Abstract
Serine is a both a proteinogenic amino acid and the source of one-carbon units essential for de novo purine and deoxythymidine synthesis. In the canonical glucose-derived serine synthesis pathway, Homo sapiens phosphoglycerate dehydrogenase (PHGDH) catalyzes the first, rate-limiting step. Genetic loss of PHGDH is toxic towards PHGDH-overexpressing breast cancer cell lines even in the presence of exogenous serine. Here, we use a quantitative high-throughput screen to identify small molecule PHGDH inhibitors. These compounds reduce the production of glucose-derived serine in cells and suppress the growth of PHGDH-dependent cancer cells in culture and in orthotopic xenograft tumors. Surprisingly, PHGDH inhibition reduced the incorporation into nucleotides of one-carbon units from glucose-derived and exogenous serine. We conclude that glycolytic serine synthesis coordinates the use of one-carbon units from endogenous and exogenous serine in nucleotide synthesis, and suggest that one-carbon unit wasting may contribute to the efficacy of PHGDH inhibitors in vitro and in vivo.
Introduction
One-carbon metabolism uses the coenzyme tetrahydrofolate to carry reactive one-carbon units, which are essential for the synthesis of the dTMP and purines ultimately incorporated into DNA and RNA1,2. Antifolates, such as methotrexate, target the enzymes responsible for tetrahydrofolate synthesis and have a long record of efficacy in the treatment of malignancies3,4. The proteinogenic amino acids serine and glycine are also the source of the one-carbon units carried by tetrahydrofolate (reviewed in 2,5) and incorporated into nucleotides6. It is well appreciated that proliferating cells not only obtain serine exogenously7 but also synthesize serine from glucose8–10 via the canonical serine synthesis pathway, in which 3-phosphoglycerate dehydrogenase (PHGDH), which converts the glycolytic intermediate 3-phosphoglycerate (3-PG) to phosphohydroxypyruvate (P-Pyr), catalyzes the first, often rate limiting step8,10. Recent work demonstrating that PHGDH loss is selectively toxic to tumor cell lines with high PHGDH expression or flux through the serine synthesis pathway11–15 has contributed to interest in understanding serine synthesis and downstream one-carbon metabolism16–20. Unlike for the tetrahydrofolate synthesis pathway, there are no small molecule tools for interrogating the serine synthesis pathway.
Here we report small molecule probes of PHGDH and demonstrate the utility of these compounds in studying the biological consequences of PHGDH inhibition. We find that PHGDH inhibitors reduce the production of glucose-derived serine, and that these compounds attenuate the growth of PHGDH-dependent cell lines both in culture and in orthotopic xenograft tumors. Surprisingly, PHGDH inhibitors reduce the incorporation into nucleotides of one-carbon units derived not only from glucose-derived serine, but also from exogenous serine present in the cell medium. We trace this to PHGDH inhibitor-induced wasting of serine-derived one-carbon units. We conclude that glucose-derived serine synthesis coordinates the availability of one-carbon units from both endogenously produced and exogenous, imported serine for nucleotide synthesis, and hypothesize that this wasting of one-carbon units may contribute to the efficacy of PHGDH inhibitors in vitro and in vivo.
Results
Identification and characterization of PHGDH inhibitors
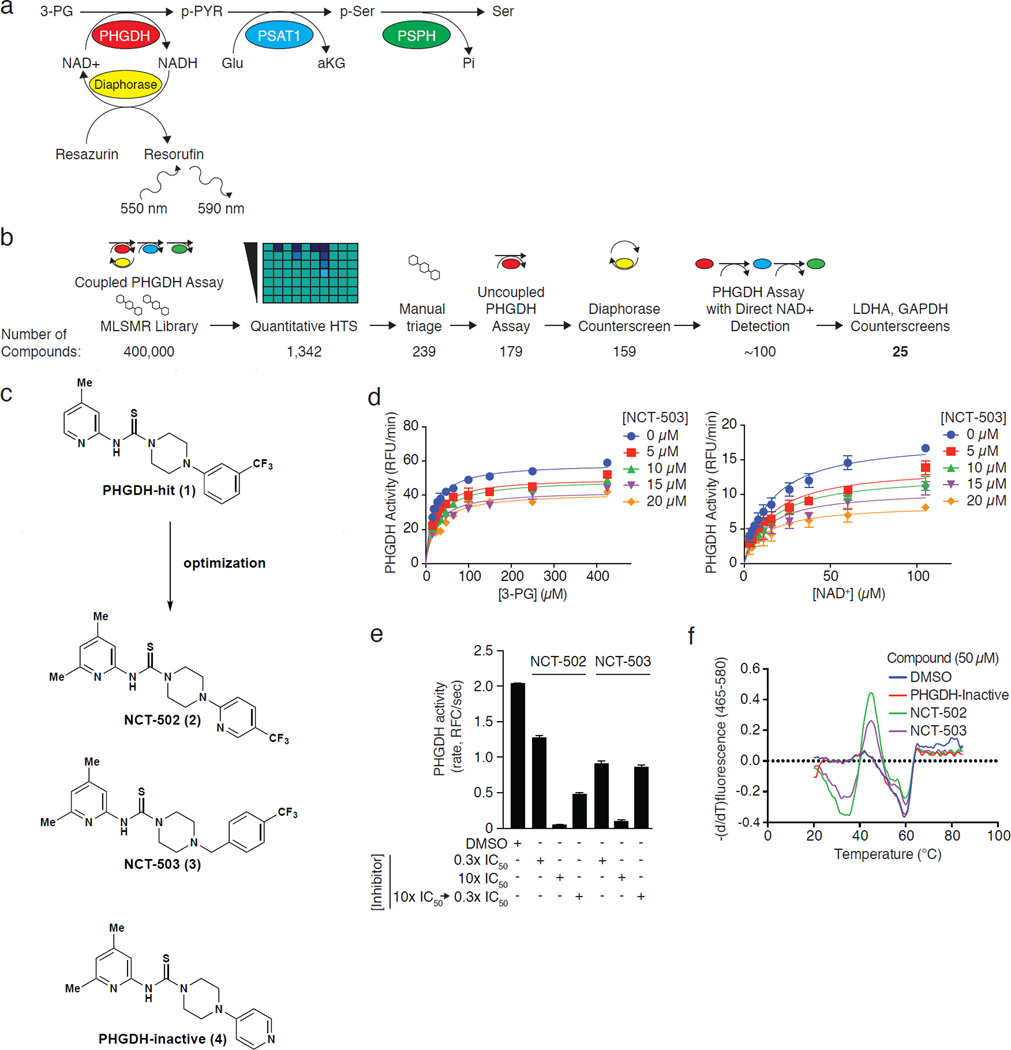
Because the PHGDH-catalyzed reaction in isolation is at near thermodynamic equilibrium21, we developed a coupled primary screening assay in which phosphoserine aminotransferase (PSAT1) and phosphoserine phosphatase (PSPH), the enzymes downstream of PHGDH, are included to minimize P-Pyr-mediated feedback inhibition of PHGDH and to pull the reaction forward (Fig. 1a). In this endogenous pathway coupled assay we measured the production of NADH by PHGDH through the diaphorase-mediated reduction of resazurin22. We screened the 400,000 compound NIH Molecular Libraries Small Molecule Repository (MLSMR) library in four-point dose-response and ranked the hits by curve class, IC50 values, and maximum inhibition obtained from the primary screen23 (Fig. 1b; Supplementary Results, Supplementary Table 1). Following removal of synthetically intractable and promiscuous hits (defined as compounds active in >15% of NCATS-run assays), confirmatory PHGDH assays validated the remaining hits and counterscreening assays eliminated pan-dehydrogenase inhibitors (Fig. 1b). Most of the remaining compounds contained a piperazine-1-carbothioamide scaffold, among which PHGDH-Hit (1) (Fig. 1c) was the best hit.
Figure 1. Identification and characterization of small molecule PHGDH inhibitors.
a, Coupled PHGDH assay with diaphorase/resazurin readout used for the primary screen. b, Screening pipeline for PHGDH inhibitors. Following HTS, manual triage selected synthetically tractable compounds and eliminated promiscuous inhibitors. Remaining compounds were confirmed and counterscreened to eliminate false positives and pan-dehydrogenase inhibitors. The number of compounds remaining is listed beneath each step. c, Piperazine-1-carbothioamide PHGDH inhibitors. PHGDH-hit (1) was the initial hit in the screen; NCT-502 (2) was a derivative with improved potency, and NCT-503 (3) has improved solubility and in vivo characteristics. The structurally related inactive compound (PHGDH-inactive; 4) had no activity against PHGDH and served as a negative control. d, NCT-503 exhibits noncompetitive inhibition with respect to both 3-PG and NAD+. Data are average of three experiments and error bars represent standard deviations. e, Dilution data demonstrating in vitro reversibility of NCT-502 and NCT-503. Data are average of 96 experiments and error bars represent standard deviations. f, Melting temperature curves demonstrating NCT-502 and NCT-503-induced destabilization of PHGDH. Curves are representative of 3 experiments.
Ultimately, PHGDH-hit was validated as a PHGDH inhibitor (IC50 = 15.3 µM; Table 1). In an effort to improve potency, we undertook a brief chemistry optimization effort. Attempts to replace the thiourea with urea, thioamide or replace the pyridine with a phenyl derivative resulted in considerable loss of activity (Supplementary Fig. 1a, entries 1–4). Addition of a methyl group to the 6-position of the pyridine ring slightly improved potency (Supplementary Results, Supplementary Fig. 1a, entry 5). Subsequently moving the trifluoromethyl group to the para-position and incorporating a nitrogen into the aromatic ring gave the first considerable improvement in potency (NCT-502 (2), IC50 = 3.7±1 µM, Fig. 1c; Table 1) with reasonable in vitro ADME (Supplementary Fig. 1b). Replacing the 2-pyridine-4-trifluoromethyl substituent with a 4-pyridinyl group resulted in a soluble compound (114 µM in PBS buffer) that did not inhibit PHGDH (PHGDH-Inactive (4); IC50>57 µM; Fig. 1c; Table 1), and was a key inactive control for subsequent experiments. Next, it was discovered that the piperazine N-aryl bond could be replaced with an N-benzyl group, resulting in a slight improvement in potency, solubility and microsomal stability yielding NCT-503 (3) (IC50 = 2.5±0.6 µM, Fig. 1c; Table 1; Supplementary Fig. 1b). NCT-502 and NCT-503 are more soluble in assay buffer than in PBS, which may explain the discrepancy between the reported solubility and IC50 for NCT-502 (Table 1)24.
Table 1.
IC50, solubility and microsomal stability of PHGDH inhibitors. IC50s are averages±standard deviations and the number of replicates is provided in the table. Compound solubility was determined in assay buffer containing 0.05% BSA (kinetic solubility) and in PBS pH 7.4 (aqueous solubility). IC50s were measured in assay buffer.
| PHGDH IC50 (µM) |
Kinetic Solubility in Assay Buffer (µM) |
Aqueous Solubility (µM) |
Rat Microsome Stability (t1/2, min.) |
|
|---|---|---|---|---|
| PHGDH-Hit (1) | 15.3 | - | 19 | 24 |
| NCT-502 (2) | 3.7±1.0 (n=8) | 662.2±39.8 (n=3) | 1.2±1.2 (n=8) | >30 (n=10) |
| NCT-503 (3) | 2.5±0.6 (n=4) | 61.5±2.0 (n=3) | 25.3±2.6 (n=8) | >30 (n=8) |
| PHGDH-Inactive (4) | >57 (n=4) | 251.4±24.9 (n=3) | >148 (n=2) | >30 (n=2) |
NCT-502, NCT-503 and inactive compound were inactive against a panel of other dehydrogenases (Supplementary Fig. 1c), and showed minimal cross-reactivity (<30% modulation of activity) in a panel of 168 GPCRs (Supplementary Results, Supplementary Dataset 1). Compounds in this class are known to inhibit bacterial phosphopantetheinyl transferase, but are inactive against the human ortholog25. Competition studies of NCT-503 against 3-phosphoglycerate (3-PG) and the co-substrate NAD+ revealed a non-competitive mode of inhibition with respect to both 3-PG and NAD+ (Fig. 1d). Dilution experiments demonstrated reversible inhibition (Fig. 1e). NCT-502 and NCT-503 decreased the Tm of PHGDH as measured by differential scanning fluorimetry, while the inactive compound did not, consistent with decreased stability of PHGDH induced by binding of active PHGDH inhibitors (Fig. 1f; Supplementary Fig. 1d). Although this is not typical, destabilization has been previously observed in the specific binding of small molecules to their protein targets26. NCT-503 had reasonable aqueous solubility, and both NCT-502 and NCT-503 exhibited favorable absorption, distribution, metabolism and excretion (ADME) properties (Supplementary Fig. 1b). For comparison, ML265, a well-characterized PKM2 activator, has an aqueous kinetic solubility of 79 µM (comparable to NCT-503 solubility in assay buffer in Table 1), stability of >90% after 30 minute exposure to mouse, rat, and human liver microsomes, and an efflux ratio of 1.3527,28. Taken together, this and subsequent data validate these compounds as useful tools to study PHGDH biology.
PHGDH inhibitors engage their target in cells
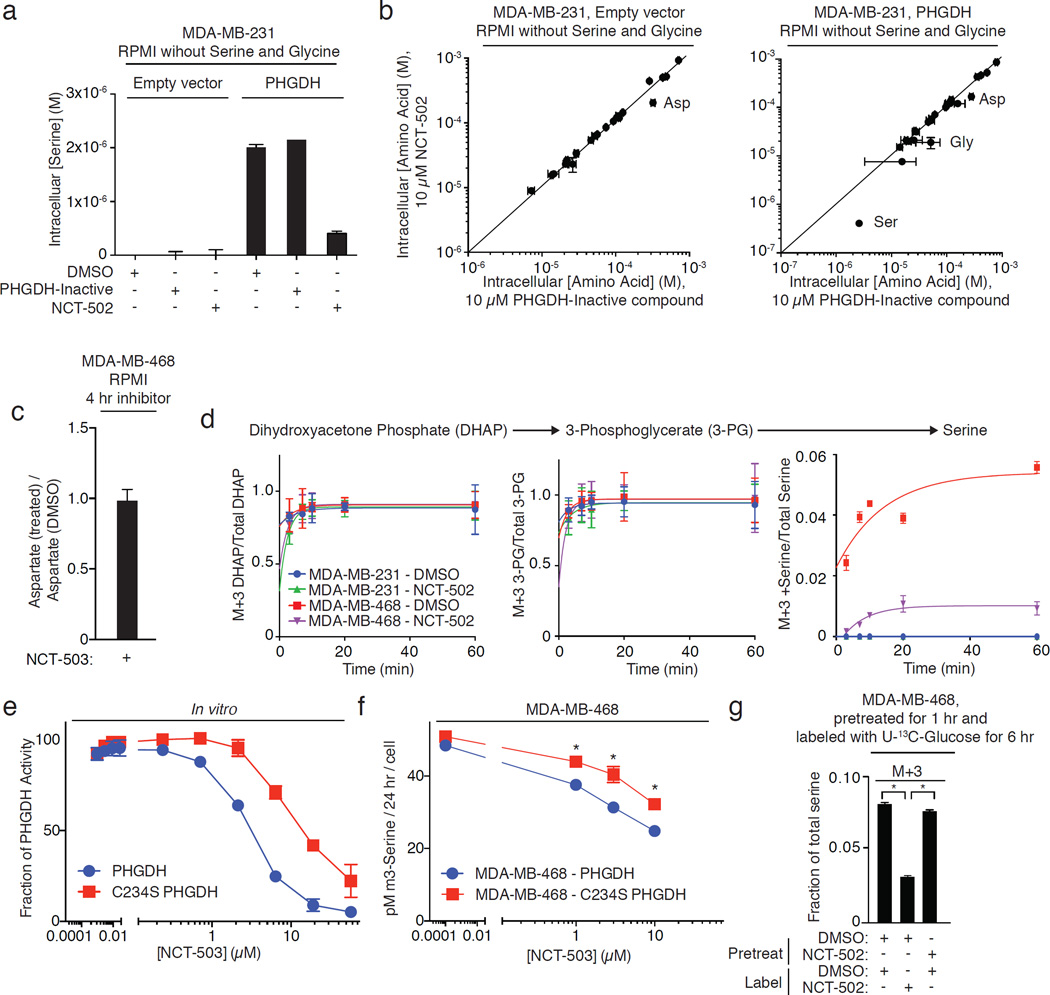
To examine target engagement of PHGDH inhibitors in cells, MDA-MB-231 cells that lack detectable expression of PHGDH and MDA-MB-231 cells engineered to stably express full-length human PHGDH (MDA-MB-231-PHGDH) were treated with PHGDH inhibitors in RPMI lacking serine and glycine (Supplementary Results, Supplementary Fig. 2a). In this medium, MDA-MB-231 cells had low intracellular serine concentrations, while MDA-MB-231-PHGDH cells generated significant quantities of intracellular serine (Fig. 2a). In MDA-MB-231-PHGDH cells, NCT-502 treatment decreased intracellular serine and glycine concentrations (Fig. 2a) and did not change the concentration of any other amino acid except for aspartate, which also decreased in parental MDA-MB-231 cells (Fig. 2b). However, MDA-MB-468 cells treated with NCT-503 did not exhibit a decrease in aspartate (Fig. 2c), suggesting that this effect is greater with NCT-502.
Figure 2. Target engagement and efficacy of PHGDH inhibitors.
All data points are the average of 3 biological replicates. Error bars represent standard deviations. a, NCT-502 reduces intracellular serine concentrations in MDA-MB-231 cells expressing PHGDH in medium lacking serine and glycine. Inactive compound (PHGDH-inactive) has no effect on intracellular serine concentrations. b, NCT-502 treatment in media lacking serine and glycine decreases the concentrations of serine and glycine only in MDA-MB-231 cells expressing PHGDH, while sparing all other amino acids except for aspartate. c, NCT-503 does not affect the intracellular concentration of aspartate in MDA-MB-468 cells in complete RPMI. d, PHGDH inhibitors reduce M+3 serine produced from U-13C glucose while sparing the labeling of the glycolytic intermediates M+3 dihydroxyacetone phosphate (DHAP) and M+3 3-phosphoglycerate. e, C234S PHGDH is less sensitive to NCT-503 inhibition than wild type PHGDH in vitro. f, Expression of C234S PHGDH in MDA-MB-468 cells increases glucose-mediated serine flux in the presence of NCT-503. g, Intracellular synthesis of M+3-serine from U-13C glucose following washout of NCT-502 demonstrates PHGDH inhibitor reversibility. *, p<0.05, Student’s t-test.
Intracellular aspartate levels are influenced by electron transport chain activity in proliferating cells29,30. To test the possibility that our compounds might decrease intracellular [aspartate] by inhibiting the electron transport chain, we measured oxygen consumption in MDA-MB-468 cells following treatment with our PHGDH inhibitors and inactive compound. Both the active and inactive compounds decreased oxygen consumption (Supplementary Fig. 2b), consistent with electron transport chain inhibition. At 50 µM, the inactive and active compounds equally inhibited oxygen consumption, (Supplementary Fig. 2b), but the active compounds inhibited the production of glucose-derived serine substantially more than the inactive compound (Supplementary Fig. 2c). NCT-503 treatment also did not change intracellular glucose concentration (Supplementary Fig. 2d). Use of our structurally related inactive compound as a control should separate the effect of our compounds on electron transport chain activity and on serine synthesis pathway activity.
To demonstrate that inhibition of serine synthesis was due to PHGDH inhibition rather than inhibition of glycolysis, we treated MDA-MB-231 and PHGDH-dependent MDA-MB-468 cells in complete RPMI with glucose labeled at all six carbons with carbon-13 (U-13C-glucose) in the presence or absence of PHGDH inhibitors, and measured the fraction of glycolytic intermediates and glucose-derived serine. PHGDH inhibition did not affect the incorporation of 13C into the glycolytic intermediates dihydroxyacetone phosphate (DHAP) and 3-PG, the substrate of PHGDH, but decreased the production of M+3 serine (Fig. 2d).
Finally, we also demonstrated PHGDH target engagement by mutating cysteine 234 to serine (C234S) near the PHGDH active site (PDB: 2G76; Supplementary Fig. 2e), which reduced the potency of NCT-503-mediated PHGDH inhibition by approximately 3-fold (Fig. 2e). Expression of C234S PHGDH in MDA-MB-468 partially restores serine flux in these cells in the presence of NCT-503 (Fig. 2f) in spite of slightly decreased expression of PHGDH in these cells (Supplementary Fig. 2f).
NCT-502-mediated inhibition of serine synthesis was reversible in cells, as evidenced by resumption of the production of M+3 serine from U-13C-glucose following washout of the inhibitor (Fig. 2g). Consistent with prior observations11, NCT-502 also reduced the PSAT1-catalyzed production of M+5-α-ketoglutarate from U-13C glutamate and 15N-serine from α-15N-glutamate (generated from U-13C-glutamine and α-15N-glutamine; Supplementary Figs. 2g and 2h). Therefore, PHGDH inhibitors reversibly reduce serine synthesis in cells by engagement of PHGDH.
In vitro and in vivo effects of PHGDH inhibitors
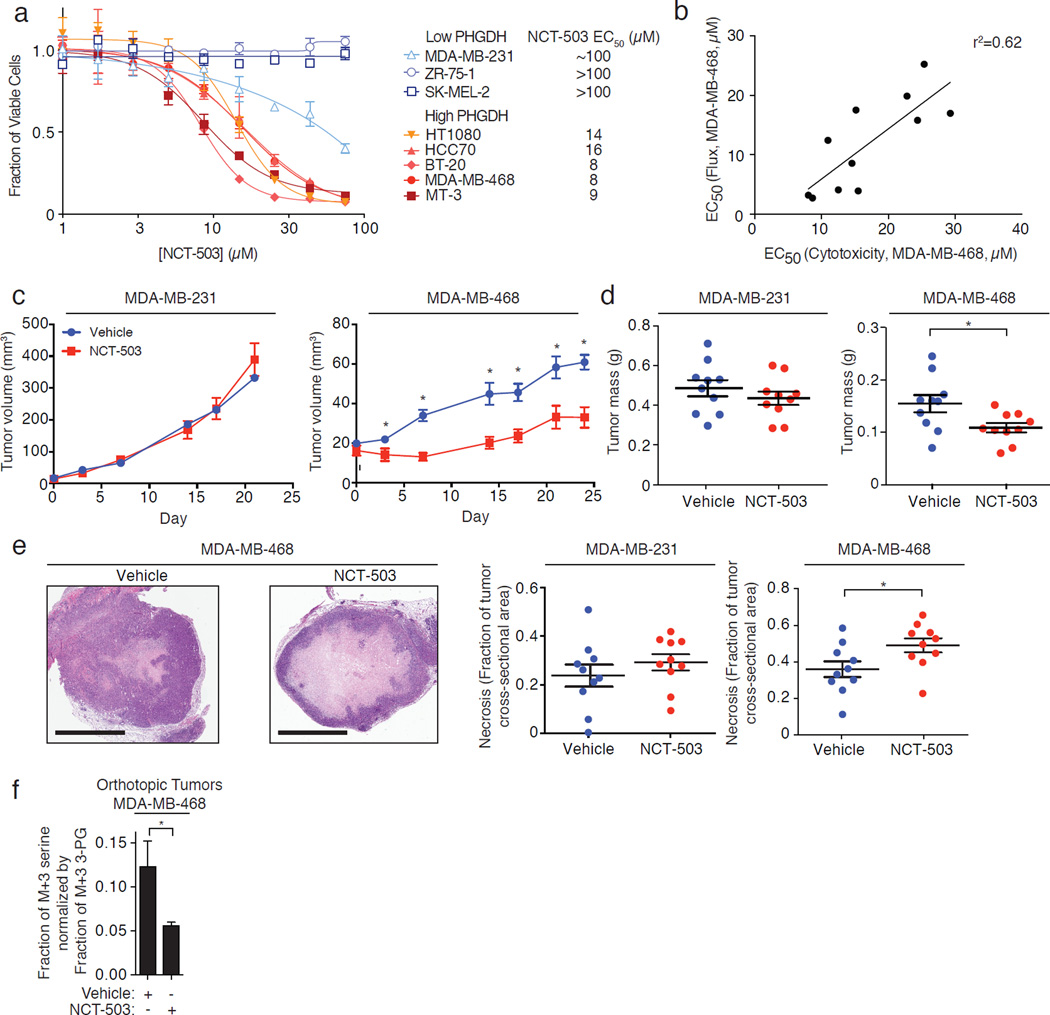
Knockdown of PHGDH is selectively toxic towards PHGDH-dependent cell lines, and minimally toxic towards PHGDH-independent cell lines11–13. Treatment of three PHGDH-independent cell lines (MDA-MB-231, ZR-75-1, and SK-MEL-2), and five PHGDH-dependent cell lines (MDA-MB-468, BT-20, HCC70, HT1080, and MT-3; Supplementary Fig. 3a) in dose-response with NCT-503 demonstrated that PHGDH inhibitors had EC50s of 8–16 µM for the PHGDH-dependent cell lines, a 6- to 10-fold higher EC50 for MDA-MB-231 cells, and no toxicity towards other PHGDH-independent cell lines (Fig. 3a). The inactive compound was not toxic towards any of these cell lines (Supplementary Fig. 3b). We hypothesized that more potent PHGDH inhibitors should be more cytotoxic towards PHGDH-dependent cells. Accordingly, the EC50s for M+3 serine production from U-13C-glucose of a set of piperazine-1-carbothioamides showed a strong positive correlation with their EC50 values for cytotoxicity in MDA-MB-468 cells (Fig. 3b; Supplementary Fig. 3c).
Figure 3. In vitro and in vivo efficacy of PHGDH inhibitors.
a, Selective toxicity of NCT-503 towards five cell lines that overexpress PHGDH relative to three cell lines with low PHGDH expression. Data points are the average of three independent biological experiments and error bars represent standard deviations. b, Compound cytotoxicity towards PHGDH-expressing MDA-MB-468 cells correlates with inhibition of M+3 serine production. Each data point represents an EC50 that is the average of three independent experiments and a single IC50 flux experiment comprised of 6 data points. c, NCT-503 reduces the volume of MDA-MB-468 orthotopic xenografts while sparing the growth of MDA-MB-231 xenografts. Data points are the mean of ten animals, and error bars represent standard error of the mean. *, p<0.05, Student’s t-test. d, NCT-503 reduces the weight of MDA-MB-468 xenografts but not the weight of MDA-MB-231 xenografts. Each data point is a single animal, n=10 in each arm. Horizontal bar indicates the mean of ten animals and error bars represent standard error of the mean. *, p<0.05, Student’s t-test. e, NCT-503 increases the fraction of necrosis in MDA-MB-468 orthotopic xenografts but not in MDA-MB-231 orthotopic xenografts. Scale bars, 2 mm. Images are representative of ten animals. Each data point is a single animal, n=10 in each arm. Horizontal bar indicates mean of ten animals and error bars represent standard error of the mean. *, p<0.05, Student’s t-test. f, Following infusion of U-13C glucose, NCT-503 reduces the fraction of M+3 serine (normalized to M+3 3-PG) in MDA-MB-468 orthotopic xenografts. Data are the mean of three independent experiments (3 animals in each arm) and error bars represent standard deviations. *, p<0.05, Student’s t-test.
Pharmacokinetics to evaluate the utility of NCT-503 as an in vivo probe determined that the compound had good exposure (AUClast=14,700 hr*ng/mL), half-life (2.5 hr) and Cmax (~20 µM in plasma) following intraperitoneal administration with significant partitioning into the liver and brain (Supplementary Figs. 3d and 3e). To evaluate NCT-503 activity in vivo, NOD.SCID mice bearing MDA-MB-231 and MDA-MB-468 orthotropic xenografts were treated with vehicle or NCT-503 (40 mg/kg daily, IP, Supplementary Fig. 3f). PHGDH inhibitor treatment reduced the growth and weight of PHGDH-dependent MDA-MB-468 xenografts but did not affect the growth or weight of PHGDH-independent MDA-MB-231 xenografts (Fig. 3c–d). PHGDH inhibition also selectively increased necrosis in MDA-MB-468 xenografts, but not in MDA-MB-231 xenografts (Fig. 3e). Importantly, mice treated with the compound did not lose weight during the 24-day treatment (Supplementary Fig. 3g) in spite of the potential systemic toxicities of inhibiting serine biosynthesis. Levels of NCT-503 in tumors were ~3 µM at the conclusion of the experiment, validating exposure of the tumor to compound. We evaluated target engagement in NOD.SCID mice by treating mice bearing MDA-MB-468 tumors with vehicle or NCT-503 and infusing the animals with U-13C-glucose. MDA-MB-468 tumors in NCT-503-treated mice exhibited decreased production of glucose-derived serine (Fig. 3f), while intratumoral serine concentrations did not change with NCT-503 treatment (Supplementary Fig. 3h). Although we cannot completely exclude off-target contributions to the cytotoxic effects that we observe, our small molecule PHGDH inhibitors engage PHGDH in tumors and recapitulate the selective toxicity of PHGDH knockdown in vivo.
PHGDH inhibition affects the fate of serine
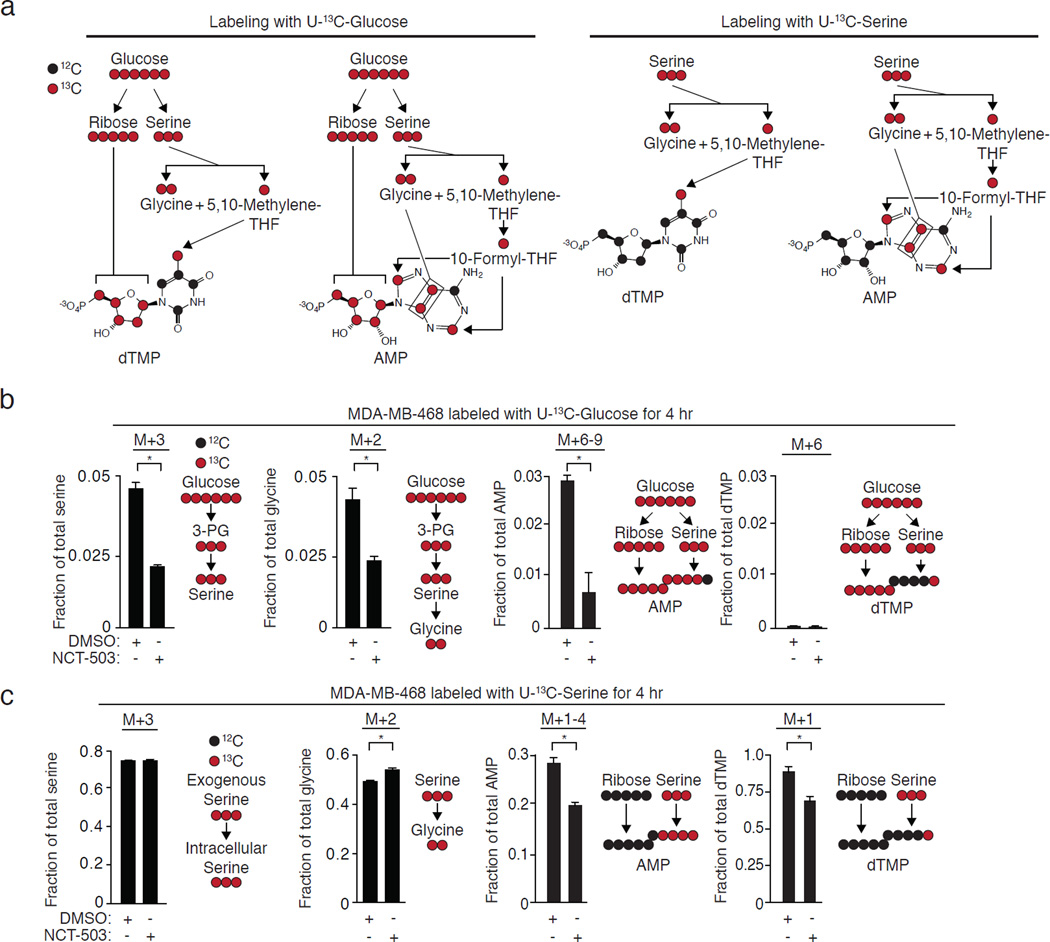
PHGDH knockdown11,12 and small molecule PHGDH inhibitors are selectively toxic towards PHGDH-dependent cells and tumors even when serine is present (all cells in Fig. 3a were treated in RPMI containing 288 µM serine; see Supplementary Fig. 3h for intratumoral serine concentrations). For this reason, we examined the fates of both glucose-derived serine and exogenous serine in PHGDH-dependent cells following acute PHGDH inhibition. Serine is incorporated into the purine ring of AMP via 10-formyl-tetrahydrofolate and glycine, and into the methyl group of the pyrimidine moiety of dTMP via 5,10-methylene-tetrahydrofolate (Fig. 4a). We measured the production of M+3 serine and labeled nucleotides in cells fed U-13C-glucose. As expected, PHGDH inhibition reduced the production of M+3-serine and its product, M+2 glycine, from U-13C-glucose (Fig. 4b, Supplementary Results; Supplementary Fig. Fig. 5a), resulting in the incorporation of less 13C into AMP and dTMP via M+3 serine (Fig. 4b). The loss of incorporation of glucose-derived serine carbons into both AMP and dTMP persisted at 24 hours (Supplementary Fig. 4a, Supplementary Fig. 5b).
Figure 4. PHGDH inhibition in a PHGDH-dependent cell line unexpectedly reduces the incorporation of exogenous serine into dTMP and AMP.
All data are the mean of three biological replicates. Error bars represent standard deviations. *, p<0.05, Student’s t-test. a, Incorporation of 13C from glucose, glucose-derived serine, and exogenous serine into nucleotides. b, 10 µM NCT-503 treatment for four hours reduces the synthesis of glucose-derived serine and decreases the incorporation of 13C from glucose via serine into AMP. c, 10 µM NCT-503 treatment for four hours in the presence of exogenous U-13C-serine does not increases the proportion of labeled serine but increases the fraction of labeled glycine, consistent with decreased synthesis of unlabeled serine. Unexpectedly, NCT-503 reduces the incorporation of one-carbon units from exogenous U-13C-serine into AMP and dTMP.
To determine the fate of exogenous serine, we fed cells U-13C-serine instead of unlabeled serine in the medium. PHGDH inhibition did not change the uptake of labeled serine at 4 hours (Supplementary Figs. 4b, 5c) but increased the pool size of exogenous, labeled serine at 24 hours, resulting in an increase in the size of the total serine pool at 24 hours (Supplementary Results; Supplementary Figs. 6a–b) and increased incorporation of exogenous, labeled serine into glycine (Fig. 4c). Unexpectedly, NCT-503 treatment for 4 hours decreased the incorporation of 13C from exogenous U-13C-serine into AMP and dTMP (Fig. 4c, Supplementary Fig. 5c), effects that persisted at 24 hours of treatment as a reduction in labeled pool size (Supplementary Figs. 5d and 6a–b). NCT-503 treatment does not decrease the total pool size of AMP due to an increase in unlabeled AMP, but decreases the dTMP pool size relative to untreated cells at 24 hours (Supplementary Fig. 6a, 6b), in spite of an increase in the pool of labeled ribose and ribulose-5-phosphate generated by the pentose phosphate pathway (Supplementary Fig. 6c). The inhibitor did not have these effects on MDA-MB-231 cells that lack PHGDH (Supplementary Fig. 4c).
Serine synthesis regulates one-carbon unit availability
Exogenous and endogenous serine are incorporated into AMP via glycine and 10-formyl-tetrahydrofolate, and into dTMP via 5,10-methylene tetrahydrofolate (5,10-CH2-THF) generated by the mitochondrial serine hydroxymethyl transferase (SHMT2) or the cytosolic serine hydroxymethyl transferase (SHMT1)6. SHMT1 is also capable of synthesizing serine from 5,10-CH2-THF and glycine1,31. As we did not observe a defect in serine import (Fig. 4c; Supplementary Figs. 5c and 5d), we hypothesized that increased consumption of serine-derived 5,10-CH2-THF by SHMT1 might generate serine at the expense of dTMP synthesis when the serine synthesis pathway is inhibited. Moreover, attenuation of SHMT1 activity might redirect one-carbon units from both exogenous and endogenous serine towards nucleotide synthesis.
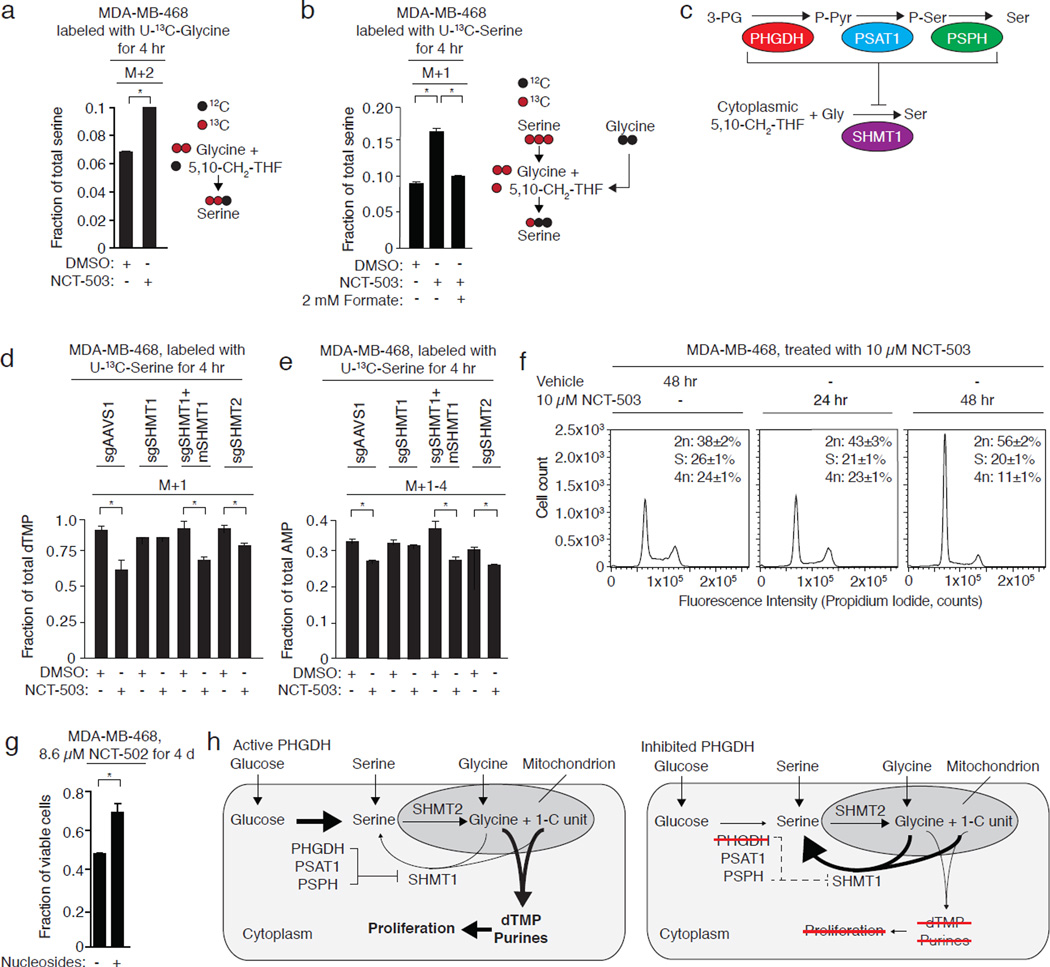
In agreement with this, MDA-MB-468 cells fed exogenous U-13C-glycine exhibit increased production of M+2 serine in the presence of NCT-503 (Fig. 5a; Supplementary Results, Supplementary Fig. 7a). This did not occur in MDA-MB-231 cells lacking PHGDH (Supplementary Figs. 7b–c). Increased SHMT1 activity in the presence of U-13C-serine should increase the amount of serine containing a single 13C derived from 5,10-13CH2-THF and unlabeled glycine from the cytosolic pool. Consistent with this hypothesis, NCT-503 treatment increased the fraction of M+1 serine in MDA-MB-468 cells fed with U-13C-serine (Fig. 5b). The addition of unlabeled formate reduced M+1 serine production from U-13C-serine, which demonstrates that the M+1 serine arose from unlabeled glycine and 5,10-13CH2-THF (Fig. 5b, Supplementary Fig. 7d). Cells lacking PHGDH did not exhibit increased M+1 serine production from M+3 serine following NCT-503 treatment (Supplementary Fig. 7e). These data support a hypothesis in which the glucose-derived serine biosynthesis pathway inhibits SHMT1 (Fig. 5c) to prevent wasting of 5,10-CH2-THF while serine synthesis is ongoing.
Figure 5. SHMT1 mediates the loss of nucleotide labeling induced by PHGDH inhibition.
All data are the mean of three biological replicates. Error bars represent standard deviations. *, p<0.05, Student’s t-test. a, NCT-503 induces increased synthesis of M+2 serine from M+2 glycine and unlabeled 5,10-CH2-THF in a PHGDH-dependent cell line. b, Probable SHMT1-catalyzed synthesis of M+1-serine from unlabeled glycine and 13C-serine-derived 5,10-methylene THF (5,10-CH2-THF) increases with PHGDH inhibition (10 µM NCT-503) and is suppressed by exogenous unlabeled formate. c, Serine synthesis pathway activity, or a serine synthesis pathway intermediate, represses SHMT1 activity. SHMT1 catalyzes serine synthesis from glycine and 5,10-CH2-THF. d, SHMT1 deletion restores incorporation of carbon from U-13C serine into dTMP in the presence of PHGDH inhibitor. Mouse SHMT1 expression restores decreased dTMP labeling induced by PHGDH inhibition. SHMT2 knockout does not block PHGDH inhibitor-mediated loss of dTMP labeling. e, SHMT1 deletion restores incorporation of carbon from U-13C serine into AMP in the presence of a PHGDH inhibitor. Mouse SHMT1 restores PHGDH inhibitor-mediated loss of AMP labeling by U-13C serine. f, NCT-503 treatment induces G1/S cell cycle arrest in MDA-MB-468 cells, consistent with a defect in nucleotide synthesis. g, Nucleoside supplementation partially rescues PHGDH inhibitor toxicity. h, Model of one-carbon unit wasting induced by PHGDH inhibition. Suppression of PHGDH activity increases the activity of SHMT1, which consumes one-carbon units to resynthesize serine but reduces the availability of one-carbon units needed for purine and dTMP synthesis.
To address this hypothesis, we generated MDA-MB-468 cells in which cytosolic SHMT1 or mitochondrial SHMT2 was deleted with CRISPR/Cas9 (Supplementary Fig. 7f). Loss of SHMT1, but not SHMT2, restored the incorporation of serine-derived carbon into AMP and dTMP in the presence of a PHGDH inhibitor (Fig. 5d, Fig. 5e). Expression of an sgRNA-resistant mouse SHMT1 (Supplementary Fig. 7f) restored the loss of U-13C-serine incorporation into AMP and dTMP induced by PHGDH inhibition (Fig. 5d, Fig. 5e). We conclude that serine synthesis-mediated inhibition of SHMT1 directs the incorporation of serine-derived one-carbon units into AMP and dTMP. Consistent with this hypothesis, NCT-503 caused G1/S arrest after 48 hours (Fig. 5f) and supplementation of RPMI with nucleosides reduced the toxicity of PHGDH inhibition (Fig. 5g).
Discussion
Here, we identify PHGDH inhibitors that inhibit the synthesis of glucose-derived serine through direct target engagement. Counter-screens of these compounds against dehydrogenases, steady state metabolite profiling, and 13C labeling studies eliminated hits that inhibited glycolytic labeling and confirmed the effectiveness of our PHGDH probes in cells and in vivo. Although counter-screening and metabolomics cannot exclude all off-target effects or metabolic vulnerabilities due to media composition or cell density, we anticipate that these techniques will greatly assist the development of improved PHGDH inhibitors and other novel antimetabolites.
Our small molecule PHGDH inhibitors demonstrate that the serine synthesis pathway not only generates serine, but also ensures that one-carbon units derived from both endogenous and exogenous serine are available for nucleotide synthesis by reducing SHMT1 activity (Fig. 5h). One-carbon unit wasting induced by PHGDH inhibition and SHMT1 activation reduces one-carbon unit incorporation into nucleotides even in the presence of ample serine, and may contribute to the inability of exogenous serine to rescue PHGDH inhibition or knockdown. We anticipate that our PHGDH inhibitors will assist in deciphering the role of serine synthesis and one-carbon metabolism in a variety of additional contexts.
Substantial genetic evidence supports the importance of the serine synthesis pathway in the survival and proliferation of breast11–14, melanoma, and non-small cell lung cancer cells15. The selective toxicity and tolerability of these compounds in vitro and in vivo suggest that PHGDH inhibitors may be therapeutically useful.
Online Methods
Materials
The following antibodies were used: antibodies to PHGDH (HPA021241) and SHMT2 (HPA020549) from Sigma, an antibody to SHMT1 (12612) from Cell Signaling Technologies, an antibody to GAPDH (GT239) from GeneTex, and an antibody to actin (sc-1616) from Santa Cruz Biotechnologies. Antibodies were used at 1:1000 dilution except for GAPDH (1:5000) and actin (1:10000). Western blots are shown in Supplementary Results; Supplementary Fig. 8. BT-20 (HTB-19), HCC70 (CRL-2315), HT1080 (CCL-121), MDA-MB-468 (HTB-132), MDA-MB-231 (HTB-26), SK-MEL-2 (HTB-68), and ZR-75-1 (CRL-1500) cells were from ATCC and MT-3 (ACC 403) cells were from DSMZ. Cell lines were directly obtained from authenticated sources and were not STR profiled. The BT-20 cell line has previously been misidentified but this PHGDH-amplified cell line has previously been used to demonstrate selective toxicity of PHGDH knockdown11. Cell lines were verified to be free of mycoplasma contamination by PCR32. 3-Phosphoglycerate (P8877), NAD+ (N0632), glutamate (49621), DL-glyceraldehyde 3-phosphate (G5251), dihydroxyacetone phosphate (D7137), glycerol-3-phosphate (G7886), TCEP (646547), diaphorase (D5540), and resazurin (R7017) were from Sigma. U-13C-glucose (CLM-1396-1), U-13C-serine (CLM-1574-H-0.25), U-13C-glycine (CLM-1017-1), Phe-d8 (DLM-372) and Val-d8 (DLM-311) were from Cambridge Isotope Laboratories. Matrigel (536230) was from BD Biosciences. Protein concentrations were determined using Bio-Rad Protein assay (Bio-Rad 500-0006). Amino acid-free and glucose-free RPMI was from US Biological. All LC-MS reagents were Optima grade (Fisher).
Compound Synthesis
Please see Supplementary Note on Compound Synthesis.
Protein overexpression and purification
cDNAs to human PHGDH, PSAT1, PSPH, GAPDH, GPD1, and GPD1L were PCR amplified from human liver cDNA prepared from human liver mRNA using SuperScript III (Life Technologies) and cloned into pET30-2 with an N-terminal 6×His Tag. Proteins were expressed in Rosetta (DE3)pLysS E. coli (EMD Millipore) grown to an OD of 0.6 and induced with 1 mM IPTG for 16 hours at 16 °C. Bacteria were lysed at 4 °C in a French press and purified by Ni2+ affinity chromatography on a 5 mL HiTrap chelating HP column (GE Healthcare) attached to an AktaPURE FPLC system (GE Healthcare) using a gradient of 0–500 mM imidazole in 50 mM Na-Phosphate pH 8 and 300 mM NaCl. Peak fraction purity was assessed by SDS gel electrophoresis (Supplementary Results; Supplementary Fig. 9a). Pure fractions were combined, concentrated in 15 mL UltraFree 30 concentrators (EMD Millipore) and loaded onto a HiLoad Superdex 200 prep grade 16/60 column equilibrated in 20 mM Tris pH 7.4, 100 mM NaCl, and 1 mM TCEP. Peak fractions were concentrated to [protein] ≥ 5 mg/mL, flash frozen in liquid nitrogen, and stored at −80 °C prior to use (Supplementary Fig. 9a).
Enzyme assays
PHGDH assay buffer contained 50 mM TEA pH 8.0, 10 mM MgCl2, 0.05% BSA, and 0.01% Tween-20. PHGDH enzyme buffer consisted of assay buffer with 20 nM PHGDH and 0.2 mg/mL diaphorase. PHGDH substrate buffer contained 0.3 mM NAD+, 1.25 mM glutamate, 0.1 mM 3-phosphoglycerate, 0.2 mM resazurin, 1 µM PSAT1, and 1 µM PSPH. qHTS was performed in 1536-well plates dispensed with a BioRAPTR FRD. Each well contained equal volumes of substrate buffer and assay buffer. Plates were read at 0 minutes and 20 minutes at room temperature with a ViewLux uHTS Microplate Imager (PerkinElmer). Follow-up assays were performed in black 384-well plates (Greiner) in 20 µL of enzyme buffer to which compounds were added in dose-response with an HP D300 digital dispenser (Hewlett-Packard), followed by addition of 20 µL of substrate buffer. Plates were incubated at room temperature (25 °C) and read at 0 and 20 minutes with a Spectramax M5 plate reader (Molecular Devices) in fluorescence intensity mode with a λex=550 nm and λem=600 nm (emission cutoff=590 nm). Inhibition models were identified using DynaFit 33. The enzyme concentration was fixed at 10 nM and the Km, Ki, and Vmax were allowed to vary. Data were fit to competitive, uncompetitive, noncompetitive, and mixed inhibition models of complex equilibria. The Km values were not greatly affected, and by F-test, the probability of fit to a noncompetitive model was higher than all other models (91.5% for 3-PG and 84.6% for NAD+).
GAPDH substrate buffer contained 210 mM Tris pH 7.4, 2.5 mM NaH2PO4 pH 7.4, 2 mM DL-Glyceraldehyde 3-phosphate, 1.75 mM MgCl2, 0.01 mM NAD+, 0.11 mM resazurin, 0.2 mg/mL BSA, and 0.01% Tween-20. GAPDH enzyme buffer contained 50 mM Tris pH 7.4, 100 mM NaCl, 0.02 mM TCEP, 0.01 mM EDTA, 0.1 mg/mL BSA, 0.42 mg/mL diaphorase, and 2.5 nM GAPDH. The GAPDH, GPD1, and GPD1L assays were run in 384-well plates using the same protocol and readout as the PHGDH assay. GPD1 substrate buffer contained 114 mM Tris pH 7.4, 0.25 mM DHAP, 1.14 mM MgCl2, 0.011 mM NADH, 0.06 mg/mL BSA, and 0.011% Tween-20. GPD1 enzyme buffer contained 50 mM Tris pH 7.4, 100 mM NaCl, 0.08 mM TCEP, 0.4 mg/mL BSA, 0.03 µM EDTA, and 0.8 nM GPD1. 70 µL of substrate buffer were mixed with 10 µL enzyme buffer.
GPD1L substrate buffer contained 200 mM Tris pH 7.4, 0.05 mM sn-G3-P, 2 mM MgCl2, 0.04 mM NAD+, 0.11 mM resazurin, 0.1 mg/mL BSA, and 0.02% Tween-20. GPD1L enzyme buffer contained 50 mM Tris pH 7.4, 100 mM NaCl, 0.02 mM TCEP, 0.42 mg/mL diaphorase, 0.1 mg/mL BSA, 0.008 µM EDTA, and 16 nM GPD1L. 40 µL of substrate buffer were mixed with 40 µL of enzyme buffer.
The GAPDH and GPD1L assays were read using the same protocol as the PHGDH assay. The GPD1 assay was read using loss of NADH fluorescence (λex=340 nm and λem=460 nm).
PHGDH inhibition data was analyzed by calculating delta RFUs (increase in resorufin fluorescence between T=0 and T=30 mins) and normalizing to the delta RFU values of 0× and 1× PHGDH enzyme controls as 100% and 0% PHGDH inhibition, respectively. 32 wells of each 1536-well assay plate were dedicated to each of these 0× and 1× PHGDH controls. These normalized dose-response values were plotted in GraphPad Prism and fit using a Sigmoidal dose response (variable slope) equation. IC50s for each replicate were then averaged to determine average IC50s with standard deviations.
Thermal shift assays
Differential scanning fluorimetry assays were carried out in 20 mM TEA pH 8, 100 mM NaCl, 1× SYPRO Orange (Sigma), and 2 µM PHGDH in a volume of 10 µL. Unfolding was monitored using a LightCycler 480 II real-time PCR instrument (λex=465 nm and λem=580 nm) (Roche) over a linear 20 to 85°C gradient. Compounds were dispensed with an HP D300 digital dispenser and DMSO concentration was normalized to 1% in all samples. Plots of the first derivative of fluorescence vs. temperature were generated in LightCycler software.
Cell culture
All cells were grown as adherent cell lines in RPMI supplemented with 10% IFS and penicillin/streptomycin. Media for metabolite profiling experiments utilized dialyzed inactivated fetal serum (IFS), prepared by dialyzing IFS for 72 hours using SnakeSkin 3.5K MWCO dialysis tubing (ThermoFisher) against a 10-fold higher volume of phosphate-buffered saline (PBS) with a complete PBS exchange every 12 hours.
Overexpression of PHGDH
Full length human PHGDH was cloned into pMXS-IRES-BLAST which was used to generate retrovirus in supernatants using transient transfection34. MDA-MB-231 cells were transduced with retrovirus by spin infection (2250 rpm for 30 minutes) in polybrene. After 24 hours, cells were selected with 2 µg/mL puromycin.
CRISPR-Cas9 mediated gene knockout
We used CRISPR/Cas-9 mediated genome editing to achieve gene knockout, using pLentiCRISPR (Addgene Plasmid #49535) in which the sgRNA and Cas9 are delivered on a single plasmid. Editing of the PHGDH locus in MDA-MB-231 cells was accomplished by transfection of cells with the pLENTICRISPR plasmid into which an sgRNA targeting the PHGDH locus had been cloned. Transfected cells were subjected to single cell cloning by limiting dilution in 96 well plates. Editing of the PHGDH locus was confirmed by Sanger sequencing of the targeted locus. PHGDH null clones exhibited biallelic insertion or deletion of a single "A" at the targeted site and were compared to unedited control clones.
Editing of the SHMT1 and SHMT2 loci in MDA-MB-468 cells was accomplished by infection of cells with lentivirus delivering sgRNA and Cas9. Lentiviruses were generated in supernatants using transient transfection34 and MDA-MB-468 cells were transduced by spin infection at 2250 rpm for 30 minutes in the presence of polybrene followed by overnight incubation. Lentiviruses with a sgRNA against AAVS1 served as a negative control, and uninfected cells were used as negative controls for transduction.
After transduction, infected MDA-MB-468 cells were selected with puromycin for three days. Loss of SHMT1 and SHMT2 expression was confirmed by Western blotting. For addback experiments, mouse SHMT1 was cloned into pLJM5 (Addgene Plasmid #61614). Lentivirus production and MDA-MB-468 infection were as above. Following spin infection at 2250 rpm and incubation for 24 hours, cells were selected with hygromycin (Sigma) for one week and re-selected with puromycin for 24 hours. All cells were seeded into medium lacking antibiotics for 24 hours prior to further experiments. The following target site sequences35 were used:
AAVS1: GGGGCCACTAGGGACAGGAT
PHGDH: AAAGCAGAACCTTAGCAAAG
SHMT1: GAACGGGGCGTATCTCATGG
SHMT2: GAGAAGGACAGGCAGTGTCG
Cytotoxicity experiments
Cells were seeded in white 96-well plates (Greiner) at a density of 2000 cells/well (MDA-MB-468, BT-20, MT-3) or 1000 cells/well (all other cell lines) and allowed to attach for 24 hours. Compounds were prepared in DMSO and dispensed using an HP D300 compound dispenser. Cell viability was assessed with Cell Titer-Glo (Promega) at four days following treatment and luminescence measured with a SpectraMax M5 Plate Reader (Molecular Devices). Luminescence was normalized to an untreated control in identical medium. For rescue experiments, RPMI was supplemented with 40 µM adenosine, uridine, guanosine, cytidine, deoxyadenosine, thymidine, deoxyguanosine, and deoxycytidine and the medium was replaced daily.
Oxygen consumption measurements
Oxygen consumption of intact MDA-MB-468 cells was measured using an XF24 Extracellular Flux Analyzer (Seahorse Bioscience). 85,000 cells were plated in RPMI media and exposed to compounds at 10 and 50 µM. Each measurement represents the average of six independent wells.
Metabolite profiling: Steady-state and labeling experiments
Cells were evenly seeded at 400,000 cells per well of a 6-well plate and allowed to attach for 24 hours. Prior to all labeling experiments, cells were pretreated with 10 µM compound or an equivalent volume of DMSO in RPMI for 1 hour. For steady-state metabolite concentrations, cells were washed with PBS prior to pretreatment and treatment in RPMI lacking serine and glycine. For labeling experiments, U-13C-glucose, U-13C-serine, or U-13C-glycine replaced the corresponding unlabeled RPMI component. Cells were washed in 4 °C 0.9% (w/v) NaCl in LCMS-grade water and extracted in 1 mL/well of 80:20 (v/v) methanol:water with 0.01 ng/mL Val-d8 and Phe-d8 as internal extraction standards. The extraction solvent was dried under nitrogen gas and metabolite samples were stored at −80 °C until analysis. Triplicate identically seeded and treated wells were trypsinzed and analyzed with a Multisizer Coulter Counter (Beckman Coulter) to obtain cell counts and total cell volumes for normalization.
Liquid Chromatography-Mass Spectrometry
Dried metabolites were resuspended in 100 µL water, centrifuged at 13,000 × g at 4 °C for 10 minutes, and the supernatant recovered for analysis. Chromatographic separation was achieved by injecting 1 µl of sample on a SeQuant ZIC-pHILIC Polymeric column (2.1 × 150 mm, 5 µM, EMD Millipore). Flow rate was set to 0.1 ml per minute, column compartment was set to 25 °C, and autosampler sample tray was set to 4 °C. Mobile Phase A consisted of 20 mM ammonium carbonate, 0.1% ammonium hydroxide. Mobile Phase B was 100% acetonitrile. The mobile phase gradient (%B) was as follows: 0 min 80%, 30 min 20%, 31 min 80%, 42 min 80%. All mobile phase was introduced into the ionization source set with the following parameters: sheath gas = 40, auxiliary gas = 15, sweep gas = 5, spray voltage = −3.1 kV or + 3.0kV, capillary temperature = 275 °C, S-lens RF level = 40, probe temperature = 350 °C. Metabolites were monitored using a polarity-switching full-scan method and identified by accurate mass (± 20 ppm) and retention time within 15 seconds of a previously run pure standard. Metabolite peaks were identified and integrated with Xcalibur v.2.2 software (Thermo Fisher Scientific) and normalized to internal standards and to total cell volume. m/z ratios for stable isotopically labeled metabolites were obtained from IsoMETLIN36 and corrected for natural abundance.
Mouse orthotopic xenografts
Female NOD.CB17-Prkdcscid/J mice, 6–8 weeks old, were obtained from Jackson Laboratories. All animals were provided with food ad libitum for the duration for the duration of the experiment. The animals were allocated randomly for induction with MDA-MB-231 or MDA-MB-468 tumors and tumor group was assigned blindly. 500,000 MDA-MB-231 or MDA-MB-468 cells were injected into the 4th mammary fat pad of each mouse. After 30 days, the tumors were palpable, and the mice were pooled by tumor type and divided randomly to two groups, which were assigned blindly to vehicle or NCT-503 treatment. Each arm contained 10 mice for a total of forty mice in all arms. NCT-503 was prepared in a vehicle of 5% ethanol, 35% PEG 300 (Sigma), and 60% of an aqueous 30% hydroxypropyl-β-cyclodextrin (Sigma) solution, and injected intraperitoneally once daily. Dose was adjusted to mouse weight, and the volume of injection did not exceed 150 µL. Caliper measurements were obtained twice weekly and tumor volumes were calculated with the modified ellipsoid formula: volume = 0.5 × width2 × length.
For quantitation of necrotic regions, fixed tumors were embedded and sections stained with hematoxylin and eosin. Slides were scanned with a Leica Aperio AT2 brightfield scanner. Tumor and necrotic cross-sectional regions were manually delineated and measured using Leica ImageScope software to calculate the percentage of necrosis.
Glucose infusions in mice
Chronic catheters were surgically implanted into the jugular veins of normal or tumor bearing animals 3–4 days prior to infusions. Animals were fasted for 6 hours (morning fast) and infusions were performed in free-moving, conscious animals at 1:00pm for all studies to minimize metabolic changes associated with circadian rhythm. Following administration of either vehicle or NCT-503 at 30 mg/kg, a constant infusion of U-13C-glucose (30 mg/kg/min) (Cambridge Isotope Laboratories) was administered for a 3-hour duration. Animals were terminally anesthetized with sodium pentobarbital and all tissues were fully harvested in less than 5 minutes to preserve the metabolic state. Tumors and adjacent lung tissue were carefully dissected and rapidly frozen using a BioSqueezer (BioSpec Products) to ensure rapid quenching of metabolism throughout the tissue section. Tissues were stored at −80C and extracted with 80:20 (v/v) methanol:water in the same manner as cells prior to LCMS analysis.
High-Throughput PAMPA Protocol
The stirring double-sink PAMPA method patented by Pion Inc. (Billerica, MA) was employed to determine the permeability of compounds via PAMPA passive diffusion. The artificial membrane contained a proprietary lipid mixture and dodecane, optimized to predict gastrointestinal tract (GIT) passive diffusion permeability. Membranes were immobilized on a plastic matrix of a 96 well “donor” filter plate placed above a 96 well “acceptor” plate. A pH 7.0 solution was used in both “donor” and “acceptor” wells. A 10 mM stock of compound in DMSO was diluted to 0.05 mM in aqueous buffer (pH 7.4), with a final concentration of DMSO was 0.5. During the 30-minute permeation period at room temperature, the test samples in the “donor” compartment were stirred. Compound concentrations in the “donor” and “acceptor” compartments were measured using an UV plate reader (Nano Quant, Infinite® 200 PRO, Tecan Inc., Männedorf, Switzerland). Permeability calculations were performed using Pion Inc. software and were expressed in the unit of 10−6cm/s. All samples were tested in duplicate, and control compounds (ranitidine, dexamethasone and verapamil) were included in each run.
High-Throughput Kinetic Solubility Test Protocol
Compounds were evaluated using a µSOL assay for kinetic solubility determination (Pion) that adapts classical saturation shake-flask solubility method to a 96-well microtiter plate format with a co-solvent method using n-propanol. Test compounds were prepared in 10 mM DMSO solutions, and diluted with the co-solvent before being spiked to the aqueous solution (pH 7.4). The final drug concentration in the aqueous solution was 150 µM. Samples were incubated at room temperature for 6 hours and filtered to remove precipitate. The drug concentration in the filtrate was determined by direct UV measurement (λ: 250–498 nm). The reference drug concentration of 17 µM in a co-solvent was used for quantitation of unknown drug concentration in filtrate. Spectroscopically pure n-propanol used as the co-solvent suppressed precipitation in the reference solutions. Kinetic solubility (µg/mL) was calculated using µSOL Evolution software. All samples were tested in duplicate and control compounds (albendazole, phenazolpyridine and furosemide) were included in each run.
High-Throughput Rat Liver Microsomal Stability Determination Protocol
Microsomal stability of test articles was determined in a 96-well plates at a single time point. Compound concentrations were determined by LC/MS/MS (Waters Xevo TQ-S) and used to calculate in vitro half-life. All samples were tested in duplicate, and six standard controls were tested in each run: buspirone and propranolol (short half-life), loperamide and diclofenac (short to medium half-life), and carbamazepine and antipyrine (long half-life). The assay incubation system consisted of 0.5 mg/mLmicrosomal protein, 1.0 µM drug concentration, and NADPH regeneration system (containing 0.650 mM NADP+, 1.65 mM glucose 6-phosphate, 1.65 mM MgCl2, and 0.2 unit/mL G6PDH) in 100 mM phosphate buffer at pH 7.4. Incubation was carried out at 37 °C for 15 min and quenched by adding 555 µL of acetonitrile (~1:2 ratio) containing 0.28 µM albendazole (internal standard). After a 20 min centrifugation at 3000 rpm, 30 µL of the supernatant was transferred to an analysis plate and was diluted 5-fold using 1:2 v/v acetonitrile/water before the samples were analyzed by LC/MS−MS.
Statistics and animal-model statements
All experiments consisted of at least three biological replicates unless otherwise stated, with the exception of the xenograft based experiments, which were performed once with groups of ten mice. All center values shown in graphs refer to the mean. Error bars represent standard deviations unless otherwise stated. Asterisks in figure legends represent p<0.05. t-tests were heteroscedastic to allow for unequal variance and distributions assumed to follow a Student’s t distribution, and these assumptions are not contradicted by the data. All t-tests were two-sided. No statistical methods were used to predetermine sample size. No samples or animals were excluded from analyses. Animals were blindly and randomly assigned to tumor type and to the vehicle or treatment groups. All experiments involving mice were carried out with approval from the Committee for Animal Care at MIT and under the supervision of the MIT Division of Comparative Medicine in accordance with the MIT Policy on the use of animals in research and teaching.
Deposition of data
Compounds will be deposited into PubChem (http://pubchem.ncbi.nlm.nih.gov) with the following accession codes and URLs:
NCT-502: NCGC00242267 (https://pubchem.ncbi.nlm.nih.gov/compound/49853368)
NCT-503: NCGC00351958 (http://pubchem.ncbi.nlm.nih.gov/compound/118796328)
PHGDH-inactive: NCGC00242266 (https://pubchem.ncbi.nlm.nih.gov/compound/49853203)
Supplementary Material
Acknowledgments
We thank Tim Wang and Ellen Edenberg for critical reading of the manuscript, Samatha Murphy for assistance with mouse experiments, and Joseph Pacold of the Lawrence Berkeley National Laboratory for assistance in interpreting Tm data. This research is supported by the Sally Gordon Fellowship of the Damon Runyon Cancer Research Foundation (DRG-112-12), a Department of Defense Breast Cancer Research Program Postdoctoral Fellowship (BC120208), and an ASTRO Resident Seed Grant (RA-2011-1) (all to M.E.P.), by Susan G. Komen for the Cure (to R.L.P.), by an EMBO Long-Term Fellowship (to M.A.-R.), by the NIH (R03 DA034602-01A1, R01 CA129105, R01 CA103866, and R37 AI047389 to D.M.S.), the U.S. Department of Defense (W81XWH-14-PRCRP-IA to D.M.S.) and by the Stewart Trust (to D.M.S.). D.M.S. is an investigator of the Howard Hughes Medical Institute.
Footnotes
URLs and Accession codes
Compounds will be deposited into PubChem (http://pubchem.ncbi.nlm.nih.gov) with the following accession codes and URLs:
NCT-502: NCGC00242267 (https://pubchem.ncbi.nlm.nih.gov/compound/49853368)
NCT-503: NCGC00351958 (http://pubchem.ncbi.nlm.nih.gov/compound/118796328)
PHGDH-inactive: NCGC00242266 (https://pubchem.ncbi.nlm.nih.gov/compound/49853203)
Author Contributions
M.E.P. and D.M.S. conceived of the study and designed most of the experiments with advice from N.G. M.E.P. performed most of the experiments (in vitro assays, cell viability and proliferation, western blots, xenografts, knockdowns, and metabolomics) with assistance from L.J.Y.M.S., S.H.C., R.P., S.W.C., M.Z., E.F., K.B., M.A.-R., Y.D.S., C.M.L., H.C., M.J.K., W.W.C., and K.D.W. and in discussion with C.A.L., B.P.F, L.B.S., and M.G.V.H. K.R.B. and M.B.B. helped design and carried out the quantitative high throughput screen. J.M.R., L.L. G.R., and M.B.B. designed and carried out SAR and synthesis of all compounds. A.Y. assisted with additional in vitro assays, and A.Q.W. and X.X. designed and carried out pharmacokinetic analyses. M.S. was responsible for chemoinformatics during the screen and SAR. S.M.D., A.L, and M.G.V.H. designed and carried out in vivo isotope tracing experiments. M.E.P. and D.M.S. wrote and all authors edited the manuscript.
Competing Financial Interests
D.M.S. is a founder and holds equity in Raze Therapeutics, which has interest in targeting one-carbon metabolism in cancer. M.E.P. is a consultant and holds equity in Raze Therapeutics.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Tibbetts AS, Appling DR. Compartmentalization of Mammalian folate-mediated one-carbon metabolism. Annu. Rev. Nutr. 2010;30:57–81. doi: 10.1146/annurev.nutr.012809.104810. [DOI] [PubMed] [Google Scholar]
- 2.Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer. 2013;13:572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farber S, Diamond LK, Mercer R, Sylvester R, Wolff J. Temporary remissions in acute leukemia in children produced by folic acid antagonist, 4-aminopteroyl-glutamic acid. New England Journal of Medicine. 1948;238:787–793. doi: 10.1056/NEJM194806032382301. [DOI] [PubMed] [Google Scholar]
- 4.Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov. 2011;10:671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 5.Cantor JR, Sabatini DM. Cancer cell metabolism: one hallmark, many faces. Cancer Discov. 2012;2:881–898. doi: 10.1158/2159-8290.CD-12-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Labuschagne CF, van den Broek NJF, Mackay GM, Vousden KH, Maddocks ODK. Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep. 2014;7:1248–1258. doi: 10.1016/j.celrep.2014.04.045. [DOI] [PubMed] [Google Scholar]
- 7.Maddocks ODK, et al. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2013;493:542–546. doi: 10.1038/nature11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Snell K, Natsumeda Y, Eble JN, Glover JL, Weber G. Enzymic imbalance in serine metabolism in human colon carcinoma and rat sarcoma. Br J Cancer. 1988;57:87–90. doi: 10.1038/bjc.1988.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snell K, Weber G. Enzymic imbalance in serine metabolism in rat hepatomas. Biochem J. 1986;233:617–620. doi: 10.1042/bj2330617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fell DA, Snell K. Control analysis of mammalian serine biosynthesis. Feedback inhibition on the final step. Biochem J. 1988;256:97–101. doi: 10.1042/bj2560097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Possemato R, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Locasale JW, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J, et al. Phosphoglycerate dehydrogenase is dispensable for breast tumor maintenance and growth. Oncotarget. 2013;4:2502–2511. doi: 10.18632/oncotarget.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mattaini KR, et al. An epitope tag alters phosphoglycerate dehydrogenase structure and impairs ability to support cell proliferation. Cancer Metab. 2015;3:5. doi: 10.1186/s40170-015-0131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeNicola GM, et al. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat Genet. 2015;47:1475–1481. doi: 10.1038/ng.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang WC, et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell. 2012;148:259–272. doi: 10.1016/j.cell.2011.11.050. [DOI] [PubMed] [Google Scholar]
- 17.Kim D, et al. SHMT2 drives glioma cell survival in ischaemia but imposes a dependence on glycine clearance. Nature. 2015;520:363–367. doi: 10.1038/nature14363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaneton B, et al. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature. 2012;491:458–462. doi: 10.1038/nature11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan J, et al. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510:298–302. doi: 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nilsson R, et al. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nature Communications. 2014;5:3128. doi: 10.1038/ncomms4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lund K, Merrill DK, Guynn RW. The reactions of the phosphorylated pathway of L-serine biosynthesis: thermodynamic relationships in rabbit liver in vivo. Arch Biochem Biophys. 1985 doi: 10.1016/0003-9861(85)90268-1. [DOI] [PubMed] [Google Scholar]
- 22.Chakraborty S, Sakka M, Kimura T, Sakka K. Characterization of a dihydrolipoyl dehydrogenase having diaphorase activity of Clostridium kluyveri. Biosci. Biotechnol. Biochem. 2008;72:982–988. doi: 10.1271/bbb.70724. [DOI] [PubMed] [Google Scholar]
- 23.Inglese J, et al. Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci USA. 2006;103:11473–11478. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di L, Kerns EH. In: Solvent Systems and their Selection in Pharmaceutics and Biopharmaceutics. Augustijns P, Brewster ME, editors. New York: Springer; 2007. [Google Scholar]
- 25.Foley TL, et al. 4-(3-Chloro-5-(trifluoromethyl)pyridin-2-yl)-N-(4-methoxypyridin-2-yl)piperazine-1-carbothioamide (ML267), a potent inhibitor of bacterial phosphopantetheinyl transferase that attenuates secondary metabolism and thwarts bacterial growth. J Med Chem. 2014;57:1063–1078. doi: 10.1021/jm401752p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamiaux C, et al. DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr Biol. 2012;22:2032–2036. doi: 10.1016/j.cub.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 27.Walsh MJ, et al. ML265: A potent PKM2 activator induces tetramerization and reduces tumor formation and size in a mouse xenograft model. National Center for Biotechnology Information (US); 2010. [PubMed] [Google Scholar]
- 28.Anastasiou D, et al. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat Chem Biol. 2012;8:839–847. doi: 10.1038/nchembio.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Birsoy K, et al. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell. 2015;162:540–551. doi: 10.1016/j.cell.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sullivan LB, et al. Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell. 2015;162:552–563. doi: 10.1016/j.cell.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Narkewicz MR, Sauls SD, Tjoa SS, Teng C, Fennessey PV. Evidence for intracellular partitioning of serine and glycine metabolism in Chinese hamster ovary cells. Biochem J. 1996;313(Pt 3):991–996. doi: 10.1042/bj3130991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 32.Young L, Sung J, Stacey G, Masters JR. Detection of Mycoplasma in cell cultures. Nat Protoc. 2010;5:929–934. doi: 10.1038/nprot.2010.43. [DOI] [PubMed] [Google Scholar]
- 33.Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 34.Luo B, et al. Highly parallel identification of essential genes in cancer cells. Proceedings of the National Academy of Sciences. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cho K, et al. isoMETLIN: a database for isotope-based metabolomics. Anal Chem. 2014;86:9358–9361. doi: 10.1021/ac5029177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.