Abstract
Two plant growth promoting rhizobacteria (PGPR) Pseudomonas putida NBRIRA and Bacillus amyloliquefaciens NBRISN13 with ability to tolerate abiotic stress along with multiple PGP traits like ACC deaminase activity, minerals solubilisation, hormones production, biofilm formation, siderophore activity were evaluated for their synergistic effect to ameliorate drought stress in chickpea. Earlier we have reported both the strains individually for their PGP attributes and stress amelioration in host plants. The present study explains in detail the possibilities and benefits of utilizing these 2 PGPR in consortium for improving the chickpea growth under control and drought stressed condition. In vitro results clearly demonstrate that both the PGPR strains are compatible to each other and their synergistic growth enhances the PGP attributes. Greenhouse experiments were conducted to evaluate the effect of inoculation of both strains individually and consortia in drought tolerant and sensitive cultivars (BG362 and P1003). The growth parameters were observed significantly higher in consortium as compared to individual PGPR. Colonization of both PGPR in chickpea rhizosphere has been visualized by using gfp labeling. Apart from growth parameters, defense enzymes, soil enzymes and microbial diversity were significantly modulated in individually PGPR and in consortia inoculated plants. Negative effects of drought stress has been ameliorated and apparently seen by higher biomass and reversal of stress indicators in chickpea cultivars treated with PGPR individually or in consortia. Findings from the present study demonstrate that synergistic application has better potential to improve plant growth promotion under drought stress conditions.
Keywords: biofertilizers, consortia, PGPR, rhizosphere, synergism
Abbreviations
- PGPR
plant growth promoting rhizobacteria
- IAA
Indole 3 acetic acid
- ACC
1 amino cyclopropane 1 carboxylate.
Introduction
Plant remains in close interaction with soil microbes. Generally, these microbes are mutualistic and beneficial for plants. The root system produces exudates which affect the growth of soil microorganism. Bacterial associations with plants have been well studied1-6 and known that communication between interacting partners involves physiological and molecular processes. There are several examples of beneficial interactions between plant growth promoting microbes and plants.1,7 These rhizospheric competent and beneficial bacterial species mainly comprises group of plant growth promoting rhizobacteria (PGPR) such as Azospirillum, Azotobacter, Acetobacter, Bacillus, Burkholderia, Paenibacillus, Pseudomonas, Rhizobium and Serratia.2,8,9 Bacteria help plants directly by promoting growth via nitrogen fixation, nutrient channelization by solubilizing in absorbable forms,10 growth hormones production, production of 1- aminocyclopropane, 1- carboxylate (ACC) deaminase, and indirectly by producing siderophores, chitinases, fluorescent pigment molecules, antibiotics, β-1-3-glucanase and sometimes also by some poisonous compounds like cyanide.8
The PGPR are known to adhere or colonize in the rhizoshpere and grow in close association to stimulate the growth of plants. Prime requirement of good PGPR is its ability to colonize the plant root and survive in its vicinity and failing to survive and colonize in rhizosphere reduces the PGP effects.11 It is proposed that use of more than single PGPR in biofertilizer preparation could be better choice over a single bacterium to bring synergistic effect of nutrient mobilisation, enhanced efficacy, stability and uniformity when applied to different fields.12 In general, it has been reported by several researchers that inoculation with consortia have better plant growth promotion as compared to individual inoculations because individual strain is supplementing to each other for their beneficial traits.13,14
We have used 2 well proven PGPR Pseudomonas putida NBRIRA (RA) and Bacillus amyloliquefaciens NBRISN13 (SN13) in our laboratory.15,16 The strain RA is Gram negative and displayed multiple PGP activities like auxin production, phosphate solubilisation and tolerance to drought and salt stresses.17 Other PGPR is Gram positive SN13, with property of salt stress amelioration under hydroponic and soil condition as it showed increased colonization with increase ACC deaminase activity, reduced reduction in chlorophyll content, more proline accumulation.16 SN13 has also shown the osmoprotectant properties in rice rhizosphere together with both up regulation and down regulation of transcripts in leaves. In this paper we have studied in detail, ability of 2 PGPR to be utilized as consortia to improve chickpea growth promotion under control and drought stress conditions. Begin with compatibility assay, several PGP attributes in vitro conditions and physical parameters like growth parameters i.e. effect on shoot and root length, fresh and dry weight in different combinations were studied. Modulation in defense enzymes, physiological parameters, soil enzymes and functional microbial diversity of individually PGPR or in consortia was also evaluated. Results in this study will justify the utilization of 2 PGPR with multiple synergistic effects as better option than individual application in context.
Results
Compatibility assay
Initially compatibility assay was performed among 12 potential PGP strains of our laboratory by using well diffusion method and results showed 2 strains RA and SN13 were compatible to each other (Figs. 1A, 1B and 1C). The PGP traits like biofilm formation, motility, IAA production, phosphate solubilisation, siderophore production and ACC deaminase activity were evaluated individually and in consortia of RA and SN13 (Table 1), and all the attributes were positive in RA, SN13 and in consortia. All the PGP attributes were either comparable to individual strains or higher in consortia as compared to individual strains.
Figure 1.
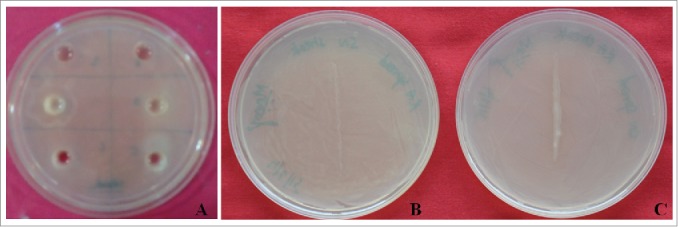
In vitro compatibility assay of Pseudomonas putida RA and Bacillus amyloliquefaciens SN13. (A) Well diffusion method for compatibility assay in NA plate. (B) RA (spread) and SN13 (streaked) to determined compatibility by the plate confrontation culture method. (C) SN13 (spread) and RA (streaked) to determined compatibility by the plate confrontation culture method.
Table 1.
Plant growth promoting attributes for Pseudomonas putida RA and Bacillus amyloliquefaciens SN13 and consortium
| PGP traits | RA | SN13 | RS |
|---|---|---|---|
| IAA* | 48.69±1.66 µg/ml | 24.45±0.85 µg/ml | 59.52±1.40 µg/ml |
| Phosphate solubilization* | 117.68±2.61 µg/ml | 110.43±1.47 µg/ml | 120.62±1.14 µg/ml |
| Siderophore | + | – | + |
| Biofilm formation | + | + | + |
| ACC deaminase | + | + | + |
| Motility | + | + | + |
RA (P. putida); SN13 (B. amyloliquefaciense); RS (RA+SN13);
IAA and P solubilization was checked after 48 h of incubation.
Growth patterns were observed for RA and SN13 to evaluate the effect of different concentrations of 48h grown culture supernatant of other compatible bacteria, and results (Fig. 2) revealed that the absorbance at 600 nm and viable CFU were not found significantly different from the control.
Figure 2.
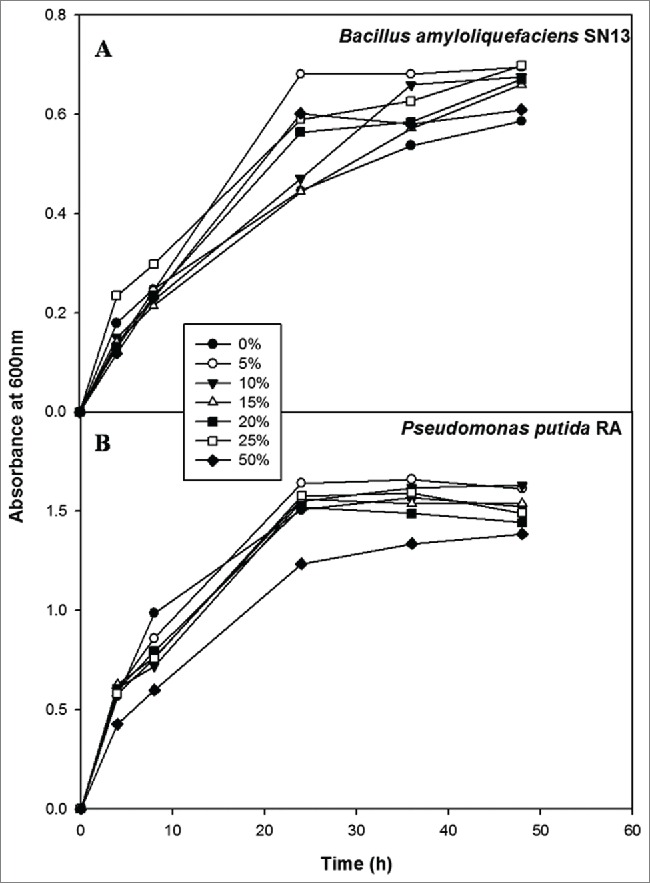
Competitive growth assessment of Pseudomonas putida RA and Bacillus amyloliquefaciens SN13. (A) Filtered supernatant of RA supplemented at 0% to 50% in NB medium to check the growth of SN13 at different time interval (B) Filtered supernatant of SN13 supplemented at 0% to 50% in NB medium to check the growth of RA at different time interval.
Effect of RA, SN13 and consortia on chickpea growth in control and drought conditions
Drought tolerant and sensitive cultivars were evaluated for their growth promotion in presence and absence of individual PGPR and in consortia under control and drought conditions. Results clearly revealed that inoculation with consortia has significantly enhanced the plant biomass of both chickpea cultivars as compared to uninoculated control and inoculated with individual strains. Moreover inoculation with individual strains were also improved the plant biomass as compared to uninoculated control. Thirty days old plants were subjected for drought conditions by withdrawing water for 15 d resulted in early drought responsive symptoms in sensitive cultivars and delayed or no symptoms in tolerant cultivars. Prominent results (Table 2) in consortia inoculated drought sensitive cultivars were observed by ameliorating the drought like symptoms as compared to uninoculated control under drought stress. While in tolerant variety visual effects of drought stress amelioration were not much significant due to consortia inoculation. Inoculations with individual strains are showing comparable improvements as compared to uninoculated control, but inoculation with consortia has better response under drought stress. It has been reported that inoculation of PGPR has ability to induce the nodulation in leguminous crops which has been clearly seen in PGPR inoculated roots of both cultivars, moreover as compared to individual strains consortia has significantly enhanced the number of nodules.
Table 2.
Plant growth promotion in chickpea cultivars treated with Pseudomonas putida RA and Bacillus amyloliquefaciens SN13 individually and consortium under control and drought stress conditions
| Treatments | Root length (cm) | Shoot length (cm) | Fresh weight root (mg) | Fresh weight shoot (mg) | Dry weight root (mg) | Dry weight shoot (mg) | No. of Root nodules | R/S DW ratio |
|---|---|---|---|---|---|---|---|---|
| BC | 16.7 ± 1.3de | 12.3±0.7def | 321.66±19.56abcd | 808.00 ± 31.05a | 90.00 ± 12.60d | 80.00 ± 5.10a | 44.67 ± 3.05ef | 1.13 |
| BC+D | 13.3 ± 1.3cd | 11.8 ± 1.0d | 270.66 ± 26.65abc | 707.34 ± 74.45a | 27.57 ± 2.24a | 75.13 ± 8.64a | 40.67 ± 3.79e | 0.37 |
| BRA | 20.1 ± 3.4ef | 13.8 ± 0.3fg | 595.33 ± 64.39de | 899.67 ± 64.01ab | 110.90 ± 13.28e | 110.00 ± 10.13bc | 42.00 ± 5.29e | 1.01 |
| BRA+D | 18.7 ± 3.1e | 12.2 ± 0.9de | 537.00 ± 41.60cde | 818.67 ± 84.28a | 40.60 ± 4.68ab | 88.54 ± 11.01ab | 35.00 ± 5.00d | 0.46 |
| BSN | 24.1 ± 3.8gh | 14.6 ± 1.1gh | 457.66 ± 91.15bcde | 809.34 ± 51.98a | 130.40 ± 6.42f | 100.34 ± 8.90ab | 48.00 ± 3.60fg | 1.30 |
| BSN+D | 22.4 ± 3.4fg | 12.6 ± 0.4ef | 440.66 ± 93.30bcde | 793.34 ± 43.31a | 31.63 ± 6.66a | 79.81 ± 3.99a | 44.34 ± 4.93ef | 0.40 |
| BRS | 26.5 ± 1.8h | 16.3 ± 0.7i | 573.00 ± 48.53de | 962.34 ± 18.47ab | 180.90 ± 14.35g | 129.69 ± 13.20cd | 50.34 ± 1.52g | 1.39 |
| BRS+D | 26.3 ± 1.1h | 16.0 ± 0.8hi | 560.33 ± 60.45cde | 934.34 ± 61.58ab | 46.63 ± 6.76b | 128.34 ± 12.74cd | 48.34 ± 1.53fg | 0.36 |
| PC | 8.2 ± 1.6ab | 6.1 ± 1.2a | 174.33 ± 37.07ab | 1270.00 ± 101.48abc | 48.33 ± 9.07b | 99.60 ± 6.35ab | 18.67 ± 3.05b | 0.49 |
| PC+D | 4.9 ± 0.5a | 4.8 ± 1.1a | 83.67 ± 5.50a | 610.00 ± 85.44a | 27.33 ± 6.42a | 90.00 ± 9.64ab | 11.34 ± 1.52a | 0.30 |
| PRA | 11.6 ± 0.4bc | 9.0 ± 0.4bc | 206.67 ± 15.27ab | 1893.34 ± 120.5cde | 63.33 ± 4.72c | 139.00 ± 5.29b | 31.34 ± 1.52d | 0.46 |
| PRA+D | 9.9 ± 1.1bc | 7.8 ± 0.9b | 180.00 ± 12.12ab | 1653.34 ± 177.85bcd | 47.00 ± 2.64b | 224.00 ± 42.55e | 24.00 ± 1.00c | 0.21 |
| PSN | 10.1 ± 0.5bc | 9.5 ± 1.1c | 550.00 ± 30.00cde | 2346.67 ± 395.13de | 110.00 ± 3.78e | 148.67 ± 9.16b | 30.00 ± 2.00d | 0.74 |
| PSN+D | 8.6 ± 0.5bc | 7.9 ± 0.5bc | 445.33 ± 63.90bcde | 2176.67 ± 70.23de | 87.67 ± 5.13d | 140.67 ± 18.02d | 21.67 ± 1.52bc | 0.62 |
| PRS | 12.2 ± 1.8bc | 11.5 ± 0.8de | 661.33 ± 81.39e | 2540.00 ± 210.00e | 180.67 ± 3.00g | 150.33 ± 5.56d | 34.67 ± 1.52d | 1.20 |
| PRS+D | 11.3 ± 2.1bc | 10.9 ± 0.8d | 321.66 ± 19.56e | 808.00 ± 31.05de | 117.50 ± 11.60ef | 84.87 ± 4.58d | 44.67 ± 3.05d | 1.30 |
values of root and shoot length taken 30 DAS, dry weight after drying at 45°C for 3 days.
RA (P. putida); SN13 (B. amyloliquefaciens); RS (consortia of RA and SN13).
“ ± : Standard errors (n = 6)”. Different letters within column represents significant difference at (P = 0.05) by using DMRT.
Modulation in defense enzymes
Defense enzymes like APX, Catalase, LPX, PAL and SOD were analyzed using enzyme extract from leaves. In general plants subjected to drought, showed that all defense enzymes were elevated compared to their controls (Fig. 3). With the inoculation of PGPR, these defense enzyme levels in drought stressed condition minimize the stress in plant by lowering the stress molecules. Moreover, both varieties when treated with bacterial consortium showed defense enzymes level in drought comparable to control. It indicates that RA and SN13 used in consortia have synergistic effect on drought stress amelioration. For example in SOD, the value of drought sensitive cultivar P1003 was doubled in drought condition while in inoculated samples with RA, SN13 and consortia, the activities were significantly reduced and best results were observed in consortia inoculated plants. Consortia showed value of SOD approximately equal to the control, indicating the complete drought amelioration of both the PGPR in greenhouse conditions. Similar results were also observed for APX, Catalase, LPX and PAL activities under drought stress conditions (Fig. 3). Bar diagram indicated the presence of drought stress amelioration activity due to PGPR inoculation. Total chlorophyll content was significantly reduced due to drought stress in both cultivars while inoculation with PGPR ameliorates the negative effect (Fig. 4). Another stress indicator i.e., proline was significantly enhanced under drought stress conditions as compared to control, which was also mitigated by PGPR inoculation. Similar to drought sensitive cultivar, tolerant variety has also showed induced defense enzymes under drought stress which were ameliorated by inoculation of PGPR individually or in consortium which confer the defense enzyme management by these bacteria in a synergistic manner. Results clearly demonstrated by assessing defense enzymes, stress indicators and PGP attributes that inoculation of PGPR in consortia has better response toward minimizing the deleterious effect of drought stress.
Figure 3.
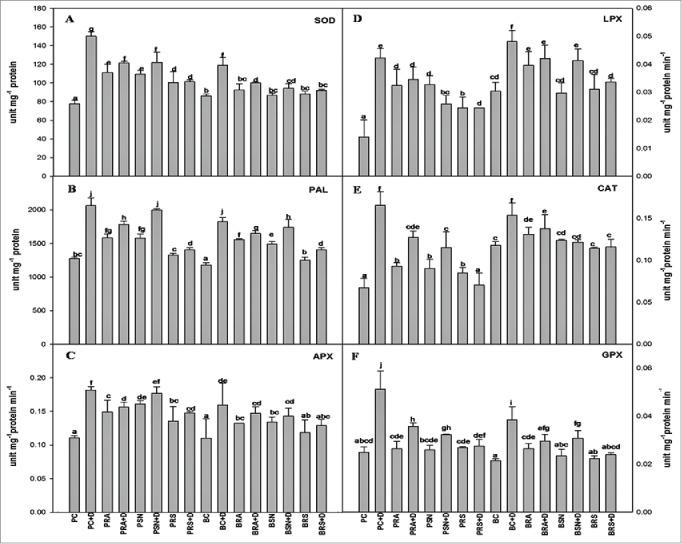
Defense enzymes activities in chickpea leaves treated with Pseudomonas putida RA and Bacillus amyloliquefaciens SN13 individually and consortium under control and drought stress conditions. (A) Superoxide dismutase (SOD). (B) Phenylalainine ammonia lyase (PAL). (C) Ascorbate peroxidase (APX). (D) Lipid peroxidase (LPX). (E) Catalase (F) Glutathione peroxidase (GPX). PC (control of P1003). PRA (P1003 treated with RA). PSN (P1003 treated with SN13). PRS (P1003 treated with Consortium of RA and SN13). BC (Control of BG362). BRA (BG362 treated with RA). BSN (BG362 treated with SN13). BRS (BG362 treated with Consortium of RA and SN13). D represents for drought; ±: Standard errors (n = 3). Different letters within column represents significant difference at (P = 0.05) by using DMRT.
Figure 4.
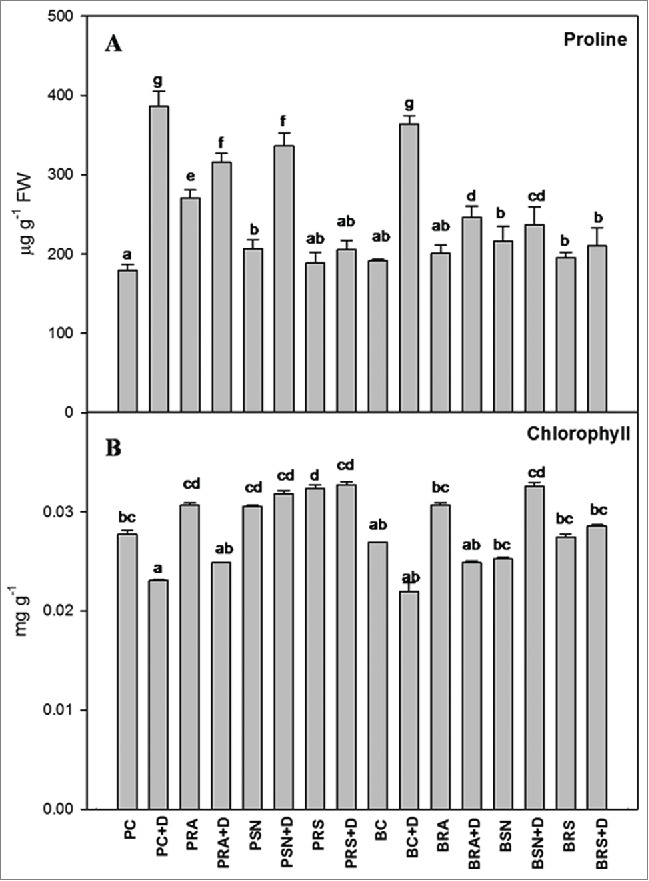
Physiological parameters in chickpea leaves treated with Pseudomonas putida RA and Bacillus amyloliquefaciens SN13 individually and consortium under control and drought stress conditions. (A) Proline (B) Chlorophyll PC (control of P1003). PRA (P1003 treated with RA). PSN (P1003 treated with SN13). PRS (P1003 treated with Consortium of RA and SN13). BC (Control of BG362). BRA (BG362 treated with RA). BSN (BG362 treated with SN13). BRS (BG362 treated with Consortium of RA and SN13). D represents for drought; ±: Standard errors (n = 3). Different letters within column represents significant difference at (P = 0.05) by using DMRT.
Soil enzymes
Soil enzymes were performed to determine the soil health in terms of microbial activity in rhizosphere soil samples of plants treated with RA, SN13 and consortia under control and drought conditions. Soil enzymes dehydrogenase, acid/alkaline phosphatase, protease, urease and β-glucosidase were determined and results (Table 3) showed the activity of soil enzymes in rhizosphere of 2 chickpea cultivars enhanced due to PGPR inoculation. As compared to individual strains RA and SN13, consortium inoculation has higher soil enzymes activities as compared to uninoculated control under normal and drought conditions. Higher soil enzyme activities due to consortium inoculation under drought conditions indicated synergistic effect of both bacteria.
Table 3.
Soil enzymes activities in rhizosphere of chickpea cultivars treated with Pseudomonas putida RA and Bacillus amyloliquefaciens SN13 individually and consortium under control and drought stress conditions
| Treatments | DHA (ug.g−1 soil h−1) | AP (g g−1 dwt h−1) | ALP (g g−1 dwt h−1) | Urease (g NH4-N g−1 dwt 2h−1) | Protease (g tyrosine g−1 dwt 2h−1) | -glucosidase (g g−1 dwt h−1) |
|---|---|---|---|---|---|---|
| PC | 3474.71 ± 51.20h | 308.04 ± 35.01bcd | 392.50 ± 31.94def | 2435.72 ± 152.86fg | 5.10 ± 0.09gh | 240.71 ± 6.59f |
| PC+D | 2255.74 ± 63.57c | 233.44 ± 7.95a | 291.99 ± 16.18a | 1824.27 ± 223.59d | 3.84 ± 0.14b | 141.73 ± 8.61a |
| PRA | 2457.72 ± 76.66d | 264.15 ± 9.52ab | 364.85 ± 36.78bcd | 2079.08 ± 106.67e | 4.09 ± 0.06bc | 195.91 ± 12.04e |
| PRA+D | 2456.70 ± 55.86d | 327.07 ± 41.28bcde | 418.29 ± 9.87fg | 2122.84 ± 168.65e | 5.17 ± 0.07gh | 191.70 ± 2.42e |
| PSN | 2851.29 ± 23.89f | 335.14 ± 65.54cdef | 382.51 ± 20.04cde | 2067.53 ± 47.12e | 3.33 ± 0.07a | 209.26 ± 6.42e |
| PSN+D | 2090.78 ± 28.12b | 355.94 ± 39.34defg | 429.23 ± 7.72g | 2146.56 ± 192.48e | 4.67 ± 0.05ef | 203.64 ± 1.83e |
| PRS | 3788.85 ± 28.96i | 416.63 ± 31.68gh | 412.97 ± 12.58efg | 2621.52 ± 163.30g | 5.22 ± 0.21gh | 245.69 ± 10.08f |
| PRS+D | 4011.45 ± 36.76j | 332.62 ± 35.90cdef | 411.96 ± 16.21efg | 2390.31 ± 42.56f | 4.26 ± 0.14cd | 233.80 ± 4.21f |
| BC | 2599.54 ± 20.63e | 380.77 ± 42.43efdh | 428.64 ± 13.70g | 1646.58 ± 89.43cd | 5.31 ± 0.14h | 155.56 ± 1.10bcd |
| BC+D | 1961.08 ± 15.40a | 289.05 ± 12.91abc | 348.07 ± 10.66b | 1028.18 ± 56.43a | 4.53 ± 0.39de | 125.52 ± 19.42a |
| BRA | 3166.77 ± 9.622g | 374.36 ± 22.55efgh | 392.35 ± 7.37def | 1450.53 ± 177.50b | 5.72 ± 0.21i | 165.52 ± 9.40cd |
| BRA+D | 3103.71 ± 107.5g | 431.57 ± 37.90h | 528.03 ± 14.22i | 1381.81 ± 122.12b | 6.24 ± 0.17j | 179.63 ± 6.66bc |
| BSN | 2861.99 ± 91.13f | 295.67 ± 34.76bcd | 354.86 ± 1.51bc | 1378.31 ± 78.30b | 4.88 ± 0.17fg | 173.20 ± 6.86d |
| BSN+D | 2089.63 ± 14.80b | 319.99 ± 35.89bcde | 404.20 ± 3.74efg | 1289.62 ± 103.19b | 6.14 ± 0.44j | 162.40 ± 9.02cd |
| BRS | 3470.63 ± 13.91h | 395.12 ± 18.52fgh | 434.55 ± 20.67g | 1620.15 ± 66.43cd | 5.27 ± 0.14h | 159.48 ± 7.20bcd |
| BRS+D | 3454.52 ± 84.70h | 426.27 ± 29.76h | 469.78 ± 27.05h | 1622.10 ± 84.40gd | 5.38 ± 0.15h | 159.23 ± 4.58bcd |
DHA (Dehydrogenase activity); AP (Acid phosphatase); ALP (Alkaline phosphatase); PC (control of P1003); PRA (P1003 treated with RA); PSN (P1003 treated with SN13); PRS (P1003 treated with Consortium of RA and SN13); BC (Control of BG362); BRA (BG362 treated with RA); BSN (BG362 treated with SN13); BRS (BG362 treated with Consortium of RA and SN13); D represents drought.
“ ± : Standard errors (n = 6)”. Different letters within column represents significant difference at (P=0.05) by using DMRT.
Microbial diversity assessment based on carbon source utilization pattern
Microbial richness, diversity and evenness of rhizosphere soil of both cultivars under control and drought conditions were estimated in order to check the effects of PGPR inoculation on native microbial population. Carbon source utilization pattern on Biolog Eco plates was determined to calculate Shannon, McIntosh and Simpson diversity and their related evenness for rhizosphere region of chickpea. Results (Table 4) clearly demonstrate that diversity and evenness indices have been influenced significantly by inoculation of PGPR individually and in consortia. Drought treatment has significantly reduced the microbial diversity in the rhizosphere samples of both the cultivars moreover inoculation of PGPR has ameliorated the negative effect drought stress on native microflora. Principal component (PC) score plot distributed at 71.33% and 11.86% on factor 1 and 2, respectively. Treatment of drought has more prominent effects as compared to cultivars and inoculation of PGPR, which is clearly reflected in the PCA plot (Fig. 5) where 2 groups formed and separated by drought treatment.
Table 4.
Rhizosphere functional diversities and evenness based on carbon source utilization pattern for rhizosphere chickpea cultivars treated with Pseudomonas putida RA and Bacillus amyloliquefaciens SN13 individually and consortium under control and drought stress conditions
| Samples | ShD | ShE | McD | McE | SimpD |
|---|---|---|---|---|---|
| BC | 4.41 ± 0.00i | 0.969 ± 0.001g | 0.983 ± 0.00i | 0.983 ± 0.00g | 0.996 ± 0.00i |
| BC+D | 4.11 ± 0.01b | 0.939 ± 0.002b | 0.963 ± 0.01b | 0.974 ± 0.00b | 0.992 ± 0.00b |
| BRA | 4.47 ± 0.00n | 0.982 ± 0.002l | 0.990 ± 0.00n | 0.990 ± 0.00l | 0.998 ± 0.00n |
| BRA+D | 4.25 ± 0.00c | 0.951 ± 0.001c | 0.974 ± 0.01c | 0.979 ± 0.00d | 0.995 ± 0.00c |
| BSN | 4.47 ± 0.00n | 0.982 ± 0.001l | 0.990 ± 0.00o | 0.990 ± 0.00m | 0.998 ± 0.00o |
| BSN+D | 4.28 ± 0.00f | 0.958 ± 0.000f | 0.977 ± 0.00f | 0.981 ± 0.00f | 0.995 ± 0.00f |
| BRS | 4.44 ± 0.00j | 0.975 ± 0.001h | 0.986 ± 0.00j | 0.986 ± 0.00h | 0.997 ± 0.00j |
| BRS+D | 4.28 ± 0.01e | 0.949 ± 0.001c | 0.976 ± 0.00d | 0.978 ± 0.00c | 0.995 ± 0.00d |
| PC | 4.45 ± 0.00k | 0.977 ± 0.000i | 0.988 ± 0.00k | 0.988 ± 0.00i | 0.997 ± 0.00k |
| PC+D | 4.08 ± 0.00a | 0.937 ± 0.001a | 0.960 ± 0.01a | 0.971 ± 0.00a | 0.991 ± 0.00a |
| PRA | 4.47 ± 0.00m | 0.981 ± 0.000e | 0.989 ± 0.00m | 0.989 ± 0.00k | 0.998 ± 0.00m |
| PRA+D | 4.27 ± 0.00d | 0.952 ± 0.001d | 0.976 ± 0.00e | 0.980 ± 0.00e | 0.995 ± 0.00e |
| PSN | 4.47 ± 0.02m | 0.981 ± 0.001k | 0.990 ± 0.00n | 0.990 ± 0.00l | 0.998 ± 0.00n |
| PSN+D | 4.30 ± 0.00g | 0.949 ± 0.000c | 0.977 ± 0.00g | 0.978 ± 0.00c | 0.995 ± 0.00g |
| PRS | 4.46 ± 0.00l | 0.979 ± 0.000j | 0.989 ± 0.00l | 0.989 ± 0.00j | 0.997 ± 0.00l |
| PRS+D | 4.36 ± 0.01h | 0.957 ± 0.000e | 0.981 ± 0.00h | 0.981 ± 0.00f | 0.996 ± 0.00h |
ShD (Shannon diversity); ShE (Shannon evenness); McD (McIntosh diversity); McE (McIntosh evenness); SimpD (Simpson diversity); PC (control of P1003); PRA (P1003 treated with RA); PSN (P1003 treated with SN13); PRS (P1003 treated with Consortium of RA and SN13); BC (Control of BG362); BRA (BG362 treated with RA); BSN (BG362 treated with SN13); BRS (BG362 treated with Consortium of RA and SN13); D represents drought.
“ ± : Standard errors (n=3)”. Different letters within column represents significant difference at (P=0.05) by using DMRT.
Figure 5.
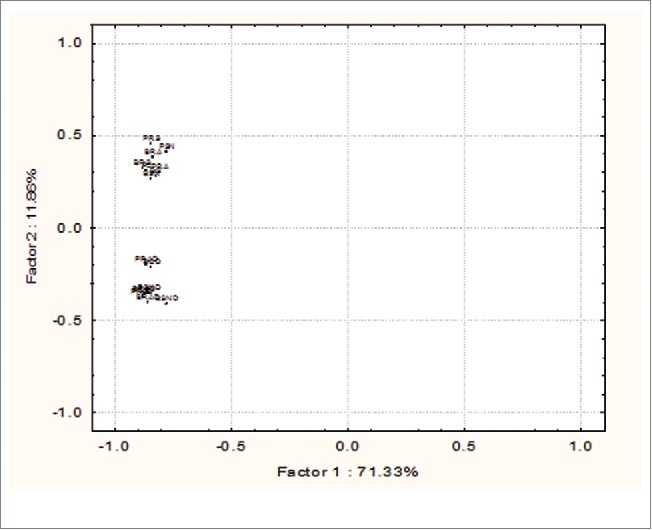
Principal component analysis (PCA) based on carbon source utilization pattern of rhizospheric soil treated with Pseudomonas putida RA and Bacillus amyloliquefaciens SN13 individually and consortium under control and drought stress conditions. PC (Control of P1003). PRA (P1003 treated with RA). PSN (P1003 treated with SN13). PRS (P1003 treated with consortium). BC (Control of BG362). BRA (BG362 treated with RA). BSN (BG362 treated with SN13). BRS (BG362 treated with consortium). D represents drought.
Tracking and visualization of PGPR in the root
Competence in the rhizosphere of host plant is the most important trait for any successful PGPR. Population of strains SN13 and RA individually tracked as described earlier by Nautiyal (1997), at the time of harvesting i.e. 45 d after sowing, in both cultivars population of SN13 and RA were more than 106 CFU/g rhizosphere soil alone or in consortia. Population with 107 CFU/g soil in the rhizosphere gives clear indication that these strains have ability to compete, grow and survive under the control and drought stress conditions. To visualize the SN13 and RA on the roots for both the cultivars, strains were labeled with green fluorescent protein and results (Fig. 6) from roots of both cultivars and florescence was observed form the roots surfaces.
Figure 6.
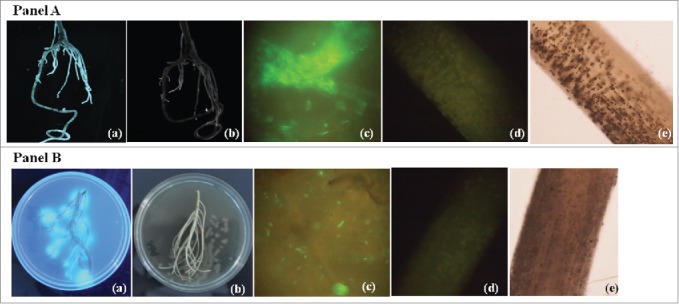
Visualization of gfp tagged Pseudomonas putida RA and Bacillus amyloliquefaciens SN13 on chickpea roots. Panel A shows root adherence of gfp transformed in Pseudomonas putida (a), (c) and (d) showing gfp adherence on root whereas (b) and (e) are control without gfp label bacteria. Panel B shows Bacillus amyloliquefaciens (a), (d) showing gfp adherence on root whereas (b) and (e) are control without gfp label bacteria.
Discussion
Compatibility between 2 PGPR, their performance in synergistic manner without altering the native rhizosphere microbial population and drought stress amelioration could leads to prepare a bioinoculant formulation for chickpea. In presented study we have selected 2 PGPR P. putida and B. amyloliquefaciens based on their multiple PGP attributes and stress ameliorating abilities in host crops. These both strains are compatible and performing better under in vitro and in vivo experiments when applied in consortia as compared to individual strain or control. Several mechanisms have been proposed to understand the mechanism behind microbial mediated stress amelioration in plants e.g. production of growth hormones, resulted in increased root biomass, which leads to higher uptake of nutrients thereby improving plant growth under stress conditions.1 Experiments conducted in live soil conditions colonization ability of both strains have been monitored and their performance under controlled and drought conditions can be considered as a consequence of it. Many researchers have been reported that colonization and competence is the most important factor for any bacterial inoculation as PGPR or as biofertilizers.18-21 We have selected 2 chickpea cultivars, which are drought tolerant (BG362) and sensitive (P1003) and our results clearly demonstrated that inoculation of PGPR strains alone or in consortia has significantly not only enhanced the plant growth under controlled conditions but also ameliorates the negative effect of drought stress. We have evaluated defense enzymes to check the possible mechanism of action to ameliorate drought stress due to inoculation of theses strains. Under drought conditions, complex interactions among native microorganisms, roots and water content in the rhizosphere of host plant alter the physiological properties of associated soil. In general, proline protects plant membranes against the adverse effects of high salt concentration, extreme drought and temperature. Proline accumulation can act as buffers cellular redox potential under different stresses.40 Higher accumulation of proline helps the plants to maintain osmotic balance when growing under drought conditions. It has been reported earlier that proline protects higher plants against salt/osmotic stresses.41 Some PGPR strains with ability to produce cytokinin and antioxidants, resulted in abscisic acid (ABA) accumulation followed by degradation of reactive oxygen species. Antioxdant enzymes are linked with oxidative stress tolerance,42 in our results we have also reported that when chickpea plants subjected to drought stress cause induction in defense enzymes. While inoculation of PGPR alone or in consortia have tendency to reduce these enzymes that results in less reactive oxygen species (ROS) formation, which ameliorate the damages to cell membranes and proteins. Plant cells must retain water potential lower than soil to absorb the water and when soil dries higher solutes concentrations causes the cell damage, even less moisture has negative effect on soil microorganism. Drought conditions not only affects the bacterial physiological functions, mobility, chemotaxis and adsorption to soil particles but also has adverse effect on root exudates, required for the plant-microbes interaction. From our results inoculation of stress tolerant PGPR induced the growth under drought conditions clearly attributed their role by plant hormones production that enhanced lateral roots growth and consequently nutrients and water uptake. The study suggested that an improved tolerance of plants to drought, with higher growth and yields, and synergistic effects from the use of consortia were found. In conclusion, the results achieved till now in utilizing microbial strains selected for a specific function or a specific plant/microorganism interaction are encouraging to apply these strategies. The improvement of plant response to drought or nutritional stresses when specifically selected microbial strains are used is a successful example. Nevertheless, more knowledge about the processes and the regulation of the plant-rhizosphere microorganism interaction is needed to standardize a practice which is supporting the sustainability of agricultural productions.
Materials and Methods
Bacterial strains and their culture preparations
In the experiments, microbial strains B. amyloliquifaciens (SN13) was isolated from the alkaline soil of DRC, Banthra, Lucknow, Uttar Pradesh, India whereas P. putida (RA) from chickpea rhizospheric soil of Dholpur, Rajasthan India. These 2 PGPR strains were grown separately at 28°C for 16 h in nutrient broth (NB) on a rotary shaker at 180 rpm. To check their compatibility under in vitro conditions 50 µl culture of one bacterium was spreaded on NA plate and 50 µl cultures of different PGPR were filled in the wells formed on plates (Well diffusion method). Another experiment was conducted by streaking of 1 PGPR on the NA plate spreaded with overnight grown other PGPR (plate confrontation method).
PGP traits evaluation test
For PGP traits evaluation both physiological and biochemical assays including motility, IAA production, ACC deaminase activity, phosphate solubilisation, siderophore activity and biofilm formation were assayed using standard protocols.16-20 These properties were screened for SN13, RA and consortia of RA and SN13 (RS).
Competitive growth assessments
Cultures of both bacteria grown for 48 h (at 30°C at 180 rpm) were filtered using 0.22 µm filters (PALL, Corporation, USA). The filtered supernatant from grown bacterial culture was mixed at concentration of 5, 10, 15, 20, 25 and 50% (v/v) to the fresh NB medium. The growth of one bacterium in presence of filtered supernatant of the other was plotted at different time intervals of 4, 8, 24, 36 and 48 h by taking absorbance at 600 nm in spectrophotometer (Thermo Scientific Pvt. Ltd.) and colony forming units.
Greenhouse experiments
Two contrasting drought tolerant and sensitive chickpea varieties BG362 and P1003 were selected for evaluation of individual PGPR and in consortia inoculation effects under control and drought stress conditions. Seed bacterization was carried out as described by Nautiyal (1997)21 for individual strains and for consortium 1:1 ratio of both strains were mixed before seed bacterization. Uninoculanted and bacterized plants were grown for 30 d under greenhouse conditions and for drought stress 4 weeks old plants were subjected to progressive drought by no further addition of water, whereas normal plants were regularly watered to maintain the 20% moisture. Plants were harvested after 2 weeks of progressive drought stress and data on root length, shoot length, root, shoot fresh and dry biomass were recorded.
Plant defense enzymes and soil enzymes assays
Total soluble protein was extracted from the leaves of the control, drought stressed, bacterized and drought with bacterized in both the varieties of chickpea varying in drought sensitivity. Protein quantification was done according to Bradford assay22 at 595nm. Estimation of defense enzymes of leaf samples were done using established standard protocols like for Superoxide dismutase (SOD),23 Phenylalainine ammonia lyase (PAL),24 Catalase,25 Ascorbate peroxidae (APX),26 Lipid peroxidase (LPX),27 Glutathione peroxidase (GPX),28 Proline,29 Chlorophyll and Carotenoid.30
Soil enzymes estimations were also done using previous published protocols like for Dehydrogenase (DHA),31 Acid/Alkaline phosphatases,32,33 Urease,34 Protease35 and β-Glucosidase.36
Functional microbial diversity
Rhizosphere microbial diversity was assessed by carbon source utilization pattern using Biolog Eco plates (Biolog, Inc.., Hayward, CA, USA) in chickpea. Rhizosphere soil samples (1.0 g) from all the treatments of both varieties were collected and suspended in 9.0 ml sterile MQW and mixed them on a rotary shaker for 4 h at 28°C, 10−3 final dilution of suspension was further used. Samples were coated in each well of Biolog plates (150 µl) and kept at 30°C for incubation. Carbon source utilization was measured by the rate of reduction of tetrazolium, a redox indicator dye. Data was recorded for day 1-7 at interval of 24 hours at 590 nm. Average well-color development (AWCD) was determined which is microbial activity in each microplates. Richness, McIntosh, Shannon and Simpson functional diversity and related evenness were calculated and Principal component analysis (PCA) was performed on AWCD normalized data.37,38
Green fluorescent protein visualization
Both SN13 and RA were transformed with plasmid harboring gfp gene using heat shock method.39 PGPR adheres to root surface and forms biofilm around the roots. They form the majority population in rhizosphere community. For verification of root adhering property overnight grown tagged bacteria culture individually and in consortia were pelleted and resuspended in saline (0.85%) to make 0.8 OD (600 nm). The roots of 25 d old chickpea plants were dipped for 2 h in the culture and washed with sterile MQW. These roots were visualized under UV illumination at 10 and 100X lens with microscope (Olympus CX41). Statistical analyses. Results were subjected for statistical analysis by using Microsoft excel 2007, SPSS 16.0 and Statistica 7.0 software.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
The study was supported by New Initiative (as a Cross Flow Technology project) “Root Biology and Its Correlation to Sustainable Plant Development and Soil Fertility” (Root SF; BSC0204) from the Council of Scientific and Industrial Research (CSIR), New Delhi, India.
References
- 1.Glick BR. The enhancement of plant growth by free-living bacteria. Can J Microbiol 1995; 41:109-17; http://dx.doi.org/ 10.1139/m95-015 [DOI] [Google Scholar]
- 2.Glick BR, Bashan Y. Genetic manipulation of plant growth promoting bacteria to enhance biocontrol of phytopathogens. Biotechnol Adv 1997; 15:353-78; PMID:14538716; http://dx.doi.org/ 10.1016/S0734-9750(97)00004-9 [DOI] [PubMed] [Google Scholar]
- 3.Glick BR, Cheng Z, Czarny J, Duan J. Promotion of plant growth by ACC deaminase-containing soil bacteria. Eur J Plant Pathol 2007; 119:329-39; http://dx.doi.org/ 10.1007/s10658-007-9162-4 [DOI] [Google Scholar]
- 4.Kloepper JW, Leong J, Teintze M, Schroth MN. Enhanced plant growth by siderophores produced by plant growth-promoting rhizobacteria. Nature 1980; 286:885-6; http://dx.doi.org/ 10.1038/286885a0 [DOI] [Google Scholar]
- 5.Kloepper JW, Lifshitz R, Zablotowicz RM. Free-living bacterial inocula for enhancing crop productivity. Tre Biotechnol 1989; 7:39-43; http://dx.doi.org/ 10.1016/0167-7799(89)90057-7 [DOI] [Google Scholar]
- 6.Kloepper JW, Ryu CM, Zhang S. Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology 2004; 94:1259-66; PMID:18944464; http://dx.doi.org/ 10.1094/PHYTO.2004.94.11.1259 [DOI] [PubMed] [Google Scholar]
- 7.James K, Olivares FL. Infection and colonization of sugar cane and other Graminaceous plants by endophytic diazotrophs. Crit Rev Plant Sci 1997; 17:77-119; http://dx.doi.org/ 10.1016/S0735-2689(98)00357-8 [DOI] [Google Scholar]
- 8.Kloepper JW. Plant-growth-promoting rhizobacteria as biological control agents, in: Soil Microbial Ecology, Metting F.B., Jr., (ed.), Marcel Dekker inc., N.Y. 1993; pp.255-73. [Google Scholar]
- 9.Khalid A, Arshad M, Zahir ZA. Screening plant growth promoting rhizobacteria for improving growth and yield of wheat. J Appl Microbiol 2004a; 96:473-80; http://dx.doi.org/ 10.1046/j.1365-2672.2003.02161.x [DOI] [PubMed] [Google Scholar]
- 10.Rodrı´guez H, Fraga R. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol Adv 1999; 17:319-39; http://dx.doi.org/ 10.1016/S0734-9750(99)00014-2 [DOI] [PubMed] [Google Scholar]
- 11.Normander B, Prosser JI. Bacterial origin and community composition in the barley phytosphere as a function of habitat and pre-sowing conditions. Appl Environ Microbiol 2000; 66:4372-7; PMID:11010885; http://dx.doi.org/ 10.1128/AEM.66.10.4372-4377.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stockwell VO, Johnson KB, Sugar D, Loper JE. Mechanistically compatible mixtures of bacterial antagonists improve biological control of fire blight of pear. Phytopathology 2011; 101:113-23; PMID:20839962; http://dx.doi.org/ 10.1094/PHYTO-03-10-0098 [DOI] [PubMed] [Google Scholar]
- 13.Singh A, Jain A, Sarma BK, Upadhyay RS, Singh HB. Rhizosphere competent microbial consortium mediates rapid changes in phenolic profiles in chickpea during Sclerotium rolfsii infection. Microbiol Res 2014; 169:353-60; PMID:24168925; http://dx.doi.org/ 10.1016/j.micres.2013.09.014 [DOI] [PubMed] [Google Scholar]
- 14.Panwar M, Tewari R, Nayyar H. Microbial consortium of plant growth-promoting rhizobacteria improves the performance of plants growing in stressed soils: An overview In: Phosphate solubilizing microorganisms, Principles and application of microphos technology. Khan MS, Zaidi A, Musarrat J (Eds.) Springer, Germany: 2014; pp 257-85. [Google Scholar]
- 15.Chaudhry V, Asif MH, Bag S, Goel R, Mantri SS, Singh SK, Chauhan PS, Sawant SV, Nautiyal CS. Draft Genome Sequence of Pseudomonas putida Strain MTCC5279. Gen Ann 2013; 1: e00560-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nautiyal CS, Srivastava S, Chauhan PS, Seem K, Mishra A, Sopory SK. Plant growth-promoting bacteria Bacillus amyloliquefaciens NBRISN13 modulates gene expression profile of leaf and rhizosphere community in rice during salt stress. Plant Physiol Biochem 2013; 66:1-9; PMID:23454292; http://dx.doi.org/ 10.1016/j.plaphy.2013.01.020 [DOI] [PubMed] [Google Scholar]
- 17.Srivastava S, Chaudhry V, Mishra A, Chauhan PS, Rehman A, Yadav A, Tuteja N, Nautiyal CS. Gene expression profiling through microarray analysis in Arabidopsis thaliana colonized by Pseudomonas putida MTCC5279, a plant growth promoting rhizobacterium. Plant Sig Behav 2012; 7:2, 235-45; http://dx.doi.org/ 10.4161/psb.18957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khan N, Mishra A, Chauhan PS, Nautiyal CS, Induction of Paenibacillus lentimorbus biofilm by sodium alginate and CaCl2 alleviates drought stress in chickpea, Ann App Biol 2011; 159:372-86; http://dx.doi.org/ 10.1111/j.1744-7348.2011.00502.x [DOI] [Google Scholar]
- 19.Siddikee MA, Glick BR, Chauhan PS, Yima WJ, Sa T. Enhancement of growth and salt tolerance of red pepper seedlings (Capsicum annuum L.) by regulating stress ethylene synthesis with halotolerant bacteria containing 1- aminocyclopropane-1-carboxylic acid deaminase activity, Plant Physiol Biochem 2011; 49:427-34; PMID:21300550; http://dx.doi.org/ 10.1016/j.plaphy.2011.01.015 [DOI] [PubMed] [Google Scholar]
- 20.Mishra S, Nautiyal CS, Reducing the allelopathic effect of Parthenium hysterophorus L. on wheat (Triticum aestivum L.) by Pseudomonas putida, Plant Growth Regul 2012; 66:155-65; http://dx.doi.org/ 10.1007/s10725-011-9639-1 [DOI] [Google Scholar]
- 21.Nautiyal CS. Selection of Chickpea-Rhizosphere-Competent Pseudomonas fluorescens NBRI1303 Antagonistic to Fusarium oxysporum f. sp. ciceri, Rhizoctonia bataticola and Pythium sp. Curr Microbiol 1997; 35:52-8; http://dx.doi.org/ 10.1007/s002849900211 [DOI] [Google Scholar]
- 22.Bradford MM. Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976; 72:248-54; PMID:942051; http://dx.doi.org/ 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 23.Beauchamp C, Fridovich I. Superoxide dismutase: Improved assays and an assay applicable to acrylamide gels. Anal Biochem Rev 1971; 44:276-87; http://dx.doi.org/ 10.1016/0003-2697(71)90370-8 [DOI] [PubMed] [Google Scholar]
- 24.Fritz RR, Hodgins DS, Abell C. Phenylalainine ammonia lyase induction and purification from yeast and clearance in mammals. J Biol Chem 1976; 251:15, 4646-50; PMID:985816 [PubMed] [Google Scholar]
- 25.Aebi H. Catalase Methods of enzymatic analysis In: Bergmeyer HU. (Ed.). Vol. 2, Academic Press Inc., New York, USA: 1974; ISBN: 352725370X, pp: 673-85. [Google Scholar]
- 26.Nakano Y, Asada K. Hydrogen peroxide scavenged by Ascorbate- specific peroxidase in spinach chloroplast. Plant Cell Physiol 1981; 22:867-80 [Google Scholar]
- 27.Heath RL, Packer L. Photoperoxidation in isolated chloroplast. I. kinetics and stoichiochemistry of fatty acid peroxidation. Arch Biochem Biophys 1968; 125:189-98; PMID:5655425; http://dx.doi.org/ 10.1016/0003-9861(68)90654-1 [DOI] [PubMed] [Google Scholar]
- 28.Hemeda HM, Klein BP. Effect of naturally occurring antioxidants on peroxidase activity of vegetable extracts. J Food Sci 1990; 55:184-5; http://dx.doi.org/ 10.1111/j.1365-2621.1990.tb06048.x [DOI] [Google Scholar]
- 29.Bates L, Waldren RP, Teare ID. Rapid determination of free proline for water-stress studies. Plant Soil 1973; 39, 205-7; http://dx.doi.org/ 10.1007/BF00018060 [DOI] [Google Scholar]
- 30.Arnon DI. Copper enzyme polyphenoloxides in isolated chloroplast in Beta vulgaris. Plant Physiol 1949; 24:1-15; PMID:16654194; http://dx.doi.org/ 10.1104/pp.24.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alef K. Dehydrogenase activity In: Alef K., Nannipieri P (Eds.), Methods in Applied Soil Microbiology and Biochemistry. Academic Press, San Diego, California: 1995; pp. 228-31. [Google Scholar]
- 32.Tabatabai MA, Bremner JM. Use of p-nitro phenyl phosphate for assay of soil phosphatase activity. Soil Biol Biochem 1969; 1:301-7; http://dx.doi.org/ 10.1016/0038-0717(69)90012-1 [DOI] [Google Scholar]
- 33.Eivazi F, Tabatabai MA. Phosphatase in soils. Soil Biol Biochem 1977; 9:167-72; http://dx.doi.org/ 10.1016/0038-0717(77)90070-0 [DOI] [Google Scholar]
- 34.Kandeler E, Gerber H. Short-term assay of soil urease activity using colorimetric determination of ammonium. Biol Fert Soils 1988; 6:68-72; http://dx.doi.org/ 10.1007/BF00257924 [DOI] [Google Scholar]
- 35.Tabatabai MA, Bremner J. Assay of urease, catalase in soil. Soil Biol Biochem 1972; 4:479-82; http://dx.doi.org/ 10.1016/0038-0717(72)90064-8 [DOI] [Google Scholar]
- 36.Eivazi F, Tabatabai MA. Glucosidases and galactosidases in soils. Soil Biol Biochem 1988; 20:601-6; http://dx.doi.org/ 10.1016/0038-0717(88)90141-1 [DOI] [Google Scholar]
- 37.Nautiyal CS. Self-Purificatory Ganga Water Facilitates Death of Pathogenic Escherichia coli O157: H7. Curr Microbiol 2009; 58:25-9; PMID:18810537; http://dx.doi.org/ 10.1007/s00284-008-9260-3 [DOI] [PubMed] [Google Scholar]
- 38.Nautiyal CS, Rehman A, Chauhan PS. Environmental Escherichia coli occur as natural plant growth-promoting soil bacterium. Arch Microbiol 2010; 192:185-93; PMID:20084366; http://dx.doi.org/ 10.1007/s00203-010-0544-1 [DOI] [PubMed] [Google Scholar]
- 39.Froger A, Hall JE. Transformation of plasmid DNA into E. coli using the heat shock method. J Vis Exp 2007; 6:253; PMID:18997900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ashraf M, Foolad MR. Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ Exp Bot 2007; 59:206-16; http://dx.doi.org/ 10.1016/j.envexpbot.2005.12.006 [DOI] [Google Scholar]
- 41.Hare PD, Cress WA. Metabolic implications of stress-induced proline accumulation in plants. Plant Growth Reg 1997; 21:79-102; http://dx.doi.org/ 10.1023/A:1005703923347 [DOI] [Google Scholar]
- 42.Patel PK, Hemantaranjan A. Antioxidant defense system in chickpea (Cicer arietinum L.): Influence by drought stress implemented at pre and post-anthesis stage. Ame J Plant Physiol 2012; 7(4):164-73; http://dx.doi.org/ 10.3923/ajpp.2012.164.173 [DOI] [Google Scholar]


