Abstract
Background: Platycodon grandiflorum is the only species in the genus Platycodon of the family Campanulaceae, which has been traditionally used as a medicinal plant for its lung-heat-clearing, antitussive, and expectorant properties in China, Japanese, and Korean. Oleanane-type triterpenoid saponins were the main chemical components of P. grandiflorum and platycodin D was the abundant and main bioactive component, but little is known about their biosynthesis in plants. Hence, P. grandiflorum is an ideal medicinal plant for studying the biosynthesis of Oleanane-type saponins. In addition, the genomic information of this important herbal plant is unavailable.
Principal findings: A total of 58,580,566 clean reads were obtained, which were assembled into 34,053 unigenes, with an average length of 936 bp and N50 of 1,661 bp by analyzing the transcriptome data of P. grandiflorum. Among these 34,053 unigenes, 22,409 unigenes (65.80%) were annotated based on the information available from public databases, including Nr, NCBI, Swiss-Prot, KOG, and KEGG. Furthermore, 21 candidate cytochrome P450 genes and 17 candidate UDP-glycosyltransferase genes most likely involved in triterpenoid saponins biosynthesis pathway were discovered from the transcriptome sequencing of P. grandiflorum. In addition, 10,626 SSRs were identified based on the transcriptome data, which would provide abundant candidates of molecular markers for genetic diversity and genetic map for this medicinal plant.
Conclusion: The genomic data obtained from P. grandiflorum, especially the identification of putative genes involved in triterpenoid saponins biosynthesis pathway, will facilitate our understanding of the biosynthesis of triterpenoid saponins at molecular level.
Keywords: Platycodon grandiflorum, transcriptome, triterpenoid saponins, platycodin D, biosynthesis
Introduction
Platycodon grandiflorum (Jacq.) A. DC. is a perennial flowering plant of the Campanulaceae family and the only species of the genus Platycodon. It is a well-known medicinal plant in China and other East Asian countries and has been traditionally used as a medicine and food additive for various respiratory diseases, including bronchitis, asthma, tonsillitis, pulmonary tuberculosis and other inflammatory diseases (Takagi and Lee, 1972; Kim et al., 1995; Shin and Lee, 2002). Oleanane-type triterpenoid saponins are the main chemical components of P. grandiflorum, mainly including platycodin D, D2, D3, deapioplatycodin D, D2, polygalacin D and platyconic acid A (Kim J.W. et al., 2013). In addition to their natural effects, these triterpenoid saponins have various pharmacological activities, such as anti-inflammatory, anti-cancer, immune enhancing effects and preventing chemicals-induced hepatotoxicity (Lee et al., 2004, 2008; Kim et al., 2008, 2012a; Khanal et al., 2009). Especially, chemical investigation of P. grandiflorum has revealed that platycodin D is the most abundant and the main bioactive component (Shin et al., 2009; Xie et al., 2009; Kim et al., 2012b).
Triterpenoid saponins are a group of mostly studied compounds in plants, and their biosynthesis has been extensively studied and described (Haralampidis et al., 2002; Yendo et al., 2010; Augustin et al., 2011; Moses et al., 2014a). The direct precursor of triterpenoid saponins is 2, 3-oxidosqualene which is synthesized via the mevalonic acid (MVA) pathway (Haralampidis et al., 2002). Three key enzymes are involved in the biosynthesis of these saponins: oxidosqualene cyclases (OSCs), cytochrome P450 monooxygenases (P450s) and uridine diphosphate-dependent glycosyltransferases (UGTs; Figure 1, Supplementary Table S4). The most important progress in the biosynthesis of triterpenoid saponins is achieved in Panax species (Araliaceae family), which contains a special group of triterpenoid saponins, i.e., ginsenosides. Three P450s in Panax ginseng have been functionally characterized, they are protopanaxadiol synthase (PPDS, CYP716A47), which catalyzes the conversion of dammarenediol-II to protopanaxadiol (Han et al., 2011), protopanaxatriol synthase (PPTS, CYP716A53v2) catalyzing the conversion of protopanaxadiol to protopanaxatriol (Han et al., 2012), and β-A28O (CYP716A52v2) catalyzing the conversion of β-amyrin to oleanolic acid (Han et al., 2013). Recently, two UGTs (PgUGT74AE2 and PgUGT94Q2) have also been characterized in P. ginseng which are involved in the biosynthesis of ginsenoside Rg3 and Rd (Jung et al., 2014). Even though the biosynthesis of some ginsenosides or their aglycones have been well-documented and can be conducted in a yeast fermentation system (Dai et al., 2014; Jung et al., 2014), the biosynthesis of triterpenoid saponins in different plant species is far from conclusive.
FIGURE 1.
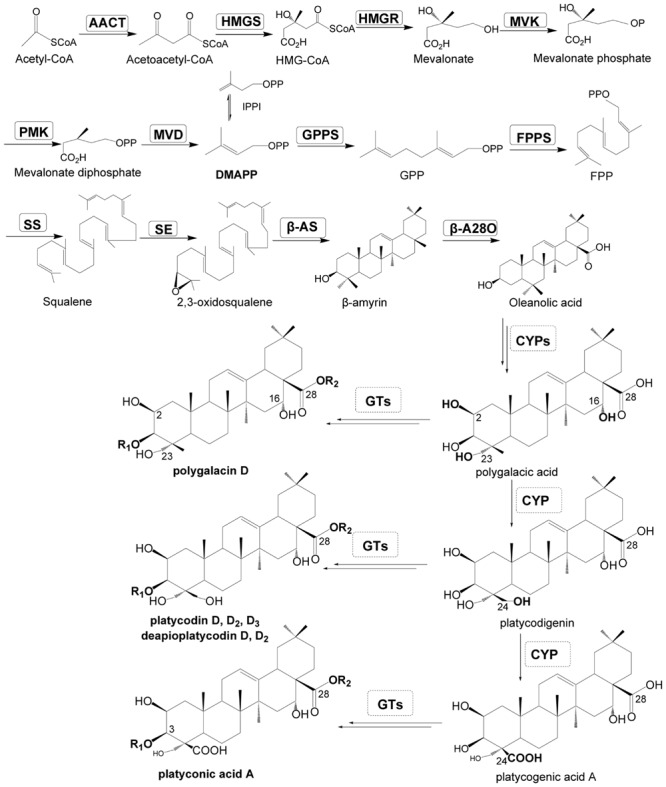
Putative pathway for triterpenoid saponin biosynthesis in Platycodon grandiflorum. Enzymes found in this study are boxed. AACT, acetyl-CoA acetyltransferase; HMGS, HMG-CoA synthase; IPPI, IPP isomerase; HMGR, HMG-CoA reductase; MVK, mevalonate kinase; PMK, phosphomevalonate kinase; MVD, mevalonate diphosphate decarboxylase; GPPS, geranylgeranyl pyrophosphate synthase; FPPS, farnesyl diphosphate synthase; SS, squalene synthase; SE, squalene epoxidase; β-AS, β-amyrin synthase; β-A28O, β-amyrin 28-oxidase; HMG-CoA, 3-hydroxy-3-methylglutaryl coenzyme A; DMAPP, dimethylallyl diphosphate; FPP, farnesyl diphosphate; GPP, geranyl pyrophosphate; IPP, isopentenyl diphosphate; GT, glycosyltransferase; CYPs, cytochrome P450.
Despite many genes encoding enzymes involved in the biosynthesis of the triterpenoid saponins have been identified from Panax species (Sun et al., 2010; Chen et al., 2011; Luo et al., 2011; Li et al., 2013), information about those genes in P. grandiflorum is still lacking (Kim Y.K. et al., 2013). Although the pharmacological activity of platycodin D has been investigated (Kim et al., 2012a,b; Chun et al., 2013; Chun and Kim, 2013; Hwang et al., 2013; Li et al., 2014), a complete biosynthesis pathway of platycodin D has not been elucidated, especially the last two steps. At present, the genomes or transcripts of about 46 species of medicinal plants have been sequenced, which will lead to an efficient way of deciphering novel gene functions involved in specific metabolic pathways in medicinal plants (Misra, 2014). Characterization of these novel genes will be useful for investigating the synthesis of platycodins in P. grandiflorum. The objective of the present study was to characterize the transcriptome of P. grandiflorum using Illumina HiSeqTM2000 sequencing platform in order to uncover the candidate genes encoding enzymes involved in the triterpene saponin biosynthetic pathway, especially in oleanane-type saponins biosynthesis, and to screen molecular markers of SSRs for facilitation the marker-assisted breeding of this species.
Results and Discussion
Illumina Sequencing and De Novo Assembly
The root tissue of P. grandiflorum was used for transcriptome sequencing and analysis because roots have traditionally been used for medicinal purpose. A cDNA library was constructed from total RNA of P. grandiflorum roots, and sequenced using Illumina paired-end sequencing technology. After removal of adaptor sequences, ambiguous reads and low-quality reads (Q20 < 20), a total of 58,580,566 clean reads were obtained. The Q20 percentage (sequencing error rate < 1%) and GC percentage were 97.04 and 45.51%, respectively. An overview of the sequencing and assembly statistics is shown in Table 1. The high quality reads obtained in this study have been deposited in the NCBI SRA database (accession number: SRA226668).
Table 1.
Summary of Illumina paired-end sequencing and assembly for Platycodon grandiflorum.
| Database | Number | Total length (bp) |
|---|---|---|
| Total clean reads | 58,580,566 | 58,580,56600 |
| Q20 percentage | 97.04% | |
| GC percentage | 45.51% | |
| Number of transcripts | 50,408 | 55,568,306 |
| Average length of transcripts (bp) | 1,102 | |
| Max length of transcripts (bp) | 15,684 | |
| Min length of transcripts (bp) | 201 | |
| Transcript size N50 (bp) | 1,796 | |
| Number of unigenes | 34,053 | 31,887,854 |
| Average length of unigenes (bp) | 936 | |
| Max length of unigenes (bp) | 15,684 | |
| Min length of unigenes (bp) | 2,01 | |
| Unigene size N50 (bp) | 1,661 |
All the clean reads (58,580,566) were de novo assembled using the Trinity program into 50,408 transcripts consisting of 55,568,306 bp. The size of the transcripts ranged from 201 to 15,684 bp, with an average length of 1,102 bp and N50 length of 1,796 bp. Among these transcripts, 20,939 (41.54%) were longer than 1000 bp, and 19,808 (39.30%) were shorter than 500 bp (Figure 2). Using paired-end joining and gap-filling methods, these contigs were further assembled into 34,053 unigenes with an average length of 936 bp and an N50 length of 1,661 bp. There were 11,291 unigenes (33.16%) longer than 1,000 bp, and 4,202 unigenes (12.34%) longer than 2,000 bp (Figure 2).
FIGURE 2.
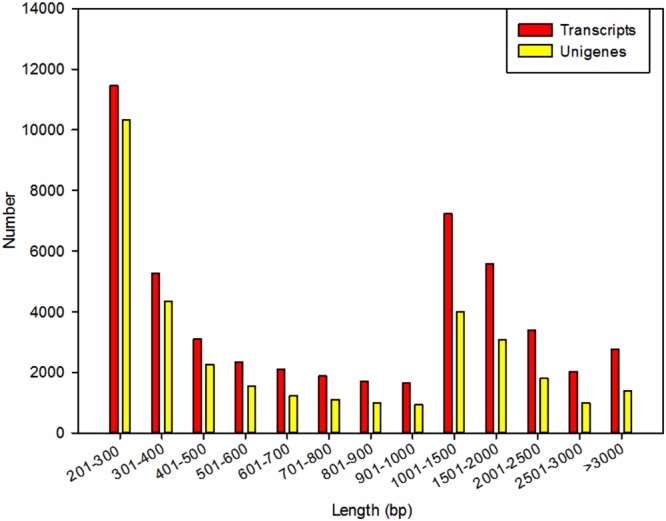
Overview of the P. grandiflorum transcriptome assembly.
Functional Annotation
In our study, we used the Nr, Nt, KEGG, SwissProt, PFAM, GO, and KOG publicly available databases to annotate the unigenes. The overall function annotation is depicted in Table 2. Altogether, 22,409 unigenes (65.80%) were annotated in the public databases. There were 21,310 unigenes (62.57%) matched in the Nr databases, and 11,877 unigenes (34.87%) matched with known proteins in the Nt databases. A total of 6,998 unigenes (20.55%) matched to the KEGG database and 15,870 unigenes (46.60%) matched to the SwissProt. The number of unigenes matched to the PFAM, GO and KOG databases was 14,877 (43.68%), 16,677 (48.97%), and 8,779 (25.78%), respectively.
Table 2.
Summary of the annotation percentage of P. grandiflorum compared with public databases.
| Database | Number of unigenes | Annotation percentage (%) |
|---|---|---|
| Nr | 21,310 | 62.57 |
| Nt | 11,877 | 34.87 |
| KEGG | 6,998 | 20.55 |
| SwissProt | 15,870 | 46.60 |
| PFAM | 14,877 | 43.68 |
| GO | 16,677 | 48.97 |
| KOG | 8,779 | 25.78 |
| All annotated unigenes | 22,409 | 65.80 |
| Total unigenes | 34,053 |
Gene Ontology Classification
A total of 16,677 unigenes were characterized using GO analysis based on Nr annotation, including biological process, cellular component, and molecular function. There were 31,810 unigenes were grouped under cellular component, 21,705 unigenes under molecular function, 44,810 unigenes under biological process. Under the cellular component category, the majority of unigenes were involved in cell (6,586 unigenes, 20.29%) and cell part (6,579 unigenes, 20.27%). For the biological process class, the cellular process (10,127 unigenes, 22.50%) and metabolic process (9,737 unigenes, 21.63%) were the most abundant classes. In the molecular function category, binding (9,999, 46.07%) and catalytic activities (8,438, 38.88%) were predominant (Figure 3).
FIGURE 3.
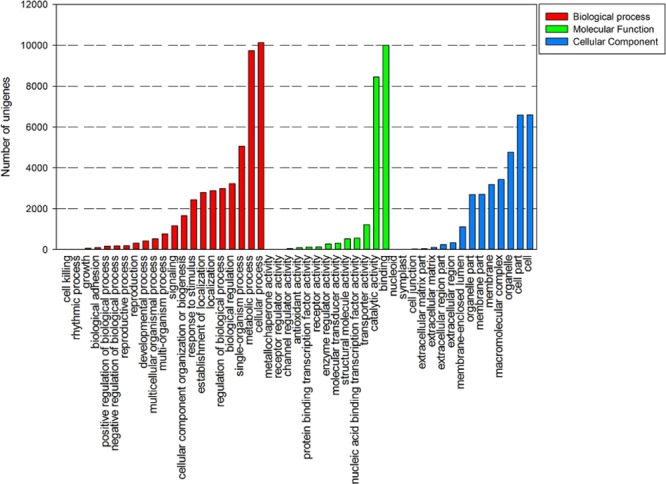
Gene ontology classification of assembled unigenes.
KOG Classification
All unigenes were subjected to a search against the KOG database for functional prediction and classification. Totally, 8,779 unigenes were clustered into 26 functional categories. The general function prediction only (1,444 unigenes, 16.45%) was the major KOG category, followed by post-translational modification, protein turnover, chaperones (1,215 unigenes, 13.84%), signal transduction mechanisms (763 unigenes, 8.7%), translation, ribosomal structure and biogenesis (683 unigenes, 7.78%), transcription (534 unigenes, 6.08%), intracellular trafficking, secretion, and vesicular transport (513 unigenes, 5.84%), energy production and conversion (512 unigenes, 5.83%; Figure 4).
FIGURE 4.

Eukaryotic Orthologous Groups (KOGs) function classification of P. grandiflorum.
Functional Classification by KEGG
In our study, 6,998 unigenes (20.55%) were annotated and assigned to 258 pathways by the KEGG, including metabolism, genetic information processing, environmental information processing, cellular processes, organismal systems and human diseases. The category with the largest number of unigenes was metabolism, which included carbohydrate metabolism (718 unigenes, 19.43%), energy metabolism (480 unigenes, 12.99%), amino acid metabolism (443 unigenes, 11.99%), lipid metabolism (360 unigenes, 9.74%), metabolism of cofactors and vitamins (226 unigenes, 6.11%), nucleotide metabolism (209 unigenes, 5.65%), metabolism of other amino acids (196 unigenes, 5.30%), biosynthesis of other secondary metabolites (179 unigenes, 4.84%), metabolism of terpenoids and polyketides (173 unigenes, 4.68%), glycan biosynthesis and metabolism (123 unigenes, 3.33%), xenobiotics biodegradation and metabolism (98 unigenes, 2.65%; Figure 5A).
FIGURE 5.
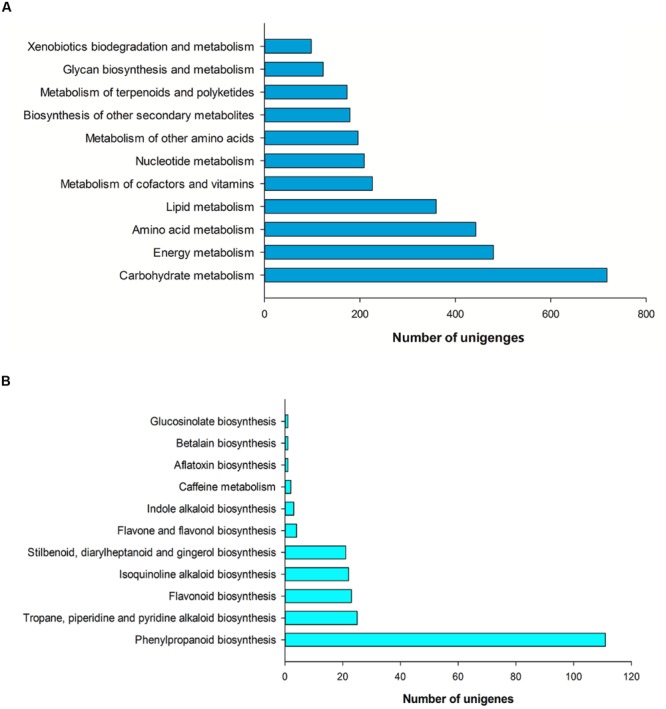
Pathway assignment based on KEGG. (A) Classification based on metabolism categories. (B) Classification based on biosynthesis of other secondary metabolites.
In the other secondary metabolites, the most represented category was phenylpropanoid biosynthesis (111 unigenes, 62.01%), followed by tropane, piperidine and pyridine alkaloid biosynthesis (25 unigenes, 13.97%), flavonoid biosynthesis (23 unigenes, 12.85%), isoquinoline alkaloid biosynthesis (22 unigenes, 12.29%), stilbenoid, diarylheptanoid and gingerol biosynthesis (21 unigenes, 11.73%), flavone and flavonol biosynthesis (4 unigenes, 2.23%; Figure 5B).
Candidate Genes Encoding Enzymes Involved in Triterpenoid Saponin Biosynthesis
The transcripts encoding all the known enzymes involved in triterpenoid saponin biosynthesis were discovered in this Illumina dataset, including AACT, HMGS, HMGR, MVK, PMK, MVD, GGPPS, FPPS, IPPI, SS, SE, β-AS, and β-A28O (Table 3). These findings were in accordance with the fact that P. grandiflorum contains high contents of oleanane-type saponins. Platycodin D is the main triterpenoid saponin in P. grandiflorum, the β-AS (seven unigenes) and β-A28O (one unigenes) were the key enzymes in the biosynthesis of platycodin D. Functional characterization of these unigenes will help us to understand the molecular mechanism of the biosynthesis of oleanane-type saponins in P. grandiflorum.
Table 3.
Transcripts involved in triterpene saponin biosynthesis pathway in P. grandiflorum.
| Gene name | EC number | Unigene number |
|---|---|---|
| AACT, acetyl-CoA acetyltransferase | 2.3.1.9 | 2 |
| HMGS, hydroxymethylglutaryl-CoA synthase | 2.3.3.10 | 5 |
| HMGR, hydroxymethylglutaryl-CoA reductase | 1.1.1.34 | 4 |
| MVK, mevalonate kinase | 2.7.1.36 | 3 |
| PMK, phosphomevalonate kinase | 2.7.4.2 | 2 |
| MVD, mevalonate diphosphate decarboxylase | 4.1.1.33 | 1 |
| GPPS, geranylgeranyl pyrophosphate synthase | 2.5.1.29 | 6 |
| FPPS, farnesyl diphosphate synthase | 2.5.1.10 | 1 |
| IPPI, isopentenyl diphosphate isomerase | 5.3.3.2 | 1 |
| SS, squalene synthase | 2.5.1.21 | 1 |
| SE, squalene epoxidase | 1.14.99.7 | 10 |
| β-AS, β-amyrin synthase | 5.4.99.39 | 7 |
| β-A28O, β-amyrin 28-oxidase | 1.14.13.- | 1 |
The Cytochrome P450 Monooxygenases and UDP-Glycosyltransferase Genes
The CYP450 enzymes, which catalyze the oxidations of β-amyrin, especially at C-2, C-16, C-23, C-24 and C-28, are required for the biosynthesis of the main triterpenoid saponins in P. grandiflorum (Figure 1). In the transcriptomic data of P. grandiflorum, 87 unigenes were annotated to CYP450 (Supplementary Table S1). Among them, unigene comp13745 c0 was annotated to P. ginseng CYP716A52v2 (Figure 6), and com13950 c0 was highly homologous to P. ginseng CYP716A52v2, Medicago truncatula CYP716A12, Vitis vinifera CYP716A15 and CYP716A17 (Carelli et al., 2011; Fukushima et al., 2011; Han et al., 2013), strongly suggesting that both of them might encode β-A28O catalyzing conversion of β-amyrin to oleanolic acid (Figure 6). Bupleurum falcatum CYP716Y1 catalyzes conversion of β-amyrin to 16α hydroxyl β-amyrin (Moses et al., 2014b), no homologous gene was found in this study, and only one unigene (comp21656 c0) was of some similarity (Figure 6). Two unigenes (comp21069 c0 and comp63723 c0) were homologous to M. truncatula CYP72A68v2 which catalyze the hydroxylation of oleanolic acid at C-23 (Fukushima et al., 2013), suggesting that both of them have the same catalytic activities in P. grandiflorum. We also found that five unigenes (comp7080 c0, comp17806 c0, comp10382 c0, comp17206 c0, and comp9845 c0) were highly homologous to CYP93E1 of Glycine max, M. truncatula, and Glycyrrhiza uralensis (Seki et al., 2008), which catalyzes the C-24 hydroxylation of β-amyrin and sophoradiol in soyasaponin biosynthesis (Shibuya et al., 2006; Li et al., 2007; Seki et al., 2008; Fukushima et al., 2013), thus proteins encoded by these unigenes might be also responsible for the C-24 hydroxylation in P. grandiflorum.
FIGURE 6.

Phylogenetic tree of the P. grandiflorum CYP450s. Phylogenetic tree is constructed based on the deduced amino acid sequences for the P. grandiflorum CYP450s (bold letters) and other plant CYP450s involved in triterpenoid biosynthesis. Protein sequences are retrieved from NCBI GenBank using the following accession numbers: Vitis vinifera VvCYP716A15 (BAJ84106.1) and VvCYP716A17 (BAJ84107.1); Medicago truncatula MtCYP716A12 (ABC59076.1), MtCYP93E2 (ABC59085), MtCYP72A63 (H1A981.1), MtCYP72A65v2 (BAL45202), MtCYP72A67v2 (BAL45203) and MtCYP72A67v2 (BAL45203), and MtCYP72A61v2 (BAL45199); Panax ginseng PgCYP716A52v2 (AFO63032.1), PgCYP716A53v2 (I7CT85.1) and PgCYP716A47 (H2DH16.2); Arabidopsis thaliana AtCYP708A2 (NP_001078732.1) and AtCYP705A5 (EFH40098); Glycyrrhiza uralensis GuCYP88D6 (B5BSX1.1), GuCYP93E3 (BAG68930) and GuCYP72A154 (H1A988.1); Avena strigosa AsCYP51H10 (ABG88965.1); Glycine max GmCYP93E1 (NP_001236154.1); BfCYP716Y1.
Surprisingly, two unigenes (comp22091 c0 and comp64057 c0), which were highly homologous to P. ginseng CYP716A47 (Han et al., 2011) and CYP716A53v2 (Han et al., 2012), were also found in these transcriptomic data, suggesting that trace amount of protopanaxadiol-type and protopanaxatriol-type ginsenosides might also be synthesized in the root of P. grandiflorum. Moreover, some unigenes homologous to G. uralensis CYP88D6 (β-amyrin 11-oxidase, Seki et al., 2008), Avena strigosa CYP51H10 (C-12, 13 epoxy and C-16 β-oxidase, Qi et al., 2006; Kunii et al., 2012; Geisler et al., 2013) and Arabidopsis CYP708A2 (thalianol hydroxylase) and CYP705A5 (thaliana-diol desaturase) were also found in the transcriptomic data of P. grandiflorum (Field et al., 2011). In the putative pathway, we proposed that the carboxylation at C-28 is before the hydroxylation reactions at other carbon atoms (Figure 1); actually it is more likely to occur in the opposite order. Even though some unigenes are homologous to the known CYP450s in other plants, further studies are needed to characterize their functions in the biosynthesis pathway of triterpenoid saponins, including those key intermediates in P. grandiflorum.
Uridine diphosphate-dependent glycosyltransferases catalyze the glucosylation of C3- and C28-carboxyl for the biosynthesis of triterpenoid saponins in P. grandiflorum (Figure 1). In the present study, 106 unigenes encoding UGTs were obtained (Supplementary Table S2), the phylogenetic relationship between these UGTs and characterized UGTs from other plants is depicted in Figure 7. Two unigenes (comp18634 c0 and comp20876 c0) were highly homologous to Barbarea vulgaris UGT73C11 and UGT73C10, which catalyze sapogenin 3-O-glucosylation (Augustin et al., 2012), suggesting that both of them have the same function in P. grandiflorum. Two unigenes (comp18634 c0 and comp20876 c0) were closely related to Saponaria vaccaria UGT74M1, which is a triterpene carboxylic acid glucosyltransferase (Meesapyodsuk et al., 2007), suggesting that these two unigenes might catalyze the glucosylation of C28-carboxyl for the biosynthesis of triterpenoid saponins. Further studies are required to characterize functionally the aforementioned four unigenes in the biosynthesis of triterpenoid saponins in P. grandiflorum.
FIGURE 7.
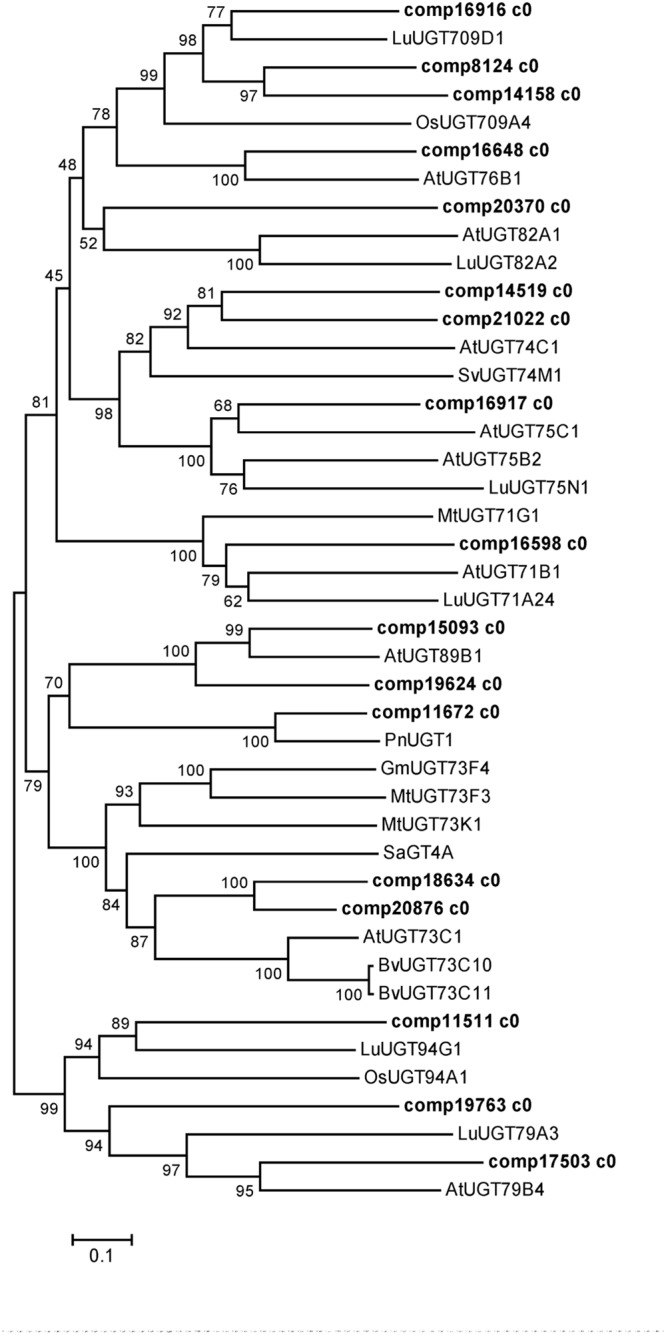
Phylogenetic tree constructed based on the deduced amino acid sequences for the P. grandiflorum UGTs (bold letters) and other plant UGTs. Accession numbers in the NCBI GenBank database are as follows: Barbarea vulgaris BvUGT73C11 (AFN26667) and BvUGT73C10 (AFN26666); Arabidopsis thaliana AtUGT73C1 (NP_181213.1), AtUGT82A1 (NP_188864.1), AtUGT76B1 (NP_187742.1), AtUGT71B1 (NP_188812.1), AtUGT89B1 (NP_177529.2), AtUGT75B2 (NP_172044.1), AtUGT75C1 (NP_193146.1), AtUGT74C1 (NP_180738.1), AtUGT79B4 (Q9LJA6.1) and AtUGT79B1 (Q9LVW3.1); Solanum aculeatissimum SaGT4A (BAD89042); M. truncatula MtUGT73K1 (AAW56091), MtUGT73F3 (ACT34898) and MtUGT71G1 (AAW56092); G max GmUGT73F4 (BAM29363); Panax notoginseng PnUGT1 (JX018210); Oryza sativa OsUGT709A4 (Q7XHR3); Saponaria vaccaria SvUGT74M1 (ABK76266); Linum usitatissimum LuUGT71A24 (AFJ52909), LuUGT82A2 (AFJ52979), LuUGT709D1 (AFJ53007), LuUGT75N1 (AFJ52962), LuUGT94G1 (AFJ53037.1), LuUGT79A3 (AFJ52973.1).
Tissue-Specific Expression of Genes Involved in the Biosynthesis of Triterpenoid Saponins
The qPCR analysis was used to investigate the tissue-specific expression patterns of 19 unigenes related to the triterpenoid saponin biosynthesis in this species. The expression pattern of these genes is shown in Figure 8. The unigenes encoding AACT, HMGS, MVK, PMK, MVD, FPPS, and SS were expressed at much higher level in leaves than in roots, young stems, and flowers (P < 0.05). The HMGR, IPPI, and SE genes showed very high expression in the flower tissue (P < 0.05). All genes mentioned above play a role in upstream biochemical reactions of the triterpenoid saponin pathway, and showed high expression at mRNA level in leaves and flowers, indicating that leaves are the factories for synthesizing the precursors of triterpenoid saponins. A high expression of β-A28O was observed in young stems (P < 0.05), but PD accumulated mainly in roots, indicating that young stems were the modification site of triterpenoid saponins before storage. UGT1 and UGT5 were expressed at much higher level in roots than in other tissues (P < 0.05), whereas the expression level of UGT1 and UGT2 was higher in P. grandiflorum as compared to that of UGT3, UGT4, UGT5, and UGT6. These results demonstrated that the expression of several genes involved in the biosynthesis of triterpenoid saponins in P. grandiflorum was in a tissue-specific manner.
FIGURE 8.
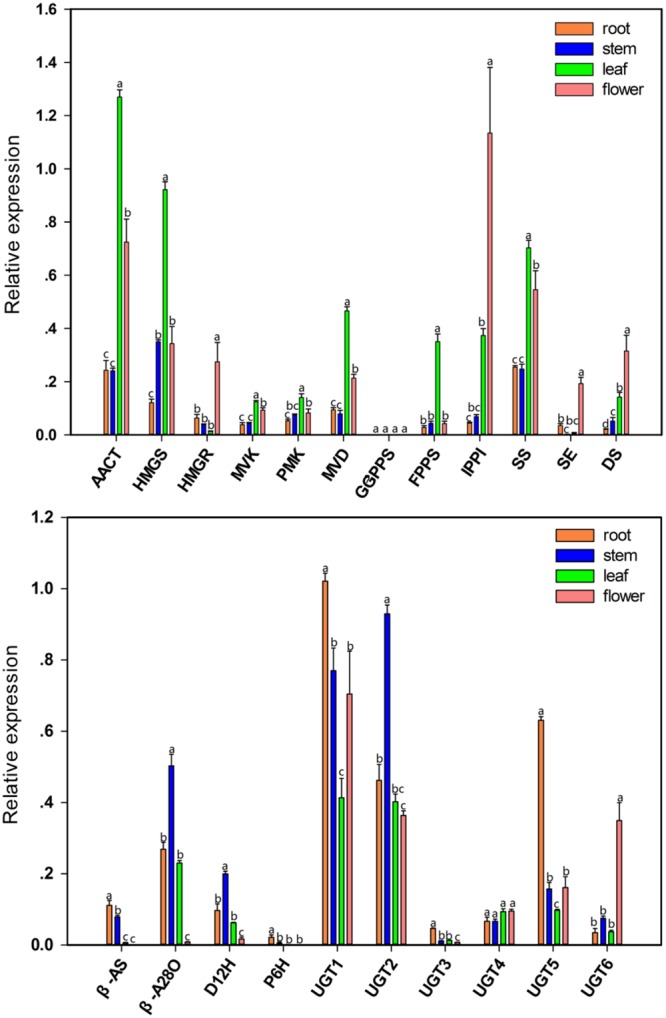
Validation of candidate unigenes involved in triterpene saponin biosynthesis in P. grandiflorum by qPCR. Bars represent the mean (± SD) of four experiments. Statistical analysis is performed with one way ANOVA with Tukey’s test to compare the difference in the mean expression level of a given gene among different tissues. P ≤ 0.05 was considered statistically significant.
SSR Marker Analysis
In order to develop SSR markers in P. grandiflorum, MISA software was used to detect the SSRs in 34,053 unigenes. A total of 10,626 SSRs were identified in 8,185 unigenes. Among them, 1,916 sequences contained more than one SSR and 807 SSRs were found in compound formation. On average, 3.33 SSRs per 10 Kb were found. In 10,626 SSRs identified the di-nucleotide repeat motifs were the most abundant types (46.05%), followed by mono (33.99%), tri- nucleotide (17.79%), tetra-nucleotide (1.77%), penta-nucleotide (0.24%), and hexa-nucleotide tandem repeats (0.16%; Tables 4 and 5).
Table 4.
Summary of SSR markers.
| Item | Number |
|---|---|
| Total number of sequences examined | 34,053 |
| Total size of examined sequences (bp) | 31,887,854 |
| Total number of identified SSRs | 10,626 |
| Number of SSR containing Sequences | 8,185 |
| Average number of SSRs per 10 kb | 3.33 |
| Number of sequences containing more than 1 SSR | 1,916 |
| Number of SSRs present in compound formation | 807 |
Table 5.
Distribution of identified SSRs using the MISA software.
| Motif | Repeat numbers |
Total | % | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 5–8 | 9–12 | 13–16 | 17–20 | 21–24 | 25–28 | 29–33 | |||
| Mono- | 0 | 1860 | 835 | 697 | 220 | 0 | 0 | 3612 | 33.99 |
| Di- | 2866 | 2027 | 0 | 0 | 0 | 0 | 0 | 4893 | 46.05 |
| Tri- | 1885 | 2 | 2 | 0 | 0 | 0 | 1 | 1890 | 17.79 |
| Tetra- | 185 | 3 | 0 | 0 | 0 | 0 | 0 | 188 | 1.77 |
| Penta- | 25 | 1 | 0 | 0 | 0 | 0 | 0 | 26 | 0.24 |
| Hexa- | 14 | 2 | 0 | 1 | 0 | 0 | 0 | 17 | 0.16 |
| Total | 4975 | 3895 | 837 | 698 | 220 | 0 | 1 | 10626 | 100.00 |
| % | 46.82 | 36.66 | 7.88 | 6.57 | 2.07 | 0.00 | 0.01 | 100.00 | |
Conclusion
Transcriptome sequencing of P. grandiflorum was performed for the first time using Illumina next-generation sequencing technologies and a total of 34,053 unigenes were obtained. Particularly, 19 unigenes involved in the biosynthesis of triterpenoid saponins were identified, the expression of which was in a tissue-specific manner. These findings will not only provide valuable information for our complete understanding of the biosynthesis pathway of triterpenoid saponins in P. grandiflorum, but also provide opportunities for the de novo production of active ingredients by engineering microorganisms. Furthermore, this study will also contribute to the improvements on this species through marker-assisted breeding or genetic engineering.
Materials and Methods
Ethics Statement
No specific permits were required for the described field studies. No specific permissions were required for these locations and activities. The location was not privately owned or protected in any way and the field studies did not involve endangered or protected species.
Plant Materials
Two-years-old P. grandiflorum plants were collected from Jianchuan County, Yunnan province, southwest of China (Latitude: 26° 16′ 13″ N, Longitude: 99° 32′ 4″ E, Altitude: 2900 m). After morphological and molecular identification according to the reference (Kim et al., 2012a), the root tissues were collected, frozen immediately in liquid nitrogen, and stored at -80°C until use.
RNA Library Preparation and Sequencing
Total RNA was extracted from roots by using Trizol reagent (Invitrogen), following by purification with RNeasy MiniElute Cleanup Kit (Qiagen) according to the manufacture’s protocol. For mRNA library construction and deep sequencing, at least 20 μg of total RNA samples were prepared by using the NEBNext® UltraTM RNA Library Prep Kit for Illumina sequencing on Hiseq 2000 platform at Novogene Bioinformatics Technology, Co. Ltd., (Beijing, China). The high quality reads obtained in this study have been deposited in the NCBI SRA database.
Transcriptome Data Processing and Assembly
The raw data processing was the same as described previously (Zhang et al., 2015). In brief, raw reads with adaptors and unknown nucleotides above 5% or those that were of low quality (containing more than 50% bases with Q-value ≤ 20) were firstly removed to obtain clean reads using a custom Perl script. Then the clean reads were de novo assembled using Trinity program (K-mer = 25, group pairsdistance = 300) with default parameters (Grabherr et al., 2011). Firstly, clean reads with a certain length of overlap were combined to form longer fragments without N, which were called contigs. These clean reads were then mapped back to the corresponding contigs with paired-end reads to detect contigs from the same transcript as well as the distances between contigs, and their paired-end information was also used to fill gaps or extend the sequences. Finally, these resultant sequences were clustered to remove redundant sequences using the TIGR gene Indices clustering tools (TGICL) to form longer sequences without N and cannot be extended on either end. Such sequences are defined as unigenes.
Functional Annotation and Predicted CDS
Functional annotations were performed as described previously (Zhang et al., 2015). Briefly, functional annotations were performed by sequence comparison with public databases, including the NCBI non-redundant nucleotide database1, non-redundant protein database, Swiss-Prot database2 and the KOG database using BLASTN and BLASTX3, with an e-value of 1e-5. A Perl script was written to assign the functional class to unigenes. Unigenes were also compared with KEGG (Kanehisa et al., 2006) using BLASTX with an e-value of less than 1e-10. A Perl script was used to retrieve KEGG Orthology (KO) information from blast result and then established pathway associations between unigenes and database. Based on the results of Nr database annotation, we used Blast2GO program (Conesa et al., 2005) to perform GO annotation of unigenes. After achieving GO annotation for every unigene, WEGO (Ye et al., 2006) software was used to perform GO classification and draw GO tree. Moreover, the conserved domains/families of the assembled unigenes encoding proteins were searched against the Pfam database (version 26.0; Finn et al., 2014) using Pfam_Scan script.
The CDS for unigene was predicted by BlastX and ESTscan. The unigene sequences were searched against the Nr, KOG, KEGG, and Swiss-Prot protein databases using BLASTX (e-value < 10-5). Unigenes aligned to a higher priority database would not be aligned to lower priority database. The best alignment results were used to determine the sequence direction of unigenes. When a unigene could not be aligned to any database, ESTScan (Iseli et al., 1999) program was used to predict coding regions and determine sequence direction.
EST-SSR Detection and Primer Design
Potential SSR markers were detected among the 34,053 unigenes using the MISA tool4 as described previously (Jiang et al., 2014). We searched for SSRs with motifs ranging from mono- to hexa-nucleotides in size. The minimum of repeat units were set as follows: 10 repeat units for mono-nucleotide, six for di-nucleotides, and five for tri-, tetra-, penta-, and hexa-nucleotides. Primer pairs were designed using Primer35 with default parameters.
Phylogenetic Analysis
Phylogenetic analysis was performed based on the deduced amino acid sequences of CYP450 and UGT from P. grandiflorum and other plants. All of the deduced amino acid sequences were aligned with Clustal X with a gap opening penalty of 10, a gap extension penalty of 0.1, a delay divergent cutoff of 25%, and the other default parameters as described previously (Jiang et al., 2014). The evolutionary distances were computed using MEGA5.10 with the Poisson correction method. For the phylogenetic analysis, a neighbor-joining tree was constructed using MEGA5.0. Bootstrap values obtained after 1000 replications are indicated on the branches. The scale represents 0.1 amino acid substitutions per site.
Quantitative Real-Time PCR (qPCR) Analysis
Nineteen unigenes with potential roles in ginsenoside biosynthesis were chosen for validation using qPCR with gene specific primers designed with Primer3 software, as described previously (Zhang et al., 2015). All the primer sequences used for the qPCR analysis are shown in Supplementary Table S3. Total RNA from different organs (roots, stems, leaves, and flowers) of P. grandiflorum were extracted individually using Trizol Kit (Promega, USA) following the manufacturer’s protocol. Subsequently, RNA was treated with 4 × g DNA wiperMix at 42°C for 2 min to remove DNA. The purified RNA (1 μg) was reverse transcribed to cDNA using HiScript QRT SuperMix for qPCR (Vazyme, Nanjing, China). The qPCR reactions were performed in a 20 μl volume composed of 2 μl of cDNA, 0.4 μl of each primer, and 10 μl 2 × SYBR Green Master mix (TaKaRa) in Roche LightCycler 2.0 system (Roche Applied Science, Branford, CT, USA). 574 PCR amplifications were performed under the following conditions: 30 s at 94°C, followed by 45 cycles of 94°C for 20 s, 55°C for 20 s, and 72°C for 30 s. Three technical replications were performed for all qPCRs. The PMK gene, which was found in our transcriptome database, was chosen as reference control for normalization after the expression of three reference genes (actin, GAPDH, and PMK) was compared in different tissues. The relative changes in gene expression levels were calculated using the 2-ΔΔCt method. For a given gene, the relative expression level was expressed as mean ± standard deviation (SD) of three determinations after normalization with the mRNA level of reference gene PMK. One way ANOVA with Tukey’s test was used to compare the difference in the mean expression level of a given gene among different organs. P ≤ 0.05 was considered statistically significant.
Author Contributions
This study was conceived by G-HZ and S-CY. The plant material preparation were carried out by M-RH and J-HS. Z-JG, J-JZ, and WZ analyzed the RNA-Seq data. C-HM and G-HZ drafted the manuscript. J-WC and C-HM revised the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was funded by the project of young and middle-aged talent of Yunnan province (Grant No. 2014HB011).
Abbreviations
- β-A28O
β-amyrin 28-oxidase
- β-AS
β-amyrin synthase
- AACT
acetyl-CoA acetyltransferase
- Api
apiose
- Ara
arabinose
- BLAST
Basic Local Alignment Search Tool
- bp
base pair
- cDNA
complementary DNA
- CDS
coding sequence
- CYPs
cytochrome P450
- DMAPP
dimethylallyl diphosphate
- FPP
farnesyl diphosphate
- FPPS
farnesyl diphosphate synthase
- Gen
gentiobiose
- Glc
glucose
- GO
Gene Ontology
- GPP
geranyl pyrophosphate
- GPPS
geranylgeranyl pyrophosphate synthase
- GT
glycosyltransferase
- HMG-CoA
3-hydroxy-3-methylglutaryl coenzyme A
- HMGR
HMG-CoA reductase
- HMGS
HMG-CoA synthase
- IPP
isopentenyl diphosphate
- IPPI
IPP isomerase
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- KOGs
Eukaryotic Orthologous Groups
- Lam
lipoarabinomannan
- MVD
mevalonate diphosphate decarboxylase
- MVK
mevalonate kinase
- NCBI
National Center for Biotechnology Information
- Nr
non-redundant protein
- PMK
phosphomevalonate kinase
- Rha
Rhamnose
- SE
squalene epoxidase
- SS
squalene synthase
- SSRs
simple sequence repeats
- Xyl
xylose
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00673
References
- Augustin J. M., Drok S., Shinoda T., Sanmiya K., Nielsen J. K., Khakimov B., et al. (2012). UDP-glycosyltransferases from the UGT73C subfamily in Barbarea vulgaris catalyze sapogenin 3-O-glucosylation in saponin-mediated insect resistance. Plant Physiol. 160 1881–1895. 10.1104/pp.112.202747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustin J. M., Kuzina V., Andersen S. B., Bak S. (2011). Molecular activities, biosynthesis and evolution of triterpenoid saponins. Phytochemistry 72 435–457. 10.1016/j.phytochem.2011.01.015 [DOI] [PubMed] [Google Scholar]
- Carelli M., Biazzi E., Panara F., Tava A., Scaramelli L., Porceddu A., et al. (2011). Medicago truncatula CYP716A12 is a multifunctional oxidase involved in the biosynthesis of hemolytic saponins. Plant Cell 23 3070–3081. 10.1105/tpc.111.087312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Luo H., Li Y., Sun Y., Wu Q., Niu Y., et al. (2011). 454 EST analysis detects genes putatively involved in ginsenoside biosynthesis in Panax ginseng. Plant Cell Rep. 30 1593–1601. 10.1007/s00299-011-1070-6 [DOI] [PubMed] [Google Scholar]
- Chun J., Ha I. J., Kim Y. S. (2013). Antiproliferative and apoptotic activities of triterpenoid saponins from the roots of Platycodon grandiflorum and their structure-activity relationships. Planta Med. 79 639–645. 10.1055/s-0032-1328401 [DOI] [PubMed] [Google Scholar]
- Chun J., Kim Y. S. (2013). Platycodin D inhibits migration, invasion, and growth of MDA-MB-231 human breast cancer cells via suppression of EGFR-mediated Akt and MAPK pathways. Chem. Biol. Interact. 205 212–221. [DOI] [PubMed] [Google Scholar]
- Conesa A., Gotz S., Garcia-Gomez J. M., Terol J., Talon M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21 3674–3676. 10.1093/bioinformatics/bti610 [DOI] [PubMed] [Google Scholar]
- Dai Z., Wang B., Liu Y., Shi M., Wang D., Zhang X., et al. (2014). Producing aglycons of ginsenosides in bakers’ yeast. Sci. Rep. 4:3698 10.1038/srep03698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field B., Fiston-Lavier A. S., Kemen A., Geisler K., Quesneville H., Osbourn A. E. (2011). Formation of plant metabolic gene clusters within dynamic chromosomal regions. Proc. Natl. Acad. Sci. U.S.A. 108 16116–16121. 10.1073/pnas.1109273108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Bateman A., Clements J., Coggill P., Eberhardt R. Y. (2014). Pfam: the protein families database. Nucleic Acids Res. 42 D222–D230. 10.1093/nar/gkt1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima E. O., Seki H., Ohyama K., Ono E., Umemoto N., Mizutani M., et al. (2011). CYP716A subfamily members are multifunctional oxidases in triterpenoid biosynthesis. Plant Cell Physiol. 52 2050–2061. 10.1093/pcp/pcr146 [DOI] [PubMed] [Google Scholar]
- Fukushima E. O., Seki H., Sawai S., Suzuki M., Ohyama K., Saito K., et al. (2013). Combinatorial biosynthesis of legume natural and rare triterpenoids in engineered yeast. Plant Cell Physiol. 54 740–749. 10.1093/pcp/pct015 [DOI] [PubMed] [Google Scholar]
- Geisler K., Hughes R. K., Sainsbury F., Lomonossoff G. P., Rejzek M., Fairhurst S., et al. (2013). Biochemical analysis of a multifunctional cytochrome P450 (CYP51) enzyme required for synthesis of antimicrobial triterpenes in plants. Proc. Natl. Acad. Sci. U.S.A. 110 E3360–E3367. 10.1073/pnas.1309157110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J. Y., Hwang H. S., Choi S. W., Kim H. J., Choi Y. E. (2012). Cytochrome P450 CYP716A53v2 catalyzes the formation of protopanaxatriol from protopanaxadiol during ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 53 1535–1545. 10.1093/pcp/pcs106 [DOI] [PubMed] [Google Scholar]
- Han J. Y., Kim H. J., Kwon Y. S., Choi Y. E. (2011). The Cyt P450 enzyme CYP716A47 catalyzes the formation of protopanaxadiol from dammarenediol-II during ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 52 2062–2073. 10.1093/pcp/pcr150 [DOI] [PubMed] [Google Scholar]
- Han J. Y., Kim M. J., Ban Y. W., Hwang H. S., Choi Y. E. (2013). The involvement of β-amyrin 28-oxidase (CYP716A52v2) in oleanane-type ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 54 2034–2046. 10.1093/pcp/pct141 [DOI] [PubMed] [Google Scholar]
- Haralampidis K., Trojanowska M., Osbourn A. E. (2002). Biosynthesis of triterpenoid saponins in plants. Adv. Biochem. Eng. Biotechnol. 75 31–49. [DOI] [PubMed] [Google Scholar]
- Hwang Y. P., Choi J. H., Kim H. G., Khanal T., Song G. Y., Nam M. S., et al. (2013). Saponins, especially platycodin D, from Platycodon grandiflorum modulate hepatic lipogenesis in high-fat diet-fed rats and high glucose-exposed HepG2 cells. Toxicol. Appl. Pharmacol. 267 174–183. 10.1016/j.taap.2013.01.001 [DOI] [PubMed] [Google Scholar]
- Iseli C., Jongeneel C. V., Bucher P. (1999). ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 138–148. [PubMed] [Google Scholar]
- Jiang N. H., Zhang G. H., Zhang J. J., Shu L. P., Zhang W., Long G. Q., et al. (2014). Analysis of the transcriptome of Erigeron breviscapus uncovers putative scutellarin and chlorogenic acids biosynthetic genes and genetic markers. PLoS ONE 9:e10035 10.1371/journal.pone.0100357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S. C., Kim W., Park S. C., Jeong J., Park M. K., Lim S., et al. (2014). Two ginseng UDP-glycosyltransferases synthesize ginsenoside Rg3 and Rd. Plant Cell Physiol. 55 2177–2188. 10.1093/pcp/pcu147 [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Hattori M., Aoki-Kinoshita K. F., Itoh M. (2006). From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 34 D354–D357. 10.1093/nar/gkj102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanal T., Choi J. H., Hwang Y. P., Chung Y. C., Jeong H. G. (2009). Saponins isolated from the root of Platycodon grandiflorum protect against acute ethanol-induced hepatotoxicity in mice. Food Chem. Toxicol. 47 530–535. 10.1016/j.fct.2008.12.009 [DOI] [PubMed] [Google Scholar]
- Kim J. W., Park S. J., Lim J. H., Yang J. W., Shin J. C., Lee S. W., et al. (2013). Triterpenoid saponins isolated from Platycodon grandiflorum inhibit hepatitis C virus replication. Evid Based Complement. Alternat. Med. 2013 560417 10.1155/2013/560417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. S., Ezaki O., Ikemoto S., Itakura H. (1995). Effects of Platycodon grandiflorum feeding on serum and liver lipid concentrations in rats with diet-induced hyperlipidemia. J. Nutr. Sci. Vitaminol. (Tokyo) 41 485–491. 10.3177/jnsv.41.485 [DOI] [PubMed] [Google Scholar]
- Kim M. O., Moon D. O., Choi Y. H., Shin D. Y., Kang H. S., Choi B. T., et al. (2008). Platycodin D induces apoptosis and decreases telomerase activity in human leukemia cells. Cancer Lett. 261 98–107. 10.1016/j.canlet.2007.11.010 [DOI] [PubMed] [Google Scholar]
- Kim T. W., Lee H. K., Song I. B., Kim M. S., Hwang Y. H., Lim J. H., et al. (2012a). Protective effect of the aqueous extract from the root of Platycodon grandiflorum on cholestasis-induced hepatic injury in mice. Pharm. Biol. 50 1473–1478. 10.3109/13880209.2012.680973 [DOI] [PubMed] [Google Scholar]
- Kim T. W., Song I. B., Lee H. K., Lim J. H., Cho E. S., Son H. Y., et al. (2012b). Platycodin D, a triterpenoid sapoinin from Platycodon grandiflorum, ameliorates cisplatin-induced nephrotoxicity in mice. Food Chem. Toxicol. 50 4254–4259. 10.1016/j.fct.2012.05.022 [DOI] [PubMed] [Google Scholar]
- Kim Y. K., Kim J. K., Kim Y. B., Lee S., Kim S. U., Park S. U. (2013). Enhanced accumulation of phytosterol and triterpene in hairy root cultures of Platycodon grandiflorum by overexpression of Panax ginseng 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J. Agric. Food Chem. 61 1928–1934. 10.1021/jf304911t [DOI] [PubMed] [Google Scholar]
- Kunii M., Kitahama Y., Fukushima E. O., Seki H., Muranaka T., Yoshida Y., et al. (2012). β-Amyrin oxidation by oat CYP51H10 expressed heterologously in yeast cells: the first example of CYP51-dependent metabolism other than the 14-demethylation of sterol precursors. Biol. Pharm. Bull. 35 801–804. 10.1248/bpb.35.801 [DOI] [PubMed] [Google Scholar]
- Lee K. J., Choi C. Y., Chung Y. C., Kim Y. S., Ryu S. Y., Roh S. H., et al. (2004). Protective effect of saponins derived from roots of Platycodon grandiflorum on tert-butyl hydroperoxide-induced oxidative hepatotoxicity. Toxicol. Lett. 147 271–282. 10.1016/j.toxlet.2003.12.002 [DOI] [PubMed] [Google Scholar]
- Lee K. J., Choi J. H., Kim H. G., Han E. H., Hwang Y. P., Lee Y. C., et al. (2008). Protective effect of saponins derived from the roots of Platycodon grandiflorum against carbon tetrachloride induced hepatotoxicity in mice. Food Chem. Toxicol. 46 1778–1785. 10.1016/j.fct.2008.01.017 [DOI] [PubMed] [Google Scholar]
- Li C., Zhu Y., Guo X., Sun C., Luo H., Song J., et al. (2013). Transcriptome analysis reveals ginsenosides biosynthetic genes, microRNAs and simple sequence repeats in Panax ginseng C. A. Meyer. BMC Genomics 14:245 10.1186/1471-2164-14-245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Cheng H., Gai J., Yu D. (2007). Genome-wide identification and characterization of putative cytochrome P450 genes in the model legume Medicago truncatula. Planta 226 109–123. 10.1007/s00425-006-0473-z [DOI] [PubMed] [Google Scholar]
- Li T., Xu W. S., Wu G. S., Chen X. P., Wang Y. T., Lu J. J. (2014). Platycodin D induces apoptosis, and inhibits adhesion, migration and invasion in HepG2 hepatocellular carcinoma cells. Asian Pac. J. Cancer Prev. 15 1745–1749. 10.7314/APJCP.2014.15.4.1745 [DOI] [PubMed] [Google Scholar]
- Luo H., Sun C., Sun Y., Wu Q., Li Y., Song J., et al. (2011). Analysis of the transcriptome of Panax notoginseng root uncovers putative triterpene saponin-biosynthetic genes and genetic markers. BMC Genomics 12:S5 10.1186/1471-2164-12-S5-S5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meesapyodsuk D., Balsevich J., Reed D. W., Covello P. S. (2007). Saponin biosynthesis in Saponaria vaccaria. cDNAs encoding β-amyrin synthase and a triterpene carboxylic acid glucosyltransferase. Plant Physiol. 143 959–969. 10.1104/pp.106.088484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra B. B. (2014). An updated snapshot of recent advances in transcriptomics and genomics of phytomedicinals. J. Postdoc. Res. 2 1–15. [Google Scholar]
- Moses T., Papadopoulou K. K., Osbourn A. (2014a). Metabolic and functional diversity of saponins, biosynthetic intermediates and semi-synthetic derivatives. Crit. Rev. Biochem. Mol. Biol. 49 439–462. 10.3109/10409238.2014.953628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses T., Pollier J., Almagro L., Buyst D., Van Montagu M., Pedreño M. A., et al. (2014b). Combinatorial biosynthesis of sapogenins and saponins in Saccharomyces cerevisiae using a C-16α hydroxylase from Bupleurum falcatum. Proc. Natl. Acad. Sci. U.S.A. 111 1634–1639. 10.1073/pnas.1323369111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X., Bakht S., Qin B., Leggett M., Hemmings A., Mellon F., et al. (2006). A different function for a member of an ancient and highly conserved cytochrome P450 family: from essential sterols to plant defense. Proc. Natl. Acad. Sci. U.S.A. 103 18848–18853. 10.1073/pnas.0607849103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki H., Ohyama K., Sawai S., Mizutani M., Ohnishi T., Sudo H., et al. (2008). Licorice β-amyrin 11-oxidase, a cytochrome P450 with a key role in the biosynthesis of the triterpene sweetener glycyrrhizin. Proc. Natl. Acad. Sci. U.S.A. 105 14204–14209. 10.1073/pnas.0803876105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya M., Hoshino M., Katsube Y., Hayashi H., Kushiro T., Ebizuka Y. (2006). Identification of β-amyrin and sophoradiol 24-hydroxylase by expressed sequence tag mining and functional expression assay. FEBS J. 273 948–959. 10.1111/j.1742-4658.2006.05120.x [DOI] [PubMed] [Google Scholar]
- Shin C. Y., Lee E. B. (2002). Platycodin D and D3 increase airway mucin in vivo and in vitro in rates and hamsters. Planta Med. 68 221–225. 10.1055/s-2002-23130 [DOI] [PubMed] [Google Scholar]
- Shin D. Y., Kim G. Y., Li W., Choi B. T., Kim N. D., Kang H. S., et al. (2009). Implication of intracellular ROS formation, caspase-3 activation and Egr-1 induction in platycodon D-induced apoptosis of U937 human leukemia cells. Biomed. Pharmacother. 63 86–94. 10.1016/j.biopha.2008.08.001 [DOI] [PubMed] [Google Scholar]
- Sun C., Li Y., Wu Q., Luo H., Sun Y., Song J., et al. (2010). De novo sequencing and analysis of the American ginseng root transcriptome using a GS FLX Titanium platform to discover putative genes involved in ginsenoside biosynthesis. BMC Genomics 11:262 10.1186/1471-2164-11-262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi K., Lee E. B. (1972). Pharmacological studies on Platycodon grandiflorum A. DC. III. Activities of crude platycodin on respiratory and circulatory systems and its other pharmacological activities. Yakugaku Zasshi 92 969–973. [DOI] [PubMed] [Google Scholar]
- Xie Y., Sun H. X., Li D. (2009). Platycodin D is a potent adjuvant of specific cellular and humoral immune responses against recombinant hepatitis B antigen. Vaccine 27 757–764. 10.1016/j.vaccine.2008.11.029 [DOI] [PubMed] [Google Scholar]
- Ye J., Fang L., Zheng H., Zhang Y., Chen J. (2006). WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 34 293–297. 10.1093/nar/gkl031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yendo A. C., de Costa F., Gosmann G., Fett-Neto A. G. (2010). Production of plant bioactive triterpenoid saponins: elicitation strategies and target genes to improve yields. Mol. Biotechnol. 46 94–104. 10.1007/s12033-010-9257-6 [DOI] [PubMed] [Google Scholar]
- Zhang G. H., Ma C. H., Zhang J. J., Chen J. W., Tang Q. Y., He M. H., et al. (2015). Transcriptome analysis of Panax vietnamensis var. fuscidicus discovers putative ocotillol-type ginsenosides biosynthesis genes and genetic markers. BMC Genomics 2015:159 10.1186/s12864-015-1332-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


