14. doi: 10.1016/j.virusres.2013.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashina LN, Patrushev NA, Ivanov LI, Slonova RA, Mishin V, Kompanez GG, Zdanovskaya NI, Kuzina II, Safronov PF, Chizhikov VE, Schmaljohn C, Netesov SV. Genetic diversity of hantaviruses associated with hemorrhagic fever with renal syndrome in the Far East of Russia. Virus Res. 2000;70:31–44. doi: 10.1016/s0168-1702(00)00203-3. [DOI] [PubMed] [Google Scholar]
- Yashina LN, Slonova RA, Oleinik OV, Kuzina II, Kushnareva TV, Kompanets GG, Simonov SV, Simonova TL, Netesov SV, Morzunov SP. A new genetic variant of the PUUV virus from the Maritime Territory and its natural carrier red-grey vole Clethrionomys rufocanus. Vopr Virusol. 2004;49:34–37. in Russian. [PubMed] [Google Scholar]
- Yashina LN, Abramov SA, Gutorov VV, Dupal TA, Krivopalov AV, Panov VV, Danchinova GA, Vinogradov VV, Luchnikova EM, Hay J, Kang HJ, Yanagihara R. Seewis virus: phylogeography of a shrew-borne hantavirus in Siberia, Russia. Vector Borne Zoonotic Dis. 2010;10:585–591. doi: 10.1089/vbz.2009.0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashina LN, Danchinova GA, Seregin SV, Khasnatinov MA, Yanagihara R. Genetic analysis of Hokkaido hantavirus among Myodes rufocanus in the Baikal Lake area. Bull East Sib Sci Cent. 2013;4:147–152. in Russian. [Google Scholar]
- Zhang YZ, Zou Y, Yan YZ, Hu GW, Yao LS, Du ZS, Jin LZ, Liu YY, Li MH, Chen HX, Fu ZF. Detection of phylogenetically distinct Puumala-like viruses from red-grey vole Clethrionomys rufocanus in China. J Med Virol. 2007;79:1208–1218. doi: 10.1002/jmv.20871. [DOI] [PubMed] [Google Scholar]