Abstract
MicroRNA (miRNA) are a class of noncoding RNA involved in regulating gene expression by a posttranscriptional mechanism. Based on work from our laboratory, this review explores the hypothesis that a recently described muscle-specific miRNA, myomiR, network has a central role in the regulation of skeletal muscle plasticity by coordinating changes in fiber type and muscle mass in response to altered contractile activity.
Keywords: exercise, hypertrophy, atrophy, endurance, miRNA, miR
INTRODUCTION
MicroRNA (miRNA) represent a family of noncoding RNA of approximately 23 nucleotides that regulate gene expression through a posttranscriptional mechanism. Originally described as “short temporal RNA” in Caenorhabditis elegans, miRNA emerged from a decade of relative obscurity with the observation that let-7 was expressed in a wide range of animals and, then a year later, with the identification of 30 to 50 new miRNA in the fly, worm, and human (3,15,17,18,29). Since then, there has been an explosion in the number of identified miRNA, such that the latest version of miRBase contains more than 17,000 distinct mature miRNA (miR) sequences from 142 species with 1048 validated miR in humans (14).
The most recent analysis has revealed that more than 60% of all human protein coding genes harbor conserved miRNA target sites within their 3′-untranslated region (UTR), a finding consistent with the emerging notion that miRNA are involved in most developmental and cellular processes (9). Whereas the precise molecular details remain to be worked out, it is well established that miRNA regulate gene expression through a posttranscriptional mechanism involving inhibition of translation and/or degradation of the mRNA transcript (8). Somewhat contrary to what has been thought historically, Guo and colleagues (11) used ribosome profiling to show that the vast majority (>80%) of miRNA repression of gene expression is accounted for by changes in transcript levels resulting from mRNA destabilization. Although this is potentially a very important finding, it will need to be confirmed in vivo, given, at least in human skeletal muscle, that miRNA appear to function primarily through inhibition of translation (10). Moreover, beyond the mechanistic implications, this finding has important relevance to experimental design as it indicates that expression profiling is in fact a useful tool for identifying potential miRNA target genes.
Based on work from our laboratory, the purpose of this review was to examine the hypothesis that members of the muscle-specific miRNA family, designated myomiR, are involved in regulating the phenotypic changes that occur in fiber type and mass in response to altered patterns of muscle activity. The review provides a brief introduction on myomiR and an overview on what is known currently about the role of myomiR in skeletal muscle adaptation. It finishes with a presentation of our findings and those of others, suggesting that the myomiR network may have a role in coordinating changes in fiber type and muscle mass in response to exercise.
MyomiR: Muscle-Specific MiRNA
In addition to discovering that miRNA were conserved phylogenetically, there also was the realization that some miRNA seemed to be expressed in a tissue-specific manner. The mature form of miRNA-1 (miR-1) was found to be expressed only in the human heart, but not in the brain, kidney, liver, lung, or HeLa cells (15,18). The tissue specificity of some miRNA was later confirmed by the finding that miR-1, miR-122a, and miR-124a were expressed only in the muscle, liver, and brain, respectively (16). Sempere and coworkers (31) provided the first description of striated muscle–specific miRNA, miR-1, miR-133a, and miR-206, which were designated myomiR subsequently (23,31). The myomiR family has expanded to include miR-208a, miR-208b, miR-499, and most recently, miR-486 (Table 1) (32,35,36). Most myomiR family members are expressed in both the heart and skeletal muscle, except for miR-208a, which is cardiac specific, and miR-206, which is skeletal muscle specific and enriched in slow-twitch muscles such as the soleus (Table 1) (25).
TABLE 1.
MyomiR: muscle-specific microRNA.
| MyomiR | Host Gene | Expression Pattern | Knockout Phenotype | Study |
|---|---|---|---|---|
| MiR-1-1 | Mib1 | Heart, skeletal muscle | No knockout | — |
| MiR-1-2 | Intergenic | Heart, skeletal muscle | 50% lethal, cardiac defect | Zhao et al., 2007 (38) |
| MiR-133a-1 | Mib1 | Heart, skeletal muscle | No overt phenotype | Liu et al., 2008 (22) |
| MiR-133a-2 | Intergenic | Heart, skeletal muscle | No overt phenotype | Liu et al., 2008 (22) |
| MiR-206 | Intergenic | Skeletal muscle (Type I) | No overt phenotype | Williams et al., 2009 (37) |
| MiR-208a | Myh6 | Heart | Blunted stress response | van Rooij et al., 2007 (36) |
| Conduction defects | Callis et al., 2009 (5) | |||
| MiR | Myh7 | Heart (low), skeletal muscle (Type I) | No overt phenotype | van Rooij et al., 2009 (35) |
| MiR | Ank1 | Heart, skeletal muscle | No knockout | — |
| MiR | Myh7b/14 | Heart, skeletal muscle (Type I) | No overt phenotype | van Rooij et al., 2009 (35) |
Since their initial description, a great deal of effort has gone into determining the function of myomiR in striated muscle (reviewed in (21)). The finding that overexpression of miR-1 caused a shift toward a myogenic profile in HeLa cells suggested that myomiR may have a fundamental role in promoting a muscle cell identity (20). The importance of myomiR in myogenesis was first demonstrated by the finding of Sokol and Ambros (33), who reported that the deletion of miR-1 in the fly resulted in premature death from a failure of skeletal muscle to grow properly during the second instar. Table 1 lists the known myomiR, their host genes, expression patterns, and whether they have been knocked out in the mouse. Interestingly, myomiR seem to have uniform expression throughout the muscle (miR-1 and miR-133a), independent of fiber type, or are enriched in Type I muscles (miR-206, miR-208b, and miR-499); to date, no miRNA has been reported to be enriched specifically in fast-twitch Type II muscle. As detailed in Table 1, most of the myomiR have been deleted in the mouse with surprisingly little impact on skeletal muscle phenotype (5,22,35–38). For example, deletion of skeletal muscle-specific miR-206 resulted in no obvious phenotype, as reflected by no change in soleus muscle weight, morphology, or fiber-type distribution; however, recovery from denervation was delayed in the muscle of the miR-206 knockout (37). This finding is consistent with earlier work by the Olson laboratory, showing that miR-208a is necessary for the stress response involved in cardiac hypertrophy (36). Collectively, these findings provide credence to the notion that a primary function of miRNA is to mediate the stress response of the cell by helping to restore homeostasis through regulating gene expression (19).
Regulation of MyomiR Expression by Changes in Muscle Activity
It is well known that changes in the contractile activity of a muscle can induce a number of different stresses, the most obvious being mechanical and metabolic. Accordingly, it is reasonable to speculate a priori that miRNA may have a role in the stress response of muscle to changes in use. Table 2 summarizes the results of published studies that have examined the expression of miRNA in response to changes in contractile activity, with an emphasis on myomiR (1,2,7,24,26, 28,30). To aid in identifying a potential pattern in myomiR expression in response to different types of activity, Table 2 has been organized into resistance exercise, unloading, and endurance exercise (acute and chronic). Most of the studies have focused on the canonical myomiR (miR-1, miR-133a, and miR-206), with miR-1 appearing to be the most responsive to changes in contractile activity, regardless of the modality. With resistance exercise, independent of the species or duration, miRNA expression was down-regulated, miR-1 in particular. Similarly, prolonged unloading of muscle, whether by casting, spaceflight, or hind limb suspension, resulted in a decrease in miRNA expression. It seems that unloading for less than a week brings about little to no change in miRNA expression, whereas longer time points (12 and 28 days) lead to a more significant decrease (40%–60%) in the myomiR expression.
TABLE 2.
MyomiR expression in response to changes in contractile activity.
| MyomiR | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Modality | Duration | Species | Muscle | 1 | 133a | 206 | 208b | 486 | 499 | Study |
| Resistance exercise | ||||||||||
| Mechanical overload | 14 d | Mouse | Plantaris | ↓50% | ↓50% | No Δ | — | — | — | McCarthy, 2007 (24) |
| Leg extension + essential amino acids | 8 × 10 | Human | Vastus lateralis | ↓38% | No Δ | No Δ | — | — | — | Drummond et al., 2008 (7) |
| Unloading | ||||||||||
| Immobilization | 5 d | Mouse | Gastrocnemius | ↓22% | No Δ | ↓13% | Low | No Δ | Low | Aoi et al., 2010 (2) |
| Spaceflight | 12 d | Mouse | Gastrocnemius | No Δ | No Δ | ↓50% | — | — | — | Allen et al., 2009 (1) |
| Hind limb unloading | 7 and 28 d | Rat | Soleus | No Δ | No Δ | No $ | ?60% | — | ↓40% | McCarthy et al., 2009 (26) |
| Endurance exercise | ||||||||||
| Treadmill running | 90 min | Mouse | Quadriceps | 40% | No Δ | — | — | — | — | Safdar et al., 2009 (30) |
| Cycle ergometer | 60 min | Human | Vastus lateralis | ↑30% | ↑37% | No Δ | — | — | — | Nielsen et al., 2010 (28) |
| Treadmill running | 4 wk | Mouse | Gastrocnemius | ↑71% | No Δ | ↑24% | Low | ↑24% | Low | Aoi et al., 2010 (2) |
| Cycle ergometer | 12 wk | Human | Vastus lateralis | ↓32% | ↓23% | ↓19% | — | — | — | Nielsen et al., 2010 (28) |
Down arrow (↓), decreased expression; em-dash (—), expression not measured; low, low expression; myomiR, muscle-specific microRNA; no Δ, no change in expression; up arrow (↑), increased expression.
The results from studies investigating the impact of endurance exercise on miRNA expression are less clear than the results from the resistance exercise and unloading studies. In humans, a single bout of cycling caused an approximately 40% increase in miR-1 and miR-133a expressions, whereas a bout of treadmill running in the mouse resulted in a similar increase in the miR-1 expression, but no change in the miR-133a expression (Table 1). After training, Nielsen and coworkers (28) observed a consistent decrease in myomiR expression, whereas Aoi et al. (2) reported the opposite — a uniform increase in myomiR expression after endurance exercise training. The reason for the discrepancy in the results of these two studies is not known but may be related to a difference in species (human vs mouse), exercise modality (cycling vs treadmill running), length of training (12 wk vs 4 wk), and/or muscle (vastus lateralis vs gastrocnemius). Interestingly enough, in trained individuals, a single bout of cycling no longer induced an increase in myomiR expression as observed in untrained persons (28). Collectively, the results of the studies summarized in Table 2 provide the first evidence suggesting that myomiR have a role in skeletal muscle plasticity.
Given that changes in muscle use can induce muscle injury, one question that comes to mind immediately is the possible role of myomiR in muscle repair. If so, do the changes reported in myomiR expression after exercise reflect their involvement in the injury/repair process? Although no studies to date have addressed this important issue directly, there are studies examining muscle regeneration after cardiotoxin injection or laceration (6,27). Together, these studies indicate that myomiR can enhance muscle regeneration by promoting satellite cell differentiation through the down-regulation of Pax7 expression. It will be of interest to see if a similar mechanism is operative during muscle repair in response to exercise.
MyomiR Network
Although the field has yet to explore fully the role of the new members of the myomiR family, our laboratory reported changes in miR-208b and miR-499 expression during muscle atrophy that portends a role for these family members in skeletal muscle plasticity (26). Specifically, we found that the expressions of miR-208b and miR-499 were decreased significantly by 60% and 40%, respectively, in the rat soleus muscle after 28 days of hind limb unloading (26). In contrast, there was no change in the canonical myomiR after 2 and 7 days of hind limb loading, although it remains to be determined if the expression of these miRNA are changed after a longer period of unloading. Our finding that miR-208b and miR-499 expression was down-regulated during muscle atrophy was exciting but took on new meaning when it was realized that these miRNA were part of a recently described myomiR network (34).
The idea of a myomiR network was proposed originally by the Olson laboratory in a review article discussing the emerging role of miRNA in muscle (34). The first clue of a myomiR network appeared in a study by van Rooij and coworkers (36), in which they showed that inactivation of miR-208 (now referred to as miR-208a) blunted induction of the Myh7 gene in response to a hypertrophic stimulus or hyperthyroidism in the heart; Myh7 encodes the β-myosin heavy-chain (β-MHC) protein. The discovery that a miRNA derived from an intron of a myosin gene (miR-208a is encoded by an intron of Myh6, which encodes the α-MHC protein) regulated the expression of a second myosin gene, the β-MHC, was an intriguing revelation and suggested that other such interactions might exist. In a tour de force study, the Olson laboratory provided proof of the myomiR network recently and laid out in detail the components and interactions of the network (35).
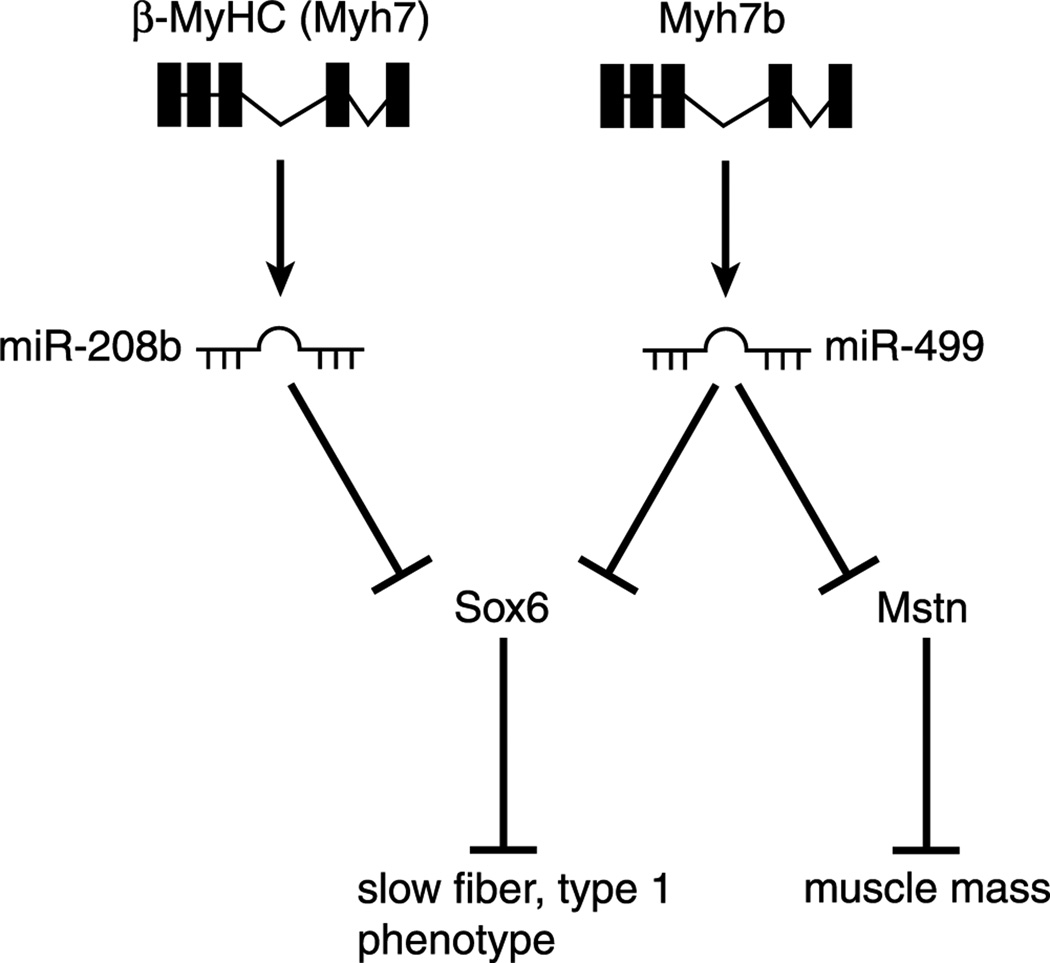
The portion of the myomiR network that is operative in skeletal muscle is presented in the Figure; the full myomiR network includes Myh6/miR-208a, which is not expressed in skeletal muscle under normal conditions (35). As depicted, the miR-208b is encoded by intron 31 of Myh7, and miR-499 is derived from intron 19 of the Myh7b gene. The expression of these myomiR parallels the expression of their respective host genes, such that miR-208b is enriched in slow-twitch Type I muscles with a very low expression in the heart, whereas miR-499 is expressed in the heart and enriched in Type I muscles (Table 1). These miRNA have been shown to regulate the expression of transcription factors (Sox6, Purβ, and Sp3) that are known to repress the expression of the slow-twitch phenotype, as assessed by β-MHC expression (12,13,35). In particular, studies have provided strong evidence that Sox6 is a potent repressor of β-MHC expression (12,35).
Figure.
The muscle-specific microRNA (myomiR) network in skeletal muscle. A schematic diagram of the myomiR network in skeletal muscle. MiRNA-208b and miR-499 are each encoded by an intron (31 and 19, respectively) within myosin heavy-chain (MHC) (Myh) genes 7 and 7b, respectively. Myh7 encodes the β-MHC protein that is expressed highly in slow-twitch skeletal muscle. Both miR-208b and miR-499 have been shown experimentally to regulate the expression of transcriptional repressors (Sox6, Purβ, and Sp3) of β-MHC expression. Multiple lines of evidence indicate that Sox6 is a major repressor of β-MHC expression in skeletal muscle. In addition to regulating fiber type, miR-208b and miR- 499 have been shown to regulate myostatin (Mstn) expression, suggesting that the myomiR network may coordinate changes in fiber type and muscle mass.
The study by van Rooij and colleagues (35) demonstrated that the myomiR network sets fiber type in skeletal muscle by repressing the expression of transcription factors, that is, Sox6, known to inhibit the expression of slow-twitch genes such as the β-MHC gene; in essence, miR-208b and miR-499 repress the repressors, thereby promoting the slow-twitch phenotype (35). These molecular interactions establish a feedback circuit in which miR-208b regulates the expression of Sox6, which in turn represses Myh7 expression, the host gene of miR-208b. As proof of this feedback mechanism, inactivation of both miR-208b and miR-499 resulted in a significant loss of Type I fibers in the soleus muscle that was associated with a 60% decrease in β-MHC expression concomitant with increased expression of Sox6 (35). Conversely, overexpression of miR-499 caused the complete conversion of all fibers in the soleus muscle to Type I from about 55% Type I in the wild-type mouse soleus. The conversion of skeletal muscle to a Type I phenotype by overexpression of miR-499 increased running performance as the miR-499 transgenic strain ran 50% longer than the wild-type strain in a run to exhaustion bout (35). Interestingly, overexpression of Sox6 caused almost complete loss of β-MHC and slow troponin I (Tnni1) expression but did not alter the expression of the fast troponins (Tnni2 and Tnnt3), indicating that target genes other than Sox6 are involved in the regulation of fast-twitch fiber gene expression (35). Together, the results of this study show that the myomiR network is a master regulator of fiber type in adult skeletal muscle (35).
MyomiR Network Regulation During Fast to Slow Fiber-Type Transition
What role does the myomiR network have in skeletal muscle plasticity? Whereas this question remains to be explored fully, results from our laboratory provided the first evidence suggesting that the myomiR network may be operative during periods of muscle plasticity. A hallmark of the fiber-type transition associated with skeletal muscle atrophy is the down-regulation of the slow-twitch myosin heavy-chain gene, the β-MHC. Accordingly, the decreases in miR-208b and miR-499 expressions we observed with muscle atrophy resulted in a greater than twofold increase in Sox6 expression, which, in turn, was associated with repression of β-MHC expression by 28% (26). Although these results are highly suggestive, we need to perform the same experiments using the miR-208b- and miR-499-knockout mice to confirm the involvement of the myomiR network in the fiber-type transition associated with muscle atrophy. Furthermore, it will be of great interest to determine if the myomiR network is involved in regulating the phenotypic transition known to occur with skeletal muscle hypertrophy.
MyomiR Network Regulation of Muscle Mass
One aspect of the myomiR network that has not been investigated is the possibility that the network also will be involved in regulating skeletal muscle mass. A predicted target gene of miR-208 and miR-499 is myostatin (Mstn), a major regulator of skeletal muscle mass. Callis and colleagues (5) reported that miR-208 can repress Mstn expression; first, by an in-vitro 3′-UTR reporter gene assay and then in vivo, showing increased myostatin protein in the miR-208a knockout and decreased myostatin protein in transgenic miR-208a overexpression, which was associated with cardiac hypertrophy. Bell et al. (4) extended these findings by showing that miR-499 also was capable of repressing Mstn expression. The miR-208b- and miR-499-knockout mice will be an invaluable resource for determining if, in fact, the myomiR network does have a role in regulating skeletal mass in addition to fiber type.
SUMMARY
The MiRNA have emerged as important players in the regulation of gene expression. The notion that they are involved primarily in the stress response of the cell makes them ideal candidates for mediating the response of skeletal muscle to changes in contractile activity. Although it is still early, the findings of the studies presented in Table 2 suggest that muscle-specific miRNA, the myomiR, will have a central role in skeletal muscle plasticity. This optimism, however, is tempered by the realization that the findings to date, which support a role for myomiR in muscle plasticity, are correlative. The challenge for the field will be to move beyond these descriptive studies by performing gain- and loss-of-function studies using the genetic mouse models described in Table 1. Moreover, to understand fully the molecular mechanism by which myomiR regulate skeletal muscle plasticity, it will be essential to identify and validate by microarray analysis and 3′-UTR assay, respectively, target genes. It will be exciting to see if the myomiR network is involved truly in the acute response to changes in contractile activity as well as coordinating the change in fiber type and muscle mass that are known to occur with extended training.
Acknowledgments
J.J. McCarthy is supported by a National Institutes of Health grant funding (no. RAR053641A; no. RAR060701A).
References
- 1.Allen DL, Bandstra ER, Harrison BC, et al. Effects of spaceflight on murine skeletal muscle gene expression. J. Appl. Physiol. 2009;106:582–595. doi: 10.1152/japplphysiol.90780.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoi W, Naito Y, Mizushima K, et al. The microRNA miR-696 regulates PGC-1{alpha} in mouse skeletal muscle in response to physical activity. Am. J. Physiol. Endocrinol. Metab. 2010;298:E799–E806. doi: 10.1152/ajpendo.00448.2009. [DOI] [PubMed] [Google Scholar]
- 3.Arasu P, Wightman B, Ruvkun G. Temporal regulation of lin-14 by the antagonistic action of two other heterochronic genes lin-4 and lin-28. Genes Dev. 1991;5:1825–1833. doi: 10.1101/gad.5.10.1825. [DOI] [PubMed] [Google Scholar]
- 4.Bell ML, Buvoli M, Leinwand LA. Uncoupling of expression of an intronic microRNA and its myosin host gene by exon skipping. Mol. Cell. Biol. 2010;30:1937–1945. doi: 10.1128/MCB.01370-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callis TE, Pandya K, Seok HY, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J. Clin. Invest. 2009;119:2772–2786. doi: 10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dey BK, Gagan J, Dutta A. MiR-206 and-486 induce myoblast differentiation by downregulating Pax7. Mol. Cell. Biol. 2011;31:203–214. doi: 10.1128/MCB.01009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drummond MJ, McCarthy JJ, Fry CS, Esser KA, Rasmussen BB. Aging differentially affects human skeletal muscle microRNA expression at rest and after an anabolic stimulus of resistance exercise and essential amino acids. Am. J. Physiol. Endocrinol. Metab. 2008;295:E1333–E1340. doi: 10.1152/ajpendo.90562.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 9.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallagher IJ, Scheele C, Keller P, et al. Integration of microRNA changes in vivo identifies novel molecular features of muscle insulin resistance in type 2 diabetes. Genome Med. 2010;2:9. doi: 10.1186/gm130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagiwara N, Yeh M, Liu A. Sox6 is required for normal fiber type differentiation of fetal skeletal muscle in mice. Dev. Dyn. 2007;236:2062–2076. doi: 10.1002/dvdy.21223. [DOI] [PubMed] [Google Scholar]
- 13.Ji J, Tsika GL, Rindt H, et al. Puralpha and Purbeta collaborate with Sp3 to negatively regulate beta-myosin heavy chain gene expression during skeletal muscle inactivity. Mol. Cell Biol. 2007;27:1531–1543. doi: 10.1128/MCB.00629-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kozomara A, Griffiths-Jones S. MiRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 16.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 17.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 18.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 19.Leung AK, Sharp PA. MicroRNA functions in stress responses. Mol. Cell. 2010;40:205–215. doi: 10.1016/j.molcel.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 21.Liu N, Olson EN. MicroRNA regulatory networks in cardiovascular development. Dev. Cell. 2010;18:510–525. doi: 10.1016/j.devcel.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu N, Bezprozvannaya S, Williams AH, et al. MicroRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCarthy JJ. MicroRNA-206: the skeletal muscle-specific myomiR. Biochim. Biophys. Acta. 2008;1779:682–691. doi: 10.1016/j.bbagrm.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCarthy JJ, Esser KA. MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. J. Appl. Physiol. 2007;102:306–313. doi: 10.1152/japplphysiol.00932.2006. [DOI] [PubMed] [Google Scholar]
- 25.McCarthy JJ, Esser KA, Andrade FH. MicroRNA-206 is overexpressed in the diaphragm but not the hindlimb muscle of mdx mouse. Am. J. Physiol. Cell. Physiol. 2007;293:C451–C457. doi: 10.1152/ajpcell.00077.2007. [DOI] [PubMed] [Google Scholar]
- 26.McCarthy JJ, Esser KA, Peterson CA, Dupont-Versteegden EE. Evidence of myomiR network regulation of beta-myosin heavy chain gene expression during skeletal muscle atrophy. Physiol. Genomics. 2009;39:219–226. doi: 10.1152/physiolgenomics.00042.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakasa T, Ishikawa M, Shi M, Shibuya H, Adachi N, Ochi M. Acceleration of muscle regeneration by local injection of muscle-specific micro-RNAs in rat skeletal muscle injury model. J. Cell Mol. Med. 14:2495–2505. doi: 10.1111/j.1582-4934.2009.00898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nielsen S, Scheele C, Yfanti C, et al. Muscle specific microRNAs are regulated by endurance exercise in human skeletal muscle. J. Physiol. 2010;588:4029–4037. doi: 10.1113/jphysiol.2010.189860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pasquinelli AE, Reinhart BJ, Slack F, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 30.Safdar A, Abadi A, Akhtar M, Hettinga BP, Tarnopolsky MA. MiRNA in the regulation of skeletal muscle adaptation to acute endurance exercise in C57Bl/6J male mice. PLoS One. 2009;4:e5610. doi: 10.1371/journal.pone.0005610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Small EM, O’Rourke JR, Moresi V, et al. Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4218–4223. doi: 10.1073/pnas.1000300107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sokol NS, Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes Dev. 2005;19:2343–2354. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Rooij E, Liu N, Olson EN. MicroRNAs flex their muscles. Trends Genet. 2008;24:159–166. doi: 10.1016/j.tig.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 35.van Rooij E, Quiat D, Johnson BA, et al. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev. Cell. 2009;17:662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 37.Williams AH, Valdez G, Moresi V, et al. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326:1549–1554. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao Y, Ransom JF, Li A, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1–2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]