Abstract
Arabidopsis thaliana cryptochrome 2 (AtCRY2), a light-sensitive photosensory protein, was previously adapted for use controling protein-protein interactions through light-dependent binding to a partner protein, CIB1. While the existing CRY2/CIB dimerization system has been used extensively for optogenetic applications, some limitations exist. Here, we set out to optimize function of the CRY2/CIB system, to identify versions of CRY2/CIB that are smaller, show reduced dark interaction, and maintain longer or shorter signaling states in response to a pulse of light. We describe minimal functional CRY2 and CIB1 domains maintaining light-dependent interaction and new signaling mutations affecting AtCRY2 photocycle kinetics. The latter work implicates a α13-α14 turn motif within plant CRYs where perturbations alter signaling state lifetime. Using a long-lived L348F photocycle mutant, we engineered a second generation photoactivatable Cre recombinase, PA-Cre2.0, that shows five-fold improved dynamic range allowing robust recombination following exposure to a single, brief pulse of light.
Introduction
Optical dimerizers are a powerful new class of optogenetic tools that allow light-inducible control of protein-protein interactions. As protein-protein interactions are the predominant facilitator of cellular behaviors, such tools allow exquisite spatial, temporal, and dose-dependent control of biological events. The basis of these tools is an interaction between two proteins or domains where one of the interacting partners is a photosensory protein or domain that exists in a ‘ground’ or unexcited state, but undergoes a conformational change with light excitation. The second protein or domain selectively binds either the ground or photoexcited state of the photosensory protein. If photostimulation is not maintained, all photosensory proteins naturally revert to their ground states over time (which varies from seconds to hours depending on the system), and thus the binding interactions are naturally reversible.
A variety of optical dimerizer systems have been described based on different light-sensing domains: phytochromes, cryptochromes, LOV domains, and UVR81–10. One of the most widely used optical dimerizers is the CRY2/CIB system, based on a light-dependent interaction between Arabidopsis cryptochrome 2 (AtCRY2) and an interacting partner, CIB1. This system has been used in a variety of cell lines and model systems to optically regulate transcription4,11–14, recombinase activity4,15, phosphoinositide levels16–18, signaling19,20, cytoskeletal dynamics21,22, and other cellular functions. While the CRY2/CIB system has been easily adapted for use with diverse targets, limitations include the large size of the constructs, the lack of known dark reversion mutants that can be used to alter the lifetime of CRY2/CIB1 interaction, and dark baseline interaction of CRY2 and CIB1 versions in some applications.
In this work, we set out to improve the versatility and tunability of the CRY2/CIB dimerizer system for specific applications. We first generated CRY2 truncations that show improved dynamic range in response to light, and minimal CIB1 truncations. Using genetic screens, we then identified CRY2 signaling mutants with altered photocycles, resulting in longer or shorter half-lives for CIB1 binding. These mutations, near the flavin and ATP binding sites, provide insight into the molecular mechanism of CRY2 photoactivation, which is still under debate. We tested these mutations in two previously-validated systems: a photoactivatable transcriptional system, and a photoactivatable Cre recombinase (PA-Cre). With further engineering, we generated a second-generation photoactivatable Cre recombinase that shows 5-fold enhanced dynamic range. We expect these second generation CRY2, CIB1, and PA-Cre modules will have a wealth of new uses in optogenetic experiments.
Results
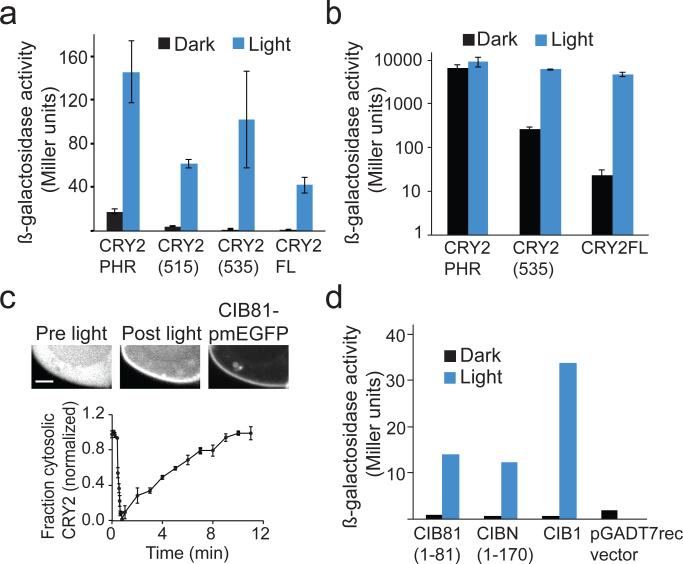
To generate improved second generation CRY2/CIB1 optical dimerizer modules, we first focused on functional differences between CRY2PHR (residues 1–498 of CRY2) and full length CRY2. Previously, we found that CRY2PHR shows improved expression over full-length CRY2, but in most cases, such as when regulating transcription4, this construct shows enhanced background interaction with CIB1 in the dark. We sought to identify a truncation of CRY2 that would express well and have a smaller size, but also maintain tight light control when used with CIB1. We tested CRY2 truncations for interaction with CIB1 using a transcriptional assay or by visualizing recruitment to membrane-associated CIB in mammalian cells4. Two truncations, CRY2(515) (residues 1–515) and CRY2(535) (residues 1–535), interacted well with CIB1 and showed improved dynamic range compared with CRY2PHR in transcriptional studies reconstituting a split Gal4 protein (Fig. 1a and Supplementary Results, Supplementary Fig. 1), however CRY2(515) retained higher background interaction with CIB1 in dark compared with CRY2(535). As CRY2 has been found to cluster and self-associate23–25, We also examined self-interaction of truncation mutants. We observed substantial self-interaction of CRY2PHR and CRY2(515) in both light and dark (Supplementary Fig. 2). In constrast, while still interacting substantially in light, CRY2(535) showed greatly reduced self-association in dark. We further examined CRY2(535) in a more sensitive assay, in which the CRY2/CIB interaction was used to reconstitute a split LexA-VP16 transcription factor26 (Fig. 1b). Use of a LexABD-CRY2(535) fusion resulted in a 26-fold reduction in dark activity compared to LexABD-CRY2PHR, indicating this module, only 37 residues larger, may show improved function compared with CRY2PHR in many applications. Full length CRY2 used in this assay still showed the highest dynamic range and lowest background dark activity compared to either truncation (Fig. 1b), and thus may still be preferable for applications with stringent background requirements.
Figure 1. Truncations of of CRY2/CIB1 modules.
(a) β-galactosidase activity of Gal4BD-CRY2 (full length and truncation constructs) and Gal4AD-CIB1 expressed in AH109/Y187 yeast and tested for interaction in the dark or blue light (1s pulse every 3 min, 461 nm, 5.8 mW/cm2) for 2.5 hrs. Data represent mean values ± s.e.m. (n=3 independent experiments) (b) β-galactosidase reporter activity of W303-1A yeast expressing different length LexABD-CRY2 constructs, VP16-CIB1, and a pSH18-34 reporter plasmid after 3 h treatment with blue light or dark. Data represent mean values ± s.e.m. for three independent experiments. (c) Fluorescence images of a HEK293T cell expressing CRY2PHR-mCh and membrane localized CIB81-pmEGFP (containing a CaaX motif at C-terminus of EGFP) pre and 1 min post blue light exposure. Graph depicted below shows quantification of the change in cytoplasmic CRY2PHR-mCh fluorescent signal over time after exposure to a single 488 nm pulse (mean values ± s.d., n=3 cells). Scale bar, 2 μm. (d) β-galactosidase activity of Gal4BD-CRY2 and indicated Gal4AD-CIB1 full length or truncation mutants expressed in AH109/Y187 yeast and tested for interaction in the dark or in blue light (1s pulse every 3 min, 461 nm, 5.8 mW/cm2) for 2.5 hrs. Data represent mean values of two independent experiments (3 replicates each).
We next examined whether we could identify smaller domains of CIB1 that would retain robust light-dependent binding to CRY2. A truncated version of CIB1, ‘CIBN’ (residues 1–170) was previously found to promote robust light-dependent interaction with CRY2 and CRY2PHR in a variety of applications4, but this tag is still fairly large. A further truncation of CIB1 containing only the first 81 amino acids, ‘CIB81’, showed light-dependent interaction with CRY2 similar to that observed with CIBN (Fig. 1c,d and Supplementary Video 1). Residues 1–81 of CIB1 contain homology with other CIB family proteins found to also interact with CRY227. A smaller truncation, containing residues 16–43 of CIB1, retained minimal binding to CRY2 and showed some promise in recruitment experiments with cytosolic CRY2PHR-mCherry, however, recruitment was substantially weaker than using either CIBN or CIB81 and yeast two-hybrid experiments showed strong interaction with full length CRY2 even in dark (Supplementary Fig. 3).
Characterization of CRY2 photocycle mutants
We next turned our efforts to improving the versatility of the CRY2/CIB1 modules by identifying CRY2 variants with altered photocycle kinetics. When photostimulated, AtCRY2 undergoes a conformational change allowing binding to CIB128, but does not remain stably in this photoactivated state. When expressed in mammalian cells (at 34°C), CRY2 spontaneously dissociates from CIB1 with a half-life of ~5.5 min as it returns to the ground (dark) state4. While this time frame is sufficient for many optogenetic applications, it would be useful for many experiments to have versions of CRY2 that maintain the photoactivated state for longer or shorter durations. We reasoned that such mutations would also provide insight into the photocycle of plant CRYs.
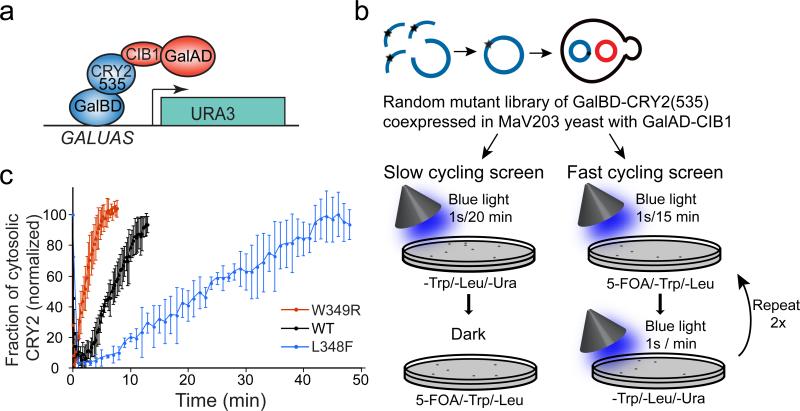
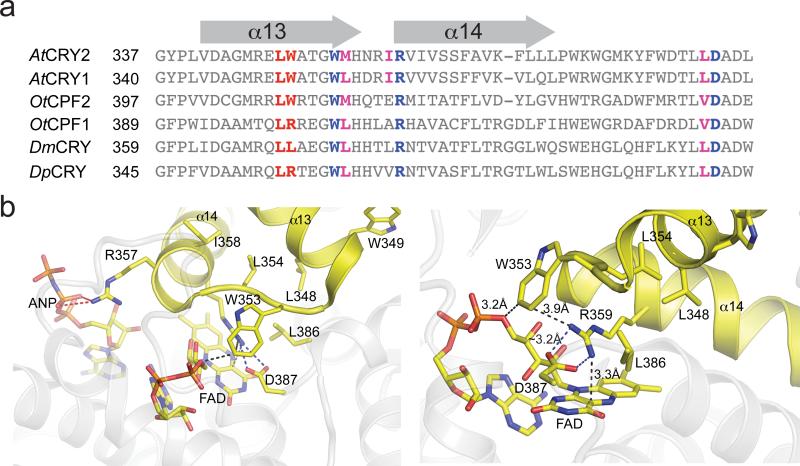
Despite detailed study, the mechanism of signaling of AtCRY2 and related CRYs, including the identity of the ground and excited states of the flavin cofactor, remains controversial29–38. In plant and animal CRYs, the ground state is proposed to be an oxidized FAD that undergoes photoreduction to an anionic semiquinone (animal CRYs)39 or a neutral semiquinone (plant CRYs)40. Previous studies of residues affecting photocycle kinetics and signal transduction in plant and animal CRYs identified three key loci that alter function: a conserved salt bridge over the si-face of the FAD isoalloxazine ring (R359-D387) 41, a residue adjacent to the N5 position of FAD42, and a remote cleft near the FAD binding pocket that engages ATP (plants) or the CCT (insects)30. To identify photocycle mutations, we targeted residues 290–498 of AtCRY2, containing these and flavin binding regions, using a mutagenesis library screening approach based on yeast-two hybrid (Fig. 2a,b). Yeast expressing GalBD-CRY2(535) and GalAD-CIB1 were subjected to rounds of screening optimized to identify either long-lived or short-lived photocycle mutants (Fig. 2b). Each screen included a positive growth selection to identify mutants that retain interaction under specific light conditions (described in Methods), where interaction resulted in activation of a Ura3 reporter allowing growth on plates lacking uracil. Yeast were also subjected to a negative selection, with 5-Fluoroorotic Acid (5-FOA), to eliminate yeast showing interaction under alternate light conditions (5-FOA is toxic to cells expressing the Ura3 reporter). In independent screens optimized for either long-lived or short-lived mutations, we identified mutations on adjacent residues on CRY2 with opposite effects on photocycle kinetics: a long-cycling L348F variant, and a short-cycling W349R variant. When tested in a membrane recruitment assay4, monitoring recruitment and subsequent dissociation rate of CRY2PHR-mCherry to a membrane-tethered CIBN protein following delivery of a pulse of blue light, we obtained half-lives of ~24 min for the long-cycling L348F variant, and ~2.5 min for the W349R variant (Fig. 2c). In the highly conserved AtCRY1 structure29, Leu348 and Trp349 are located ~10 Å away from the flavin-binding pocket, at the C-terminal end of the α13 helix (Fig. 3a,b).
Figure 2. Identification of CRY2 photocycle mutants.
(a) Growth selection assay used for screening. Interaction of Gal4BD-CRY2(535) and Gal4AD-CIB1 results in reconstitution of Gal4 and activation of a URA3 reporter. (b) Schematic showing slow and fast cycling selection screens testing interaction of a GalBD-CRY2(535) mutant library with GalAD-CIB1. (c) Quantification of cytosolic levels of wild-type (wt) or indicated mutants in CRY2PHR-mCh upon recruitment to CIBN-pmEGFP at the plasma membrane after delivery of a pulse of 488nm light at time 0. The data represent mean values ± s.d., n=3 cells.
Figure 3. Sequence alignment and modeling of photocycle mutants.
(a) Sequence alignment of CRYs from Arabidopsis thaliana (AtCRY1, AtCRY2), Ostreococcus tauri (OtCPF1, OtCPF2), Drosophila melanogaster (DmCRY), and Danaus plexippus (DpCRY). The amino acids resulting in photocycle alteration in AtCRY2 are indicated in red. Conserved hydrophobic residues and the residues involved in stabilizing electrostatic interactions are indicated in magenta and blue. (b) Location of mutations in AtCRY2 structure at end of helix α13. Key structural contacts are mediated by the α13-α14 turn motif (yellow helix-turn-helix) that positions conserved Trp353 adjacent to a salt bridge (Arg359-Asp387) required for signal transduction in DmCRY2. Arg357 of α14 forms a salt bridge to ANP in AtCRY1 crystal structures. Leu348 is buried in a hydrophobic pocket (Leu354, Ile358, Leu386) that anchors contacts between FAD, Arg359 (salt bridge, blue-dash) and Trp353, whereas Trp349 is partially solvent-exposed. The surrounding protein (grey) is transparent for clarity.
While L348 and W349 are strictly conserved in AtCRY1 and AtCRY2, and L348 is conserved in all CRYs as well as DNA photolyase, the W349 site is more variable (Fig. 3a). In fact, a related algal CRY from Ostreococcus tauri (OtCPF1) contains a Arg residue at the equivalent position (R401), as do some other animal CRYs, including DpCRY from D. plexippus (monarch butterfly), which shows significantly faster rates of flavin reoxidation compared with DmCRY (Drosophila melanogaster), which has Leu at this position42. To assess the effect of Arg/Trp substitution at this residue, we generated the Trp401 mutation in OtCPF1 and examined effects on photocycle kinetics in vitro. As shown in Table 1, substitution of Arg401 to Trp extended the rate of flavin reoxidation from the neutral semiquinone to the oxidized FAD by two-fold. These in vitro data are consistent with the faster reversion rate we observe in vivo, and suggest that the Trp349 site affects CRY photocycling in a generalizable manner in related plant and algal CRYs.
Table 1.
Kinetics of neutral semiquinone oxidation for OtCPFI
| Construct | t1/2 (s) | k (s−1) | Error (s−1) |
|---|---|---|---|
| WT OtCPFI | 1100 | 9.0E-4 | 8E-5 |
| R401W OtCPFI | 2350 | 4.3E-4 | 8E-5 |
1k is reported as the first order rate constant for oxidation of the FAD neutral semiquinone to the oxidized FAD.
Functional analysis of L348F slow-cycling mutant
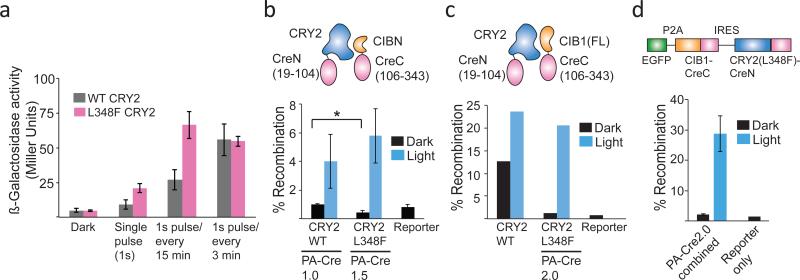
We reasoned the L348F slow-cycling mutant, which permits extended CRY2-CIB interaction in response to a single pulse of light, would be useful in applications where fast turnoff is not desired, such as transcription or DNA recombination. We examined light response of CRY2 L348F in the photoactivatable Gal4 transcriptional system used in our original screen (reconstituting split fragments of Gal4BD-CRY2(535) and Gal4AD-CIB1). Compared to wild-type CRY2, substitution of L348F resulted in extended activity with brief or infrequent light exposure but similar activity with frequent light exposure, consistent with a slow-cycling mutant (Fig. 4a).
Figure 4. Functional analysis of CRY2 L348F.
(a) CRY2 L348F allows extended transcriptional activation in response to a light pulse. Yeast expressing wt or L348F (also K329M) GalBD-CRY2(535) and Gal4AD-CIB1 were assayed for β-galactosidase reporter activity after 2.5 hrs in dark or under indicated light pulse conditions (1s pulse every 3 min, 461 nm, 5.8 mW/cm2). Data represent mean values ± s.d. (n=3 independent experiments) (b) Comparison of Cre recombinase activity generated using PA-Cre1.0 (CIBN-CreC/CRY2-CreN) or PA-Cre1.5 (same constructs, with L348F instead of wild-type CRY2). Constructs were transfected along with a fluorescent loxP-STOP-loxP-EGFP reporter in HEK293 cells, and exposed to a single 4s pulse of light 24 hr after transfection (461 nm, 5.8 mW/cm2). Reporter activity was quantified by flow cytometry 48 hours after transfection. Data represent mean values ± s.d. (n=3 independent experiments). *, p-value < .05 (c) Comparison of Cre recombinase activity of CRY2-CreN(wt or L348F) and CIB1-CreC(N1). Constructs were transfected and assayed as in (b). Data represent mean values of two independent experiments (triplicate replicates) analyzed by flow cytometry. Similar results were obtained by manual count in two independent experiments. (d) Cre recombinase activity of combined PA-Cre2.0 construct expressed in HEK293 cells. The combined PA-Cre2.0 construct was transfected along with a loxP-STOP-loxP-dsRed reporter, and exposed to a single 4s pulse of light 24 hr after transfection. Reporter activity was quantified by manual count 48 hrs after transfection. Data represent mean values ± s.d. (n=3) from one experiment, and experiments were repeated two times with similar results.
To further examine the properties of L348F, we examined this variant in a completely different system, in which CRY2 and CIBN bring together split N- and C-terminal fragments of Cre recombinase4. While a first generation photoactivatable Cre recombinase (PA-Cre1.0) showed tight light dependence, it showed poor recombination efficiency following exposure to brief light inputs4,15,43. We reasoned that the L348F variant would allow increased Cre recombination activity with minimal light illumination times, facilitating use in model organisms and for focal stimulation in individual cells. Substitution of L348F in this system (termed PA-Cre1.5) resulted in ~35% improved recombinase activity in response to a brief light flash (single 4 s flash, 461 nm light, 5.8 mW/cm2) (Fig. 4b). Surprisingly, while the levels of recombination in the dark using wild-type CRY2 were already quite low, use of L348F in this system resulted in a significant 50% reduction in dark background levels.
Second generation photoactivatable Cre recombinase
Given the extended lifetime (and potential reduced background) provided by use of the L348F mutation in PA-Cre, we examined whether this mutation could be combined with other alterations to generate a second generation PA-Cre with greater dynamic range. We first examined the effect of different Cre split sites. Our initial studies split the protein at amino acid 104, a site validated previously with chemical dimerizers44. Cre recombinase fragments split at residues 60/61, 66/67, 101/102, 109/110, and 279/280 all showed light-dependent reconstitution of activity but variable dark activity (Supplementary Fig. 4), with no improvement in dynamic range. We next examined use of different CRY2 and CIB1 truncations and expression levels. Substitution of full length CIB1 for CIBN in the CIBN-CreC fusion (CIB1-CreC(N1)) yielded significantly higher levels of recombination in response to a single flash of light (23.6 ± 1.25%), but also much higher levels of recombination in dark (12.7 ± 4.8%) (Fig. 4c). Use of CRY2(L348F) (CRY2 L348F-CreN) maintained high recombination levels in response to a single flash of light (20.7 +/− 4%), but significantly reduced levels of dark recombination (1.3 ± 0.8%) to nearly that of reporter only control cells (0.8 ± 0.3%) (Fig 4c). With extended 24 hour illumination, this system achieved recombination rates of over 50% (Supplementary Fig. 5). Placement of CIB1-CreC immediately after the CMV promoter of the mCherryN1 vector (CIB1-CreC(N1)), or downstream of an IRES element after mCherry (CIB1-CreC(IRES)) had no effect on activity (Supplementary Fig. 6), indicating expression levels of CIB1-CreC did not require precise tuning. To test if we could reduce the construct size, we tested functionality of the shorter CRY2(535)-L348F-CreN, which behaved similar to full length CRY2-L348F-CreN (Supplementary Fig. 6). For tracking expression in cells, we also tested myc and HA-tagged versions of the split Cre: attachment of an HA tag at the C-terminus of CIB1-CreC greatly reduced light dependent activity, while a myc-tag attached at the N-terminus of CRY2 retained function (Supplementary Fig. 6). We named the optimized CRY2(L348F)-CreN/CIB1-CreC combination achieving high levels of light activation and low dark background ‘PA-Cre2.0’.
To package PA-Cre2.0 into a more compact format, we generated a combined construct containing both Cre halves. We fused a EGFP reporter at the N-terminus of CIB1-CreC, separated from CIB1 by a P2A cleavable peptide sequence. The GFP-P2A-CIB1-CreC ORF was followed by a IRES element, then the coding sequence of CRY2FL(L348F)-CreC. When tested with a LoxP-STOP-LoxP-dsRed reporter in transient transfections, the combined construct behaved similar to the split PA-Cre2.0 version, showing >20% recombination rates with a single pulse of light and low background (Fig. 4d).
Finally, to follow up on whether the effects we observed with L348F on dark background activity in the Cre recombinase system were generalizable to other functional assays using CRY2 and CIB1, we turned to a functional assay showing high dark background with wild-type CRY2, the LexA-VP16 transcriptional system described in Fig. 1b. Using CRY2(535)-LexA and CIB1-VP16, we observed enhanced activity in response to a single brief light exposure with CRY2(535) L348F, but no reduction in dark background levels of activation, which remained high (Supplementary Fig. 7). Thus, we conclude that this mutation does not universally reduce dark background interaction of CRY2 and CIB1, but may have more contextual functional effects depending on the fused proteins.
Discussion
Here we present new CRY2 and CIB1 variants and fusion constructs with improved properties useful for specific dimerization applications. We identified an 81 residue fragment of CIB1, CIB81, that retains interaction with CRY2 and can be used to control protein localization in cells. CIB81 is expected to be useful with protein targets that cannot tolerate a larger fusion tag, or for packaging the dimerization system into size-restricted viral vectors. The CRY2 truncation CRY2(535) shows tighter light control than CRY2PHR, is shorter than full length CRY2, and may be preferable for use regulating many cellular activities.
We identified mutants of AtCRY2 that prolong (L348F) or shorten (W349R) the CRY2 photocycle, as measured by functional interaction with CIB1. Despite intensive study, only a few regions have been previously identified to affect cryptochrome photocycle kinetics, and we expect these mutations may help provide new insight into the CRY signaling mechanism. Intriguingly, the L348/W349 mutations are adjacent to the α13-α14 turn motif, which links to regions recognized as important for signal transduction: the FAD binding pocket and the ATP binding site (Fig. 3). The turn motif positions Trp353 adjacent to a salt bridge (Arg359-Asp387) that is required for signal transduction in dCRY. In the AtCRY1 structure, R357 of α14 also forms a salt bridge to ANP, a region implicated in signaling in plant CRYs. In combination with the active site FAD, the FAD-Arg359-Trp353 triad forms a π-cation-π interaction, linking these regions and possibly stabilizing the FAD radical. We hypothesize local perturbations in structure at L348/W349 sites may alter positioning of the α13-α14 turn motif and the π-cation-π interaction, ultimately perturbing the local FAD environment. However, since the signaling mechanism of cryptochromes is still under debate35–38,45–47 and these residues lie in critical structural regions implicated in signal propagation, we cannot rule out the possibility that the mutations perturb structural relaxation time of the CRY2-CIB1 interaction independent of any effects on flavin redox state.
Our studies on CRY2 and the conserved OtCPF1 (26% identity in PHR domain with CRY2) link functional in vivo measurements of CRY photocycle with direct kinetic measurements of FAD oxidation state. We demonstrate that the substitution of an Arg residue for Trp at 349 shortens CRY2 photocycle, as measured by in vivo binding to CIB1. Likewise, the same amino acid substitutions, measured in vitro with the related OtCPF1 CRY, alter the rate of flavin oxidation from the neutral semiquinone to the oxidized FAD. This result supports prior studies indicating that the signaling state of AtCRY1/2 is the neutral semiquinone with a ground state corresponding to the oxidized FAD48–50, and suggests the amino acid specificity of this site, which naturally occurs as Arg in some CRYs, may be important for conferring differences in CRY photocycle rate.
We expect the fast-cycling mutant, W349R, and the slow cycling mutant, L348F, to be useful substitutes for wild-type AtCRY2 in a variety of optogenetic applications. W349R should be helpful in achieving better local control of protein recruitment. In turn, the slow-cycling L348F mutant should allow less light input in applications where extended photoactivation is preferable. In future studies, we are exploring the significance of the L348F variant on dark state interaction of CRY2 and CIB1. Surprisingly, use of L348F significantly reduced background interaction of CRY2 and CIB1 in the context of a split Cre, but did not affect background in a split transcriptional system. However, as we find background levels using CRY2/CIB dimerizers to be highly contextual based on target proteins fused26, it is possible this mutant may be useful in reducing dark state binding in other contexts.
Using the L348F variant, we generated a dramatically improved second generation photoactivatable Cre DNA recombinase, PA-Cre2.0. The PA-Cre2.0 system shows minimal dark background but generates a robust response even to a single flash of blue light. Indeed, we were able to achieve 5-fold higher levels of activity in response to light than the original PA-Cre1.04, with little difference in dark background levels. The increased efficacy of PA-Cre2.0 with shorter light inputs will facilitate use for local control of Cre recombination in user-specified cells using focal illumination. The increased efficacy will also facilitate use in vivo in model organisms: shorter light stimulation regimes will reduce the time an animal is exposed to surgery, anesthesia, and/or to immobilized positioning under microscopes. For more extended light treatments, the long lifetime of the L348F mutant allows for longer intervals between light exposure, thus reducing the potential for light-mediated toxicity. Altogether, PA-Cre2.0 should provide an improved system allowing precise spatial and temporal control of gene editing in targets ranging from single cells to whole organisms.
Methods
Strains and plasmids
Yeast strains used for two-hybrid assays were AH109 (MATa, trp1-901, leu2-3, 112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2::GAL1UAS-GAL1TATA-HIS3, GAL2UAS-GAL2TATA-ADE2, URA3::MEL1UAS-MEL1TATA-lacZ, MEL1), Y187 (MATα, ura3-52, his3-200, ade2-101, trp1-901, leu2-3, 112, gal4Δ, gal80Δ, met-, URA3::GAL1UAS-GAL1TATA-lacZ, MEL1) (Clontech), W3031A (MATa, leu2-3,112, trp1-1, can1-100, ura3-1, ade2-1, his3-11,15), and MaV203 (MATα; leu2-3,112; trp1-901; his3Δ200; ade2-101; cyh2R; can1R; gal4Δ; gal80Δ; GAL1::lacZ; HIS3UASGAL1::HIS3@LYS2; SPAL10::URA3). Sequences of new constructs generated for this manuscript and oligos used are provided in Supplementary Dataset 1. CIBN-pmEGFP, CRY2PHR-mCh, pDBTrp-CRY2PHR, pDBTrp-CRY2(FL,ΔNLS), and pGADT7rec-CIB1 were described previously4. The EGFP and dsRed (pCALVL-dsRed) Cre recombinase reporters were described previously4,51. pDBTrp-CRY2(515) and pDBTrp-CRY2(535) were generated by homologous recombination of CRY2 fragments (residues 1-515 using oligos 1054F/1113R; residues 1-535 using 1054F/1082R), into pDBTrp cut with SmaI. GalAD-CRY2PHR, GalAD-CRY2(515), and GalAD-CRY2(535) were generated by homologous recombination of CRY2 truncation fragments (PHR, residues 1-498, using 490F/644R; CRY2(515), residues 1-515, using 490F/1842R; or CRY2(535), residues 1-535, using 490F/1841R) into pGADT7rec cut with NcoI and BamHI. GalAD-CIB81 was generated by homologous recombination of CIB1 (residues 1-81, oligos 933F/935R) into pGADT7rec cut with BamHI/EcoRI. CIB81-pmGFP and CIB(16-43)-pmEGFP (CIB28-pmEGFP) were generated by replacing CIB1 in CIB1-pmEGFP with CIB(1-81) or CIB(16-43) at NheI and AgeI sites. To generate LexA-CRY2(535) and LexA-CRY2(535)L348F, we used homologous recombination in yeast into LexA-CRY2PHR26 cut with MscI and NotI, along with a first PCR fragment using oligos 977f and 1145r to amplify LexA-CRY2(535), and a second PCR fragment using oligos 1144f and 1082r with template CRY2(535)L348F-mCherry.
For mammalian expression studies, L348F or W349R were generated in CRY2PHR-mCh using sequence overlap extension PCR with mutagenic primers. To generate CRY2(535)-mCherry and CRY2(535)L348F-mCherry, we added XhoI and XmaI sites using PCR, then cloned into XhoI/XmaI on CRY2-mCherry, replacing CRY2. To generate CRY2(L348F)-CreN, a first fragment containing residues 1-400 (including the L348F mutation) was amplified from CRY2PHR(L348F)-mCherry using PCR, and a second fragment beginning at residue 400 of CRY2 through the end of CreN was amplified from CRY2-CreN. The two fragments were joined using sequence overlap extension and cloned into SacI and XmaI sites in CRY2-CreN. CIB1-CreC was generated by replacing CIBN in CIBN-CreC4 with full length CIB1 using homologous recombination in yeast. Then, the CIB1-CreC open reading frame was inserted into mCherryN1 (Clontech) at Xho I and Xma I sites, designated CIB1-CreC(N1), or at SacI and XmaI sites into the IRES-containing vector CIBN-CreC described previously4 (replacing CIBN with CIB1), designated CIBN-CreC(IRES).
To generate the split Cre fragments at different sites, mCherry-IRES-CRY2-CreN and mCherry-IRES-CIBN-CreC were digested at NotI and XmaI sites to remove existing Cre fragments. Fragments containing split N-terminal Cre fragments were amplified from pCIG2-Cre using oligos 750F/1212R and 1213F/753R (split at residue 60/61); 750F/1214R and 1215F/753R (split at 66/67); 750F/1216F and 1217F/753R (split at 101/102); 750F/1218R and 1219F/753R (split at 109/110); 750F/1220R and 1221F/753R (split at 279/280). To generate CRY2(535)L348F-CreN, we used oligos 1272f and 1273r to add a SacI site and a ‘MPKKKRKV’ SV40 large T antigen nuclear localization sequence at the N-terminus of CRY2, and a GGGSGGGS linker and BspEI site at the C-terminus of CRY2(535). The insert was cloned into CRY2-CreN digested with SacI and BspEI, in place of the original SacI-BspEI fragment. The myc-tagged myc-CRY2(L348F)-CreN construct was generated by adding the myc tag and SacI and XmaI restriction sites using nested PCR using oligo 1683f /1685r followed by 1684f /1685r and cloning into the CIBN-CreC IRES vector at SacI/XmaI (after removing the CIBN-CreC insert). The HA-tagged CIB1-CreC-2xHA construct was generated using PCR to add HA and SacI/XmaI sites using 1686f / 1687r, then 1686f/1688r, and inserting at Sac I and Xma I in the same IRES vector. To generate pEGFPC1-PA-Cre2.0, we assembled the following sequence using PCR with overlapping ends and homologous recombination in yeast, in vector p416ADH: EGFP-P2A cleavable peptide-CIB1-CreC-IRES-CRY2(L348F)-CreN-STOP. The CIB1-CreC-IRES-CRY2(L348F)-CreN-STOP fragment was cut out of p416ADH with BsrGI and XmaI and cloned into EGFP-C1 (Clontech) at BsrGI and XmaI sites.
Statistical analysis
Statistical significance for experiments comparing two populations was determined using a two-tailed unpaired Student's t-test.
Directed evolution in yeast
Error-prone PCR was performed on CRY2 between amino acid 290 and 498 using a MnCl2/unbalanced nucleotide protocol52. The reaction used Taq enzyme and included a final concentration of 0.125 MnCl2, 1 mM dTTP, 1mM dCTP, 0.1mM dGTP, and 0.1mM dATP, giving an average mutation rate of 0.6%. The selection screen was carried out using pDBTrp-CRY2(535), as initial tests of CRY2PHR/CIB1 showed too high background on 5-FOA plates, and CRY2(FL)/CIB1 required frequent light illumination for growth on Ura-plates. To insert the mutagenic library, we first made a non-functional pDBTrp-CRY2(535) using a mCherry stuffer. Thus, pDBTrp-CRY2(535) was digested with Sma I and Stu I (using dam-/dcm- strain GM2163), and mCherry was amplified with 1109F/1110R, and CRY2 was amplified with 1111F/1082R, allowing homologous recombination of mCherry between residue 329 and residue 474 of CRY2. The library of mutagenic fragments was inserted by homologous recombination into pDBTrp-CRY2-mChstuffer-CRY2(535) digested with SbfI and XmaI, and transformed into MaV203 yeast already containing pGADT7rec-CIB14, allowing growth selection based on the CRY2-CIB1 interaction. Approximately 300,000 transformants were initially obtained on SC-Trp/-Leu plates.
To identify slow-cycling mutants, approximately 100,000 yeast cells from the pooled library were incubated on SC -Trp/-Leu/-Ura plates. Plates were placed in a 30°C incubator and illuminated with 461 nm LED light (infrequent pulses, 1s every 15 min, 5.8 mW/cm2) under conditions where yeast cells expressing wild type pDBTrp-CRY2(535)/pGADT7rec-CIB1 did not grow. Yeast colonies that could grow under these conditions were pooled and plated on SC-Trp/-Leu + 0.1% 5-Fluoroorotic Acid (5-FOA) plates and incubated in the dark. Because 5-FOA is toxic to cells expressing the URA3 gene product, this screen provides a counterselection to eliminate constitutive variants of CRY2 (that bind to CIB1 in dark and thus induce URA3 expression in dark). For the fast-cycling screen, 100,000 yeast were initially plated on SC-Trp/-Leu +0.1% 5-FOA plates, and subjected to conditions (infrequent frequency pulses, 1s every 15 min) where yeast cells expressing wild type pDBTrp-CRY2(535)/ pGADT7rec-CIB1 did not grow (as they generate Ura3 at too high levels due to interaction half-life of CRY2 and CIB1). Surviving colonies were then plated on SC-Trp/-Leu/-Ura treated with frequent blue light flashes (1s every min) for 48 hours, to eliminate mutations resulting in complete loss of light-dependent CRY2-CIB1 interaction. In both screens, colonies surviving after two rounds of positive and negative selection were sequenced for mutations within CRY2. Multiple mutations were identified in each positive clone: L348F was originally identified in conjunction with a second mutation, K329M, while W349R was found in conjunction with A451T. Each mutation was generated individually in pDBTrp-CRY2(535) and retested in the original assay for slow-cycling or fast-cycling phenotypes.
Live cell imaging to monitor CRY2-CIB1 interaction lifetime
HEK293 cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) with 10% fetal bovine serum. Cells were plated on 35-mm glass bottom imaging dishes and transfected using Lipofectamine 2000 (Life Technologies) according to the manufacturer's protocol. Cells were incubated in dark (wrapped with foil) and manipulations were carried out under dim light or using a red safelight. Sixteen to twenty-four hours after transfection, cells were moved to Hepes-buffered Saline (HBS) with 1 mM CaCl2 for imaging. Live cell imaging was performed at 33.5 °C on an Olympus IX71 microscope equipped with a spinning disc scan head (Yokogawa Corporation of America) with a × 60/NA 1.4 objective. Excitation illumination was delivered from an AOTF-controlled laser launch (Andor Technology) and images were collected on a 1,024 × 1,024 pixel EM-CCD camera (iXon; Andor Technology).
Yeast two-hybrid interaction assays
For GalBD fused baits, BD-fusions were initially expressed in strain AH109 and AD-fusions expressed in strain Y187 (Clontech). These strains were mated and selected on SD -Trp/-Leu to generate AH109 x Y187 strains containing both AD and BD plasmids. For LexA fused baits, we used strain W303-1A, transformed with a pSH18-34 reporter plasmid and AD and BD fusions. For quantitative interaction testing, we assayed ONPG activity. Strains were grown overnight in dark, then diluted to 0.2 OD600 in the next morning. Following an initial 3 hour growth period in the dark, cultures were either kept in the dark or exposed to a 461 nm LED light source for 2-3 hours (1s pulse every 3 min) as indicated in figures. After light treatment, cultures were harvested and lysed with Y-PER reagent (Thermo Scientific) and assayed for β-galactosidase activity using a standard protocol (Clontech Laboratories, protocol #PT3024-1) using ONPG (Sigma-Aldrich) as a substrate.
Cre recombinase assays
Cre recombination experiments were performed similar to those previously described4, using either FACS analysis or manual counting to quantify fluorescence, as indicated in figures. For testing the split Cre constructs, HEK293 cells were transiently transfected using Lipofectamine 2000 (Life Technologies) with N- and C-terminal Cre constructs and a LoxP-Stop-LoxP-EGFP reporter, containing a stop codon flanked by LoxP sites that is removed by active Cre recombinase. 24 hr after transfection, samples were either kept in the dark or treated with light as indicated, then cells were assayed after an additional 24 hours incubation. Rates of recombination were determined by quantifying the percent of mCherry expressing cells also expressing EGFP. For FACS analysis, trypsinized cells were fixed 48 hr after transfection in 4% paraformaldehyde. After washing twice with PBS, samples were analyzed with a Beckman Coulter Gallios 561 flow cytometer at the University of Colorado Cancer Center Flow Cytometry Shared Resource center. Data was analyzed using Kaluza flow analysis software (Beckman Coulter). Rates of recombination were quantified for mCherry positive cells. For testing the combined vector, PA-Cre2.0 (in EGFP-C1 backbone) was transfected similar to above, along with a loxP-STOP-loxP-dsRed reporter (pCALVL-dsRed). Rates of recombination were determined by quantifying the percent of EGFP expressing cells also expressing dsRed by manual cell count.
OtCPH1 studies
Full-length OtCPF1 was codon optimized for expression in E. coli and cloned into the pGST-Parallel vector using NcoI/XhoI restriction sites. A R401W variant was introduced using the QuickChange Protocol (Stratagene). Both WT and R401W OtCPF1 were expressed in E. coli BL21(DE3) cells. Cells were initially grown in LB media to an OD600 of 0.6 prior to induction with 0.2 mM IPTG. OtCPF1 was then expressed for 22 hours at 18°C. Cells were pelleted and stored at −80°C in buffer containing 50 mM Tris pH 7.4, 10% glycerol and 100 mM NaCl (Purification Buffer). Immediately prior to use, OtCPF1 cells were lysed and applied to glutathione affinity resin (Qiagen). OtCPF1 was eluted via on column cleavage of the GST tag by addition of 2 mg of TEV protease. Eluted protein was subsequently purified on a Superdex S200 sizing column in purification buffer prior to kinetics measurements. All kinetics measurements were conducted on an Agilent 8453 spectrometer. Protein was concentrated to 20 μM and illuminated on ice with a 150 W broad spectrum light source in the presence of 10 mM DTT and 3 mM EDTA for 5 minutes. It is important to note that the degree of photoexcitation and kinetics is dependent upon the presence of DTT and the illumination protocol. Rates of flavin oxidation were determined by measuring the absorbance at 450, 478 and 638 nm and fit with a monoexponential function. Kinetics were measured for three independent samples and averaged. The reported values represent the average rate constant with error representing the standard deviation.
Supplementary Material
Acknowledgements
We thank Dr. Constance Cepko for the pCALVL-dsRed Cre reporter (13769), obtained through Addgene, Dr. Matthew Kennedy for critical reading of the manuscript, and Jessica Spiltoir and Qi Liu for experimental assistance. This work was supported by grants from the National Institutes of Health (GM100225) and the McKnight Endowment Fund for Neuroscience (Technological Innovations in Neuroscience Award) to C.L.T.
Footnotes
Author Contributions
A.T., B.Z., J.G.M., G.P., R.M.H., and C.L.T. carried out experiments. B.Z. carried out protein structural characterization and in vitro studies with OtCPF1. A.T., B.Z., and C.L.T. analyzed data and wrote the manuscript. C.L.T. conceived the project and edited the manuscript.
Competing Financial Interests Statement
The authors declare no competing financial interests.
References
- 1.Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. A light-switchable gene promoter system. Nat Biotechnol. 2002;20:1041–1044. doi: 10.1038/nbt734. [DOI] [PubMed] [Google Scholar]
- 2.Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yazawa M, Sadaghiani AM, Hsueh B, Dolmetsch RE. Induction of protein-protein interactions in live cells using light. Nat Biotechnol. 2009;27:941–945. doi: 10.1038/nbt.1569. [DOI] [PubMed] [Google Scholar]
- 4.Kennedy MJ, et al. Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods. 2010;7:973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strickland D, et al. TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat Methods. 2012;9:379–384. doi: 10.1038/nmeth.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen D, Gibson ES, Kennedy MJ. A light-triggered protein secretion system. J. Cell Biol. 2013;201:631–40. doi: 10.1083/jcb.201210119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crefcoeur RP, Yin R, Ulm R, Halazonetis TD. Ultraviolet-B-mediated induction of protein- protein interactions in mammalian cells. Nat. Commun. 2013;4:1779. doi: 10.1038/ncomms2800. [DOI] [PubMed] [Google Scholar]
- 8.Müller K, et al. Multi-chromatic control of mammalian gene expression and signaling. Nucleic Acids Res. 2013;41:e124. doi: 10.1093/nar/gkt340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guntas G, et al. Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc. Natl. Acad. Sci. U. S. A. 2015;112:112–7. doi: 10.1073/pnas.1417910112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawano F, Suzuki H, Furuya A, Sato M. Engineered pairs of distinct photoswitches for optogenetic control of cellular proteins. Nat. Commun. 2015;6:6256. doi: 10.1038/ncomms7256. [DOI] [PubMed] [Google Scholar]
- 11.Hughes RM, Bolger S, Tapadia H, Tucker CL. Light-mediated control of DNA transcription in yeast. Methods. 2012;58:385–391. doi: 10.1016/j.ymeth.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konermann S, et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–6. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Polstein LR, Gersbach CA. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat. Chem. Biol. 2015;11:198–200. doi: 10.1038/nchembio.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nihongaki Y, Yamamoto S, Kawano F, Suzuki H, Sato M. CRISPR-Cas9-based Photoactivatable Transcription System. Chem. Biol. 2015;22:169–74. doi: 10.1016/j.chembiol.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 15.Boulina M, Samarajeewa H, Baker JD, Kim MD, Chiba a. Live imaging of multicolor- labeled cells in Drosophila. Development. 2013;140:1605–1613. doi: 10.1242/dev.088930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Idevall-Hagren O, Dickson EJ, Hille B, Toomre DK, De Camilli P. Optogenetic control of phosphoinositide metabolism. Proc Natl Acad Sci U S A. 2012;109:E2316–23. doi: 10.1073/pnas.1211305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giordano F, et al. PI(4,5)P2-Dependent and Ca(2+)-Regulated ER-PM Interactions Mediated by the Extended Synaptotagmins. Cell. 2013;153:1494–509. doi: 10.1016/j.cell.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kakumoto T, Nakata T. Optogenetic Control of PIP3: PIP3 Is Sufficient to Induce the Actin- Based Active Part of Growth Cones and Is Regulated via Endocytosis. PLoS One. 2013;8:e70861. doi: 10.1371/journal.pone.0070861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aoki K, et al. Stochastic ERK activation induced by noise and cell-to-cell propagation regulates cell density-dependent proliferation. Mol. Cell. 2013;52:529–40. doi: 10.1016/j.molcel.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 20.O'Neill PR, Gautam N. Subcellular optogenetic inhibition of G proteins generates signaling gradients and cell migration. Mol. Biol. Cell. 2014;25:2305–14. doi: 10.1091/mbc.E14-04-0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maiuri P, et al. Actin Flows Mediate a Universal Coupling between Cell Speed and Cell Persistence. Cell. 2015;161:374–86. doi: 10.1016/j.cell.2015.01.056. [DOI] [PubMed] [Google Scholar]
- 22.Duan L, et al. Optogenetic Control of Molecular Motors and Organelle Distributions in Cells. Chem. Biol. 2015;22:671–82. doi: 10.1016/j.chembiol.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bugaj LJ, Choksi AT, Mesuda CK, Kane RS, Schaffer DV. Optogenetic protein clustering and signaling activation in mammalian cells. Nat Methods. 2013;10:249–52. doi: 10.1038/nmeth.2360. [DOI] [PubMed] [Google Scholar]
- 24.Lee S, et al. Reversible protein inactivation by optogenetic trapping in cells. Nat. Methods. 2014;11:633–6. doi: 10.1038/nmeth.2940. [DOI] [PubMed] [Google Scholar]
- 25.Taslimi A, et al. An optimized optogenetic clustering tool for probing protein interaction and function. Nat. Commun. 2014;5:4925. doi: 10.1038/ncomms5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pathak GP, Strickland D, Vrana JD, Tucker CL. Benchmarking of optical dimerizer systems. ACS Synth. Biol. 2014;3:832–8. doi: 10.1021/sb500291r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, Li X, Li K, Liu H, Lin C. Multiple bHLH proteins form heterodimers to mediate CRY2- dependent regulation of flowering-time in Arabidopsis. PLoS Genet. 2013;9:e1003861. doi: 10.1371/journal.pgen.1003861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu H, et al. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science. 2008;322:1535–1539. doi: 10.1126/science.1163927. [DOI] [PubMed] [Google Scholar]
- 29.Brautigam CA, et al. Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2004;101:12142–12147. doi: 10.1073/pnas.0404851101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Müller P, et al. ATP binding turns plant cryptochrome into an efficient natural photoswitch. Sci. Rep. 2014;4:5175. doi: 10.1038/srep05175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang HQ, et al. The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell. 2000;103:815–827. doi: 10.1016/s0092-8674(00)00184-7. [DOI] [PubMed] [Google Scholar]
- 32.Zeugner A, et al. Light-induced electron transfer in Arabidopsis cryptochrome-1 correlates with in vivo function. J. Biol. Chem. 2005;280:19437–40. doi: 10.1074/jbc.C500077200. [DOI] [PubMed] [Google Scholar]
- 33.Zoltowski BD, et al. Structure of full-length Drosophila cryptochrome. Nature. 2011;480:396–399. doi: 10.1038/nature10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Partch CL, Sancar A. Photochemistry and photobiology of cryptochrome blue-light photopigments: the search for a photocycle. Photochem. Photobiol. 2005;81:1291–304. doi: 10.1562/2005-07-08-IR-607. [DOI] [PubMed] [Google Scholar]
- 35.Engelhard C, et al. Cellular metabolites enhance the light sensitivity of Arabidopsis cryptochrome through alternate electron transfer pathways. Plant Cell. 2014;26:4519–31. doi: 10.1105/tpc.114.129809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.El-Esawi M, et al. Cellular metabolites modulate in vivo signaling of Arabidopsis cryptochrome-1. Plant Signal. Behav. 2015;10:e1063758. doi: 10.1080/15592324.2015.1063758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao J, et al. Trp triad-dependent rapid photoreduction is not required for the function of Arabidopsis CRY1. Proc. Natl. Acad. Sci. U. S. A. 2015;112:9135–40. doi: 10.1073/pnas.1504404112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zoltowski BD. Resolving cryptic aspects of cryptochrome signaling. Proc. Natl. Acad. Sci. U. S. A. 2015;112:8811–2. doi: 10.1073/pnas.1511092112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vaidya AT, et al. Flavin reduction activates Drosophila cryptochrome. Proc. Natl. Acad. Sci. U. S. A. 2013;110:20455–60. doi: 10.1073/pnas.1313336110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Müller P, Bouly J-P. Searching for the mechanism of signalling by plant photoreceptor cryptochrome. FEBS Lett. 2015;589:189–92. doi: 10.1016/j.febslet.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 41.Stanewsky R, et al. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell. 1998;95:681–92. doi: 10.1016/s0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- 42.Oztürk N, Song S-H, Selby CP, Sancar A. Animal type 1 cryptochromes. Analysis of the redox state of the flavin cofactor by site-directed mutagenesis. J. Biol. Chem. 2008;283:3256–63. doi: 10.1074/jbc.M708612200. [DOI] [PubMed] [Google Scholar]
- 43.Schindler SE, et al. Photo-activatable Cre recombinase regulates gene expression in vivo. Sci. Rep. 2015;5:13627. doi: 10.1038/srep13627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jullien N, Sampieri F, Enjalbert A, Herman JP. Regulation of Cre recombinase by ligand- induced complementation of inactive fragments. Nucleic Acids Res. 2003;31:e131. doi: 10.1093/nar/gng131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X, et al. Arabidopsis cryptochrome 2 (CRY2) functions by the photoactivation mechanism distinct from the tryptophan (trp) triad-dependent photoreduction. Proc Natl Acad Sci U S A. 2011;108:20844–9. doi: 10.1073/pnas.1114579108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thöing C, Oldemeyer S, Kottke T. Microsecond Deprotonation of Aspartic Acid and Response of the α/β Subdomain Precede C-Terminal Signaling in the Blue Light Sensor Plant Cryptochrome. J. Am. Chem. Soc. 2015;137:5990–9. doi: 10.1021/jacs.5b01404. [DOI] [PubMed] [Google Scholar]
- 47.Solov'yov IA, Domratcheva T, Moughal Shahi AR, Schulten K. Decrypting cryptochrome: revealing the molecular identity of the photoactivation reaction. J. Am. Chem. Soc. 2012;134:18046–52. doi: 10.1021/ja3074819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herbel V, et al. Lifetimes of Arabidopsis cryptochrome signaling states in vivo. Plant J. 2013;74:583–92. doi: 10.1111/tpj.12144. [DOI] [PubMed] [Google Scholar]
- 49.Bouly JP, et al. Cryptochrome blue light photoreceptors are activated through interconversion of flavin redox states. J Biol Chem. 2007;282:9383–9391. doi: 10.1074/jbc.M609842200. [DOI] [PubMed] [Google Scholar]
- 50.Banerjee R, et al. The signaling state of Arabidopsis cryptochrome 2 contains flavin semiquinone. J Biol Chem. 2007;282:14916–14922. doi: 10.1074/jbc.M700616200. [DOI] [PubMed] [Google Scholar]
- 51.Matsuda T, Cepko CL. Controlled expression of transgenes introduced by in vivo electroporation. Proc. Natl. Acad. Sci. U. S. A. 2007;104:1027–32. doi: 10.1073/pnas.0610155104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cadwell RC, Joyce GF. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.