ABSTRACT
Plants have developed highly efficient and remarkable mechanisms to survive under frequent and extreme environmental stress conditions. Exposure of plants to various stress factors is associated with coordinated changes in gene expression at the transcriptional level and hence transcription factors, such as those belonging to the MYB family play a central role in triggering the right responses. MYB transcription factors have been extensively studied in regard of their involvement in the regulation of a number of such stress responses in plants. Genetic and molecular biological studies, primarily in Arabidopsis, have also begun to unravel the role of MYB transcription factors in the epigenetic regulation of stress responses in plants. This review focuses on the role of MYB transcription factors in the regulation of various stress responses in general, highlighting on recent advances in our understanding of the involvement of this class of transcription factors in epigenetic regulation of stress response in plant genome.
KEYWORDS: Abiotic stress, crop improvement, epigenetic regulation, MYB transcription factor, plant stress response
Introduction
Understanding the mechanisms of plant responses, tolerance and adaptation toward abiotic stress is crucial for the development of stress tolerance varieties in crop plants. Therefore, in depth knowledge on plant stress response mechanisms and transfer of gained information from the coordinated experimental and additional in silico approaches have received much attention in plant biology research for last couple of years. Plants have developed various efficient strategies to respond, adapt and survive under stress situations. Plant response toward various stresses, particularly abiotic stress such as UV-B radiation, high soil salinity, drought and low temperature involve rapid and coordinated changes at the transcriptional level of the genome. Several transcription factors, belonging to diverse families, have been found to play crucial role in stress signaling, either by acting as positive or negative regulators of stress responsive genes. Therefore, understanding the transcriptional response of plants under stress has remain the subject of extensive investigation for better understanding plant growth and developmental pattern in the context of global climate change.
Transcription factors (TFs) generally act as key regulators of gene expression. In general, the transcription factors with 2 distinct domains, a DNA binding domain and a transcriptional activation/repression domain, regulate diverse cellular processes via governing the transcriptional rates of target genes. Arabidopsis genome sequence (Arabidopsis Genome Initiative, 2000) have led to the identification of more than 1600 transcription factor genes, contributing approximately up to 6%; of the total number of genes (Gong et al. 2004). Based on the DNA binding domains, genes for several transcription factors, such as MYC, MYB, MADS, bZIP, BHLH etc. have been characterized and assigned into different families and superfamilies. Previous studies have demonstrated important role of different families of transcription factors, including AP2/ERF, bZIP, Zn-finger, NAC, MYB, and WRKY in the regulation of in abiotic stress tolerance in plants (Fig. 1).14
Figure 1.
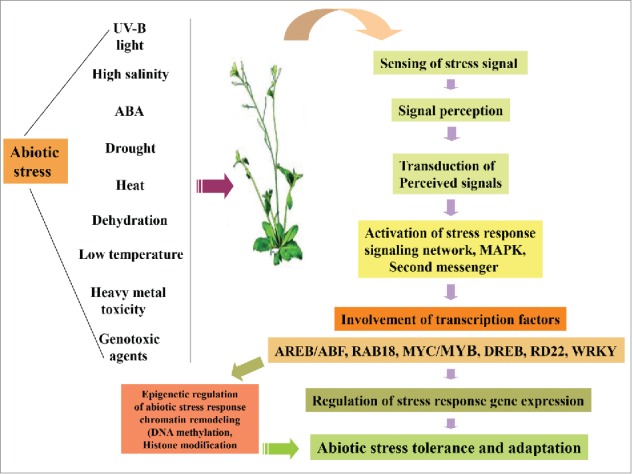
Abiotic stress response and transcriptional regulation in plants. Schematic representation illustrating exposure of plants toward different abiotic stress factors and the subsequent signal sensing, perception and transduction through sensors and associated signaling networks which result in the transcriptional activation of stress response genes through the involvement of various transcription factors including the MYB domain proteins. The epigenetic regulation of abiotic stress response via the activity of transcription factors has been indicated.
MYB domain proteins act as DNA-binding transcription factors
The MYB family represents one of the large, functionally diverse classes of proteins, found in all eukaryotes. In general, most of the MYB proteins function as transcription factors and are characterized by the presence of variable numbers of N-terminus conserved MYB repeats (R), mainly associated with DNA-binding and protein-protein interactions. The variable C-terminal region is responsible for modulating the regulatory activity of the protein. Several members of this family have been identified in Arabidopsis, rice, maize, and soybean and shown to be involved in regulating various cellular processes, including cell cycle and cell morphogenesis, biotic and abiotic stress responses11; Jin and Martin 1999.38
Since the initial finding of the first MYB-like gene CI from Zea mays, involved in anthocyanin biosynthesis in kernels,24 plant genome has been found to contain large MYB gene families. The classification criterion used for MYB proteins is based on the number of repeats present in their sequences, varying from one to 4. Each repeat generally comprises of about 52 amino acid residues, forming 3 α-helices. The second and third helices in each repeat are involved in the formation of a helix-turn-helix (HTH) fold with 3 tryptophan residues spaced regularly, producing a hydrophobic core in the 3-dimensional HTH structure.22 Therefore, based on the number of MYB domains, the MYB protein family has been classified into 4 different groups, such as 1R-, R2R3-, 3R- and 4RMYB proteins, respectively.3,11,38
In plants, majority of the MYB proteins belong to the R2R3-MYB subfamily, involved in regulation of multiple responses, such as biotic and abiotic stresses,32,18 hormone signaling,2 phenylpropanoid biosynthesis,13 determination of cell shape21 and regulation of differentiation,45 More than over 100 R2R3-MYB members have been reported in dicots and monocots.44 Based on the conservation of the DNA binding domain and the amino acid motifs at the C-terminal domain, the members of R2R3-type MYB domain proteins have been classified into 23 subgroups.11 In plants, whereas the 3R-MYB class constitutes a very small group with only 5 members, MYB transcription factors with 4 R1/R2-like repeats are generally not commonly found. Only one member of such type has been reported from different plant species. Furthermore, MYB proteins with a single or a partial repeat, known as ‘MYB-related’ have been recognized and grouped into different subclasses, depending on the presence of particular repeats. The MYB-related proteins are involved in various processes, including control of cellular and organ morphogenesis, secondary metabolism and circadian rhythm, respectively.5
MYB transcription factors regulate abiotic stress response in plants
Genetic and molecular approaches have facilitated extensive functional characterization of MYB domain proteins, particularly the R2R3-type members in various plant species, including the crop plants like rice, maize, soybean etc. A genome wide comparative analysis of MYB genes and their expression in Arabidopsis and rice have indicated potential role of several MYB domain proteins in plant stress responses.16 Several members of R2R3-type MYB transcription factors are involved in the regulation of phenylpropanoid pathway which produces various secondary metabolic compounds involved in abiotic stress response in plants. Among the various secondary metabolites produced in plants, the sinapate esters and flavonoids act as key UV-B absorbing sunscreen compounds to protect plants against the harmful effects of UV-radiation. Plants produce higher levels of UV-B absorbing compounds under low doses of UV-B to compromise the initial damage of the major UV-B targets like nucleic acids, proteins and lipids. Molecular and genetic analysis in Arabidopsis mutants impaired in UV-B response have revealed key role of flavonoids and phenolics in UV-B absorption, facilitating enhanced UV-B tolerance (Bieza and Lois 2001). Recent studies in Arabidopsis have indicated important role of MYB transcription factors in the regulation of biosynthesis of secondary metabolites involved in UV-B absorption in plants. Arabidopsis MYB4, a member of R2R3 subgroup, represses the transcription of the gene encoding cinnamate 4-hydroxylase, involved in hydroxycinnamate ester biosynthesis. The MYB4 loss-of-function mutant showed UV-B tolerance due to increased accumulation level of hydroxycinnamate esters, while MYB4 overexpression caused reduced level of UV-B absorbing compounds, resulting in UV-B hypersensitivity.15 Another R2R3 MYB protein in Arabidopsis, AtMYB7 has been shown to be involved in regulating accumulation of UV-B absorbing phenylpropanoid compounds. The atmyb7 mutants showed induction of several flavonoids biosynthetic genes. Interestingly, under UV-B stress, along with its own transcriptional repression, AtMYB4 was also found to inhibit AtMYB7 expression, which was consistent with the reduced flavonoids contents in atmyb4 mutant, indicating repression of flavonoids biosynthesis by AtMYB7 and functional involvement of both AtMYB4 and AtMYB7 in maintaining the balance of accumulation of UV-B absorbing compounds in plants.
In plants, the phytohormone ABA plays the central role in regulating the drought stress response through integrated coordination of a complicated gene regulatory system, facilitating plants to adjust under water deficit condition.10 For regulating stress adaptation, the ABA-dependent signaling pathways have been shown to be functional via the involvement of MYC and MYB transcription factors.1,30 Various MYB transcription factors, mainly in Arabidopsis and also in some other crop species have been characterized in recent years for their involvement in drought response. The expression of several MYB genes has been found to be drought regulated. Arabidopsis thaliana transcriptomic data analyses have revealed increased expression of about 51%; of MYB genes in response to drought while 41%; gene were found to be down-regulated.5,16,53 In Arabidopsis, various MYB transcription factor genes like AtMYB2, AtMYB74 and AtMYB102 showed enhanced expression under drought stress.2 Previous studies have revealed involvement of several MYB genes in drought stress and ABA signaling network, governing various plant specific responses under abiotic stress. The Arabidopsis R2R3-MYB transcription factor MYB96 has been shown to be involved in regulation of lateral root growth in response to drought stress via the ABA-auxin signaling network.34 Furthermore, overexpression of MYB96 caused enhanced drought tolerance. BcMYB1, a member of R2R3-type MYB protein family has been shown to be strongly induced under drought stress in Boea crassifolia. BcMYB1 message level was shown to be substantially increased following drought stress and also in presence of high salinity and polyethylene glycol (PEG), while marginal induction was observed under low temperature stress. Interestingly, exogenous application of ABA and methyl jasmonate showed only negligible effect on BcMYB1 transcript induction, suggesting role of BcMYB1 in regulation of drought-responsive genes via ABA-independent pathway and also probably do not participate in wound signaling.6 Another MYB transcription factor from sugarcane (Saccharum officinarum), ScMYBAS1–3 showed induced response under water-deficit condition and salinity stress.26
The MYB transcription factors AtMYB60 and AtMYB96 in Arabidopsis were shown to be functional through the ABA signaling network, regulating many plant specific responses like drought stress, disease resistance,35 and stomatal movement,9 respectively. AtMYB96, an R2R3-type MYB protein in Arabidopsis, regulates lateral root meristem activation under drought stress through an ABA-auxin signaling crosstalk pathway.34 In sugarcane (Saccharum officinarum), the stress-related MYB transcription factor gene, ScMYBAS1–3 showed enhanced expression in response to water-deficit and salt stress.26 In soyabean (Glycine max), 156 MYB genes (GmMYB) have been identified and 43 of such genes showed altered expression level following treatment with ABA, salt, dehydration and cold stress.17 More recent studies in rice (Oryza sativa) have established role of MYB gene OsMYBS3 in conferring cold tolerance (Su et al. 2010), while the R2R3-MYB gene, OsMYB2, was shown to be involved in salt, cold, and dehydration tolerance in rice.50 ABA, PEG, dehydration and H2O2 have been shown to distinctly induce the expression of a MYB-related transcription factor gene OsMYB48-1 in rice, while only marginal induction was detected under high salinity and low temperature stress. OsMYB48-1 overexpression in rice was found to confer enhanced tolerance toward drought and salinity stresses. Enhanced tolerance to abiotic stresses was suggested to be accomplished via regulation of stress-mediated ABA biosynthesis.46
Plants have developed elaborate and complex mechanisms to survive under abiotic stress conditions. Several line of evidences have implicated role of MYB protein in regulation of abiotic stress tolerance in plant genome. The MYB genes OsMYB2, OsMYB3R-2, OsMYB4, OsMYBS3, TaMYB2, TaMYB32, TaMYB56, TaMYB30 and TaMYB73 from rice and wheat genome have been identified as important component of plant abiotic stress response pathways.19,39,50,20 A wheat R2R3-MYB transcription factor gene, TaMYB19, has recently been characterized and shown to be involved in enhanced tolerance to abiotic stress in Arabidopsis via regulation of expression levels of a number of abiotic stress-related genes.51 SbMYB44, a member of R2R3 class of MYB transcription factor, has recently been isolated from the succulent halophyte, Salicornia brachiata. SbMYB44 message level showed increased accumulation in response to salinity, desiccation, high temperature, abscisic acid and salicylic acid treatments, while recombinant SbMYB44 protein showed binding to dehydration responsive elements like RD22 and MBS-1, indicating role of SbMYB44 in stress signaling.36 Several salt response genes, including 76 MYB family genes have been identified using high throughput sequencing technologies in sheepgrass, a close relative of wheat. Recent studies have led to the characterization of a MYB-related gene LcMYB1 which appeared to be involved in salt stress response through activation of the pathways independent of the classical DREB1A- and MYB2-mediated signaling pathway. In addition, LcMYB1 overexpression conferred improved salt tolerance in Arabidopsis.7
Interestingly, in addition to stress signals, recent studies have indicated the role of mircoRNA in the regulation of endogenous message levels of MYB transcription factors. In Arabidopsis, the MYB transcription factors MYB33 and MYB101 act as positive regulators of ABA responses during seed germination. However, in germinating seeds, ABA induces the accumulation of the microRNA 159 (miR159) via the activity of ABI3 (abscisic acid-insensitive 3) transcription factor. MYB33 and MYB101 transcripts were found to be cleaved by miR159. These observations were also supported by the ABA hypersensitivity in myb33 and myb101 null mutant lines and transgenic plants overexpressing miR159.29
Epigenetic regulation of stress response and involvement of MYB transcription factors
In eukaryotes, including plants, epigenetic regulation is accomplished through a complex signaling network, involving regulatory small RNA species and chromatin remodeling activities. The chromatin remodeling activities are mainly associated with DNA methylation and histone modifications.48 Epigenetic control of stress-induced phenotypic response in plants mainly involves regulation of gene expression. Multiple evidences have accumulated to indicate that along with the normal developmental processes and functions, plants have evolved complex gene regulatory mechanisms for responding to various environmental stresses.
Although several studies have indicated role of MYB transcription factors in regulation of abiotic stress responses in plant genome, its link with the epigenetic control remains largely unknown (Fig. 1). The v-myb oncogene of avian myeloblastosis virus (AMV) encodes the v-Myb transcription factor which has been shown to transform myelomonocytic cells via deregulation of the expression of specific target genes. The chicken mim-1 gene, a target of c-Myb gene, is not activated by v-Myb and contains a strong cell type-specific and Myb-inducible enhancer element in addition to a Myb-responsive promoter. Recent studies have shown that the Myb-dependent activation of the mim-1 gene is associated with extensive remodeling of the nucleosomal architecture at the enhancer region, indicating direct involvement of Myb induced remodeling of the chromatin organization at a pertinent target site.42 Joe Lipsick's group at the Stanford University has carried out much of the current work on epigenetic regulation by Myb protein and its interacting protein partners in Drosophila. Studies from this group have indicated that in Drosophila, absence of functional Myb gene causes death of larvae. However, Drosophila flies lacking a functional Myb gene and a functional Mip130 gene sometimes were found to survive to adulthood. Such type of viability was also observed in case of Myb/Mip120 and Myb/Mip40 double mutants. On the other hand, abnormal mitosis has been found in Drosophila Myb null mutant with irregular chromatin condensation, suggesting role of Myb protein in epigenetic regulation. More recent studies in Drosophila have shown that the Myb transcription factor, RBF (Retinoblastoma-family protein) and Mip130/LIN-9 tumor suppressor proteins exist in a conserved Myb-MuvB (MMB)/dREAM complex. The MYB transcription factor has been shown to be involved in regulating the in vivo expression of Polo kinase, a regulator of spindle pole assembly and function and key component of the spindle assembly checkpoint (SAC). The absence of both MYB and Mip130, or of both MYB and E2F2 transcriptional repressor function resulted in variegated phenotype where high or low levels of Polo kinase were found to be stably transmitted through successive cell divisions in the imaginal wing discs. Interestingly, restoration of MYB activity caused consistently high level of Polo expression as like in wild-type tissue. However, restoration of Mip130 or E2F2 compromised Polo expression. These observations have suggested epigenetic regulation of gene expression by MYB transcription factor and Mip130 or E2F2 in Drosophila in vivo.41
Plants have developed various mechanisms to survive under environmental stresses which are accomplished by means of alteration of expression level of some genes through the introduction of epigenetic modifications, such as DNA methylation. DNA methylation plays an important role in gene expression by increasing the RNA-directed DNA methylation (RdDM) of genes and by inducing some histone modifications.49 In maize, the flavonoid biosynthetic pathway associated genes encoding the structural enzymes and transcription regulators have been characterized at the genetic and molecular level. Multiple natural alleles and epialleles of these genes have also been identified. An R2R3 Myb transcription factor, encoded by pericarp color1 (p1) gene has been shown to control the biosynthesis of phlobaphenes, brick red flavonoids which accumulate in floral organs including pericarp, tassel and cob glumes, and silk in maize (Styles and Ceska 1989). The p1 comprises of approximately 100 natural alleles and epialleles and many of them may be identified based on their expression profiles in the floral organs. Studies based on expression pattern analysis and transgenic plants carrying promoter and coding region of p1 alleles have suggested involvement of epigenetic mechanism of regulation for the unique expression pattern of p1.8 However, the nature of such epigenetic mechanism is not well defined. In maize, gradual loss of DNA methylation has been shown to reduce epigenetic gene silencing from a tandemly arranged Myb gene,33 indicating epigenetic regulation of MYB gene function. In Mimulus guttatus ((yellow monkey-flower) differential regulation of a MYB transcription factor MYB MIXTA-like 8 has been correlated with the transgenerational epigenetic inheritance of trichome density.31 In pines (Pinus pinaster), transcriptional and epigenetic regulation during the zygotic embryo development was shown to associated with ˜10-fold and 5-fold up-regulated expression of MYB3 and MYB4-type transcription factors.40
Recent studies in soybean have indicated importance of epigenetic modification in the regulation of gene expression and plant development under salinity stress. Microarray analyses have identified 49 transcription factors in soybean. These transcription factors were found to be inducible by salinity stress. Further expression based analyses have demonstrated salinity stress mediated inducibility of 45 out of 49 transcription factors, and 10 of them showed upregulated expression in seedlings in response to the demethylation agent 5-aza-2-deoxycytidine. Salinity stress was found to affect the methylation level of 4 of these 10 transcription factors, including one MYB transcription factor, along with one b-ZIP and 2 AP2/DREB family members, respectively. ChIP analysis further revealed that the activation of 3 of the 4 DNA methylated transcription factors was associated with a higher level of histone H3K4 trimethylation and H3K9 acetylation, and a decreased level of H3K9 demethylation in the various regions of the promoter or coding regions of the corresponding genes, indicating key role of epigenetic modification in the activation or suppression of these transcription factors in the context of salinity stress tolerance in soybean.37
In plants, transcription factors and RNA-directed DNA methylation (RdDM) play important role in regulation of gene expression under abiotic stress. The Arabidopsis R2R3-MYB gene family member AtMYB74 is primarily regulated by RdDM at the transcriptional level in response to salt stress. Bisulphite sequencing studies have identified a target site of 24-nucleotides long siRNAs located in the region about 500 bp upstream of the transcription initiation site of AtMYB74. This target site for siRNA has been shown to be heavily methylated. Whereas the DNA methylation level in this region was found to be considerably reduced in wild type plants under salt stress, no change in the methylation level was detected in case of RdDM mutants. Furthermore, promoter deletion studies have indicated the importance of the siRNA target region for maintaining AtMYB74 expression patterns and also suggested that changes in the levels of the 5 24-nucleotides long siRNAs modulate the expression of AtMYB74 transcription factor through RdDM under salt stress.47
Plant development involves epigenetic regulation for modification of developmental fate. Genomic imprinting is one type of epigenetic regulation which causes monoallelic gene expression in a parent-of-origin dependent manner and is established by the differential epigenetic marking of parental alleles. Imprinting has been shown to be regulated via genome wide DNA demethylation in the central cell before fertilization and intend for repression of individual loci with the polycomb repressive complex 2 (PRC2). Imprinted loci are involved in processes associated with the developmental programs. In maize, among the 5 recently described potentially imprinted genes, transcripts from only one parental allele have been identified in the endosperm, encoding the homeodomain and MYB family of transcription factors. However information is still limited regarding the function of such imprinted genes in endosperm development.28 Genome-wide analyses have identified several nuclear proteins, like transcription factors and chromatin-related proteins as imprinted genes, which are commonly expressed in plant seed endosperms and exhibit preferential uniparental allelic expression. Several members of the MYB transcription factor family have also been identified as maternally expressed genes (MEGs), indicating their link with epigenetic regulation.43,12,25,4
Genome wide sequence analysis in Arabidopsis thaliana, yeast (Saccharomyces cerevisiae), Caenorhabditis elegans and Drosophila have indicated the presence of 3 families of histone deacetylase (HDAC) and 3 families of histone acetyltransferase (HAT) proteins. Plants, animals and fungi were found to possess a single member of each of 3 subfamilies of HATs.23 In silico analyses (http://string-db.org/) have indicated potential interaction of Arabidopsis Myb4 subgroup of transcription factor with other regulatory proteins, such as histone acetyltrasferase (HAC2, E1A/-CREB binding protein), histone acetyltrasferase CBP family 4 (HAC4) and p300/CBP acetyltrasferase-related protein. Active DNA demethylation plays an important role in epigenetic regulation in plants. Recent studies have identified a histone H3 acetyltransferase, IDM1 in Arabidopsis. IDM1 has been shown to regulate active DNA demethylation and found to be required for inhibiting DNA hypermethylation in highly homologous multicopy genes and other repetitive sequences which are generally targeted by Repressor of Silencing 1 and related 5-methylcytosine DNA glycosylases for active DNA demethylation. IDM1 appears to bind to methylated DNA at chromatin sites lacking histone H3K4 di- or trimethylation and acetylates H3, generating a chromatin environment permissible for 5-methylcytosine DNA glycosylases to become functional.27 In Arabidopsis, the SWI2/SNF2 chromatin remodeling ATPase BRAHMA (BRM) protein has been shown to be involved in regulating gene expression in plant development processes through interaction with different transcription factors including MYB,52 suggesting potential functional involvement of MYB transcription factors in epigenetic regulation through interaction with other protein partners.
Conclusions and outlook
Understanding epigenetic mechanisms in response to environmental stress and its probable implication in the genetic management of plants is one of the most important challenges for plant scientists. In plants, the MYB transcription factor family has been expanded, mainly through the large family of R2R3-MYB. Members of this family are key factors in regulatory networks controlling development, metabolism and responses to biotic and abiotic stresses in plant genome. Although, at present information is limited to understand role of MYB proteins in the epigenetic control of abiotic stress response in plants, some exciting observations have already started accumulating to shed new light on this field. Future work will lead to a better understanding of the epigenetic regulation by MYB genes and how this important family of transcription factors and their interacting partners are integrated in the network for epigenetic regulation of stress response in plants.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
I sincerely thank Prof. K.P. Das, Dept. of Chemistry, Bose Institute, Kolkata, India and Dr. Swarup Roy Choudhury, Donald Danforth Plant Science Center, St.Louis, Missouri, USA for the critical reading and corrections of the Manuscript. I apologize to those whose work could not be cited due to space limitations.
References
- 1.Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 1997; 9:1859-68; PMID:9368419; http://dx.doi.org/ 10.1105/tpc.9.10.1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003; 15:63-78; PMID:12509522; http://dx.doi.org/ 10.1105/tpc.006130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambawat S, Sharma P, Yadav NR, Yadav RC. MYB transcription factor genes as regulators for plant responses: an overview. Physiol Mol Biol Plants 2013; 19:307-21; PMID:24431500; http://dx.doi.org/ 10.1007/s12298-013-0179-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bai F, Settles AM. Imprinting in plants as a mechanism to generate seed phenotypic diversity. Front Plant Sci 2015; http://dx.doi.org/ 10.3389/fpls.2014.00780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldoni E, Genga A, Cominelli E. Plant MYB transcription factors: their role in drought response mechanisms. Int J Mol Sci 2015; 16:15811-51; PMID:26184177; http://dx.doi.org/ 10.3390/ijms160715811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen B-J, Wang Y, Hu Y-L, Wu Q, Lin Z-P. Cloning and characterization of a drought-inducible MYB gene from Boea crassifolia. Plant Sci 2005; 168:493-500; http://dx.doi.org/ 10.1016/j.plantsci.2004.09.013 [DOI] [Google Scholar]
- 7.Cheng L, Li X, Huang X, Maa T, Liang Y, Maa X, Peng X, Jia J, Chen S, Chen Y, et al.. Overexpression of sheepgrass R1-MYB transcription factor LcMYB1 confers salt tolerance in transgenic Arabidopsis. Plant Physiol Biochem 2013; 70:252-60; PMID:23800660; http://dx.doi.org/ 10.1016/j.plaphy.2013.05.025 [DOI] [PubMed] [Google Scholar]
- 8.Cocciolone SM, Nettleton D, Snook ME, Peterson T. Transformation of maize with the p1 transcription factor directs production of silk maysin, a corn earworm resistance factor, in concordance with a hierarchy of floral organ pigmentation. Plant Biotechnol J 2005; 3:225-35; PMID:17173622; http://dx.doi.org/ 10.1111/j.1467-7652.2005.00120.x [DOI] [PubMed] [Google Scholar]
- 9.Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, Leonhardt N, Dellaporta SL, Tonelli C. A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol 2005; 15:1196-200; PMID:16005291; http://dx.doi.org/ 10.1016/j.cub.2005.05.048 [DOI] [PubMed] [Google Scholar]
- 10.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Ann Rev Plant Biol 2010; 61:651-79; PMID:20192755; http://dx.doi.org/ 10.1146/annurev-arplant-042809-112122 [DOI] [PubMed] [Google Scholar]
- 11.Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci 2010; 15:573-81; PMID:20674465; http;//dx.doi.org/ 10.1146/annurev-arplant-042809-112122 [DOI] [PubMed] [Google Scholar]
- 12.Gehring M, Missirian V, Henikoff S. Genomic analysis of parent-of-origin allelic expression in Arabidopsis thaliana seeds. PLoS One 2011; 6:e23687; PMID:21858209; http://dx.doi.org/ 10.1371/journal.pone.0023687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hichri I, Barrieu F, Bogs J, Kappel C, Delrot S, Lauvergeat V. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J Exp Bot 2011; 62:2465-83; PMID:21278228; http://dx.doi.org/ 10.1093/jxb/erq442 [DOI] [PubMed] [Google Scholar]
- 14.Hirayama T, Shinozaki K. Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J 2010; 61:1041-52; PMID:20409277; http://dx.doi.org/ 10.1111/j.1365-313X.2010.04124.x [DOI] [PubMed] [Google Scholar]
- 15.Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C. Transcriptional repression by AtMYB4 controls production of UV- protecting sunscreens in Arabidopsis. EMBO J 2000; 19:6150-61; PMID:11080161; http://dx.doi.org/ 10.1093/emboj/19.22.6150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katiyar A, Smita S, Keshari S, Rajwanshi LR, Chinnusamy V, Bansal KC. Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genomics 2012; 13:544-63; PMID:23050870; http://dx.doi.org/ 10.1186/1471-2164-13-544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liao Y, Zou HF, Wang HW, Zhang WK, Ma B, Zhang JS. Soybean GmMYB76, GmMYB92, and GmMYB177 genes confer stress tolerance in transgenic Arabidopsis plants. Cell Res 2008; 18:1047-60; PMID:18725908; http://dx.doi.org/ 10.1038/cr.2008.280 [DOI] [PubMed] [Google Scholar]
- 18.Lippold F, Sanchez DH, Musialak M, Schlereth A, Scheible WR, Hincha DK, Udvardi MK. AtMyb41 regulates transcriptional and metabolic responses to osmotic stress in Arabidopsis. Plant Physiol 2009; 149:1761-72; PMID:19211694; http://dx.doi.org/ 10.1104/pp.108.134874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma QB, Dai XY, Xu YY, Guo J, Liu YJ, Chen N, Xiao J, Zhang D, Xu Z, Zhang X, et al.. Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol 2009; 150:244-56; PMID:19279197; http://dx.doi.org/ 10.1104/pp.108.133454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mao X, Jia D, Li A, Zhang H, Tian S, Zhang X, Jia J, Jing R. Transgenic expression of TaMYB2A confers enhanced tolerance to multiple abiotic stresses in Arabidopsis. Funct Integrat Genomics 2011; 11:445-65; PMID:21472467; http://dx.doi.org/ 10.1007/s10142-011-0218-3 [DOI] [PubMed] [Google Scholar]
- 21.Noda KI, Glover BJ, Linstead P, Martin C. Flower colour intensity depends on specialized cell shape controlled by a Myb related transcription factor. Nature 1994; 369:661-4; PMID:8208293; http://dx.doi.org/ 10.1038/369661a0 [DOI] [PubMed] [Google Scholar]
- 22.Ogata K, Kanei-Ishii C, Sasaki M, Hatanaka H, Nagadoi A, Enari M, Nakamura H, Nishimura Y, Ishii S, Sarai A. The cavity in the hydrophobic core of Myb DNA binding domain is reserved for DNA recognition and transactivation. Nat Struct Biol 1996; 3:178-818; PMID:8564545; http://dx.doi.org/ 10.1038/nsb0296-178 [DOI] [PubMed] [Google Scholar]
- 23.Pandey R, Müller A, Napoli CA, Selinger DA, Pikaard CS, Richards EJ, Bender J, Mount DW, Jorgensen RA. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res 2002; 30:5036-55; PMID:12466527; http://dx.doi.org/ 10.1093/nar/gkf660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paz-Ares J, Ghosal D, Wienand U, Peterson PA, Saedler H. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J 1987; 6:3553-8; PMID:3428265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pignatta D, Erdmann RM, Scheer E, Picard CL, Bell GW, Gehring M. Natural epigenetic polymorphisms lead to intraspecific variation in Arabidopsis gene imprinting. Elife 2014; 3:e03198; PMID:24994762; http://dx.doi.org/ 10.7554/eLife.03198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prabu G, Prasad DT. Functional characterization of sugarcane MYB transcription factor gene promoter (PScMYBAS1) in response to abiotic stresses and hormones. Plant Cell Rep 2012; 4:661-9; PMID:22083650; http://dx.doi.org/ 10.1007/s00299-011-1183-y [DOI] [PubMed] [Google Scholar]
- 27.Qian W, Miki D, Zhang H, Liu Y, Zhang X, Tang K, Kan Y, La H, Li X, Li S, et al.. A histone acetyltransferase regulates active DNA demethylation in arabidopsis. Science 2012; 336:1445-8; PMID:22700931; http://dx.doi.org/ 10.1126/science.1219416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raissig MT, Baroux C, Grossniklaus U. Regulation and flexibility of genomic imprinting during seed development. The Plant Cell 2011; 23:16-26; PMID:21278124; http://dx.doi.org/ 10.1105/tpc.110.081018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reyes JL, Chua N-H. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J 2007; 49:592-606; PMID:17217461; http://dx.doi.org/ 10.1111/j.1365-313X.2006.02980.x [DOI] [PubMed] [Google Scholar]
- 30.Saibo NJM, Lourenco T, Oliveira MM. Transcription factors and regulation of photosynthetic and related metabolism under environmental stresses. Ann Botany 2009; 103:609-23; PMID:19010801; http://dx.doi.org/ 10.1093/aob/mcn227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scoville AG, Barnett LL, Bodbyl-Roels S, Kelly JK, Hileman LC. Differential regulation of a MYB transcription factor is correlated with transgenerational epigenetic inheritance of trichome density in Mimulus guttatus. New Phytologist 2011; 91:251-63; PMID:21352232; http://dx.doi.org/ 10.1111/j.1469-8137.2011.03656.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Segarra G, Van der Ent S, Trillas I, Pieterse CMJ. MYB72, a node of convergence in induced systemic resistance triggered by a fungal and a bacterial beneficial Microbe. Plant Biol 2009; 11:90-6; PMID:19121118; http://dx.doi.org/ 10.1111/j.1438-8677.2008.00162.x [DOI] [PubMed] [Google Scholar]
- 33.Sekhon RS, Chopra S. Progressive loss of DNA methylation releases epigenetic gene silencing from a tandemly repeated maize Myb gene. Genetics 2009; 181:81-91; PMID:19001287; http://dx.doi.org/ 10.1534/genetics.108.097170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seo PJ, Park CM. Auxin homeostasis during lateral root development under drought condition. Plant Signaling Behav 2009; 4:1002-4; PMID:19826230; http://dx.doi.org/ 10.4161/psb.4.10.9716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seo PJ, Park CM. MYB96-mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis. New Phytologist 2010; 186:471-83; PMID:20149112; http://dx.doi.org/ 10.1111/j.1469-8137.2010.03183.x [DOI] [PubMed] [Google Scholar]
- 36.Shukla PS, Agarwal P Gupta K, Agarwal PK. Molecular characterisation of a MYB transcription factor from a succulent halophyte involved in stress tolerance. AoB Plants 2015; 17;7; pii: plv054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song Y, Ji D, Li S, Wang P, Li Q, Xiang F. The dynamic changes of DNA methylation and histone modifications of salt responsive transcription factor genes in soybean. PLoS One 2012; 7(7):e41274; PMID:22815985; http://dx.doi.org/ 10.1371/journal.pone.0041274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stracke R, Werber M, Weisshaar B. The R2R3–MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 2001; 4:447-56; PMID:11597504; http://dx.doi.org/ 10.1016/S1369-5266(00)00199-0 [DOI] [PubMed] [Google Scholar]
- 39.Vannini C, Locatelli F, Bracale M, Magnani E, Marsoni M, Osnato M, Mattana M, Baldoni E, Coraggio I. Overexpression of the rice OsMYB4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. Plant J 2004; 37:115-27; PMID:14675437; http://dx.doi.org/ 10.1046/j.1365-313X.2003.01938.x [DOI] [PubMed] [Google Scholar]
- 40.Vega-Bartol JJ de, Simões M, Lorenz WW. Transcriptomic analysis highlights epigenetic and transcriptional regulation during zygotic embryo development of Pinus pinaster. BMC Plant Biol 2013; 13:123-43; PMID:23987738; http://dx.doi.org/ 10.1186/1471-2229-13-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wen H, Andrejka L, Ashton J, Karess R, Lipsick JS. Epigenetic regulation of gene expression by Drosophila Myb and E2F2-RBF via the Myb-MuvB/dREAM complex. Genes Dev 2008; 22:601-14; PMID:18316477; http://dx.doi.org/ 10.1101/gad.1626308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilczek C, Chayka O, Plachetka A, Klempnauer KH. Myb-induced chromatin remodeling at a dual enhancer/promoter element involves non-coding rna transcription and is disrupted by oncogenic mutations of v-myb. J Biol Chem 2009; 284:35314-24; PMID:19841477; http://dx.doi.org/ 10.1074/jbc.M109.066175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolff P, Weinhofer I, Seguin J, Roszak P, Beisel C, Donoghue MT, Spillane C, et al.. High-resolution analysis of parent-of-origin allelic expression in the Arabidopsis endosperm. PLoS Genetics 2011; 7:e1002126; PMID:21698132; http://dx.doi.org/ 10.1371/journal.pgen.1002126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilkins O, Nahal H, Foong J, Provart NJ, Campbell MM. Expansion and diversification of the Populus R2R3–MYB family of transcription factors. Plant Physiol 2009; 149:981-93; PMID:19091872; http://dx.doi.org/ 10.1104/pp.108.132795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie Z, Lee EK, Lucas JR, Morohashi K, Li D, Murray JAH, Sack FD, Grotewold E. Role of the stomatal development regulators FLP/MYB88 in abiotic stress responses. The Plant Cell 2010; 22:2306-21; PMID:20675570; http://dx.doi.org/ 10.1105/tpc.110.074609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiong H, Li J, Liu P, Duan J, Zhao Y, Guo X, Li Y, Zhang H, Ali J, Li Z. Overexpression of OsMYB48-1, a novel MYB-related transcription factor, enhances drought and salinity tolerance in rice. PLoS One 2014; 9(3):e92913; PMID:24667379; http://dx.doi.org/ 10.1371/journal.pone.0092913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu R, Wang Y, Zheng H, Lu W, Wu C, Huang J, Yan K, Yang G, Zheng C. Salt-induced transcription factor MYB74 is regulated by the RNA-directed DNA methylation pathway in Arabidopsis. J Exp Botany 2015; 66(19):5997–6008; PMID:26139822; http://dx.doi.org/19419532 10.1093/jxb/erv312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yaish MWF, Peng M, Rothstein SJ. AtMBD9 modulates Arabidopsis development through the dual epigenetic pathways of DNA methylation and histone acetylation. Plant J 2009; 59:123-35; PMID:19419532; http://dx.doi.org/ 10.1111/j.1365-313X.2009.03860.x [DOI] [PubMed] [Google Scholar]
- 49.Yaish MWF. DNA methylation-associated epigenetic changes in stress tolerance of plants Rout GR and Das AB (eds.) Molecular Stress Physiology of Plants, 2013; doi: 10.1007/978-81-322-0807-5-17 [DOI] [Google Scholar]
- 50.Yang A, Xiaoyan DX, Zhang WH. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J Exp Botany 2012; 63:2541-56; PMID:22301384; http://dx.doi.org/ 10.1093/jxb/err431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang L, Liu G, Zhao G, Xia C, Jia J, Liu X, Kong X. Characterization of a wheat R2R3-MYB transcription factor gene, TaMYB19, involved in enhanced abiotic stresses in Arabidopsis. Plant Cell Physiol 2014; 55:1802-12; PMID:25146486; http://dx.doi.org/ 10.1093/pcp/pcu109 [DOI] [PubMed] [Google Scholar]
- 52.Zhao M, Yang S, Chen C-Y, Li C, Shan W, Lu W, Cui Y, Liu X, Wu K. Arabidopsis BREVIPEDICELLUS interacts with the SWI2/SNF2 chromatin remodeling ATPase BRAHMA to regulate KNAT2 and KNAT6 expression in control of inflorescence architecture. PLoS Genet 2015; 11(3):e1005125; PMID:25822547; http://dx.doi.org/ 10.1371/journal.pgen.1005125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. Genevestigator. Arabidopsis microarray database and analysis toolbox. Plant Physiol 2004; 136:2621-32; PMID:15375207; http://dx.doi.org/ 10.1104/pp.104.046367 [DOI] [PMC free article] [PubMed] [Google Scholar]


