Abstract
Corynebacterium ulcerans is an emerging pathogen transmitted by a zoonotic pathway with a very broad host spectrum to humans. Despite rising numbers of infections and potentially fatal outcomes, data on the molecular basis of pathogenicity are scarce. In this study, the interaction of 2 C. ulcerans isolates - one from an asymptomatic dog, one from a fatal case of human infection - with human macrophages was investigated. C. ulcerans strains were able to survive in macrophages for at least 20 hours. Uptake led to delay of phagolysosome maturation and detrimental effects on the macrophages as deduced from cytotoxicity measurements and FACS analyses. The data presented here indicate a high infectious potential of this emerging pathogen.
Keywords: Corynebacterium ulcerans, FACS, host-pathogen interaction, necrotic cell death, THP-1 cells
Abbreviations
- CFU
colony forming units
- G-CSF
granulocyte-colony stimulating factor
- GFP
green fluorescent protein
- IL-6
Interleukin 6
- JAK
Janus kinase
- MOI
multiplicity of infection
- STAT3
signal transducers and activators of transcription-3
Introduction
Corynebacterium ulcerans is a pathogenic member of the genus Corynebacterium, which is part of the family Corynebacteriaceae, the order Actinomycetales and the phylum Actinobacteria.1 Within the taxon Actinobacteria, the genera Corynebacterium, Nocardia and Mycobacterium form a monophyletic branch, the CMN group, based on their unusual cell envelope composition.2 The genus Rhodococcus shares this features and is sometimes also regarded as a member of this group, then designated as CMNR group.3,4 C. ulcerans was first described by Gilbert and Stewart, who isolated the bacteria from the throat of a patient with respiratory diphtheria-like illness.5 In fact, when lysogenized by a tox gene-carrying corynephage, C. ulcerans can – as Corynebacterium diphtheriae – produce diphtheria toxin and during the past decade, diphtheria-like infections with toxigenic C. ulcerans have outnumbered those caused by toxigenic C. diphtheriae in many industrialized countries.6 Moreover, during the last years, human infections associated with C. ulcerans appear to be increasing in various countries and can most often be ascribed to zoonotic transmission.7,8 The range of mammals that may serve as a reservoir for human infections is extremely broad. C. ulcerans was e.g. isolated from cattle, goats, pigs, wild boars, dogs, cats, ground squirrels, otters, camels, monkeys, orcas and water rats.9-22
In 2011, 2 C. ulcerans strains from the metropolitan area of Rio de Janeiro, Brazil, were sequenced: BR-AD22, isolated from a nasal swab of an asymptomatic dog, and 809, isolated from a bronchoalveolar lavage (BAL) sample of an 80-year-old woman with fatal pulmonary infection.23-25 Based on these genome sequences and comparative genomics approaches, a number of putative virulence factors were annotated, while functional data were scarce. Up to now, data on adhesion and invasion of epithelial cells, fibrinogen, fibronectin and collagen binding, antimicrobial profiles and arthritogenic potential of isolates were published.26-28 In the study presented here, C. ulcerans 809 and BR-AD22 were characterized for the first time to our knowledge in respect to their interaction with macrophages as important part of the innate immune system in order to get deeper insight into the pathogenicity of this emerging pathogen. The interaction with macrophages was of special interest, since the closely related animal pathogen Corynebacterium pseudotuberculosis, a facultative intracellular bacterium, is able to survive and grow in macrophages in order to disseminate within the host,3,29 and also the human pathogen C. diphtheriae is able to survive within human macrophages.30
Results
Internalization of C. ulcerans by human macrophages
As a first approach, the interaction of human macrophages with C. ulcerans was analyzed by fluorescence microscopy, together with Corynebacterium glutamicum used as control. This bacterium is non-pathogenic and was expected to be eliminated quickly by the macrophages. GFP-labeled bacteria were readily internalized by human THP-1 macrophages without opsonization or additional external primining e.g., by lipopolysaccharide. The fate of the bacteria was monitored for 2, 8 and 20 hours. The number of fluorescent bacteria within the macrophages appeared to be constant or declining within the 20 hours for C. glutamicum ATCC 13032. In contrast, a time-dependent increase of C. ulcerans 809 and BR-AD22 within macrophages was observed by fluorescence microscopy within this time interval (Fig. 1). Similar results were obtained with the murine macrophage cell line RAW 264.7 (data not shown).
Figure 1.
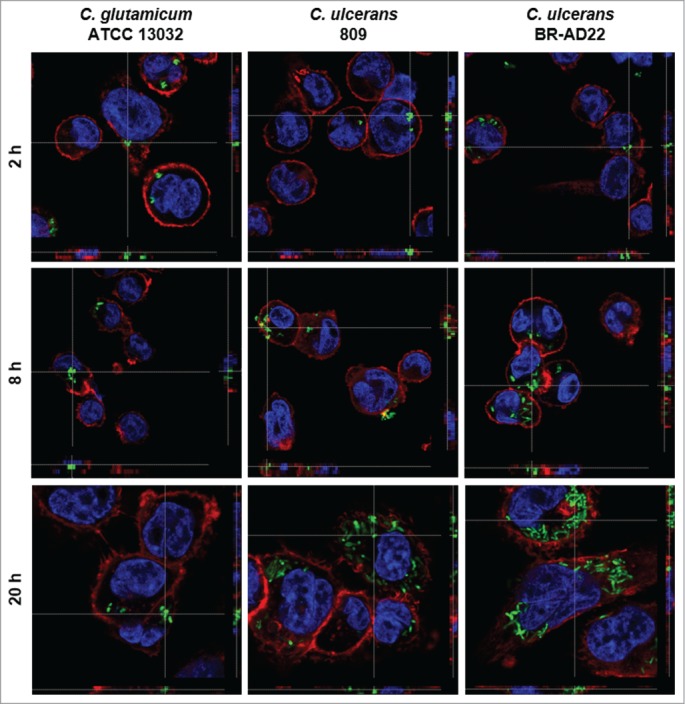
Fluorescence microscopy of C. ulcerans and THP-1 cells. THP-1 cells were infected with C. glutamicum ATCC 13032 pEPR1p45gfp, C. ulcerans 809 pEPR1p45gfp and C. ulcerans BR-AD22 pEPR1p45gfp at an MOI of 10 for 30 min. Extracellular bacteria were killed by the addition of gentamicin and after different time points, cells were fixed. Nuclei were stained with DAPI, the cytoskeleton with Alexa Fluor® 647 Phalloidin and z-stack micrographs were taken using the confocal laser-scanning microscope Leica SP5 II and analyzed with the LAS software suite to proof that bacteria are located inside of the macrophages. Representative pictures are shown.
Survival of C. ulcerans after internalization by human macrophages
For a more quantitative analysis of C. ulcerans – macrophage interaction, a replication assay was carried out and CFU were determined after adhering bacteria were killed by gentamicin addition. When the number of bacteria within THP-1 cells was analyzed, already after 2 hours almost no viable C. glutamicum were detectable (Fig. 2A), indicating that the fluorescent bacteria observed at the same and later time points by microscopy were already irreversibly inactivated or killed by the macrophages. When the low number of viable C. glutamicum at 2 hours post-infection were set to 100 %, a constant decline of CFU could be observed at the later time points (Fig. 2B). For C. ulcerans strains 809 and BR-AD22 already after 2 hours post-infection higher CFU were detected compared to the non-pathogenic C. glutamicum strain. After 8 hours of macrophage infection, even a strong increase of CFU was observed both, in relation to the number of bacteria used for infection and in relation to the number of intracellular CFU determined at 2 hours post infection (Fig. 2A, B). This result indicates that – in contrast to C. glutamicum - C. ulcerans is able to proliferate in the macrophages within the first hours of internalization. After 20 hours, the number of CFU declined for the C. ulcerans strains, indicating successful inactivation of bacteria at this late time point by the macrophages. Interestingly, with longer incubation time, cells challenged by C. ulcerans in this experimental set-up began to detach from the wells and showed signs of cell death, which was analyzed in more detail in subsequent approaches (see below). Taken together, in contrast to C. glutamicum, C. ulcerans strains seem to be able to interfere with macrophage function independent of their origin from animal or human sources. Similar results were obtained with murine RAW 264.7 and J774E macrophages indicating that these findings are not limited to a single macrophage cell line or exclusively to human macrophages (data not shown).
Figure 2.
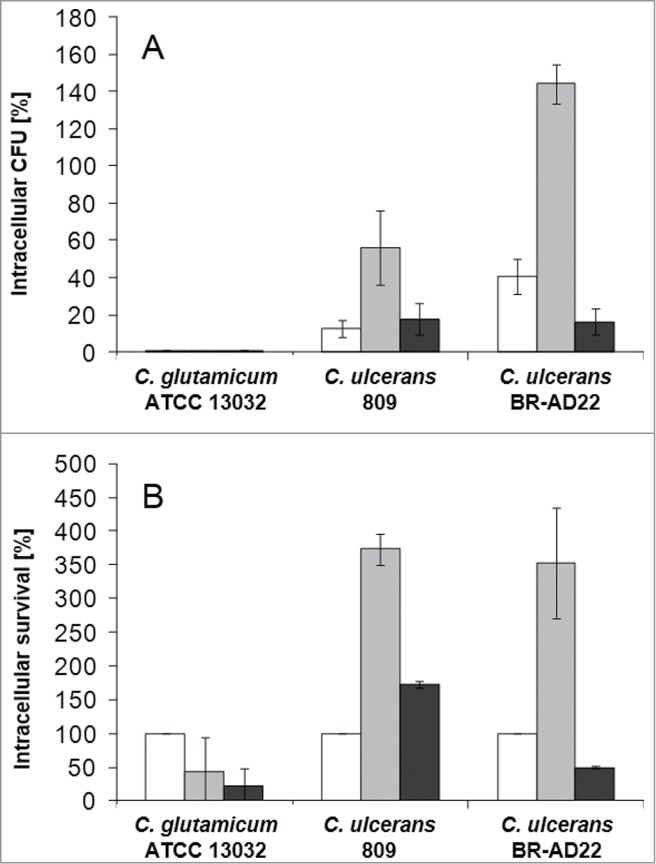
Quantitative analysis of viable intracellular C. ulcerans in THP-1 cells. THP-1 cells were infected with C. ulcerans wild type strains 809 and BR-AD22 at an MOI of 1 for 30 min. To kill extracellular bacteria, cells were incubated with medium containing gentamicin and after 2 (white bars), 8 (gray bars) and 20 h (black bars), cells were harvested, lysed and lysates were plated on blood agar plates to recover intracellular CFU. A) Intracellular CFU in percent referred to the inoculum, B) intracellular survival in percent referred to the bacteria that were taken up after 2 h. Data shown are mean values of 4 independent biological replicates each performed in triplicates ± standard deviation.
Delay of phagolysosome maturation by C. ulcerans
A hallmark of macrophage function after phagocytosis is the formation of phagolysosomes by fusion of phagosomes and acidic lysosomes. In order to investigate the reason for the observed growth and delay of degradation of C. ulcerans, formation of acidic compartments within macrophages was monitored using a LysoTracker dye. In parallel, Alexa Fluor® 647 Phalloidin and DAPI staining visualized cytoskeleton and nuclei, respectively, and corynebacteria were labeled by GFP (Fig. 3). In case of C. glutamicum ATCC 13032, a clear co-localization of bacteria and acidic compartments was observed already after 2 hours. After 8 and 20 hours, this co-localization was even more pronounced with almost all bacteria located together with acidic compartments. This situation differed strongly in case of C. ulcerans isolates. After 2 hours, no co-localization of strain 809 or BR-AD22 with lysosomes was detected. After 8 hours, only about half of the bacteria detected were co-localizing with lysosomes, and even after 20 hours, a considerable number of GFP-labeled bacteria were not co-localizing with acidic compartments. The data indicate a delay of phagolysosome formation by C. ulcerans. In accordance with the CFU determined (Fig. 2), the delaying effect of 809 seemed to be stronger than in case of BR-AD22.
Figure 3.
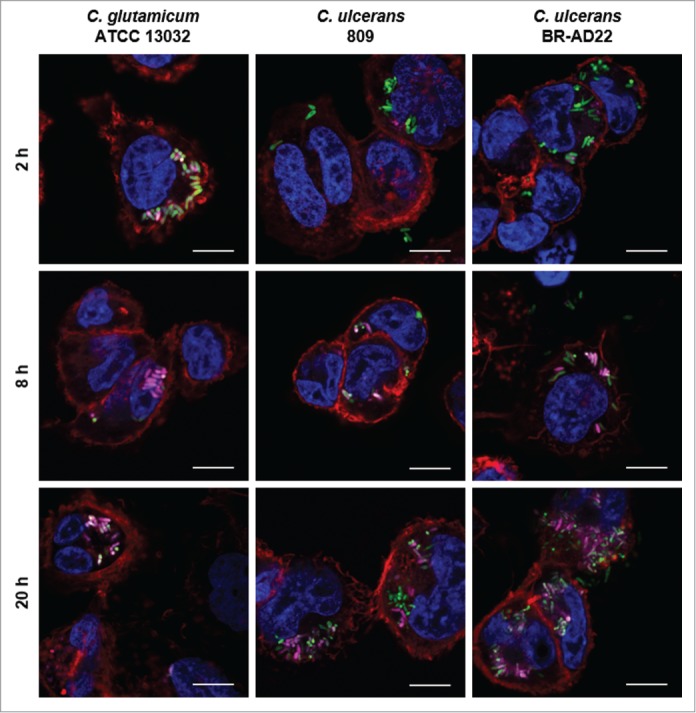
Labeling and tracking of acidic organelles in THP-1 cells infected with C. ulcerans. THP-1 cells were incubated with LysoTracker® Red DND-99 for 120 min before cells were infected with C. glutamicum ATCC 13032 pEPR1p45gfp, C. ulcerans 809 pEPR1p45gfp and C. ulcerans BR-AD22 pEPR1p45gfp at an MOI of 10 for 30 min. Extracellular bacteria were killed by the addition of gentamicin and after 2, 8 and 20 h, cells were fixed. Nuclei were stained with DAPI, the cytoskeleton with Alexa Fluor® 647 Phalloidin and micrographs were taken using the confocal laser-scanning microscope Leica SP5 II and analyzed with the LAS software suite. Non-pathogenic C. glutamicum immediately co-localize with acidic compartments (2 h) whereas pathogenic C. ulcerans only show co-localization after longer incubation time (8 h to 20 h). Representative pictures are shown. Scale bars: 10 µm.
For an unbiased and more quantitative approach, pictures were automatically evaluated either on the level of pixels or bacterial cells. Both methods showed consistently that C. ulcerans impairs phagosome maturation. In contrast to the non-pathogenic C. glutamicum, which already shows approximately 60 % co-localization after 2 hours post-infection and at least 80 % after 20 hours, the C. ulcerans strains applied show lower co-localization at all time points investigated with less than half of the bacteria co-localized with acidic compartments after 20 hours (Table 1).
Table 2.
Strains, cell lines and plasmids used in this study.
| Strain, cell line or plasmid | Description/genotype | Reference/source |
|---|---|---|
| C. glutamicum | ||
| Strains | ||
| ATCC 13032 | type-strain, non-pathogenic | 50 |
| Corynebacterium ulcerans | ||
| 809 | isolated from an 80-year-old woman with fatal pulmonary infection | 24 |
| BR-AD22 | isolated from an asymptomatic dog | 23 |
| ELHA1 | BR-AD22 pld::pK18mob-pld’, phospholipase D-deficient mutant | 35 |
| ELHA3 | BR-AD22 CULC22_00609::pK18mob-CULC22_00609’ | 27 |
| Cell lines | ||
| THP-1 | human leukemic monocytic cells | 51 |
| THP1-Blue NFκB | THP-1 cells with stable integrated NFκB inducible SEAP reporter construct | Invivogen |
| RAW 246.7 | murine leukemic macrophages | 52 |
| J774E | mannose receptor-expressing clone of the J774 mouse macrophage cell line | 53 |
| Plasmids | ||
| pEPR1-p45gfp | P45, gfpuv, KmR, rep, per, T1, T2 | 54 |
Table 1.
Automated analysis of co-localization of bacteria with acidic compartments. At least 19 fluorescence microscopy pictures were analyzed for each data set as described in the Materials and Methods section.
| % co-localization after evaluated on pixel level |
||||
|---|---|---|---|---|
| 2 h | 8 h | 20 h | ||
| C. glutamicum ATCC 13032 | 59 | 82 | 91 | |
| C. ulcerans 809 | 8 | 35 | 49 | |
| C. ulcerans BR-AD22 | 19 | 29 | 47 | |
|
% co-localization after evaluated on bacterium level |
||||
| 2 h | 8 h | 20 h | ||
| C. glutamicum ATCC 13032 | 54 | 80 | 79 | |
| C. ulcerans 809 | 11 | 34 | 49 | |
| C. ulcerans BR-AD22 | 20 | 29 | 47 | |
Response of human macrophages to C. ulcerans contact
The observed delay in phagolysosome maturation gave rise to the question whether macrophage function is impaired in general or specifically. To address this, NFκB induction and cytokine secretion were analyzed in response to infection with C. ulcerans. Cells of the reporter cell line THP1-Blue NF-κB were incubated for 20 hours with viable (Fig. 4A) and UV-killed (Fig. 4B) bacteria of non-pathogenic C. glutamicum ATCC 13032 and pathogenic C. ulcerans strains 809 and BR-AD22. Viable corynebacteria led to strong NF-κB activation when MOI 1 and 10 were tested independent of their pathogenicity (Fig. 4A). When an MOI of 100 was applied, NF-κB activation by C. ulcerans strains was decreased (data not shown) most likely due to detrimental effects of these pathogenic bacteria on the reporter cells (see below). Infection with dead bacteria led to a weaker NF-κB activation, which reached the values obtained for viable cells only in case of 10-fold higher MOIs were applied. The activation by dead bacteria was dose-dependent; detrimental effects of UV-inactivated pathogens were not observed (Fig. 4B), indicating that macrophage damage is the result of an active process induced by C. ulcerans.
Figure 4.
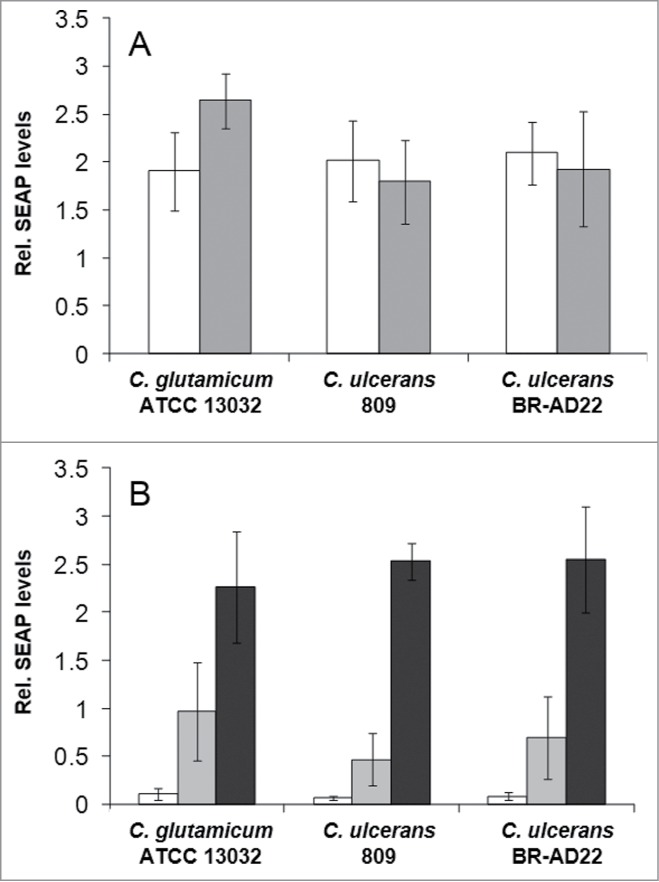
NF-κB activation in THP1-Blue™ NF-κB reporter cells after C. ulcerans infection. THP1-Blue™ NF-κB cells were incubated for 20 h with (A) viable and (B) UV-killed bacteria of the non-pathogenic C. glutamicum ATCC 13032 and pathogenic C. ulcerans strains 809 and BR-AD22 at an MOI of 1 (white bars), 10 (gray bars) and 100 (black bars). For viable C. ulcerans, MOI 100 led to detrimental effects on cells and data are not shown. Subsequently, supernatants were taken and mixed with QuantiBlue SEAP detection solution leading to a change in color upon NF-κB activation. Living C. glutamicum led to high SEAP levels in all MOIs, whereas living C. ulcerans showed lower levels at higher MOIs. Infection with dead bacteria led to a dose-dependent NF-κB activation in all cases. Data shown are mean values of 3 independent biological replicates each performed in triplicates ± standard deviation.
Additionally, supernatants of THP-1 cells infected with C. ulcerans with an MOI of 1 and 10, respectively, were collected at 2, 8 and 20 hours post-infection and used for determination of IL-6 (Fig. 5A) and G-CSF (Fig. 5B) secretion. Upon receptor binding, IL-6 and G-CSF activate JAK kinases, which then activate STAT3 signaling leading to increased expression of the anti-apoptotic proteins Bcl-xL and Bcl-2.31,32 C. ulcerans 809 and BR-AD22 led to rising cytokine levels with infection time and reached concentrations of 1,000 to 1,400 pg ml−1 for IL-6 and 4,500 to 6,500 pg ml−1 for G-CSF, respectively.
Figure 5.
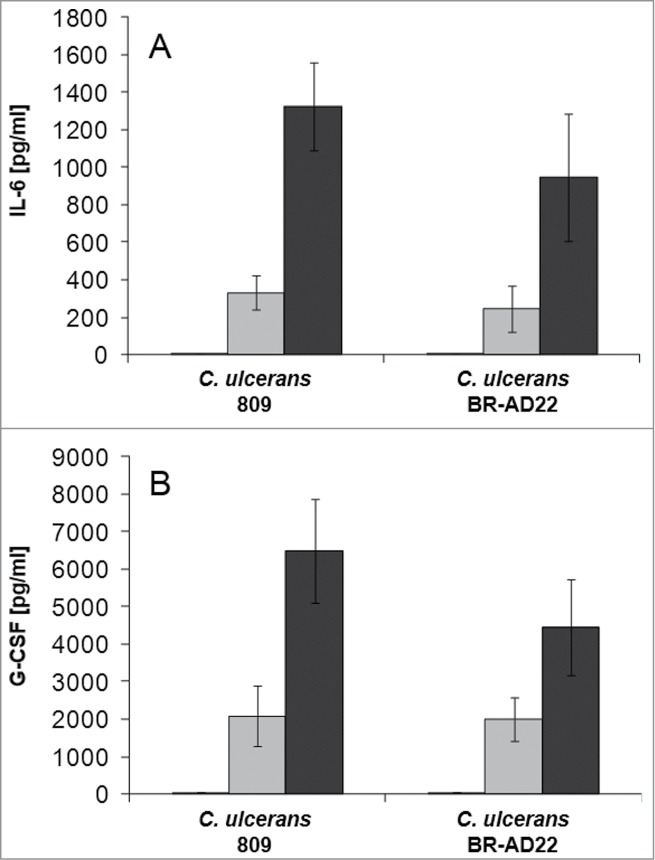
Cytokine ELISA of THP-1 cells after infection with C. ulcerans. Supernatants of THP-1 cells infected with C. ulcerans were collected at 2 (white bars), 8 (gray bars) and 20 h (black bars) post-infection and used as samples for determination of (A) IL-6 and (B) G-CSF concentrations. Data shown are mean values of 3 independent biological replicates each performed in triplicates ± standard deviation.
Taken together, these data indicate functional NF-κB and STAT3 signal transduction pathways in the macrophages, indicating that delay of phagolysosome formation is caused by a specific mechanism and not a general detrimental effect of C. ulcerans.
Killing of macrophages by C. ulcerans
As described above, a partial detachment of cells from the surface of cell culture wells was observed in response to C. ulcerans contact. This phenomenon was analyzed in more detail in subsequent experiments. First, different MOIs and times of infection were tested. Detachment of cells was not observed with untreated cells and those infected with an MOI of 1. In contrast, C. ulcerans strains 809 and BR-AD22 applied with an MOI of 10 led to malformation and detachment of cells after 8 hours. An effect, which was not observed with C. glutamicum ATCC 13032 (Fig. 6).
Figure 6.
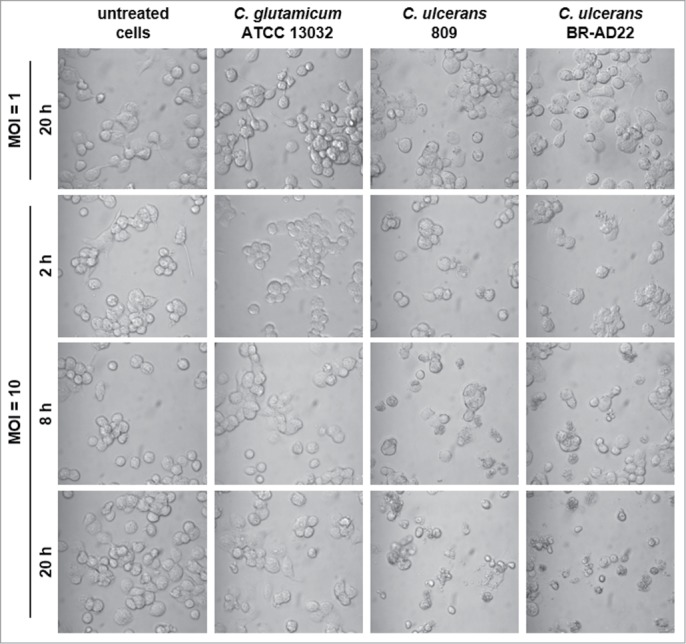
Detachment of THP-1 cells infected with C. ulcerans. Cells infected with non-pathogenic C. glutamicum or pathogenic C. ulcerans at an MOI of 10 and 1 were analyzed microscopically at 2, 8 and 20 h post-infection, untreated cells served as negative control. Both C. ulcerans wild type strains caused detachment of cells at MOI 10 after 8 h, and an increasing effect with longer incubation time. Cells treated with C. glutamicum looked healthy even after 20 h, as well as cells incubated with a lower amount of bacteria (MOI 1).
Microscopic analysis of DAPI stained fragmented nuclei as a sign of apoptotic cell death of THP-1 cells after infection with C. ulcerans revealed only single apoptotic cells independent of the time point analyzed, which is in accordance with the activation of anti-apoptotic cell signaling as described above. To further analyze the mode of cell death, FACS experiments were carried out. First, the SubG1 technique was used as assay to measure apoptosis rates. However, no typical SubG1 fractions were detectable for THP-1 cells infected with C. ulcerans at MOI 50 further supporting the idea that apoptosis is not involved in macrophage death (data not shown).
As second approach, lysis of cells was tested. To quantify the number of lytic cells upon C. ulcerans infection, cells were harvested at different time points, stained with 7-AAD and analyzed by flow cytometry after gating and exclusion of cell debris (Fig. 7A). Seven-AAD is a dye that is excluded from viable cells, but can penetrate damaged cell membranes. In the DNA histogram, 7-AAD positive cells show a shift to the right and can be easily separated from viable cells (Fig. 7B). Uninfected cells show a background level of around 10 % 7-AAD positive cells over all points in time investigated. Handling and infection of cells increased this background to about 30 % at 2 hours post-infection. At 8 hours post-infection, the count of positive cells increased for all samples (Fig. 7B); however, when all events are considered (Fig. 7A), the amount of small fragments detected was much higher for cells infected with C. ulcerans compared to cells infected with C. glutamicum, indicating that many cells were already fragmented due to C. ulcerans infection. These were gated out and not considered in the count of 7-AAD positive cells, resulting in a relatively low difference between the strains. After 20 hours, cells infected with C. glutamicum recover and drop down to background level of uninfected cells, whereas cells infected with C. ulcerans strains show about 50 % positive cells. Furthermore, again a high amount of cell debris was observed. In summary, a lytic cell death mechanism resembling necrosis upon infection with C. ulcerans was indicated by this approach (Fig. 7B, C).
Figure 7.
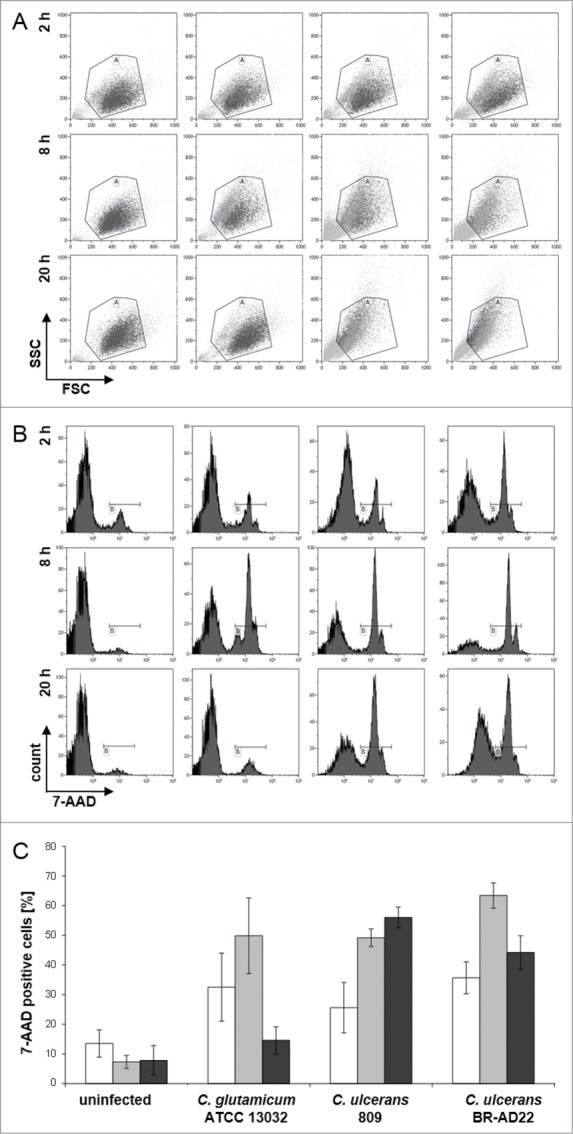
7-AAD staining and FACS analysis of THP-1 cells infected with C. ulcerans. THP-1 cells were infected with the non-pathogenic C. glutamicum ATCC 13032 and pathogenic C. ulcerans strains 809 and BR-AD22 at MOI 50, uninfected cells were carried along as negative control. After different points in time, cells were harvested and stained with 7-AAD, a dye that only penetrates dead cells, and analyzed by flow cytometry. (A) Dot plots of representative samples show forward scatter versus side scatter. Gate A excludes cell debris and small fragments like bacteria. (B) Histograms (7-AAD signal vs. cell count) of 10,000 cells after gating. Gate B represents 7-AAD positive, dead cells (light gray in gate A). (C) Quantitative analysis of 7-AAD positive cells at 2 (white bars), 8 (gray bars) and 20 h (black bars). Mean values and standard deviations of at least 2 independent assays performed in triplicate are depicted.
In addition to the FACS assays, the release of cytosolic lactate dehydrogenase into the supernatant as a sign of host cell damage was measured during infection of THP1-Blue NF-κB cells with C. ulcerans. Cells were infected for 20 hours with viable and UV-killed C. glutamicum ATCC 13032 and C. ulcerans strains 809 and BR-AD22 at an MOI of 1 and 10. The non-pathogenic C. glutamicum as well as dead C. ulcerans had no damaging effect, while viable C. ulcerans strains 809 and BR-AD22 induced cell lysis as indicated by strong LDH release already at an MOI of 1 (Fig. 8).
Figure 8.
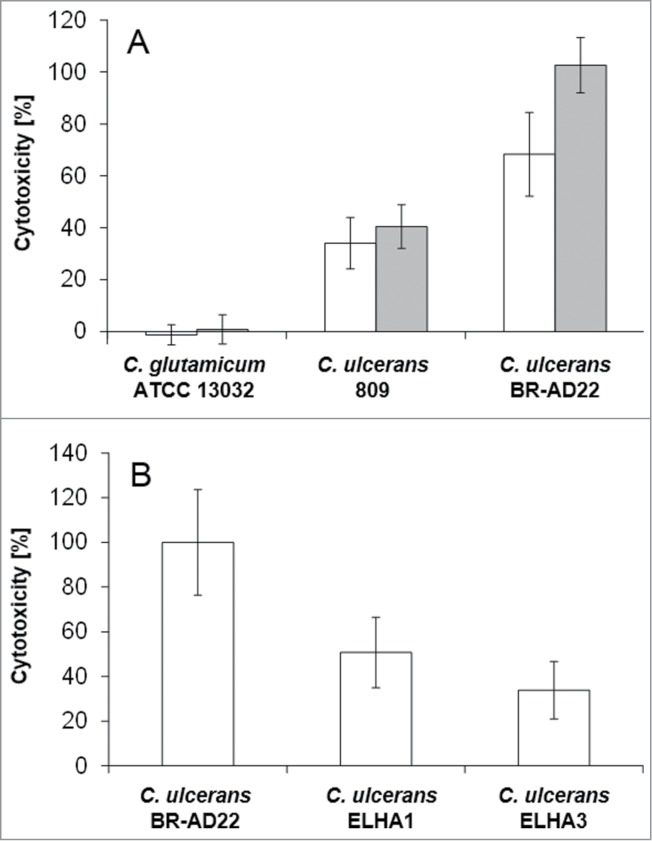
LDH release of THP1-Blue cells infected with C. ulcerans. The release of lactate dehydrogenase (LDH) as a sign of host cell damage during infection of THP1-Blue™ NF-κB cells with C. ulcerans was measured using the cytotoxicity detection kit (Roche). (A) Cells were infected for 20 h with the non-pathogenic C. glutamicum ATCC 13032 and pathogenic C. ulcerans wild type strains 809 and BR-AD22 at an MOI of 1 (white bars) and 10 (gray bars). The non-pathogenic C. glutamicum had no damaging effect. However, C. ulcerans led to LDH release at MOI 1 and 10. (B) Cells were infected with C. ulcerans mutant strains ELHA1 and ELHA3 at MOI 1 and the LDH release was compared to the corresponding wild type strain BR-AD22 which was set to 100 %. Data shown are mean values of 3 independent biological replicates each performed in triplicates ± standard deviation.
Factors involved in the killing of macrophages by C. ulcerans were not characterized before. However, based on homology to closely related pathogenic corynebacteria, different C. ulcerans proteins have been annotated as putative virulence factors.25 Phospholipase D (PLD) is the most important virulence factor of the closely related C. pseudotuberculosis.33 This so-called ovis toxin is involved in dissemination of this animal pathogen from the initial site of infection to other parts of the body. Another example of a putative virulence factor encoded in the C. ulcerans genome is CULC22_00609, a homolog of C. diphtheriae DIP0733, which was found recently to be involved in adhesion to and invasion of epithelial cells and in the survival inside macrophages.34 BR-AD22 strains carrying a pld and CULC22_00609 gene disruption, respectively, were generated and designated ELHA1 and ELHA3.27,35 When cells were infected with these mutant strains and LDH release was measured, at MOI 1 strain ELHA1 showed 51 ± 16 % and ELHA3 34 ± 13 % cytotoxicity compared to the wild type, indicating a multifactorial mechanism of macrophage damage, which has to be analyzed in more detail in future. A complex interaction is also indicated by the fact that the defect of mutant strains observed at MOI 1 could be overcome by higher MOIs (data not shown).
Discussion
C. ulcerans is an emerging pathogen, which is transmitted by a zoonotic pathway from a wide number of mammals to the human host.7,8 In fact, in the last decade cases of respiratory diphtheria caused by C. diphtheriae were outnumbered by C. ulcerans infections in Western Europe.6,36,37 Despite increasing numbers of infections and the occurrence of fatal cases, basic mechanisms of host-pathogen interaction and specific virulence factors of C. ulcerans are widely uncharacterized.
In the study presented here, C. ulcerans BR-AD22, isolated from an asymptomatic dog, and 809, isolated from an 80-year-old woman with fatal pulmonary infection were characterized for the first time to our knowledge in respect to their interaction with macrophages. Interestingly, the human and animal isolate did not differ significantly in respect to internalization and survival within macrophages and showed higher uptake and survival rates compared to C. diphtheriae strains in U937 macrophages,30 supporting the high infectious potential of this emerging pathogen.
In contrast to non-pathogenic C. glutamicum strain ATCC 13032, the C. ulcerans strains were able to multiply within macrophages at least 8 hours post-infection and were able to delay phagolysosome formation, which might be an effective mechanism for immune evasion and spreading of C. ulcerans within the body supporting the establishment and progress of infections. Corynebacterium species are closely related to the genera Mycobacterium, Nocardia and Rhodoccoccus, forming the CMNR group of Actinobacteria. The members of this group are characterized by a complex cell wall architecture with an additional layer of mycolic acids. Interaction with macrophages and the influence of mycolic acids is best investigated for Mycobacterium tuberculosis. Here, trehalose dimycolate, the cord factor, was shown to inhibit fusion events between phospholipid vesicles inside the host macrophage and to be involved in macrophage activation.38 Furthermore, Rhodococcus equi, a facultative intracellular pathogen, is able to arrest phagosome maturation in macrophages before the late endocytic stage.39 For corynebacteria, data on macrophage interaction and the role of mycolic acids are scarce. As shown previously, outer membrane lipids of C. pseudotuberculosis have a lethal effect on murine and caprine macrophages.40,41 It might therefore be worth to investigate the role of mycolic acids in the interaction of C. ulcerans with host macrophages, besides the influence of PLD and CULC22_00609.
Flow cytometry analysis revealed a high amount of cell debris when THP-1 cells were infected with C. ulcerans 809 and BR-AD22 and a strong lytic effect was found when LDH release was tested. Together with the absence of evidence for nucleus fragmentation a lytic cell death mechanism is favored by these results rather than apoptosis. In the past few years, several mechanisms of programmed necrotic cell death besides apoptosis have been described. These include besides others pyroptosis and necroptosis, which result in plasma membrane rupture. In contrast to classical necrosis, they are initiated in response to various intrinsic or extrinsic signals and underlie specific signaling cascades resulting finally in cell death (for review see42,43). To unravel the molecular background and exact mode of cell death induction by C. ulcerans in THP-1 cells, further investigations need to be carried out.
A host cell damaging effect of C. ulcerans to eukaryotic cells was already described for the epithelial cell line Detroit562. Here, the transepithelial resistance of cell monolayers was rapidly reduced in response infection with C. ulcerans 809 and in a less severe mode with C. ulcerans BR-AD22. The more aggressive behavior of C. ulcerans 809 toward epithelial cells might be due to the additional Shiga-like toxin this strain carries.27
Taken together, the observations made in this study hint to the existence of more than one active mechanism of C. ulcerans to cope with macrophage function. In line with this idea, initial sequencing of strains 809 and BR-AD2225 as well as next generation sequencing of 9 C. ulcerans isolates8 revealed a number of different virulence factors, which might influence survival of this pathogen besides the Shiga-like toxin of strain 809 as well as PLD and DIP0733 homolog CULC22_00609 of BR-AD22. Mutations in the genes encoding PLD and CULC22_00609 led reduced cytotoxicity toward THP1-Blue™ NF-κB cells compared to the wild type strain BR-AD22. To further investigate the effect of these putative virulence factors, studies analyzing the effect of purified proteins on macrophage survival would be of high interest. Further proteins found for C. ulcerans were e.g. neuraminidase H, endoglycosidase E, a Fic toxin homolog and a RhuM homolog.7,8,25
In conclusion, the emerging pathogen C. ulcerans is a potent pathogen, which comprises a broad and varying set of virulence factors.25 Furthermore, by acquisition of new genes, this repertoire can change quickly,8 supporting the necessity of a constant surveillance of this pathogen.44 The contribution of putative virulence proteins identified so far and of other components of the cell, like glycolipids, which play an important role in virulence of C. diphtheriae, M. tuberculosis or R. equi,38,39,45,46 has to be elucidated at the molecular level by genetic experiments with defined mutants in future.
Materials and Methods
Bacterial strains and growth
Strains and plasmids used in this study are shown in Table 2. C. glutamicum and C. ulcerans strains were grown in Heart Infusion (HI) broth at 37°C. If appropriate, 50 μg ml−1 kanamycin was added. For generation of GFP expressing strains used in fluorescence microscopy studies, electrocompetent C. ulcerans cells were transformed with the plasmid pEPR1-p45gfp and positive clones were selected on HI agar with kanamycin.
Replication assay
THP-1 human monocytic cells cultures in 10 % FCS supplemented RPMI medium 1640 (containing 100 U penicillin ml−1 and 0.1 mg streptomycin ml−1) at 37°C in 5 % CO2 in a humified cell culture incubator. For replication assays, cells were seeded in 24-well plates (Nunc) at a density of 2 × 105 and differentiated by addition of 10 ng ml−1 phorbol 12-myristate 13-acetate (PMA) 24 h prior to infection. Overnight cultures of C. ulcerans grown in HI were re-inoculated to an OD600 of 0.1 in fresh medium and grown to an OD600 of 0.4 to 0.6. An inoculum with an MOI of 1 or 10 was prepared in RPMI without antibiotics and 500 µl per well were used to infect the cells. The plates were centrifuged for 5 min at 350 × g to synchronize infection and incubated for 30 min (37°C, 5 % CO2, 90 % humidity) to allow phagocytosis of bacteria. Subsequently, the supernatant containing non-engulfed bacteria was aspirated, cells were washed once with PBS and remaining extracellular bacteria were killed by addition of 100 µg ml−1 gentamicin in cell culture medium. After 2 h, cells were either lysed and intracellular bacteria were recovered or further incubated with medium containing 10 µg ml−1 gentamicin for analysis at later time points (8 h and 20 h). To recover intracellular bacteria, the medium was aspirated and cells were lysed by adding 500 µl of 0.1 % Triton X-100 in PBS. Serial dilutions of the lysate and the inoculi were plated on blood agar plates (Oxoid) using an Eddy Jet Version 1.22 (IUL Instruments). After incubation at 37°C for 2 days, the number of colony forming units (CFU) was determined. The ratio of bacteria used for infection (number of colonies on inoculum plates) and bacteria in the lysate (number of colonies on the lysate plates) multiplied with 100 gave the percentage of viable intracellular bacteria at different time points. When the survival of intracellular bacteria in THP-1 cells was analyzed over the time, the number of CFU at 2 h was set to 100 % and later time points were calculated based on this value. The assay was performed in 4 biological replicates each performed in triplicates and means and standard deviations were calculated.
Fluorescence microscopy
For qualitative analysis of intracellular CFU, THP-1 cells were infected with GFP-expressing bacteria and analyzed by fluorescence microscopy. THP-1 cells were seeded one day prior to infection in a density of 1 × 105 cells on sterile coverslips in 24-well plates. Overnight cultures of C. glutamicum or C. ulcerans strains transformed with plasmids encoding gfp cultivated in HI medium containing kanamycin were re-inoculated to an OD600 of 0.1 in fresh medium, harvested at the beginning of the exponential growth phase and used to infect macrophages as described above. After different time points, the medium was aspirated and cells were fixed by addition of 500 µl 4 % paraformaldehyde in PBS and incubated for 20 min at 37°C. Until further staining, cells were stored in PBS at 4°C. For subsequent analysis by microscopy, coverslips were incubated with 30 µl of Alexa Fluor® 647 Phalloidin diluted 1:200 in Image-iT™ FX Signal Enhancer (Molecular Probes, Life Technologies) for 45 min in the dark to stain the cytoskeleton of THP-1 cells. After washing twice with PBS, the coverslips were dried and embedded on glass slides in ProLong® Gold antifade mountant with DAPI (Molecular Probes, Life Technologies) and samples were stored in the dark at 4°C. Micrographs were taken with the confocal laser scanning microscope Leica SP5 CLSM-1P (Leica Microsystems) and analyzed with the LAS software suite.
Staining of acidic compartments in infected THP-1 cells
To analyze if C. ulcerans co-localizes with acidic compartments, THP-1 cells we were treated with 200 nM LysoTracker® Red DND-99 (Molecular Probes, Life Technologies), a red fluorescent dye that stains acidic compartments in live cells, 2 h before infection. Then, cells were infected and further treated as described for fluorescence microscopy above.
Automated analysis of fluorescence microscopic images
The analysis of bacteria on fluorescence images poses a demanding challenge as the bacteria tend to stick together and form clusters. To circumvent this problem, i.e. to avoid the problem of segmentation of single bacteria, data were analyzed with the software tool CaeT47 in 2 ways. In the first approach the area of all bacteria was analyzed on pixel level. For this purpose, an adaptive threshold algorithm48 based on k-means clustering49 was applied to the images of the GFP channel to determine pixels, which belong to bacteria. If intensities of corresponding pixels in the LysoTracker® Red DND-99 channel exceeded an empirically determined threshold, the pixels were regarded as co-localized. For all images the proportion between the number of co-localizing pixels and the total number of pixels of bacteria was determined. The second method was based on the evaluation of images at bacterial cell level. For this purpose, pixels belonging to bacteria which had been calculated in the first method, were grouped into regions. To prevent the analysis of cell clusters, these regions were filtered according to a statistical shape model to analyze exclusively single bacteria. The single bacteria had to reach a minimum average intensity in the LysoTracker® Red DND-99 channel to be positive for co-localization with acidic compartments. Then, the proportion between the number of positive bacteria and the total number of bacteria was determined.
FACS apoptosis assay (SubG1 analysis)
For analysis of apoptotic cell death induced by infection with C. ulcerans, cells were stained with PI and the SubG1 peak was analyzed by FACS. Four × 105 THP-1 cells were seeded 24 h prior to infection in 12-well plates and differentiated by addition of 10 ng ml−1 PMA. Cells were infected with C. ulcerans at an MOI of 50 as described above. After different time points, cells were harvested and fixed. To collect all cells, the supernatant with potentially detached cells was saved. Cells were washed once with 50 mM EDTA in PBS, detached by incubation for 5 to 10 min with trypsin at 37°C, and washed with 10 % FCS in PBS. Then everything was combined and centrifuged at 350 × g for 10 min. The supernatant was carefully removed and the cell pellet was washed once with 10 % FCS in PBS. The cell pellets were resuspended in 500 µl ice-cold PBS and 3 ml of 70 % ethanol (−20°C) were added dropwise under vortexing. Fixed cells were stored up to 2 weeks at 4°C. For staining, cells were harvested by centrifugation (500 × g, 10 min), and cell pellets were resuspended in 1 ml extraction buffer (9 parts 50 mM Na2HPO4, 1 part 25 mM citric acid, 0.1 % Triton X-100, 0.01 % NaN3, pH 7.8) to extract low molecular weight DNA, transferred to a FACS tube and centrifuged again (500 × g, 10 min). Finally, cells were stained by adding 400 µl of the staining buffer (10 mM PIPES, 0.1 N NaCl, 2 mM MgCl2, 0.1 % Triton X-100, 0.02 % NaN3, pH 6.8), 25 µl 10 mg ml−1 RNase in PBS and 20 µl 1 mg ml−1 PI dissolved in ddH2O. Until flow cytometric analysis on a BD FACS Canto II, samples were incubated in the dark.
FACS necrosis assay (7-AAD staining)
For analysis of necrotic cell death forms in response to infection with C. ulcerans, cells were stained with 7-AAD. Four × 105 THP-1 cells were seeded 24 h prior to infection in 12-well plates and differentiated by addition of 10 ng ml−1 PMA. Cells were infected with C. ulcerans at an MOI of 50 as described above. At different time points after infection, cells were harvested as described above for the SubG1 analysis, the resulting pellet was resuspended in 250 µl PBS but then not fixed but immediately stained with 5 µl of the 7-AAD staining solution (BD Bioscience). Uninfected cells served as negative control and cells treated with 0.01 % Triton X-100 as positive control. Analysis was performed within 10 minutes by flow cytometry on a BD FACS Canto II after gating and excluding cell debris. Data were analyzed using Kaluza™ flow cytometry analysis software V1.1 (Beckmann Coulter).
NF-κB reporter assay
THP1-Blue™ NF-κB cells (InvivoGen) carrying a stable integrated NF-κB-inducible secreted embryonic alkaline phosphatase (SEAP) reporter construct were used to analyze NF-κB induction by C. ulcerans. C. ulcerans strains were inoculated from an overnight culture to an OD600 of 0.1 in fresh medium and grown to an OD600 of 0.4 to 0.6. An inoculum with an OD600 of 1.25 in 1,000 µl PBS was prepared and 20 µl of this inoculum or of the 10−1 and 10−2 dilutions were mixed with 180 µl of a suspension with 5 × 105 THP1-Blue™ NF-κB cells in cell culture medium resulting in an MOI of 100, 10 or 1. UV-killed bacteria in the same concentrations were also carried along. After incubation for 20 h at cell culture conditions, the 96-well plates were centrifuged (350 × g, 5 min) and 20 µl of the cell free supernatant was mixed with 180 µl prewarmed SEAP detection reagent QUANTI-Blue™ (InvivoGen). After further incubation at cell culture conditions for 3 h, the levels of NF-κB-induced SEAP resulting in a color change from pink to blue was measured in a microplate reader (TECAN Infinite 200 PRO) at 620 nm.
Determination of cytokine excretion
For determination of cytokine activation through C. ulcerans, supernatants of infected THP-1 cells were collected after different time points and stored at −20°C. IL-6 and G-CSF concentrations were measured using the DuoSet ELISA Kits according to the manufacturer's recommendations (R&D systems). Briefly, the ELISA plates were coated over night with a capture antibody at room temperature, washed 3 times with 0.05 % Tween20 in PBS, blocked for 1 h at room temperature with 1 % BSA in PBS and washed again 3 times. Subsequently, 100 µl supernatant of infected cells or standard dilutions were added and the plates were incubated for 2 h, washed again 3 times and further incubated for 2 h at room temperature. After another washing step, a streptavidin-HRP solution was added and the plates were stored for 20 min under light exclusion, washed again and incubated for another 20 min in the dark with substrate solution. To stop the color reaction, 2 N H2SO4 was added to the wells and the optical density was determined using a microplate reader (TECAN Infinite 200 PRO) set to 450 nm with wavelength correction at 550 nm.
LDH release
The release of cytosolic lactate dehydrogenase (LDH) as a sign of host cell damage during infection was measured using the cytotoxicity detection kit according to the supplier (Roche). Briefly, 100 µl supernatant of infected cells were mixed with 2.5 µl of the provided catalyst solution and 112.5 µl of the provided dye solution in 96 well plates, incubated in the dark for 30 minutes and the absorbance was measured at 490 nm and wavelength correction at 620 nm in a multiplate reader (TECAN Infinite 200 PRO). Cells treated with 2 % Triton X-100 served as positive control for maximal LDH release and were set to 100 %, untreated cells served as negative control.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
The project was supported by the Deutsche Forschungsgemeinschaft in frame of SFB796 (A4, B8 and MGK).
References
- 1.Tauch A, Sandbote J. The family Corynebacteriaceae In: Rosenberg E, DeLong E, Lory S, Stackebrandt E, Thompson F, eds. The Prokaryotes. Berlin Heidelberg: Springer; 2014:239-77 [Google Scholar]
- 2.Burkovski A. Cell envelope of corynebacteria: structure and influence on pathogenicity. ISRN Microbiol 2013; 2013:935736; PMID:23724339; http://dx.doi.org/ 10.1155/2013/935736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dorella FA, Pacheco LG, Oliveira SC, Miyoshi A, Azevedo V. Corynebacterium pseudotuberculosis: microbiology, biochemical properties, pathogenesis and molecular studies of virulence. Vet Res 2006; 37:201-18; PMID:16472520; http://dx.doi.org/ 10.1051/vetres:2005056 [DOI] [PubMed] [Google Scholar]
- 4.Silva AS, Baraúna RA, de Sá PC, das Gracas DA, Carneiro AR, Thouvenin M, Azevedo V, Badell E, Guiso N, da Silva AL, Ramos RT. Draft genome sequence of Corynebacterium ulcerans FRC58, isolated from the bronchitic aspiration of a patient in France. Genome Announc 2014; 2:e01132-13; http://dx.doi.org/ 10.1128/genomeA.01132-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilbert R, Stewart FC. Corynebacterium ulcerans; a pathogenic microorganism resembling Corynebacterium diphtheriae. J Lab Clin Med 1927; 12:756-61 [Google Scholar]
- 6.Zakikhany K, Efstratiou A. Diphtheria in Europe: current problems and new challenges. Future Microbiol 2012; 7:595-607; PMID:22568715; http://dx.doi.org/ 10.2217/fmb.12.24 [DOI] [PubMed] [Google Scholar]
- 7.Meinel DM, Konrad R, Berger A, König C, Schmidt-Wieland T, Hogardt M, Bischoff H, Ackermann N, Hörmansdorfer S, Krebs S, et al.. Zoonotic transmission of toxigenic Corynebacterium ulcerans strain, Germany, 2012. Emerg Infect Dis 2015; 21:356-8; PMID:25625779; http://dx.doi.org/ 10.3201/eid2102.141160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meinel DM, Margos G, Konrad R, Krebs S, Blum H, Sing A. Next generation sequencing analysis of nine Corynebacterium ulcerans isolates reveals zoonotic transmission and a novel putative diphtheria toxin-encoding pathogenicity island. Genome Med 2014; 6:113; PMID:25587356; http://dx.doi.org/ 10.1186/s13073-014-0113-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berger A, Huber I, Merbecks SS, Ehrhard I, Konrad R, Hörmansdorfer S, Hogardt M, Sing A. Toxigenic Corynebacterium ulcerans in woman and cat. Emerg Infect Dis 2011; 17:1767-9; PMID:21888821; http://dx.doi.org/ 10.3201/eid1709.110391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergin IL, Chien CC, Marini RP, Fox JG. Isolation and characterization of Corynebacterium ulcerans from cephalic implants in macaques. Comp Med 2000; 50:530-5; PMID:11099137 [PubMed] [Google Scholar]
- 11.Contzen M, Sting R, Blazey B, Rau J. Corynebacterium ulcerans from diseased wild boars. Zoonoses Public Health 2011; 58:479-88; PMID:21824349; http://dx.doi.org/ 10.1111/j.1863-2378.2011.01396.x [DOI] [PubMed] [Google Scholar]
- 12.Eisenberg T, Mauder N, Contzen M, Rau J, Ewers C, Schlez K, Althoff G, Schauerte N, Geiger C, Margos G, et al.. Outbreak with clonally related isolates of Corynebacterium ulcerans in a group of water rats. BMC Microbiol 2015; 15:42; PMID:25887321; http://dx.doi.org/ 10.1186/s12866-015-0384-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster G, Patterson T, Howie F, Simpson V, Davison N, Efstratiou A, Lai S. Corynebacterium ulcerans in free-ranging otters. Vet Rec 2002; 150:524; PMID:12017529 [PubMed] [Google Scholar]
- 14.Hirai-Yuki A, Komiya T, Suzaki Y, Ami Y, Katsukawa C, Takahashi M, Yamamoto A, Yamada YK. Isolation and characterization of toxigenic Corynebacterium ulcerans from 2 closed colonies of cynomolgus macaques (Macaca fascicularis) in Japan. Comp Med 2013; 63:272-8; PMID:23759530 [PMC free article] [PubMed] [Google Scholar]
- 15.Hogg RA, Wessels J, Hart J, Efstratiou A, De Zoysa A, Mann G, Allen T, Pritchard GC. Possible zoonotic transmission of toxigenic Corynebacterium ulcerans from companion animals in a human case of fatal diphtheria. Vet Rec 2009; 165:691-2; PMID:19966333 [PubMed] [Google Scholar]
- 16.Hommez J, Devriese LA, Vaneechoutte M, Riegel P, Butaye P, Haesebrouck F. Identification of nonlipophilic corynebacteria isolated from dairy cows with mastitis. J Clin Microbiol 1999; 37:954-7; PMID:10074508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morris WE, Uzal FA, Cipolla AL. Pyogranulomatous meningoencephalitis in a goat due to Corynebacterium ulcerans. Vet Rec 2005; 156:317-8; PMID:15786922; http://dx.doi.org/ 10.1136/vr.156.10.317 [DOI] [PubMed] [Google Scholar]
- 18.Olson ME, Goemans I, Bolingbroke D, Lundberg S. Gangrenous dermatitis caused by Corynebacterium ulcerans in Richardson ground squirrels. J Am Vet Med Assoc 1988; 193:367-8; PMID:3182392 [PubMed] [Google Scholar]
- 19.Schuhegger R, Schoerner C, Dlugaiczyk J, Lichtenfeld I, Trouillier A, Zeller-Peronnet V, Busch U, Berger A, Kugler R, Hörmansdorfer S, et al.. Pigs as source for toxigenic Corynebacterium ulcerans. Emerg Infect Dis 2009; 15:1314-5; PMID:19751602; http://dx.doi.org/ 10.3201/eid1508.081568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seto Y, Komiya T, Iwaki M, Kohda T, Mukamoto M, Takahashi M, Kozaki S. Properties of corynephage attachment site and molecular epidemiology of Corynebacterium ulcerans isolated from humans and animals in Japan. Jpn J Infect Dis 2008; 61:116-22; PMID:18362399 [PubMed] [Google Scholar]
- 21.Sykes JE, Mapes S, Lindsay LL, Samitz E, Byrne BA. Corynebacterium ulcerans bronchopneumonia in a dog. J Vet Intern Med 2010; 24:973-6; PMID:20337919; http://dx.doi.org/ 10.1111/j.1939-1676.2010.0491.x [DOI] [PubMed] [Google Scholar]
- 22.Tejedor MT, Martin JL, Lupiola P, Gutierrez C. Caseous lymphadenitis caused by Corynebacterium ulcerans in the dromedary camel. Can Vet J 2000; 41:126-7; PMID:10723599 [PMC free article] [PubMed] [Google Scholar]
- 23.Dias AA, Silva FC Jr., Pereira GA, Souza MC, Camello TC, Damasceno JA, Pacheco LG, Miyoshi A, Azevedo VA, Hirata R Jr., et al.. Corynebacterium ulcerans isolated from an asymptomatic dog kept in an animal shelter in the metropolitan area of Rio de Janeiro, Brazil. Vector Borne Zoonotic Dis 2010; 10:743-8; PMID:20055577; http://dx.doi.org/ 10.1089/vbz.2009.0132 [DOI] [PubMed] [Google Scholar]
- 24.Mattos-Guaraldi AL, Sampaio JL, Santos CS, Pimenta FP, Pereira GA, Pacheco LG, Miyoshi A, Azevedo V, Moreira LO, Gutierrez FL, et al.. First detection of Corynebacterium ulcerans producing a diphtheria-like toxin in a case of human with pulmonary infection in the Rio de Janeiro metropolitan area, Brazil. Mem Inst Oswaldo Cruz 2008; 103:396-400; PMID:18660996; http://dx.doi.org/ 10.1590/S0074-02762008000400014 [DOI] [PubMed] [Google Scholar]
- 25.Trost E, Al-Dilaimi A, Papavasiliou P, Schneider J, Viehoever P, Burkovski A, Soares SC, Almeida SS, Dorella FA, Miyoshi A, et al.. Comparative analysis of two complete Corynebacterium ulcerans genomes and detection of candidate virulence factors. BMC Genomics 2011; 12:383; PMID:21801446; http://dx.doi.org/ 10.1186/1471-2164-12-383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dias AA, Silva FC Jr., Santos LS, Ribeiro-Carvalho MM, Sabbadini PS, Santos CS, Filardy AA, Myioshi A, Azevedo VA, Hirata R Jr., et al.. Strain-dependent arthritogenic potential of the zoonotic pathogen Corynebacterium ulcerans. Vet Microbiol 2011; 153:323-31; PMID:21742447; http://dx.doi.org/ 10.1016/j.vetmic.2011.06.007 [DOI] [PubMed] [Google Scholar]
- 27.Hacker E, Ott L, Hasselt K, Mattos-Guaraldi AL, Tauch A, Burkovski A. Colonization of human epithelial cell lines by Corynebacterium ulcerans from human and animal sources. Microbiology 2015; 161:1582-91; PMID:26066797; http://dx.doi.org/ 10.1099/mic.0.000121 [DOI] [PubMed] [Google Scholar]
- 28.Simpson-Louredo L, Ramos JN, Peixoto RS, Santos LS, Antunes CA, Ladeira EM, Santos CS, Vieira VV, Boas MH, Hirata R Jr., et al.. Corynebacterium ulcerans isolates from humans and dogs: fibrinogen, fibronectin and collagen-binding, antimicrobial and PFGE profiles. Antonie Van Leeuwenhoek 2014; 105:343-52; PMID:24281735; http://dx.doi.org/ 10.1007/s10482-013-0080-5 [DOI] [PubMed] [Google Scholar]
- 29.McKean S, Davies J, Moore R. Identification of macrophage induced genes of Corynebacterium pseudotuberculosis by differential fluorescence induction. Microbes Infect 2005; 7:1352-63; PMID:16046166; http://dx.doi.org/ 10.1016/j.micinf.2005.05.002 [DOI] [PubMed] [Google Scholar]
- 30.dos Santos CS, dos Santos LS, de Souza MC, dos Santos Dourado F, de Souza de Oliveira Dias AA, Sabbadini PS, Pereira GA, Cabral MC, Hirata R Jr., Mattos-Guaraldi AL. Non-opsonic phagocytosis of homologous non-toxigenic and toxigenic Corynebacterium diphtheriae strains by human U-937 macrophages. Microbiol Immunol 2010; 54:1-10; PMID:20055937; http://dx.doi.org/ 10.1111/j.1348-0421.2009.00179.x [DOI] [PubMed] [Google Scholar]
- 31.Hirano T, Ishihara K, Hibi M. Roles of STAT3 in mediating the cell growth, differentiation and survival signals relayed through the IL-6 family of cytokine receptors. Oncogene 2000; 19:2548-56; PMID:10851053; http://dx.doi.org/ 10.1038/sj.onc.1203551 [DOI] [PubMed] [Google Scholar]
- 32.Solaroglu I, Cahill J, Jadhav V, Zhang JH. A novel neuroprotectant granulocyte-colony stimulating factor. Stroke 2006; 37:1123-8; PMID:16514095; http://dx.doi.org/ 10.1161/01.STR.0000208205.26253.96 [DOI] [PubMed] [Google Scholar]
- 33.Hodgson AL, Krywult J, Corner LA, Rothel JS, Radford AJ. Rational attenuation of Corynebacterium pseudotuberculosis: potential cheesy gland vaccine and live delivery vehicle. Infect Immun 1992; 60:2900-5; PMID:1612756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Antunes CA, Sanches Dos Santos L, Hacker E, Köhler S, Bösl K, Ott L, de Luna M, Hirata R Jr., Azevedo VA, Mattos-Guaraldi AL, et al.. Characterization of DIP0733, a multi-functional virulence factor of Corynebacterium diphtheriae. Microbiology 2015; 161:639-47; PMID:25635272; http://dx.doi.org/ 10.1099/mic.0.000020 [DOI] [PubMed] [Google Scholar]
- 35.Ott L, McKenzie A, Baltazar MT, Britting S, Bischof A, Burkovski A, Hoskisson PA. Evaluation of invertebrate infection models for pathogenic corynebacteria. FEMS Immunol Med Microbiol 2012; 65:413-21; PMID:22443092; http://dx.doi.org/ 10.1111/j.1574-695X.2012.00963.x [DOI] [PubMed] [Google Scholar]
- 36.Bonmarin I, Guiso N, Le Fleche-Mateos A, Patey O, Patrick AD, Levy-Bruhl D. Diphtheria: A zoonotic disease in France? Vaccine 2009; 27:4196-200; PMID:19393707; http://dx.doi.org/ 10.1016/j.vaccine.2009.04.048 [DOI] [PubMed] [Google Scholar]
- 37.Wagner KS, White JM, Crowcroft NS, De Martin S, Mann G, Efstratiou A. Diphtheria in the United Kingdom, 1986-2008: the increasing role of Corynebacterium ulcerans. Epidemiol Infect 2010; 138:1519-30; PMID:20696088; http://dx.doi.org/ 10.1017/S0950268810001895 [DOI] [PubMed] [Google Scholar]
- 38.Indrigo J, Hunter RL Jr., Actor JK. Cord factor trehalose 6,6'-dimycolate (TDM) mediates trafficking events during mycobacterial infection of murine macrophages. Microbiology 2003; 149:2049-59; PMID:12904545; http://dx.doi.org/ 10.1099/mic.0.26226-0 [DOI] [PubMed] [Google Scholar]
- 39.Sydor T, von Bargen K, Hsu FF, Huth G, Holst O, Wohlmann J, Becken U, Dykstra T, Söhl K, Lindner B, et al.. Diversion of phagosome trafficking by pathogenic Rhodococcus equi depends on mycolic acid chain length. Cell Microbiol 2013; 15:458-73; PMID:23078612; http://dx.doi.org/ 10.1111/cmi.12050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hard GC. Comparative toxic effect of the surface lipid of Corynebacterium ovis on peritoneal macrophages. Infect Immun 1975; 12:1439-49; PMID:1205621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tashjian JJ, Campbell SG. Interaction between caprine macrophages and Corynebacterium pseudotuberculosis: an electron microscopic study. Am J Vet Res 1983; 44:690-3; PMID:6869967 [PubMed] [Google Scholar]
- 42.Aki T, Funakoshi T, Uemura K. Regulated necrosis and its implications in toxicology. Toxicology 2015; 333:118-26; PMID:25865964; http://dx.doi.org/ 10.1016/j.tox.2015.04.003 [DOI] [PubMed] [Google Scholar]
- 43.Sridharan H, Upton JW. Programmed necrosis in microbial pathogenesis. Trends Microbiol 2014; 22:199-207; PMID:24565922; http://dx.doi.org/ 10.1016/j.tim.2014.01.005 [DOI] [PubMed] [Google Scholar]
- 44.Both L, Collins S, de Zoysa A, White J, Mandal S, Efstratiou A. Molecular and epidemiological review of toxigenic diphtheria infections in England between 2007 and 2013. J Clin Microbiol 2015; 53:567-72; PMID:25502525; http://dx.doi.org/ 10.1128/JCM.03398-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Axelrod S, Oschkinat H, Enders J, Schlegel B, Brinkmann V, Kaufmann SH, Haas A, Schaible UE. Delay of phagosome maturation by a mycobacterial lipid is reversed by nitric oxide. Cell Microbiol 2008; 10:1530-45; PMID:18363878; http://dx.doi.org/ 10.1111/j.1462-5822.2008.01147.x [DOI] [PubMed] [Google Scholar]
- 46.Moreira LO, Mattos-Guaraldi AL, Andrade AF. Novel lipoarabinomannan-like lipoglycan (CdiLAM) contributes to the adherence of Corynebacterium diphtheriae to epithelial cells. Arch Microbiol 2008; 190:521-30; PMID:18575847; http://dx.doi.org/ 10.1007/s00203-008-0398-y [DOI] [PubMed] [Google Scholar]
- 47.Wiesmann V, Reimer D, Franz D, Hüttmayer H, Mielenz D, Wittenberg T. Automated high-throughput analysis of B cell spreading on immobilized antibodies with whole slide imaging. Curr Directions in Biomed Engineering 2015; 1:224-7; http://dx.doi.org/ 10.1515/cdbme-2015-0056 [DOI] [Google Scholar]
- 48.Held C. Towards increased efficiency and automation in fluorescence micrograph analysis based on hand-labeled data Bielefeld: Universität Bielefeld, 2013. [Google Scholar]
- 49.Hartigan JA, Wong MA. Algorithm AS 136: A K-means Clustering Algorithm. J Royal Statistical Soc Series C (applied Statistics) 1979; 28:100-8 [Google Scholar]
- 50.Abe S, Takayama K, Kinoshita S. Taxonomical studies on glutamic acid producing bacteria. J Gen Microbiol 1967; 13:279-301; http://dx.doi.org/ 10.2323/jgam.13.279 [DOI] [Google Scholar]
- 51.Tsuchiya S, Yamabe M, Yamaguchi Y, Kobayashi Y, Konno T, Tada K. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1). Int J Cancer 1980; 26:171-6; PMID:6970727; http://dx.doi.org/ 10.1002/ijc.2910260208 [DOI] [PubMed] [Google Scholar]
- 52.Ralph P, Nakoinz I. Antibody-dependent killing of erythrocyte and tumor targets by macrophage-related cell lines: enhancement by PPD and LPS. J Immunol 1977; 119:950-54; PMID:894031 [PubMed] [Google Scholar]
- 53.Fiani ML, Beitz J, Turvy D, Blum JS, Stahl PD. Regulation of mannose receptor synthesis and turnover in mouse J774 macrophages. J Leukoc Biol 1998; 64:85-91; PMID:9665280 [DOI] [PubMed] [Google Scholar]
- 54.Knoppová M, Phensaijai M, Veselý M, Zemanová M, Nesvera J, Pátek M. Plasmid vectors for testing in vivo promoter activities in Corynebacterium glutamicum and Rhodococcus erythropolis. Curr Microbiol 2007; 55:234-9; http://dx.doi.org/ 10.1007/s00284-007-0106-1 [DOI] [PubMed] [Google Scholar]


