Abstract
The biomedical field has greatly benefited from the discovery of bioluminescent proteins. Currently, scientists employ bioluminescent systems for numerous biomedical applications, ranging from highly sensitive cellular assays to bioluminescence-based molecular imaging. Traditionally, these systems are based on Firefly and Renilla luciferases; however, the applicability of these enzymes is limited by their size, stability, and luminescence efficiency. NanoLuc (NLuc), a novel bioluminescence platform, offers several advantages over established systems, including enhanced stability, smaller size, and >150-fold increase in luminescence. In addition, the substrate for NLuc displays enhanced stability and lower background activity, opening up new possibilities in the field of bioluminescence imaging. The NLuc system is incredibly versatile and may be utilized for a wide array of applications. The increased sensitivity, high stability, and small size of the NLuc system have the potential to drastically change the field of reporter assays in the future. However, as with all such technology, NLuc has limitations (including a non-ideal emission for in vivo applications and its unique substrate) which may cause it to find restricted use in certain areas of molecular biology. As this unique technology continues to broaden, NLuc may have a significant impact in both preclinical and clinical fields, with potential roles in disease detection, molecular imaging, and therapeutic monitoring. This review will present the NLuc technology to the scientific community in a non-biased manner, allowing the audience to adopt their own views of this novel system.
Keywords: NanoLuc Luciferase (NLuc), luciferase, bioluminescence, bioluminescence imaging, bioluminescence resonance energy transfer (BRET)
1. Introduction
The bioluminescence phenomenon has intrigued both scientists and the general population alike since the beginning of ancient civilization. Bioluminescence is the product of a chemical reaction occurring in certain organisms that result in the production of visible light.1 In nature, bioluminescence is produced by organisms for several reasons, including interspecies communication, hunting and locating food, attracting prey, and self-defense. While bioluminescence has been observed for thousands of years, it was only within the last century that advances in molecular biology made it possible to investigate the underlying mechanism of bioluminescence.2 Now, it is known that bioluminescence is a chemical process that relies upon on the interaction of an enzyme and substrate for the production of light. The required enzyme is called luciferase, and the substrate varies upon the type of luciferase (e.g. luciferin). The bioluminescence reaction will be discussed in Section 2.
While light produced from fire and electricity result in significant release of heat, the “cold light” or light produced from bioluminescence is highly efficient, resulting in minimal energy loss via heat. While the biomedical community has already employed bioluminescence for various applications, researchers are currently developing bioluminescence-based products that may assist the general population in everyday life. For example, the potential of utilizing bioluminescent trees and plants to replace traditional street lights is being explored.3 Also, several governmental agencies have provided funding for researchers to investigate new ways the armed forces may benefit from bioluminescence.
While several luciferase enzymes and corresponding substrates have been available to researchers for over a decade, a few novel luciferases have only recently become commercially available. NanoLuc luciferase (NLuc) is the newest commercially available luciferase enzyme. NLuc is a 19.1 kDa luciferase enzyme that relies on the substrate furimazine to produce high intensity, glow-type luminescence.4 Recently, NLuc was shown to exhibit several properties superior to traditional luciferases. In this review, we examine how NLuc has altered the field of bioluminescence in the biomedical sciences by discussing the initial discovery and history of bioluminescence since its initial discovery, examining the luciferase enzymes, and introducing the NLuc system. NLuc has been utilized in biomedical research for several applications, including studying protein-protein interactions, investigating genetic regulation and cell signaling, monitoring protein stability, BRET-based sensors, and molecular imaging. Section 3 examines the applicability of NLuc for each of these applications with a short discussion of how NLuc was beneficial in these applications. While NLuc is believed to be superior to other luciferases, Section 4 investigates the improvements and limitations of the NLuc platform in comparison to traditional luciferase systems. Lastly, Section 5 summarizes the key findings, discusses the biomedical impact of the NLuc system, and provides future perspectives.
2. Evolution of bioluminescence for biomedical research
The field of bioluminescence has grown tremendously in the past decade as researchers are effectively harnessing the benefits of this unique phenomenon for biomedical purposes. It was less than a century ago that the first luciferase enzyme was discovered.5, 6 Since the discovery of the luciferase enzyme and substrate, there have been significant achievements in the field of bioluminescence.
2.1 Discovery of luciferase enzymes
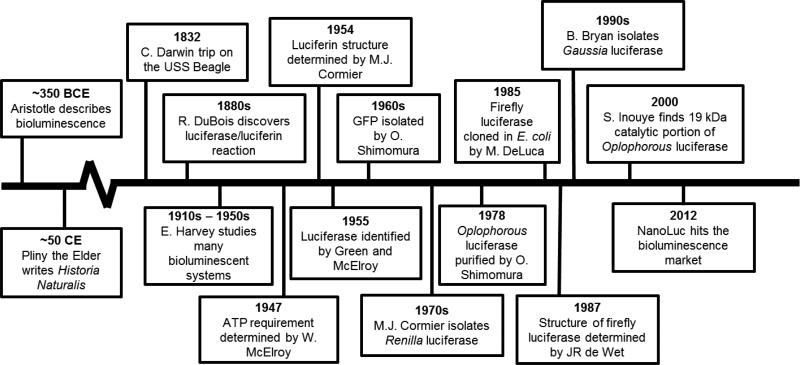
Bioluminescence is a naturally occurring phenomenon that occurs in many living terrestrial and aquatic organisms, including insects, bacteria, fungi, and marine animals.7 While bioluminescence has been observed for thousands of years (Figure 1), humans only recently began to utilize this unique occurrence for biomedical purposes. While the first reference of bioluminescence can be traced back to the folklore of ancient civilizations, the first historical reference of bioluminescence came from the Greek philosopher Aristotle (384-322 BCE), who was fascinated by how organisms could produce light.2 It was almost three centuries later when Pliny the Elder (23-79 CE) became the first person to report the bioluminescence of several species of animal, providing in-depth descriptions of the luminous mollusk, purple jellyfish, glowworm, fireflies, and others. His findings were published in an encyclopedia called Historia Naturalis or Natural History. In 1667, it was discovered by Robert Boyle that bioluminescence required air. Bioluminescence was also noted by Charles Darwin, who termed the process phosphorescence during his memorable trip aboard the Beagle.
Figure 1.
Timeline illustrating several significant events that occurred in the field of bioluminescence.
It was the French pharmacologist Raphael Dubois (1849 – 1929) who first investigated the components required for the bioluminescent reaction from the click beetle.8 Using only cold water and the abdomens of the Elateridae click beetle, Raphael Dubois was able to produce luminescence in the laboratory. Eventually, he named the two extracted components, calling the molecule that was consumed in the reaction “luciferine” and the enzyme responsible for the reaction was termed “luciferase.” As history continues, E. Newton Harvey tested various combinations of luciferase enzymes and substrates to find that both luciferases and luciferins were not interchangeable between species.
While DuBois discovered the reaction between luciferin and luciferase in 1885, it was not until the late 1940s when the luciferase protein was first extracted and purified firefly lanterns by Drs. Green and McElroy.6 Using this process, they isolated the enzyme and determined its conformational structure. The ATP requirement for bioluminescence from the Firefly was elucidated by W. McElroy through in vitro studies of firefly luminescence in 1947.5 In the 1960's, Osamu Shimomura and colleagues discovered the calcium-activated photo-protein aequorin from the jellyfish Aequorea victoria.9 The next vital discovery occurred in 1985 when Marlene DeLuca reported the cloning of firefly luciferase (FLuc) in Escherichia coli, effectively paving the way for this technique to be widely utilized in the future with many luciferase systems.10
During the thirty years following cloning of FLuc, several essential discoveries and advancements in the field of molecular biology transpired that revolutionized the field of bioluminescence. Eventually, this led to the discovery of the luciferase and substrate from Oplophorus gracilirostris (OLuc) and Gaussia princeps (GLuc).11 In recent years, research in the area of bioluminescence has emphasized the development of highly stable luciferase enzymes. To this end, some luciferases have been discovered and modified that have allowed bioluminescence to gain considerable attention in the biomedical field. For more details regarding the history of bioluminescence, readers are directed to the detailed review by John Lee and references within.7
While it was initially thought that only a few organisms were capable of producing bioluminescence, further investigations have revealed that numerous organisms possess this unique ability. The most commonly employed luciferases for biomedical purposes are listed in Table 1. One of the first bioluminescent systems explored for biomedical purposes was that of Pyrophorus plagiophthalamus, a click beetle.2, 7 The click beetle luciferase catalyzes an ATP-dependent reaction that utilizes the substrate D-luciferin, along with magnesium and molecular oxygen, making this luciferase similar to other ATP-dependent luciferases like FLuc. The large size of the click beetle luciferase may greatly impact the behavior of smaller investigative compounds. Derivatives of the click beetle luciferase have subsequently been developed with a range of light emissions from green to red.
Table 1.
Common luciferase enzymes with their origin, required substrates and cofactors, molecular weight, and maximum emission wavelength.
| Luciferase | Organism | Substrate | Cofactor(s) | Size (kDa) | Emission wavelength (nm) |
|---|---|---|---|---|---|
| North American Firefly (FLuc) | Photinus pyralis | D-luciferin | ATP and Mg | 61 | 560 |
| Click Beetle | Pyrophorus plagiophthalamus | D-luciferin | ATP and Mg | 64 | ~600 |
| Renilla (RLuc) | Renilla reniformis | Coelenterazine | N/A | 36 | 480 |
| Renilla Mutant (RLuc8) | Renilla reniformis | Coelenterazine | N/A | 36 | 535 |
| Gaussia (GLuc) | Gaussia princeps | Coelenterazine | N/A | 20 | 470 |
| OLuc | Oplophorus gracilirostris | Coelenterazine | N/A | 19 | 460 |
| NanoLuc (NLuc) | Oplophorus gracilirostris | Furimazine | N/A | 19 | 460 |
In the 1970s, M.J. Cormier and colleagues purified the Renilla reniformis luciferase from the sea pansy.12 This 36 kDa enzyme is ATP-independent and uses coelenterazine and oxygen as its cofactors. This system provides a so-called long–flash emission, an approximately 30-second burst of 480 nm light. Perhaps the most widely-known luciferase for biomedical purposes is known as Photinus pyralis, or the American firefly. Its structure was determined in 1987 by J.R. de Wet et al., who also expressed the gene in mammalian cells.8 Unlike the luciferases from marine organisms, this enzyme requires ATP and magnesium, along with D-luciferin, to produce luminescence. The combination of FLuc and click beetle luciferases have allowed for the development of dual luciferase reporter assays, in which one may have both an experimental reporter and an internal control reporter present.13 Dual luciferase systems have also been developed with other platforms, including FLuc and RLuc which is commercially available.14
Currently, many researchers have switched to smaller luciferases. GLuc is a luciferase that was originally extracted from Gaussia princeps, a marine copepod.7, 8 This luciferase is distinctive as it is a secreted luciferase, thus assays using this luciferase system will use cell media for analysis. Similar to other marine organisms, this small luciferase (20 kDa) produces light at 470 nm and is ATP-independent. Dr. Bryan isolated this flash-type enzyme in the 1990s, and it has since found unique applications in the cellular secretory pathway, assays for viral infectivity, and chemokine receptor activation. Next, Dr. Shimomura identified the bioluminescent properties of the deep-sea shrimp Oplophorus gracilirostris in 1978.15 The intact luciferase contains two regions composed of a 35 kDa and 19 kDa subunit each, and, in its natural state, uses coelenterazine as a substrate. However, this enzyme has undergone extensive development, with the 19 kDa subunit being identified as the catalytic portion for bioluminescence in 2000.4 Since that time, this subunit has been modified and a novel substrate, furimazine, developed, to create the system known as NLuc.
2.2 Luciferases: Mechanism of action
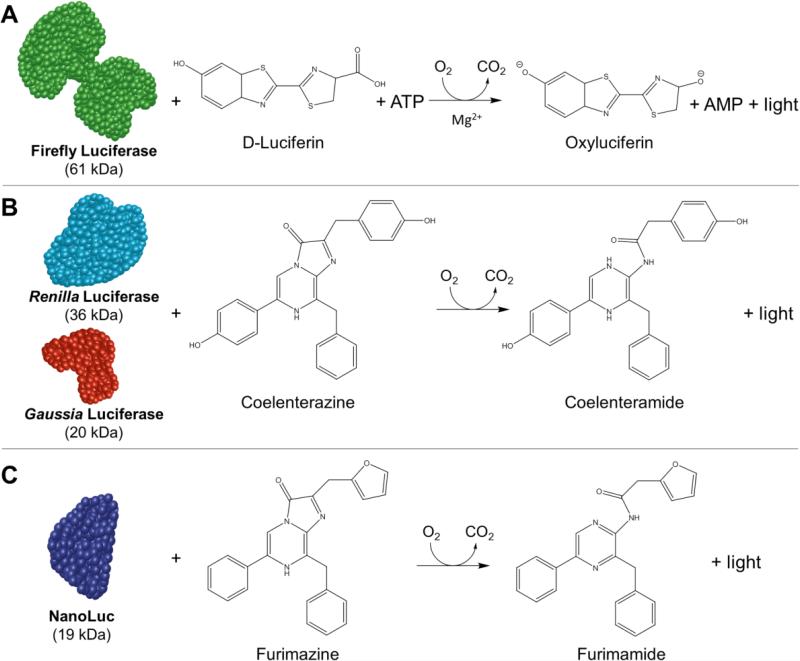
Luciferase is a general name for enzymes that produce light in living organisms. There are two key requirements for the production of bioluminescence, including the enzyme responsible for catalyzing the reaction and producing light (luciferase) and the substrate for this enzyme (luciferin). Many such systems have been characterized, with several famous examples outlined in Figure 2. For an extensive review of luciferase mechanisms, the interested reader is referred to the excellent explanation by Frank McCapra.16 Briefly, the reaction between FLuc and its substrate (D-Luciferin) produces bioluminescence in the presence of ATP (Figure 2A). Next, RLuc and GLuc react with coelenterazine to produce bioluminescence, while NLuc reacts with a coelenterazine derivative, known as furimazine (Figure 2B-C).
Figure 2.
Bioluminescence is based upon a chemical reaction that occurs between the luciferase enzyme and corresponding substrate. (A) Firefly luciferase (FLuc) reacts with D-luciferin in the presence of adenosine triphosphate (ATP), molecular oxygen, and magnesium to produce light. (B) Both Renilla (RLuc) and Gaussia luciferase (GLuc) only require coelenterazine and oxygen to produce light. (C) Bioluminescence from the NanoLuc (NLuc) system occurs when the optimized substrate called furimazine reacts with NLuc in the presence of molecular oxygen. This reaction yields furimamide and luminescence output.
2.3 Development of the NanoLuc system
NLuc is the newest addition to the family of luciferase enzyme systems commercially available for bioluminescence applications. This small luciferase enzyme was derived from deep-sea shrimp Oplophorus gracilirostris, during which three rounds of mutagenesis were performed to optimize the luminescence output of the enzyme.4 While the Oplophorus gracilirostris luciferase (OLuc) consists of two heterodimeric regions with a 35 kDa and 19 kDa subunit each, its bioluminescence properties have been mapped to a single 19 kDa subunit of the intact luciferase, called OLuc-19. While this subunit is poorly expressed and unstable, structural optimization of the small catalytic subunit with mutagenesis and the addition of a novel substrate were found to create a very effective bioluminescent system. Also, Mary Hall and her colleagues were able to optimize the stability of the enzyme.4 Overall, they found that the novel NLuc system displayed a specific activity over 150-fold higher than both FLuc and RLuc. In addition, the luminescence output remained stable and lasted longer than previous luciferases. This subsection investigates the engineering of the NLuc enzyme and its corresponding substrate for improved bioluminescence. Advantages of the NLuc system will be further discussed in Section 4.
While the NLuc technology is relatively straightforward, choosing a vector for a specific application may seem tedious at first. As a short review, expression vectors are plasmid constructs designed for protein expression in cells by introducing a specific gene into a target cell.17 For more details on vectors, readers are directed to an extensive review of this topic in Gene Therapy.18 Currently, there are several NLuc protein fusion vectors available for generation of N- or C-terminal fusions of NLuc with a protein of interest. They have been divided into two formats, including the pNLF vector series uses the traditional cloning system with multiple cloning sites and the pF series using the Flexi vector cloning system, which is based on directional cloning with two rare-cutting restriction enzymes and eliminates the need for resequencing if the protein-coding regions are transferred between other Flexi vectors.19
There are three main forms of NLuc available for specific applications, including an unfused form of NLuc, a form that is secreted from the cells (secNLuc), and a destabilized form of NLuc called NLuc-PEST (NLucP). Each form of NLuc was configured for different applications.19 The unfused form of NLuc provides the highest sensitivity and light output and can actively accumulate in cells due to its intracellular stability.19 Next, secNLuc is an excellent choice for those applications requiring the enzyme to be secreted into the media, as secNLuc remains stable in cell culture media for more than 4 days at 37 °C. NLucP is a unique version of the protein as it has a protein degradation signal to destabilize the enzyme, thus providing a close coupling of NLuc to alterations in transcriptional activity and protein expression with increased signal-to-noise background ratios.19 Due to the short lifetime of NLucP (10-30 minutes), expression levels are rapidly changed in response to transcriptional changes. Also, Promega offers other NLuc-based technologies, such as NanoBRET that can be used to study protein interaction dynamics20, 21 and stability sensors for studying signaling proteins. 22 For more information about the NanoLuc platform offerings, readers are directed to the Promega website.19
3. Applications of the NanoLuc technology
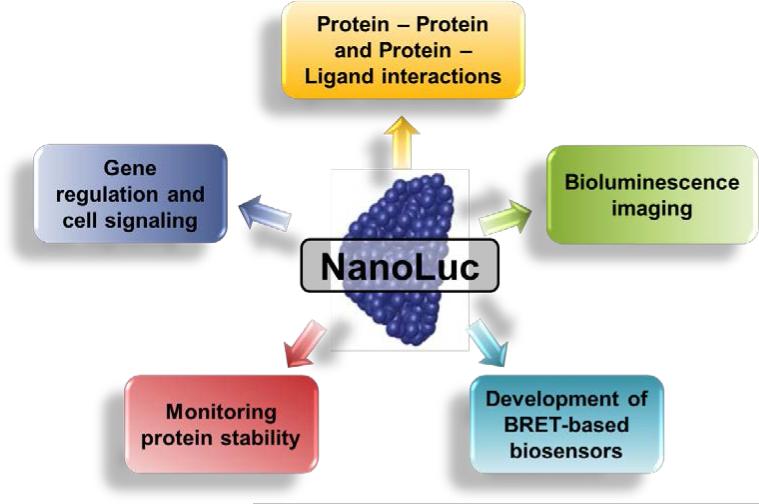
NLuc is an extremely versatile technology that may be employed for several applications, including the investigation of protein – protein and protein – ligand interactions, exploring gene regulation and cell signaling, monitoring protein stability, development of novel BRET-based biosensors, and for bioluminescence imaging (Figure 3). This section examines the current uses and advantages of NLuc in each of these applications.
Figure 3.
The NanoLuc luciferase (NLuc) technology has been successfully utilized for several applications, including the investigation of protein – protein and protein – ligand interactions, exploring gene regulation and cell signaling, monitoring protein stability, utilization as BRET-based biosensors, and bioluminescence imaging.
3.1 NanoLuc for investigating protein – protein and protein – ligand interactions
Ligand-receptor binding assays are the cornerstone of molecular biology. While effective, most traditional binding assays rely on the use of radioactive tracers, which pose health risks to users, and many radionuclides have short half-lives. To overcome these limitations, scientists have searched for novel non-radioactive receptor-binding assays, with bioluminescent ligand-receptor binding assays showing excellent potential. In particular, bioluminescent tracers display higher sensitivity than radioactive tracers, easier preparation, longer shelf life, and better safety.23
Development of novel receptor – binding assays is often tedious and time-consuming, yet Song et al. developed a simplified method for rapid preparation of a novel NLuc-based protein tracer using erythropoietin (Epo) as a model.23 The glycosylated cytokine Epo binds to the cell membrane receptor EpoR, thus promoting erythropoiesis or red blood cell formation. In this study, the secreted form of NLuc was fused to C-terminus of Epo and overexpressed in human embryonic kidney 293 cells (HEK-293T). The Epo-NLuc protein maintained a high binding affinity for EpoR with a dissociation constant (Kd) of 0.59 ± 0.06 nM, which was similar to the values measured with 125I-labeled Epo. Also, overexpression of Epo-NLuc resulted in large quantities of tracer that could be obtained within four days of transfection. Using these procedures, it was possible to prepare and test novel NLuc-based tracers within 1 to 2 weeks.
Two characteristics of NLuc that make it an optimal tracer for cell-based assays are its enhanced luminescence output and the individualized substrate. Since NLuc utilizes a substrate different from other luciferases, it is possible to investigate more than one protein – protein interaction concurrently through multiplexing with other luciferases. Recently, Verhoef et al. verified these properties by generating a split NLuc reporter that could be multiplexed with FLuc.24 Both mouse double minute 2 (MDM2) homolog and MDM4 are negative regulators of the p53 tumor suppressor. Thus, simultaneous measurement of these interactions may provide new insight into complex interactions. The NLuc fragments were genetically fused to p53 and MDM2. The two NLuc fragments would come into proximity when MDM2 and p53 interacted, allowing for reactivation of the enzyme and the production of bioluminescence in the presence of furimazine. As NLuc and FLuc use different substrates and display different emission spectra, it would be possible to study the interaction of p53 with both MDM2 and MDM4.
In a study conducted by the laboratory of Ping Wang, the process of ubiquitination was investigated using NLuc that was fused to the C-terminus of ubiquitin which was bound to agarose beads.25 Ubiquitination is a post-translational process that regulates protein degradation, protein trafficking, transcriptional signaling, and oncogenesis. In the presence of deubiquitinating enzymes, NLuc was released from ubiquitin beads into the supernatant. The luminescence signal correlated with the activity of the deubiquitinating enzymes. Furthermore, they applied NLuc for measuring other ubiquitin-like isopeptidases and investigating OTULIN activity.
In 2015, Liu et al. successfully monitored the internalization of the G protein-coupled relaxin family peptide receptor (RXFP3) using NLuc.26 A stable HEK293T cell line was generated that co-expressed NLuc-tagged RXFP3 and an enhanced green fluorescent protein (EGFP)-tagged RXFP3. While EGFP was used to visualize the receptor internalization process using microscopy, NLuc was used to quantify the measurements via bioluminescence. As both fused proteins were encoded on a single plasmid, an inducible bi-directional promoter was used to ensure that expression of both proteins was equivalent and controllable. As expected, NLuc-tagged RXFP3 was internalized into acidic lysosomes, causing a decreased bioluminescence signal. Also, ligand-induced internalization of RXFP3 was quantitatively assessed, as cells exposed to 100 nM of its endogenous ligand, relaxin-3, resulted in rapid receptor internalization and diminished bioluminescence signal. In a similar study, Wu et al. investigated the interaction of human relaxin with RXFP1.27 With a similar goal in mind, they conjugated NLuc to relaxin by replacing three lysines of human relaxin with arginine. After expression in Pichia pastoris and in vitro enzymatic maturation, a disulfide linkage was made between the B-chain N-terminus of relaxin with a C-terminus cysteine of NLuc. After construction of the assay, they showed that NLuc-conjugated relaxin retained its high affinity for RXFP1 (Kd=1.11 ± 0.08 nM), thus making the assay suitable for screening of novel antagonists or agonists of RXFP1.
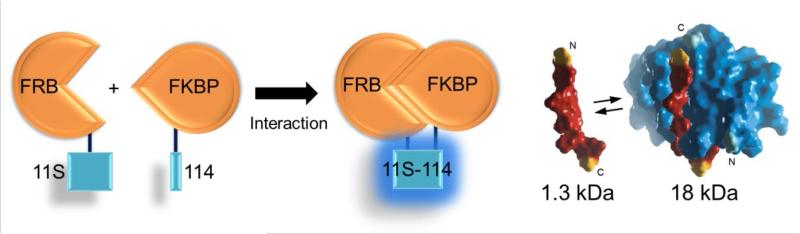
Protein interactions are often investigated using traditional protein-fragment complementation assays, yet these fragments are often non-optimized and structurally compromised. Dixon and colleagues engineered a novel complementation reporter (NanoBiT) using NLuc that would allow for quantitative investigation of protein interactions under physiological conditions.28 This was accomplished by splitting NLuc into two components, either of which would be fused to two proteins of interest. One of the NanoBiT components was only 11 amino acids long (114) while the other component was two-thirds the size of GFP (11S). Unlike other complementation reporters, NanoBiT was structurally optimized to increase conformational stability and minimize steric hindrance. Optimized stability of 11S resulted in enhanced expression and performance at physiological temperatures while the small size of 114 ensured that it would not influence the interaction of the target protein. Next, the rapamycin-inducible FKBP rapamycin binding protein (FRB)/FK506 binding protein (FKBP) model was used to ensure that NanoBiT would function in mammalian cells, using FRB-11S and FKBP-114 fusion proteins (Figure 4). Luminescence was measured in live cells and cell lysates, after transfection with equal amounts of NanoBiT or NLuc DNA and showed a linear trend in luminescence at high DNA concentrations with NanoBiT and NLuc lysates in the presence of rapamycin while luminescence was significantly reduced in the absence of rapamycin.
Figure 4.
Quantitative assessment of protein interactions under physiological conditions using NanoBiT. NanoBiT was used to study the rapamycin-inducible FKBP rapamycin binding protein (FRB)/FK506 binding protein (FKBP) interaction mammalian cells, using FRB-11S and FKBP-114 fusion proteins. Adapted with permission from ref. 28. Copyright 2015 American Chemical Society.
Two studies have investigated the iron efflux transporter, known as human ferroportin (Fpn), using the NLuc system. First, Song at al. attached NLuc to the C-terminus of Fpn to monitor the internalization of the receptor in mammalian cells. 29 In addition, this fusion protein was used to quantitatively assess the interaction of Fpn with the peptide hormone, known as hepcidin. Luminescence measurements were made with both intact and lysed cells expressing Fpn-NLuc with and without hepcidin treatment. A slight decrease in the bioluminescence signal of lysed cells was seen after hepcidin treatment, attributed to the high stability of NLuc. However, intact cells showed a significant 50% decrease of luminescence after hepcidin treatment, which was linked to the low pH values found in lysosomes. This was confirmed by measuring the activity of NLuc at pH 4-5, which is similar to the acidity found in lysosomes. Next, the same research group further utilized NLuc for studying insulin-like peptide 3 (INSL3), whose interaction with the relaxin family peptide receptor 2 (RXFP2) plays a significant role in fertility.30 In this work, the authors established a simple procedure for chemically linking NLuc with the protein of interest by first creating a fully active 6×histidine-cysteine-NLuc (6×His-Cys-NLuc) that would react with sulfhydryl protein residues. Similarly, 6× His-Cys-NLuc was later employed by He et al. studied the binding of human leukemia inhibitory factor (LIF) to its native receptor (LIFR).31 As expected, LIF-NLuc showed an unaltered binding affinity with a detection limit of fewer than ten receptors per cell, making this platform an ultrasensitive bioluminescent tracer.
In a similar study, NLuc was employed for investigating inositol-requiring enzyme 1 (IRE1) activity in vitro.32 Activation of IRE1 is caused by abnormal conditions of the endoplasmic reticulum (ER), which has been linked to several neurodegenerative diseases. When this occurs, the unspliced form of X-box binding protein 1 is cleaved to produce sXBP1. A NLuc reporter construct was designed with the mouse XBP1-splice region upstream, thus, expression of XBP1-NLuc could be induced by different endoplasmic reticulum stress inducers. To assess the efficiency of the reporter protein in vitro, XBP1-NLuc was transfected into four cell lines which were later treated with an ER stress inducer. It was shown that NLuc activity increased upon exposure to the ER stress inducers. In another study by the same group, NLuc was used to monitor the transcriptional regulation of mesencephalic astrocyte-derived neurotrophic factor (MANF) in HEK293 cells.33 Secretion of NLuc-tagged MANF from HEK293 cells was shown to be time-dependent and could be slowed by two proteins known to decrease wild-type MANF secretion. In addition, the cells were stimulated to produce more MANF, which caused significant increases in the NLuc activity measured in the cell media. This method can be applied to study other ER stress-induced diseases, including amyotrophic lateral sclerosis, ischemia, and diabetes.
The potential applications of NLuc extend beyond the study of endogenous proteins, including the study of viral infection. Recently, Seay et al. utilized NLuc for developing a human immunodeficiency virus (HIV) reporter construct.34, 35 While RLuc was originally developed for this study; the authors wanted to improve the sensitivity and stability of the assay. The enhanced sensitivity of NLuc is important for this application because it allows for quantification of low-level HIV replication during initial viral infection. As the researchers wanted to study acute HIV infection and the efficacy of pre-exposure prophylaxis (PrEP) for inhibiting HIV infection via the vaginal mucosa, an R5-tropic env gene from an African woman recently infected with HIV was employed (C.Du151.2). After generating the virus in 293T cells, they found high levels of NLuc activity in the cells, suggesting that NLuc expression was stable and highly sensitive. Also, they were able to monitor the initial stages of viral infection. Furthermore, this group later developed a novel strain of mice termed hCD4/R5/cT1, a transgenic strain carrying the essential genes for HIV-1 infection, including human cluster of differentiation 4 (CD4), C-C chemokine receptor type 5 (CCR5), and cyclin T1.35 This mouse model was infected with HIV-1-expressing NLuc, which made it possible to study how substance abuse and other factors increase the risk of HIV-1 infection via mucosal transmission.
NLuc has also been used to study other viral infections, including hepatitis B virus (HBV). Nishitsuji et al. developed a recombinant hepatitis B virus (HBV) called HBV/NLuc to monitor viral replication in vitro.36 This was accomplished by inserting the NLuc gene downstream of the HBV control region. It was imperative that the HBV DNA size remained unchanged, as replication is significantly hindered if the genome size was more than 0.7 kilobase pairs (kbp) over the original genome size. It was shown that NLuc activity accurately reflected HBV RNA levels in transfected cells, as both increased for more than 6 days post HBV infection. Next, the novel HBV/NLuc reporter was used to screen HBV inhibitors, including entecavir and IFNB, both of which resulted in dose-dependent decreases in NLuc activity. While qPCR and qRT-PCR are common procedures for evaluating HBV infection, the HBV/NLuc system was shown to be simpler and more sensitive. This assay, utilizing the unique properties of NLuc for monitoring viral replication, may be applied for identifying novel anti-HBV agents and antibodies in the future.
In addition to viruses, the utilization of NLuc has extended to other pathogens like Plasmodium falciparum, the parasite that causes malaria. While advancements in the treatment and patient control have effectively reduced the number of malaria cases, more than 400,000 people still die from malaria each year according to the World Health Organization.37 To develop a new high throughput screening assay for screening anti-malarial compounds, Azevedo et al. described a method to express NLuc in Plasmodium falciparum.38 As a proof-of-concept, parasites expressing NLuc were shown to act as functional screening assays for measuring the effects of inhibitors of anti-malarial compounds. Similar procedures have been employed to express NLuc in bacteria as well. In a study conducted by the laboratory of Dr. Thomas Proft, NLuc was used for generating bioluminescent A Streptococcus (GAS) strains.39 It was shown that NLuc-modified bacteria were more than 15-times brighter than FLuc-modified GAS. However, NLuc-GAS could not be used to differentiate between metabolically active and inactive bacteria, as NLuc is ATP-independent. For this reason, NLuc may be restricted in potential future applications of this type, in which ATP-dependent FLuc will function better.
3.2 NanoLuc for exploring gene regulation and cell signaling
Gene delivery is a significant advancement from the field of molecular biology, which has been rapidly gaining attention in the biomedical community. To ensure the success of gene delivery, certain markers may be utilized. In a recent study, NLuc was used to confirm the success of mitochondrial gene delivery via hydrodynamic limb vein injection (quickly injecting a large volume of plasmid DNA in the tail vein) in mice without using a virus for transfection.40 A genetic construct was designed with the DNA from an endogenous mitochondrial protein, known as the human NADH-ubiquinone oxidoreductase subunit 4 (ND4). The authors expected that the mRNA transcribed from the ND4-nLuc vector would efficiently bind to the mitochondrial ribosomes for translation into detectable protein. While Western blot lacked the sensitivity to detect the exogenous protein from the mitochondrial transfection, NLuc allowed for validation of gene delivery in both liver and skeletal muscle with this novel construct
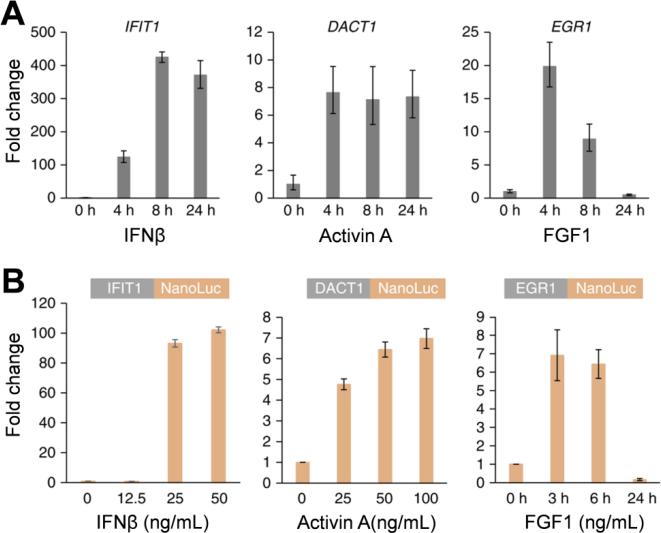
The small size and brightness of NLuc were also utilized by Lackner et al. in choosing a system that would be compatible with the low signal intensities that arise from single events of genomic integration.41 This study examined a novel strategy for tagging genes at both the N- and C- terminus, using NLuc to demonstrate the approach. NLuc tagged cell lines were used to monitor endogenous gene expression. The cell lines contained NLuc integrations in three cytokine-inducible genes (DACT1, IFIT1, and EGR1). First, the upregulation of mRNA to cytokine stimulation was assessed in wild-type HAP1 cells using quantitative PCR (Figure 5A). To test the functionality of NLuc, cell lines bearing NLuc integrations in DACT1, IFIT1, and EGR1 were stimulated with the cognate cytokine ligands (specific for each gene), which resulted in cytokine-induced upregulation of NLuc (Figure 5B). Importantly, it was shown that NLuc data regarding cytokine stimulation and kinetics correlated well with qPCR data. Also, the tagged alleles remained fully functional regarding cellular localization and gene expression regulation. Ultimately, NLuc reporter cell lines were shown to allow for monitoring of gene expression at endogenous levels in live cells.
Figure 5.
Monitoring of gene expression with cell lines expression NanoLuc (NLuc) integrations. (A) HAP1 cells were stimulated with three cytokines, IFN-β, activin A, and FGF1 for 4, 8, or 24 h with a final concentration of 50 ng/mL. RNA was isolated and analyzed for IFIT1, DACT1, and EGR1 signatures. (B) Clonal cell lines bearing the NLuc integrations in IFIT1, DACT1, and EGR1 were stimulated with three cytokines. NLuc levels were measured at 24 h for IFIT1 and DACT1and after the noted time points for EGR1 from collected cell lines. Reproduced with permission from ref. 41. Copyright 2015 Nature Publishing Group.
Monitoring of gene expression is crucial to understanding a variety of phenomena from viral infection to gene delivery. In addition to studying gene expression, the development of novel genetic reporters has made it possible to study cellular events coupled to gene expression. Bioluminescent reporters have been used to study RNA interference (RNAi), the structure of various promoters, and the functional analysis of promoter single nucleotide polymorphisms (SNPs); thus, we predict future studies will employ NLuc for similar applications.
3.3 NanoLuc for monitoring protein stability
Protein aggregation has been found to be a crucial factor in several human diseases, ranging from Huntington's to Alzheimer's disease. While the role of protein aggregation has been well documented, there are limited assays for investigating protein aggregation in vitro and in vivo. To successfully investigate protein aggregation and enhance the development of novel therapeutic strategies targeting protein aggregation, highly sensitive agents are required. The NLuc construct is a possible platform for exploring this biological occurrence, demonstrating yet another biomedical application that may benefit from this new technology.
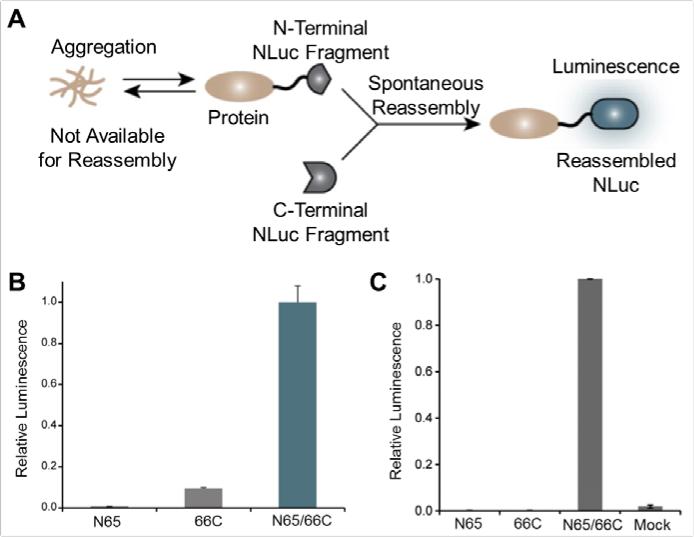
With these limitations in mind, Zhao et al. monitored protein aggregation in living cells using a conditional split – NLuc system.42 A portion of NLuc (N65) was fused to a protein known to aggregate while the other portion of NLuc was used to measure the level of protein aggregation (Figure 6A). If the protein – NL fusion aggregated, then the addition of the second portion of NLuc would not result in bioluminescence upon the addition of substrate. If the protein did not aggregate, the addition of the second portion of NLuc would produce a bioluminescent signal upon addition of furimazine. The two fragments of NLuc were characterized in vitro to ensure that both fragments (N65 and 66C) were required for the production of a luminescence signal (Figure 6B). Lastly, the authors transferred this technique from bacteria to mammalian cells as a proof-of-principle, showing that luminescence was only observed when NIH-3T3 cells were co-transfected to express both NLuc fragments (Figure 6C), further validating the use of NLuc for monitoring protein aggregation in vitro.
Figure 6.
NanoLuc (NLuc) fragments for determining protein aggregation. (A) Schematic of the aggregation assay in which a fusion of a protein to the N-terminus of NLuc was used to monitor protein aggregation, with decreased luminescent signal signifying protein insolubility. (B) Luminescence output from individual NLuc fragments and combined fragments 50 min after addition of furimazine. (C) As a proof-of-concept, individual or both NLuc fragments were expressed in mammalian cells. Luminescence required the presence of both NLuc fragments, N65, and 66C. Adapted with permission from ref. 42. Copyright 2016 American Chemical Society.
3.4 NanoLuc for BRET-based biosensors
Bioluminescence resonance energy transfer (BRET) makes it possible to investigate molecular interactions in real-time in living cells at physiological temperatures.43 In BRET, the luciferase enzyme acts as a resonance energy donor, while the fluorescent protein is the resonance energy acceptor.44 Upon addition of the luciferase substrate, the bioluminescence energy from the luciferase excites the fluorophore, avoiding the need for excitation light and preventing possible photobleaching of the fluorophore. Optimized BRET platforms utilize donors with high bioluminescence quantum yield and excellent spectral resolution.45 Also, successful translation of BRET into small animal imaging requires red-shifted signal due to tissue attenuation. As an example, this was previously accomplished using the mutated RLuc8 (λex/em = 480 nm with coelenterazine) with mOrange (λex/em = 514/530 nm), as this BRET reaction shifted the energy to 530 nm.46
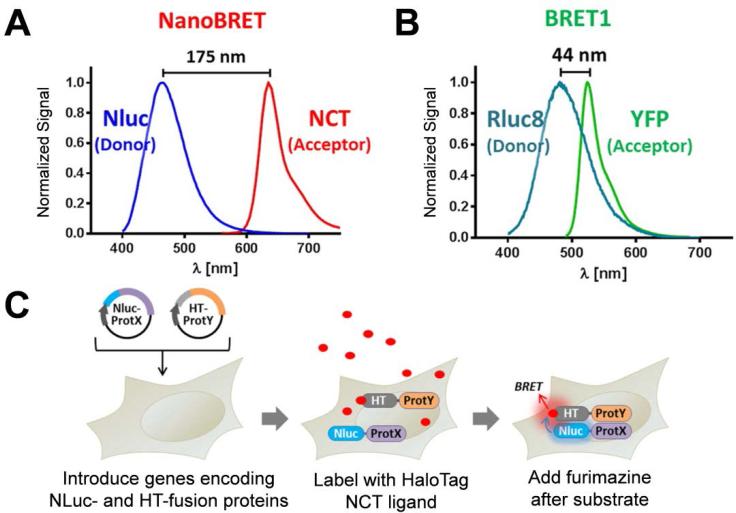
RLuc is a common energy donor used in BRET studies, matched with a yellow fluorescent protein (YFP). While effective, this method is limited by high background noise and low sensitivity. In comparison to RLuc, the bioluminescence spectrum of NLuc is much narrower, making it easier to discriminate the acceptor fluorophore. Another novel technology called the HaloTag fusion tag, allowed for rapid evaluation of several potential fluorophores.47 The HaloTag system is based on the formation of a stable fusion protein that carries an active site for binding of application-specific ligands. This results in a covalent interaction between the HaloTag-tagged protein and specific ligand. Both of these technologies were utilized to investigate the BRET efficiency of the rapamycin-induced interaction between FKBP12 and Frb, as this simple model is commonly employed for investigating new protein – protein interactions.20 First, the optimal HaloTag ligand was determined to be nonchloroTOM (NCT), having an excitation and emission maxima at 595/635 nm, which is more than 175 nm red shifted from that of NLuc (Figure 7A). For comparison, the BRET efficiency between RLuc and YFP was also investigated (Figure 7B). Two constructs were created, and the genes were transfected into cells (Figure 7C). Next, the cells were incubated with NCT, which formed a covalent linkage with the HaloTag-modified protein. Cells were then incubated with furimazine, which reacts with NLuc to produce luminescence with the energy being effectively transferred to NCT. This validates the potential employment of NLuc-based BRET for monitor protein interactions in live cells.
Figure 7.
NanoBRET for detecting protein – protein interactions between Frb and FKBP. (A) In NanoBRET, there is a difference of 175 nm from NLuc to NCT. (B) BRET with RLuc8 and YFP was used as a control system and showed a 44 nm separation in signals. (C) Schematic of the NanoBRET system, in which two proteins were fused with NLuc or HaloTag and expressed in mammalian cells for in vitro BRET imaging. Adapted with permission from ref. 47. Copyright 2015 American Chemical Society.
Recently, Robers and colleagues developed an approach that utilized a cell permeable fluorescent protein, in conjunction with an NLuc-fused intracellular target protein in live cells.48 Using a compound that binds to the target protein, it was shown that BRET signal correlated with the binding efficiency of the interacting molecule. Thus, binding of the interacting molecule to the target protein would lead to less target protein available for binding of the fluorescent tracer, resulting in decreased BRET signal. A similar study by Schaub et al. led to the development of a novel class of BRET imaging reporters for in vivo use, called LumiFluors.49 Two LumiFluors were constructed, eGFP-NLuc (GpNLuc) and LSSmOrange-NanoLuc (OgNLuc) that were connected together by a flexible peptide linker between the C-terminus of the fluorophore and N-terminus of NLuc. The intramolecular BRET signal gained from these two LumiFluors resulted in the brightest bioluminescent signal known to date in A549 (non-small cell lung cancer) subcutaneous and orthotopic mice injected with furimazine. Also, the novel system was sensitive enough to detect micrometastases in regional lymph nodes.
NLuc has also been used to study the binding of insulin to the insulin receptor. The two insulin-interacting domains of the insulin receptor (αCT and L1 domain) were genetically fused with NLuc or the fluorescent protein YPet, respectively. 50 Upon binding of insulin, these two insulin-interacting domains come into proximity of each other, effectively bringing NLuc and YPet into close contact for BRET signal production. BRET signal was found to be proportional to the amount of insulin in the sample. The use of NLuc for BRET has allowed researchers to develop highly sensitive constructs for visualization of processes from insulin binding to cancer detection. While in vitro testing is a crucial tool in the biomedical field, further investigations in animal models are necessary for preclinical translation.
3.5 NanoLuc for in vivo bioluminescence imaging
Bioluminescence has become a widely exploited approach for in vivo optical imaging over the last decade, as bioluminescent techniques allow for non-invasive molecular imaging with low background noise and high sensitivity. For these reasons, bioluminescence imaging has become increasingly popular in the preclinical setting for biomedical research.51 Traditionally, bioluminescence imaging has been performed with FLuc, as its strong emission at 560 nm allows for imaging of tissue several centimeters deep. The use of other traditional luciferases for in vivo remains limited to date as their spectral emission profiles are not optimal for imaging tissue at depth. While NLuc does not have an optimal spectral emission for in vivo imaging, it has been employed for several imaging applications, including tracking of viral infection and monitoring disease progression.
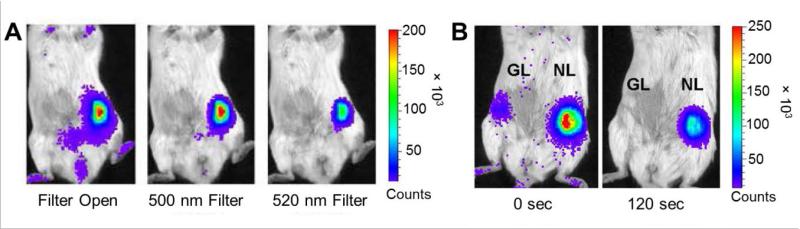
In a study conducted by Stacer et al., NLuc was used in the dual optical imaging of breast cancer by transiently transfecting MDA-MB-231 human breast cancer cells with plasmids for secreted NLuc or GLuc.52 NLuc-transfected cells were implanted into the mammary fat pad of mice. After mice had received an injection of furimazine, imaging showed that no filter showed the highest signal, followed by the 500 nm filter (Figure 8A). Next, mice were implanted with both NLuc and GLuc-transfected tumor xenografts to provide a comparison between the two bioluminescent agents. Injection of 5 μg furimazine provided an intense optical signal from the MDA-MB-231-NLuc tumors, while the signal from the MDA-MB-231-GLuc tumor remained at background levels (Figure 8B), further demonstrating the substrate specificity of furimazine for NLuc and indicating the potential of this dual-imaging system for coincidental reporting. Also, NLuc displayed an enhanced signal in comparison to GLuc at each imaging time point. This also confirmed that NLuc can be used for in vivo imaging of superficial tumors and internal organs. To determine the multiplexing capabilities, tumor progression was also monitored using MDA-MB-231-NLuc/FLuc cells co-expressing both NLuc and FLuc. While this bioluminescence system was able to monitor effectively tumor growth, the emission wavelength of NLuc (~460 nm) was less suited for deep tissue imaging than the red-shifted FLuc with an emission greater than 600 nm.
Figure 8.
Comparison of NLuc and GLuc for imaging of breast cancer. MDA-MB-231 cells were transfected with NLuc or GLuc. (A) Bioluminescence imaging of mice implanted in the mammary fat pad with MDA-MB-231 cells transfected with NLuc. After injecting furimazine, images were obtained with an open filter, 500 nm filter, and 520 nm filter. (B) Mice implanted with either NLuc or GLuc-expressing MDA-MB-231 tumor xenografts were injected with coelenterazine and imaged for two minutes. Adapted with permission from ref. 52. Copyright 2013 SAGE Publications.
Germain-Genevois et al. also investigated NLuc for dual reporter imaging with FLuc, specifically in deep brain tumor and systemic metastases.53 U87-FRT-CMV-NL-IRES-FL tumors were imaged using substrates for NLuc (furimazine), FLuc (D-luciferin), and also using coelenterazine, a substrate suitable for both. The use of coelenterazine produced signals from both luciferases, showing that sequential determination of the activity of both NLuc and FLuc is possible with a single substrate injection, even within the same tumor. While NLuc showed higher activity ex vivo, FLuc signal was greater in vivo due to the more suitable emission for tissue penetration. In a similar study, viral vectors expressing either NLuc or FLuc were injected into the footpads of mice and imaged to track the spread of the virus.54 Increased signal from FLuc was evident in individual positive tissues, but the whole-body signal from NLuc was found to persist out to 48 h after injection and was approximately two logs higher than the whole-body FLuc signal at the same time point. As both NLuc and GLuc produce light in the presence of coelenterazine, dual NLuc and GLuc reporter imaging does require some optimization. Luckily, this setback was solved by Heise et al. who showed that the background signal from NLuc with coelenterazine may be calculated by determining the activity of NLuc in the presence of its modified substrate furimazine.55 In return, the activity of GLuc can be calculated, as it does not react with furimazine. This discovery opens up new avenues for highly sensitive dual luciferase systems, which may be useful for high throughput screening.
Bioluminescent reporters have long been utilized in the imaging of virus spread in animals. However, many viral agents, including influenza, are particularly sensitive to changes in their genome, making the insertion of a large traditional reporter impossible. NLuc, with its small 19kDa size, overcomes this obstacle and made the first in vivo tracking of influenza spread possible in a murine model by the generation of a reporter virus encoding a PA-NLuc fusion.56 An increase in the flux produced by the NLuc virus was seen as the mouse's weight decreased and the infection became more advanced. As ferrets are generally accepted as the closest models to the human immune system, Karlsson et al. demonstrated the capabilities of NLuc to image influenza spread within this model as well.57 Bioluminescence from the nasal passages was significantly correlated with the viral titers and was easily visible on all scans of infected ferrets. Respiratory tract infection was much less readily seen, with the presence of overlying tissue reducing the flux from the lungs by a factor of up to 4.5.
4. Improvements and limitations of the NanoLuc technology
Nearly all of the applications discussed can use other luciferase systems, so what advantages does the NLuc system provide over traditional luciferase systems? The benefits of the NLuc platform include its enhanced brightness, thermal stability, pH stability, and unbiased distribution in cells. Also, NLuc does not require post-translational modifications in mammalian cells, unlike other reporter genes like green fluorescent protein (GFP).58 These advantages have made it possible to study biology under more physiologically relevant conditions where reporters can be expressed at low levels.41,59 Furthermore, NLuc has shown specific advantages in several applications. More specifically, the enhanced brightness of NLuc and ability to assay live cells has made cellular imaging using luminescence microscopy more feasible. While this area has been dominated by confocal microscopy with fluorescent proteins (xFP), NanoLuc allows for excellent macroscopic detection and cellular imaging to confirm and study biology.28 In addition, the NLuc has improved upon previous similar technologies as NanoBiT and NanoBRET can be used for in vitro and in vivo assays that can reversibly measure the interactions of proteins.60 Since these systems are reversible, they can also be used to monitor the displacement of interacting proteins when incubated with competitive inhibitors.
When considering the luciferase platforms currently available on the market, GLuc has the most similarities with NLuc. To review, both of these luciferase systems and others (e.g. RLuc) are derived from marine species, thus, these enzymes are normally smaller than the luciferase enzymes from other organisms (Table 1). Also, the luciferase enzymes derived from marine animals are ATP-independent and utilize coelenterazine as their substrate, except for NLuc. Several studies have shown that coelenterazine is less stable, less soluble, more toxic, and more expensive than luciferin. Also, GLuc is a secreted luciferase, thus, it is limited in potential applications, whereas both non-secreted and secreted NLuc platforms have been established. As reported by Hall et al., GLuc suffers from the rapid decay of light intensity.4 Under most conditions, light produced from the GLuc reaction rapidly decays, requiring illuminometers equipped with injectors for measurements. NLuc can be tagged with the PEST degradation signal, which allows for rapid changes in luciferase expression that is longer lasting in response to transcriptional dynamics. While there are no reports of GLuc-PEST at this time, GLuc and other luciferases could be regulated in a similar manner. There have been significant efforts to improve upon these limitations, as scientists seek out novel and genetically manipulated luciferase systems.
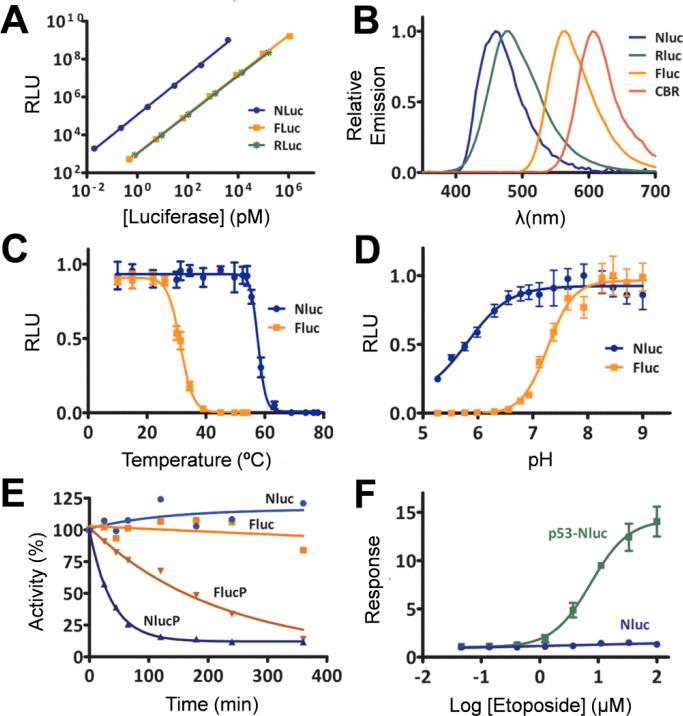
Development of novel luciferase systems is not a simple task, as the optimal situation requires that the enzyme is small, monomeric, and highly stable in both acidic/basic conditions and at high/low temperatures. It was the genetic manipulation of OLuc-19 that resulted in the formation of the NLuc system. In combination with the enzyme-specific substrate furimazine, this novel technology surpassed most expectations, as it provided a highly versatile luciferase platform for numerous bioluminescence applications. During synthesis of NLuc, several studies were performed to compare several luciferases regarding bioluminescence brightness (Figure 9A), maximum wavelength emission (Figure 9B), temperature stability (Figure 9C), pH stability (Figure 9D).4 Also, the destabilization properties were determined for NLucP and were compared to the destabilized form of FLuc, showing faster response rates (Figure 9E). Lastly, cells expressing p53-NLuc showed that it was possible to monitor the accumulation of p53 in cells after treatment with etoposide, with increasing etoposide concentration resulting in higher signal (Figure 9F).
Figure 9.
Comparison of luminescence intensity of NLuc, FLuc, and RLuc at 10 min. Spectral profile of NLuc, RLuc, FLuc, and click beetle red luciferase (CBR), showing a broad range of emission peaks. NLuc and FLuc were compared for sensitivity to (C) temperature and (D) pH. (E) Luminescence measured over time after treatment with cycloheximide to inhibit protein synthesis, showing that NLuc-PEST (NLucP) signal quickly decreased, followed by FLucP. (F) NLuc was used to investigate regulated changes in p53 stability, by expressing p53 –NLuc in HEK293 cells. The addition of etoposide caused an accumulation of p53, which could be measured with furimazine. Adapted with permission from ref. 4. Copyright 2012 American Chemical Society.
While NLuc has several advantages over traditional luciferase systems, this system has some limitations that may need to be addressed for further implementation of this technology. The first limitation of the NLuc platform is its emission spectra, which is not optimal for in vivo investigations. The spectral profile of NLuc exhibits an emission maximum of 460 nm, which is approximately 20 and 25 nm blue-shifted relative to RLuc and GLuc, respectively. Several luciferase platforms have been genetically manipulated to shift their maximum emission spectra to the red region, including RLuc8 (520 nm) and another genetically-modified RLuc known as Green-RLuc (560 nm). In addition, the click beetle luciferase platform is well known for its red-shifted spectral emission at 610 nm. By shifting these luciferase systems to the red spectral region, their potential applications broaden as in vivo use becomes available. While still not optimal for in vivo imaging, which works best in the NIR spectral region, these are significant improvements in the field of bioluminescence imaging. As NLuc is not an optimal candidate for in vivo imaging, several studies described in Section 3 utilized the BRET capabilities of NLuc to effectively overcome this limitation. Another potential benefit that should be considered is the multiplexing capacity of the NLuc platform with other luciferase systems, allowing for dual-luciferase assays with spectral emission profiles adequately separated for greater dynamic range and improved sensitivity.
Another potential limitation of the NLuc platform is the requirement for furimazine, a substrate that is specific for only NLuc and is not generically available. Thus, the cost of the NLuc platform is higher than traditional luciferase systems, which may effectively limit its availability to many laboratories. As the system becomes well established and more commonly utilized in the scientific community, we believe the cost will become less of a hindrance. With these concepts in mind, the use of NLuc is continually growing in the biomedical field. For example, a recent protocol was published in Nature Methods describing how to monitor ligand binding to GPCRs using the NLuc platform.61 In this protocol, the authors provided a direct comparison between NLuc and RLuc8 and showed that NLuc produced substantially greater bioluminescence signal than RLuc8 in mammalian cells. Also, NLuc exhibited no adverse effects on normal activity or cellular tracking of the receptor, while RLuc8 caused alterations in the trafficking of the receptor. Direct comparisons between the luciferase systems are invaluable, as they provide researchers with the facts required to make an informed decision on which luciferase system may be best suited for a particular application.
5. Conclusions and future perspectives
The field of bioluminescence is continually growing as novel luciferases are being developed. As NLuc is the newest member of the luciferase family, researchers are still investigating the potential uses of the system. In this review, several applications of the NLuc technology were discussed, including the investigation of protein – protein and protein – ligand interactions. While several studies have utilized NLuc for this application, there is still a need for new high-throughput screening assays. While NLuc showed promise for this purpose, it remains difficult to monitor more than one interaction occurring in vitro or in vivo. Since there are thousands of interactions occurring every minute in cells, the development of novel bioluminescent multiplexing techniques will significantly improve our understanding of disease progression and therapeutic intervention.
NLuc was also shown to be an effective tool for monitoring cell signaling and gene regulation. As gene delivery is a central theme for molecular biologists, the NLuc system may be further extended to monitor the success of gene delivery in vivo. While NLuc is currently limited by its blue spectral emission, scientists may be able to genetically manipulate the enzyme to produce a far-red shifted NLuc in the future that maintains its enhanced stability and superior brightness. Next, NLuc was used to monitor protein stability; yet, NLuc has not been extensively investigated for this application. As several diseases are directly linked to protein aggregation, such as Alzheimer's, the NLuc system may hold future promise in detecting novel protein instability pathways. The fourth application for NLuc was its use as BRET-based biosensors.
In addition to the applications mentioned here, the versatility of NLuc extends beyond mammals. Several studies utilized NLuc for monitoring viruses and bacteria. As bioluminescence assays are relatively fast and inexpensive, future studies may employ NLuc-based assays for detecting or monitoring certain infections in patient samples. Also, the enhanced brightness of NLuc makes it feasible to detect diseases early, including HIV or some bacterial infections.
As the NLuc technology is still new, many researchers remain cautious about switching from traditional luciferase systems. In some cases, NLuc may not be an optimal choice for specific applications. However, NLuc may be an excellent alternative for those researchers with studies that have been halted due to low bioluminescence signal. As the field of bioluminescence continues to grow, the development of novel luciferases will continue to expand the range of possibilities.
Acknowledgements
This work was supported, in part, by the University of Wisconsin - Madison, the National Institutes of Health (NIBIB/NCI 1R01CA169365, P30CA014520, T32CA009206, and T32GM008505), and the American Cancer Society (125246-RSG-13-099-01-CCE).
References
- 1.Widder EA, Falls B. Review of Bioluminescence for Engineers and Scientists in Biophotonics. IEEE J. Sel. Top. Quantum Electron. 2014;20 [Google Scholar]
- 2.Roda A. Chemiluminescence and Bioluminescence: Past, Present and Future. The Royal Society of Chemistry; 2011. hapter 1 A History of Bioluminescence and Chemiluminescence from Ancient Times to the Present; pp. 1–50. [Google Scholar]
- 3.Cui B, Zhang L, Song Y, Wei J, Li C, Wang T, Wang Y, Zhao T, Shen X. Engineering an enhanced, thermostable, monomeric bacterial luciferase gene as a reporter in plant protoplasts. PLoS One. 2014;9:e107885. doi: 10.1371/journal.pone.0107885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, Otto P, Zimmerman K, Vidugiris G, Machleidt T, et al. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem. Biol. 2012;7:1848–1857. doi: 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McElroy WD. The Energy Source for Bioluminescence in an Isolated System. Proc. Natl. Acad. Sci. U. S. A. 1947;33:342–345. doi: 10.1073/pnas.33.11.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McElroy WD, Ballentine R. The Mechanism of Bioluminescence. Proc. Natl. Acad. Sci. U. S. A. 1944;30:377–382. doi: 10.1073/pnas.30.12.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee J. Bioluminescence: the First 3000 Years. J. Sib. Fed. Univ., Chem. 2008;3:194–205. [Google Scholar]
- 8.Fraga H. Firefly luminescence: a historical perspective and recent developments. Photochem Photobiol Sci. 2008;7:146–158. doi: 10.1039/b719181b. [DOI] [PubMed] [Google Scholar]
- 9.Shimomura O. Discovery of green fluorescent protein. Methods Biochem. Anal. 2006;47:1–13. [PubMed] [Google Scholar]
- 10.Kricka LJ, Leach FR. In memoriam Dr Marlene DeLuca. 1987 O. M. Smith Lecture. Firefly luciferase: mechanism of action, cloning and expression of the active enzyme. J. Biolumin. Chemilumin. 1989;3:1–5. doi: 10.1002/bio.1170030102. [DOI] [PubMed] [Google Scholar]
- 11.Yamaguchi I. Oplophorus oxyluciferin and a model luciferin compound biologically active with Oplophorus luciferase. Biochem. J. 1975;151:9–15. doi: 10.1042/bj1510009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matthews JC, Hori K, Cormier MJ. Purification and properties of Renilla reniformis luciferase. Biochemistry. 1977;16:85–91. doi: 10.1021/bi00620a014. [DOI] [PubMed] [Google Scholar]
- 13.Villalobos V, Naik S, Bruinsma M, Dothager RS, Pan MH, Samrakandi M, Moss B, Elhammali A, Piwnica-Worms D. Dual-color click beetle luciferase heteroprotein fragment complementation assays. Chem. Biol. 2010;17:1018–1029. doi: 10.1016/j.chembiol.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McNabb DS, Reed R, Marciniak RA. Dual luciferase assay system for rapid assessment of gene expression in Saccharomyces cerevisiae. Eukaryot. Cell. 2005;4:1539–1549. doi: 10.1128/EC.4.9.1539-1549.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimomura O, Masugi T, Johnson FH, Haneda Y. Properties and reaction mechanism of the bioluminescence system of the deep-sea shrimp Oplophorus gracilorostris. Biochemistry. 1978;17:994–998. doi: 10.1021/bi00599a008. [DOI] [PubMed] [Google Scholar]
- 16.Mccapra F. Chemical Mechanisms in Bioluminescence. Acc. Chem. Res. 1976;9:201–208. [Google Scholar]
- 17.Rosano GL, Ceccarelli EA. Recombinant protein expression in Escherichia coli: advances and challenges. Front. Microbiol. 2014;5:172. doi: 10.3389/fmicb.2014.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gill DR, Pringle IA, Hyde SC. Progress and prospects: the design and production of plasmid vectors. Gene Ther. 2009;16:165–171. doi: 10.1038/gt.2008.183. [DOI] [PubMed] [Google Scholar]
- 19.Promega, editor. Promega Corporation; Madison, WI: 2016. URL: https://www.promega.com/products/reporter-assays-and-transfection/reporter-assays/nanoluc-luciferase-redefining-reporter-assays/ [Google Scholar]
- 20.Machleidt T, Woodroofe CC, Schwinn MK, Mendez J, Robers MB, Zimmerman K, Otto P, Daniels DL, Kirkland TA, Wood KV. NanoBRET--A Novel BRET Platform for the Analysis of Protein-Protein Interactions. ACS Chem. Biol. 2015;10:1797–1804. doi: 10.1021/acschembio.5b00143. [DOI] [PubMed] [Google Scholar]
- 21.Bradley WD, Arora S, Busby J, Balasubramanian S, Gehling VS, Nasveschuk CG, Vaswani RG, Yuan CC, Hatton C, Zhao F, et al. EZH2 inhibitor efficacy in non-Hodgkin's lymphoma does not require suppression of H3K27 monomethylation. Chem. Biol. 2014;21:1463–1475. doi: 10.1016/j.chembiol.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 22.Robers M, Eggers C, Binkowski B, Hartnett J, Wilkinson J, Zimprich C, Stecha P, Cong M. Protein Stability Exposes Cellular Stress: Measure Dynamic Protein Regulation with a Luminescent Reporter. Drug Discovery Tutorial. 2014;34 [Google Scholar]
- 23.Song G, Wu QP, Xu T, Liu YL, Xu ZG, Zhang SF, Guo ZY. Quick preparation of nanoluciferase-based tracers for novel bioluminescent receptor-binding assays of protein hormones: Using erythropoietin as a model. J. Photochem. Photobiol. B. 2015;153:311–316. doi: 10.1016/j.jphotobiol.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 24.Verhoef LG, Mattioli M, Ricci F, Li YC, Wade M. Multiplex detection of protein-protein interactions using a next generation luciferase reporter. Biochim. Biophys. Acta. 2016;1863:284–292. doi: 10.1016/j.bbamcr.2015.11.031. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y, Wang L, Cheng X, Ge X, Wang P. An ultrasensitive system for measuring the USPs and OTULIN activity using Nanoluc as a reporter. Biochem. Biophys. Res. Commun. 2014;455:178–183. doi: 10.1016/j.bbrc.2014.10.139. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, Song G, Shao XX, Liu YL, Guo ZY. Quantitative measurement of cell membrane receptor internalization by the nanoluciferase reporter: Using the G protein-coupled receptor RXFP3 as a model. Biochim. Biophys. Acta. 2015;1848:688–694. doi: 10.1016/j.bbamem.2014.11.026. [DOI] [PubMed] [Google Scholar]
- 27.Wu QP, Zhang L, Shao XX, Wang JH, Gao Y, Xu ZG, Liu YL, Guo ZY. Application of the novel bioluminescent ligand-receptor binding assay to relaxin-RXFP1 system for interaction studies. Amino Acids. 2016;48:1099–1107. doi: 10.1007/s00726-015-2146-3. [DOI] [PubMed] [Google Scholar]
- 28.Dixon AS, Schwinn MK, Hall MP, Zimmerman K, Otto P, Lubben TH, Butler BL, Binkowski BF, Machleidt T, Kirkland TA, et al. NanoLuc Complementation Reporter Optimized for Accurate Measurement of Protein Interactions in Cells. ACS Chem. Biol. 2016;11:400–408. doi: 10.1021/acschembio.5b00753. [DOI] [PubMed] [Google Scholar]
- 29.Song G, Jiang Q, Xu T, Liu YL, Xu ZG, Guo ZY. A convenient luminescence assay of ferroportin internalization to study its interaction with hepcidin. FEBS J. 2013;280:1773–1781. doi: 10.1111/febs.12192. [DOI] [PubMed] [Google Scholar]
- 30.Zhang L, Song G, Xu T, Wu QP, Shao XX, Liu YL, Xu ZG, Guo ZY. A novel ultrasensitive bioluminescent receptor-binding assay of INSL3 through chemical conjugation with nanoluciferase. Biochimie. 2013;95:2454–2459. doi: 10.1016/j.biochi.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 31.He SX, Song G, Shi JP, Guo YQ, Guo ZY. Nanoluciferase as a novel quantitative protein fusion tag: Application for overexpression and bioluminescent receptor-binding assays of human leukemia inhibitory factor. Biochimie. 2014;106:140–148. doi: 10.1016/j.biochi.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 32.Hikiji T, Norisada J, Hirata Y, Okuda K, Nagasawa H, Ishigaki S, Sobue G, Kiuchi K, Oh-hashi K. A highly sensitive assay of IRE1 activity using the small luciferase NanoLuc: Evaluation of ALS-related genetic and pathological factors. Biochem. Biophys. Res. Commun. 2015;463:881–887. doi: 10.1016/j.bbrc.2015.05.132. [DOI] [PubMed] [Google Scholar]
- 33.Norisada J, Hirata Y, Amaya F, Kiuchi K, Oh-hashi K. A sensitive assay for the biosynthesis and secretion of MANF using NanoLuc activity. Biochem. Biophys. Res. Commun. 2014;449:483–489. doi: 10.1016/j.bbrc.2014.05.031. [DOI] [PubMed] [Google Scholar]
- 34.Seay K, Khajoueinejad N, Zheng JH, Kiser P, Ochsenbauer C, Kappes JC, Herold B, Goldstein H. The Vaginal Acquisition and Dissemination of HIV-1 Infection in a Novel Transgenic Mouse Model Is Facilitated by Coinfection with Herpes Simplex Virus 2 and Is Inhibited by Microbicide Treatment. J. Virol. 2015;89:9559–9570. doi: 10.1128/JVI.01326-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas T, Seay K, Zheng JH, Zhang C, Ochsenbauer C, Kappes JC, Goldstein H. High-Throughput Humanized Mouse Models for Evaluation of HIV-1 Therapeutics and Pathogenesis. Methods Mol. Biol. 2016;1354:221–235. doi: 10.1007/978-1-4939-3046-3_15. [DOI] [PubMed] [Google Scholar]
- 36.Nishitsuji H, Ujino S, Shimizu Y, Harada K, Zhang J, Sugiyama M, Mizokami M, Shimotohno K. Novel reporter system to monitor early stages of the hepatitis B virus life cycle. Cancer Sci. 2015;106:1616–1624. doi: 10.1111/cas.12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.World Health Organization; 2015. URL: http://www.who.int/topics/malaria/en/ [Google Scholar]
- 38.Azevedo MF, Nie CQ, Elsworth B, Charnaud SC, Sanders PR, Crabb BS, Gilson PR. Plasmodium falciparum transfected with ultra bright NanoLuc luciferase offers high sensitivity detection for the screening of growth and cellular trafficking inhibitors. PLoS One. 2014;9:e112571. doi: 10.1371/journal.pone.0112571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loh JM, Proft T. Comparison of firefly luciferase and NanoLuc luciferase for biophotonic labeling of group A Streptococcus. Biotechnol. Lett. 2014;36:829–834. doi: 10.1007/s10529-013-1423-z. [DOI] [PubMed] [Google Scholar]
- 40.Yasuzaki Y, Yamada Y, Ishikawa T, Harashima H. Validation of Mitochondrial Gene Delivery in Liver and Skeletal Muscle via Hydrodynamic Injection Using an Artificial Mitochondrial Reporter DNA Vector. Mol. Pharm. 2015;12:4311–4320. doi: 10.1021/acs.molpharmaceut.5b00511. [DOI] [PubMed] [Google Scholar]
- 41.Lackner DH, Carre A, Guzzardo PM, Banning C, Mangena R, Henley T, Oberndorfer S, Gapp BV, Nijman SM, Brummelkamp TR, et al. A generic strategy for CRISPR-Cas9-mediated gene tagging. Nat. Commun. 2015;6:10237. doi: 10.1038/ncomms10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao J, Nelson TJ, Vu Q, Truong T, Stains CI. Self-Assembling NanoLuc Luciferase Fragments as Probes for Protein Aggregation in Living Cells. ACS Chem. Biol. 2016;11:132–138. doi: 10.1021/acschembio.5b00758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Y, Piston DW, Johnson CH. A bioluminescence resonance energy transfer (BRET) system: application to interacting circadian clock proteins. Proc. Natl. Acad. Sci. U. S. A. 1999;96:151–156. doi: 10.1073/pnas.96.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lohse MJ, Nuber S, Hoffmann C. Fluorescence/bioluminescence resonance energy transfer techniques to study G-protein-coupled receptor activation and signaling. Pharmacol. Rev. 2012;64:299–336. doi: 10.1124/pr.110.004309. [DOI] [PubMed] [Google Scholar]
- 45.Sapsford KE, Berti L, Medintz IL. Materials for fluorescence resonance energy transfer analysis: beyond traditional donor-acceptor combinations. Angew. Chem. Int. Ed. Engl. 2006;45:4562–4589. doi: 10.1002/anie.200503873. [DOI] [PubMed] [Google Scholar]
- 46.Loening AM, Wu AM, Gambhir SS. Red-shifted Renilla reniformis luciferase variants for imaging in living subjects. Nat. Methods. 2007;4:641–643. doi: 10.1038/nmeth1070. [DOI] [PubMed] [Google Scholar]
- 47.England CG, Luo H, Cai W. HaloTag technology: a versatile platform for biomedical applications. Bioconjug. Chem. 2015;26:975–986. doi: 10.1021/acs.bioconjchem.5b00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robers MB, Dart ML, Woodroofe CC, Zimprich CA, Kirkland TA, Machleidt T, Kupcho KR, Levin S, Hartnett JR, Zimmerman K, et al. Target engagement and drug residence time can be observed in living cells with BRET. Nat. Commun. 2015;6:10091. doi: 10.1038/ncomms10091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schaub FX, Reza MS, Flaveny CA, Li W, Musicant AM, Hoxha S, Guo M, Cleveland JL, Amelio AL. Fluorophore-NanoLuc BRET Reporters Enable Sensitive In Vivo Optical Imaging and Flow Cytometry for Monitoring Tumorigenesis. Cancer Res. 2015;75:5023–5033. doi: 10.1158/0008-5472.CAN-14-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shigeto H, Ikeda T, Kuroda A, Funabashi H. A BRET-based homogeneous insulin assay using interacting domains in the primary binding site of the insulin receptor. Anal. Chem. 2015;87:2764–2770. doi: 10.1021/ac504063x. [DOI] [PubMed] [Google Scholar]
- 51.O'Neill K, Lyons SK, Gallagher WM, Curran KM, Byrne AT. Bioluminescent imaging: a critical tool in pre-clinical oncology research. J. Pathol. 2010;220:317–327. doi: 10.1002/path.2656. [DOI] [PubMed] [Google Scholar]
- 52.Stacer AC, Nyati S, Moudgil P, Iyengar R, Luker KE, Rehemtulla A, Luker GD. NanoLuc reporter for dual luciferase imaging in living animals. Mol. Imaging. 2013;12:1–13. [PMC free article] [PubMed] [Google Scholar]
- 53.Germain-Genevois C, Garandeau O, Couillaud F. Detection of Brain Tumors and Systemic Metastases Using NanoLuc and Fluc for Dual Reporter Imaging. Mol. Imaging Biol. 2016;18:62–69. doi: 10.1007/s11307-015-0864-2. [DOI] [PubMed] [Google Scholar]
- 54.Sun C, Gardner CL, Watson AM, Ryman KD, Klimstra WB. Stable, high-level expression of reporter proteins from improved alphavirus expression vectors to track replication and dissemination during encephalitic and arthritogenic disease. J. Virol. 2014;88:2035–2046. doi: 10.1128/JVI.02990-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heise K, Oppermann H, Meixensberger J, Gebhardt R, Gaunitz F. Dual luciferase assay for secreted luciferases based on Gaussia and NanoLuc. Assay Drug. Dev. Technol. 2013;11:244–252. doi: 10.1089/adt.2013.509. [DOI] [PubMed] [Google Scholar]
- 56.Tran V, Moser LA, Poole DS, Mehle A. Highly sensitive real-time in vivo imaging of an influenza reporter virus reveals dynamics of replication and spread. J. Virol. 2013;87:13321–13329. doi: 10.1128/JVI.02381-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karlsson EA, Meliopoulos VA, Savage C, Livingston B, Mehle A, Schultz-Cherry S. Visualizing real-time influenza virus infection, transmission and protection in ferrets. Nat. Commun. 2015;6:6378. doi: 10.1038/ncomms7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Coralli C, Cemazar M, Kanthou C, Tozer GM, Dachs GU. Limitations of the reporter green fluorescent protein under simulated tumor conditions. Cancer Res. 2001;61:4784–4790. [PubMed] [Google Scholar]
- 59.Hasson SA, Fogel AI, Wang C, MacArthur R, Guha R, Heman-Ackah S, Martin S, Youle RJ, Inglese J. Chemogenomic profiling of endogenous PARK2 expression using a genome-edited coincidence reporter. ACS Chem. Biol. 2015;10:1188–1197. doi: 10.1021/cb5010417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Demont EH, Bamborough P, Chung CW, Craggs PD, Fallon D, Gordon LJ, Grandi P, Hobbs CI, Hussain J, Jones EJ, et al. 1,3-Dimethyl Benzimidazolones Are Potent, Selective Inhibitors of the BRPF1 Bromodomain. ACS Med. Chem. Lett. 2014;5:1190–1195. doi: 10.1021/ml5002932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stoddart LA, Johnstone EK, Wheal AJ, Goulding J, Robers MB, Machleidt T, Wood KV, Hill SJ, Pfleger KD. Application of BRET to monitor ligand binding to GPCRs. Nat. Methods. 2015;12:661–663. doi: 10.1038/nmeth.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]