Abstract
Usher syndrome type III (USH3) characterized by progressive deafness, variable balance disorder, and blindness is caused by destabilizing mutations in the gene encoding the clarin-1 protein (CLRN1). Here we report a novel strategy to mitigate hearing loss associated with a common USH3 mutation CLRN1N48K that involved a cell-based high-throughput screening of small molecules capable of stabilizing CLRN1N48K, a secondary screening to eliminate general proteasome inhibitors, and finally an iterative process to optimize structure activity relationships. This resulted in the identification of BF844. To test the efficacy of BF844, a mouse model was developed that mimicked the progressive hearing loss of USH3. BF844 effectively attenuated progressive hearing loss and prevented deafness in this model. Because the human CLRN1N48K mutation causes both hearing and vision loss, BF844 could in principle prevent both sensory deficiencies in USH3. Moreover, the strategy described here could help identify drugs for other protein-destabilizing monogenic disorders.
INTRODUCTION
Recessively inherited diseases are most frequently caused by point mutations which result in either attenuation or loss of gene function. In such diseases, unstable gene products are dramatically down-regulated and often prone to proteasome-mediated degradation. An example is Usher syndrome type III (USH3) characterized by progressive loss of vision and hearing, with variable difficulty in maintaining balance1–3. This is caused by loss-of-function mutations in the clarin-1 (CLRN1) gene which encodes a putative four transmembrane protein (CLRN1) closely related to tetraspanins4,5.
The CLRN1N48K missense mutation is the most common USH3 causative mutation in both North America and among those of Ashkenazi Jewish descent2. Replacement of the conserved N-glycosylation site residue, Asn at position 48 to Lys, is thought to affect proper folding and stability of the CLRN1N48K mutant protein6. Clarin-1 N48K knockin (Clrn1N48K/N48K) mice develop hearing but rapidly become profoundly deaf associated with the degeneration of mechanosensory hair cells by postnatal day 24 (P24)7, suggesting that CLRN1-mediated function is not sustained in the Clrn1N48K/N48K background. Moreover, studies with HEK293 cells indicate that N-linked glycosylation is required for the stability and plasma membrane localization of human CLRN16,7, and studies with mouse and zebrafish hair cells expressing human CLRN1N48K confirmed mislocalization of the mutant protein in vivo7,8. These observations demonstrate that the function of the pathogenic variant, CLRN1N48K, is compromised by reduced protein levels and localization defects.
No treatment is available to stop or slow the progression of USH3 disease. Thus, a generally applicable strategy is needed to develop therapeutic agents for treating USH3 and other diseases caused by protein-destabilizing point mutations. We hypothesized that increasing the stability of CLRN1N48K would help the mutant protein reach the proper site at the plasma membrane and restore CLRN1-mediated function, mitigating the CLRN1N48K-associated hearing loss. Here we describe a novel strategy to identify and optimize small molecules that specifically stabilize CLRN1N48K. Moreover, attenuation of hearing loss in our new USH3 mouse model provides proof of concept that such an approach is effective in identifying pre-clinical candidates for treatment of the USH3 sensory deficit associated with the CLRN1N48K mutation.
RESULTS
A high-throughput screen identifies a CLRN1N48K stabilizer
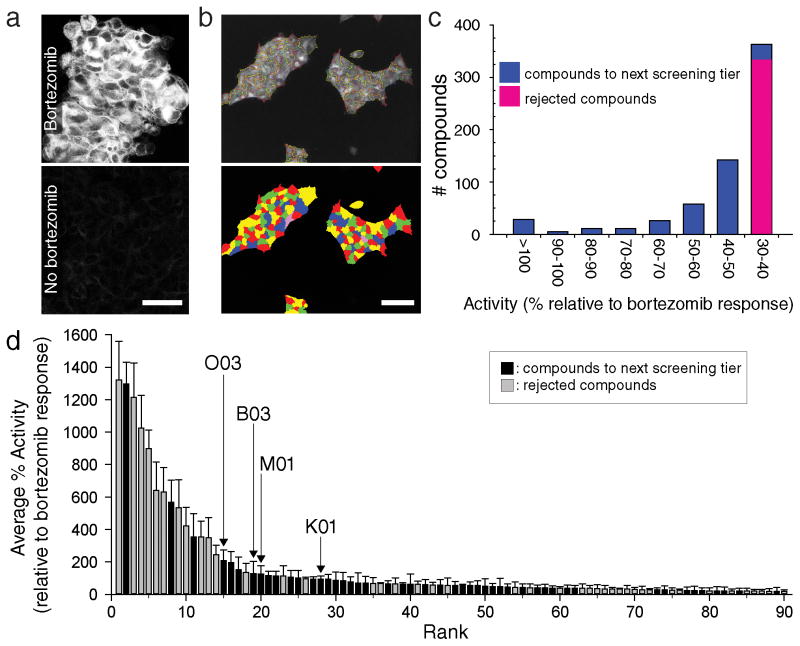
To identify molecules that stabilize CLRN1N48K, we conducted a high-throughput screening (HTS) of ~50,000 small molecules with HEK293 cells stably expressing human CLRN1N48K fused to the influenza hemagglutinin (HA) and FLAG™ epitope tags (D6 cell line) (Fig. 1a, b). As CLRN1N48K is effectively degraded by proteasomes, addition of the proteasome inhibitor bortezomib (25 nM) resulted in an increased amount of CLRN1N48K in the D6 cell line (Fig. 1a). Based on assessments of the cell’s responses to 25 nM bortezomib and DMSO, the z′-value was determined to be 0.43, which is suitable for HTS9. Out of ~50,000 molecules, 644 at 16.8 μM showed activities relative to bortezomib-treated cells equal to or greater than 30% (Fig. 1c). Among them, the top 320 compounds with high activities (Fig. 1c) were selected and re-tested six times for reproducibility. Molecules then were ranked based on their average percent activities (percent activities of the top 90 molecules are shown in Fig. 1d and Supplementary Data Set). Molecules were eliminated (Fig. 1d) due to undesirable chemical attributes10,11, including but not limited to high sulfur content, the presence of a planar polycyclic structure, dye-like structure, or a structure similar to those already represented by other high-tier compounds. In total, 48 molecules were selected for further characterization (Fig. 1d, black, Supplementary Data Set highlighted structures).
Figure 1. High-throughput screening identifies compounds that stabilize human CLRN1N48K.
(a) Inhibition of proteasomes by bortezomib increased CLRN1N48K levels in a D6 cell line stably expressing CLRN1N48K mRNA. CLRN1N48K was tagged with an HA epitope that was detected by immunofluorescence microscopy. (b) Cell-containing areas were segmented to measure relative concentrations of CLRN1N48K. Cells were outlined in the top image and colored in the bottom image. (c) Approximately 50,000 compounds were tested by high-throughput screening for stabilization of CLRN1N48K, and the measured efficacies of these compounds were normalized to 25 nM bortezomib assayed on the same plate. The top 320 compounds (blue) were selected for further analysis. (d) The top 320 compounds were subjected to the same assay 6 times. Of these, 90 compounds with highest average percentage (%) activities are shown. Among them, 48 compounds (black) were selected for secondary screening but 42 compounds (grey) were eliminated due to unfavorable properties such as autofluorescence, the formation of dye-like structures, or chemical structures unsuitable for further pharmaceutical development10,11. Data on the y-axis are presented as means ± SEMs (n = 6). Compounds O03, B03, M01, and K01 are labeled. Scale bars = 50 μm.
A dual-reporter assay for monitoring proteasome activity
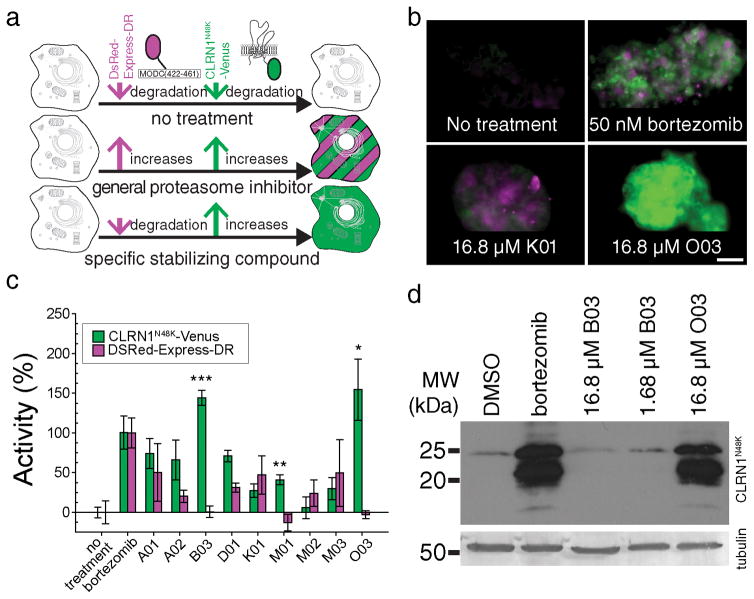
DsRed-Express-DR was used as a reporter for proteasome-mediated protein degradation, as this is a fusion of DsRed-Express with a portion of ornithine decarboxylase that is prone to proteasome-mediated degradation12 (Fig. 2a). Human CLRN1N48K-Venus fluorescent protein was used as a second reporter which also is prone to proteasome-mediated degradation (Fig. 2a). General inhibition of proteasomes causes increased fluorescence of both DsRed and Venus (Fig. 2a, b), whereas the specific stabilization of CLRN1N48K would be expected to result in increased fluorescence of Venus but not DsRed (Fig. 2a).
Figure 2. A dual-reporter assay eliminates pan-proteasome inhibitors.
(a) Cells were engineered to co-express human CLRN1N48K fused to Venus fluorescent protein (green) and DsRed-Express-DR (magenta). CLRN1N48K-Venus and DsRed-Express-DR are both degraded by proteasome (top row). Thus, a proteasome inhibitor will cause both increased Venus fluorescence and DsRed fluorescence (middle row). A molecule specifically stabilizing CLRN1N48K will cause increased Venus fluorescence but will minimally affect DsRed fluorescence (bottom row). (b) Dual-color assays conducted for 0 nM bortezomib (no treatment), 50 nM bortezomib, 16.8 μM K01, and 16.8 μM O03. O03 specifically increased CLRN1N48K-Venus as compared to bortezomib and K01 which increased both Venus and DsRed. Scale bar = 50 μm. (c) Fluorescence intensities from Venus and DsRed normalized to intensity values obtained for 50 nM bortezomib. The specific and most significant increase in fluorescence with characteristics of Venus was observed with compounds B03, M01, and O03 (***P = 1.22 × 10−5, **P = 3.70 ×10−4, *P = 2.27 × 10−3, respectively, compared to no treatment; two-sided t-test). Results are expressed as means ± SDs (all images are taken in triplicate and 15 different areas from each replicate were picked for quantification). (d) Immunoblotting analysis of human CLRN1N48K tagged with an HA epitope. Cells were treated with either bortezomib (50 nM), B03 (16.8 and 1.68 μM) or O03 (16.8 μM). CLRN1N48K expression levels increased dramatically in cells treated with bortezomib and O03 but did not increase in cells treated with B03. Tubulin loading controls are indicated. See also Supplementary Figure 13.
The 48 compounds selected from the initial screen (Fig. 1d) were investigated in this secondary screen with the dual-reporter cell line (Fig. 2b, c and Supplementary Results, Supplementary Fig. 1). For quantification of fluorescence, the average relative fluorescence intensities of Venus and DsRed after a 50 nM bortezomib treatment were used to standardize the fluorescence changes caused by each small molecule (Fig. 2b, c). Three potential hits, B03, M01, and O03 (Fig. 2c) did not show statistically significant increases of DsRed fluorescence (P > 0.1 by t-test, vs. no treatment,) but showed an increase of fluorescence consistent with Venus (P < 0.003 by t-test, vs. no treatment). Among them, M01 was excluded as this compound was fluorescent when bound to control HEK293 cells not expressing CLRN1N48K-Venus and DsRed-Express-DR. B03 and O03 were further validated by immunoblotting with the D6 cell line. Here O03 demonstrated a specific increase of CLRN1N48K whereas B03 failed to show an appreciable increase (Fig. 2d). Thus B03 is a false positive possibly caused by an increased Venus level in the secondary screen (Fig. 2c). O03 was similarly effective in enhancing levels of mouse CLRN1N48K in NIH/3T3 cells (Fig. 3a). Based on these results, O03 was used as a lead compound for further characterization.
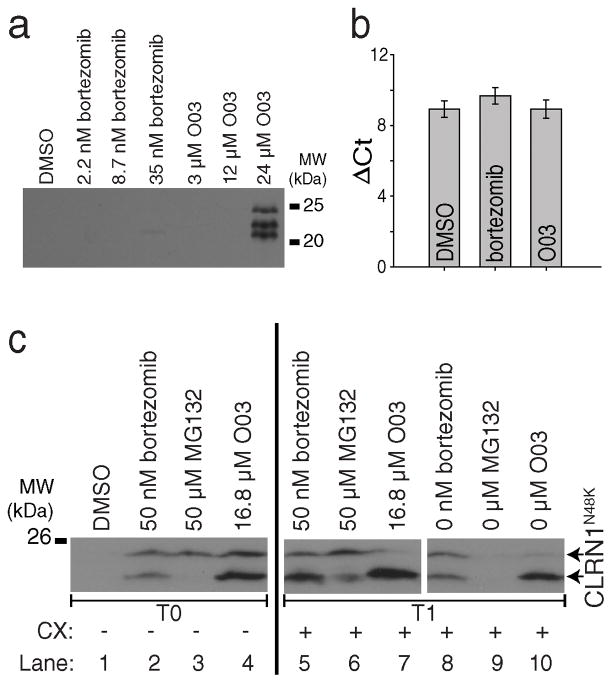
Figure 3. The lead compound O03 stabilizes CLRN1N48K by a post-translational mechanism.
(a) Mouse CLRN1N48K expressed in NIH/3T3 cells was stabilized by O03 treatment. (b) Quantitative RT-PCR of HEK293 cells stably expressing human CLRN1N48K. CLRN1 mRNA levels were not affected by either bortezomib or O03 treatment (O03 vs. DMSO, P = 0.9995, two-sided t-test, n=4, means ± SDs). (c) Protein synthesis is not required for the effect of O03. Cells treated with proteasome inhibitors bortezomib and MG132 showed increased levels of CLRN1N48K as did O03 treatment when compared to DMSO (lanes 1–4, T0). CLRN1N48K levels remained similar after treatment for an additional 6 h in the presence of 100 μM cycloheximide (CX) (lanes 5–7, T1), indicating that O03 stabilized CLRN1N48K in the absence of protein synthesis. To confirm that CLRN1N48K can be effectively degraded in the absence of protein synthesis, cells were treated with bortezomib, MG132, or O03 for 6 h. Compounds then were washed out, and cells were further incubated for 6 h in the presence of CX (lanes 8 – 10, T1). CLRN1N48K was undetectable in cells after removing the reversible proteasome inhibitor MG132 (compare lanes 3 and 9). Effects of O03 and bortezomib persisted 6 h after the washout. Note: Multiple immunopositive bands were detected by the HA-antibody which recognizes the Ct-tail of CLRN1N48K (a and c). Based on the analysis of the primary structure39, CLRN1 contains a few signal peptide cleavage sites; incomplete cleavage leads to a few differently sized bands. See also Supplementary Figure 13.
O03 acts through a post-translational mechanism
To understand the mechanism of action for O03, we examined three different processes that can increase cellular levels of CLRN1N48K: (i) increased transcription, (ii) increased translation, and (iii) greater stabilization of CLRN1N48K. We found that increased CLRN1N48K is not due to altered transcription because O03 did not increase the level of the CLRN1N48K mRNA (Fig. 3b). To investigate if protein translation is required for the effect of O03, we introduced cycloheximide (CX) which has been confirmed to inhibit the synthesis of CLRN1N48K (Supplementary Fig. 2). Cells were initially allowed to synthesize and accumulate CLRN1N48K for 6 h (Fig. 3c), and then incubated with CX for an additional 6 h in the presence of O03 (Fig. 3c). In the absence of protein translation, we were able to confirm that O03 increased the stability of CLRN1N48K.
In subsequent experiments, we compared the effects of O03 and proteasome inhibitors, MG132 and bortezomib. After 6 h of incubating the cells with O03, a condition known to increase the stability of CLRN1N48K, cells were washed and then incubated in the absence of O03 and in the presence of CX for 6 h. CLRN1N48K remained stable even after this procedure (Fig. 3c). MG132, a reversible inhibitor of proteasomes, did not stabilize CLRN1N48K after a similar washout protocol (Fig. 3c). Thus, the experiment with MG132 confirms that CLRN1N48K is prone to degradation in the absence of stabilizing reagents. Similar to O03 but unlike MG132, bortezomib was effective in stabilizing and protecting CLRN1N48K from degradation after 6 h following the washout (Fig. 3c). Bortezomib binds tightly to its targets and is known for its extremely slow ‘off rate’ kinetics13. A long lasting effect similar to that noted with bortezomib suggests that O03 is also tightly bound to its respective target protein.
Medicinal chemistry of O03 and its analogues
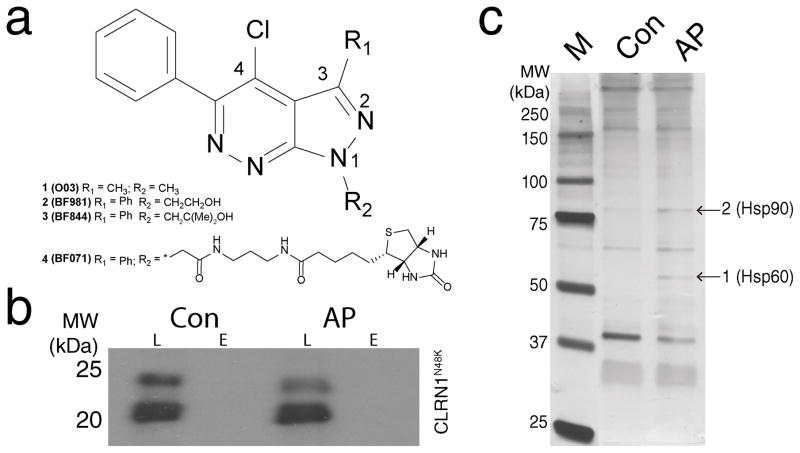
O03 was a low potency compound (compound 1, EC50 of 2 μM, Supplementary Table 1), and therefore we optimized the potency and physicochemical properties of O03 (Supplementary Note). Potencies of O03 derivatives were monitored by the same cell-based assay used for the HTS. Initially, different substitutions on position 4 of the pyrazolo[3,4-c]pyridazine core were explored (Fig. 4a). The chloro-substituent yielded the highest efficacy, and therefore was maintained during subsequent optimizations of substituents on C3 and N1. These initial results suggested that the electrophilic character of these compounds is essential for efficacy. Replacement of the methyl moiety at C3 with a phenyl group and replacement of the methyl moiety at N1 with a hydroxyethyl group resulted in compound 2 (BF981) with improved solubility and potency (EC50 = 0.31 μM, Supplementary Table 1). BF981 was metabolically unstable (Supplementary Table 1), and consistently a metabolite identification study in mouse hepatocytes indicated that BF981 underwent both oxidation and glucuronidation of the primary alcohol (Supplementary Table 2). To improve the metabolic stability of BF981, we introduced steric hindrance in the proximity of the primary hydroxyl group, leading to compound 3 (BF844) (Fig. 4a). BF844 retained the improved solubility and potency observed for BF981 (Supplementary Table 1). Further, BF844 had a higher area under the curve (AUC represented by concentration × hours (μM.h) derived from plasma concentration data) than BF981 in P20 mice, suggesting a lower in vivo clearance due to a reduction in oxidation, glucuronidation or both (Supplementary Table 3). AUC was higher in mice at P6 than at P20 (Supplementary Table 3), indicating that an escalating dosage regimen would be needed to achieve constant exposure in young mice as they age.
Figure 4. Design of BF844; identification of proteins bound to its pharmacophore.
(a) Chemical structures of the lead compound (1, O03), the optimized compounds (2, BF981; 3, BF844) and biotinylated compound (4, BF071). BF844, BF981 and BF071 share the same core structure and BF071’s biotin moiety was used for avidin affinity purification. (b) CLRN1N48K does not bind to the core structure of BF071. BF071 was used for affinity purification (AP) of binding proteins from cell homogenates which contained CLRN1N48K. As a negative control (Con), target proteins in cell homogenates were blocked with BF981 prior to incubation and affinity purification with BF071. L, whole cell lysates prior to purification; E, the fraction eluted from avidin agarose. (c) BF071 binding proteins were analyzed by SDS-PAGE; protein bands (arrows) at the sizes of ~ 60 and 90 kDa were enriched by BF071 (AP) as compared to the negative control (Con) for which samples were blocked with BF981 prior to affinity purification. Based on quantitative mass spectroscopy, HSP60 was enriched 28 times in position 1 and HSP90 was enriched 10 times in position 2, compared to the control. Positions of the protein molecular mass markers (in kDa) are indicated (lane M). See also Supplementary Figure 13.
Prediction of effective dosages for systemic administration
For systemic administration, candidate drugs for USH3 need to cross blood-cochlear and blood-retina barriers to reach affected cells. To predict their ability to cross these barriers, we used an MDR1-MDCK cell assay14 as a surrogate to assess passive permeability and P-glycoprotein-mediated efflux. The latter transporter plays an important role in physiological barriers15. BF981 demonstrated a good apical to basolateral apparent permeability and a low effective efflux ratio (Supplementary Table 3), indicative of low in vivo efflux. This was subsequently confirmed by measurements of brain and retinal concentrations of BF981, relative to those in plasma, following in vivo compound administration (Supplementary Table 3). Likewise, BF844 showed similarly good in vitro permeability and low efflux potential (Supplementary Table 3), predictive of good penetration into the retina and cochlea in vivo.
To estimate the in vivo exposure required to achieve CLRN1N48K stabilization, we incubated D6 cells with BF981 or BF844 for periods between 0.5 h and 2 h. Cells then were washed and incubated with fresh compound-free media for 22 – 23.5 h prior to the analysis of CLRN1N48K expression. Following such treatment, BF981 and BF844 displayed significant potency after the cells were exposed to each compound for a 0.5 h period (Supplementary Table 3). Because exposure at 15 μM for 0.5 h, corresponding to an AUC of 7.5 μM.h, is sufficient for near maximal (~90%) rescue of CLRN1N48K in vitro (EC90 = 15 μM), we estimated that an in vivo AUC of 7.5 μM.h for either compound might be sufficient to rescue the majority of CLRN1N48K in vivo. Following intraperitoneal (i.p.) administration of BF844 into P6 and P20 mice (3 mg/kg and 10 mg/kg doses, respectively), in vivo AUC values were determined to be 1.76 μM.h and 1.98 μM.h, respectively. Assuming that AUC and dose are linearly correlated, we estimated that BF844 would achieve an AUC of 7.5 μM.h with doses of 10 and 30 mg/kg at P6 and P20, whereas higher doses of BF981 would be needed to achieve the same exposure (100 mg/kg at P20). Hence, BF844 was selected for further in vivo studies.
The BF844 core structure interacts with HSP60/90
To identify targets of our newly developed compounds, we conducted an affinity purification of proteins using the core structure of BF844 linked with a biotin moiety (Fig. 4a). The biotinylated molecule, BF071 (4), demonstrated reasonable potency (~1.6 μM, Supplementary Table 1) and efficacy (~1.1 times higher than that of 25 nM bortezomib) in the cell-based assay described above. The affinity purification procedure failed to yield CLRN1N48K (Fig. 4b) suggesting that there is no direct interaction between the BF071 and CLRN1N48K. We found upon further analysis that two particular protein bands were enriched by binding to the core structure of BF844 (Fig. 4c). Through quantitative mass spectrometry analyses, the major constituents of these bands were identified as HSP90 and HSP60 (Fig. 4c). Further evidence supporting these interactions came from the finding that ~2% of the HSP60 and HSP90 peptides was covalently conjugated with BF071 (Supplementary Fig. 3), wherein the Cys residues responsible for the activity of HSP90 and HSP60 constituted the binding sites.
To test if these interactions are biologically significant, we measured the chaperone activities of HSP60 and HSP90 toward a model substrate, thermally denatured citrate synthase (CS), in the presence and absence of BF844 (Supplementary Fig. 4). At submicromolar concentration, BF844 effectively inhibited HSP60 activity (87.07±27.70% inhibition, n=3). When applied at 20 fold molar excess, BF844 only moderately inhibited HSP90 (40.06±19.10% inhibition, n=3), suggesting attenuation of HSP90 chaperone activity is not the primary mechanism for BF844 associated stabilization of CLRN1N48K. To further test the involvement of HSP90, 17-AAG, a potent inhibitor of HSP9016, was applied to D6 cells (Supplementary Fig. 5). When applied alone, 17-AAG did not alter the amount of CLRN1N48K. When applied in combination with BF844, however, 17-AAG caused ~5-fold enhancement of the BF844-mediated stabilization (Supplementary Fig. 5), providing evidence that the HSP90 pathway is indirectly involved in the stabilization of CLRN1N48K.
If HSP60 and HSP90 are the on-targets of BF844, changes in the potency of O03 analogs by medicinal chemistry efforts should correlate with changes in the interactions between the analogs and the two HSPs. To study the correlation between cell-based and HSP activity readouts, we introduced two analogs, BF066 (5) and BF136 (6), which are inactive according to the cell-based assay (Supplementary Fig. 4 and Supplementary Table 1). At submicromolar concentration (0.846 μM), BF066 and BF136 had no significant effect on HSP60 chaperone activity. At the same concentration, BF844 inhibited HSP60 almost completely. Both BF066 and BF136 demonstrated either little or no significant effect on HSP90 activities. Therefore, our medicinal chemistry efforts were ultimately directed toward improving functional interaction with HSPs, especially HSP60.
Effects of BF844 on non-glycosylated CLRN1
CLRN1 is present in the plasma membrane when expressed in HEK293 cells, whereas CLRN1N48K is not effectively trafficked to this location6. We tested if BF844 can enhance the plasma membrane representation of CLRN1N48K in HEK293 cells. For this assay, plasma membrane proteins were conjugated by EZ-Link® Sulfo-NHS-SS-Biotin and fractionated to compare the amounts of plasma membrane and intracellularly localized proteins. BF844 treatment induced about 6% of total CLRN1N48K to be transported to the plasma membrane compared to 49–60% of wild type CLRN1 (P < 0.0006 by two-tailed t-test, Supplementary Fig. 6). Thus in the presence of BF844, CLRN1N48K is delivered to the plasma membrane eight to ten fold less efficiently than CLRN1. Successful fractionation of plasma membrane proteins was confirmed by the presence of Na+/K+ ATPase and the absence of cytoplasmic Golgi-associated protein GRASP55 in the plasma membrane fraction. The increase in the plasma membrane level of CLRN1N48K indicates that BF844 can partially ameliorate the localization defects observed for the N48K mutant of CLRN1.
Glycosylation is part of the ER protein quality control system. To clarify the quality control step at which BF844 is involved, we tested the effect of BF844 on CLRN1, using two stable cell lines with high and low expression levels of CLRN1, C1 and D1 cell lines, respectively. In both cell lines, BF844 effectively increased the amount of non-glycosylated CLRN1 (Supplementary Fig. 7). Although the amount of non-glycosylated CLRN1 observed in the presence of BF844 was small relative to the glycosylated form of the protein (Supplementary Fig. 7), this finding is consistent with the proposal that BF844 is able to stabilize a fraction of the CLRN1 polypeptide which otherwise is degraded because of improper folding. Protein glycosylation is initiated in the ER, and thus stabilization of non-glycosylated CLRN1 suggests that BF844 is acting on the ER where nascent CLRN1 polypeptides are synthesized. Similar to CLRN1N48K, non-glycosylated CLRN1 was effectively transported to the plasma membrane after BF844 treatment. Similar stabilization effects also were observed for BF981 (Supplementary Fig. 6 and 7). Taken together, non-glycosylated CLRN1 polypeptides, both wild type and the N48K mutant, are capable of passing through the ER protein quality control mechanism. We hypothesize that BF844-mediated prevention of endoplasmic reticulum-associated degradation (ERAD) allowed the protein more time to acquire proper folding so that a fraction was able to exit the ER for plasma membrane-directed transport. Such findings are consistent with the modulation of HSPs by BF844.
Selectivity of BF844-mediated protein stabilization
We tested the effect of BF844 on two inherently unstable membrane proteins of rod photoreceptors, the P23H rhodopsin mutant (RHOP23H)17 (Supplementary Fig. 8a) and the rod cGMP-gated channel β-subunit (CNGB1)18 (Supplementary Fig. 8b). The effect of BF844 was tested with an NIH3T3 cell line stably expressing RHOP23H. RHOP23H is improperly folded and actively degraded through ERAD19–21. However, BF844 failed to stabilize RHOP23H (Supplementary Fig. 8a). The effect of BF844 also was tested on CNGB1 fused to a fluorescent protein Dendra2 (Dend2-CNGB1)22 expressed in telomerase-immortalized human retinal pigmented epithelium (hTERT-RPE1) cells. Under normal conditions, CNGB1 forms a stable oligomer with CNGA1 in rod photoreceptors. However CNGB1 is prone to proteasome-mediated degradation when expressed alone in hTERT-RPE1cells (Supplementary Fig. 8b). Under the latter condition, BF844 failed to stabilize CNGB1 (Supplementary Fig. 8b). The lack of an effect on CNGB1 further supports the concept that BF844 does not act as a general inhibitor of proteasomes. Taken together, BF844 is a protein stabilizer with selectivity toward CLRN1N48K (Supplementary Fig. 8c), and does not have an effect on photoreceptor-specific GPCR or channel.
A new mouse model of USH3
Clrn1N48K/N48K (KI/KI) mice develop early onset, profound hearing loss7. A model that shows progressive hearing loss in the KI/KI background would parallel more closely the progression of hearing loss in CLRN1N48K/N48K subjects2. Such a model would be useful for testing compounds like BF844, as it would permit treating animals after complete maturation of their hair cells but before the onset of degenerative events. We hypothesized that conditional and transient expression of Clrn1 in hair cells in the KI/KI background during pre- to perinatal periods would delay the onset of hearing loss, and the ensuing decrease in hearing sensitivity would be more gradual relative to the phenotype evidenced by KI/KI mice. To test this hypothesis, we generated a transgenic line expressing Clrn1 under control of the Atoh1 gene enhancer (Supplementary Fig. 9), which is known to drive hair-cell specific expression from the embryonic to perinatal stage and then down regulate around P523. Mice carrying Transgene Atoh1-enhacer-Clrn1 cDNA (TgAC1) and two copies of the Knock-In allele Clrn1N48K are designated as “Tg;KI/KI”.
Hearing in Tg;KI/KI mice (n=10) along with KI/KI mice (n=10) and wild-type mice (n=5) was monitored from P22 to P70 (Supplementary Fig. 10) using auditory evoked brainstem responses (ABRs)24,25. The median ABR threshold (‘threshold’) in control mice was 30±5 dB SPL (decibel sound pressure level) (Supplementary Fig. 10). Compared to controls, KI/KI mice evidenced early onset (P22) profound hearing loss (threshold ≥90 dB SPL). In contrast, at P22 most Tg;KI/KI mice had thresholds closer to those of control mice at all frequencies tested. Tg;KI/KI mice were predicted to display hearing loss associated with elevated (above normal) thresholds by P30 and continue to experience gradual hearing loss over the next 40 days. Around P70, 2 out of 10 Tg;KI/KI mice showed a 70–80 dB SPL threshold (severe hearing loss), whereas 8 out of 10 displayed ≥90 dB SPL threshold (profound hearing loss) at all frequencies tested (Supplementary Fig. 10). One-way ANOVA analysis confirmed the statistical significance of these results (P≤0.0001). However, in the KI/KI model reported previously, mutants showed moderate to severe hearing loss (60–70 dB SPL) as early as P18, and profound hearing loss by P247. In summary, we successfully generated a new model for USH3 that displays delayed-onset, progressive hearing loss. Tg;KI/KI mice closely mimics the hearing loss profile manifested by USH3 patients with the CLRN1N48K/N48K mutation.
BF844 mitigates hearing loss linked to CLRN1N48K
To test the efficacy of BF844 in mitigating hearing loss, two dosage regimens were tested. Regimen I was designed to test the efficacy of BF844 in Tg;KI/KI mice soon after they are predicted to display elevated thresholds (P30) (Supplementary Fig. 11). Regimen II was designed to intervene at a point (P10) before the onset of hearing loss in Tg;KI/KI mice to prevent earlier changes that culminate in hair cell degeneration. As P10 mice are less likely to tolerate the same dose as P30 mice, an i.p. dose escalation starting at 10 mg/kg was implemented (for details, see Supplementary Fig. 11). Weights of the injected mice were comparable to those of controls (Supplementary Fig. 12) suggesting that BF844 did not negatively affect the growth of the animals.
Regimen I
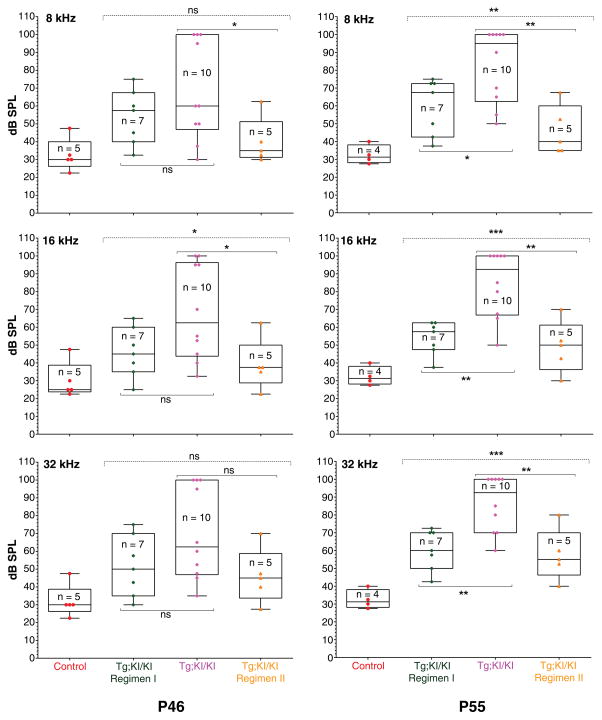
At P46, the median threshold of treated animals was lower than their untreated counterparts at all frequencies tested, but the difference was greater at 16 kHz and 32 kHz than at 8 kHz (Fig. 5, Table 1). Notably, a 10–20 dB SPL lower threshold translates into 10–100 times more sensitive hearing in treated as compared to untreated mice. At P55, the effect of BF844 on halting the progression of hearing loss in Tg;KI/KI mice was statistically significant across all tested frequencies. The median threshold was 92.5–95 dB SPL for untreated Tg;KI/KI mice, whereas the median thresholds for BF844 treated mice were 57.5–67.5 dB SPL (Table 1). This is 27.5–35.0 dB SPL lower, signifying about 1,000 times more sensitive hearing compared to untreated controls at P55.
Figure 5. BF844 significantly preserves hearing in Tg;KI/KI mice.
The y-axis indicates sound intensity in log scale (dB SPL). Each color symbol represents the average ABR threshold (“threshold”) from both ears of an individual mouse. Boxes represent the interquartile range. Horizontal lines within each box indicate the median values whereas the outermost bars present the extremes of the data. In each group, thresholds were recorded from the same mice at P46 and P55. Hearing at P46 (left column): The median threshold is 27.5 dB SPL for control mice and 60–62.5 dB SPL for Tg;KI/KI mice. For Regimen I, the median thresholds at 8, 16 and 32 kHz are 57.5, 45 and 50 dB SPL, respectively. For Regimen II, the median thresholds at 8, 16 and 32 kHz are 35, 37.5 and 45 dB SPL, respectively. ABR data at P55 (right column): The median threshold is 31.25 dB SPL for control mice and 92.5–95 dB SPL for Tg;KI/KI mice. For Regimen I, the median thresholds at 8, 16 and 32 kHz are 67.5, 57.5 and 60 dB SPL, respectively. For Regimen II, the median thresholds at 8, 16 and 32 kHz are 40, 50 and 55 dB SPL, respectively. At P55, Tg;KI/KI mice treated under Regimens I or II show a 30 to 40 dB SPL lower threshold compared to that of untreated Tg;KI/KI mice, indicating 1,000- to 10,000-fold more sensitive hearing across frequencies at P55 in BF844-treated mice. Statistics: ns (not significant) P≥0.05, * P≤0.05, **P≤0.01, and ***P≤0.001
Table 1.
Median ABR thresholds in BF844 treated versus untreated Tg;KI/KI mice
| Groups | 8 kHz | 16 kHz | 32 kHz | |||
|---|---|---|---|---|---|---|
| Threshold* at P46 | Threshold* at P55 | Threshold* at P46 | Threshold* at P55 | Threshold* at P46 | Threshold* at P55 | |
| Control (B6 WT) | 30.00 | 31.25 | 25.00 | 31.25 | 30.00 | 31.25 |
| KI/KI | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 |
| Tg; KI/KI | 60.00 | 95.00 | 62.50 | 92.50 | 62.50 | 92.50 |
| Tg; KI/KI, Regimen I | 57.50 | 67.50 | 45.00 | 57.50 | 50.00 | 60.00 |
| Tg; KI/KI, Regimen II | 35.00 | 40.00 | 37.50 | 50.00 | 45.00 | 55.00 |
ABR threshold in dB SPL (sound intensity in a log scale).
Regimen II
At P46, the median thresholds for treated Tg;KI/KI mice across stimulus frequencies were 17–25 dB SPL lower than those of untreated controls (Table 1), achieving statistical significance at 8 and 16 kHz (Fig. 5). Moreover, at P46 the median thresholds of mice in Regimen II were 7.5–22.5 dB SPL lower than the median thresholds of mice in Regimen I (Table 1). At P55, the effect of BF844 in halting the progression of hearing loss in Tg;KI/KI mice was not only statistically significant across all frequencies tested, but the difference for Regimen II was more dramatic than that for Regimen I. The median thresholds for Tg;KI/KI mice at P55 in the treated group were 55, 42.5 and 37.5 dB SPL lower at 8, 16 and 32 kHz, respectively, compared to untreated mice (Table 1); these data correspond to an average of 45 dB SPL lower across the three frequencies, signifying about 10,000 times more sensitive hearing than untreated controls at P55. This means that untreated Tg;KI/KI mice progressed to ‘severe-to-profound’ hearing loss, although most of their treated counterparts were protected from rapid deterioration of hearing.
DISCUSSION
Here we developed BF844 which mitigates progressive hearing loss in a USH3 mouse model. HTS enabled us to identify O03, a lead molecule which increased the stability of CLRN1N48K. Different synthetic modifications of O03 were then tested to improve the potency as well as the pharmacokinetic properties of the compound. The product of these studies, BF844 not only stabilized CLRN1N48K and increased its plasma membrane localization in HEK293 cells, but also improved hearing by more than three orders of magnitude in a CLRN1N48K USH3 mouse model. BF844 did not act as a general inhibitor of proteasome activity. Although general proteasome inhibition can increase the cellular levels of CLRN1N48K, that approach cannot be recommended for treating USH3 disorder, since protein homeostasis is especially critical for the maintenance and survival of auditory hair cells and photoreceptor neurons26,27. To our knowledge, BF844 constitutes the first targeted therapy for Usher syndrome, successfully delaying hearing loss.
To evaluate investigational new drugs including BF844, we made improvements to the KI/KI mouse model. The new model allowed us to observe the hair cell deficiencies caused by CLRN1 defects only in mature hair cells, a condition more closely resembling USH3 in humans. In this model, BF844 effectively mitigated hearing loss when applied before the onset of hearing loss (Regimen II) and during the early stages in the progression of hearing loss (Regimen I). Our data demonstrate that BF844 effectively crosses the blood-labyrinth barrier and protects hearing in the low, mid, and high frequency ranges. CLRN1N48K/N48K USH3 patients display post-lingual progressive hearing loss, which indicates that they acquire hair cell function but fail to maintain it2,3. The onset of hearing loss varies from 3–6 years of age to more than 35 years2. Thus BF844 can provide treatment options at different stages of USH3, since genetic testing28 can identify CLRN1N48K/N48K individuals before the onset of hearing loss.
We believe that the phenotype observed in humans and mice with the N48K mutation is due to diminished surface expression of CLRN1N48K, resulting in the loss of hair cell function over time. Human CLRN1N48K expressed in HEK293 cells accumulated in the endoplasmic reticulum (ER)7. Though accumulation of misfolded protein in the ER triggers ERAD and ER stress signals29, several cell types can activate a compensatory mechanism that delivers a fraction of the accumulated proteins to the cellular surface or other normal destinations29–31. Such a mechanism is well documented for RHOP23H and the cystic fibrosis transmembrane regulator mutant (CFTRΔ F508), both of which are prone to misfold and accumulate in the ER32–34. The RHOP23H mutant is dramatically down-regulated due to ERAD, but some of the mutated protein can escape degradation and reach the photoreceptive disk membranes where it can trigger the phototransduction cascade17,20. CFTRΔF508 can be translocated to the surface of airway epithelial cells and conduct chloride ions to a certain extent, but patients with the CFTRΔF508 mutation eventually succumb to disease33,34. For its cell surface expression, CFTRΔF508 escapes ERAD through an unconventional secretory pathway33,34. We hypothesize that a nominal amount of ER-retained CLRN1N48K in developing hair cells can escape ERAD and reach the plasma membrane to mediate its function at early stages of life. However, the amount of CLRN1N48K reaching the plasma membrane diminishes over time resulting in progressive loss of CLRN1-mediated function and hair bundle integrity. Based on our in vitro and in vivo studies, we propose that BF844 stabilizes CLRN1N48K in hair cells, enhancing its chances to escape ERAD and to reach the plasma membrane. Consistent with this hypothesis, BF844 was effective in mitigating hearing loss in our USH3 mouse model. CLRN1N48K/N48K mouse models do not develop visual impairments35–37. Therefore, the efficacy of BF844 in attenuating vision loss linked to the CLRN1N48K mutation could not be tested. However, because the same mutation causes both vision and hearing loss in humans, BF844-based therapy could preserve both hearing and vision in those with USH3.
In summary, we successfully identified the lead molecule O03 and designed an improved small molecule derivative, BF844, a compound that mitigates hearing loss associated with CLRN1N48K in vivo. BF844 stabilized CLRN1N48K without directly interacting with the mutated protein, rather the compound associates with HSP90 and HSP60. As demonstrated in other studies, interference of HSP-co-chaperone interaction can stabilize proteins by preventing ERAD38. Consistent with this concept, BF844 effectively prevented ERAD of CLRN1N48K and the non-glycosylated form of CLRN1. BF844 appears to preferentially bind to and inhibit HSP60. However, the observed inhibition of HSP60 chaperone activity (Supplementary Fig. 3) may not be attributable to the stabilization of CLRN1N48K. Because molecular components involved in HSP-assisted ERAD have not been fully characterized, it is not clear how the HSP60-co-chaperone network affects the ER-mediated protein quality control mechanism. Thus, a future direction of this study will be to address more specifically how BF844 affects the HSP60/90-co-chaperone network in vivo to stabilize CLRN1. The stabilization effect of BF844 thus far appears selective toward CLRN1. In this regard, it would be beneficial to further assess BF844’s selectivity and therapeutic potential for other genetic disorders35,36 caused by misfolded membrane proteins.
ONLINE METHODS
Preparation of expression constructs and stable cell lines
A stable cell line (D6), expressing human CLRN1N48K fused to HA and FLAG epitopes was cloned from HEK293 stable cells established previously6. The D6 cell line was tested for mycoplasma contamination using MycoAlert™ Mycoplasma Detection Kit according to the manufacturer’s instructions (Lonza, Allendale, NJ), and found to be negative. A second stable cell line co-expressing CLRN1N48K-Venus-1D4 and DsRed-Express-DR was originated by the following procedure. The coding region of Ds-Red-Express-DR (Clontech, Mountain View, CA, #632423) was cloned into the pMSCV-puro vector (Clontech). To express human CLRN1N48K-Venus-1D4, the C-terminus of the coding region of human CLRN1N48K was fused to the coding sequence of Venus40, followed by the addition of a 1D4 epitope41. The entire sequence (CLRN1N48K-Venus-1D4) then was cloned into the pMSCV-hygro vector (Clontech). The two vectors were used to establish a stable HEK293 cell line co-expressing CLRN1N48K-Venus-1D4 and Ds-Red-Express-DR. Mouse NIH/3T3 cells stably expressing mouse CLRN1N48K fused to the 1D4 epitope (CLRN1N48K-1D4) were established employing the following procedure. cDNA encoding CLRN1N48K-1D4 was cloned into the pMSCV-puro vector, and the expression construct for mouse CLRN1N48K was generated by introducing a point mutation into mouse Clrn1 via QuikChange site-directed mutagenesis (Qiagen, Valencia, CA). Stable NIH/3T3 cells were established in the manner described above.
Automated HTS
Approximately 50,000 compounds representing the chemical diversity space of pharmacologically relevant compounds contained in the University of Cincinnati Drug Discovery Center chemical library were screened with a D6 cell-based immunofluorescence assay. Ten thousand cells were plated together with culture media (DMEM + L-Glutamine + 14.5 g/L D-Glucose (Invitrogen, Grand Island, NY, #11965092), 10% fetal bovine serum (FBS) (Sigma-Aldrich, St. Louis, MO, #F4135, Lot #116K8402), and 1% penicillin/streptomycin (Fisher Scientific, Pittsburgh, PA, #30002CI, lot#3002123)) into each well of a 384 microwell plate (PerkinElmer, Waltham, MA, #6007430) using a Multidrop reagent dispenser (Thermo Scientific, Waltham, MA) and then incubated overnight at 37 °C, in 5% CO2 and 95% humidity. On day 2, cells were treated for 24 h with either 16.8 μM of test compound, 25 nM of bortezomib (LC Laboratories, Woburn, MA), or DMSO (vehicle control). The concentration of 16.8 μM was selected to cover low potency lead compounds. The DMSO concentration was 0.2% (v/v). These compounds were transferred with a CybiWell pipettor (Analytik Jena AG, Jena, Germany). On day 3, cells were fixed in 4% formaldehyde (EMS, Hatfield, PA, #19208) for 20 min. Cells were washed 3 times with PBST (0.01 M PO43−, 0.138 M NaCl, 2.7 mM KCl, pH 7.4, 1% (v/v) Triton X-100) and incubated at room temperature with anti-HA primary antibody (HA.11 clone 16B12 monoclonal antibody, Covance, Princeton, NJ, #MMS-101P) for 90 min. Cells then were washed 3 times with PBST, and incubated in secondary anti-mouse IgG conjugated with Cy3 (goat anti-mouse IgG- Cy3 (1.5 mg/ml), Jackson ImmunoResearch, West Grove, PA, #115165003) for 45 min. After incubation, cells were washed again and imaged with the Opera High Content screening system (PerkinElmer). Six independent images from each well were obtained for each compound tested. With Acapella software (PerkinElmer), cells were outlined and segmented after which the fluorescence intensity per area of the cells was obtained. Average fluorescence intensities per area of cells were loaded into a HTS database and the analysis software Genedata (Genedata, Basel, Switzerland) was used to normalize the results based on control samples (bortezomib and no treatment) for each microtiter plate, and yield a percent activity for each compound. To obtain data for 50,071 compounds, 157 plates were assayed in 15 separate runs. The 320 compounds exhibiting the highest percentage activities were tested at 24.8 μM in triplicate with a final DMSO concentration of 0.5% (v/v) to determine reproducibility. Additional information about the HTS is described in Supplementary Table 4.
Dual color assay to identify compounds with activity specific to CLRN1N48K
A cell line stably expressing CLRN1N48K-Venus-1D4 and DsRed–Express-DR was grown to 70% confluency and treated with 16.8 μM of each test compound in triplicate. Cells treated with 50 nM bortezomib or vehicle (DMSO) served as positive and negative controls, respectively. After treatment for 24 h, cells were fixed with 4% formaldehyde, washed with PBS, and imaged with a Leica DMI6000B inverted fluorescence microscope. Bright field illumination was used to verify the presence of cells prior to capturing images. Venus and DsRed fluorescence intensities were quantified by ImageJ software to draw a series of regions of interest (ROIs) with identical sizes encompassing cell-containing areas (15 ROIs per well, three wells per treatment). Average integrated pixel densities were obtained for each cell replicate. The background signal measured from an area without cells was subtracted from each measurement. Finally, the average pixel intensities for each treatment were normalized both to DMSO and bortezomib to yield the percent activity for each treatment.
Measurements of protein stability in HEK293 cells
The stabilities of human CLRN1N48K-Venus, DsRed-Express-DR, and human CLRN1-Venus were tested in HEK293 cell lines. To stabilize CLRN1N48K-Venus and DsRed-Express-DR, a cell line (1C4) co-expressing CLRN1N48K-Venus and DsRed-Express-DR was treated with MG132 (5 μM) in DMEM containing 10% FBS. After 14 h, cells were allowed to degrade protein by replacing MG132-DMEM with fresh DMEM medium containing 10% FBS. Cells were fixed with 4% paraformaldehyde at different time points (0, 4, and 8 h), and Venus and DS-Red were imaged and quantified by fluorescence microscopy following the methods described in the previous section. Half-lives of CLRN1N48K-Venus and DsRed-Express-DR were estimated by non-liner regression analysis using SigmaPlot v12.0 software. To test the stability of CLRN1-Venus, a cell line expressing CLRN1-Venus (C3 cell line) was treated with MG132. As control, 1C4 and C3 cells were treated with 0.1% DMSO for 14 h, and Venus and DsRed were imaged by the methods described above.
Accommodation of HTS assay to support medicinal chemistry optimization of O03
D6 cells were cultured in DMEM containing 10% FBS in a humidified incubator at 37 °C and 5% CO2. Cells were seeded on collagen-coated 96-well plates at a density of 20,000 cells per well in DMEM containing 10% FBS in a humidified incubator at 37 °C and 5% CO2. Following an overnight incubation, compounds were added for 24 h and tested in triplicate. DMSO was used as a negative control at 0.25% final concentration. Cells were fixed by the addition of 10% buffered formaldehyde to each well to achieve a final concentration of 4%. After 20 min, wells were washed three times with PBS (Gibco, Grand Island, NY) containing Triton X-100 (0.1%). HA-tagged CLRN1N48K was detected with anti-HA primary antibody at a dilution of 1:1000 in PBS containing Triton X-100 (0.1%, v/v). After a 90 min incubation, wells were washed three times with PBS containing Triton X-100 (0.1%, v/v), and a secondary antibody (goat anti-mouse IgG- Cy3) was added at a dilution of 1:250 in PBS containing Triton X-100 (0.1%, v/v) for 45 min. Wells were subsequently washed three times with PBS containing Triton X-100 (0.1%, v/v), and nuclear staining was accomplished by the addition of Hoechst 33342 (Life Technologies) at a dilution of 1:10,000. Imaging of the stained cells was performed with an InCell 2000 High Content Imager (GE Healthcare, Buckinghamshire, UK), reading the Cy3 channel for CLRN1N48K and the Hoechst channel for nuclei. Images were analyzed and quantitated using an algorithm that measured the CLRN1N48K staining for each cell, which was identified through the nuclear segmentation derived from the Hoechst signal. The algorithm measured the intensity of staining per cell, and thus was not sensitive to variations in cell number. Approximately 2,000 cells per well were analyzed to achieve an average density per cell measurement.
Immunoblotting
Cells were suspended and then sonicated in RIPA buffer (150 mM NaCl, 50 mM Tris (pH 8.0), 1% Igepal-630 (Sigma-Aldrich), 0.5% sodium deoxycholate, 1 tablet Complete Mini protease inhibitor cocktail (Roche, Basel, Switzerland)) and mixed end-over-end for 1 h at 4 °C. Lysates were cleared by centrifugation, and the total protein was quantified with a Bradford assay kit according to the manufacturer’s instructions (BioRad, Hercules, CA). Equal amounts of protein were mixed with Laemmli buffer and loaded onto 12% gels and separated by SDS-PAGE. Proteins then were transferred to PVDF membranes using a semi-dry method. Membranes were blocked with PBSTween (PBS with 0.1% Tween-20 (v/v)) containing 5% dry milk (w/v), incubated in primary antibody (either 1:2,000 mouse HA.11 clone 16B12 (BioLegend, San Diego, CA, #MMS-101R) or 1:6,000 mouse anti-β tubulin (Hybridoma Bank, Iowa City, IA, #E7)), washed with PBSTween, incubated in secondary antibody (1:6,000 anti-mouse IgG AP-linked (Promega, Madison, WI) or anti-mouse IgG HRP-linked (Cell Signaling Technology, Beverly, MA)), and washed again with PBSTween. Antibody binding was detected using either BCIP/NBT substrate (Promega) or ECL substrate (HyBlot, Denville Scientific, Hollison, MA). For ECL detection, membranes were exposed to photosensitive film (Denville Scientific). All antibodies were diluted in PBSTween containing 5% dry milk.
Characterizing the effect of B03 and O03 on human CLRN1N48K stabilization
D6 cells were grown to 70% confluency and treated with either bortezomib (50 nM), O03 (16.8 μM, University of Cincinnati), or B03 (1.68 or 16.8 μM, Enamine Ltd., Kiev, Ukraine). Compounds were diluted with DMSO to a final concentration of 0.25% (v/v). After 24 h, cells were collected and subjected to immunoblotting as described above.
Reverse transcription and quantitative PCR
D6 cells were grown to 70% confluency and then treated in triplicate with either 50 nM bortezomib, 16.8 μM O03, or 0.25% DMSO for 24 h. Total RNA was isolated using TRIzol (Invitrogen) according to the manufacturer’s instructions. RNA (5 μg) was treated with DNase I, and first strand cDNA was synthesized using M-MLV reverse transcriptase (Life Technologies, Grand Island, NY, #28025-013) with an oligo (dT) primer mixture containing 10 μM each of dT12, dT15, and dT18. cDNA was subjected to real-time PCR using the TaqMan Fast Universal Mix system (Life Technologies #4352042) and a probe for human CLRN1 (Life Technologies, Hs00602425_m1). A probe for 18s rRNA (Life Technologies #4319413E) served as an internal control.
Examining the effect of O03 on mouse CLRN1N48K stabilization
An NIH/3T3 cell line was established by methods described above to stably express mouse CLRN1N48K fused to the 1D4 antibody epitope41. Cells were grown to 70% confluency and treated with either bortezomib (2.2, 8.7, or 35 nM), O03 (3, 12, or 24 μM), or vehicle (0.25% (v/v) DMSO). After 24 h, cells were washed and processed for SDS-PAGE and immunoblotting with 1D4 monoclonal antibody41 (0.8 μg/ml) (see above for details).
Combined treatment of cells with cycloheximide and other test reagents
D6 cells were grown to 70% confluency and treated for 6 h with either 50 nM bortezomib, 50 μM MG132, or 16.8 μM O03. Untreated cells served as a negative control. Cells then were collected for immunoblot analyses or cultured further for additional treatment. To inhibit protein translation, cells were cultured with 100 μM cycloheximide. These cells were cultured in the presence or absence of bortezomib (50 nM), MG132 (50 μM), or O03 (16.8 μM). After an additional 6 h of culture, cells were collected for immunoblotting as described above.
Proteomics analysis of proteins affinity purified with a biotinylated compound
D6 cells were collected in lysis buffer (150 mM NaCl, 50 mM Tris, pH 8.0, and Complete protease inhibitor (Roche) containing 1% (v/v) Triton X-100), sonicated, incubated at 4 °C for 1 h, and finally subjected to centrifugation at 22,000 × g for 15 min at 4 °C. The supernatant was incubated with biotinylated compound, BF071 (10 μM). A control group was incubated with non-biotinylated compound, BF981 (500 μM) for 4 h prior to incubation with biotinylated-BF071 (10 μM) at 4 °C for 4 h. Mixtures then were incubated with Monomeric Avidin Agarose (Thermo Scientific) at 4 °C overnight. Samples were subjected to centrifugation, washed four times with lysis buffer containing Triton X-100, and then washed with lysis buffer without Triton X-100. Bound proteins were eluted using biotin (2 mM) in lysis buffer without Triton X-100. Eluted fractions were combined and concentrated with an Amicon ultra filter unit (3 kDa cut off). Affinity-purified proteins and proteins non-specifically bound to the resin (control) were separated by SDS-PAGE and then polyacrylamide gels were stained either with Bio-Safe Coomassie G-250 Stain (BioRad) (preparative gel) or silver stain (BioRad) (analytical gel). Enriched protein bands from the preparative gel were excised and subjected to in-gel protein digestion with porcine trypsin (sequencing grade, Promega). Peptides derived from samples incubated with BF071 were then labeled with 18O, whereas peptides from the control group were labeled with 16O as previously described42. 18O and 16O labeled peptides were mixed at a 1:1 ratio, and desalted using a C18 column (2–100 μL loading, 3–30 μg capacity, The Nest Group, Inc., Southborough, MA) according to the manufacturer’s instructions. Mixed peptides were dissolved in 0.1% formic acid and analyzed by liquid chromatography-tandem mass spectrometry with an Orbitrap Elite hybrid mass spectrometer (Thermo Scientific). Acquired data were subjected to Mascot search 2.4 (Matrix Science, London, UK) against a Swiss-Prot v56 human data base to identify specific proteins. Oxidation of Cys residues (to cysteic acid) and Met residues (to methionine sulfone) were set as fixed modifications with one or two C-terminal 18O labels set as variable modifications. Mass tolerance was set at 8 ppm for the precursor ion and 0.5 Da for product ions. Up to three incomplete cleavages of trypsin were allowed. Peptides with at least six amino acid residues and a minimum mascot score of 20 were considered significant. Proteomics Tools 3.4.3 developed by Quanhu Sheng43 were used to calculate the quantitative ratios of 18O/16O-labeled peptides.
To determine if CLRN1N48K is a potential target for BF844, D6 cells prior to affinity purification were treated overnight with bortezomib (50 nM) and then subjected to the purification procedures just described. Eluted fractions were concentrated, and CLRN1 N48K was identified by immunoblotting with anti-HA antibody (see above for details).
Citrate synthase aggregation assay
The chaperon activity of HSP60 and HSP90 was evaluated by monitoring thermal-induced aggregation of citrate synthase (CS) from porcine heart (Sigma-Aldrich) as previously described44. Briefly, CS was dialyzed overnight against 40 mM HEPES buffer (pH 7.4) and incubated at 0.16 mg/ml with or without HSP90β (StressMarq Biosciences, Vitoria, BC, Canada; 42 μg/ml) or HSP60 and its co-chaperone HSP10 (R&D Systems, Boston, USA; 28 μg/ml and 5 μg/ml, respectively) in a buffer containing 1 mM ATP at 43 °C. To test the effect of the compounds, HSP60/10 and HSP90β mixtures were pretreated either with BF844 (at 2 and 20 times molar ratio), BF066, or BF136 (each at 2 times and 20 times molar ratio). The total reaction volume was 250 μl. CS aggregation was monitored by measuring light scattering at 360 nm in 96-microwell plates (96-Well UV Microplate, Thermo Scientific) using a micro plate reader FlexStation 3 (Molecular Devices, Sunnyvale, CA). Control conditions included assays conducted without the addition of either HSP or HSPs without the addition of compounds, and all measurements were conducted in triplicate.
Effect of HSP90 inhibiton on CLRN1N48K expression
D6 cells were treated with either 2.90 μM BF844, 50 nM bortezomib, 20 μM 17-N-allylamino-17-demethoxygeldanamycin (17-AAG, LC Laboratories), or vehicle (DMSO) for 24 h. In one condition cells were pre-incubated with 17-AAG for 6 h followed by the addition of BF844 for 24 h. Cells were then harvested and immunoblotting analyses were performed (see above for details).
Effect of BF844 on CLRN1
Two HEK293 cell lines, C1 and D1, stably expressing HA tagged WT human CLRN16 were treated with bortezomib (50 nM), BF844 (2.9 μM), BF982 (2.2 μM) or vehicle (DMSO) for 24 h. Protein samples were prepared from C1 and D1 cells following the methods described above, and then treated with PNGase F (New England Biolabs, Ipswich, MA, USA) according to the manufacturer’s procedures. Briefly, 7 μl of protein sample were mixed with 1 μl of 10× G7 reaction buffer, 1 μl of 10× glycoprotein denaturing buffer, 1 μl of 10% Nonidet P-40, and 1 μl of PNGase F enzyme, and the resulting mixture was incubated at 37 °C for 1 h. As a control, 1 μM RIPA buffer was added instead of PNGase F. After incubation, samples were analyzed by immunoblotting as described above.
Cell surface protein labeling by biotin
C1 and D1 cell lines expressing HA tagged CLRN1 or D6 cells expressing HA tagged CLRN1N48K were treated with either 2.90 μM BF844 or vehicle (DMSO) for 24 h. Labeling of proteins at the plasma membrane was performed as previously described6 with the following modifications. First, cells washed with PBS were incubated in a PBS solution containing 1 mg/ml Sulfo-NHS-SS- Biotin (ProteoChem, Loves Park, IL) with gentle mixing at room temperature for 30 min. The biotin solution was aspirated and excess biotin was quenched with 100 mM glycine dissolved in PBS at room temperature for 30 min. Cells were collected by centrifugation. Pellets were lysed in RIPA buffer and total protein was determined using the Bradford assay. Lysates containing equal amounts of protein were added to NeutrAvidin agarose beads (Thermo Scientific), equilibrated in RIPA buffer, and incubated overnight at 4 °C with end-over-end mixing. Samples were subjected to centrifugation, supernatants isolated, and beads then washed 4 times with RIPA buffer. Avidin-bound proteins were eluted with Laemmli buffer and separated by SDS-PAGE on a 12% polyacrylamide gel, side-by-side with the cytosolic protein fraction. Immunoblotting analyses were performed as described in a previous section.
Testing BF844 and BF981 on unstable opsin mutant RHOP23H and CNGB1
The NIH/3T3 cell line stably expressing RHOP23H was a generous gift from Yuanyuan Chen (Case Western Reserve University)19. CNGB1 fused to a fluorescent protein Dendra222 (Dend2-CNGB1) was cloned into the pcDNA3.3-TOPO vector (Life Technologies). hTERT-RPE1 cells (ATCC, Manassas, VA), grown in DMEM/F12 with 10% FBS, 0.01 mg/ml hygromycin B (Sigma-Aldrich), and 1% penicillin/streptomycin at 37 °C with 5% CO2, were first transfected with pCDNA3.3 Dend2-CNGB1 using the FuGENE reagent (Promega) according to the manufacturer’s instructions. After 24 h post-transfection, cells were treated with G418 (500 μg/ml, Life Technologies). After 2 weeks of selection, the hTERT-RPE1 Dend-2-CNGB1 cell line was successfully obtained. Stable cells (NIH/3T3 cells expressing RHOP23H or CLRNN48K and hTERT-RPE1 cells expressing Dend2-CNGB1) were treated with either DMSO, bortezomib (50 nM), BF844 (2.9 μM) or BF981 (2.5 μM) for 24 h. Cells were then harvested and immunoblotting analyses were performed as described previously. 1D4 monoclonal antibody, rabbit Dendra2 antiserum45 and HA monoclonal antibody were used for immuno-detection of RHOP23H, Dend2-CNGB1 and CLRNN48K, respectively.
Mice
All animal procedures described in this report were approved by the Institutional Animal Care and Use Committee (IACUC) at Case Western Reserve University. Founder transgenic mice (TgAC1) were generated with the assistance of the Case Transgenic and Targeting Facility under IACUC protocol number 2011-0094. The testing of drug efficacy in mice was conducted under IACUC protocol number 2012-0068. Mice were maintained under standard care and housing conditions provided by the Animal Resource Center of Case Western Reserve University.
Mice with appropriate genotypes were setup for breeding and all mice from a litter were used without any bias or pre-selection. A litter consisted of age-matched male and female mice of various genotypes. All mice used in this study were maintained in the C57BL/6J genetic background. Genotypes and gender were documented after the experiments were completed. Results from mice with wild-type (Clrn1+/+), KI/KI and Tg;KI/KI genotype were used to generate the graphs shown in the results and supplementary information section. For all studies reported here, a sample size of 5 (minimum) to 10 (maximum) mice per group was chosen, based on hearing evaluation in USH3 mouse models reported previously7,35.
Generation of Transgenic line Tg;KI/KI
To obtain a mouse USH3 model with slow progressive hearing loss, we first generated a construct in which the ‘Atoh1 enhancer-β globin basal promoter’ was fused to cDNA representing a full length transcript of mouse Clrn1 (Supplementary Fig. 9). Mice carrying Transgene Atoh1-enhacer-Clrn1 (TgAC1) were produced by pronuclear microinjection of TgAC1 into the male pronucleus of fertilized eggs from wild-type mice of a desired genetic background (C57BL/6J mice). Details regarding the generation, identification and propagation of mice can be found elsewhere (http://ko.cwru.edu/services/transgenics.shtml). Injected transgenes integrate into the host genome by non-homologous recombination or random, non-targeted insertion. The endogenous Clrn1 was intact in the founder generation. These ‘TgAC1’ founders (Tg; Clrn1+/+) were crossed with Clrn1N48K/N48K (KI/KI) mice7 (also in the C57BL/6J genetic background) to generate Tg;KI/KI mice. A PCR-based approach was used to identify mice carrying TgAC1. Genomic DNA was isolated as described previously35. The primers used for genotyping were 5′-CCCTCTCTCACACCCCATTA-3 ′ (KA1109) and 5′-TGAGAACCGGAAAGGCCTTGC-3 ′ (KA1085). The expected size of the PCR product is 1938 base pairs (bp). A PCR-based approach was used to identify mice carrying Clrn1 or Clrn1N48K alleles as described previously7,35. TgAC1; Clrn1N48K/N48K mice are denoted as “Tg;KI/KI” mice in this report. Mice carrying the wild-type allele Clrn1+/+ were used as controls, unless otherwise stated.
BF844 administration to mice
To test the efficacy of BF844 in preserving hearing of Tg;KI/KI mice, two regimens were implemented. Two stock concentrations of BF844 were prepared at 1mg/ml and 3 mg/ml. The solvent solution consisted of the following: 10% DMSO, 10% Cremaphor® EL (Sigma-Aldrich) and 80% sterile distilled H2O. Further descriptions of Regimens I and II can be found in Supplementary Fig. 11. Summaries of the procedures are presented here.
Regimen I
BF844 was delivered i.p. at 30 mg/kg of body weight (bw) daily from P30 to P45. Hearing was tested at P22 (before drug administration) to make sure the Tg;KI/KI mice displayed ABR thresholds in the expected range, as shown in Supplementary Fig. 10. After BF844 treatment was completed on P45, hearing was tested at P46 and P55.
Regimen II
This regimen was designed to determine whether earlier intervention with BF844 (starting at P10) was more effective in preserving hearing in the Tg;KI/KI mouse model. Because P10 mice did not tolerate 30 mg/kg of BF844 (data not shown), we started at 10 mg/kg every other day and gradually escalated the dose up to 20 mg/kg at P28. From P30 to P45, mice received 30 mg/kg daily (see schematic in Supplementary Fig. 11). Hearing tests were performed at P46 and P55.
ABR recording
ABR recording was performed as described previously7,35. Auditory function in mice reaches adult levels of sensitivity about 3 weeks after birth. Briefly, P22 and older mice were anesthetized with an i.p. injection of diluted ketamine, xylazine, and acepromazine at doses of 40, 5, and 1 mg/kg, respectively. Body temperatures were maintained at 37–38 °C by placing mice on a homeothermic heating pad (Harvard Apparatus, Holliston, MA). Mice then were placed in a sound proof chamber during recording. ABRs were recorded with a SmartEP system from Intelligent Hearing Systems (Miami, FL). Platinum subdermal electrodes were inserted at the vertex and ventrolaterally into the right and left ears. To test hearing function, mutant mice were presented with 16 kHz pure tone stimuli at a stimulus intensity starting at a 100 decibel sound pressure level (dB SPL) that decreased in 10 dB steps; this sequence was repeated in 5 dB steps until the lowest intensity that evoked a reproducible ABR waveform (peaks I–IV) was detected. A plastic tube connected to the transducer at one end and the opening of the tube at the other end was placed into the external ear to enable closed-field measurements. One hundred millisecond stimuli were presented for at least 500 sweeps to the ear through high-frequency transducers. To obtain more robust data, the left and right ears were tested separately, and ABRs were recorded from both ears of a mouse in all cases. ABR thresholds from the left and right were averaged and used for statistical analyses and graphic presentation. Thus, if n = 10, it means the data from 20 ears were averaged.
Statistical methods used to analyze ABR data from Tg; KI/KI mice
ABR threshold values of left and right ears were averaged (Excel, Microsoft®, Redmond, VA) for each mouse belonging to Clrn1+/+ (Control), KI/KI, or Tg; KI/KI mice at five time points (P22, P35, P46, P55 and P70). The ABR threshold value from each mouse was displayed as a single data point (color dot/symbol) on a scatter plot. The ABR threshold data are presented in a box and whisker plot format superimposed on the scatter plot. Boxes in the plot represent values from the 25th to 75th percentile in each group and the middle line in each box is the median ABR threshold. A line extends from the box to the minimum and maximum values. One-way ANOVA analyses were used to determine whether the differences in ABR thresholds observed between the groups were significant. P values of <0.05 were considered statistically significant. The same statistical approach was used to analyze ABR threshold data from Tg:KI/KI mice after treatment (Regimens I and II). The hearing threshold is represented in log scale. A 20 dB SPL lower or reduction in ABR threshold in treated mice would imply that hearing sensitivity in the treated mice is 100 times more sensitive than untreated mice. Therefore, a 20 dB SPL mean difference in ABR threshold following treatment was considered significant (efficacious).
Measurements of kinetic solubility
Using a 10 mM stock solution of each test compound in 100% DMSO, sample dilutions were prepared to a theoretical concentration of 200 μM both in PBS (pH 7.4, containing 2% DMSO) and in 100% DMSO. An aliquot of the 200 μM DMSO solution was then further diluted to 10 μM (n = 2 for each compound) and equilibrated at room temperature on an orbital shaker for 2 h. PBS dilutions were filtered using a Multiscreen HTS solubility filter plate (Millipore, Billerica, MA), and the filtrates were analysed by LC (1100 binary LC system, Agilent Technologies Ltd, Cheshire, UK) with a variable wavelength UV detector (Agilent Technologies Ltd) and a MS detector (Quattro Micro MS, Waters Ltd, Hertfordshire, UK). The concentration of a test compound in the PBS filtrate was determined by comparing its UV absorbance peak (λ = 254 nm) with that of the two DMSO dilutions. Mass spectrometry was used to confirm the presence of the expected molecular ion in the measured UV peak. The effective concentration range of the assays was 5 – 200 μM.
Measurements of mouse hepatic microsome stability (half-life)
Incubations of test compounds (1 μM initial concentration, n=2) were carried out with microsomes (0.25 mg protein/mL). Assays were performed at 37 °C with 100 μL samples removed from the incubation mixtures at 0, 5, 10, 20 and 40 min. Reactions were terminated by the addition of 100 μL of acetonitrile containing carbamazepine as an internal standard.
Samples were subjected to centrifugation, and supernatants were analysed by LC-MS/MS (TQD, Waters Ltd.). Parameters used for electrospray mass spectrometry were as follows: capillary voltage, 3.5 kV; extractor voltage, 3.0 V; source temp., 150 °C; desolvation gas temp, 500 °C; desolvation gas flow, 900 L/h; cone gas flow, 50 L/h; collision gas flow, 0.20 mL/min. Parameters used for multiple reaction monitoring (MRM) were as follows for each compound.
1 (O03): Ionization mode ES+; Transition (parent ion > fragment ion (m/z)) 260.12 > 149.96; Cone voltage 55 V; Collision energy 28 eV. 2: Ionization mode ES+; Transition (parent ion > fragment ion (m/z)) 322.14 > 77.06; Cone voltage 64 V; Collision energy 46 eV. 3 (BF981): Ionization mode ES+; Transition (parent ion > fragment ion (m/z)) 351.17 > 77.10; Cone voltage 70 V; Collision energy 64 eV. 4 (BF844): Ionization mode ES+; Transition (parent ion > fragment ion (m/z)) 379.23 > 307.15; Cone voltage 46 V; Collision energy 28 eV. Carbamazepine (internal standard): Ionization mode ES+; Transition (parent ion > fragment ion (m/z)) 237.11 > 194.18; Cone voltage 46 V; Collision energy 19 eV.
Chromatographic conditions were as follows: Column: Kinetex XB-C18 100A 50 × 2.1 mm, 2.6 μm (Phenomenex, Torrance, CA). Column temperature, 40 °C; Flow rate, 0.8 mL/min; Injection volume, 2 μL; Mobile phase A, 0.01% formic acid in water; Mobile phase B, 0.01% formic acid in acetonitrile. The gradient used ran from 5% Mobile phase B at T = 0 min to 95% at T=0.5 min. This percentage was held until T=0.85 min. The gradient then ran to 5% Mobile phase B at T=0.90 min and this percentage was held until T = 1.1 min. The instrument responses (i.e. chromatographic peak heights) were referenced to the zero time-point samples (as 100%) to determine the percentage of test compound remaining. Natural logarithm (ln) plots of the % remaining vs time, for each compound, were used to determine the half-lives for the microsomal incubations. Half-life values were calculated from the relationship: T1/2 (min) = −0.693/λ, where λ was the exponential decay constant calculated from the slope of the ln concentration vs time curve. The in vitro intrinsic clearance, Clint (μL/min/mg microsomal protein), was calculated by the following formula: Clint = 0.693 × 1/T½ (min) × incubation volume (μL)/mg of microsomal protein.
Metabolite identification
To generate metabolites, a test compound (10 μM initial concentration) was incubated with mouse cryopreserved hepatocytes (0.25 million cells/mL) for 90 min at 37 °C. Samples were then prepared from the incubation mixture by solvent precipitation (1:1 addition of acetonitrile). Extracted samples were mixed by vortex and subjected to centrifugation; the supernatant was removed and placed in a clean polypropylene plate for analysis. A Waters Xevo-ToF G2 mass spectrometer (S/N: YCA258, Waters Ltd, 730–740 Centennial Court, Centennial Park, Hertfordshire) was used for metabolite identification. Settings of the electrospray ion source were selected for compatibility with a wide range of compounds and metabolites, whereas the MSE collision energy settings were fixed at 4 eV for low energy scans and ramped up to 20 to 30 eV for high energy scans. To ensure accurate mass measurements of ions, calibration of the instrument was verified with a sodium formate calibration standard (500 μM sodium formate in 80:20 acetonitrile:water containing 1% formic acid) immediately prior to use. An internal accurate mass reference (lockmass) was used to further stabilize mass accuracy over the course of an analysis. A solution of leucine-enkephalin (Sigma-Aldrich, L9133) at 2 ng/mL in a 1:1 mixture of water and acetonitrile containing 0.1% formic acid was run in parallel with all analytical samples, using Waters’ LockSpray independent ion source. All spectra gathered were adjusted to maintain the lockmass at mass charge ratios (m/z) of 556.27 and 278.11 Da in a positive ion mode.
Samples were loaded onto a cooled Waters Acquity UPLC autosampler (Waters Ltd.), held at 10 °C (± 1 °C). A 5 μL aliquot of each sample was loaded onto an UPLC BEH C18 column (2.1 × 100 mm with a 1.7 μm particle size) (Acquity, Waters Ltd.) under starting conditions of 5% acetonitrile in water containing 0.1% formic acid at a flow rate of 0.2 mL/min; the injection triggered a gradient rise in acetonitrile from 5% at 0.2 min to 95% at 4.2 min, holding until 4.7 min before falling to 5% and equilibrating until 5.0 min. Mass to charge ratio (m/z) chromatograms were generated for study sample extracts over a 50–1200 Da range. Two collision energy levels were employed for mass spectrometry in the electrospray mode to generate fragment ions for additional structural elucidation. When significant chromatographic peaks were observed in the samples derived from the compound mixtures but were absent in vehicle control samples, both high and low energy mass spectra were extracted and the elemental composition of the underlying metabolites and fragments was determined from the accurate monoisotopic mass.
Cellular EC50 measurements
The D6 cells were cultured in DMEM containing 10% FBS in a humidified incubator at 37 °C and 5% CO2. For the CLRN1N48K expression assay, cells were seeded on collagen coated 96-well plates at a density of 20,000 cells per well in DMEM containing 10% FBS in a humidified incubator at 37 °C and 5% CO2. After incubation overnight, test compounds in triplicate were added for a 24 h incubation. DMSO (0.25%) was used as a negative control. The standard protocol involved continued exposure to compounds for 24 h. Alternatively, cells were exposed to a test compound for 30 min or 2 h before the medium was replenished and the incubation continued without the compound for 23.5 or 22 h, respectively. Cells were fixed with 10% buffered formaldehyde introduced into the wells to achieve a final concentration of 4%. Cells were processed and imaged as described above (see Accommodation of HTS assay to support medicinal chemistry optimization of O03).
MDR1-MDCK effective efflux assay
The MDR1-MDCK cell line, which was engineered to over-express the efflux transporter MDR1 (P-glycoprotein), was obtained from Solvo Biotechnology (Boston, MA). Both MDCK and MDR1-MDCK cells were seeded onto 24-well Transwell plates at 2.35 × 105 cells per well and used in confluent monolayers after a 3-day culture at 37 °C under 5% CO2. For both the cell lines, test and control (propranolol, vinblastine) compounds were added (10 μM, 0.1% DMSO final concentration, n=2) to donor compartments of the Transwell plate assembly in assay buffer (Hanks balanced salt solution supplemented with 25 mM HEPES, adjusted to pH 7.4) for apical to basolateral (A>B) and basolateral to apical (B>A) measurements. Incubations were performed at 37 °C, with samples removed from both donor and acceptor chambers at T=0 and 1 h. Samples containing an internal standard then were analyzed by mass spectrometry (LC-MS/MS). Results were calculated as follows: Apparent permeability Papp = [CompoundAcceptor T=end]× VAcceptor/([CompoundDonor T=0]×VDonor)/incubation time×VDonor/Area×60 × 10−6 cm/s, where V is the volume of each Transwell compartment (apical 125 μL, basolateral 600 μL), and concentrations are the relative MS responses for the test compound (normalized to internal standard) in the donor chamber before incubation and in the acceptor chamber at the end of the incubation. Area = area of cells exposed for drug transfer (0.33 cm2). Efflux ratios (Papp B>A/Papp A>B) were calculated for each compound from the mean Papp values in each direction for both MDCK and MDR1-MDCK cells. The finding of a good basolateral to apical permeability (B>A) but a poor apical to basolateral permeability (A>B), suggests that a test compound is a substrate for the efflux transporter MDR1. To confirm the involvement of MDR1 in any observed efflux, an “effective efflux ratio” (EER) was calculated by comparing compound efflux ratios (ER) in the two cell types, i.e. EER = ER (MDR1-MDCK)/ER (MDCK). This ratio illustrates the effect of the over-expressed MDR1 normalized for the background movement of compound through wild-type cells. Lucifer Yellow (LY) was added to the apical buffer in all wells to assess viability of the cell layer. As LY cannot freely permeate lipophilic barriers, a high degree of LY transport indicates poor integrity of the cell layer. Therefore wells with a LY Papp > 10 × 10−6 cm/s were rejected.
Measurements of brain to plasma ratios
Plasma proteins were precipitated from plasma samples and test compounds extracted by the addition of three volumes of acetonitrile containing analytical internal standard (carbamazepine). Samples were subjected to centrifugation for 10 min at 14,462 g in a benchtop centrifuge and the supernatant fractions removed for MS analysis. Brain tissue was homogenized in 2:1 (w/v) volumes of 0.1 M PBS, pH 7.4, using a Precellys® homogenizer (Bertin Corp., Rockville, MD) (5,000 rpm for 15 s). Compound BF981 was extracted from brain homogenates by the addition of three volumes of acetonitrile containing analytical internal standard (carbamazepine). Samples were centrifuged for 30 min at 2,773 g in a benchtop centrifuge and the supernatant fractions removed for MS analysis. Samples of brain homogenates were quantified against an external calibration curve generated in control brain homogenates (final concentrations of 0.5, 1.0, 2.0, 5.0, 10, 50, 100, 500, 1,000 and 2,000 ng eq/mL), alongside QC samples prepared at 8, 80 and 800 ng eq/mL in control rat brain.
At a minimum, 75% of all the calibration standards and at least one calibration standard per concentration met the accuracy and precision criteria of ±20%. There was no bias in the accuracy or precision of the calibration standards. The coefficient of variation (CV%) of the internal standard signal/area response for the entire run was within the criteria of ±20%, and there was no bias or trend in the internal standard signal/area response.
Concentrations of BF981 in plasma and brain samples from mice given single i.p. doses of 30 mg/kg were measured at T=15, 30 and 60 min (n=3). Brain to plasma compound concentration ratios were calculated for each animal, and an overall average brain to plasma ratio (B/P) was calculated from all time points combined.
Measurements of retina to plasma ratios
Retinas were dissected from rat eyes and weighed. Plasma proteins were precipitated from plasma samples and test compounds extracted by the addition of three volumes of acetonitrile containing analytical internal standard (carbamazepine) and processed for MS analysis as described in a previous section.
Plasma samples were quantified against an external calibration curve generated in control plasma (final concentrations of 0.5, 1.0, 2.0, 5.0, 10, 50, 100, 500, 1,000 and 2,000 ng eq/mL). Retina samples were quantified against an external calibration standard curve generated in control retina tissue from the same species and strain used in the study (final concentrations of 0.5, 1.0, 2.0, 5.0, 10, 50, 100, 500, 1,000 and 2,000 ng eq/mL). QC samples were prepared at 8, 80 and 800 ng eq/mL in rat plasma, sheep retina and rat brain and at 80 ng eq/mL in rat retina.
Concentrations of BF981 in plasma and retina samples from rats given single i.p. doses of 10 mg/kg were measured at T=15 and 30 min (n=3). Retina to plasma compound concentration ratios were calculated for each animal, and an overall average retina to plasma ratio (R/P) was calculated from both time points combined.
Reaction temperature
Unless otherwise noted, all the procedures were carried out at room temperature.
Statistical Analysis
Procedures used for statistical analyses are described throughout the manuscript.
Supplementary Material
Acknowledgments
This work was supported by the Usher III Initiative (Y.I., K.N.A., and BioFocus group), Hope for Vision (Y.I.), the Prince Family Foundation (Y.I.), start-up funding from CWRU (Y.I.), the National Institutes of Health Grants R01-DC010816 (K.N.A.), R01-EY020826 (Y.I.) and R24 EY021126 (K.P.) and the Arnold and Mabel Beckman Foundation (K.P.). We thank the Elden family and The Prince Family Foundation for their generous support of our research via the Usher III Initiative, Dr. David Saperstein for his leadership of the Initiative’s research efforts and Dr. Alexander R. Moise for useful suggestions and discussions prior to the inception of this project. We also thank Drs. Ruben Papoian, Sandra Nelson, and William Seibel at the Drug Discovery Center (Univ. of Cincinnati) for conducting the HTS. The monoclonal antibody E7 developed by Dr. Michael Klymkowsky was obtained from the Developmental Studies Hybridoma Bank created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242. Fluorescence microscopy was conducted to optimize the HTS assay at the light microscopy imaging core facility of CWRU. We thank Dr. Yuanyuan Chen at Case Western Reserve University for providing the RHOP23H NIH/3T3 cell line. We thank Dr. Leslie T. Webster, Jr., for insightful comments on this manuscript. K.P. is John H. Hord Professor of Pharmacology.
Abbreviations
- 17-AAG
17-N-allylamino-17-demethoxygeldanamycin
- ABR
auditory brainstem response
- AUC
area under the drug concentration-time curve
- BF prefix
synthesized at BioFocus
- B/P
brain to plasma ratio
- C1 cell line
HEK293 stable cell line expressing higher amounts of human CLRN1
- CFTRδF508
cystic fibrosis transmembrane regulator mutant in which Phe residue at position 508 is deleted
- CLRN1
human clarin-1 gene
- Clrn1
mouse clarin-1 gene
- CLRN1
clarin-1 protein
- CLRN1N48K
clarin-1 protein with Asn at position 48 replaced with Lys
- CS
citrate synthase
- D1 cell line
HEK293 stable cell line expressing lower amounts of human CLRN1
- CX
cycloheximide
- D6 cell line
HEK293 stable cell line expressing human CLRN1N48K-HA-FLAG™
- CNGB1
cGMP-gated channel β-subunit
- dB SPL
decibel sound pressure level
- DEAD
diethyl azodicarboxylate
- DIPEA
N,N-diisopropylethylamine
- DMEM
Dulbecco’s Modified Eagle Medium
- DMF
N,N-dimethylformamide
- DMSO
dimethyl sulfoxide
- EER
effective efflux ratio
- ERAD
endoplasmic reticulum-associated degradation
- FBS
fetal bovine serum
- HA
hemagglutinin
- HATU
1-[bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxid hexafluorophosphate
- HPLC
high performance liquid chromatography
- HSP
heat shock protein
- hTERT-RPE
telomerase immortalized human retinal pigmented epithelium
- HTS
high-throughput screening
- i.p.
intraperitoneal/ly
- LC-MS
liquid chromatography and mass spectrometry
- LY
Lucifer Yellow
- MRM
multiple reaction monitoring
- NMR
nuclear magnetic resonance
- PBS
phosphate buffered saline (1.05 mM KH2PO4, 155 mM NaCl, and 2.97 mM Na2HPO4, pH 7.4)
- PBSTween
phosphate buffered saline containing 0.1% Tween-20 (v/v)
- PVDF
polyvinylidene fluoride
- PX
post-natal day X
- RHOP23H
rhodopsin mutant (protein) in which Pro at position 23 is substituted with His
- RIPA buffer
radio-immunoprecipitation assay buffer
- ROIs
regions of interest
- R/P
retina to plasma ratio
- Rt
retention time
- TFA
trifluoroacetic acid
- TgAC1
transgene consisting of a regulatory element of the atonal homolog 1 enhancer-β globin basal promoter fused to mouse Clrn1
- THF
tetrahydrofuran
- Tg;KI/KI
the previously noted transgene (TgAC1) coupled with the N48K knockin mutation in Clrn1
- USH3
Usher syndrome type III
Footnotes
Author contributions
Y.I., and K.P. conceived the small molecule screening experiments and designed the small molecule HTS assay, and G.T. and R.L. contributed to the establishment of the assay; G.T. I. N., and R.L. conducted the experiments characterizing small molecules; I.N. and M.M. performed mass spectroscopy and associated experiments; K.N.A. conceived the transgenic mouse models TgAC1 and TgAC1;KI/KI; K.N.A., S.R.G., R.G. and D.H-C., were involved in the generation TgAC1;KI/KI mice; S.R.G. characterized the hearing loss profile in TgAC1; KI/KI mice; K.N.A., S.R.G. and D.H-C., conceived the dosing scheme for Regimen II; S.R.G. and D.H-C. performed efficacy testing associated with Regimens I and II; K.F.M., C.J.L., W.R.E., and A.P.O. performed the medicinal chemistry experiments. N.A.L. accommodated the small molecule screening assay at BioFocus; K.O. and F.J.A. conducted the small molecule screening assay at BioFocus; K.F.M., N.A.L., D.F.F., R.W.B., A.M.M. and W.E.H. provided project supervision at BioFocus. Y.I. and K.N.A. provided project supervision at CWRU. I.N. and K.F.M. wrote the results section associated with Medicinal Chemistry and target identification. K.N.A., K.P., and Y.I. wrote the manuscript.
Competing financial interests
Usher III Initiative contracted K.F.M, C.J.L, W.R.E., A.P.O., N.A.L., K. O., F.J.A., D.F.F., R.W.B. and A.M.M. from BioFocus to carry out hit-to-lead optimization of small molecules described in this report. W.E.H. serves as the Director of Pharmaceutical Development for the Usher III Initiative and Chief Translational Officer for the School of Medicine at CWRU. W.E.H., R.W.B., K.F.M, C.J.L, W.R.E. and A.P.O. are named on patents WO 2014066835, WO 2014066836 and WO 2012148994. The mouse Clrn1 cDNA used to generate transgenic mouse TgAC1 and the human ortholog are protected by a provisional US patent # 62/076,114 to K.N.A.
S.R.G., R.G., D.H-C., I.N., R.L., G.T., M.M., K.P., and Y.I. have no conflicts of interest.
Additional information
BF844 patent information: WO 2012148994
Preparation of pyrazolopyridazines and methods for treating retinal-degenerative diseases and hearing loss associated with Usher Syndrome; Burli, Roland Werner; Esmieu, William Rameshchandra Krishna; Lock, Christopher James; Malagu, Karine Fabienne; Owens, Andrew Pate; Harte, William Edward.
References
- 1.Pakarinen L, Karjalainen S, Simola KO, Laippala P, Kaitalo H. Usher’s syndrome type 3 in Finland. Laryngoscope. 1995;105:613–617. doi: 10.1288/00005537-199506000-00010. [DOI] [PubMed] [Google Scholar]
- 2.Ness SL, et al. Genetic homogeneity and phenotypic variability among Ashkenazi Jews with Usher syndrome type III. J Med Genet. 2003;40:767–772. doi: 10.1136/jmg.40.10.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plantinga RF, et al. Serial audiometry and speech recognition findings in Finnish Usher syndrome type III patients. Audiol Neurootol. 2005;10:79–89. doi: 10.1159/000083363. [DOI] [PubMed] [Google Scholar]
- 4.Joensuu T, et al. Mutations in a novel gene with transmembrane domains underlie Usher syndrome type 3. Am J Hum Genet. 2001;69:673–684. doi: 10.1086/323610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adato A, et al. USH3A transcripts encode clarin-1, a four-transmembrane-domain protein with a possible role in sensory synapses. Eur J Hum Genet. 2002;10:339–350. doi: 10.1038/sj.ejhg.5200831. [DOI] [PubMed] [Google Scholar]
- 6.Tian G, et al. Clarin-1, encoded by the Usher Syndrome III causative gene, forms a membranous microdomain: possible role of clarin-1 in organizing the actin cytoskeleton. J Biol Chem. 2009;284:18980–18993. doi: 10.1074/jbc.M109.003160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geng R, et al. The mechanosensory structure of the hair cell requires clarin-1, a protein encoded by Usher syndrome III causative gene. J Neurosci. 2012;32:9485–9498. doi: 10.1523/JNEUROSCI.0311-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gopal SR, et al. Zebrafish Models for the Mechanosensory Hair Cell Dysfunction in Usher Syndrome 3 Reveal That Clarin-1 Is an Essential Hair Bundle Protein. J Neurosci. 2015;35:10188–10201. doi: 10.1523/JNEUROSCI.1096-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 10.Baell JB, Holloway GA. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 11.Cumming JG, Davis AM, Muresan S, Haeberlein M, Chen H. Chemical predictive modelling to improve compound quality. Nat Rev Drug Discov. 2013;12:948–962. doi: 10.1038/nrd4128. [DOI] [PubMed] [Google Scholar]
- 12.Li X, et al. Generation of destabilized green fluorescent protein as a transcription reporter. J Biol Chem. 1998;273:34970–34975. doi: 10.1074/jbc.273.52.34970. [DOI] [PubMed] [Google Scholar]
- 13.Kisselev AF, van der Linden WA, Overkleeft HS. Proteasome inhibitors: an expanding army attacking a unique target. Chem Biol. 2012;19:99–115. doi: 10.1016/j.chembiol.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polli JW, et al. Rational use of in vitro P-glycoprotein assays in drug discovery. J Pharmacol Exp Ther. 2001;299:620–628. [PubMed] [Google Scholar]
- 15.Schinkel AH, et al. Disruption of the mouse mdr1a P-glycoprotein gene leads to a deficiency in the blood-brain barrier and to increased sensitivity to drugs. Cell. 1994;77:491–502. doi: 10.1016/0092-8674(94)90212-7. [DOI] [PubMed] [Google Scholar]
- 16.Schulte TW, Neckers LM. The benzoquinone ansamycin 17-allylamino-17-demethoxygeldanamycin binds to HSP90 and shares important biologic activities with geldanamycin. Cancer Chemother Pharmacol. 1998;42:273–279. doi: 10.1007/s002800050817. [DOI] [PubMed] [Google Scholar]
- 17.Sakami S, et al. Probing mechanisms of photoreceptor degeneration in a new mouse model of the common form of autosomal dominant retinitis pigmentosa due to P23H opsin mutations. J Biol Chem. 2011;286:10551–10567. doi: 10.1074/jbc.M110.209759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korschen HG, et al. A 240 kDa protein represents the complete beta subunit of the cyclic nucleotide-gated channel from rod photoreceptor. Neuron. 1995;15:627–636. doi: 10.1016/0896-6273(95)90151-5. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y, et al. A High-Throughput Drug Screening Strategy for Detecting Rhodopsin P23H Mutant Rescue and Degradation. Invest Ophthalmol Vis Sci. 2015;56:2553–2567. doi: 10.1167/iovs.14-16298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakami S, Kolesnikov AV, Kefalov VJ, Palczewski K. P23H opsin knock-in mice reveal a novel step in retinal rod disc morphogenesis. Hum Mol Genet. 2014;23:1723–1741. doi: 10.1093/hmg/ddt561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiang WC, et al. Robust Endoplasmic Reticulum-Associated Degradation of Rhodopsin Precedes Retinal Degeneration. Mol Neurobiol. 2015;52:679–695. doi: 10.1007/s12035-014-8881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nemet I, Tian G, Imanishi Y. Organization of cGMP sensing structures on the rod photoreceptor outer segment plasma membrane. Channels (Austin) 2014;8:528–535. doi: 10.4161/19336950.2014.973776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lumpkin EA, et al. Math1-driven GFP expression in the developing nervous system of transgenic mice. Gene Expr Patterns. 2003;3:389–395. doi: 10.1016/s1567-133x(03)00089-9. [DOI] [PubMed] [Google Scholar]
- 24.Zhou X, Jen PH, Seburn KL, Frankel WN, Zheng QY. Auditory brainstem responses in 10 inbred strains of mice. Brain Res. 2006;1091:16–26. doi: 10.1016/j.brainres.2006.01.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burkard RF, Eggermont Jso J, Don Manuel, editors. Auditory evoked potentials: basic principles and clinical application. Lippincott Williams & Wilkins; Philadelphi: 2007. [Google Scholar]
- 26.Lobanova ES, Finkelstein S, Skiba NP, Arshavsky VY. Proteasome overload is a common stress factor in multiple forms of inherited retinal degeneration. Proc Natl Acad Sci U S A. 2013;110:9986–9991. doi: 10.1073/pnas.1305521110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JN, et al. Proteasome inhibitors induce auditory hair cell death through peroxisome dysfunction. Biochem Biophys Res Commun. 2015;456:269–274. doi: 10.1016/j.bbrc.2014.11.070. [DOI] [PubMed] [Google Scholar]
- 28.Shearer AE, et al. Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc Natl Acad Sci U S A. 2010;107:21104–21109. doi: 10.1073/pnas.1012989107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiseman RL, Haynes CM, Ron D. SnapShot: The unfolded protein response. Cell. 2010;140:590–590. e592. doi: 10.1016/j.cell.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 30.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 31.Claessen JH, Kundrat L, Ploegh HL. Protein quality control in the ER: balancing the ubiquitin checkbook. Trends Cell Biol. 2012;22:22–32. doi: 10.1016/j.tcb.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ward CL, Omura S, Kopito RR. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 33.Luo Y, McDonald K, Hanrahan JW. Trafficking of immature DeltaF508-CFTR to the plasma membrane and its detection by biotinylation. Biochem J. 2009;419:211–219. 212–219. doi: 10.1042/BJ20081869. [DOI] [PubMed] [Google Scholar]
- 34.Heon TG, Noh SH, Tang BL, Kim KH, Le MG. Rescue of delta F508-CFTR trafficking via a GRASP-Dependent Unconventional Secretion Pathway. Cell. 2011;146:746–760. doi: 10.1016/j.cell.2011.07.021. [DOI] [PubMed] [Google Scholar]
- 35.Geng R, et al. Usher syndrome IIIA gene clarin-1 is essential for hair cell function and associated neural activation. Hum Mol Genet. 2009;18:2748–2760. doi: 10.1093/hmg/ddp210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geller SF, et al. CLRN1 is nonessential in the mouse retina but is required for cochlear hair cell development. PLoS Genet. 2009;5:e1000607. doi: 10.1371/journal.pgen.1000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian G, Lee R, Ropelewski P, Imanishi Y. Impairment of vision in a mouse model of Usher syndrome type III. Invest Ophthalmol Vis Sci. 2016;57:866–875. doi: 10.1167/iovs.15-16946. [DOI] [PubMed] [Google Scholar]
- 38.Wang X, et al. Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell. 2006;127:803–815. doi: 10.1016/j.cell.2006.09.043. [DOI] [PubMed] [Google Scholar]
- 39.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 40.Nagai T, et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 41.MacKenzie D, Arendt A, Hargrave P, McDowell JH, Molday RS. Localization of binding sites for carboxyl terminal specific anti-rhodopsin monoclonal antibodies using synthetic peptides. Biochemistry. 1984;23:6544–6549. doi: 10.1021/bi00321a041. [DOI] [PubMed] [Google Scholar]
- 42.Hajkova D, et al. Proteomic changes in the photoreceptor outer segment upon intense light exposure. J Proteome Res. 2010;9:1173–1181. doi: 10.1021/pr900819k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo Y, Miyagi M, Zeng R, Sheng Q. O18Quant: a semiautomatic strategy for quantitative analysis of high-resolution 16O/18O labeled data. Biomed Res Int. 2014;2014:971857. doi: 10.1155/2014/971857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santhoshkumar P, Sharma KK. Phe71 is essential for chaperone-like function in alpha A-crystallin. J Biol Chem. 2001;276:47094–47099. doi: 10.1074/jbc.M107737200. [DOI] [PubMed] [Google Scholar]
- 45.Lodowski KH, et al. Signals governing the trafficking and mistrafficking of a ciliary GPCR, rhodopsin. J Neurosci. 2013;33:13621–13638. doi: 10.1523/JNEUROSCI.1520-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.