Abstract
Strain specific mouse brain magnetic resonance imaging (MRI) atlases provide coordinate space linked anatomical registration. This allows longitudinal quantitative analyses of neuroanatomical volumes and imaging metrics for assessing the role played by aging and disease to the central nervous system. As NOD/scid-IL-2Rγcnull (NSG) mice allow human cell transplantation to study human disease, these animals are used to assess brain morphology. Manganese enhanced MRI (MEMRI) improves contrasts amongst brain components and as such can greatly help identifying a broad number of structures on MRI. To this end, NSG adult mouse brains were imaged in vivo on a 7.0 Tesla MR scanner at an isotropic resolution of 100 µm. A population averaged brain of 19 mice was generated using an iterative alignment algorithm. MEMRI provided sufficient contrast permitting 41 brain structures to be manually labeled. Volumes of 7 humanized mice brain structures were measured by atlas-based segmentation and compared against non-humanized controls. The humanized NSG mice brain volumes were smaller than controls (p<0.001). Many brain structures of humanized mice were significantly smaller than controls. We posit that the irradiation and cell grafting involved in the creation of humanized mice were responsible for the morphological differences. Six NSG mice without MnCl2 administration were scanned with high resolution T2-weighted MRI and segmented to test broad utility of the atlas.
Keywords: In vivo mouse brain atlas, MEMRI, Mouse brain morphology, Atlas-based segmentation, NSG mice
Introduction
Advancements in genetic engineering enabled wide spread use of transgenic mice for biomedical research. These mice are extensively used in studies of cell, tissue and organism growth, differentiation and disease. Immune deficiency induced by affecting the integrity of the adaptive immune system in NOD/scid-IL-2Rγcnull (NSG) mice permitted the efficient and sustained engraftment of human immunocytes in mice (Ito et al. 2002; Saito et al. 2002). As a result, these mice are used for studies of a broad range of human diseases covering the disciplines of oncology, hematology, infectious disease and regenerative medicine. In particular, our laboratories and others have pursued investigation of human immunodeficiency virus type one (HIV-1) pathobiology including the studies of viral reservoirs and direct tissue injuries including the lung and the central nervous system (CNS) (Janus and Welzl 2010; Trancikova et al. 2011; Gorantla et al. 2012).
Apropos to studies of end organ diseases associated with HIV-1 infection, magnetic resonance imaging (MRI) has provided critical insights into the mechanisms of virus-induced damage as well as repair following antiretroviral therapy (ART). We posit that such investigations can be substantively improved if specific mouse atlases are generated. Such an atlas could permit broad longitudinal investigation of brain morphology under conditions that mimic aspects of human neurologic disease. Specifically, brain parcellation can automate analyses of structure-wise MRI based metrics (e.g., T1 and T2 relaxation times, diffusion tensor imaging (DTI) measures, metabolites concentrations, pharmacokinetics and pharmacodynamics (PK and PD), and drug biodistribution of magnetically labeled cells and nanomaterials). These works would serve to complement and extend analyses of morphological aberrations seen during progressive infection. Such measures could also be harnessed as biomarkers of disease as well as to determine drug efficacy.
Mouse brain atlases were developed by others (Kovacevic et al. 2005; Ma et al. 2005; Chan et al. 2007; Dorr et al. 2008; Aggarwal et al. 2009; Chuang et al. 2011; Nie and Shen 2013; Sunkin et al. 2013). Such works were heralded through the need to integrate gene expression with neuroanatomical data and now available as an online public resource (Lein et al. 2007). Indeed, recent studies have focused on generating developmental and functional brain atlases (Chuang et al. 2011). The realization of multi-dimensional (multi-modality and/or multi-parametric) data is notable (MacKenzie-Graham et al. 2004; Aggarwal et al. 2009).
Acquisition of high resolution data with high signal-to-noise ratios (SNR) involves long MRI scanning times that are difficult in a live mouse. To overcome such difficulties, most researchers have performed ex vivo imaging on brain-in-skull or fixed brains and created mouse brain atlases. However, tissue deformations that are common and are linked to the type and duration of brain preparation and fixation methods (de Guzman et al. 2013) may affect the atlases generated. Thus, in vivo MRI data based atlases can help improving accuracy of brain atlases constructed and can be used for longitudinal analyses of individual mice. In vivo atlases on C57BL/6J mouse brain were generated before by others (Ma et al. 2008; Bai et al. 2012). We wished to take this idea a step further in sensitivity through the use of manganese enhanced MRI (MEMRI). Administration of MnCl2 shortens T1 relaxation times in most brain structures, improving signal to noise per unit time and providing excellent contrast between many brain substructures including hippocampus, olfactory bulb, cerebellum, and cerebral cortical layers as noticed in the present study and previous studies (Aoki et al. 2004; Silva et al. 2008). This allows T1-weighted brain MRI at high field strength to be used to acquire high resolution in vivo images while providing enhanced contrast for brain structure identifications. As high-resolution 3D MRI show significant neuroanatomical differences between mouse strains (Chen et al. 2006), generation of a brain atlas on the same genetic background as used for a disease model serves to enhance accuracy of brain tissue segmentation on MRI.
To these ends, the current study developed a 3D in vivo MEMRI atlas of NSG mouse and used this atlas to compare brain morphological changes between normal and humanized mice. Our overarching goal is to analyze morphological and MRI based parametric changes that follow HIV-1 disease progression. This can be used to identify disease biomarkers and to assess treatment strategies. We posit that such investigations will allow a more complete elucidation of the broad effects that HIV-1 infection has on the nervous system. In addition, mapping of MEMRI atlas labels onto T2-weighted brain images without MnCl2 administration demonstrated the atlas utility for longitudinal studies.
Materials and Methods
Experimental Animals
Nineteen NSG mice (male, weight=28.5±2.4 g, age~1 year) from a University of Nebraska Medical Center (UNMC) breeding colony were used in study. Animals were maintained in sterile microisolator cages under pathogen-free conditions in accordance with ethical for care of laboratory animals at UNMC set forth by the National Institutes of Health. All procedures were approved by the University’s Institutional Animal Care and Use Committee. Seven human CD34+ hematopoietic stem cells (HSC) reconstituted (humanized) NSG mice (male, weight=22.1±5.3 g, age~1 year) were scanned using MEMRI to study brain morphology. Additional 6 NSG mice (male, weight=30.6±2.9 g, age~1.5 years) were included in the study for whole brain T2-weighted MRI data acquisition without MnCl2 administration.
Human CD34+ HSC Reconstitution (Humanization) of NSG Mice
CD34-NSG mice were generated as described in (Gorantla et al. 2012). Human CD34+ HSC were obtained from cord blood (Department of Gynecology and Obstetrics, UNMC) and enriched to high purity by magnetic bead selection (Miltenyi Biotech Inc., Auburn, CA). The purity of CD34+ cells was >90 % by flow cytometry. Cells were transplanted into newborn mice irradiated at 1Gy using a C9 cobalt 60 source (Picker Corporation). CD34+ cells were injected intrahepatically at 105 cells/mouse in 20 µl of PBS using a 30 gauge needle. The levels of engraftment and number of human cells in peripheral blood were analyzed by flow cytometry (Dash et al. 2011).
MnCl2 Administration
MnCl2.4H2O (Sigma-Aldrich, St Louis, MO) was added to saline (0.9 % w/v of NaCl solution) to make 120 mM MnCl2 solution. MnCl2 was administered at a dose of 125 mg/kg bodyweight using intravenous (i.v.) injections through the tail vein. MnCl2 was injected using a syringe pump (Harvard Apparatus, MA) at the rate of 125 µL/h. The dosing scheme was designed based on our experience in MEMRI and several previous studies (Koretsky and Silva 2004; Silva et al. 2004, 2008; Lee et al. 2005; Kuo et al. 2006). Mice were placed on an electrically heated tail vein injection platform (Braintree Scientific, MA), and were anesthetized by inhalation of isoflurane in 100 % oxygen. Breathing rate, cardiac rate and blood oxygen saturation were continuously monitored. Anesthesia level was varied from 0.3 to 1.5 % isoflurane to maintain the breathing rate between 40 and 100 breaths per minute. Immediately after injection, the mouse was placed on a heating pad in the cage, and its behavior was observed up to 4 h to detect any side effects of MnCl2. The animal was then returned to animal facility and scanned 24 h later.
MRI Data Acquisition
MRI of the 19 NSG mice used for atlas generation were scanned 24 h after MnCl2 administration on Bruker Biospec 7T (Bruker, Billerica, MA) operating Paravision 4.0 with a custom-built 18 mm birdcage volume coil. The humanized mice that were used to study the effect of humanization on brain volume were scanned using the same MRI scanner operating Paravision 5.1. An 82 mm actively decoupled volume resonator was used for signal transmission and a four-channel phase array coil was used for reception.
Mice were anesthetized by inhalation of isoflurane in 100 % oxygen and maintained 40–80 breaths/min. Three-dimensional T1-weighted data were acquired using a Rapid Acquisition with Relaxation Enhancement (RARE) sequence with the following parameters: Repetition time (TR)=400 ms, Effective echo time (TEeff)=7.2 ms, RARE factor=4, number of averages=1, image matrix=176×128×128 with 100 µm isotropic pixel size, total scan time=27 min, anterior-posterior as the readout direction. MRI data were acquired from both normal and humanized mice. Three-dimensional T2-weighted MRI were obtained from six NSG mice without MnCl2 administration using the same scanning parameters as for 3D T1-weighted data except: TR/TEeff=1500/36 ms, RARE factor = 8, number of averages=1, total scan time=1 h 55 m.
Population Averaged MRI Mouse Brain
All MR brain images were manually brain extracted by separating brain from extracranial tissue using Analyze 10.0v software (www.analyzedirect.com). All brains were registered to median size brain in the group using rigid image registration. Population average brain was created by averaging all registered individual brain images. Then all individual brain images were iteratively (3 times) affine registered to population average brain and average was updated at each iteration (Kovacevic et al. 2005). Finally, nonlinear registration of individual brain images to the average was performed using Large Deformation Diffeomorphic Metric Mapping (LDDMM) to align differences (Beg et al. 2005). To minimize the interpolation errors, transformation matrices from individual registrations were combined and applied in one step to each original MRI to generate the final average. All the registration procedures were performed using Diffeomap 1.6v as implemented in DTIStudio software (www.mristudio.org). The final step was to sharpen the boundaries between anatomic features (enhanced brain) by applying the Laplacian as:
where g(x, y) and f(x, y) represent enhanced and input images respectively, and ∇2 represents the Laplacian operator.
Humanized Mouse Brain Data Processing
Humanized mouse images acquired using the volume-phase array coil system were first corrected for intensity inhomogeneity using a nonparametric nonuniform normalization (N3) of intensity method (Sled et al. 1998). After the correction, brains were manually extracted using Analyze10.0v software. Individual brain images were registered to the averaged brain image of the 19 NSG mice first using linear transformations followed by LDDMM as described previously. Laplacian operation was applied on these images as well.
Structures Delineation and Labeling
Paxinos atlas (Paxinos and Franklin 2001) and Allen brain digital atlas (Lein et al. 2007) (http://mouse.brain-map.org/) were followed as reference for identifying and naming different structures on the averaged MEMRI brain images. Amira® 5.21v VSG software (www.amira.com) was used for generating colored labels of brain structures. A three dimensional view with connected cursor was used for accurate identification of various structures.
Atlas Based Segmentation
Volumes of individual brain structures from NSG (n=19) and humanized mice (n=7) were measured by performing atlas based segmentation (Nie and Shen 2013). Here, image transformation matrices were obtained by registering the population averaged MRI to individual mouse brain. These matrices were then applied on atlas to transform atlas labels onto individual brains. The transformed color coded labels provided the volume of each brain structure. A Student t-test was carried out to evaluate the statistical significance of differences in measurements between control and humanized mice. Brain structures on T2-weighted images were segmented in similar manner.
Results
Brain Structures Labeling
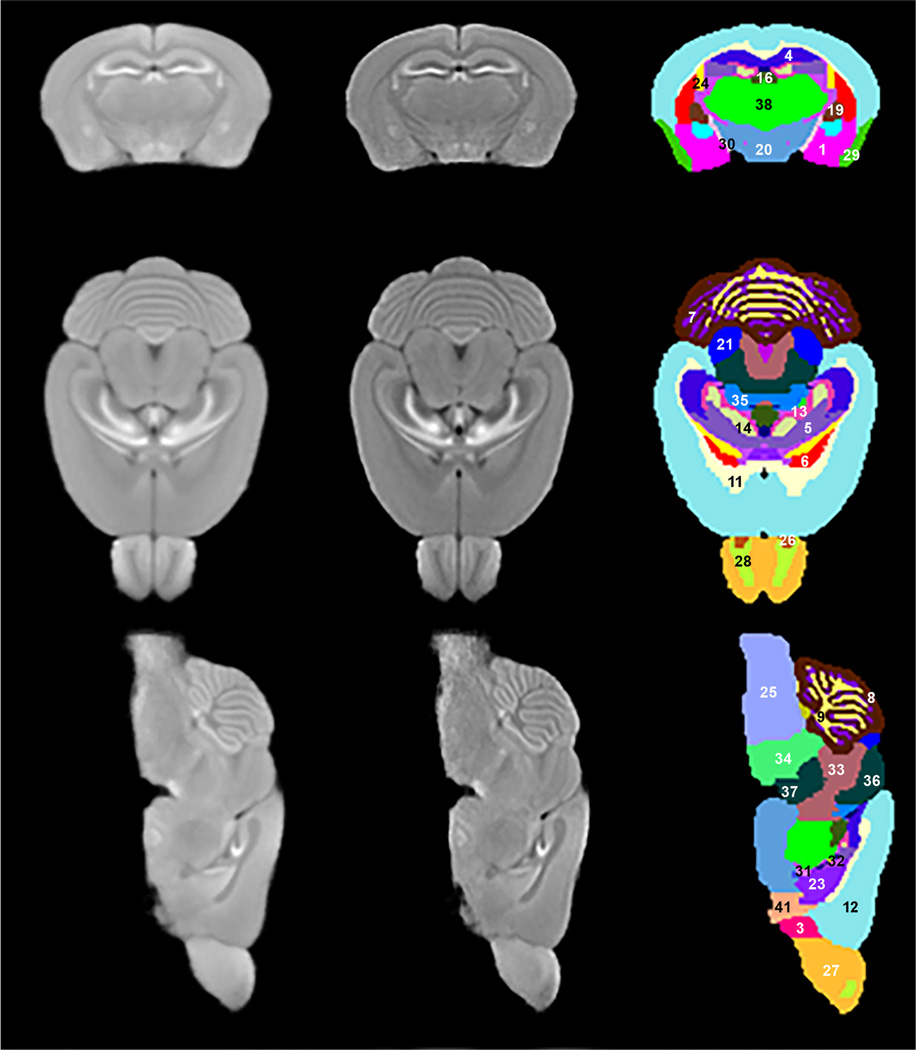
Representative slices from three orthogonal cross-sections of the population averaged MRI are shown in the first column of Fig. 1. The second displays the same slices with Laplacian edge enhancement. Improved contrast between structures is realized. The third column shows the manually labeled structures using Amira software. Significant image contrast seen is due to MnCl2 and allowed the identification and delineation of 41 brain structures from the cerebrum (CH), brain stem (BS), cerebellum (CB), fiber tracts (FB), and ventricular systems (VS). All the identified structures’ names are listed in Table 1. To the best of our knowledge this is the highest number of structures identified on in vivo mouse brain MRI.
Fig. 1.
Columns: First: Three orthogonal planes of population average MRI. Second: Edge enhancement of images in first column. Third: Identified structures on these planes: 


 (Note: Structures (2) ac, (10) AQ, (15) DN, (17) RFB, (18) FN (22) IP, (39) V3, and (40) V4 are not presented in the third column of the figure)
(Note: Structures (2) ac, (10) AQ, (15) DN, (17) RFB, (18) FN (22) IP, (39) V3, and (40) V4 are not presented in the third column of the figure)
Table 1.
Volumes of brain structures from normal (n=19) and humanized (n=7) mice.
| Region | Structure | Normal Mean ± SD (mm3) (n=19) |
Humanized Mean ± SD (mm3) (n=7) |
|---|---|---|---|
| CH: Cerebrum | |||
| Isocortex: Isocortex | Isocortex: Isocortex | 116.64±4.49 | 97.46±6.32 |
| OLF: Olfactory areas | MOBgl: Main olfactory bulb, glomerular layer | 15.44±1.37 | 12.11±1.06 |
| MOBgr: Main olfactory bulb, granule layer | 3.36±0.35 | 2.60±0.26 | |
| AOB: Accessory olfactory bulb | 0.79±0.08 | 0.66±0.05 | |
| AON: Anterior olfactory nucleus | 4.77±0.28 | 4.13±0.19 | |
| PIR: Piriform area | 12.56±0.48 | 10.43±0.65 | |
| HPF: Hippocampal formation | CA1_CA2_SUB: Field CA1+Field CA2+Subiculum | 17.32±0.55 | 13.68±1.38 |
| CA3: field CA3 of hippocampus | 7.87±0.23 | 6.51±0.59 | |
| DG-mo: Dentate gyrus_molecular layer | 3.35±0.13 | 2.75±0.31 | |
| DG-(po+sg): Dentate gyrus_(polymorph layer+granular layer) | 3.06±0.13 | 2.54±0.29 | |
| STR: Striatum | CP: Caudoputamen | 23.30±0.84 | 19.95±1.24 |
| STRv: Striatum ventral region | 12.06±0.52 | 10.00±0.61 | |
| LSX: Lateral septal complex | 4.41±0.16 | 3.99±0.17 | |
| PAL: Pallidum | PALc: Pallidium, caudal region | 2.44±0.12 | 2.15±0.11 |
| GP: Globus pallidus | 3.15±0.12 | 2.74±0.16 | |
| MS: Medial septal nucleus | 0.35±0.02 | 0.32±0.01 | |
| AMY: Amygdala | AMY: Amygdala | 18.1±0.72 | 14.55±0.92 |
| FB: Fiber tracts | |||
| cc: corpus callosum | 11.85±0.58 | 9.84±0.76 | |
| opt: optic tract | 0.85±0.04 | 0.72±0.05 | |
| ac: anterior commissure | 0.82±0.03 | 0.71±0.04 | |
| RFB: Rest of fiber tracts | 9.13±0.3 | 7.9±0.5 | |
| BS: Brain stem | |||
| TH: Thalamus | 19.67±0.58 | 17.37±1.12 | |
| EPI: Epithalamus | 0.87±0.04 | 0.79±0.06 | |
| HY: Hypothalamus | 19.02±0.69 | 16.51±0.57 | |
| IC: Inferior colliculus | 6.50±0.28 | 5.58±0.36 | |
| PAG: Periaqueductal gray | 7.59±0.24 | 6.71±0.32 | |
| PRT: Pretectal region | 2.59±0.08 | 2.26±0.21 | |
| SN: Substantia nigra | 2.63±0.10 | 2.36±0.12 | |
| RMB: Rest of midbrain | 23.95±0.56 | 20.82±1.36 | |
| P: Pons | 19.68±0.88 | 17.46±0.85 | |
| MY: Medulla | 52.51±3.12 | 43.55±2.62 | |
| CB: Cerebellum | |||
| CBXmo: Cerebellar cortex, molecular layer | 44.35±1.57 | 36.06±3.19 | |
| CBXgr: Cerebellar cortex, granular layer | 11.65±0.42 | 9.24±0.92 | |
| CBwm: Cerebellar white matter | 14.20±0.48 | 11.46±1.18 | |
| FN: Fastigial nucleus | 0.64±0.04 | 0.54±0.06 | |
| IP: Interpose nucleus | 1.07±0.05 | 0.88±0.12 | |
| DN: Dentate nucleus | 0.72±0.05 | 0.58±0.07 | |
| VS: Ventricular system | |||
| VL: Lateral ventricles | 2.47±0.09 | 2.16±0.16 | |
| V3: Third ventricle* | 0.52±0.03 | 0.47±0.04 | |
| AQ: Cerebral aqueduct* | 0.123±0.01 | 0.121±0.01 | |
| V4: Fourth ventricle | 1.21±0.07 | 1.00±0.09 | |
| Brain volume | 503.63±13.05 | 421.66±25.62 |
‘*’ denotes NOT significant (p>0.01) volume differences
Volume Comparison of Individual Brain Structures
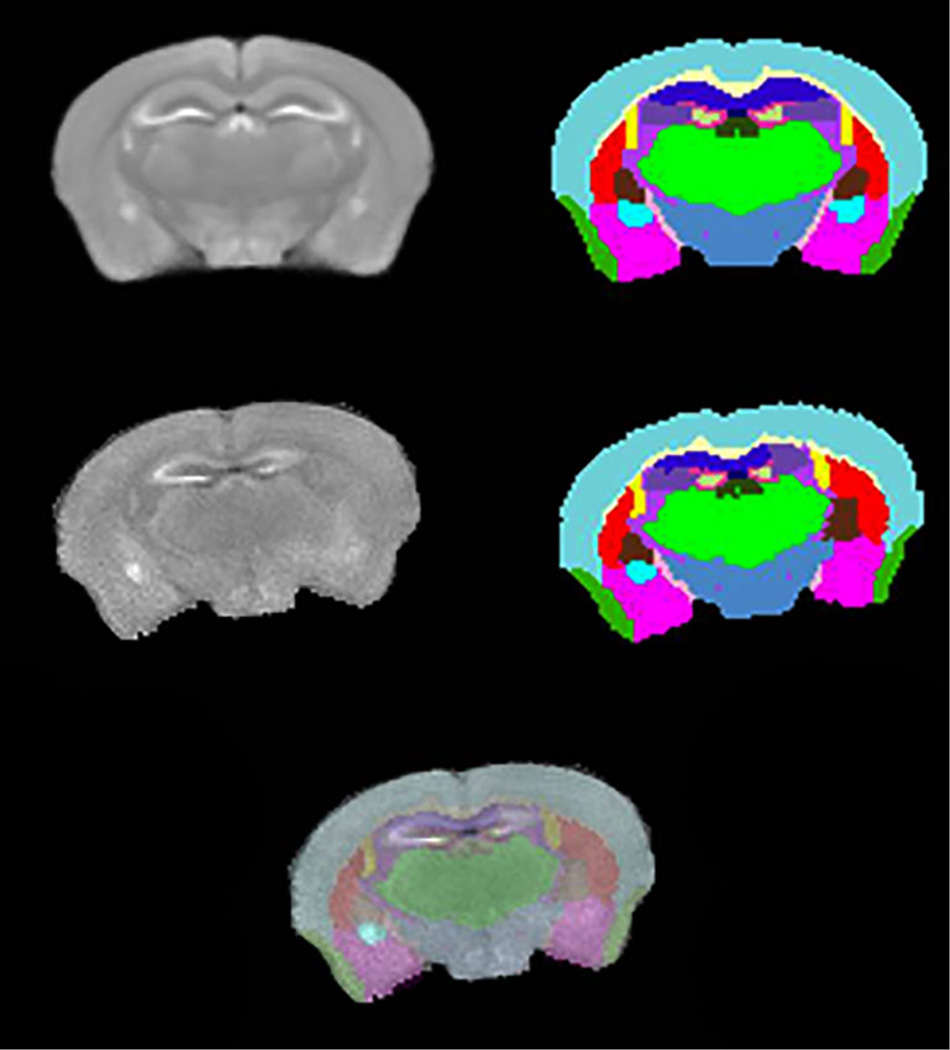
Image segmentations were performed using the developed mouse brain atlas and the corresponding population average MRI to identify structures of individual brains. For visual assessment of the segmentation quality, a segmented cross-section is shown in Fig. 2. The first row of Fig. 2 has one slice of average MRI and the corresponding atlas labels. In the second row, the first image is a slice from one of the humanized mice and the second image is the corresponding transformed labels from atlas. The quality of segmentation is shown in third row by overlaying the transformed labels of humanized mouse onto the corresponding MR image. The average and standard deviations of volumes for all identified structures are shown in Table 1. Almost all structures of humanized mice are significantly smaller than the corresponding volumes in unaltered NSG mice. In addition, the total brain sizes of humanized mice are significantly smaller than the NSG mice (p<0.001). Structure volumes were normalized with respect to whole brain volumes to remove the effect of brain size on comparison of individual structures. After normalization, 12 structures were found to be significantly different (p<0.01) for humanized compared to control mice. The results are shown in Table 2.
Fig. 2.
Rows: First: A cross-section from population averaged MRI and the corresponding labeled slice. Second: A similar location MR slice from a humanized mouse and the corresponding transformed labels. Third: Overlay of transformed labels on the corresponding MRI
Table 2.
Significantly different (p<0.01) normalized brain sub-region volumes (%) in control and humanized NSG mice
| S. No. | Structure | Unaltered (mean ± SD) |
Humanized (mean ± SD) |
|---|---|---|---|
| 1 | TH | 3.905±0.089 | 4.142±0.112 |
| 2 | VL | 0.491±0.019 | 0.512±0.014 |
| 3 | PAG | 1.508±0.045 | 1.592±0.038 |
| 4 | LSX | 0.877±0.037 | 0.948±0.042 |
| 5 | SN | 0.522±0.017 | 0.561±0.020 |
| 6 | P | 3.908±0.139 | 4.145±0.103 |
| 7 | RMB | 4.756±0.068 | 4.494±0.085 |
| 8 | PALc | 0.486±0.021 | 0.512±0.017 |
| 9 | RFB | 1.814±0.047 | 1.873±0.033 |
| 10 | CA1_CA2_Sub+opt | 3.440±0.082 | 3.239±0.134 |
| 11 | EPI | 0.173±0.009 | 0.187±0.006 |
| 12 | AQ | 0.025±0.001 | 0.029±0.002 |
Atlas Registration to T2-weighted Images
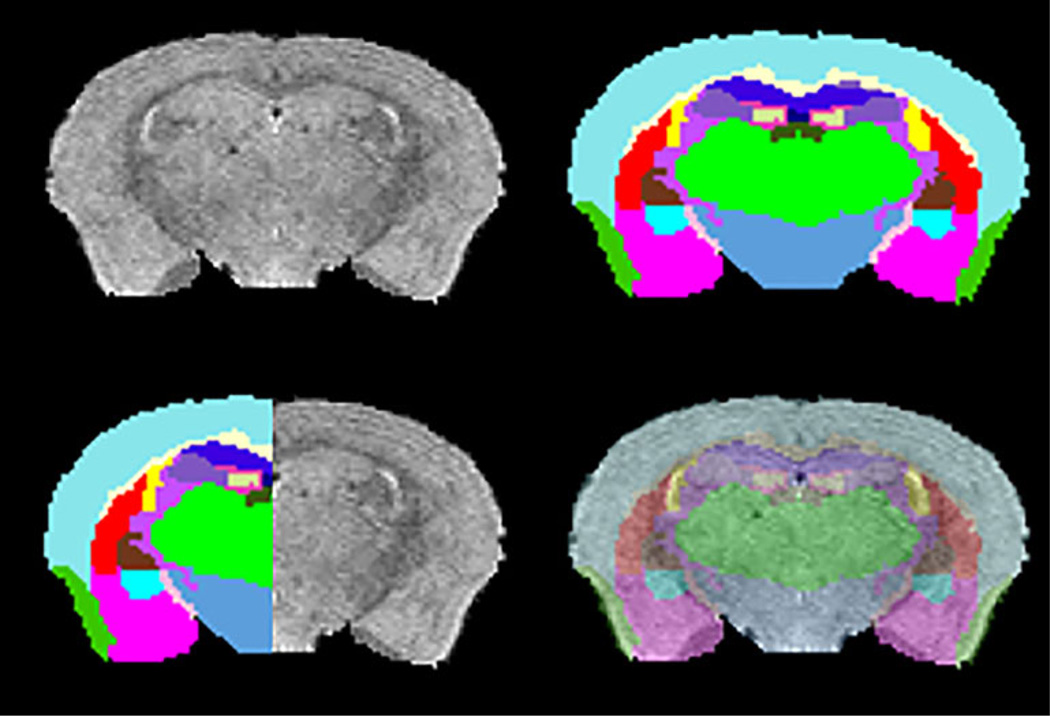
Neurotoxicity and long washout times associated with the administration of MnCl2 limits the use of MEMRI for longitudinal studies of individual mice. In order to overcome this limitation, we have used the MEMRI atlas to segment high resolution T2-weighted MRI of 6 NSG mice without injecting MnCl2. A good agreement between MRI cross-section and the corresponding registered labels is shown in first row of Fig. 3. For visual assessment, half image with MRI and the other half with transformed labels of the same cross-section and overlay of labels on the MRI in transparent mode are shown in second row. It can be clearly seen that the atlas was registered well onto the T2-weighted image, and thus the structures were identified accurately. The quality of parcellation was similar in all 6 mice.
Fig. 3.
Rows: First: A cross section from a T2-weighted brain image and the corresponding brain labels obtained by registering with MEMRI atlas. Second: Qualitative assessment of segmentation accuracy by matching labels and MRI image from the same cross section and overlaying labels onto T2-weight slice in transparent mode
Discussion
We have developed a 3D in vivo MEMRI brain atlas for NSG mice containing 41 sub regions. Noted volumetric comparison of these brain regions was made following humanization. We acknowledge that although this is not the first mouse brain atlas made, it is the sole one constructed by MEMRI for NSG mice analyses. Majority of previous such constructions were made on fixed or post-mortem in situ brains (Ma et al. 2005; Dorr et al. 2008; Aggarwal et al. 2009). However, such prior works may not provide accurate assessment of in vivo volumetric and geometrical changes amongst brain regions (Ma et al. 2008; Aggarwal et al. 2011). Indeed, fixation protocols cause alterations in analyses for brain morphology even when MRI data with high resolution and SNR are employed (de Guzman et al. 2013). To this end, the present in vivo MEMRI brain atlas allows longitudinal quantitative morphological studies.
There are some advantages of the present approach. First, due to MnCl2 ability to selectively reduce local T1 relaxation times, the MEMRI provided increased contrast to noise ratio. Second, boundaries between brain structures are enhanced by Laplace Transform image processing. Third, specific molecular and granular layers in the brain regions such as the olfactory bulb and cerebellum were identified (Fig. 1). Through such an approach, 41 structures on averaged in vivo MRI were delineated and then labeled. This is a significant improvement from prior 3D in vivo MRI atlases that enabled only half of the MEMRI-identified brain structures to be processed (Ma et al. 2008; Bai et al. 2012).
The use of humanized mice in the present study is of particular interest as they are used to study various human diseases. For instance, humanized mice can readily be infected by HIV-1 and have been used successfully to develop new treatment regimens to combat disease (Guo et al. 2013, 2014; Puligujja et al. 2013). As mouse brain morphology is strain dependent (Chen et al. 2006), the current brain atlas can be specifically used to assess how the virus can alter specific brain structures and in regards to specific morphological and parametric changes that occur over time. It is also recently shown that the brain pathology during the progressive HIV-1 infection in humanized mice alters the MEMRI signal in specific brain regions (Bade et al. 2015). The developed atlas is made available to researchers through Neuroimaging Informatics Tools and Resources Clearinghouse (NITRC) website (https://www.nitrc.org/projects/memribrainatlas/).
All together the current in vivo MEMRI based NSG mouse brain atlas can assess how humanization of lymphoid tissues in NSG mice can alter brain structural integrity. For example, we found significant brain volume reductions (p<0.01) during the humanization procedure with a number of specific brain structures showing significant volume reductions (Table 1). In support of such observations, prior studies have shown that infant irradiation, as applied for our humanized mice, can affect brain development (Hossain et al. 2005; Manda et al. 2009; Rao et al. 2011; Gazdzinski et al. 2012; Park et al. 2012). Histological studies of hippocampus of adult mice show permanent brain deficits when mice were prenatally irradiated (Hossain et al. 2005). Our own observations support such prior investigations as we have seen significant volume changes at 1 year of age and indicating that the structural volume deficits continue until adulthood and have permanent effects on the brain. Such morphological results likely represent combined effect of irradiation and cell engraftment. Specifically, normalized volume changes in 12 structures suggest regional specificity (Table 2). Future assessment of irradiation and cell engraftment made over time will be necessary to assess the individual manipulations of the animals for brain development.
For longitudinal studies on individual mice, repeated MEMRI with multiple Mn2+ administrations could be problematic. Accumulation of manganese in brain as a consequence of repeated MEMRI can result in neurotoxicity. Due to potential toxicities, multiple injections could interfere with normal brain maturation, disease progression, and drug efficacy and thus limit the utility of MEMRI for longitudinal studies. Mn2+ administration is also complicated by the long half-life of Mn2+ in brain (>1.5 months), requiring extensive time for complete washout (5X half-life) (Mok et al. 2012). However, we have demonstrated the feasibility of registering MEMRI atlas to T2-weighted images acquired without Mn2+. Successful segmentation of brain structures on T2-weighted MRI extended the utility of MEMRI atlas for longitudinal studies.
Conclusion
An in vivo MEMRI-based atlas was generated for brains of NSG mice. Forty-one brain structures were identified to provide a coordinate system for spatial normalization. The atlas provides a database for studies of brain morphology, metabolomics, MR metrics, disease pathobiology, and drug pharmacokinetics in a range of infectious, inflammatory and degenerative disease of the nervous system; most notably for studies of HIV-1 infections.
Acknowledgments
This work was supported, in part, by the University of Nebraska Foundation which includes individual donations from Carol Swarts and Frances and Louie Blumkin, the Vice Chancellor’s office of the University of Nebraska Medical Center for Core Facility Developments, Shoemaker Award for Neurodegenerative Research, ViiV Healthcare and National Institutes of Health grants K25 MH08985, P01 DA028555, R01 NS36126, P01 NS31492, 2R01 NS034239, P01 MH64570, P01 NS43985, P30MH062261 and R01 AG043540. Authors thank Dr. Jorge Rodriguez-Sierra for instructive discussions on mouse brain anatomy.
Abbreviations
- NSG
NOD/scid-IL-2Rγcnull
- HIV-1
Human immunodeficiency virus type one
- ART
Antiretroviral therapy
- DTI
Diffusion tensor imaging
- PK
Pharmacokinetics
- PD
Pharmacodynamics
- MRI
Magnetic resonance imaging
- MEMRI
Manganese enhanced MRI
- LDDMM
Large deformation diffeomorphic metric mapping
- CH
Cerebrum
- OLF
Olfactory areas
- MOBgl
Main olfactory bulb, glomerular layer
- MOBgr
Main olfactory bulb, granule layer
- AOB
Accessory olfactory bulb
- AON
Anterior olfactory nucleus
- PIR
Piriform area
- HPF
Hippocampal formation
- CA1_CA2_SUB
Field CA1 + Field CA2 + Subiculum
- CA3
Field CA3 of hippocampus
- DG-mo
Dentate gyrus_molecular layer
- DG-(po + sg)
Dentate gyrus_(polymorph layer + granular cell layer)
- STR
Striatum
- CP
Caudoputamen
- STRv
Striatum ventral region
- LSX
Lateral septal complex
- PAL
Pallidum
- PALc
Pallidium, caudal region
- GP
Globus pallidus
- MS
Medial septal nucleus
- AMY
Amygdala
- FB
Fiber tracts
- cc
Corpus callosum
- opt
Optic tract
- ac
Anterior commissure
- RFB
Rest of fiber tracts
- BS
Brain stem
- TH
Thalamus
- EPI
Epithalamus
- HY
Hypothalamus
- IC
Inferior colliculus
- PAG
Periaqueductal gray
- PRT
Pretectal region
- SN
Substantia nigra
- RMB
Rest of midbrain
- P
Pons
- MY
Medulla
- CB
Cerebellum
- CBXmo
Cerebellar cortex, molecular layer
- CBXgr
Cerebellar cortex, granular layer
- CBwm
Cerebellar white matter
- FN
Fastigial nucleus
- IP
Interpose nucleus
- DN
Dentate nucleus
- VS
Ventricular system
- VL
Lateral ventricles
- V3
Third ventricle
- AQ
Cerebral aqueduct
- V4
Fourth ventricle.
Footnotes
Compliance with Ethical Standards
Conflict of Interest The authors declare that they have no conflict of interest.
References
- Aggarwal M, Zhang J, Miller MI, Sidman RL, Mori S. Magnetic resonance imaging and micro-computed tomography combined atlas of developing and adult mouse brains for stereotaxic surgery. Neuroscience. 2009;162:1339–1350. doi: 10.1016/j.neuroscience.2009.05.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal M, Zhang J, Mori S. Magnetic resonance imaging-based mouse brain atlas and its applications. Methods Mol Biol. 2011;711:251–270. doi: 10.1007/978-1-61737-992-5_12. [DOI] [PubMed] [Google Scholar]
- Aoki I, Wu YJ, Silva AC, Lynch RM, Koretsky AP. In vivo detection of neuroarchitecture in the rodent brain using manganese-enhanced MRI. Neuroimage. 2004;22:1046–1059. doi: 10.1016/j.neuroimage.2004.03.031. [DOI] [PubMed] [Google Scholar]
- Bade AN, Gorantla S, Dash PK, Makarov E, Sajja BR, Poluektova LY, Luo J, Gendelman HE, Boska MD, Liu Y. Manganese-enhanced magnetic resonance imaging reflects brain pathology during progressive HIV-1 infection of humanized mice. Molecular neurobiology. 2015 doi: 10.1007/s12035-015-9258-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J, Trinh TL, Chuang KH, Qiu A. Atlas-based automatic mouse brain image segmentation revisited: model complexity vs. image registration. Magn Reson Imaging. 2012;30:789–798. doi: 10.1016/j.mri.2012.02.010. [DOI] [PubMed] [Google Scholar]
- Beg MF, Miller MI, Trouve A, Younes L. Computing large deformation metric mappings via geodesic flows of diffeomorphisms. Int J Comput Vis. 2005;61:139–157. [Google Scholar]
- Chan E, Kovacevic N, Ho SK, Henkelman RM, Henderson JT. Development of a high resolution three-dimensional surgical atlas of the murine head for strains 129S1/SvImJ and C57Bl/6 J using magnetic resonance imaging and micro-computed tomography. Neuroscience. 2007;144:604–615. doi: 10.1016/j.neuroscience.2006.08.080. [DOI] [PubMed] [Google Scholar]
- Chen XJ, Kovacevic N, Lobaugh NJ, Sled JG, Henkelman RM, Henderson JT. Neuroanatomical differences between mouse strains as shown by high-resolution 3D MRI. Neuroimage. 2006;29:99–105. doi: 10.1016/j.neuroimage.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Chuang N, Mori S, Yamamoto A, Jiang H, Ye X, Xu X, Richards LJ, Nathans J, Miller MI, Toga AW, Sidman RL, Zhang J. An MRI-based atlas and database of the developing mouse brain. Neuroimage. 2011;54:80–89. doi: 10.1016/j.neuroimage.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash PK, Gorantla S, Gendelman HE, Knibbe J, Casale GP, Makarov E, Epstein AA, Gelbard HA, Boska MD, Poluektova LY. Loss of neuronal integrity during progressive HIV-1 infection of humanized mice. J Neurosci : Off J Soc Neurosci. 2011;31:3148–3157. doi: 10.1523/JNEUROSCI.5473-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Guzman EA, Wong MD, Gleave JA, Nieman BJ. The fixaation protocol alters brain morphology in ex-vivo MRI mouse phenotyping. ISMRM Proc. Intl Soc Mag Reson Med. 2013:0271. [Google Scholar]
- Dorr AE, Lerch JP, Spring S, Kabani N, Henkelman RM. High resolution three-dimensional brain atlas using an average magnetic resonance image of 40 adult C57Bl/6 J mice. Neuroimage. 2008;42:60–69. doi: 10.1016/j.neuroimage.2008.03.037. [DOI] [PubMed] [Google Scholar]
- Gazdzinski LM, Cormier K, Lu FG, Lerch JP, Wong CS, Nieman BJ. Radiation-induced alterations in mouse brain development characterized by magnetic resonance imaging. Int J Radiat Oncol Biol Phys. 2012;84:e631–e638. doi: 10.1016/j.ijrobp.2012.06.053. [DOI] [PubMed] [Google Scholar]
- Gorantla S, Poluektova L, Gendelman HE. Rodent models for HIV-associated neurocognitive disorders. Trends Neurosci. 2012;35:197–208. doi: 10.1016/j.tins.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Li T, McMillan J, Sajja BR, Puligujja P, Boska MD, Gendelman HE, Liu XM. Small magnetite antiretroviral therapeutic nanoparticle probes for MRI of drug biodistribution. Nanomedicine. 2013 doi: 10.2217/nnm.13.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Zhang G, Wysocki TA, Wysocki BJ, Gelbard HA, Liu XM, McMillan JM, Gendelman HE. Endosomal trafficking of nanoformulated antiretroviral therapy facilitates drug particle carriage and HIV clearance. J Virol. 2014 doi: 10.1128/JVI.01557-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M, Chetana M, Devi PU. Late effect of prenatal irradiation on the hippocampal histology and brain weight in adult mice. Int J Dev Neurosci : Off J Int Soc Dev Neurosci. 2005;23:307–313. doi: 10.1016/j.ijdevneu.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Ito M, Hiramatsu H, Kobayashi K, Suzue K, Kawahata M, Hioki K, Ueyama Y, Koyanagi Y, Sugamura K, Tsuji K, Heike T, Nakahata T. NOD/SCID/gamma(c)(null) mouse: an excellent recipient mouse model for engraftment of human cells. Blood. 2002;100:3175–3182. doi: 10.1182/blood-2001-12-0207. [DOI] [PubMed] [Google Scholar]
- Janus C, Welzl H. Mouse models of neurodegenerative diseases: criteria and general methodology. Methods Mol Biol. 2010;602:323–345. doi: 10.1007/978-1-60761-058-8_19. [DOI] [PubMed] [Google Scholar]
- Koretsky AP, Silva AC. Manganese-enhanced magnetic resonance imaging (MEMRI) NMR Biomed. 2004;17:527–531. doi: 10.1002/nbm.940. [DOI] [PubMed] [Google Scholar]
- Kovacevic N, Henderson JT, Chan E, Lifshitz N, Bishop J, Evans AC, Henkelman RM, Chen XJ. A three-dimensional MRI atlas of the mouse brain with estimates of the average and variability. Cereb Cortex. 2005;15:639–645. doi: 10.1093/cercor/bhh165. [DOI] [PubMed] [Google Scholar]
- Kuo YT, Herlihy AH, So PW, Bell JD. Manganese-enhanced magnetic resonance imaging (MEMRI) without compromise of the blood–brain barrier detects hypothalamic neuronal activity in vivo. NMR Biomed. 2006;19:1028–1034. doi: 10.1002/nbm.1070. [DOI] [PubMed] [Google Scholar]
- Lee JH, Silva AC, Merkle H, Koretsky AP. Manganese-enhanced magnetic resonance imaging of mouse brain after systemic administration of MnCl2: dose-dependent and temporal evolution of T1 contrast. Magn Reson Med. 2005;53:640–648. doi: 10.1002/mrm.20368. [DOI] [PubMed] [Google Scholar]
- Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Ma Y, Hof PR, Grant SC, Blackband SJ, Bennett R, Slatest L, McGuigan MD, Benveniste H. A three-dimensional digital atlas database of the adult C57BL/6 J mouse brain by magnetic resonance microscopy. Neuroscience. 2005;135:1203–1215. doi: 10.1016/j.neuroscience.2005.07.014. [DOI] [PubMed] [Google Scholar]
- Ma Y, Smith D, Hof PR, Foerster B, Hamilton S, Blackband SJ, Yu M, Benveniste H. In Vivo 3D digital atlas database of the adult C57BL/6 J mouse brain by magnetic resonance microscopy. Front Neuroanat. 2008;2:1. doi: 10.3389/neuro.05.001.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie-Graham A, Lee EF, Dinov ID, Bota M, Shattuck DW, Ruffins S, Yuan H, Konstantinidis F, Pitiot A, Ding Y, Hu G, Jacobs RE, Toga AW. A multimodal, multidimensional atlas of the C57BL/6 J mouse brain. J Anat. 2004;204:93–102. doi: 10.1111/j.1469-7580.2004.00264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manda K, Ueno M, Anzai K. Cranial irradiation-induced inhibition of neurogenesis in hippocampal dentate gyrus of adult mice: attenuation by melatonin pretreatment. J Pineal Res. 2009;46:71–78. doi: 10.1111/j.1600-079X.2008.00632.x. [DOI] [PubMed] [Google Scholar]
- Mok SI, Munasinghe JP, Young WS. Infusion-based manganese-enhanced MRI: a new imaging technique to visualize the mouse brain. Brain Struct Funct. 2012;217:107–114. doi: 10.1007/s00429-011-0324-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie J, Shen D. Automated segmentation of mouse brain images using multi-atlas multi-ROI deformation and label fusion. Neuroinformatics. 2013;11:35–45. doi: 10.1007/s12021-012-9163-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MK, Kim S, Jung U, Kim I, Kim JK, Roh C. Effect of acute and fractionated irradiation on hippocampal neurogenesis. Molecules. 2012;17:9462–9468. doi: 10.3390/molecules17089462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Franklin K. The mouse brain in stereotaxic coordinates. San Diego: Academic; 2001. [Google Scholar]
- Puligujja P, McMillan J, Kendrick L, Li T, Balkundi S, Smith N, Veerubhotla RS, Edagwa BJ, Kabanov AV, Bronich T, Gendelman HE, Liu XM. Macrophage folate receptor-targeted antiretroviral therapy facilitates drug entry, retention, antiretroviral activities and biodistribution for reduction of human immunodeficiency virus infections. Nanomedicine. 2013;9:1263–1273. doi: 10.1016/j.nano.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao AA, Ye H, Decker PA, Howe CL, Wetmore C. Therapeutic doses of cranial irradiation induce hippocampus-dependent cognitive deficits in young mice. J Neuro-Oncol. 2011;105:191–198. doi: 10.1007/s11060-011-0582-9. [DOI] [PubMed] [Google Scholar]
- Saito Y, Kametani Y, Hozumi K, Mochida N, Ando K, Ito M, Nomura T, Tokuda Y, Makuuchi H, Tajima T, Habu S. The in vivo development of human T cells from CD34(+) cells in the murine thymic environment. Int Immunol. 2002;14:1113–1124. doi: 10.1093/intimm/dxf087. [DOI] [PubMed] [Google Scholar]
- Silva AC, Lee JH, Aoki I, Koretsky AP. Manganese-enhanced magnetic resonance imaging (MEMRI): methodological and practical considerations. NMR Biomed. 2004;17:532–543. doi: 10.1002/nbm.945. [DOI] [PubMed] [Google Scholar]
- Silva AC, Lee JH, Wu CW, Tucciarone J, Pelled G, Aoki I, Koretsky AP. Detection of cortical laminar architecture using manganese-enhanced MRI. J Neurosci Methods. 2008;167:246–257. doi: 10.1016/j.jneumeth.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Sunkin SM, Ng L, Lau C, Dolbeare T, Gilbert TL, Thompson CL, Hawrylycz M, Dang C. Allen Brain Atlas: an integrated spatio-temporal portal for exploring the central nervous system. Nucleic Acids Res. 2013;41:D996–D1008. doi: 10.1093/nar/gks1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trancikova A, Ramonet D, Moore DJ. Genetic mouse models of neurodegenerative diseases. Prog Mol Biol Transl Sci. 2011;100:419–482. doi: 10.1016/B978-0-12-384878-9.00012-1. [DOI] [PubMed] [Google Scholar]