Abstract
Extraintestinal pathogenic Escherichia coli (ExPEC) causes a variety of acute infections in its hosts, and multidrug-resistant strains present significant challenges to public health and animal husbandry. Therefore, it is necessary to explore new drug targets to control E. coli epidemics. Previous studies have reported that ppk mutants of Burkholderia pseudomallei and Mycobacterium tuberculosis are more susceptible than the wild types (WTs) to stress. Therefore, we investigated the stress response to antibiotics mediated by polyphosphate kinase (PPK) in ExPEC strain PCN033. We observed that planktonic cells of a ppk knockout strain (Δppk) were more susceptible to antibiotics than was WT. However, biofilm-grown Δppk cells showed similar susceptibility to that of the WT and were more tolerant than the planktonic cells. During the planktonic lifestyle, the expression of genes involved in antibiotic tolerance (including resistance-conferring genes, and antibiotic influx, and efflux genes) did not change in the Δppk mutant without antibiotic treatment. However, the resistance-conferring gene bla and efflux genes were upregulated more in the WT than in the Δppk mutant by treatment with tazobactam. After treatment with gentamycin, the efflux genes and influx genes were upregulated and downregulated, respectively, more in the WT than in the Δppk mutant. The expression of genes involved in biofilm regulation also changed after treatment with tazobactam or gentamycin, and which is consistent with the results of the biofilm formation. Together, these observations indicate that PPK is important for the antibiotic stress response during the planktonic growth of ExPEC and might be a potential drug target in bacteria.
Keywords: ExPEC, antibiotic resistance, PPK, gene expression, RNA-seq, qPCR
Introduction
Extraintestinal pathogenic Escherichia coli (ExPEC) is a major cause of urinary tract infections in women, abdominal sepsis, and septicemia in elderly or immunocompromised individuals, and meningitis in newborns, with high morbidity and mortality (Gaschignard et al., 2011; Weston et al., 2011; Mellata, 2013). ExPEC strains commonly colonize domestic animals, such as pigs, chickens, and cattle, causing significant losses in animal husbandry and threating human health (Girardeau et al., 2003; Johnson et al., 2005; Bergeron et al., 2012). We previously investigated the prevalence of ExPEC in swine across China and detected ExPEC in 10.1% of porcine samples. The frequency of ExPEC isolated from pigs increased between 2004 and 2007 from 3.1 to 14.6% (Tan et al., 2012). The emergence of multidrug-resistant strains has significantly hindered the prevention and control of ExPEC epidemics (Sedláková et al., 2014; Sidrach-Cardona et al., 2014). Therefore, it is urgent that we identify new drug targets to control these increasing E. coli outbreaks.
Polyphosphate kinase (PPK) is an essential enzyme in polyphosphate (polyP) synthesis and has been implicated in many intracellular biological processes. Pseudomonas aeruginosa in which ppk was deleted showed impairments in motility, quorum sensing, and virulence (Rashid et al., 2000), compacted nucleoids, membrane distortion, extracellular polymer production, and a susceptibility to desiccation (Fraley et al., 2007). PPK also plays a prominent role in the stress response, and a Burkholderia pseudomallei ppk mutant was susceptible to hydrogen peroxide under oxidative stress conditions (Tunpiboonsak et al., 2010). A ppk1 mutant strain of Mycobacterium tuberculosis displayed a survival defect in response to nitrosative stress, and the negligible levels of polyP were associated with its increased susceptibility to certain tuberculosis drugs (Singh et al., 2013). PPK is highly conserved in bacteria, but is absent in higher mammals (Brown and Kornberg, 2004), indicating that PPK has potential utility as an antibacterial drug target.
As an opportunistic pathogen, E. coli mainly causes acute infections in immunocompromised individuals (Chaudhuri and Henderson, 2012; Mellata, 2013); further, acute infections are associated with its planktonic growth mode (Li et al., 2014). Therefore, we explored the role of PPK in antibiotic resistance in the planktonic cells of ExPEC strain PCN033. Biofilm formation contributes to chronic bacterial infections, such as the recurrent pyelonephritis caused by uropathogenic E. coli in children (Tapiainen et al., 2014). Therefore, we also studied the role of PPK in antibiotic tolerance in biofilm-grown cells.
Materials and methods
Bacterial strains and culture conditions
The wild-type (WT) strain used in this study, PCN033, was isolated from a diseased swine in Hubei Province, Central China (Liu et al., 2015). The Δppk mutant was obtained by in-frame deletion with the suicide plasmid pRE112 (He et al., 2012). Details of both these strains and the primers used in this study are listed in Table S1 (available as Supplementary Data). The antibiotic susceptibility and biofilm formation assays were performed in MOPS broth (an inorganic phosphorus [Pi]-limited medium) at 28°C (Neidhardt et al., 1974). The antibiotics used in the biofilm inhibition assay were added below the minimum inhibitory concentrations (MICs), and had no bactericidal effect on the planktonic cells.
Growth characteristics
Fresh colonies of both strains were taken from Luria–Bertani (LB) agar plates, used to inoculate LB broth, and then cultured in MOPS in a shaker incubator for 12 h. The growth characteristics were monitored turbidimetrically at 600 nm on a spectrophotometer (Eppendorf, Hamburg, Germany) and the colony-forming units (cfu) were counted at 1 h intervals. The generation times were calculated with the formula (Penfold and Norris, 1912):
where G is the generation time; T is the length of the logarithmic phase; a is the initial number of bacteria; and b is the final number of bacteria.
Susceptibility assay
Each MIC was determined with a series of two-fold dilutions of the antibiotic in MOPS broth, according to the Clinical Laboratory and Standards Institute guidelines. A pre-grown inoculum of each strain was diluted in MOPS to a final concentration of 107 cfu/mL, and the concentration of antibiotic added varied from 0.25 to 512 mg/L. The plates were incubated for 24 h, and the MICs were determined as the lowest antibiotic concentrations that produced no visible growth.
The susceptibility assay of the biofilms was performed as described previously (Benthall et al., 2015), with some modifications. The MIC on the biofilm was determined by allowing a biofilm to form in a 96-well-plate for 24 h. The unattached cells were washed off three times with 0.9% saline. The biofilm was incubated for 24 h with a range of antibiotic concentrations from 512 to 0.25 mg/L. The MIC was defined as the lowest antibiotic concentration at which no bacterial growth was detected. To determine the effect of the biofilm on the bacterial susceptibility to antibiotics, the viability of planktonic and biofilm-grown cells was calculated after the antibiotic treatments. About 107 cfu were incubated with antibiotic concentrations of 2 × MIC for 3 h, and the cfu were then counted.
To quantify the bactericidal activity of these antibiotics on the biofilm, their activity percentage was assessed according to a previous report (Sánchez-Gómez et al., 2015), with some modifications. The ability of these antibiotics to remove the biofilm attached to the microplate was determined with crystal violet (CV) staining. For this purpose, the treated biofilm was stained with CV for 30 min at room temperature. The excess stain was then rinsed off with saline and the CV remaining on the biofilm was dissolved in 33% acetic acid. The absorbance was measured at 595 nm with a Synergy HT microplate reader (BioTek, USA). The activity percentage was calculated according to Sánchez-Gómez et al., with the formula:
where C is the absorbance of the control well-containing untreated biofilm; T is the absorbance of the well-containing treated biofilm; and B is the absorbance of the blank well (i.e., no biofilm).
Biofilm formation assay
Static cultures of biofilm grown at 28°C were analyzed in flat-bottom 96-well-microtiter plates (Corning, USA) using CV. Briefly, approximately 107 cfu were inoculated with sub-MIC antibiotics and cultured for 24 h. The unattached cells were then washed off as described above, fixed with absolute ethanol for 30 min, dried, and stained with 0.1% CV solution for 1 h. The microplates were then washed three times to remove any unattached CV. The CV in the stained biofilm was then dissolved in 33% acetic acid solution and the absorbance read at 595 nm. Each experiment was repeated twice with three technical replicates.
RNA-seq assay and quantitative real-time (qRT)–PCR validation
RNA samples from each strain were prepared for RNA sequencing. Sequencing was performed on an Illumina Hiseq 2500 sequencer (Illumine Inc.) by Shanghai Hanyu Biotechnology Co., Ltd (Shanghai, China). The RNA-seq results were confirmed with qRT–PCR. Before qRT-PCR, the RNA of both strains was extracted with RNAiso Plus reagent (Takara, China). Any genomic DNA contamination was eliminated, and the RNA was reverse transcribed to cDNA with the PrimeScript™RT reagent Kit with gDNA Eraser (Takara). Quantitative real-time PCR was performed in triplicate in optical 96-well-reaction plates (Life Technologies, China) using Power SYBR Green PCR Master Mix (Life Technologies). The primers are listed in Table S1. The mRNA levels of the target genes were normalized to the internal 16S rRNA control with the ΔΔCt method (Kubista et al., 2006). The planktonic cells of the WT and Δppk mutant were cultured to an optical density at a wavelength of 600 nm (OD600) of about 0.5. Tazobactam or gentamycin was added at a concentration of 2 × MIC and the cells were incubated for 2 h. The cells were then collected for RNA extraction and qRT–PCR.
Statistical analysis
Statistical analysis was performed with the SPSS software (SPSS, Inc., Chicago, IL, USA) on a Windows XP system. Biofilm formation, differential expression, and generation times were compared with one-way analysis of variance (ANOVA). Values are expressed as means ± SD, and statistically significant differences are marked with asterisks. To identify the levels of gene expression, the RNA-seq data were analyzed with an MA-plot-based method with the random sampling model (MARS) in the DEGseq software (http://www.bioconductor.org/packages/release/bioc/html/DESeq.html). Genes showing two-fold changes in expression, a false discovery rate < 0.001, and reads per kilobase per million >20 in at least one sample were considered to be differentially expressed under the conditions used.
Results
Growth characteristics
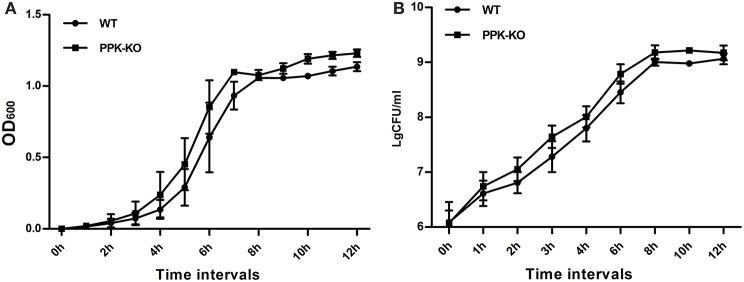
In MOPS minimal medium, the density of the Δppk cultures was slightly higher than that of the WT at each time point tested (Figure 1). However, the generation times, calculated from both the optical density and cfu, for Δppk (GOD = 56.5 ± 4.73, GCFU = 42.0 ± 1.88) were not significantly different (P > 0.05) from those for WT (GOD = 56.3 ± 3.64, Gcfu = 45.9 ± 4.35).
Figure 1.
Growth characteristics of E. coli strains PCN033 (wild type) and Δppk in MOPS broth: (A) optical density, and (B) colony-forming units.
Planktonic Δppkcells are more susceptible to antibiotics than WT cells
As shown in Table 1, the 17 antibiotics screened in this study were categorized based on their targets: cell wall biosynthesis (type A), protein biosynthesis (type B), nucleotide metabolism (type C), and cell membrane (type D). There were no significant differences in the MIC values for the Δppk strain and WT in LB broth. However, in MOPS broth, Δppk was more susceptible to antibiotics than WT. Specifically, Δppk was much more susceptible to type B antibiotics than was WT, followed by type A antibiotics. Because the WT strain can accumulate more polyP in MOPS broth than in other medi (Ault-Riché et al., 1998), we performed all further assays in MOPS.
Table 1.
Assay of the susceptibility of planktonic cells to antibiotics (mg/L).
| Target (type) | antibiotic category | compound | MIC in LB | MIC change (fold) | MIC in MOPS | MIC fold (fold) | References | ||
|---|---|---|---|---|---|---|---|---|---|
| PCN033 | Δppk | PCN033 | Δppk | ||||||
| Cell wall biosynthesis(A) | β-lactams | cefotaxime | 2 | 1 | 2 | 16 | 4 | 4 | Bush, 2012 |
| ceftazidime | 256 | 256 | 1 | 512 | 512 | 1 | |||
| ampicillin | >512 | >512 | 1 | >512 | >512 | 1 | |||
| cefazolin | >512 | >512 | 1 | >512 | >512 | 1 | |||
| tazobactam | >512 | >512 | 1 | >512 | 256 | >2 | |||
| ticarcillin | >512 | >512 | 1 | >512 | >512 | 1 | |||
| Glycopeptide | vancomycin | >512 | >512 | 1 | 128 | 64 | 2 | ||
| Protein biosynthesis (B) | Aminoglycosides | gentamicin | >512 | >512 | 1 | 256 | 32 | 8 | Davis, 1987 |
| gentamicin sulfate | >512 | >512 | 1 | 256 | 64 | 4 | |||
| amikacin | >512 | >512 | 1 | >512 | 64 | ≥4 | |||
| Macrolide | Erythromycin | 512 | 512 | 1 | 512 | 128 | 4 | Brisson-Noël et al., 1988 | |
| Nucleotide metabolism (C) | Quinolones | norfloxacin | >512 | 512 | >1 | 512 | 512 | 1 | Aldred et al., 2014 |
| levofloxacin | 128 | 64 | 2 | 64 | 16 | 4 | |||
| Sulfonamides | Trimethoprim | 128 | 128 | 1 | 256 | 128 | 2 | Pérez-Trallero and Iglesias, 2003 | |
| sulfadiazine | >512 | >512 | >1 | >512 | >512 | 1 | |||
| Nitrofurans | macrodantin | 512 | 256 | 2 | 128 | 128 | 1 | Hof, 1988 | |
| Membrane (D) | Lipopeptde | polymyxin B | 0.5 | ≤ 0.25 | ≥2 | 2 | 1 | 2 | Grau-Campistany et al., 2015 |
Biofilm-grown Δppk and WT cells are similarly tolerant and more tolerant than planktonic cells
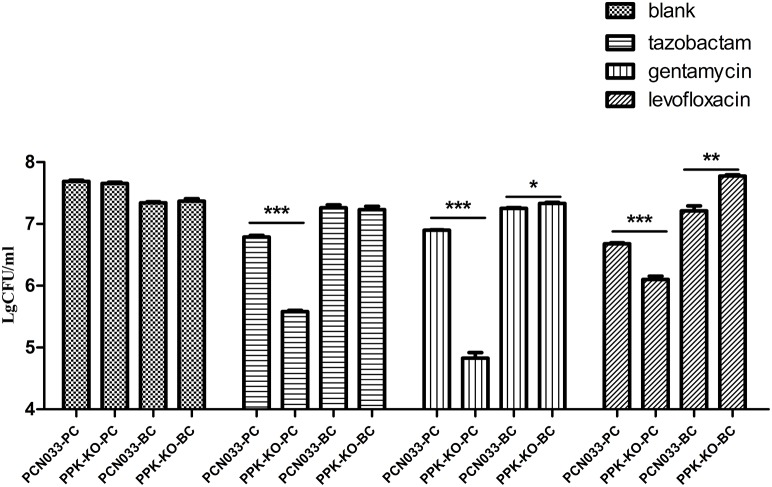
To clarify the role of biofilms in antibiotic resistance, an MIC assay using biofilm-grown cells was performed as described previously (Benthall et al., 2015). As shown in Table 2, biofilm-grown Δppk cells showed almost no difference from WT cells in their antibiotic susceptibility, and biofilm-grown cells of both strains were more tolerant than the corresponding planktonic cells. Consistent with this, there was no significant difference in the ability of antimicrobial compounds to kill biofilm-grown cells of the WT and Δppk strains (Figure S1). However, more planktonic cells of the Δppk strain were killed than WT cells (Figure 2). In a future study, we will investigate the role of PPK in antibiotic tolerance within the planktonic growth mode.
Table 2.
Assay of the susceptibility of biofilm-grown cells to antibiotics (mg/L).
| Target (type) | antibiotic category | compound | MIC for biofilm | MIC change (fold) | |
|---|---|---|---|---|---|
| PCN033 | Δppk | ||||
| Cell wall biosynthesis (A) | β-lactams | cefotaxime | 128 | 256 | 0.5 |
| ceftazidime | 512 | 512 | 1 | ||
| ampicillin | >512 | >512 | 1 | ||
| cefazolin | >512 | >512 | 1 | ||
| tazobactam | 512 | >512 | ≤ 0.5 | ||
| ticarcillin | >512 | >512 | 1 | ||
| Protein biosynthesis (B) | Aminoglycosides | gentamicin | 512 | 512 | 1 |
| gentamicin sulfate | 512 | 256 | 2 | ||
| amikacin | 512 | 512 | 1 | ||
| Macrolide | erythromycin | 512 | 512 | 1 | |
| Quinolones | norfloxacin | 512 | 512 | 1 | |
| levofloxacin | 32 | 32 | 1 | ||
| Nucleotide metabolism (C) | Sulfonamides | Trimethoprim | 512 | 512 | 1 |
| sulfadiazine | >512 | >512 | 1 | ||
| Nitrofurans | macrodantin | 512 | 512 | 1 | |
| Membrane (D) | Lipopeptde | polymyxin B | 4 | 4 | 1 |
Figure 2.
Antibiotic killing assay of planktonic and biofilm-grown cells. PCN033-PC indicates planktonic WT cells; PPK-KO-PC indicates planktonic cells of the Δppk mutant; PCN033-BC indicates biofilm-grown WT cells; PPK-KO-BC indicates biofilm-grown cells of the Δppk mutant; blank indicates no antibiotic treatment. ***p < 0.000, **p < 0.01, *p < 0.05.
Expression of antibiotic-resistance genes without antibiotic treatment
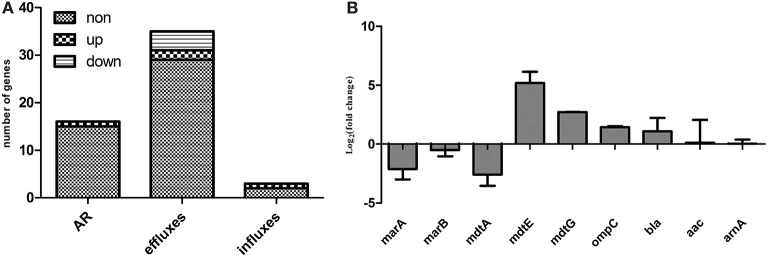
RNA-seq data for the Δppk and WT strains regarding the expression of genes involved in antibiotic resistance, including resistance-conferring genes and antibiotic efflux and influx genes, are presented in Table S2. Of 53 genes known to be involved in antibiotic resistance or multidrug resistance, the expression of one resistance-conferring gene (tetB), two efflux genes (mdtE and mdtG), and one influx gene (ompC) was upregulated, and the expression of three efflux genes (marA, marB, and mdtA) was downregulated in the mutant compared with their expression in WT (Figure 3A). The expression of some of these genes was confirmed with qRT–PCR (Figure 3B).
Figure 3.
Expression levels of genes involved in antibiotic resistance, antibiotic efflux, or antibiotic influx. (A) Expression levels determined with RNA-seq; (B) qRT–PCR confirmation. AR indicates genes that confer resistance, including beta-lactamase bla, aminoglycoside 3′N-acetyltransferase III (aac) etc.; efflux genes include acrAB–tolC, acrDEF, cusCFBA, emrAB, emrKY, mdtABCEF, etc.; and influx genes include ompC, ompF, and phoE, details are available in Table S2.
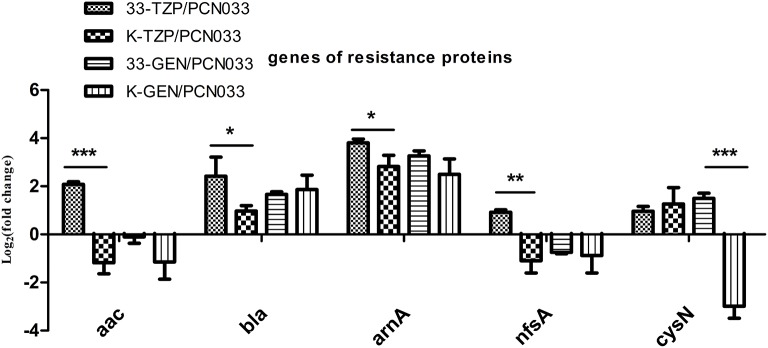
Expression of resistance-conferring genes after antibiotic treatment
Five resistance-conferring genes were selected for analysis when WT was treated with tazobactam, as shown in Figure 4. The expression of beta-lactamase (bla) was upregulated, as was that of aminoglycoside 3′N-acetyltransferase III (aac), fused UDP-L-Ara4N formyltransferase (arnA), and nitroreductase A (nfsA). The bla, arnA, and sulfate adenylyltransferase (cysN) genes were also upregulated in the Δppk mutant after tazobactam treatment, but their expression was higher in WT than in the Δppk mutant. The expression of resistance-conferring gene was not significantly altered by gentamycin treatment, except for cysN.
Figure 4.
Expression levels of genes that confer resistance, during tazobactam and gentamycin treatment. Expression levels were compared with those of WT (PCN033). 33-TZP/PCN033 indicates the expression levels in PCN033 during tazobactam treatment compared with those in untreated PCN033; K-TZP/PCN033 indicates expression levels in the Δppk mutant during tazobactam treatment compared with those in untreated PCN033; 33-GEN/PCN033 indicates expression levels in PCN033 during gentamycin treatment compared with those in untreated PCN033; K-GEN/PCN033 indicates expression levels in the Δppk mutant during gentamycin treatment compared with those in untreated PCN033. ***p < 0.000, **p < 0.01, *p < 0.05.
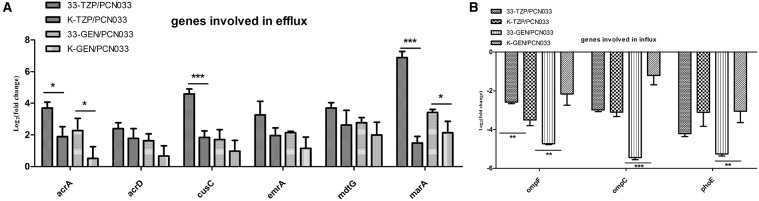
Expression of genes involved in antibiotic transport was altered more strongly in WT than in the Δppk mutant
The expression of six genes involved in antibiotic efflux was determined, as shown in Figure 5A. The efflux genes were upregulated in both the WT and Δppk mutant strains by tazobactam and gentamycin. However, the expression of acrA, cusC, and marA was higher in WT than in the Δppk mutant after treatment with tazobactam. After treatment with gentamycin, the expression of acrA and marA was also higher in WT than in the Δppk mutant. The expression of three genes involved in antibiotic influx was determined, as shown in Figure 5B. All the genes tested were downregulated by antibiotic treatment. However, ompF expression decreased more strongly in WT than in the Δppk mutant when treated with tazobactam. The expression of ompF, ompC, and phoE also decreased more strongly in WT than in the Δppk mutant after treatment with gentamycin.
Figure 5.
Expression levels of genes involved in antibiotic efflux and influx during tazobactam and gentamycin treatments. (A) Expression of efflux genes, (B) expression of influx genes. Expression levels of the genes were compared with those of WT (PCN033). 33-TZP/PCN033 indicates the expression levels in PCN033 during tazobactam treatment compared with those in untreated PCN033; K-TZP/PCN033 indicates the expression levels in the Δppk mutant during untreated PCN033.K-GEN/PCN033 indicates the expression levels in the Δppk mutant during gentamycin treatment compared with those in untreated PCN033.***p < 0.000, **p < 0.01, *p < 0.05.
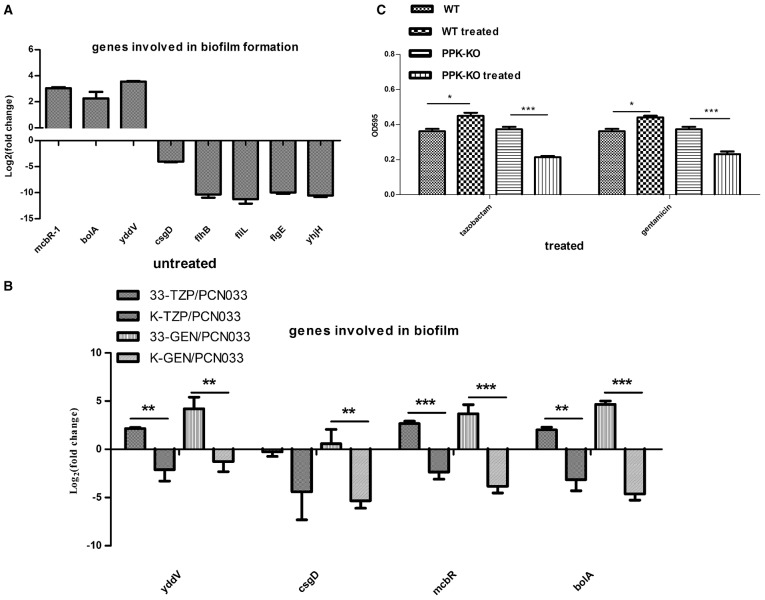
Effects of tazobactam and gentamycin in biofilm formation
The RNA-seq data showed that the transcription levels of some genes associated with biofilm formation were altered in the Δppk strain (Tables S3, S4), especially those encoding the flagella cluster, which simultaneously promotes biofilm generation and impedes biofilm maturation (Laverty et al., 2014). The expression of fimbrial and curli genes was also reduced in the Δppk mutant. The expression levels of some genes were confirmed with qRT–PCR (Figure 6A). The expression of four genes (yddV, mcbR, bolA, and csgD) involved in biofilm regulation was determined during antibiotic treatment, as shown in Figure 6B (Laverty et al., 2014; Lord et al., 2014; Dressaire et al., 2015; Wu et al., 2015). The expression of yddV, mcbR, and bolA was upregulated in WT but downregulated in the Δppk mutant when treated with tazobactam, and all four genes were upregulated in WT but downregulated in the Δppk mutant when treated with gentamycin. Biofilm formation was also evaluated in the presence of tazobactam or gentamycin, as shown Figure 6C. Biofilm formation increased in WT planktonic cells but decreased in Δppk planktonic cells when treated with antibiotics.
Figure 6.
Biofilm formation assay. (A) Validation of the expression of genes involved in biofilm formation. (B) Expression levels of genes involved in biofilm regulation during tazobactam and gentamycin treatment. (C) Biofilm formation during tazobactam or gentamycin treatment. Expression levels of genes were compared with those in WT (PCN033). 33-TZP/PCN033 indicates the expression levels in PCN033 during tazobactam treatment compared with those in untreated PCN033; K-TZP/PCN033 indicates the expression levels in the Δppk mutant during tazobactam treatment compared with those in untreated PCN033; 33-GEN/PCN033 indicates the expression levels in PCN033 during gentamycin treatment compared with those in untreated PCN033; K-GEN/PCN033 indicates the expression levels in the Δppk mutant during gentamycin treatment compared with those in untreated PCN033. WT indicates PCN033 without antibiotic treatment; WT treated indicates PCN033 with antibiotic treatment; PPK-KO indicates the Δppk mutant without antibiotic treatment; PPK-KO treated indicates the Δppk mutant with antibiotic treatment. ***p < 0.000, **p < 0.01, *p < 0.05.
Discussion
Multidrug-resistant strains of ExPEC present significant challenges to public health and animal husbandry (Girardeau et al., 2003; Johnson et al., 2005; Bergeron et al., 2012). Because pathogenic E. coli mainly causes acute infections in its planktonic growth mode (Li et al., 2014), we initially investigated the role of PPK in the antibiotic resistance of ExPEC in the planktonic growth mode. We investigated in detail its susceptibility to different types of antibiotics, mediated by PPK, in E. coli. We found that PPK is very important in aminoglycoside tolerance, regulating the expression levels of antibiotic efflux and influx genes in the planktonic growth mode. Our findings indicate that PPK could have utility as a novel antimicrobial drug target.
As reported previously, resistance-conferring proteins and antibiotic efflux and influx porins play important roles in multidrug-resistance. Efflux proteins contribute to antibiotic tolerance by transporting compounds to the extracellular environment, whereas influx proteins have the opposite effect (Wilson, 2014). The expression levels of these genes were determined with RNA-seq, and showed that without antibiotic treatment, they did not differ significantly between WT and the Δppk mutant. Because PPK is reported to play prominent roles in the stress responses elicited by other stimuli (Alcántara et al., 2014), we investigated the role of PPK in the antibiotic stress response. Gentamycin and tazobactam were selected to treatthe planktonic cells of WT and the Δppk mutant. With gentamycin treatment, the expression of the efflux genes acrA and marA increased more strongly in WT than that in the Δppk mutant, and the influx porin genes ompC and ompF decreased more strongly in WT than in the Δppk mutant. Gentamycin binds the 30S ribosomal subunit and interrupts protein synthesis, thus inhibiting bacterial multiplication (Wargo and Edwards, 2014). According to Gray et al., compounds that interrupt protein metabolism cause intracellular polyP accumulation (Gray et al., 2014). Because it is a high-energy phosphate compound, polyP can be used to phosphorylate the response regulators of two-component systems to regulate gene expression (Sureka et al., 2007). As reported previously, the two-component systems CpxR and BaeR are implicated in antibiotic resistance by regulating the efflux genes of the acr operon and mar operon (Hu et al., 2011; Weatherspoon-Griffin et al., 2014; Pletzer et al., 2015). We speculated that phosphorylation of BaeR or CpxR using polyP as phosphate donar to modulate expression of acrA and marA during gentamycin treatment. The expression of porin genes ompF and ompC is upregulated by cAMP (Dalhoff, 1983), and the level of cAMP is negatively regulated by polyP, which potently inhibits the activity of the class III adenylate cyclases (Guo et al., 2009). Therefore, we speculated that the expression of ompC and ompF was downregulated by polyP during gentamycin treatment. Therefore, polyP may influence gentamycin tolerance by regulating the expression of antibiotic efflux and influx genes.
With tazobactam treatment, the expression of the resistance gene bla and efflux genes acrA, cusC, and marA was upregulated. Tazobactam binds to the periplasmic β-lactamase, and the efflux pump is implicated in resistance to beta-lactams and beta-lactamase inhibitors (Zhanel et al., 2014). However, there are insufficient data to clarify the role of PPK in regulating the expression of efflux pump genes induced by β-lactams. It will be interesting to explore the role of PPK in this process.
Biofilms contribute to antibiotic tolerance and chronic infection; thus, we also investigated the role of PPK in the antibiotic resistance of biofilm-grown cells. We observed that biofilm formation was impaired in the Δppk mutant when treated with antibiotics. The genes involved in biofilm formation (such as those encoding the fimbriae cluster, flagella cluster, and biofilm regulators BolA and McbR), were downregulated in the Δppk mutant by both antibiotic treatments. PolyP acts as a “chemical chaperone”, stabilizing cytoplasmic proteins intracellularly, similarly to heat shock proteins (Gray et al., 2014), and chaperones are known to be involved in biofilm formation. For example, the chaperone CsgE directs the intracellular localization of CsgA, the major subunit of the extracellular amyloid protein known as curli, which is essential for biofilm formation (Andersson et al., 2013). The FliS protein acts as a chaperone for FliC, a flagellar structural protein that promotes biofilm generation (Xu et al., 2014). The universal heat shock protein chaperones are also implicated in biofilm formation by fungi, such as Candida albicans (Robbins et al., 2011; Becherelli et al., 2013), and by Gram-negative bacteria, such as E. coli (Grudniak et al., 2013). Ultimately, these different effects reduce biofilm impairment during antibiotic treatment. Interestingly, PPK did not affect the antibiotic tolerance of ExPEC in the biofilm growth mode. Biofilms manifest antibiotic tolerance through many different mechanisms, including preventing the passage of antimicrobial compounds into the cytoplasm and possessing densely adherent growth (Qu et al., 2010). As reported previously, the planktonic and biofilm modes of growth are two distinct bacterial “lifestyles” (Chua et al., 2014), so it will be interesting to explore the roles of PPK in these distinct contexts.
Author contributions
JC and CT designed and supervised the research project. JC wrote the paper, and LS, XW, HC, and TZ revised the manuscript. JC, TZ, and FL performed the experiments. JC and LS processed the data and performed the statistical analysis. All the authors have read and approved the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (grant numbers 31472201, 31030065, and 31421064) and the Programme for Introducing Talents of Discipline to Universities (grant number B12005).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank Dr. Janine Miller from the University of Tasmania for writing improvement.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00724
Percentage activity of different antibiotics in removing biofilm. (A) Targeting the cell wall, (B) targeting protein biosynthesis, and (C) targeting nucleotide metabolism.
Bacteria strains and primers.
Transcription level of genes involved in antibiotic and multi-drug resistance.
Expression of genes implicated in biofilm.
Genes involved in biofilm formation (part results of RNA-seq).
References
- Alcántara C., Blasco A., Zúñiga M., Monedero V. (2014). Accumulation of polyphosphate in Lactobacillus spp. and its involvement in stress resistance. Appl. Environ. Microbiol. 80, 1650–1659. 10.1128/AEM.03997-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldred K. J., Kerns R. J., Osheroff N. (2014). Mechanism of quinolone action and resistance. Biochemistry 53, 1565–1574. 10.1021/bi5000564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson E. K., Bengtsson C., Evans M. L., Chorell E., Sellstedt M., Lindgren A. E. (2013). Modulation of curli assembly and pellicle biofilm formation by chemical and protein chaperones. Chem. Biol. 20, 1245–1254. 10.1016/j.chembiol.2013.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ault-Riché D., Fraley C. D., Tzeng C. M., Kornberg A. (1998). Novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli. J. Bacteriol. 180, 1841–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becherelli M., Tao J., Ryder N. S. (2013). Involvement of heat shock proteins in Candida albicans biofilm formation. J. Mol. Microbiol. Biotechnol. 23, 396–400. 10.1159/000351619 [DOI] [PubMed] [Google Scholar]
- Benthall G., Touzel R. E., Hind C. K., Titball R. W., Sutton J. M., Thomas R. J. (2015). Evaluation of antibiotic efficacy against infections caused by planktonic or biofilm cultures of Pseudomonas aeruginosa and Klebsiella pneumoniae in Galleria mellonella. Int. J. Antimicrob. Agents 46, 538–545. 10.1016/j.ijantimicag.2015.07.014 [DOI] [PubMed] [Google Scholar]
- Bergeron C. R., Prussing C., Boerlin P., Daignault D., Dutil L., Reid-Smith R. J. (2012). Chicken as reservoir for extraintestinal pathogenic Escherichia coli in humans, Canada. Emerg. Infect. Dis. 18, 415–421. 10.3201/eid1803.111099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisson-Noël A., Trieu-Cuot P., Courvalin P. (1988). Mechanism of action of spiramycin and other macrolides. J. Antimicrob. Chemother. 22 (Suppl B), 13–23. [DOI] [PubMed] [Google Scholar]
- Brown M. R., Kornberg A. (2004). Inorganic polyphosphate in the origin and survival of species. Proc. Natl. Acad. Sci. U.S.A. 101, 16085–16087. 10.1073/pnas.0406909101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K. (2012). Antimicrobial agents targeting bacterial cell walls and cell membranes. Rev. Sci. Tech. 31, 43–56. 10.20506/rst.31.1.2096 [DOI] [PubMed] [Google Scholar]
- Chaudhuri R. R., Henderson I. R. (2012). The evolution of the Escherichia coli phylogeny. Infect. Genet. Evol. 12, 214–226. 10.1016/j.meegid.2012.01.005 [DOI] [PubMed] [Google Scholar]
- Chua S. L., Liu Y., Yam J. K., Chen Y., Vejborg R. M., Tan B. G. (2014). Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyles. Nat. Commun. 21, 4462 10.1038/ncomms5462 [DOI] [PubMed] [Google Scholar]
- Dalhoff A. (1983). Transport of aminoglycosides in Escherichia coli. Z. Bakteriol. Mikrobiol. Hyg. A. 254, 379–387. [PubMed] [Google Scholar]
- Davis B. D. (1987). Mechanism of bactericidal action of aminoglycosides. Microbiol. Rev. 51, 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressaire C., Moreira R. N., Barahona S., Alves de Matos A. P., Arraiano C. M. (2015). BolA is a transcriptional switch that turns off motility and turns on biofilm development. MBio 6, e02352–e02314. 10.1128/mBio.02352-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraley C. D., Rashid M. H., Lee S. S., Gottschalk R., Harrison J., Wood P. J. (2007). A polyphosphate kinase 1 (ppk1) mutant of Pseudomonas aeruginosa exhibits multiple ultrastructural and functional defects. Proc. Natl. Acad. Sci. U.S.A. 104, 3526–3531. 10.1073/pnas.0609733104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaschignard J., Levy C., Romain O., Cohen R., Bingen E., Aujard Y. (2011). Neonatal bacterial meningitis: 444 cases in 7 years. Pediatr. Infect. Dis. J. 30, 212–217. 10.1097/INF.0b013e3181fab1e7 [DOI] [PubMed] [Google Scholar]
- Girardeau J. P., Lalioui L., Said A. M., De Champs C., Le Bouguénec C. (2003). Extended virulence genotype of pathogenic Escherichia coli isolates carrying the afa-8 operon: evidence of similarities between isolates from humans and animals with extraintestinal infections. J. Clin. Microbial. 41, 218–226. 10.1128/JCM.41.1.218-226.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau-Campistany A., Manresa Á., Pujol M., Rabanal F., Cajal Y. (2015). Tryptophan-containing lipopeptide antibiotics derived from polymyxin B with activity against Gram positive and Gram negative bacteria. Biochim. Biophys. Acta 1858, 333–343. 10.1016/j.bbamem.2015.11.011 [DOI] [PubMed] [Google Scholar]
- Gray M. J., Wholey W. Y., Wagner N. O., Cremers C. M., Mueller-Schickert A., Hock N. T. (2014). Polyphosphate is a primordial chaperone. Mol. Cell. 53, 689–699. 10.1016/j.molcel.2014.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grudniak A. M., Pawlak K., Bartosik K., Wolska I. K. (2013). Physiological consequences of mutations in the htpG heat shock gene of Escherichia coli. Mutat. Res. 745–746, 1–5. 10.1016/j.mrfmmm.2013.04.003 [DOI] [PubMed] [Google Scholar]
- Guo Y. L., Mayer H., Vollmer W., Dittrich D., Sander P., Schultz A. (2009). Polyphosphates from Mycobacterium bovis–potent inhibitors of class III adenylate cyclases. FEBS J. 276, 1094–1103. 10.1111/j.1742-4658.2008.06852.x [DOI] [PubMed] [Google Scholar]
- He Y., Xu T., Fossheim L. E., Zhang X. H. (2012). FliC, a flagellin protein, is essential for the growth and virulence of fish pathogen Edwardsiella tarda. PLoS ONE. 7:e45070. 10.1371/journal.pone.0045070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hof H. (1988). Antimicrobial therapy with nitroheterocyclic compounds, for example, metronidazole and nitrofurantoin. Immun. Infekt. 16, 220–225. [PubMed] [Google Scholar]
- Hu W. S., Chen H. W., Zhang R. Y., Huang C. Y., Shen C. F. (2011). The expression levels of outer membrane proteins STM1530 and OmpD, which are influenced by the CpxAR and BaeSR two-component systems, play important roles in the ceftriaxone resistance of Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 55, 3829–3837. 10.1128/AAC.00216-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. R., Kuskowski M. A., Smith K., O'Bryan T. T., Tatini S. (2005). Antimicrobial-resistant and extraintestinal pathogenic Escherichia coli in retail foods. J. Infect. Dis. 191, 1040–1049. 10.1086/428451 [DOI] [PubMed] [Google Scholar]
- Kubista M., Andrade J. M., Bengtsson M., Forootan A., Jonák J., Lind K. (2006). The real-time polymerase chain reaction. Mol. Aspects Med. 27, 195–125. 10.1016/j.mam.2005.12.007 [DOI] [PubMed] [Google Scholar]
- Laverty G., Gorman S. P., Gilmore B. F. (2014). Biomolecular mechanisms of Pseudomonas aeruginosa and Escherichia coli biofilm formation. Pathogens 3, 596–632. 10.3390/pathogens3030596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Petrova O. E., Su S., Lau G. W., Panmanee W., Na R. (2014). BdlA, DipA and induced dispersion contribute to acute virulence and chronic persistence of Pseudomonas aeruginosa. PLoS Pathog. 10:e1004168. 10.1371/journal.ppat.1004168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Zheng H., Yang M., Xu Z., Wang X., Wei L. (2015). Genome analysis and in vivo virulence of porcine extraintestinal pathogenic Escherichia coli strain PCN033. BMC Genomics. 16:717. 10.1186/s12864-015-1890-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord D. M., Uzgoren Baran A., Soo V. W., Wood T. K., Peti W., Page R. (2014). McbR/YncC: implications for the mechanism of ligand and DNA binding by a bacterial GntR transcriptional regulator involved in biofilm formation. Biochemistry 53, 7223–7231. 10.1021/bi500871a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellata M. (2013). Human and avian extraintestinal pathogenic Escherichia coli: infections, zoonotic risks, and antibiotic resistance trends. Foodborne Pathog. Dis. 10, 916–932. 10.1089/fpd.2013.1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt F. C., Bloch P. L., Smith D. F. (1974). Culture medium for enterobacteria. J. Bacteriol. 119, 736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfold W. J., Norris D. (1912). The relation of concentration of food supply to the generation-time of bacteria. J. Hyg. 12, 527–531. 10.1017/S0022172400005180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Trallero E., Iglesias L. (2003). Tetracyclines, sulfonamides and metronidazole. Enferm. Infecc. Microbiol. Clin. 21, 520–528. 10.1016/S0213-005X(03)72999-1 [DOI] [PubMed] [Google Scholar]
- Pletzer D., Stahl A., Oja A. E., Weingart H. (2015). Role of the cell envelope stress regulators BaeR and CpxR in control of RND-type multidrug efflux pumps and transcriptional cross talk with exopolysaccharide synthesis in Erwinia amylovora. Arch. Microbiol. 197, 761–772. 10.1007/s00203-015-1109-0 [DOI] [PubMed] [Google Scholar]
- Qu Y., Daley A. J., Istivan T. S., Rouch D. A., Deighton M. A. (2010). Densely adherent growth mode, rather than extracellular polymer substance matrix build-up ability, contributes to high resistance of Staphylococcus epidermidis biofilms to antibiotics. J. Antimicrob. Chemother. 65, 1405–1411. 10.1093/jac/dkq119 [DOI] [PubMed] [Google Scholar]
- Rashid M. H., Rumbaugh K., Passador L., Davies D. G., Hamood A. N., Iglewski B. H. (2000). Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci.U.S.A. 97, 9636–9641. 10.1073/pnas.170283397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins N., Uppuluri P., Nett J., Rajendran R., Ramage G., Lopez-Ribot J. L. (2011). Hsp90 governs dispersion and drug resistance of fungal biofilms. PLoS Pathog. 7:e1002257. 10.1371/journal.ppat.1002257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Gómez S., Ferrer-Espada R., Stewart P. S., Pitts B., Lohner K., Martínez de Tejada G. (2015). Antimicrobial activity of synthetic cationic peptides and lipopeptides derived from human lactoferricin against Pseudomonas aeruginosa planktonic cultures and biofilms. BMC Microbiol. 15:137. 10.1186/s12866-015-0473-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedláková M. H., Urbánek K., Vojtová V., Suchánková H., Imwensi P., Kolář M. (2014). Antibiotic consumption and its influence on the resistance in Enterobacteriaceae. BMC Res. Notes. 7:454. 10.1186/1756-0500-7-454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrach-Cardona R., Hijosa-Valsero M., Marti E., Balcázar J. L., Becares E. (2014). Prevalence of antibiotic-resistant fecal bacteria in a river impacted by both an antibiotic production plant and urban treated discharges. Sci. Total Environ. 488, 220–227. 10.1016/j.scitotenv.2014.04.100 [DOI] [PubMed] [Google Scholar]
- Singh R., Singh M., Arora G., Kumar S., Tiwari P., Kidwai S. (2013). Polyphosphate deficiency in Mycobacterium tuberculosis is associated with enhanced drug susceptibility and impaired growth in guinea pigs. J. Bacteriol. 195, 2839–2851. 10.1128/JB.00038-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureka K., Dey S., Datta P., Singh A. K., Dasgupta A., Rodrigue S. (2007). Polyphosphate kinase is involved in stress-induced mprAB-sigE-rel signalling in mycobacteria. Mol. Microbiol. 65, 261–276. 10.1111/j.1365-2958.2007.05814.x [DOI] [PubMed] [Google Scholar]
- Tan C., Tang X., Zhang X., Ding Y., Zhao Z., Wu B. (2012). Serotypes and virulence genes of extraintestinal pathogenic Escherichia coli isolates from diseased pigs in China. Vet. J. 192, 483–488. 10.1016/j.tvjl.2011.06.038 [DOI] [PubMed] [Google Scholar]
- Tapiainen T., Hanni A. M., Salo J., Ikäheimo I., Uhari M. (2014). Escherichia coli biofilm formation and recurrences of urinary tract infections in children. Eur. J. Clin. Microbiol. Infect. Dis. 33, 111–115. 10.1007/s10096-013-1935-4 [DOI] [PubMed] [Google Scholar]
- Tunpiboonsak S., Mongkolrob R., Kitudomsub K., Thanwatanaying P., Kiettipirodom W., Tungboontina Y. (2010). Role of a Burkholderia pseudomallei polyphosphate kinase in an oxidative stress response, motilities, and biofilm formation. J. Microbiol. 48, 63–70. 10.1007/s12275-010-9138-5 [DOI] [PubMed] [Google Scholar]
- Wargo K. A., Edwards J. D. (2014). Aminoglycoside-induced nephrotoxicity. J. Pharm. Pract. 27, 573–577. 10.1177/0897190014546836 [DOI] [PubMed] [Google Scholar]
- Weatherspoon-Griffin N., Yang D., Kong W., Hua Z., Shi Y. (2014). The CpxR/CpxA two-component regulatory system up-regulates the multidrug resistance cascade to facilitate Escherichia coli resistance to a model antimicrobial peptide. J. Biol. Chem. 289, 32571–32582. 10.1074/jbc.M114.565762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston E. J., Pondo T., Lewis M. M., Martell-Cleary P., Morin C., Jewell B. (2011). The burden of invasive early-onset neonatal sepsis in the United States, 2005-2008. Pediatr. Infect. Dis. J. 30, 937–941. 10.1097/INF.0b013e318223bad2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D. N. (2014). Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat. Rev. Microbiol. 12, 35–48. 10.1038/nrmicro3155 [DOI] [PubMed] [Google Scholar]
- Wu Y., Ding Y., Cohen Y., Cao B. (2015). Elevated level of the second messenger c-di-GMP in Comamonas testosteroni enhances biofilm formation andbiofilm-based biodegradation of 3-chloroaniline. Appl. Microbiol. Biotechnol. 99, 1967–1976. 10.1007/s00253-014-6107-7 [DOI] [PubMed] [Google Scholar]
- Xu S., Peng Z., Cui B., Wang T., Song Y., Zhang L. (2014). FliS modulates FlgM activity by acting as a non-canonical chaperone to control late flagellar gene expression, motility and biofilm formation in Yersinia pseudotuberculosis. Environ. Microbiol. 16, 1090–1104. 10.1111/1462-2920.12222 [DOI] [PubMed] [Google Scholar]
- Zhanel G. G., Chung P., Adam H., Zelenitsky S., Denisuik A., Schweizer F. (2014). Ceftolozane/tazobactam: a novel cephalosporin/β-lactamase inhibitor combination with activity against multidrug-resistant gram-negative bacilli. Drugs 74, 31–51. 10.1007/s40265-013-0168-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Percentage activity of different antibiotics in removing biofilm. (A) Targeting the cell wall, (B) targeting protein biosynthesis, and (C) targeting nucleotide metabolism.
Bacteria strains and primers.
Transcription level of genes involved in antibiotic and multi-drug resistance.
Expression of genes implicated in biofilm.
Genes involved in biofilm formation (part results of RNA-seq).