Abstract
Human immunodeficiency virus (HIV) reverse transcriptase (RT) associated ribonuclease H (RNase H) remains an unvalidated antiviral target. A major challenge of specifically targeting HIV RNase H arises from the general lack of selectivity over RT polymerase (pol) and integrase (IN) strand transfer (ST) inhibitions. We report herein the synthesis and biochemical evaluations of three novel 3-hydroxypyrimidine-2,4-dione (HPD) subtypes carefully designed to achieve selective RNase H inhibition. Biochemical studies showed the two subtypes with an N-1 methyl group (9 and 10) inhibited RNase H in low micromolar range without siginificantly inhibiting RT polymerase, whereas the N-1 unsubstituted subtype 11 inhibited RNase H in submicromolar range and RT polymerase in low micromolar range. Subtype 11 also exhibited substantially reduced inhibition in the HIV-1 INST assay, and no significant cytotoxicity in the cell viability assay, suggesting that it may be amenable to further structure-activity-relationship (SAR) for identifying RNase H inhibitors with antiviral activity.
Graphical abstract
Introduction
HIV encodes three enzymes crucial for viral replicaton: RT, IN and protease (PR).1 Antivirals targeting these enzymes have successfully transformed HIV from an inevitably fatal disease into a clinically manageable chronic infection.2 One particularly successful aspect of HIV chemotherapy is the approval of multiple classes of antivirals,3 which allows effective combination therapy termed as Highly Active Antiretroviral Therapy (HAART). However, due to their long duration HAART regimens can be plagued by the emergence of resistant HIV mutants. Mechanistically novel antivirals against underexplored and unvalidated viral targets will add strategically to HAART repertoire enabling continued efficacy, especially against drug-resistant viruses that may emerge with current HAART regimens. One such novel target is RT associated RNase H activity.4–5 RT is a well established drug target with many FDA-approved nucleoside RT inhibitors (NRTIs)6 and non-nucleoside RT inhibitors (NNRTIs)7 constituting the cornerstone of HAART. However, these drugs all target the polymerase domain which carries out both RNA-dependent and DNA-dependent viral DNA polymerization. Significantly, RT also encodes an RNase H domain which selectively degrades the RNA strand from the RNA/DNA heteroduplex intermediate during reverse transcription. In stark contrast to the success of polymerase targeting antivirals, no inhibitors of RT-associated RNase H have entered clinical development. However, the critical role of RNase H in HIV replication has long been recognized and confirmed through recent experiments showing that active site mutations associated with attenuated RNase H biochemical activity conferred reduced HIV replication in cell culture.8 A similar antiviral phenotype can be expected if RNase H is selectively and potently inhibited by small molecules.
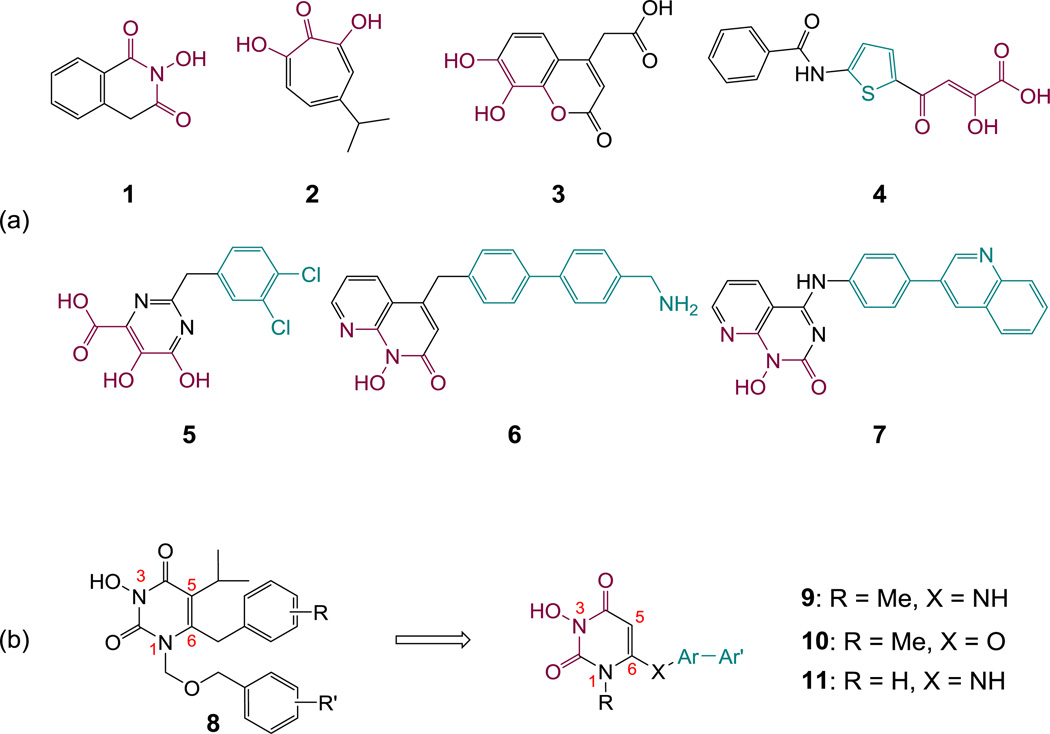
RNase H belongs to the retroviral integrase super family (RISF)9 with an active site fold and catalytic mechanism highly homologous to integrase. Accordingly, efforts in RNase H inhibition have mostly focused on targeting the active site with a pharmacophore core similar to INSTIs. The pharmacophore critically features a chelating triad (magenta) designed to bind two divalent metals (Figure 1a). Reported RNase H inhibitor types10 include 2-hydroxyisoquinolinedione (HID, 1),11 β-thujaplicinol (2),12 dihydroxycoumarin (3),13 diketoacid (DKA) 4,14 pyrimidinol carboxylic acid 5,15 hydroxynaphthyridine 616 and pyridopyrimidone 7.17 Importantly, structurally more elaborate inhibitor types 4–7 also feature a hydrophobic aromatic moiety (cyan) conferring more potent and selective RNase H inhibition. Unfortunatley, the biochemical inhibition observed with these inhibitors typically does not translate into antiviral activity in cell culture, possibly reflecting a steep biochemical barrier of competing against much larger DNA/RNA substrates.17 Recent studies by Corona et al18 on a series of DKAs demonstrated that specific interactions with highly conserved amino acid residues in the RNase H domain contributed critically to selective RNase H inhibition and effective targeting of HIV reverse transcription in cells, hence offering important insights for the rational design of RNase H inhibitors. However, the most potent antiviral activity was observed with inhibitors 6–7 presumably via tight binding to RNase H.17 Intriguingly, these inhibitors encompass a highly privileged biaryl moiety connected to the chelating core via a one atom linker, a distinct pharmacophore feature that could be key in providing tight RNase H binding. This pharmacophore hypothesis was corroborated by our redesigned HID subtype with a biaryl moiety that conferred potent RNase H inhibition and significant antiviral activity.19
Figure 1.
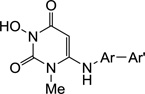
Design of active site RNase H inhibitors. (a) Major chemotypes reported as HIV RNase H active site inhibitors. All chemotypes contain a chelating triad (magenta); scaffolds 4–7 also feature an aryl or biaryl moiety (cyan) connected through a methylene or amino linker; (b) newly designed active site RNase H inhibitor chemotypes 9–11 featuring a chelating triad and a biaryl group to satisfy the pharmacophore requirements for selective RNase H inhibition.
We have previously developed an HPD chemotype (8, Figure 1, b) demonstrating exceptional antiviral activity against HIV-1 likely by dually inhibiting RT polymerase (pol) and INST.20–22 Based on the aforementioned pharmacophore model for RNase H inhibition, we have redesigned the HPD chemotype with the goal of achieving selective RNase H inhibition (Figure 1, b). Key to the redesign is the introduction of a biaryl group at C6 position through different linkers (subtypes 9–11). In addition, the new design also involves two structural simplifications: removal of the C5 isopropyl group crucial for the allosteric binding to RT pol; and substitution of the N-1 position with either a small methyl group (9 and 10) or H (11). These simplifications aim at minimizing inhibitor binding to the RT pol. We report herein the chemical synthesis and biochemical evaluations of these three subtypes.
Results and Discussion
Chemistry
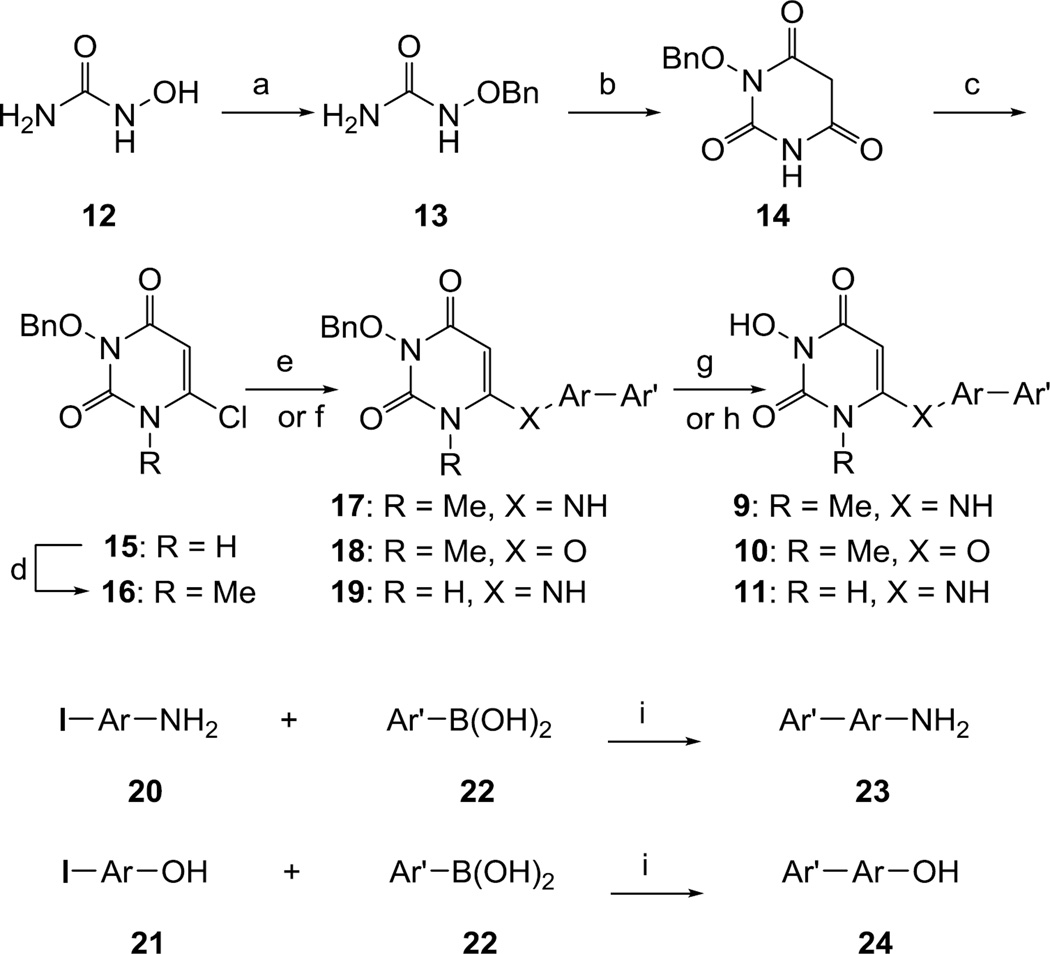
Previously reported HPD chemotypes all featured a methylene linker at C6 position and were synthesized based largely on the well documented HEPT NNRTIs.23 The key 3-OH group was typically introduced in the very last step via a base-mediated N-hydroxylation.20–22 SAR relying on this synthetic route suffers from two major drawbacks: the very early introduction of structural diversity at the C6 aromatic domain and sometimes the low efficiency of the N-hydroxylation reaction. The new design substitutes a heteroatom linker (NH or O) for the methylene group, which implicates a much more divergent synthesis where the C6 aromatic domain can be easily diversified via reacting a library of biaryl amine or alcohol nucleophiles with 6-chloropyrimidine-2,4-dione intermediates (15 and 16, Scheme 1). In addition, this new synthetic route also exploits the symmetrical nature of malonate for the ring construction (13 to 14) to allow the early introduction of a protected N-OH without the complication of regioselectivity issues. This eliminates the need for the late N-hydroxylation reaction. Overall, the concise synthesis began with the readily available hydroxyurea (12), which was selectively o-protected. The resulting benzyl protected hydroxyurea (13) was then cyclized with malonate to afford 14, which was chlorinated at C6 position to produce key intermediate 15. For final compounds with a N-1 methyl group, the methylation was conducted at this stage with methyl iodide to yield another 6-chloro intermediate 16. The synthesis of 6-chloropyrimidine-2,4-dione intermediates then allowed us to conduct two different versions of amination / etherification reactions. It turned out that for the N-1 unsubstituted chloride 15, direct amination was effected thermally via a microwave reaction with biaryl amines (23) under the influence of N,N-dimethylaniline. However, the same reaction with biaryl alcohols (24) failed to deliver desired products. On the other hand, amination of the N-1 methylated chloride 16 required a completely different set of conditions. In this event, the reaction was effected with LDA / HMPA at low temperature. Significantly, under the same conditions the biaryl alcohols also reacted to afford desired intermediates 18. The final step of the synthesis aimed at a clean benzyl deprotection and was achieved via catalytic hydrogenation (g) or TFA treatment (h). The requisite libraries of biaryl amines (23) and alcohols (24) were curated through commercial sources or quick chemical synthesis via a standard Suzuki coupling reaction.
Scheme 1a.
Synthesis of HPD analogues 9–11.
a Reagents and conditions: a) KOH, BnCl, MeOH, reflux, 6 h, 91%; b) CH2(COOEt)2, NaOEt, MW, 150 °C, 20 min, 58%; c) POCl3, BnEt3NCl, 50 °C, 6 h, 88%; d) Cs2CO3, MeI, DMF, seal tube 80 °C, 2h, 68 %; e) (for 17–18) Ar′-ArNH2/ Ar′-ArOH, LDA, HMPA, THF, −78 °C to rt, overnight; f) (for 19) Ar′-ArNH2, N,N-dimethylaniline, MW, 170 °C, 30–40 min; g) Pd/C, H2, 50 Psi, MeOH, 3–4h; h) TFA, MW, 120 °C, 30 min; i) Pd(PPh3)4 1.5 % (mol), DMF, 2M Na2CO3, MW, 160 °C, 30 min.
Biology
All final compounds of subtypes 9–11 were evaluated biochemically for inhibition against RT-associated RNase H and pol, as well as INST. To assess the multiple distinct modes of action of RNA cleavage by RNase H at multiple stages of reverse transcription, our primary biochemical assays employed three different oligonucleotide substrates19: HTS-1, which measures the random internal cleavages likely representing the majority of RNase H events; HTS-2, that gauges the DNA 3’ end directed (polymerase dependent) cleavages; and HTS-3, which evaluates the RNA 5’ end directed (polymerase independent) cleavages for degrading the recessed RNA template. These assays were performed with recombinant p66/p51 RT heterodimer and the observed inhibitory activity could be influenced by the spatial and temporal interactions between RNase H and pol domains. To confirm RNase H inhibition, a secondary assay was conducted with substrate HTS-1 using a catalytically active isolated RNase H domain fragment. The compounds were also evaluated for inhibition of RT pol activity. An INST assay was also conducted to determine the inhibitory selectivity of our new compounds for RNase H over INST. Finally, although antiviral activity against HIV-1 was not achieved with these analogues, cytoxicity was evaluated and the CC50 values could add significantly to the assessment of the suitability of compounds for further medicinal chemistry efforts.
Detailed RNase H assay results are summarized in Tables 1–3. Overall, newly synthesized HPD analogues of subtypes 9–11 all potently inhibited RT-associated RNase H activity with IC50 values ranging from nanomolar to single digit micromolar concentrations. This observation provides a strong validation to our inhibitor design. As for as SAR is concerned, the inhibition profile of the N-1 unsubstituted subtype (11) is markedly different from those of the N-1 methyl subtypes (9–10): 1) in general 11 appears to be significantly more potent, with all analogues inhibiting the RNase H in namomolar range (IC50 = 0.15–0.51 µM, HTS-1), even slightly more potent that compound 7 used as a control (table 3), whereas IC50s for subtypes 9–10 are mostly in the low micromolar range; 2) while a substantial bias towards the inhibition of the internal cleavage (HTS-1 as substrate) was observed for most analogues of subtypes 9–10 (Tables 1–2), subtype 11 tends to inhibit all three modes of RNase H cleavages nearly equally (Table 3). In addition, comparison between the two N-1 methyl substituted subtypes 9 and 10 revealed that the amino linkage at C6 (subtype 9, Table 1) conferred slightly better potency and considerably less substrate bias than the ether linkage (subtype 10, Table 2). Notably a number of subtype 10 analogues (Table 2, 10b–10f, 10i, 10l, 10o–10r, 10w–10y) did not inhibit RNase H with HTS-2 and / or HTS-3 as substrates. As for the biaryl hydrophobic moiety, although many analogues within each subtypes were synthesized, the impact of the substitution on the terminal phenyl ring (Me, CF3, OMe, OH, CN, F, Cl, etc.) turned out to be marginal. Interestingly, replacing the terminal aryl group with a saturated ring (compound 9z) completely abrogated biochemical inhibition, suggesting that the biaryl moiety may be required for activity. Nevertheless, the RNase H inhibition was further evaluated biochemically against a catalytically active RNase H domain with HTS-1 as substrate. The potency observed from this assay generally agreed with that of full-length RT with HTS-1 substrate, confirming the observed RNase H inhibition.
Table 1.
Biochemical inhibitory activity of compounds 9a–z against HIV RT RNase H and polymerase.
 | |||||||
|---|---|---|---|---|---|---|---|
| Cpd | Ar-Ar′ | Full-Length RT RNase H IC50a (µM) |
RNase H Fragmente IC50 (µM) |
RT pol IC50 (µM) |
CC50 (µM)f |
||
| HTS-1b | HTS-2c | HTS-3d | |||||
| 9a | 0.80 | 3.5 | 5.1 | 1.7 | N/Ig | 6.0 | |
| 9b | 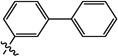 |
0.85 | 3.5 | 3.4 | NDh | N/I | ND |
| 9c | 2.7 | 6.5 | 6.6 | 1.9 | N/I | 5.0 | |
| 9d | 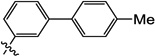 |
1.7 | 7.2 | 6.2 | ND | 17 | ND |
| 9e | 3.6 | 8.9 | 6.6 | 2.2 | N/I | 10 | |
| 9f | 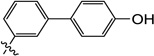 |
0.75 | 3.8 | 2.7 | ND | 12 | ND |
| 9g | 1.3 | 6.6 | 5.0 | 0.90 | N/I | 5.0 | |
| 9h | 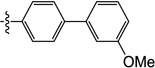 |
0.80 | 3.5 | 5.1 | 1.7 | N/I | 6.0 |
| 9i | 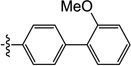 |
2.8 | 9.3 | 8.9 | ND | N/I | ND |
| 9j | 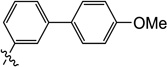 |
0.5 | 2.6 | 3.1 | 1.1 | 3.6 | 3.0 |
| 9k | 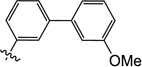 |
0.60 | 3.5 | 4.3 | 1.7 | N/I | 10 |
| 9l | 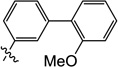 |
1.2 | 2.1 | 3.8 | 1.0 | 6.6 | 6.0 |
| 9m | 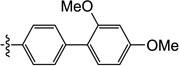 |
1.5 | 4.3 | 5.3 | ND | N/I | ND |
| 9n | 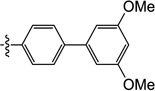 |
1.6 | 7.2 | 9.2 | 1.8 | 6.5 | 15 |
| 9o | 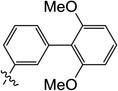 |
1.9 | 6.9 | 4.5 | 1.2 | N/I | 8.0 |
| 9p | 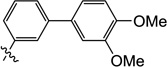 |
0.50 | 2.2 | 3.0 | 0.90 | 5.8 | 10 |
| 9q | 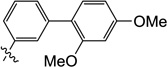 |
1.0 | 2.1 | 4.2 | 1.1 | 6.3 | 6.0 |
| 9r | 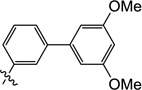 |
0.60 | 2.8 | 4.0 | 1.4 | N/I | 10 |
| 9s | 1.1 | 2.6 | 3.7 | 1.0 | 3.3 | 12 | |
| 9t | 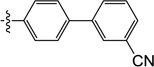 |
1.2 | 3.7 | 3.3 | 0.9 | N/I | 25 |
| 9u | 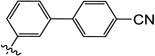 |
0.70 | 2.3 | 3.4 | 1.0 | 5.0 | 10 |
| 9v | 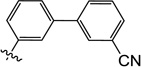 |
1.1 | 3.2 | 4.1 | 1.0 | 5.0 | >25 |
| 9w | 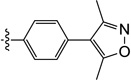 |
1.6 | 4.4 | 3.8 | ND | N/I | ND |
| 9x | 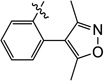 |
6.6 | N/I | N/I | ND | N/I | ND |
| 9y | 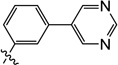 |
1.4 | 4.1 | 3.9 | 0.90 | N/I | >25 |
| 9z | 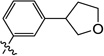 |
N/I | N/I | N/I | N/I | N/I | >25 |
| 7 | -- | 0.70 | -- | -- | -- | 0.85 | 2.0 |
IC50: concentration of a compound producing 50% inhibition, expressed as mean of at least three independent experiments (standard deviation was less than 10% and is not indicated for ease of viewing).
Substrate that measures internal cleavage.
Substrate that measures DNA 3’ end directed cleavage.
Substrate that measures RNA 5’ end directed cleavage.
Reconstituted HIV RNase H domain.
CC50: concentration of a compound causing 50% cytotoxicity.
No inhibition at concentrations up to 25 µM.
Not determined.
Table 3.
Biochemical inhibitory activity of compounds 11a–t against HIV RT RNase H and polymerase.
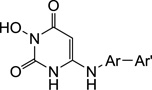 | |||||||
|---|---|---|---|---|---|---|---|
| Cpd (JT−) |
Ar-Ar′ | Full-Length RT RNase H IC50a (µM) |
RNase H Fragmente IC50 (µM) |
RT pol IC50 (µM) |
CC50 (µM)f |
||
| HTS-1b | HTS-2c | HTS-3d | |||||
| 11a | 0.25 | 0.30 | 0.30 | 0.15 | 2.0 | >25 | |
| 11b | 0.40 | 0.70 | 0.50 | 0.40 | 5.7 | >25 | |
| 11c | 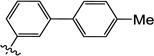 |
0.38 | 0.45 | 0.44 | NDg | 3.1 | ND |
| 11f | 0.18 | 0.45 | 0.37 | ND | 0.82 | ND | |
| 11g | 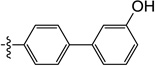 |
0.22 | 0.43 | 0.40 | ND | 0.79 | ND |
| 11h | 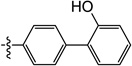 |
0.24 | 0.49 | 0.46 | ND | 0.83 | ND |
| 11i | 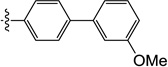 |
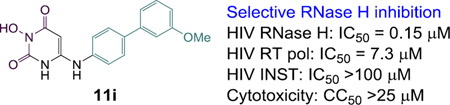
0.15 | 0.25 | 0.25 | 0.20 | 7.3 | >25 |
| 11j | 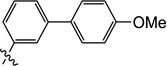 |
0.20 | 0.30 | 0.40 | 0.10 | 6.3 | >25 |
| 11k | 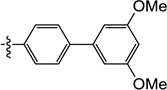 |
0.19 | 0.32 | 0.25 | ND | 0.34 | ND |
| 11l | 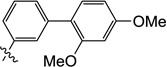 |
0.31 | 0.54 | 2.4 | ND | 3.2 | ND |
| 11m | 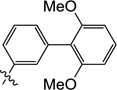 |
0.51 | 0.70 | 0.47 | ND | 1.7 | ND |
| 11o | 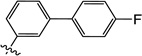 |
0.20 | 0.30 | 0.30 | 0.15 | 6.6 | >25 |
| 11p | 0.47 | 0.66 | 0.6 | ND | 0.99 | ND | |
| 11q | 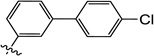 |
0.22 | 0.45 | 0.37 | ND | 0.86 | ND |
| 7 | -- | 0.70 | -- | -- | -- | 0.85 | 2.0 |
IC50: concentration of a compound producing 50% inhibition, expressed as mean of at least three independent experiments (standard deviation was less than 10% and is not indicated for ease of viewing).
Substrate that measures internal cleavage.
Substrate that measures DNA 3’ end directed cleavage.
Substrate that measures RNA 5’ end directed cleavage.
Reconstituted HIV RNase H domain.
CC50: concentration of a compound causing 50% cytotoxicity.
Not determined.
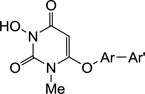
Table 2.
Biochemical inhibitory activity of compounds 10a–y against HIV RT RNase H and polymerase.
 | |||||||
|---|---|---|---|---|---|---|---|
| Cpd | Ar-Ar′ | Full-Length RT RNase H IC50a (µM) |
RNase H Fragmente IC50 (µM) |
RT pol IC50 (µM) |
CC50 (µM)f |
||
| HTS-1b | HTS-2c | HTS-3d | |||||
| 10a | 1.9 | 5.3 | 6.0 | 2.1 | N/Ig | 5.0 | |
| 10b | 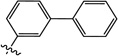 |
1.0 | N/I | 15 | 3.9 | N/I | 3.0 |
| 10c | 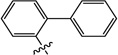 |
4.7 | N/I | N/I | NDh | N/I | ND |
| 10d | 6.2 | N/I | N/I | ND | N/I | ND | |
| 10e | 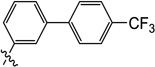 |
9.5 | N/I | N/I | N/I | N/I | >25 |
| 10f | 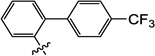 |
3.9 | N/I | 21 | 5.1 | N/I | 6.0 |
| 10g | 2.2 | 15 | 11 | ND | N/I | ND | |
| 10h | 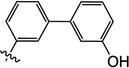 |
0.90 | 12 | 4.3 | ND | N/I | ND |
| 10i | 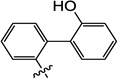 |
11 | N/I | N/I | ND | N/I | ND |
| 10j | 1.6 | 11 | 12 | 4.7 | N / I | 6.0 | |
| 10k | 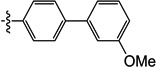 |
2.0 | 16 | 16 | 5.1 | N/I | 6.0 |
| 10l | 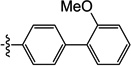 |
2.8 | N/I | 22 | 6.2 | N/I | 5.0 |
| 10m | 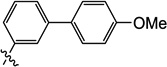 |
0.40 | 4.0 | 4.7 | 1.8 | N/I | 2.5 |
| 10n | 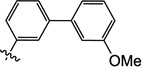 |
1.0 | 17 | 9.4 | 3.6 | N/I | 5.0 |
| 10o | 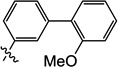 |
2.5 | N/I | 13 | 4.4 | N/I | 6.0 |
| 10p | 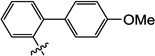 |
2.3 | N/I | 22 | 6.4 | N/I | 6.0 |
| 10q | 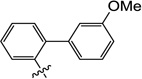 |
5.7 | N/I | N/I | 12 | N/I | 6.0 |
| 10r | 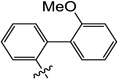 |
8.1 | N/I | N/I | 13 | N/I | 8.0 |
| 10s | 2.5 | 11 | 4.7 | ND | N/I | ND | |
| 10t | 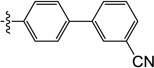 |
1.9 | 13 | 11 | ND | N/I | ND |
| 10u | 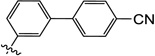 |
1.2 | 6.9 | 3.9 | ND | N/I | ND |
| 10v | 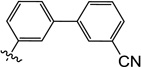 |
0.90 | 6.7 | 4.6 | ND | N/I | ND |
| 10w | 2.4 | N/I | 11 | ND | N/I | ND | |
| 10x | 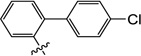 |
3.3 | N/I | 13 | ND | 14 | ND |
| 10y | 3.7 | N/I | 10 | ND | N/I | ND | |
| 7 | -- | 0.70 | -- | -- | -- | 0.85 | 2.0 |
IC50: concentration of a compound producing 50% inhibition, expressed as mean of at least three independent experiments (standard deviation was less than 10% and is not indicated for ease of viewing).
Substrate that measures DNA 3’ end directed cleavage.
Substrate that measures RNA 5’ end directed cleavage.
Reconstituted HIV RNase H domain.
CC50: concentration of a compound causing 50% cytotoxicity.
No inhibition at concentrations up to 25 µM.
Not determined.
In parallel to the RNase H assays, a classic RT pol assay was also conducted for all analogues with poly(rA) as template, oligo(dT) as primer and [3H]dTTP as nucleotide substrate. For subtype 9 (Table 1), sixteen out of twenty six analogues were completely inactive in this assay while the other ten compounds inhibited RT pol but with much reduced potency compared to RNase H inhibition (HTS-1). An even more dramatic observation was made with subtype 10 (Table 2) of which all analogues were inactive at the highest testing concentration (25 µM) with the lone exception of 10x which moderately inhibited RT pol (IC50 = 14 µM). The general lack of RT pol inhibition with these two subtypes suggests that selective inhibition of RT-associated RNase H could be achieved. In contrast, analogues with the N-1 unsubstituted chemotype 11 all inhibited RT pol at submicromolar to low micromolar concentrations (Table 3). Nevertheless, other than 11k even these compounds demonstrated considerable selectivity inhibiting RNase H over pol, particularly analogues 11a–c, 11i–j, 11l and 11o which inhibited pol 8–50 fold less potently than RNase H. This observation further confirms that our redesigned HPD chemotypes provide structural approaches towards selective RNase H inhibition. Finally, cell viability assays showed that most analogues of the two N-1 methylated subtypes 9–10 were significantly cytotoxic at low micromolar concentrations (CC50 = 3.0–10 µM), whereas cytotoxicity was not observed for any analogue of the N-1 unsubstituted subtype 11 at concentrations up to 25 µM, suggesting that 11 may be a superior chemotype for further antiviral development compared to chemotypes 9–10.
To further characterize the inhibitory selectivity towards RNase H, we also evaluated our new compounds in a biochemical assay that measures HIV INST activity. This is a critical measurement due to the high degree of homology between RNase H and IN in active site fold and catalytic mechanism. The INST assay results and the comparison between INST and RNase H inhibition are summarized in Table 4. Remarkably, six out of nine tested compounds of the two N-1 methyl subtypes 9 and 10 exhibited potent inhibition in single digit micromolar range (IC50 = 1.0–7.1µM), with the other three (9t, 9w and 10g) showing slightly lower potency (IC50 = 12–30 µM). By contrast, compounds of the N-1 unsubstituted chemotype 11 were either weakly active (11b, 11c and 11k, IC50 = 35–93 µM) or completely inactive at concentrations up to 100 µM (11i, 11j and 11o). The two exceptions are 11l and 11m which inhibited INST quite strongly at low micromolar concentrations (IC50 = 1.2–1.3 µM). These results suggest that the N-1 methyl substitution generally benefits binding to INST active site, and that the N-1 unsubstituted subtype 11 represents the preferred subtype of HPD for selective RNase H inhibition.
Table 4.
Biochemical inhibitory activity of selected compounds against HIV IN.
| Compd | RNase H IC50a (µM) |
INST IC50b (µM) |
SIc |
|---|---|---|---|
| 9b | 0.85 | 2.2 ± 0.66 | 2.6 |
| 9g | 0.75 | 4.4 ± 0.5 | 1.1 |
| 9k | 0.60 | 4.6 ± 2.1 | 7.7 |
| 9t | 1.2 | 30 ± 6.2 | 25 |
| 9v | 1.1 | 7.1 ± 2.6 | 6.5 |
| 9w | 1.6 | 25 ± 8.8 | 16 |
| 10g | 2.2 | 12 ± 3.2 | 5.5 |
| 10h | 0.90 | 7.1 ± 2.1 | 7.9 |
| 10p | 2.3 | 1.0 ± 0.28 | 0.43 |
| 11b | 0.40 | 93 ± 34 | 233 |
| 11c | 0.38 | 44 | 116 |
| 11i | 0.15 | >100 | >667 |
| 11j | 0.20 | >100 | >500 |
| 11k | 0.19 | 35 | 184 |
| 11l | 0.31 | 1.2 | 3.9 |
| 11m | 0.51 | 1.3 | 2.6 |
| 11o | 0.20 | >100 | >500 |
| L-731988d | -- | 0.40 ± 0.05 | -- |
Data with HTS-1 as substrate.
Expressed as mean ± standard deviation from at least three independent experiments.
Selectivity index defined as IC50 IN / IC50 RNase H.
Stereotypical DKA compound, reported as a selective INSTI.24
The activity and selectivity profiles of subtype 11 are manifested with a few selected analogues as shown in Table 5. Overall, these compounds demonstrated exceptional potency in inhibiting RNase H (IC50 = 0.15–0.40 µM). Although they also inhibited RT pol, the micromolar activity (IC50 = 5.7–7.3 µM) reflects a sizeable selectivity (14–49 fold) strongly favoring RNase H inhibition. Given the tendency of RNase H inhibitors to influence RT pol activity, this level of selectivity can be hard to achieve. In addition, none of these four analogues inhibited INST in a biochemical assay, amounting to a dramatic selectivity for RNase H inhibition over INST inhibition (selectivity index = 230 –>670). Furthermore, these analogues did not show appreciable cytotoxicity at concentrations up to 25 µM in cell viability assay. Although significant antiviral activity was not observed in our cell-based assays, the exceptional potency and selectivity as revealed from the detailed biochemical profiling suggest that HPD subtype 11 may present an important path towards the generation of specific inhibitors of HIV RT-associated RNase H.
Table 5.
Complete activity and selectivity profile of selected subtype 11 compounds.
| Compd | RNase H IC50a (µM) |
RT Pol IC50 (µM) (SIb) |
INST IC50 (µM) (SIb) |
CC50 (µM) |
|---|---|---|---|---|
| 11b | 0.40 | 5.7 (14) | 93 (230) | >25 |
| 11i | 0.15 | 7.3 (49) | >100 (>670) | >25 |
| 11j | 0.20 | 6.3 (32) | >100 (>500) | >25 |
| 11o | 0.20 | 6.6 (33) | >100 (>500) | >25 |
Data with HTS-1 as substrate.
Selectivity index, defined as IC50 RT pol or IN / IC50 RNase H.
Mode of Binding
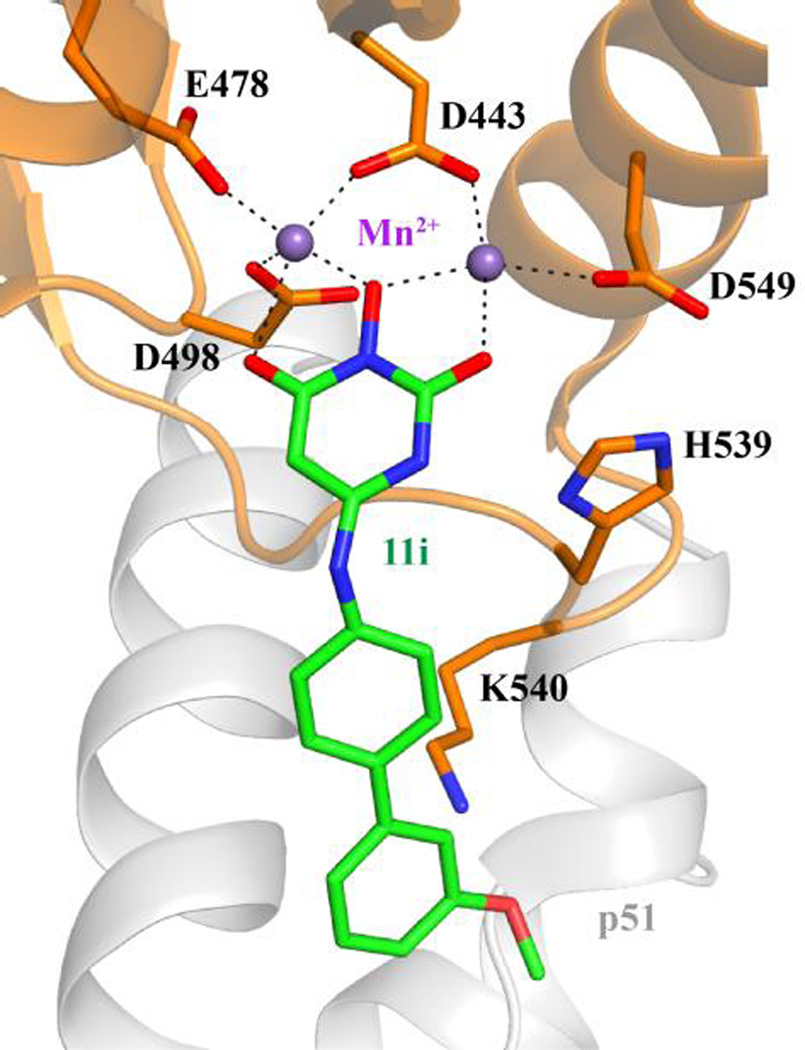
A molecular model of HIV-1 RT in complex with representative inhibitor 11i was constructed. It suggests possible interactions between this chemotype and conserved RNase H active site residues (Figure 2). Key to this binding mode is the interaction between the HPD core (the 2-C=O-3-OH-4-C=O chelating triad) and the two metal cofactors (Mn2+) which are coordinated to the active site acidic residues D443, E478, D498 and D549. The molecular model also suggests a potential interaction between the 1-NH of the HPD core and the imidazole of the highly conserved H539. In addition, the biaryl moiety is positioned to possibly interact with K540 as well as other protein residues or nucleic acid substrate near the small hydrophobic region of the RNase H. This predicted binding mode conforms to the general pharmacophore of active site RNase H inhibitors,19, 25 hence corroborating the active site inhibition mechanism.
Figure 2.
Molecular model of HIV-1 RT in complex with 11i shows possible interactions between this compound and indicated residues. RNase H active site residues are shown as orange sticks, 11i as green sticks, Mn2+ ions as purple spheres, and p51 is shown as gray cartoon.
Conclusions
New HPD chemotypes featuring a C-6 biary moiety were designed and synthesized as inhibitors of HIV RT associated RNase H domain. Detailed biochemical profiling revealed that all three newly designed subtypes can support potent RNase H inhibition, and that the N-1 unsubstituted subtype 11 is strongly favored over the N-1 methyl subtypes 9–10 due to the nanomolar potency towards RNase H inhibition, the exceptional selectivity over RT pol and INST inhibitions, and the lack of cytotoxicity at concentrations up to 25 µM. Although antiviral activity was not achieved, these highly favorable biochemical charateristics lay a solid foundation for potentially developing subtype 11 as RNase H targeting anitivirals through further SAR.
Chemistry
General Procedures
All commercial chemicals were used as supplied unless otherwise indicated. Dry solvents were either purchased (toluene and dioxane) or dispensed under argon from an anhydrous solvent system with two packed columns of neutral alumina or molecular sieves (THF and DMF). Flash chromatography was performed on a Teledyne Combiflash RF-200 with either RediSep columns (silica) or reverse-phase HP C18 Aq chromatography columns and indicated mobile phase. All moisture sensitive reactions were performed under an inert atmosphere of ultra-pure argon with oven-dried glassware. 1H and 13C NMR spectra were recorded on a Varian 600 MHz spectrometer. Mass data were acquired on an Agilent TOF II TOS/MS spectrometer capable of ESI and APCI ion sources. Analysis of sample purity was performed on a Varian Prepstar SD-1 HPLC system with a Phenomenex Gemini, 5 micron C18 column (250mm × 4.6 mm). HPLC conditions: solvent A: H2O with 0.1% TFA; solvent B: MeCN; flow rate 1.0 mL/min; compounds were eluted with a gradient of 20% MeCN/H2O with 0.1% TFA to 100% MeCN for 20 min. All tested compounds have a purity ≥ 96%.
3-(Benzyloxy)-6-hydroxypyrimidine-2,4(1H,3H)-dione (14)
To a 20 mL microwave reaction vessel were added 1-(benzyloxy)urea 13 (1.0 g, 6.0 mmol), freshly prepared sodium ethoxide (6.0 mmol, 1.0 eq) in 5.5 mL of anhydrous ethanol and diethyl malonate (6.0 mmol, 1.0 eq). The mixture was stirred at rt for 5 min, irradiated at 150 °C for 20 min and then cooled to room tempeture. After the solvent was removed under reduced pressure, the residue was dissolved in cold water, and the solution was acidified to pH = 4~5 with 2N HCl. The precipitate was filtered off, washed with Et2O and small amount of cold MeOH, and dried in vacuo to give compound 14 as light yellow solid in 58% yield. 1H NMR (600 MHz, DMSO-d6) δ 11.50 (s, 1H), 7.52 (d, J = 7.2 Hz, 2H), 7.51 (m, 3H), 4.03 (t, J = 7.2 Hz, 1H), 4.98 (s, 2H), 3.74 (s, 2H); 13C NMR (150 MHz, DMSO-d6) δ 165.8, 162.4, 148.7, 134.4, 129.3, 128.8, 128.4, 77.5, 41.0.
3-(Benzyloxy)-6-chloropyrimidine-2,4(1H,3H)-dione (15)
To a solution of 14 (2.0 g, 8.5 mmol) in POCl3 (20 mL) was added BnEt3NCl (17 mmol, 2.0 eq) and the mixture was stirred at 50 °C for 6 h. After cooled down to room temperature the reaction mixture was poured onto ice. The precipitate was filtered off, washed squentially by water, Et2O, cold MeOH, and dried in vacuo to give compound 15 as light yellow solid in 85 % yield. 1H NMR (600 MHz, DMSO-d6) δ 7.52 (d, J = 7.8 Hz, 2H), 7.40 (m, 3H), 5.97 (s, 1H), 5.00 (s, 2H); 13C NMR (150 MHz, DMSO-d6) δ 158.45, 148.1, 142.9, 134.2, 129.5, 128.9, 128.4, 99.9, 77.3.
3-(Benzyloxy)-6-chloro-1-methylpyrimidine-2,4(1H,3H)-dione (16)
To a solution of 15 (0.50 g, 2.0 mmol) in 5ml DMF was added Cs2CO3 (1.3 g, 4.0 mmmol) and MeI (0.57 g, 4.0 mmol), and the mixture was stirred at 80 °C for 3 hours when 15 was consumed completely as indicated by TLC. The mixture was cooled to room temperature and the reaction was quenched by adding 10 mL of H2O. The aqueous was extracted with EtOAc (15 mL × 3), and the combined organics were washed with brine (10 mL × 3) and dried over anhydrous Na2SO4. Solvent was removed in vacuo and the residue was purified by flash chromatography on silica gel to give the desired compound 16 (0.32 g, 60%) as offwhite solid. 1HNMR (600 MHz, CD3OD) δ 7.56 (d, J = 7.8 Hz, 2H), 7.38 (m, 3H), 6.03 (s, 1H), 5.00 (s, 2H).
General procedure for the synthesis of 3-(benzyloxy)- 1-methyl-6-aminopyrimidine-2,4(1H,3H)-dione (17)
To a solution of a biarylamine (0.20 mmol) in dry THF (3 mL) at −78 °C was added LDA (0.48 mmol). The mixture was stirred at −78 °C for 30 min followed by the addition of HMPA (1.0 mL) and compound 15 (0.40 mmol). The resulting mixture was stirred at −78 °C for 30 min, then room temperature overnight before it was quenched by adding 10 mL of water. The aqueous was extracted with EtOAc (10 mL × 3) and the combined organics were washed with brine and dried over Na2SO4. After the solvent was removed in vacuo, the residue was purified by flash column (Hexanes: EtOAc = 1:1) to give compound 17 (55–83 %).
3-(Benzyloxy)- 6-([1,1'-biphenyl]-4-ylamino)-1-methylpyrimidine-2,4(1H,3H)-dione (17a)
1H NMR: (DMSO-d6) δ 8.66 (s, 1H), 7.74 (m, 2H), 7.69 (m, 2H), 7.53 (m, 2H), 7.48 (m, 2H), 7.40 (m, 6H), 4.97 (s, 2H), 4.70 (s, 1H), 3.46 (s, 3H), 3.30 (s, 3H).
General procedure for the synthesis of 3-(benzyloxy)- 1-methyl-6-oxopyrimidine-2,4(1H,3H)-dione (18)
This chemotype was synthesized following the same procedure as described for the synthesis of 17.
6-([1,1'-Biphenyl]-4-yloxy)-3-(benzyloxy)-1-methylpyrimidine-2,4(1H,3H)-dione (18a)
1H NMR: (CDCl3) δ 7.67 (m, 2H), 7.61 (m, 2H), 7.58 (m, 2H), 7.49 (m, 2H), 7.41 (m, 4H), 7.20 (m, 2H), 5.14 (s, 2H), 4.87 (s, 1H), 3.58 (s, 3H).
General procedure for the synthesis of 3-(benzyloxy)-6-aminopyrimidine-2,4(1H,3H)-dione (19)
To a microwave reaction vessel were added compound 15 (1.0 mmol), biaryl amine (1.5 mmol) and N,N-dimethylaniline (3.0 mmol, 3.0 eq). The mixture was irradiated at 170 °C for 25 min until the completion of the reaction as monitored by both TLC and LC-MS. The reaction mixture was concentrated in vacuo. The residue was purified with Teledyne ISCO reverse-phase HP C18 Aq chromatography columns or via recrystallization (MeOH) to provide the desired compound 19 (54–85%) as offwhite solid.
3-(Benzyloxy)pyrimidine-6-([1,1'-biphenyl]-4-ylamino)-2,4(1H,3H)-dione (19a)
1H NMR: (DMSO-d6) δ 7.69 (m, 4H), 7.54 (m, 2H), 7.47 (m, 3H), 7.41 (m, 3H), 7.35 (m, 4H), 4.99 (s, 2H), 4.89 (s, 1H).
General procedure for the synthesis of 3-hydroxy-1-methyl-6-aminopyrimidine-2,4(1H,3H)-dione (9)
To a suspension of 17 (50 mg) in 5 mL MeOH in hydrogenation Parr Shaker, was added carefully 10 mg of Pd/C (20%). The reaction mixture was degassed using vacuum and refilling with H2 (40–50 psi) for three times before it was shaken under 40–50 psi H2 atmosphere at room temperature for 4 hours. The reaction mixture was filtered through a pad of celite and the solvent was removed. The residue was purified via trituration with MeOH, or Teledyne ISCO reverse-phase HP C18 Aq chromatography columns to yield the desired compound as white solid (82–90 % yield).
6-([1,1'-Biphenyl]-4-ylamino)-3-hydroxy-1-methylpyrimidine-2,4(1H,3H)-dione (9a)
1H NMR (600 MHz, DMSO-d6) δ 10.15 (s, 1H), 8.51 (s, 1H), 7.71 (d, J = 8.4 Hz, 2H), 7.67 (d, J = 7.8 Hz, 2H), 7.46 (t, J = 7.2 Hz, 2H), 7.35 (t, J = 7.8 Hz, 1H), 7.31 (d, J = 8.4 Hz, 2H), 4.71 (s, 1H), 3.42 (s, 3H); HRMS (ESI−) calcd. for C18H15N3O3 [M−H]− 308.1040, found 308.1045.
6-([1,1'-Biphenyl]-3-ylamino)-3-hydroxy-1-methylpyrimidine-2,4(1H,3H)-dione (9b)
1H NMR (600 MHz, DMSO-d6) δ 10.15 (s, 1H), 8.53 (s, 1H), 7.65 (d, J = 7.8 Hz, 2H), 7.51 (t, J = 7.8 Hz, 3H), 7.48 (t, J = 7.8 Hz, 2H), 7.39 (m, 1H), 7.24 (s, 1H), 4.70 (s, 1H), 3.84(s, 3H); HRMS (ESI−) calcd. for C17H15N3O3 [M−H]− 308.1040, found 308.1039.
3-Hydroxy-1-methyl-6-((4'-methyl-[1,1'-biphenyl]-4-yl)amino)pyrimidine-2,4(1H,3H)-dione (9c)
1H NMR (600 MHz, DMSO-d6) δ 10.14 (s, 1H), 8.50 (s, 1H), 7.68 (d, J = 7.8 Hz, 2H), 7.56 (d, J = 7.8 Hz, 2H), 7.29 (d, J = 7.8 Hz, 2H), 7.26 (d, J = 8.4 Hz, 2H), 6.73 (d, J = 8.4 Hz, 2H), 4.68 (s 1H), 3.42 (s, 3H), 3.40 (s, 3H); HRMS (ESI−) calcd. for C18H17N3O3 [M−H]− 322.1197, found 322.1200.
3-Hydroxy-1-methyl-6-((4'-methyl-[1,1'-biphenyl]-3-yl)amino)pyrimidine-2,4(1H,3H)-dione (9d)
1H NMR (600 MHz, DMSO-d6) δ 10.15 (s, 1H), 8.52 (s, 1H), 7.55 (d, J = 8.4 Hz, 2H), 7.49 (d, J = 8.4 Hz, 2H), 7.28 (d, J = 8.4 Hz, 2H), 7.21 (d, J = 9.0 Hz, 2H), 4.68 (s, 1H), 3.43 (s, 3H), 2.33 (s, 3H); HRMS (ESI−) calcd. for C18H17N3O3 [M−H]− 322.1197, found 322.1185.
3-Hydroxy-1-methyl-6-((4'-(trifluoromethyl)-[1,1'-biphenyl]-4-yl)amino)pyrimidine-2,4(1H,3H)-dione (9e)
1H NMR (600 MHz, DMSO-d6) δ 10.18 (s, 1H), 8.56 (s, 1H), 7.90 (d, J = 8.4 Hz, 2H), 7.80 (t, J = 7.8 Hz, 4H), 7.35 (d, J = 8.4 Hz, 2H), 4.77 (s, 1H), 3.42 (s, 3H); HRMS (ESI−) calcd. for C18H14F3N3O3 [M−H]− 376.0987, found 376.0990.
3-Hydroxy-6-((4'-hydroxy-[1,1'-biphenyl]-3-yl)amino)-1-methylpyrimidine-2,4(1H,3H)-dione (9f)
1H NMR (600 MHz, DMSO-d6) δ 10.14 (s, 1H), 9.58 (s, 1H), 8.49 (s, 1H), 7.47 (d, J = 6.0 Hz, 2H), 7.43 (m, 2H), 7.14 (s, 1H), 6.84 (d, J = 6.0 Hz, 2H), 4.66 (s, 1H), 3.43 (s, 3H); HRMS (ESI−) calcd. for C17H15N3O4 [M−H]− 324.0990, found 324.0993.
3-Hydroxy-6-((4'-methoxy-[1,1'-biphenyl]-4-yl)amino)-1-methylpyrimidine-2,4(1H,3H)-dione (9g)
1H NMR (600 MHz, DMSO-d6) δ 10.63 (s, 1H), 8.48 (s, 1H), 7.65 (d, J = 9.0 Hz, 2H), 7.61 (t, J = 9.0 Hz, 2H), 7.27 (d, J = 7.8 Hz, 2H), 7.01 (d, J = 8.4 Hz, 2H), 4.66 (s, 1H), 3.77 (s, 3H), 3.42 (s, 3H); HRMS (ESI−) calcd. for C18H17N3O4 [M−H]− 338.1146, found 338.1150.
3-Hydroxy-6-((3'-methoxy-[1,1'-biphenyl]-4-yl)amino)-1-methylpyrimidine-2,4(1H,3H)-dione (9h)
1H NMR (600 MHz, DMSO-d6) δ 10.15 (s, 1H), 8.52 (s, 1H), 7.71 (d, J = 7.2 Hz, 2H), 7.36 (t, J = 7.2 Hz, 1H), 7.30 (t, J = 8.4 Hz, 1H), 7.23 (d, J = 7.2 Hz, 1H), 7.18 (s, 1H), 6.92 (dd, J = 3.0, 8.4 Hz, 1H), 4.70 (s, 1H), 3.80 (s, 3H), 3.42 (s, 3H); HRMS (ESI−) calcd. for C18H17N3O4 [M−H]− 338.1146, found 338.1157.
3-Hydroxy-6-((2'-methoxy-[1,1'-biphenyl]-4-yl)amino)-1-methylpyrimidine-2,4(1H,3H)-dione (9i)
1H NMR (600 MHz, DMSO-d6) δ 10.17 (s, 1H), 8.51 (s, 1H), 7.54 (d, J = 9.0 Hz, 2H), 7.59 (m, 1H), 7.33 (m, 1H), 7.28 (d, J = 7.8 Hz, 2H), 7.13 (d, J = 8.4 Hz, 1H), 7.05 (t, J = 7.8 Hz, 1H), 4.72 (s, 1H), 3.78 (s, 3H), 3.45 (s, 3H); HRMS (ESI−) calcd. for C18H17N3O4 [M−H]− 338.1146, found 338.165.
3-Hydroxy-6-((4'-methoxy-[1,1'-biphenyl]-3-yl)amino)-1-methylpyrimidine-2,4(1H,3H)-dione (9j)
1H NMR (600 MHz, DMSO-d6) δ 10.20 (s, 1H), 8.54 (s, 1H), 7.71 (d, J = 8.4 Hz, 2H), 7.60 (d, J = 8.4 Hz, 2H), 7.47 (d, J = 6.0 Hz, 3H), 7.47 (d, J = 6.0 Hz, 1H), 7.35 (d, J =8.4 Hz, 2H), 4.69 (s, 1H), 3.78 (s, 3H); HRMS (ESI−) calcd. for C18H17N3O4 [M−H]− 338.1146, found 338.1149.
3-Hydroxy-6-((3'-methoxy-[1,1'-biphenyl]-3-yl)amino)-1-methylpyrimidine-2,4(1H,3H)-dione (9k)
1H NMR (600 MHz, DMSO-d6) δ 10.15 (s, 1H), 8.52 (s, 1H), 7.51 (m, 2H), 7.49 (t, J = 7.8 Hz, 1H), 7.38 (t, J = 7.8 Hz, 1H), 7.24 (d, J = 8.4 Hz, 1H), 7.20 (d, J = 7.8 Hz, 1H), 7.15 (s,1H), 6.94 (dd, J = 1.8, 7.8 Hz, 2H), 4.67 (s, 1H), 3.80 (s, 3H), 3.43 (s, 3H); HRMS (ESI−) calcd. for C18H17N3O4 [M−H]− 338.1146, found 338.1156.
3-Hydroxy-6-((2'-methoxy-[1,1'-biphenyl]-3-yl)amino)-1-methylpyrimidine-2,4(1H,3H)-dione (9l)
1H NMR (600 MHz, DMSO-d6) δ 10.14 (s, 1H), 8.50 (s, 1H), 7.44 (t, J = 7.8 Hz, 1H), 7.37 (s, 1H), 7.35 (t, J = 7.8 Hz, 1H), 7.30 (d, J = 7.2 Hz, 1H), 7.28 (d, J = 7.8 Hz, 1H), 7.17 (d, J = 7.2 Hz, 1H), 7.10 (d, J = 9.0 Hz, 1H), 7.03 (t, J = 7.8 Hz, 1H), 4.82 (s 1H), 3.77 (s, 3H), 3.42 (s, 3H); HRMS (ESI−) calcd. for C18H17N3O4 [M−H]− 338.1146, found 338.1149.
6-((2',4'-Dimethoxy-[1,1'-biphenyl]-4-yl)amino)-3-hydroxy-1-methylpyrimidine-2,4(1H,3H)-dione (9m)
1H NMR (600 MHz, DMSO-d6) δ 10.13 (s, 1H), 8.46 (s, 1H), 7.47 (d, J = 9.0 Hz, 2H), 7.23 (dt, J = 2.4, 8.4, 3H), 6.64 (d, J = 2.4 Hz, 1H), 6.60 (dd, J = 2.4, 9.0 Hz, 1H), 4.65 (s 1H), 3.78 (s, 3H), 3.75 (s, 3H), 3.42 (s, 3H); HRMS (ESI−) calcd. for C19H19N3O5 [M−H]− 368.1252, found 368.1272.
6-((3',5'-Dimethoxy-[1,1'-biphenyl]-4-yl)amino)-3-hydroxy-1-methylpyrimidine-2,4(1H,3H)-dione (9n)
1H NMR (600 MHz, DMSO-d6) δ 10.17 (s, 1H), 8.57 (s, 1H), 7.71 (d, J = 8.4 Hz, 2H), 7.29 (d, J = 9.0 Hz, 2H), 6.78 (m, 2H), 6.47 (s, 1H), 5.00 (s, 1H), 3.78 (s, 6H)); 13C NMR (150 MHz, DMSO-d6) δ 161.3, 159.7, 152.6, 150.3, 142.0, 138.7, 137.4, 128.2, 125.1, 105.0, 99.9, 77.7, 55.7, 30.7; HRMS (ESI−) calcd. for C19H19N3O5 [M−H]− 368.1252, found 368.1261.
6-((2',6'-Dimethoxy-[1,1'-biphenyl]-3-yl)amino)-3-hydroxy-1-methylpyrimidine-2,4(1H,3H)-dione (9o)
1H NMR (600 MHz, DMSO-d6) δ 10.11 (s, 1H), 8.45 (s, 1H), 7.39 (t, J =7.8 Hz, 1H), 7.30 (t, J =7.8 Hz, 1H), 7.11 (d, J = 8.4 Hz, 1H), 7.08 (m, 2H), 6.73 (d, J = 8.4 Hz, 2H), 4.79 (s, 1H), 3.67 (s, 6H), 3.40 (s, 3H); HRMS (ESI−) calcd. for C19H19N3O5 [M−H]− 368.1252, found 368.1255.
6-((3',4'-Dimethoxy-[1,1'-biphenyl]-3-yl)amino)-3-hydroxy-1-methylpyrimidine-2,4(1H,3H)-dione (9p)
1H NMR (600 MHz, DMSO-d6) δ 10.14 (s, 1H), 8.51 (s, 1H), 7.51 (t, J = 7.8 Hz, 2H), 7.67 (t, J = 7.2 Hz, 1H), 7.19 (m, 3H), 7.03 (t, J = 8.4 Hz, 1H), 4.64 (s, 1H), 3.82 (s, 3H), 3.77 (s, 3H); HRMS (ESI−) calcd. for C19H19N3O5 [M−H]− 368.1252, found 368.1261.
6-((2',4'-Dimethoxy-[1,1'-biphenyl]-3-yl)amino)-3-hydroxy-1-methylpyrimidine-2,4(1H,3H)-dione (9q)
1H NMR (600 MHz, CD3OD) δ 7.33 (m, 2H), 7.26 (t, J = 7.8 Hz, 1H), 7.15 (d, J = 8.4 Hz, 1H), 7.05 (d, J = 7.2 Hz, 1H), 6.51 (m, 2H), 4.96 (s, 1H), 4.72 (s, 3H), 3.74 (s, 3H), 3.72 (s, 3H); HRMS (ESI−) calcd. for C19H19N3O5 [M−H]− 368.1252, found 368.1255.
6-((3',5'-Dimethoxy-[1,1'-biphenyl]-3-yl)amino)-3-hydroxy-1-methylpyrimidine-2,4(1H,3H)-dione (9r)
1H NMR (600 MHz, DMSO-d6) δ 10.14 (s, 1H), 8.52 (s, 1H), 7.51 (m, 2H), 7.50 (t, J = 7.8 Hz, 1H), 7.24 (t, J = 7.8 Hz, 1H), 6.75 (s, 2H), 6.50 (s, 1H), 4.66 (s, 1H), 3.78 (s, 6H), 3.43 (s, 3H); HRMS (ESI−) calcd. for C19H19N3O5 [M−H]− 368.1252, found 368.1263.
4'-((1-Hydroxy-3-methyl-2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)amino)-[1,1'-biphenyl]-4-carbonitrile (9s)
1H NMR (600 MHz, DMSO-d6) δ 10.12 (s, 1H), 8.67 (s, 1H), 7.99 (d, J = 8.4 Hz, 2H), 7.97 (d, J =7.8 Hz, 2H), 7.89 (d, J = 7.8 Hz, 2H), 7.44 (t, J = 8.4 Hz, 2H), 4.88 (s, 1H), 4.01 (t, J = 7.2 Hz, 1H), 3.51 (s, 3H); HRMS (ESI−) calcd. for C18H14N4O3 [M−H]− 333.0993, found 333.0998.
4'-((1-Hydroxy-3-methyl-2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)amino)-[1,1'-biphenyl]-3-carbonitrile (9t)
1H NMR (600 MHz, DMSO-d6) δ 10.20 (s, 1H), 8.59 (s, 1H), 8.14 (s, 1H), 7.02 (d, J = 8.4 Hz, 1H), 7.79 (d, J = 7.8 Hz, 3H), 7.65 (t, J = 7.8 Hz, 1H), 7.34 (d, J = 7.8 Hz, 2H), 4.78 (s 1H), 3.43 (s, 3H), 3.42 (s, 3H); HRMS (ESI−) calcd. for C18H14N4O3 [M−H]− 333.0993, found 333.1003
3'-((1-Hydroxy-3-methyl-2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)amino)-[1,1'-biphenyl]-4-carbonitrile (9u)
1H NMR (600 MHz, DMSO-d6) δ 10.17 (s, 1H), 8.57 (s, 1H), 7.93 (t, J = 8.4 Hz, 2H), 7.86 (d, J =7.2 Hz, 2H), 7.59 (d, J = 7.2 Hz, 2H), 7.55 (t, J = 7.8 Hz, 1H), 7.31 (d, J = 7.2 Hz, 1H), 4.70 (s, 1H), 4.01 (t, J = 7.2 Hz, 1H), 3.42 (s, 3H); HRMS (ESI−) calcd. for C18H14N4O3 [M−H]− 333.0993, found 333.1004.
3'-((1-Hydroxy-3-methyl-2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)amino)-[1,1'-biphenyl]-3-carbonitrile (9v)
1H NMR (600 MHz, DMSO-d6) δ 10.18 (s, 1H), 8.59 (s, 1H), 8.12 (s, 1H), 8.00 (d, J = 7.8 Hz, 1H), 7.70 (d, J =7.2 Hz, 1H), 7.68 (t, J = 7.8 Hz, 1H), 7.58 (m, 2H), 7.53 (t, J = 7.8 Hz, 1H), 7.28 (d, J = 7.2 Hz, 1H), 4.66 (s, 1H), 3.43 (s, 3H); HRMS (ESI−) calcd. for C18H14N4O3 [M−H]− 333.0993, found 333.1009.
6-((4-(3,5-Dimethylisoxazol-4-yl)phenyl)amino)-3-hydroxy-1-methylpyrimidine-2,4(1H,3H)-dione (9w)
1H NMR (600 MHz, CD3OD) δ 7.42 (d, J = 6.0 Hz, 2H), 7.39 (d, J = 6.0 Hz, 2H), 4.93 (s, 1H), 3.57 (s, 3H), 2.42 (s, 3H), 2.27 (s, 3H); HRMS (ESI−) calcd. for C16H16N4O4 [M−H]− 327.1099, found 327.1115.
6-((2-(3,5-Dimethylisoxazol-4-yl)phenyl)amino)-3-hydroxy-1-methylpyrimidine-2,4(1H,3H)-dione (9x)
1H NMR (600 MHz, CD3OD) δ 7.47 (t, J = 6.0 Hz, 1H), 7.51 (t, J = 6.0 Hz, 1H), 7.12 (d, J = 6.0 Hz, 1H), 7.05 (d, J = 6.0 Hz, 1H), 5.39 (s, 1H), 3.33 (s, 3H), 2.18 (s, 3H), 2.04 (s, 3H) ; HRMS (ESI−) calcd. for 16H16N4O4 [M−H]− 327.1099, found 327.1088.
3-Hydroxy-1-methyl-6-((3-(pyrimidin-5-yl)phenyl)amino)pyrimidine-2,4(1H,3H)-dione (9y)
1H NMR (600 MHz, DMSO-d6) δ 10.63 (s, 1H), 10.06 (s, 1H), 9.18 (s, 1H), 9.10 (s, 2H), 8.69 (s, 1H), 7.68 (m, 1H), 7.64 (m, 1H), 7.57 (t, J = 7.8 Hz, 1H), 7.35 (d, J = 7.8 Hz, 1H), 4.72 (s, 1H), 3.44 (s, 3H); HRMS (ESI−) calcd. for C15H13N5O3 [M−H]− 310.1018, found 310.1020.
3-Hydroxy-1-methyl-6-((3-(tetrahydrofuran-3-yl)phenyl)amino)pyrimidine-2,4(1H,3H)-dione (9z)
1H NMR (600 MHz, DMSO-d6) δ 10.63 (s, 1H), 8.42 (s, 1H), 7.35 (t, J =7.8 Hz, 1H), 7.13 (m, 2H), 7.08 (d, J = 8.4 Hz, 2H), 4.47 (d, J = 1.2 Hz, 1H), 4.01 (t, J =7.2 Hz, 1H), 3.92 (m, 1H), 3.78 (q, J =7.2 Hz, 1H), 3.54 (t, J =7.2 Hz, 1H), 3.39 (m, 1H), 3.31 (s, 3H); HRMS (ESI−) calcd. for C15H17N3O4 [M−H]− 302.1219, found 302.1217.
General procedure for the synthesis of 3-hydroxy-1-methyl-6-oxopyrimidine-2,4(1H,3H)-dione (10)
This chemotype was synthesized following the same procedure as described for the synthesis of 9.
6-([1,1'-Biphenyl]-4-yloxy)-3-hydroxy-1-methylpyrimidine-2,4(1H,3H)-dione (10a)
1H NMR (600 MHz, DMSO-d6) δ 10.42 (s, 1H), 7.81 (d, J =8.4 Hz, 2H), 7.70 (d, J =7.8 Hz, 2H), 7.49 (t, J = 7.8 Hz, 2H), 7.41 (t, J = 7.8 Hz, 3H), 4.53 (s, 1H), 3.32 (s, 3H); HRMS (ESI−) calcd. for C17H14N2O4 [M−H]− 309.0954, found 309.0960.
6-([1,1'-Biphenyl]-3-yloxy)-3-hydroxy-1-methylpyrimidine-2,4(1H,3H)-dione (10b)
1H NMR (600 MHz, CDCl3) δ 8.37 (s, 1H), 7.59 (m, 4H), 7.47 (t, J = 7.8 Hz, 2H), 7.40 (t, J = 7.2 Hz, 1H), 7.33 (m, 1H), 7.10 (dd, J = 7.8, 1.8 Hz, 1H), 4.96 (s, 1H), 3.62 (s, 3H), 3.42 (s, 3H); HRMS (ESI−) calcd. for C17H14N2O4 [M−H]− 309.0954, found 309.0885.
6-([1,1'-Biphenyl]-2-yloxy)-3-hydroxy-1-methylpyrimidine-2,4(1H,3H)-dione (10c)
1H NMR (600 MHz, CDCl3) δ 7.50 (dd, J = 2.4, 6.0 Hz, 1H), 7.67 (m, 2H), 7.41 (m, 2H), 7.37 (m, 3H), 7.20 (m, 1H), 4.78 (s, 1H), 3.42 (s, 3H); HRMS (ESI−) calcd. for C17H14N2O4 [M−H]− 309.0954, found 309.0958.
3-Hydroxy-1-methyl-6-((4'-(trifluoromethyl)-[1,1'-biphenyl]-4-yl)oxy)pyrimidine-2,4(1H,3H)-dione (10d)
1H NMR (600 MHz, CDCl3) δ 8.41 (s, 1H), 7.68 (d, J = 7.8 Hz, 2H), 7.63 (t, J = 9.0 Hz, 4H), 7.19 (t, J = 7.8 Hz, 2H), 4.88 (s, 1H), 3.56 (s, 3H); HRMS (ESI−) calcd. for C18H13F3N2O4 [M−H]− 377.0755, found 377.0747.
3-Hydroxy-1-methyl-6-((4'-(trifluoromethyl)-[1,1'-biphenyl]-3-yl)oxy)pyrimidine-2,4(1H,3H)-dione (10e)
1H NMR (600 MHz, CD3OD) δ 7.77 (m, 2H), 7.73 (m, 2H), 7.67 (m, 1H), 7.62 (t, J = 7.8 Hz, 1H), 7.52 (m, 1H), 7.26 (m, 1H), 4.68 (s, 1H), 3.50 (s, 3H); HRMS (ESI−) calcd. for C18H13F3N2O4 [M−H]− 377.0755, found 377.0763.
3-Hydroxy-1-methyl-6-((4'-(trifluoromethyl)-[1,1'-biphenyl]-2-yl)oxy)pyrimidine-2,4(1H,3H)-dione (10f)
1H NMR (600 MHz, CD3OD) δ 7.75 (d, J = 7.8 Hz, 2H), 7.51 (d, J = 8.4 Hz, 2H), 7.60 (m, 2H), 7.55 (m, 1H), 7.40 (d, J = 7.8 Hz, 1H), 4.57 (s, 1H), 3.42 (s, 3H); HRMS (ESI−) calcd. for C18H13F3N2O4 [M−H]− 377.0755, found 377.0761
3-Hydroxy-6-((4'-hydroxy-[1,1'-biphenyl]-4-yl)oxy)-1-methylpyrimidine-2,4(1H,3H)-dione (10g)
1H NMR (600 MHz, CD3OD) δ 7.59 (m, 2H), 7.39 (m, 2H), 7.18 (m, 2H), 6.79 (d, J = 8.4 Hz, 2H), 4.78 (s, 1H), 3.45 (s, 3H); HRMS (ESI−) calcd. for C17H14N2O5 [M−H]− 325.0830, found 325.0835.
3-Hydroxy-6-((3'-hydroxy-[1,1'-biphenyl]-3-yl)oxy)-1-methylpyrimidine-2,4(1H,3H)-dione (10h)
1H NMR (600 MHz, CD3OD) δ 7.59 (d, J = 8.4 Hz, 2H), 7.54 (t, J = 7.8 Hz, 1H), 7.45 (s, 1H), 7.26 (t, J = 7.8 Hz, 1H), 7.19 (dd, J = 2.4, 9.0 Hz, 1H), 7.07 (d, J = 8.4 Hz, 1H), 7.03 (s, 1H), 6.80 (dd, J = 2.4, 7.8 Hz, 1H), 4.72 (s, 1H), 3.45 (s, 3H); HRMS (ESI−) calcd. for C17H14N2O4 [M−H]− 325.0830, found 325.0839.
3-Hydroxy-6-((2'-hydroxy-[1,1'-biphenyl]-2-yl)oxy)-1-methylpyrimidine-2,4(1H,3H)-dione (10i)
1H NMR (600 MHz, DMSO-d6) δ 11.08 (s, 1H), 10.39 (s, 1H), 7.57 (m, 2H), 7.46 (m, 6H), 4.45 (s, 1H), 3.18 (s, 3H); HRMS (ESI−) calcd. for C17H14N2O4 [M−H]− 325.0830, found 325.0833.
3-Hydroxy-6-((4'-methoxy-[1,1'-biphenyl]-4-yl)oxy)-1-methylpyrimidine-2,4(1H,3H)-dione (10j)
1H NMR (600 MHz, CDCl3) δ 7.63 (d, J = 9.0 Hz, 2H), 7.51 (d, J = 9.0 Hz, 2H), 7.12 (d, J = 7.8 Hz, 2H), 7.05 (d, J = 9.0 Hz, 2H), 5.00 (s, 1H), 3.87 (s, 3H), 3.62 (s, 3H); HRMS (ESI−) calcd. for C18H16N2O5 [M−H]− 339.0986, found 339.0890
3-Hydroxy-6-((3'-methoxy-[1,1'-biphenyl]-4-yl)oxy)-1-methylpyrimidine-2,4(1H,3H)-dione (10k)
1H NMR (600 MHz, CD3OD) δ 7.72 (d, J = 8.4 Hz, 2H), 7.36 (t, J = 7.8 Hz, 1H), 7.28 (d, J = 9.0 Hz, 2H), 7.17 (d, J = 6.6 Hz, 1H), 7.12 (s, 1H), 6.93 (d, J = 8.4 Hz, 1H), 4.78 (s, 1H), 3.84 (s, 3H), 3.56 (s, 3H); HRMS (ESI−) calcd. for C18H16N2O5 [M−H]− 339.0986, found 339.0989.
3-Hydroxy-6-((2'-methoxy-[1,1'-biphenyl]-4-yl)oxy)-1-methylpyrimidine-2,4(1H,3H)-dione (10l)
1H NMR (600 MHz, CD3OD) δ 7.63 (d, J = 8.4 Hz, 2H), 7.33 (t, J = 8.4 Hz, 1H), 7.39 (dd, J = 1.2, 7.2 Hz, 1H), 7.25 (d, J = 7.8 Hz, 2H), 7.08 (d, J = 7.8 Hz, 1H), 7.03 (t, J = 7.2 Hz, 1H), 4.78 (s, 1H), 3.80 (s, 3H), 3.57 (s, 3H); HRMS (ESI−) calcd. for C18H16N2O5 [M−H]− 339.0986, found 339.0991.
3-Hydroxy-6-((4'-methoxy-[1,1'-biphenyl]-3-yl)oxy)-1-methylpyrimidine-2,4(1H,3H)-dione (10m)
1H NMR (600 MHz, CDCl3) δ 7.55 (d, J =7.8 Hz, 1H), 7.51 (m, 3H), 7.29 (s, 1H), 7.05 (d, J = 7.2 Hz, 1H), 7.00 (d, J = 9.0 Hz, 2H), 4.97 (s, 1H), 3.87 (s, 3H), 3.63 (s, 3H); HRMS (ESI−) calcd. for C18H16N2O5 [M−H]− 339.09867, found 339.1004.
3-Hydroxy-6-((3'-methoxy-[1,1'-biphenyl]-3-yl)oxy)-1-methylpyrimidine-2,4(1H,3H)-dione (10n)
1H NMR (600 MHz, CDCl3) δ 7.59 (d, J = 7.8 Hz, 1H), 7.54 (d, J = 8.4 Hz, 1H), 7.39 (d, J = 8.4 Hz, 1H), 7.33 (s, 1H), 7.15 (d, J = 7.8 Hz, 2H), 7.10 (m, 2H), 6.96 (dd, J = 2.4, 7.8 Hz, 2H), 4.96 (s, 1H), 3.87 (s, 3H), 3.63 (s, 3H); HRMS (ESI−) calcd. for C18H16N2O5 [M−H]− 339.0986, found 339.0989.
3-Hydroxy-6-((2'-methoxy-[1,1'-biphenyl]-3-yl)oxy)-1-methylpyrimidine-2,4(1H,3H)-dione (10o)
1H NMR (600 MHz, CDCl3) δ 8.37 (s, 1H), 7.49 (m, 2H), 7.37 (dt, J = 1.2, 8.4 Hz, 1H), 7.33 (m, 1H), 7.30 (dd, J = 1.2, 7.2 Hz, 1H), 7.06 (m, 2H), 6.99 (d, J = 7.8 Hz, 1H), 5.04 (s, 1H), 3.82 (s, 3H), 3.61 (s, 3H);); 13C NMR (150 MHz, CDCl3) δ 160.8, 158.2, 156.3, 150.9, 146.2, 141.3, 130.5, 130.0, 129.6, 128.4, 122.2, 120.9, 119.0, 111.2, 79.3, 55.5, 29.5; HRMS (ESI−) calcd. for C18H16N2O5 [M−H]− 339.0986, found 339.0991.
3-Hydroxy-6-((4'-methoxy-[1,1'-biphenyl]-2-yl)oxy)-1-methylpyrimidine-2,4(1H,3H)-dione (10p)
1H NMR (600 MHz, CDCl3) δ 7.47 (dd, J = 3.6, 6.0 Hz, 1H), 7.42 (m, 2H), 7.30 (d, J = 9.0 Hz, 2H), 7.17 (dd, J = 3.6, 6.0 Hz, 1H), 6.92 (d, J = 9.0 Hz, 2H), 4.78 (s, 1H), 3.81 (s, 3H), 3.42 (s, 3H); HRMS (ESI−) calcd. for C18H16N2O5 [M−H]− 339.0986, found 339.0981.
3-Hydroxy-6-((3'-methoxy-[1,1'-biphenyl]-2-yl)oxy)-1-methylpyrimidine-2,4(1H,3H)-dione (10q)
1H NMR (600 MHz, CDCl3) δ 8.32 (s, 1H), 7.49 (dd, J =1.8, 7.8 Hz, 1H), 7.51 (dt, J =2.4, 7.8 Hz, 2H), 7.31 (m, 1H), 7.18 (dd, J = 1.8, 7.2 Hz, 1H), 6.93 (d, J = 8.4 Hz, 1H), 6.89 (m, 2H), 4.76 (s, 1H), 3.79 (s, 3H), 3.43 (s, 3H); HRMS (ESI−) calcd. for C18H16N2O5 [M−H]− 339.0986, found 339.0992.
3-(Benzyloxy)-6-((2'-methoxy-[1,1'-biphenyl]-2-yl)oxy)-1-methylpyrimidine-2,4(1H,3H)-dione (10r)
1H NMR (600 MHz, DMSO-d6) δ 10.34 (s, 1H), 7.51 (dt, J =1.8, 7.8 Hz, 1H), 7.48 (t, J = 7.8 Hz, 1H), 7.40 (m, 1H), 7.37 (m, 1H), 7.34 (dt, J =1.8, 7.8 Hz, 1H), 7.14 (dd, J =1.8, 7.8 Hz, 1H), 7.05 (d, J =7.8 Hz, 1H), 6.96 (t, J =1.8, 7.8 Hz, 1H), 4.43 (s, 1H), 3.64 (s, 3H), 3.10 (s, 3H); HRMS (ESI−) calcd. for C18H16N2O5 [M−H]− 339.0986, found 339.0990.
4'-((1-Hydroxy-3-methyl-2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)oxy)-[1,1'-biphenyl]-4-carbonitrile (10s)
1H NMR (600 MHz, CDCl3) δ 7.83 (m, 6H), 7.38 (d, J = 8.4 Hz, 2H), 4.76 (s, 1H), 3.56 (s, 3H); HRMS (ESI−) calcd. for C17H15N3O4 [M−H]− 324.0990, found 324.0993.
4'-((1-Hydroxy-3-methyl-2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)oxy)-[1,1'-biphenyl]-3-carbonitrile (10t)
1H NMR (600 MHz, CD3OD) δ 8.01 (s, 1H), 7.96 (d, J = 6.6 Hz, 1H), 7.80 (d, J = 7.8 Hz, 2H), 7.73 (s, 1H), 7.15 (d, J = 7.2 Hz, 1H), 7.65 (t, J = 7.8 Hz, 1H), 7.37 (d, J = 8.4 Hz, 2H), 4.74 (s, 1H), 3.55 (s, 3H), HRMS (ESI−) calcd. for C18H13N3O4 [M−H]− 334.0833, found 334.0840.
3'-((1-Hydroxy-3-methyl-2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)oxy)-[1,1'-biphenyl]-4-carbonitrile (10u)
1H NMR (600 MHz, CD3OD) δ 7.86 (d, J = 6.0 Hz, 2H), 7.83 (d, J = 6.0 Hz, 2H), 7.75 (d, J = 12.0 Hz, 1H), 7.67 (m, 2H), 7.35 (d, J = 6.0 Hz, 1H), 4.74 (s, 1H), 3.58 (s, 3H); HRMS (ESI−) calcd. for C18H13N3O4 [M−H]− 334.0833, found 334.0833.
3'-((1-Hydroxy-3-methyl-2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)oxy)-[1,1'-biphenyl]-3-carbonitrile (10v)
1H NMR (600 MHz, CDCl3) δ 7.8 (d, J = 8.4 Hz, 1H), 7.73 (d, J = 7.8 Hz, 1H), 7.64 (d, J = 7.8 Hz, 1H), 7.55 (m, 3H), 7.27 (s, 1H), 7.13 (m, 1H), 4.86 (s, 1H), 3.56 (s, 3H): HRMS (ESI−) calcd. for C18H13N3O4 [M−H]− 334.0833, found 334.0840.
6-((4'-Chloro-[1,1'-biphenyl]-4-yl)oxy)-3-hydroxy-1-methylpyrimidine-2,4(1H,3H)-dione (10w)
1H NMR (600 MHz, DMSO-d6) 10.47 (s, 1H), δ 7.80 (d, J = 9.0 Hz, 2H), 7.69 (d, J = 7.2 Hz, 2H), 7.47 (t, J = 7.8 Hz, 2H), 7.40 (d, J = 7.8 Hz, 2H), 4.51 (s, 1H), 3.41 (s, 3H); HRMS (ESI−) calcd. for C17H13ClN2O4 [M−H]− 343.0491, found 343.0483.
6-((4'-Chloro-[1,1'-biphenyl]-2-yl)oxy)-3-hydroxy-1-methylpyrimidine-2,4(1H,3H)-dione (10x)
1H NMR (600 MHz, DMSO-d6) δ 10.36 (s, 1H), 7.57 (m, 3H), 7.45 (m, 4H), 7.40 (m, 1H), 7.33 (s, 1H), 4.34 (s, 1H), 3.29 (s, 3H); HRMS (ESI−) calcd. for C17H13ClN2O4 [M−H]− 343.0491, found 343.0485.
3-Hydroxy-1-methyl-6-(4-(pyridin-4-yl)phenoxy)pyrimidine-2,4(1H,3H)-dione (10y)
1H NMR (600 MHz, CD3OD) δ 8.61 (d, J = 6.0 Hz, 2H), 7.93 (d, J = 9.0 Hz, 2H), 7.75 (d, J = 6.6 Hz, 2H), 7.43 (d, J = 9.0 Hz, 2H), 5.47 (s, 1H), 3.56 (s, 3H); HRMS (ESI−) calcd. for C16H13N3O4 [M−H]− 310.0833, found 310.0828.
General procedure for the synthesis of 3-hydroxy-6-aminopyrimidine-2,4(1H,3H)-dione (11)
This chemotype was synthesized following the same procedure as described for the synthesis of 9.
3-Hydroxy-6-((4'-methoxy-[1,1'-biphenyl]-3-yl)amino)pyrimidine-2,4(1H,3H)-dione (11a)
1H NMR (600 MHz, DMSO-d6) δ 10.56 (s, 1H), 10.07 (s, 1H), 8.36 (s, 1H), 7.66 (d, J = 9.0 Hz, 2H), 7.62 (d, J = 6.6 Hz, 2H), 7.44 (t, J = 7.8 Hz, 2H), 7.24 (t, J = 7.2 Hz, 1H), 7.26 (d, J = 8.4 Hz, 2H), 4.91 (s 1H); HRMS (ESI−) calcd. for C16H13N3O3 [M−H]− 294.0957, found 294.0961.
3-Hydroxy-6-((4'-methyl-[1,1'-biphenyl]-4-yl)amino)pyrimidine-2,4(1H,3H)-dione (11b)
1H NMR (600 MHz, CD3OD) 7.63 (d, J = 7.8 Hz, 2H), 7.50 (d, J = 7.8 Hz, 1H), 7.29 (d, J = 8.4 Hz, 2H), 7.24 (d, J = 7.8 Hz, 2H), 2.38 (s, 3H); HRMS (ESI−) calcd. for C17H15N3O3 [M−H]− 308.1035, found 308.1035.
3-Hydroxy-6-((4'-hydroxy-[1,1'-biphenyl]-3-yl)amino)-1-methylpyrimidine-2,4(1H,3H)-dione (11c)
1H NMR (600 MHz, CD3OD) δ 7.50 (d, J = 8.4 Hz, 2H), 7.46 (d, J = 8.4 Hz, 2H), 7.43 (s, 1H), 7.27 (d, J = 7.8 Hz, 2H), 7.17 (m, 1H), 5.09 (s, 1H), 2.39 (s, 3H); HRMS (ESI−) calcd. for C17H15N3O3 [M−H]− 308.1035, found 308.1042.
6-((4'-Ethyl-[1,1'-biphenyl]-4-yl)amino)-3-hydroxypyrimidine-2,4(1H,3H)-dione (11d)
1H NMR (600 MHz, DMSO-d6) δ δ 10.05 (s, 1H), 8.46 (s, 1H), 7.66 (d, J = 9.0 Hz, 2H), 7.57 (m, 2H), 7.30 (m, 4H), 4.89 (s, 1H), 2.66 (q, J = 7.8Hz, 2H), 1.22 (t, J = 7.2 Hz, 3H); HRMS (ESI−) calcd. for C18H17N3O3 [M−H]− 322.1197, found 322.1200.
3-Hydroxy-6-((4'-(trifluoromethyl)-[1,1'-biphenyl]-4-yl)amino)pyrimidine-2,4(1H,3H)-dione (11e)
1H NMR (600 MHz, DMSO-d6) δ 10.04 (s, 1H); 8.64 (s, 1H), 7.89 (m,, 2H), 7.79 m, 2H), 7.75 (m, 2H), 7.32 (m, 2H), 4.94 (s, 1H); HRMS (ESI−) calcd. for C17H12F3N3O3 [M−H]− 362.0758, found362.0761.
3-Hydroxy-6-((4'-hydroxy-[1,1'-biphenyl]-4-yl)amino)pyrimidine-2,4(1H,3H)-dione (11f)
1H NMR (600 MHz, DMSO-d6) δ 10.61 (s, 1H), 10.01 (s, 1H), 9.53 (s, 1H), 8.30 (s, 1H), 7.59-6.83 (m, 8H), 4.84 (s, 1H); 13C NMR (150 MHz, DMSO-d6) δ 160.8, 157.0, 149.6, 148.9, 136.5, 136.4, 130.2, 127.5, 126.8, 122.9, 115.7, 75.8; HRMS-ESI(−) m/z calcd for C16H13N3O4 310.0833 [M−H]−, found 310.0833.
3-Hydroxy-6-((3'-hydroxy-[1,1'-biphenyl]-4-yl)amino)pyrimidine-2,4(1H,3H)-dione (11g)
1H NMR (600 MHz, DMSO-d6) δ 10.72 (s, 1H), 10.06 (s, 1H), 9.52 (s, 1H), 8.53 (s, 1H), 7.61-6.75 (m, 8H), 4.92 (s, 1H); 13C NMR (150 MHz, DMSO-d6) δ 160.9, 157.8, 149.8, 149.3, 140.8, 137.6, 136.2, 129.9, 127.5, 122.5, 117.2, 114.3, 113.2, 76.1; HRMS-ESI(−) m/z calcd for C16H13N3O4 310.0833 [M−H]−, found 310.0838.
3-Hydroxy-6-((2'-hydroxy-[1,1'-biphenyl]-4-yl)amino)pyrimidine-2,4(1H,3H)-dione (11h)
1H NMR (600 MHz, DMSO-d6) δ 10.58 (s, 1H), 10.00 (s, 1H), 9.52 (s, 1H), 8.29 (s, 1H), 7.55-6.84 (m, 8H), 4.86 (s, 1H); 13C NMR (150 MHz, DMSO-d6): δ 160.0, 154.3, 149.7, 148.9, 136.4, 134.8, 130.1, 130.0, 128.4, 126.9, 122.0, 119.4, 116.0, 75.8; HRMS-ESI(−) m/z calcd for C16H13N3O4 310.0833 [M−H]−, found 310.0838.
3-Hydroxy-6-((3'-methoxy-[1,1'-biphenyl]-4-yl)amino)pyrimidine-2,4(1H,3H)-dione (11i)
1H NMR (600 MHz, DMSO-d6) δ 10.61 (s, 1H), 10.07 (s, 1H), 8.37 (s, 1H), 7.66 (d, J = 8.4 Hz, 2H), 7.35 (d, J = 8.4 Hz, 1H), 7.25 (d, J = 8.4 Hz, 2H), 7.20 (d, J = 8.4 Hz, 1H), 7.15(s, 1H), 6.90 (d, J = 8.4 Hz, 1H), 4.83 (s 1H), 3.80 (s, 3H); HRMS (ESI−) calcd. for C17H15N3O4 [M−H]− 324.1063, found 324.1070.
3-Hydroxy-6-((4'-methoxy-[1,1'-biphenyl]-3-yl)amino)pyrimidine-2,4(1H,3H)-dione (11j)
1H NMR (600 MHz, DMSO-d6) δ 10.62 (s, 1H), 10.17 (s, 1H), 8.36 (s, 1H), 7.56 (d, J = 8.4 Hz, 2H), 7.41 (d, J = 7.8 Hz, 2H), 7.37 (m, 1H), 7.12 (d, J = 7.8 Hz, 1H), 7.00 (d, J = 8.4 Hz, 2H), 4.83 (s 1H),;3.78 (s, 3H), HRMS (ESI−) calcd. for C17H15N3O4 [M−H]− 324.1063, found 324.1071.
6-((3',5'-Dimethoxy-[1,1'-biphenyl]-4-yl)amino)-3-hydroxypyrimidine-2,4(1H,3H)-dione (11k)
1H NMR (600 MHz, DMSO-d6) δ 10.47 (s, 1H), 9.84 (s, 1H), 8.21 (s, 1H), 7.50 (d, J = 9.0 Hz, 2H), 7.09 (d, J = 8.4 Hz, 2H), 6.59 (d, J = 8.4 Hz, 2H), 6.30 (t, J = 8.4 Hz, 1H), 4.70 (s, 1H), 3.61 (s, 6H); HRMS (ESI−) calcd. for C18H17N3O5 [M−H]− 354.1095, found 354.1089.
6-((2',4'-Dimethoxy-[1,1'-biphenyl]-3-yl)amino)-3-hydroxypyrimidine-2,4(1H,3H)-dione (11l)
1H NMR (600 MHz, DMSO-d6) δ 10.63 (s, 1H), 10.07 (s, 1H), 8.41 (s, 1H), 7.45 (t, J = 7.8 Hz, 1H), 7.39 (s, 1H), 7.30 (d, J = 9.0 Hz, 1H), 7.25 (d, J = 7.8 Hz, 1H), 7.16 (d, J = 7.8 Hz, 1H), 6.73 (s, 1H), 6.69 (d, J = 7.8 Hz, 1H), 5.04 (s, 1H), 3.86 (s, 6H); HRMS (ESI−) calcd. for C18H17N3O5 [M−H]− 354.1095, found 354.1101.
6-((2',6'-Dimethoxy-[1,1'-biphenyl]-3-yl)amino)-3-hydroxypyrimidine-2,4(1H,3H)-dione (11m)
1H NMR (600 MHz, DMSO-d6) δ 10.54 (s, 1H), 9.98 (s, 1H), 8.25 (s, 1H), 7.35 (t, J = 7.8 Hz, 1H), 7.36 (t, J = 7.8 Hz, 1H), 7.08 (m, 2H), 7.03 (d, J = 7.8 Hz, 1H), 6.74 (d, J = 7.8 Hz, 2H), 4.91 (s, 1H), 3.72 (s, 6H); HRMS (ESI−) calcd. for C18H17N3O5 [M−H]− 354.1095, found 354.1089.
4'-((1-Hydroxy-2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)amino)-[1,1'-biphenyl]-4-carbonitrile (11n)
1H NMR (600 MHz, DMSO-d6) δ 10.72 (s, 1H), 10.05 (s, 1H), 8.55 (s, 1H), 7.90 (d, J = 7.8 Hz, 2H), 7.51 (d, J = 8.4 Hz, 2H), 7.77 (d, J = 8.4 Hz, 2H), 7.31 (d, J = 7.8 Hz, 2H), 4.96 (s, 1H); HRMS (ESI−) calcd. for C17H12N4O3 [M−H]− 319.0837, found 319.0831.
6-((4'-Fluoro-[1,1'-biphenyl]-3-yl)amino)-3-hydroxypyrimidine-2,4(1H,3H)-dione (11o)
1HNMR (600 MHz, DMSO-d6) δ 10.78 (s, 1H), 10.07 (s, 1H), 8.34 (s, 1H), 7.68 (m, 2H), 7.45 (m, 1H), 7.40 (m, 1H), 7.29 (t, J = 8.4 Hz, 2H), 7.19 (d, J = 7.8 Hz, 1H), 4.84 (s 1H); HRMS (ESI−) calcd. for C16H12FN3O3 [M−H]− 312.0863, found 312.0870.
6-((4'-Chloro-[1,1'-biphenyl]-4-yl)amino)-3-hydroxypyrimidine-2,4(1H,3H)-dione (11p)
1H NMR (600 MHz, DMSO-d6) δ 10.67 (s, 1H), 10.03 (s, 1H), 8.42 (s, 1H), 7.70-7.28 (m, 8H), 4.91 (s, 1H); 13C NMR (150 MHz, DMSO-d6) δ 160.8, 149.5, 149.1, 138.2, 138.0, 134.5, 132.1, 128.9, 128.1, 127.6, 122.4, 76.4; HRMS-ESI(−) m/z calcd for C16H12ClN3O3 [M−H]−328.0494, found 328.0502.
6-((4'-Chloro-[1,1'-biphenyl]-3-yl)amino)-3-hydroxypyrimidine-2,4(1H,3H)-dione(11q)
1H NMR (600 MHz, DMSO-d6) δ 10.73 (s, 1H), 10.03 (s, 1H), 8.37 (s, 1H), 7.69-7.22 (m, 8H), 4.87 (s, 1H); 13C NMR (150 MHz, DMSO-d6) δ 160.8, 149.7, 148.9, 140.1, 138.8, 138.3, 132.6, 130.1, 128.9, 128.5,122.8, 121.8, 120.6, 76.2; HRMS-ESI(−) m/z calcd for C16H12ClN3O3 [M−H]− 328.0494, found 328.0500.
Biology
Reagents
Biologicals
Recombinant HIV-1 reverse transcriptase (RT) was expressed and purified as previously described.26 The catalytically active RNase H domain fragment of HIV-1 RT was expressed from plasmid pCSR231 (a generous gift from Dr. Daria Hazuda, Merck, West Point, PA) and purified as previously described.27 P4R5 HIV infection indicator cells were obtained from the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH (p4R5.MAGI from Dr. Nathaniel Landau). These cells express CD4, CXCR4 and CCR5 as well as a β-galactosidase reporter gene under the control of an HIV LTR promoter.
Chemicals
DNA and RNA oligonucleotides for the preparation of RNA/DNA duplexes for assay of RNase H activity were purchased from Trilink (San Diego, CA).
RNase H assay
RNase H activity was measured essentially as previously described.28 Three different RNA/DNA duplex substrates were used, each assessing a different mode of RNAse H cleavage. HTS-1 (RNA 5’-gaucugagccugggagcu -3’-fluorescein annealed to DNA 3’-CTAGACTCGGACCCTCGA -5’-Dabcyl) is a high sensitivity duplex that assesses non-specific internal cleavage. HTS-2 (RNA 5’–cugguuagaccagaucugagccugggagcu–3’-fluorescein annealed to DNA 3’-GGTCTAGACTCGGACCCTCGA–5’-Dabcyl) provides a duplex with a recessed DNA 3’-terminus and measures 3’-DNA directed or polymerase directed RNase H cleavage. HTS-3 (RNA 5’–accagaucugagccugggagcu–3-fluorescein annealed to DNA 3’-GACCAATCTGGTCTAGACTCGGACCCTCGA–5’-Dabcyl) measures 5’-RNA-directed RNase H cleavage.
RT polymerase assay
HIV RT polymerase activity was determined in the presence and the absence of inhibitor using 10 µM [3H]-TTP and 40nM poly(rA)-oligo(dT)16 (both obtained from Perkin Elmer, Waltham, MA) in 50 mM Tris-HCl, pH 7.4 (37°C) containing 60 mM KCl and 5 mM MgCl2. Reactions were initiated by the addition of 10 nM WT or mutant RT and carried out for 20 min at 37°C. Reactions were quenched by 200 µl ice cold 10% TCA containing 20 mM sodium pyrophosphate and filtered using a 1.2 µm glass fiber filter 96-well plates (Millipore, Billerica, MA) followed by sequentially wash with 10% TCA and ethanol. The extent of radionucleotide incorporation was determined by liquid scintillation spectrometry.
HIV IN assay
HIV integrase was expressed and purified as previously reported.29 Inhibition assays were performed using a modified protocol of our reported method.29 Briefly, 2.1 µL of compound suspended in DMSO was placed in duplicate into a Black 96 well non-binding plate (corning 3991). Compounds were plated in duplicate to a final concentration of 0.13 — 100 µM. To each well of the plate 186.9 µL of reaction mixture without DNA substrate was added (10 mM HEPES pH 7.5, 10 % glycerol w/v, 10 mM MnCl2, 1 mM DTT, 1 µM integrase). The enzyme was incubated with inhibitor for 10 min at 25 °C after which the reaction was initiated by the addition of 21 µL of 500 nM oligo (5’ biotin ATGTGGAAAATCTCTAGCA annealed with ACTGCTAGAGATTTTCCACAT 3’ Cy5). Reactions were incubated at 37 °C for 30 min and then quenched by the addition of 5.2 µL 500 mM EDTA. Each reaction was moved (200 µL) to a MultiScreen HTS PCR plate (Millipore MSSLBPC10) containing 20 µL streptavidin agarose beads (Life Technologies S951) and incubated with shaking for 30 min. A vacuum manifold was used to remove the reaction mixture and the beads were similarly washed 3 times with wash buffer (0.05% SDS, 1 mM EDTA in PBS). The plates were further washed 3 times with 200 µL 50 mM NaOH to denature DNA not covalently linked to the biotin modification. For each denaturation step the plate was incubated with shaking at 25 °C for 5 min and the NaOH was removed by centrifugation at 1000 g for 1 min. The reaction products were eluted from the beads by the addition of 150 µL formamide. The plate was incubated at 25 °C for 10 min and read directly at 635/675 in a SpectraMax i3 plate reader (Molecular Devices).
Antiviral assays
Antiviral assays were carried out using P4R5 indicator cells essentially as previously described.30 P4R5 cells were cultured in 96-well microplates (5×103 cells per well and maintained in DMEM/10% FBS supplemented with puromycin (0.5 µg/ml). Cells were incubated in the presence or the absence of drug for 16h then exposed to HIV followed by an additional incubation period of 48h. The extent of infection was assessed using a fluorescence-based β-galactosidase detection assay, as previously described.31
Molecular modeling
The chemical structure of compound 11i was drawn in ChemBioDraw Ultra 14.0 and minimized in ChemBio3D Ultra 14.0 (CambridgeSoft/PerkinElmer). Compound 11i was manually docked in the RNase H active site of a published crystal structure of HIV-1 RT in complex with Nevirapine and an active site-directed RNase H inhibitor (PDB ID: 3LP125) using Coot.32 No steric clashes were found upon inspection.
Supplementary Material
Acknowledgments
This research was supported by the National Institutes of Health (AI100890 to SGS, MAP and ZW) and partially by the Center for Drug Design, University of Minnesota.
ABBREVIATIONS USED
- HIV
human immunodeficiency virus
- RT
reverse transcriptase
- RNase H
ribonuclease H
- pol
polymerase
- IN
integrase
- ST
strand transfer
- HPD
3-hydroxypyrimidine-2,4-dione
- SAR
structure-activity-relationship
- PR
protease
- HAART
highly active antiretroviral therapy
- NRTIs
nucleoside RT inhibitors
- NNRTIs
nonnucleoside RT inhibitors
- DKA
diketoacid
- HID
2-hydroxyisoquinolinedione
- RISF
retroviral integrase superfamily
Footnotes
ASSOCIATED CONTENT
Supporting Information Available. Characterization data, including 1H NMR, 13C NMR and HRMS data, of intermediate chemotypes 17, 18 and 19. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hill M, Tachedjian G, Mak J. The packaging and maturation of the HIV-1 pol proteins. Curr. HIV Res. 2005;3:73–85. doi: 10.2174/1570162052772942. [DOI] [PubMed] [Google Scholar]
- 2.Richman DD. HIV chemotherapy. Nature. 2001;410:995–1001. doi: 10.1038/35073673. [DOI] [PubMed] [Google Scholar]
- 3. http://www.unaids.org/en/dataanalysis/datatools/aidsinfo/ [Google Scholar]
- 4.Beilhartz GL, Gotte M. HIV-1 Ribonuclease H: Structure, Catalytic Mechanism and Inhibitors. Viruses. 2010;2:900–926. doi: 10.3390/v2040900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sarafianos SG, Marchand B, Das K, Himmel DM, Parniak MA, Hughes SH, Arnold E. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J. Mol. Biol. 2009;385:693–713. doi: 10.1016/j.jmb.2008.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cihlar T, Ray AS. Nucleoside and nucleotide HIV reverse transcriptase inhibitors: 25 years after zidovudine. Antiviral Res. 2010;85:39–58. doi: 10.1016/j.antiviral.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 7.de Bethune MP. Non-nucleoside reverse transcriptase inhibitors (NNRTIs), their discovery, development, and use in the treatment of HIV-1 infection: a review of the last 20 years (1989–2009) Antiviral Res. 2010;85:75–90. doi: 10.1016/j.antiviral.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 8.Parniak MA. Unpublishd data. [Google Scholar]
- 9.Nowotny M. Retroviral integrase superfamily: the structural perspective. EMBO Rep. 2009;10:144–151. doi: 10.1038/embor.2008.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao L, Song W, De Clercq E, Zhan P, Liu X. Recent progress in the research of small molecule HIV-1 RNase H inhibitors. Curr. Med. Chem. 2014;21:1956–1967. doi: 10.2174/0929867321666140120121158. [DOI] [PubMed] [Google Scholar]
- 11.Klumpp K, Hang JQ, Rajendran S, Yang YL, Derosier A, PW K, Overton H, Parkes KEB, Cammack N, Martin JA. Two-metal ion mechanism of RNA cleavage by HIV RNase H and mechanism-based design of selective HIV RNase H inhibitors. Nucleic Acids Res. 2003;31:6852–6859. doi: 10.1093/nar/gkg881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Himmel DM, Maegley KA, Pauly TA, Bauman JD, Das K, Dharia C, Clark AD, Jr, Ryan K, Hickey MJ, Love RA, Hughes SH, Bergqvist S, Arnold E. Structure of HIV-1 reverse transcriptase with the inhibitor beta-Thujaplicinol bound at the RNase H active site. Structure. 2009;17:1625–1635. doi: 10.1016/j.str.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Himmel DM, Myshakina NS, Ilina T, Van Ry A, Ho WC, Parniak MA, Arnold E. Structure of a dihydroxycoumarin active-site inhibitor in complex with the RNase H domain of HIV-1 reverse transcriptase and structure-activity analysis of inhibitor analogs. J. Mol. Biol. 2014;426:2617–2631. doi: 10.1016/j.jmb.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaw-Reid CA, Munshi V, Graham P, Wolfe A, Witmer M, Danzeisen R, Olsen DB, Carroll SS, Embrey M, Wai JS, Miller MD, Cole JL, Hazuda DJ. Inhibition of HIV-1 ribonuclease H by a novel diketo acid, 4-[5-(benzoylamino)thien-2-yl]-2,4-dioxobutanoic acid. J. Biol. Chem. 2003;278:2777–2780. doi: 10.1074/jbc.C200621200. [DOI] [PubMed] [Google Scholar]
- 15.Kirschberg TA, Balakrishnan M, Squires NH, Barnes T, Brendza KM, Chen X, Eisenberg EJ, Jin W, Kutty N, Leavitt S, Liclican A, Liu Q, Liu X, Mak J, Perry JK, Wang M, Watkins WJ, Lansdon EB. RNase H active site inhibitors of human immunodeficiency virus type 1 reverse transcriptase: design, biochemical activity, and structural information. J. Med. Chem. 2009;52:5781–5784. doi: 10.1021/jm900597q. [DOI] [PubMed] [Google Scholar]
- 16.Williams PD, Staas DD, Venkatraman S, Loughran HM, Ruzek RD, Booth TM, Lyle TA, Wai JS, Vacca JP, Feuston BP, Ecto LT, Flynn JA, DiStefano DJ, Hazuda DJ, Bahnck CM, Himmelberger AL, Dornadula G, Hrin RC, Stillmock KA, Witmer MV, Miller MD, Grobler JA. Potent and selective HIV-1 ribonuclease H inhibitors based on a 1-hydroxy-1,8-naphthyridin-2(1H)-one scaffold. Bioorg. Med. Chem. Lett. 2010;20:6754–6757. doi: 10.1016/j.bmcl.2010.08.135. [DOI] [PubMed] [Google Scholar]
- 17.Beilhartz GL, Ngure M, Johns BA, Deanda F, Gerondelis P, Gotte M. Inhibition of the Ribonuclease H Activity of HIV-1 Reverse Transcriptase by GSK5750 Correlates with Slow Enzyme-Inhibitor Dissociation. J. Biol. Chem. 2014;289:16270–16277. doi: 10.1074/jbc.M114.569707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corona A, Di Leva FS, Thierry S, Pescatori L, Crucitti GC, Subra F, Delelis O, Esposito F, Rigogliuso G, Costi R, Cosconati S, Novellino E, Di Santo R, Tramontano E. Identification of Highly Conserved Residues Involved in Inhibition of HIV-1 RNase H Function by Diketo Acid Derivatives. Antimicrob. Agents Chemother. 2014;58:6101–6110. doi: 10.1128/AAC.03605-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vernekar SK, Liu Z, Nagy E, Miller L, Kirby KA, Wilson DJ, Kankanala J, Sarafianos SG, Parniak MA, Wang Z. Design, synthesis, biochemical, and antiviral evaluations of C6 benzyl and C6 biarylmethyl substituted 2-hydroxylisoquinoline-1,3-diones: dual inhibition against HIV reverse transcriptase-associated RNase H and polymerase with antiviral activities. J. Med. Chem. 2015;58:651–664. doi: 10.1021/jm501132s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang J, Maddali K, Dreis CD, Sham YY, Vince R, Pommier Y, Wang Z. N-3 Hydroxylation of Pyrimidine-2,4-diones Yields Dual Inhibitors of HIV Reverse Transcriptase and Integrase. ACS Med. Chem. Lett. 2011;2:63–67. doi: 10.1021/ml1002162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang J, Maddali K, Dreis CD, Sham YY, Vince R, Pommier Y, Wang ZQ. 6-Benzoyl-3-hydroxypyrimidine-2,4-diones as dual inhibitors of HIV reverse transcriptase and integrase. Bioorg. Med. Chem. Lett. 2011;21:2400–2402. doi: 10.1016/j.bmcl.2011.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang J, Maddali K, Metifiot M, Sham YY, Vince R, Pommier Y, Wang ZQ. 3-Hydroxypyrimidine-2,4-diones as an Inhibitor Scaffold of HIV Integrase. J. Med. Chem. 2011;54:2282–2292. doi: 10.1021/jm1014378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adrian G, Mignonac S, Lecoutteux F. Preparation of 5-(1-methylethyl)-6-(phenylmethyl)pyrimidine-2,4(1H,3H)-dione. WO 2000003999. 2000 [Google Scholar]
- 24.Hazuda DJ, Felock P, Witmer M, Wolfe A, Stillmock K, Grobler JA, Espeseth A, Gabryelski L, Schleif W, Blau C, Miller MD. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science. 2000;287:646–650. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- 25.Su H-P, Yan Y, Prasad GS, Smith RF, Daniels CL, Abeywickrema PD, Reid JC, Loughran HM, Kornienko M, Sharma S, Grobler JA, Xu B, Sardana V, Allison TJ, Williams PD, Darke PL, Hazuda DJ, Munshi S. Structural Basis for the Inhibition of RNase H Activity of HIV-1 Reverse Transcriptase by RNase H Active Site-Directed Inhibitors. J. Virol. 2010;84:7625–7633. doi: 10.1128/JVI.00353-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fletcher RS, Holleschak G, Nagy E, Arion D, Borkow G, Gu Z, Wainberg MA, Parniak MA. Single-step purification of recombinant wild-type and mutant HIV-1 reverse transcriptase. Protein Expres. Purif. 1996;7:27–32. doi: 10.1006/prep.1996.0004. [DOI] [PubMed] [Google Scholar]
- 27.Gong Q, Menon L, Ilina T, Miller LG, Ahn J, Parniak MA, Ishima R. Interaction of HIV-1 reverse transcriptase ribonuclease H with an acylhydrazone inhibitor. Chem. Biol. Drug Des. 2011;77:39–47. doi: 10.1111/j.1747-0285.2010.01052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parniak MA, Min KL, Budihas SR, Le Grice SF, Beutler JA. A fluorescence-based high-throughput screening assay for inhibitors of human immunodeficiency virus-1 reverse transcriptase-associated ribonuclease H activity. Anal. Biochem. 2003;322:33–39. doi: 10.1016/j.ab.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 29.Wang Z, Bennett EM, Wilson DJ, Salomon C, Vince R. Rationally designed dual inhibitors of HIV reverse transcriptase and integrase. J. Med. Chem. 2007;50:3416–3419. doi: 10.1021/jm070512p. [DOI] [PubMed] [Google Scholar]
- 30.Sirivolu VR, Vernekar SKV, Ilina T, Myshakina NS, Parniak MA, Wang ZQ. Clicking 3 '-Azidothymidine into Novel Potent Inhibitors of Human Immunodeficiency Virus. J. Med. Chem. 2013;56:8765–8780. doi: 10.1021/jm401232v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abram ME, Parniak MA. Virion instability of human immunodeficiency virus type 1 reverse transcriptase (RT) mutated in the protease cleavage site between RT p51 and the RT RNase H domain. J. Virol. 2005;79:11952–11961. doi: 10.1128/JVI.79.18.11952-11961.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.