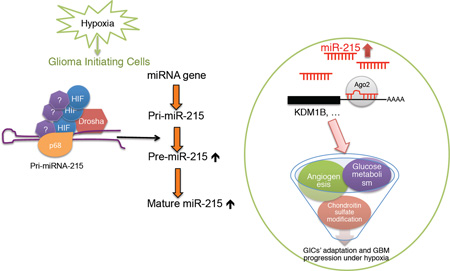
SUMMARY
The hypoxic tumor microenvironment serves as a niche for maintaining the glioma-initiating cells (GICs) that are critical for glioblastoma (GBM) occurrence and recurrence. Here we report that hypoxia-induced miR-215 is vital for reprograming GICs to fit the hypoxic microenvironment via suppressing the expression of an epigenetic regulator KDM1B and modulating activities of multiple pathways. Interestingly, biogenesis of miR-215 and several miRNAs is accelerated post-transcriptionally by hypoxia-inducible factors (HIFs) through HIF-Drosha interaction. Moreover, miR-215 expression correlates inversely with KDM1B while positively with HIF1α and GBM progression in patients. These findings reveal a direct role of HIF in regulating miRNA biogenesis and consequently activating the miR-215-KDM1B-mediated signaling required for GIC adaptation to hypoxia.
Graphical Abstract
eTOC Blurb
Hu et al. reveal a HIF-miR-215-KDM1B-mediated signaling axis in adaptation of glioma-initiating cells to hypoxia and uncover a role of HIF in post-transcriptional regulation of miRNA biogenesis through HIF-Drosha interaction. MiR-215 level in GBM correlates with HIF1α level and tumor progression in patients.
INTRODUCTION
Glioblastoma (GBM, WHO grade IV astrocytoma) is the most common and aggressive primary brain tumor in adults with an average survival of slightly more than one year past the initial diagnosis (Ostrom et al., 2014). Glioma-initiating cells (GICs), a subpopulation of cells inside GBM that could self-renew, differentiate into multi-lineages and initiate or propagate tumors, have been demonstrated to account for the fatal nature of GBM (Bao et al., 2006; Chen et al., 2012). Hypoxia is often encountered in solid tumors including GBM, with the hypoxia inducible factors (HIFs) acting as the key transcriptional modulators that promote multiple processes associated with GBM progression under hypoxic stress (Kaur et al., 2005; Majmundar et al., 2010). Notably, the hypoxic microenvironment has been revealed to serve as a niche for GICs that promotes the self-renewal and tumorigenic potential of GICs through induction of stem cell markers such as Sox2, Oct4 and Notch by HIF1α and HIF2α (Li et al., 2009; Soeda et al., 2009). In order to adapt to the hypoxic stress and even benefit from the environment to maintain their distinct biological features, multiple aspects of GICs need to be modulated.
MicroRNAs (miRNAs) are small non-coding RNAs of ∼22 nucleotides, which can regulate diverse biological processes through down-regulation of protein target expression (He and Hannon, 2004). A number of miRNAs have been demonstrated altered by hypoxia to target important oncogenes and tumor suppressors, affecting the hallmarks of tumorigenic processes (Kulshreshtha et al., 2008; Nallamshetty et al., 2013). The biogenesis of miRNAs initiates with transcription of the miRNA gene. The pri-miRNA are firstly cropped into the ∼70-nucleotide pre-miRNA by the microprocessor complex Drosha/DGCR8. Pre-miRNAs are further processed into the miRNA/miRNA* duplex by the Dicer complex. Finally, the 5’ or 3’ of the miRNA duplex is incorporated into the RNA-induced silencing complex (RISC) that suppresses the target (Winter et al., 2009). Multiple steps of miRNA biogenesis are stringently regulated during processes such as tumor progression. For example, various cofactors including Smad and p53 have been shown to associate with the Drosha complex and mediate its processing activity for specific subsets of miRNAs (Davis et al., 2010; Suzuki et al., 2009). Accumulating evidence has uncovered the significant role for the post-transcriptional modulations of miRNA processing (Ha and Kim, 2014).
In this study, we sought to examine roles of miRNAs in regulating GICs adaptation to hypoxia, as well as mechanisms mediating miRNA biogenesis under hypoxia.
RESULTS
MiR-215 is induced by hypoxia in GICs
To identify microRNAs that may mediate the responses of GICs under hypoxia, we conducted a candidate-based screen of miRNAs that have been reported to regulate the processes of cell differentiation and development. We used the cell surface marker CD133 to enrich GICs from patient-derived glioma lines maintained through serial passages in immunocompromised mice as subcutaneous xenografts (Bao et al., 2006; Wang et al., 2014b). Consistent with previous reports, the CD133+ GICs isolated from two different patient-derived lines (D456MG and 110040) could significantly promote the intracranial tumor progression compared to the CD133− cells (Figure S1A). To identify the miRNAs that may mediate responses to hypoxia, GICs isolated from these two xenografted lines were cultured in 1% O2 for 6 hrs or treated with the iron chelator desferrioxamine (DFX) for 3 hrs in the initial screen. HIF1α and HIF2α proteins were accumulated, and mRNA levels of the hypoxia-responsive genes Glut1 and ADM were induced after hypoxic treatment (Figure S1B–E), indicating a positive hypoxic response in these cells. The expression profile of 88 miRNAs in the GICs cultured under hypoxia compared to normoxia was further determined (Table S1). 11 miRNAs displayed consistent up-regulation in their expression levels under hypoxia (Table S2). Among this list, four miRNAs have been shown to play important roles in glioma progression by previous studies, such as miR-218, miR-122, miR-96 and miR-155 (Ling et al., 2013; Mathew et al., 2014; Wang et al., 2014a; Yan et al., 2014). In agreement with previous findings (Agrawal et al., 2014; Babar et al., 2011; Bruning et al., 2011), the hypoxic environment also boosted expression of miR-155, miR-129 and miR-134 in the GICs, suggesting a general regulation of these miRNAs by hypoxia.
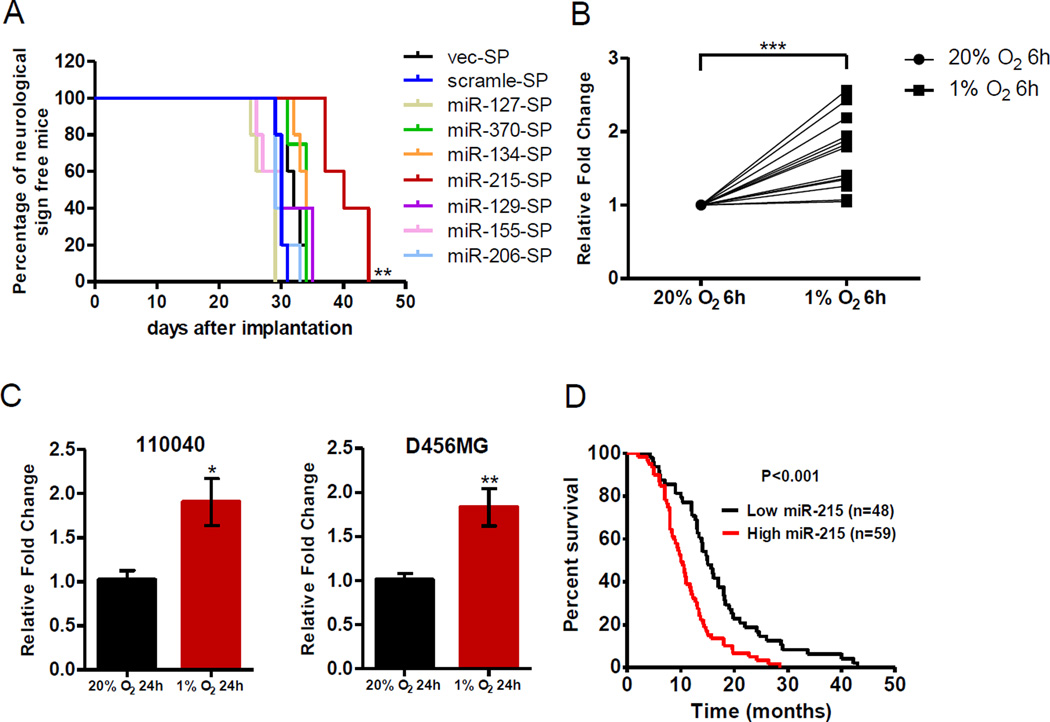
To assess the potential function of these miRNAs in the GICs cultured under hypoxia, a lentiviral-based sponge construct was generated to attenuate the function of each miRNA (Ebert et al., 2007). For the seven up-regulated miRNAs that have not been implicated in GBM progression, we successfully introduced individual sponges for five of them into the CD133+ GICs (miR-215, miR-370, miR-134, miR-127 and miR-129). As displayed in Figure 1A, antagonizing the function of miR-215, rather than other miRNAs, in GICs significantly alleviated GBM progression in mice bearing the intracranial tumor in comparison to controls. Therefore, miR-215 emerged as the top candidate for subsequent studies due to its significant impact on tumor progression in mice.
Figure 1. Identification of miR-215 as a top candidate induced by hypoxia in GICs.
(A) Kaplan-Meier curves were drawn to measure the burden of tumor progression by CD133+ D456MG cells expressing the indicated plasmids. 5000 GICs were implanted intracranially into each mouse. n=5. SP: sponge.
(B) A scatter dot plot shows the expression changes of miR-215 under hypoxia in GICs isolated from eight patient-derived xenografts and five patient specimens, as detected by qPCR.
(C) Expression of miR-215 was examined by qPCR in GICs isolated from 110040 and D456MG after cultured under 1% O2 for 24 hrs. n=4.
(D) Kaplan-Meier curves were drawn to show the correlation of miR-215 expression and clinical outcome from GBM tissue microarray.
Data are represented as the mean ± SEM. (* p<0.05, **p<0.01)
To further validate the expression pattern of miR-215 under hypoxia, we isolated GICs from six additional patient-derived xenografts by the CD133 marker, as well as five freshly isolated patient specimens by a specific culture condition that could significantly enrich the cell population displaying biological properties of GICs but containing both CD133+ and CD133− GICs (Bhat et al., 2013; Chen et al., 2010). The expression level of miR-215 was significantly elevated in majority of GIC samples exposed to hypoxia compared to normoxia (Figure 1B). In consistency with the kinetics of immediate response to hypoxia with the 6-hr treatment, expression of miR-215 was also up-regulated in GICs cultured under hypoxia for a longer period of time (Figure 1C). As an alternative to CD133 to enrich GICs, we utilized another cell surface marker, stage-specific embryonic antigen 1 or CD15 (Son et al., 2009) and detected a consistent enhancement of miR-215 expression in CD15+ GICs cultured under hypoxia (Figure S1F). Interestingly, miR-215 was induced by hypoxia in CD133+ GICs but not in paired CD133− non-GICs isolated from two xenografted lines, suggesting a GIC-specific induction of miR-215 by hypoxia (Figure S1G). Importantly, we conducted in situ hybridization (ISH) analyses to examine levels of miR-215 on GBM tissue microarrays and found that higher level of miR-215 expression was correlated with poor survival (Figure 1D). The clinical importance of miR-215 in gliomagenesis is also supported by a recent report that miR-215 expression is positively correlated with poor prognosis in glioma patients (Tong et al., 2015). In sum, these data strongly indicate that miR-215 is a hypoxia-induced miRNA in GICs, and it acts to promote GBM progression probably by mediating the hypoxic responses of GICs.
Biogenesis of miR-215 is post-transcriptionally enhanced under hypoxia
In examining the kinetics of miR-215 induction by hypoxia, we found that the expression of miR-194, which resides in the same cluster with miR-215 at chromosomal locus 1q41.1, was not boosted under hypoxia for either 6 hrs (Table S1) or 24 hrs (Figure S1H). Since miRNAs of the same clusters are usually expressed together in the same pri-miRNA transcripts (Ventura et al., 2008), the differential expression pattern between miR-215 and miR-194 in hypoxia prompted us to investigate whether the biogenesis of miR-215 might be enhanced post-transcriptionally.
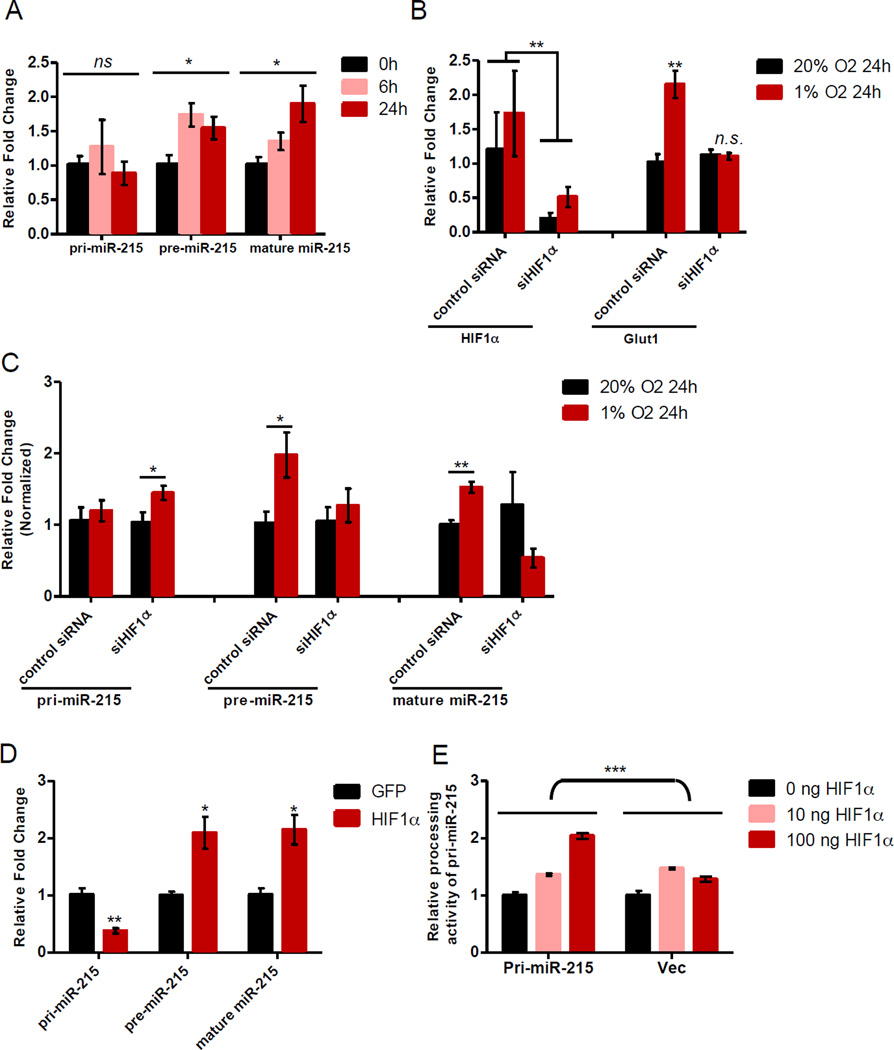
To examine which biogenesis step of miR-215 might be affected under hypoxia, we measured the expression changes of pri-miR-215, pre-miR-215 and mature miR-215 from GICs isolated from multiple xenografted lines. As shown in Figure 2A and Figure S2A–C, induction of mature miR-215 and pre-miR-215 was observed under hypoxia, while no significant change in the level of pri-miR-215 was detected, suggesting that processing of miR-215 is regulated at a post-transcriptional step likely through the action of the Drosha complex. As the key transcriptional factors mediating hypoxic response, HIFs are well known to regulate expression of a large number of genes, including miRNA genes. In order to examine the potential role of HIF in the modulation of miR-215 biogenesis, GICs was transfected with siRNA against HIF1α or scramble control and cultured in 1% O2 for 24 hrs. The mRNA level of HIF1α was knocked down dramatically and the hypoxia-responsive gene Glut1 was no longer induced by hypoxia, as expected (Figure 2B). Subsequently, expression profiles of the pri-, pre-, and mature miR-215 were measured. HIF1α-siRNA abolished the elevation of pre-miR-215 and mature miR-215 by hypoxia compared to the scramble control (Figure 2C). In contrast, the level of unprocessed pri-miR-215 was found to increase under hypoxia when HIF1α was knocked down, suggesting a blockage of the pri-miR-215 processing into pre-miR-215. These results indicate that HIF1α is required for the induction of miR-215’s biogenesis under hypoxia.
Figure 2. Biogenesis of miR-215 is enhanced by hypoxia from pri-miR-215 to pre-miR-215 through the action of Drosha and HIF.
(A) Expression profiles of pri-, pre- and mature miR-215 were examined by qPCR in 110040 GICs cultured in 1% O2 for the indicated time. n=4.
(B, C) Expression of HIF1α and Glut1 (B), as well as pri-, pre- and mature miR-215 (C) were determined by qPCR in 110040 GICs transfected with scramble control or siHIF1α when cultured in 1% O2 for 24 hrs. n=4.
(D) Expression levels of pri-, pre- and mature miR-215 were detected by qPCR in HEK293T cells transfected with pri-miR-215 and a stabilized HIF1α or GFP control. n=3.
(E) Processing activity of pri-miR-215 by Drosha in vivo was determined using a luciferase-pri-miR-215 reporter or control ectopically expressed in HEK293T cells. Different amounts of plasmid encoding the stabilized HIF1α were co-transfected as indicated. n=3.
Data are represented as the mean ± SEM. (* p<0.05, **p<0.01, n.s., non-significant)
See also Figure S2.
To further determine whether HIF1α enhances the processing of pri-miR-215 post-transcriptionally, rather than through transcriptional activation, we engineered a luciferase reporter pmirGlo-pri-miR-215, in which the transcription of luciferase/pri-miR-215 was driven by an exogenous human phosphoglycerate kinase (PGK) promoter. Since portions of pri-miR-215 are embedded in the 3’UTR of the firefly luciferase gene, cleavage of pri-miR-215 by the Drosha complex is expected to destabilize the mRNA of firefly luciferase and in turn to decrease the firefly luminescence. Consequently, the endogenous microprocessor activity could be measured by normalization of the firefly luminescence to the internal control Renilla luminescence. A similar approach has been described to monitor microprocessor activity (Mori et al., 2014; Suzuki et al., 2009). After transfection of the plasmid into HEK293T cells, expression of the pri-, pre- and mature miR-215 was measured upon overexpression of the HIF1α protein. An induction of pre-miR-215 and mature miR-215 was observed, while expression level of the pri-miR-215 was significantly reduced (Figure 2D). Using the same RNAs, induction of mature miR-215 was also detected by Northern blot (Figure S2D), indicating that cropping of the pri-miR-215 into pre-miR-215 is enhanced by HIF1α and the endogenous miR-215 promoter is not required for the up-regulation of miR-215. In agreement with the expression profiles of pri-, pre- and mature miR-215, over-expression of HIF1α accelerated the processing activity of pri-miR-215 by Drosha in a dose-dependent manner using the same luciferase reporter pmirGLo-pri-miR-215 (Figure 2E).
Taken together, these data indicate that biogenesis of miR-215 is promoted in response to hypoxia at the post-transcriptional step of pri-miR-215 to pre-miR-215 and HIF1α is required for this enhancement in the processing activity of Drosha complex.
HIF1α promotes miR-215 biogenesis through enhancing incorporation of the pri-miR-215 into the Drosha/DGCR8 complex
Hypoxia may regulate miRNA processing through multiple mechanisms. As hypoxia is known to impact gene transcription and protein synthesis (Koumenis et al., 2002; Majmundar et al., 2010), levels of the microprocessor complexes could be altered by hypoxia, leading to changes in the expression patterns of miRNAs. To test this possibility, we examined the expression profile of the Drosha/DGCR8 and Dicer complexes under hypoxia and found that levels of the Drosha or Dicer complex were not enhanced by hypoxia (Figure S3A–B), suggesting that induction of miR-215 is not due to up-regulation of Drosha or Dicer expression. This is also supported by the observation that global miRNA elevation was not detected under hypoxia from the miRNA qPCR screen (Table S1).
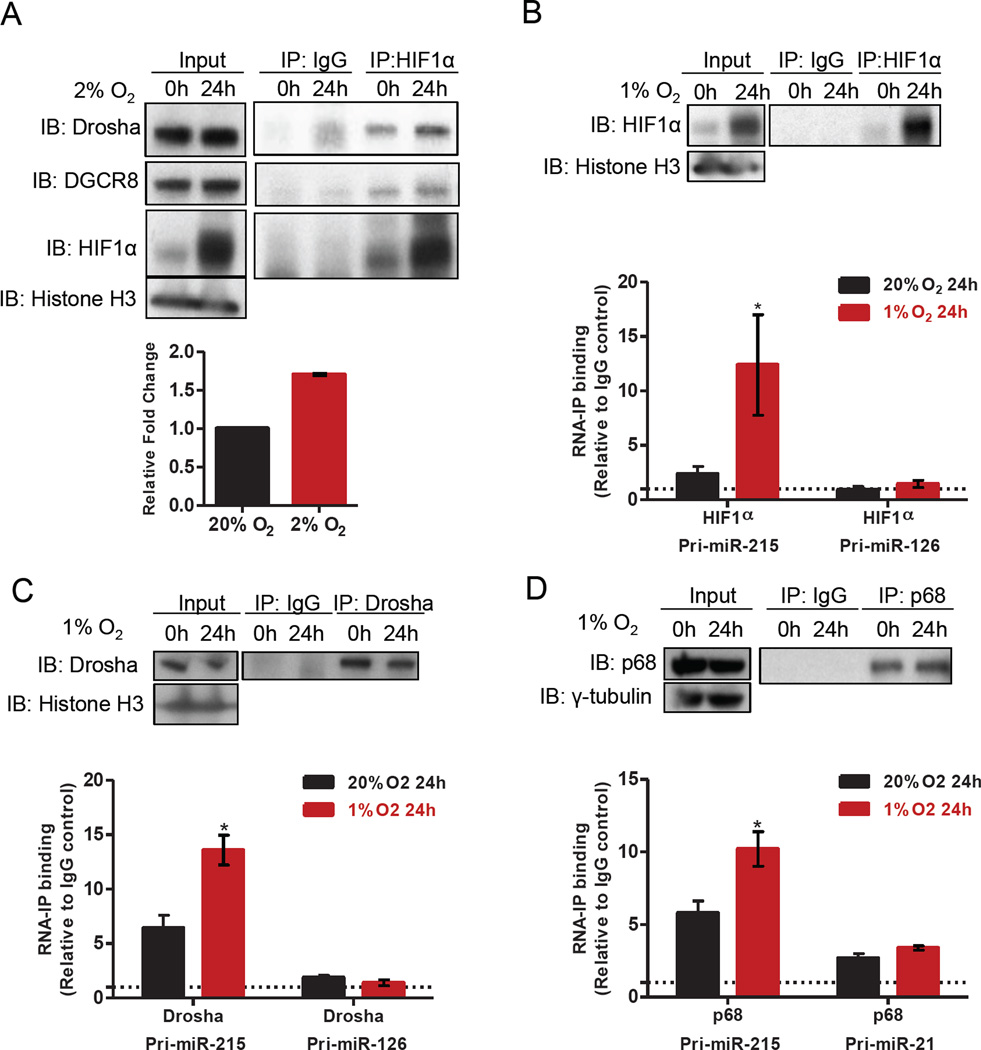
On the other hand, transcription factors such as p53 and Smads have been shown to accelerate subset of miRNAs processing through association with the Drosha complex. We examined the protein levels of p53 and p-Smads and did not detect obvious alteration of their levels in GICs cultured under hypoxia compared to normoxia, indicating that these two pathways are not responsible for the induction of miR-215 processing (Figure S3C). To test whether the enhancement of miR-215 biogenesis by HIF1α is the result of a direct interaction between HIF1α and the Drosha complex, we ectopically expressed Drosha and HIF1α in HEK293T cells and found that these proteins could be co-immunoprecipitated (Figure S3D). Consistently, HIF1α was accumulated and interacted with Drosha/DGCR8 at the endogenous level in GICs cultured under hypoxia (Figure 3A). Interestingly, at the basal level under normoxia, HIF1α was already associated with Drosha/DGCR8. Although there was a dramatic induction of HIF1α protein in cells exposed to hypoxia, the amount of Drosha associated with HIF1α did not increase proportionally, indicating that the larger pool of HIF1α may play multiple functions such as transcriptional stimulation under hypoxia.
Figure 3. HIF1α interacts with Drosha/DGCR8 complexes and enhances incorporation of pri-miR-215 into the complex to promote its processing.
(A) Co-immunoprecipitation of endogenous HIF1α was performed in 110040 GICs cultured in 2% O2 for 24 hrs followed by determination of protein levels of the associated Drosha and DGCR8 through Western blot. Quantification of Drosha levels interacted with HIF1α under hypoxia was shown in the lower panel, as measured by Image J.
(B–D) RNA-ChIP was used to detect association between pri-miR-215, but not control pri-miRNAs, and endogenous HIF1α (B), Drosha (C) or p68 (D) in 110040 GICs cultured in 1% O2 for 24 hrs. HIF1α or Drosha or p68 was immunoprecipitated using lysates from 110040 GICs (upper panels). Incorporation of pri-miR-215 and control pri-miRNAs into the protein complex was examined by qPCR (lower panels). Dotted lines indicate the background level of non-specific binding of pri-miRNAs to IgG. n=4.
Data are represented as the mean ± SEM. (* p<0.05, **p<0.01, ***p<0.001)
The interaction between HIF1α and Drosha/DGCR8 prompted us to determine whether HIF1α might modulate the incorporation of pri-miR-215 into the Drosha complex under hypoxia. RNA-Chromatin Immunoprecipitation (RNA-ChIP) analysis was performed on HEK293T cells transfected with pri-miR-215 and Flag-tagged HIF1α and an enhanced association of HIF1α with pri-miR-215 was detected (Figure S3E). Importantly, endogenous HIF1α also associated with pri-miR-215 in GICs cultured in hypoxia (Figure 3B, Figure S3F). As a control, there was no significant association between HIF1α and pri-miR-126 or pri-miR-21, of which the mature form was not altered by hypoxia in GICs. Moreover, the association between pri-miR-215 and Drosha or p68, another component of the Drosha/DGCR8 complex, was increased significantly in GICs cultured under hypoxia; whereas incorporation of other control pri-miRNAs into the Drosha complex was not affected (Figure 3C–D, Figure S3G), indicating that HIF1α specifically promotes the recruitment of the Drosha/p68 complex to pri-miR-215 to enhance its processing.
As Drosha-mediated biogenesis of miR-215 was enhanced by HIF under hypoxia, we postulated that HIF might play a general role on the processing of a specific group of miRNAs including miR-215. Since the scope of our original qPCR-based miRNA screen was limited, we conducted a small RNA deep-sequencing experiment followed by qPCR-based validation in the GICs cultured in hypoxia to identify more miRNAs that may be regulated similarly by HIF on a global scale. The expression profiles of the pri-, pre-, and mature miRNAs were further measured on the candidate miRNAs that were induced by hypoxia in GICs. While expression of most of the miRNAs, such as miR-140, miR-654 and miR-210, started to increase at the transcriptional level by hypoxia (Figure S3H–I, S3L–M), biogenesis of some miRNAs, such as let-7i and miR-1260a, were boosted post-transcriptionally at the step of Drosha-mediated cleavage, in a similar manner as miR-215 (Figure S3J–K, S3N). Also similar with miR-215, HIF1α was required for the induction of let-7i biogenesis under hypoxia (Figure S3O), through enhancing the recruitment of HIF1α and Drosha/p68 complex on pri-let-7i (Figure S3P–Q). Since HIF2α shares significant functional similarity with HIF1α, we substituted HIF1α with HIF2α and found it has a similar positive impact on the processing of pri-miR-215 (Figure S4A–D). To further distinguish the roles of HIF1α and HIF2α, we cultured D456MG GICs under 3% O2 during which only HIF2α was accumulated, but did not observe induction of miR-215 (Figure S4E). Interestingly, knocking down either HIF1α or HIF2α by shRNA could abolish the enhancement of miR-215 expression by hypoxia (Figure S4F), suggesting that a threshold of combined protein amount of HIF1α and HIF2α might be critical to promote the Drosha-mediated pri-miRNA processing. Together these results revealed a direct role of HIF1α/HIF2α in mediating processing of a subset of miRNAs via the Drosha microprocessor complex in response to hypoxia by GICs.
Inhibition of miR-215 suppressed tumorigenic abilities of GICs under hypoxia
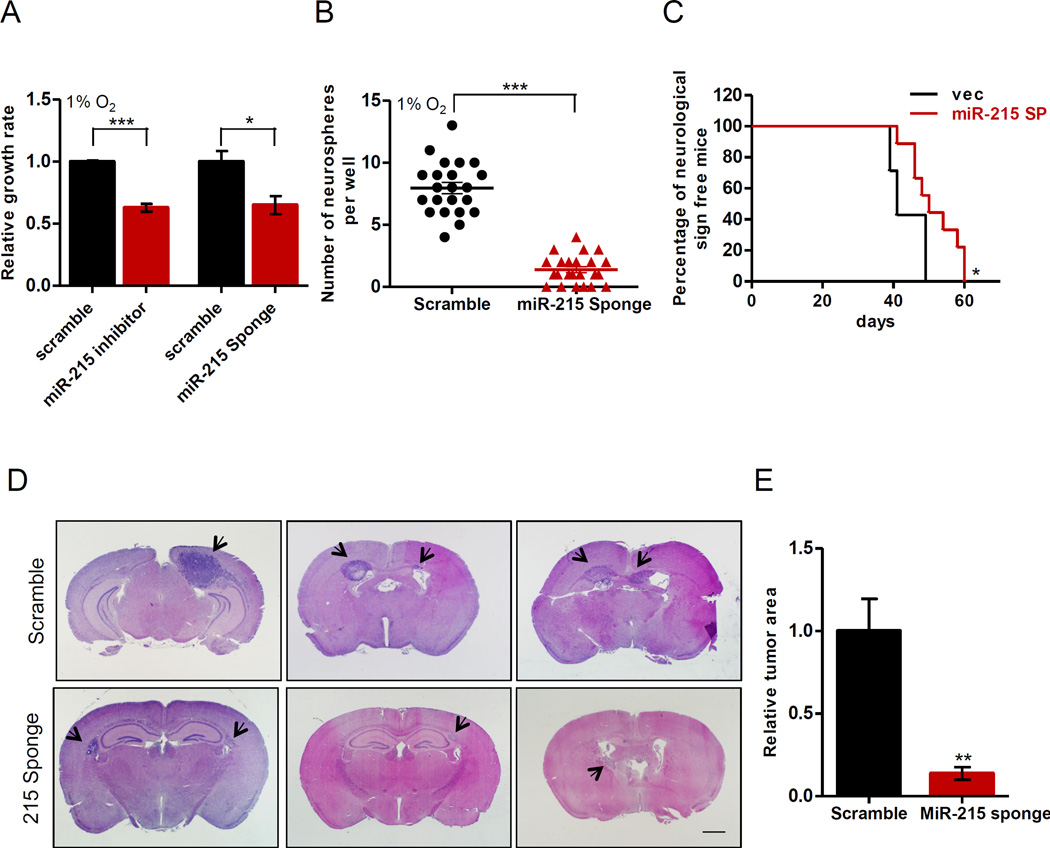
MiR-215 has been previously demonstrated to function differentially in different types of cancer. Although over-expression of miR-215 impedes the progression of colon and renal cancer (Jones et al., 2015; Senanayake et al., 2012), miR-215 is shown to serve as an onco-mir in the development of gastric cancer and hepatoma (Deng et al., 2014; Liu et al., 2014). Since miR-215 is induced in GICs under hypoxia and attenuation of its function significantly delayed tumor progression (Figure 1A), we further examined the role of miR-215 in GICs. Attenuating the function of miR-215 by miR-215-sponge or a specific LNA inhibitor in GICs significantly reduced their growth rate as well as ability to form neurospheres under hypoxia (Figure 4A–B and Figure S5A–F). In consistency with the observation in Figure 1A, miR-215-sponge also rendered a significant delay in GBM progression in mice bearing intracranial tumors derived from GICs isolated from the glioma line 110040 (Figure 4C). In another experimental setting, we inoculated equal number of GICs expressing a miR-215-sponge or scramble control intracranially into nude mice and sacrificed the two groups of mice on the same day. Histological analysis by H&E staining showed significantly smaller tumor areas in the brain of mice in the miR-215-sponge group (Figure 4D–E). Collectively, these data demonstrate that hypoxia-induced miR-215 is critical for GICs to maintain their growth in culture under hypoxic condition and generate tumors in vivo in the intracranial environment.
Figure 4. Suppression of miR-215 attenuates growth of GICs under hypoxia and their tumorigenic capability in the intracranial environment.
(A–B) Cell growth rate (A) and neurosphere formation assay (B) was measured in the 110040 GICs with the indicated modifications cultured under 1% O2. n=4 for cell growth assay.
(C) Kaplan-Meier curves were drawn to measure the burden of tumor progression by 110040 GICs expressing the indicated plasmids. 10,000 cells were implanted intracranially in each mouse. n=7 for control, n=9 for miR-215 sponge.
(D) H&E staining of brain sections showed intracranial tumors that were formed 30 days after implantation of 10,000 110040 GICs expressing the indicated plasmids. Pictures were taken at the maximum cross section of tumors from each brain. Arrows point to tumors in the sections.
(E) Tumor areas from (D) were quantified using ImageJ. n=5. Scale bar: 1.0 mm.
Data are represented as the mean ± SEM. (* p<0.05, **p<0.01, ***p<0.001)
See also Figure S5.
The epigenetic regulator KDM1B is a direct target of miR-215
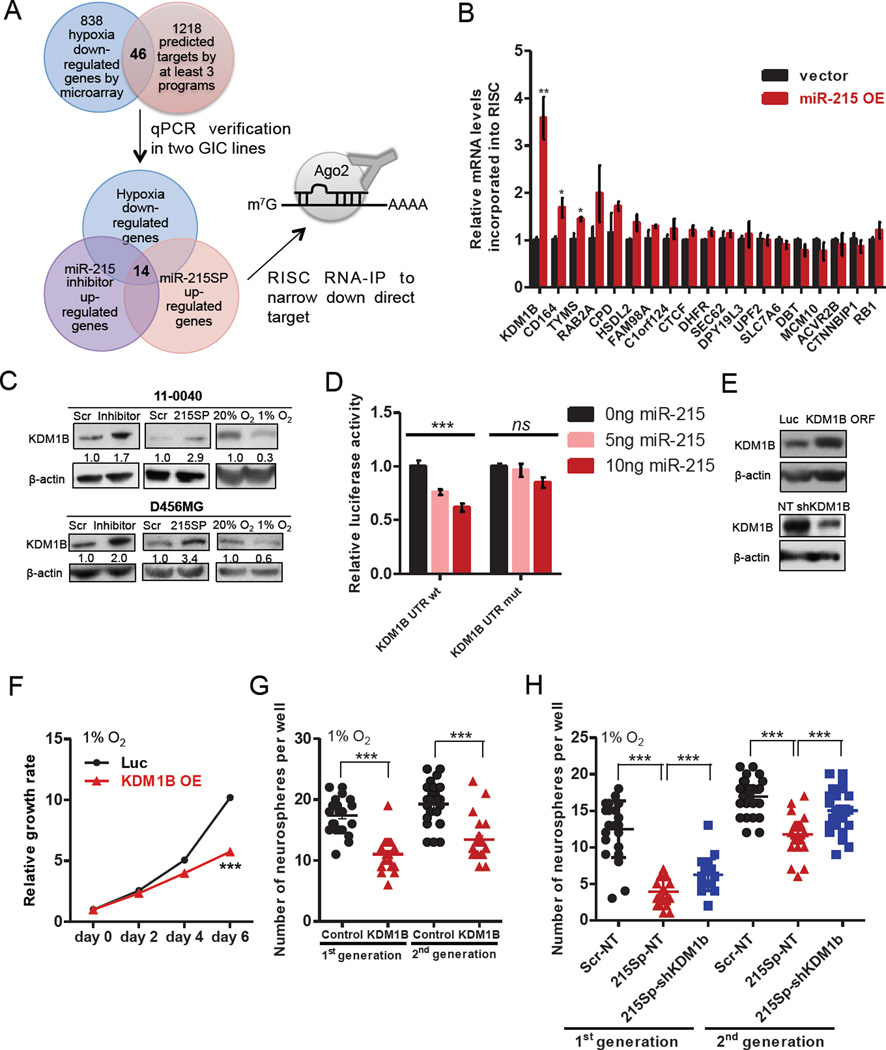
Potent effects of miR-215 in maintaining the abilities of cell growth and tumor progression in GICs prompted us to explore the downstream effectors of miR-215. To identify downstream targets, we integrated mRNA expression profiling with bioinformatics predictions. As miR-215 is induced by hypoxia, expression of its targets was postulated to be repressed in GICs under hypoxia. Through microarray analysis, we produced a list of genes that were down-regulated in GICs cultured under hypoxia compared to normoxia. Overlapping this list with the candidates generated by three prediction algorithms, we narrowed the potential targets down to 46 genes. Expression of each candidate at the mRNA level was further verified by qPCR in GICs isolated from D456MG and 110040 lines cultured under hypoxia or normoxia conditions. Both miRNA sponge and LNA inhibitor were used to antagonize the function of miR-215. This round of screen yielded 14 putative targets that were consistently down-regulated in GICs exposed to hypoxia and up-regulated when miR-215’s function was attenuated (Figure 5A and Table S3). To further narrow down the candidates that are most likely to be direct targets of miR-215, RNA-ChIP analysis was employed to identify the mRNAs selectively enriched in the Ago2/RISC complex after miR-215 over-expression (Figure S6A). Among the 14 genes, mRNA of KDM1B was detected with the most significant enrichment in miR-215-overexpressing GICs compared to the vector control group (Figure 5B). In the meantime, we assessed several targets of miR-215 that were identified previously, including TYMS, ACVR2B, CTNNBIP1 and RB1, but found little or only slight increases in the levels of these mRNAs incorporated into RISC (Figure 5B), consistent with previous notion that miRNAs suppress their targets in a context-dependent manner.
Figure 5. Identification of KDM1B as a direct target mediating the function of miR-215 in maintaining the properties of GICs under hypoxia.
(A) An illustration of strategies to identify direct targets of miR-215.
(B) RNA-ChIP analysis was conducted to detect levels of mRNAs that were incorporated into the Ago2-RISC complex from D456MG GICs expressing the indicated plasmids, as measured by qPCR. n=3.
(C) Immunoblots were performed to detect protein levels of KDM1B in D456MG and 110040 GICs with indicated modifications. Scr: scramble control.
(D) Luciferase activity of the reporter construct containing the wild-type or miR-215-binding mutant 3’UTR of KDM1B was measured after co-transfection with different amounts of pri-miR-215 in HEK293T cells. n=3.
(E) Immunoblots were used to detect the protein levels of KDM1B in 110040 GICs expressing the indicated plasmids.
(F–G) Cell growth rate (F) or Neurosphere formation assay (G) was measured in 110040 GICs expressing the luciferase control or KDM1B ORF when cultured under 1% O2. n=4 for cell growth assay.
(H) Neurosphere formation assay in 110040 GICs expressing miR-215 sponge together with control (NT) or shRNA for KDM1B as indicated.
Data are represented as the mean ± SEM. (* p<0.05, **p<0.01, ***p<0.001)
Alterations related with histone modification processes in GBM, such as mutations of KDM6A, MLL2 and SETD2 have been reported. These mutations result in disruption of the epigenetic processes and promotion of cancer progression (Sturm et al., 2014). KDM1B is a FAD-dependent histone demethylase of H3K4me1/2, which might exert its impacts on gliomagenesis through modulating the expression of a spectrum of critical genes to reprogram the cellular state to adapt to the hypoxic environment. Therefore, we focused our study on KDM1B and determined the protein level of KDM1B in GICs. Consistent with its mRNA expression profile, the protein level of KDM1B was found to be decreased under hypoxia or increased when miR-215 was suppressed in those cells (Figure 5C). In concordance with the expression pattern of miR-215, we did not observe a consistent reduction of KDM1B level by hypoxia in non-GICs isolated from multiple xenografted lines (Figure S6B). To further explore whether miR-215 suppresses KDM1B directly through the putative binding sites in the 3’UTR of KDM1B gene, a luciferase reporter was employed, in which the 3’UTR with wide-type or mutated miR-215 binding sites were embedded downstream of the firefly luciferase (Figure S6C). Indeed, the luciferase expression was repressed by miR-215 in a dose-dependent manner in HEK293T cells, whereas expression of the luciferase with mutated 3’UTR was not altered significantly (Figure 5D). In sum, these data support the notion that KDM1B serves as a direct target of miR-215.
KDM1B mediates the function of miR-215 in maintaining biological properties of GICs under hypoxia
KDM1B has not been previously implicated in GBM progression or cellular responses to hypoxia. To explore the role of KDM1B in GICs exposed to hypoxia, we ectopically expressed the KDM1B open-reading frame (ORF) which could not be suppressed by miR-215 due to lack of the miRNA binding sites. Expression of KDM1B could recapitulate phenotypes of miR-215 knockdown in GICs under hypoxia as demonstrated by the impaired cell growth and neurosphere formation (Figure 5E–G and Figure S6D–F). Next, we stably knocked down KDM1B by shRNAs and observed significant augment of GICs’ ability to form neurospheres (Figure S6D and S6G). To further determine whether properties of GICs mediated by miR-215 are attributed to the repression of KDM1B, we attenuated the expression of KDM1B in GICs with shRNAs or Cas9 and single-guide RNAs together with inhibition of miR-215 and found that co-suppression of miR-215 and KDM1B largely rescued the compromised ability of cell growth, neurosphere formation and tumor progression induced by miR-215 inhibition (Figure 5E, 5H, Figure S6H–K), suggesting that KDM1B serves as a critical effector downstream of miR-215. Together these data indicate that miR-215 functions to positively control the biological properties of GICs under hypoxia mainly through suppressing its target KDM1B.
KDM1B mediates GICs adaptation to hypoxia through modulating multiple processes
Large-scale gene profiling and ChIP-ChIP assays were performed in HeLa cells to examine the global effect on genes potentially controlled by KDM1B (Fang et al., 2010). To identify potential genes and pathways that may be regulated by KDM1B to affect the biological properties of GICs, we conducted an RNA sequencing analysis in the GICs with KDM1B knocked down and generated a list of genes that might be controlled by KDM1B. Gene ontology analysis was performed separately on this gene list and another list of altered genes under hypoxia that was produced from microarray analysis of the same GICs. The enriched pathways that are overlapped on both lists are considered to be among the likely mediators of KDM1B function during the hypoxic responses in GICs. As shown in Figure S7A–B, the analysis revealed that multiple pathways related to cellular response to hypoxia emerged in a prominent pattern, indicating that KDM1B might reprogram GICs for hypoxia-adaptation through affecting those pathways.
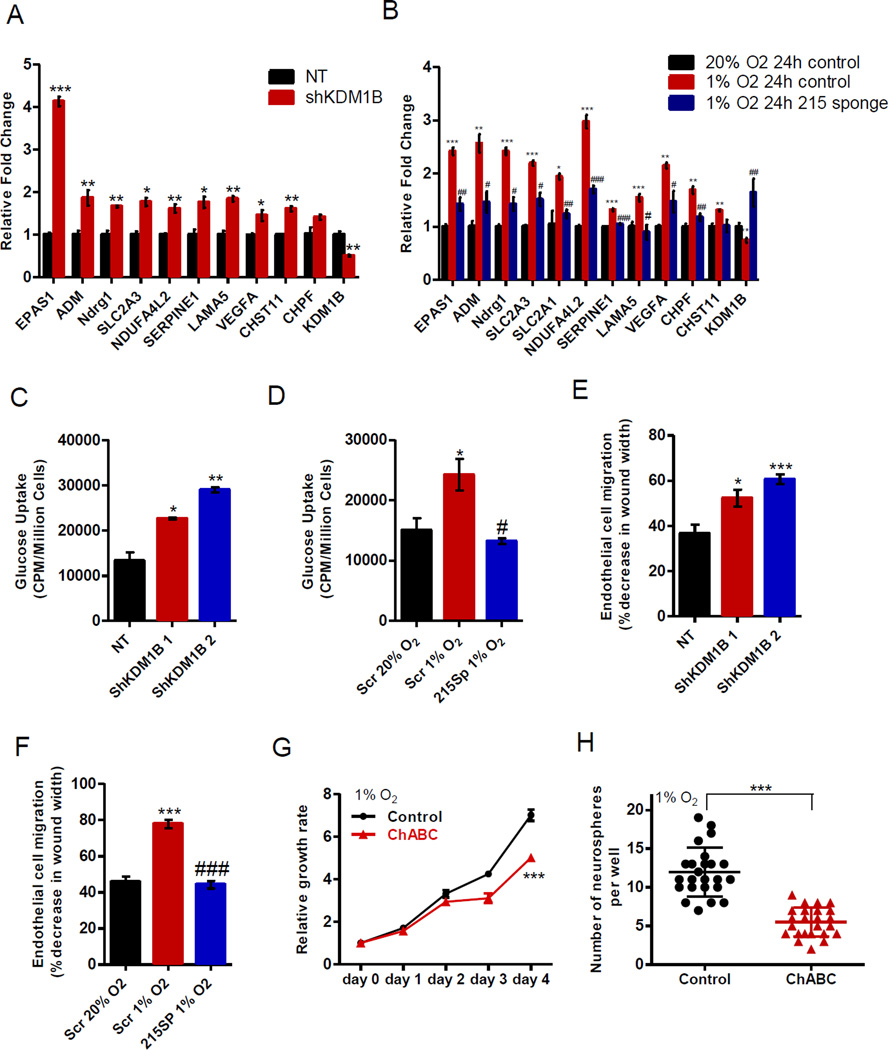
In order to confirm whether the genes enriched in those pathways were regulated by the miR-215-KDM1B axis, qPCR validation was conducted subsequently to determine the expression pattern of these genes. Our attention was drawn to three signaling pathways, mainly associated with hypoxic response and glucose metabolism (HIF2α, ADM, Ndrg1, Glut1, Glut3, NDUFA4L2), angiogenesis (VEGF, LAMA5, PAI-1), and modification of chondroitin sulfate proteoglycan (CHPF, CHST11). As shown in Figure 6A–B, expression of these genes increased when KDM1B was knocked down and no longer elevated by hypoxia when the function of miR-215 was attenuated, indicating that they serve as downstream targets of miR-215-KDM1B under hypoxia.
Figure 6. KDM1B mediates responses to hypoxia in GICs through modulating multiple processes.
(A–B) Expression profiles of genes associated with the enriched pathways downstream of miR-215-KDM1B were measured in 110040 GICs with the indicated modifications. n=3.
(C, D) Glucose uptake rate was measured in 110040 GICs with indicated modifications. n=3.
(E, F) Cell scratch assay was performed to measure the relative migration rate of immortalized human microvascular endothelial cells cultured in the conditional media derived from 110040 GICs with indicated modifications. n=3.
(G–H) Cell growth rate (G) or neurosphere formation assay (H) was measured in 110040 GICs treated with 0.05U/ml chondroitinase ABC when cultured under 1% O2. n=4 for cell growth assay.
Data are represented as the mean ± SEM. (*,# p<0.05, **,## p<0.01, ***,### p<0.001, *, compared 1% O2 24h control to 20% O2 24h control; #, compared 1% O2 24h 215 sponge to 1% O2 24h control)
See also Figure S7.
To elucidate whether the aforementioned processes contribute to the function of miR-215 and KDM1B in GICs, multiple biological aspects of those cells were assessed when the levels of miR-215 or KDM1B were manipulated. As Glut1 and Glut3 are essential glucose transporters, we firstly determined the activity of glucose uptake and found that glucose uptake by GICs with suppression of KDM1B was increased significantly (Figure 6C). On the other hand, the elevation of glucose uptake in response to hypoxia was abolished in GICs with blockage of miR-215 (Figure 6D). To further determine whether miR-215 and KDM1B regulate the process of glycolysis, we examined the extracellular acidification rate (ECAR) and observed enhanced production of acid when silencing KDM1B, but reduced acidification when repressing miR-215 in GICs (Figure S7C–D). As a consequence, these data demonstrated that miR-215 activates the anaerobic switch that favors glycolysis during hypoxic response by suppressing KDM1B.
Because VEGF, LAMA5 and PAI-1 are critical mediators in inducing tumor angiogenesis through regulation of endothelial cell activities including migration, we assessed whether manipulating miR-215 or KDM1B could impact endothelial cell migration in vitro. The cell scratch assay was performed using immortalized human microvascular endothelial cells cultured in conditional media derived from GICs with suppression of KDM1B or miR-215. As shown in Figure 6E–F and Figure S7E–F, changes in miR-215 or KDM1B expression significantly altered migratory activity of endothelial cells. Consistently, angiogenic activity in the intracranial tumors generated from GICs expressing the miR-215 sponge was significantly diminished compared to that in the scramble control group while restored when KDM1B was knocked down in the meantime, measured by CD31 immunostaining of the tumor sections (Figure S7G–H).
In addition, we found the expression of two enzymes catalyzing the biosynthesis of the chondroitin sulfate chain, chondroitin polymerizing factor (CHPF) and carbohydrate (Chondroitin 4) sulfotransferase 11 (CHST11), induced under hypoxia mediated by miR-215-KDM1B (Figure 6A–B), suggesting a potential role of the modification process of chondroitin sulfate proteoglycan (CSPG) chain during hypoxic responses. CSPG is composed of a core protein with modification of chondroitin sulfate chains, which is a main component of extracellular matrix (ECM). CHPF is known to transfer glucuronic acid (GlcUA) and N-acetylgalactosamine (GalNAc) to the non-reducing end of the chondroitin polymer. CHST11 catalyzes the transfer of sulfate to the GalNAc residue of chondroitin. To assess the potential biological impact of countering the activities of these two enzymes on the formation of chondroitin sulfate chain, we employed the enzyme chondroitinase ABC to cleave the polysaccharide chains without affecting the core proteins. As shown in Figure 6G–H, reduction in the level of the chondroitin sulfate chain significantly impaired the growth and neurosphere formation by GICs under hypoxia.
Taken together, these data illustrate that repression of KDM1B via enhancement of the miR-215 signal under hypoxia could activate multiple signaling pathways, which function together to mediate adaptation of GICs to the hypoxic stress.
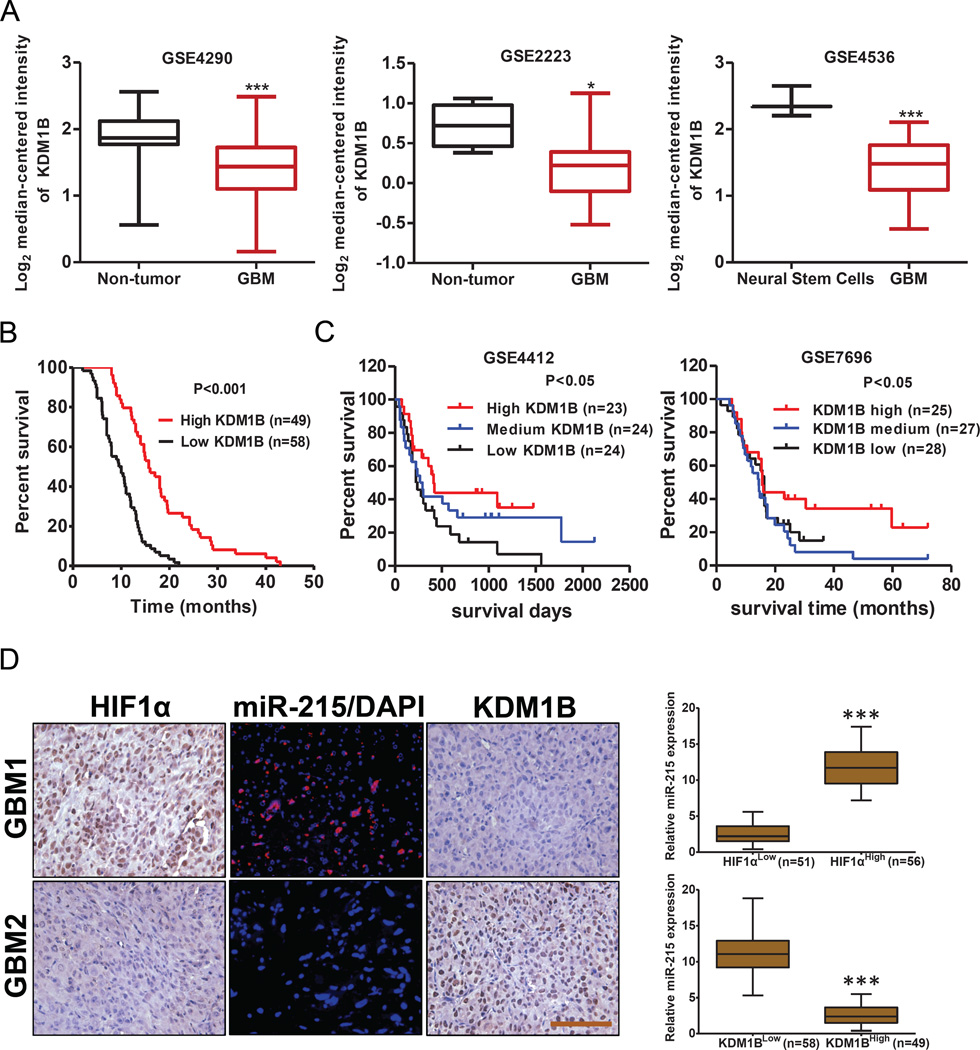
Relationship among expression profiles of HIF1α, miR-215 and KDM1B correlated with clinical prognosis of GBM patients
To evaluate the clinical relevance between KDM1B expression profiles and prognosis of GBM patients, we analyzed multiple public datasets and found that expression of KDM1B was significantly down-regulated in the GBM tissues compared to the normal tissues (Figure 7A). More importantly, low expression of KDM1B was consistently correlated with poor clinical outcome of GBM patients, based on analyses of our GBM tissue microarrays (Figure 7B) and two published datasets (Figure 7C). Finally, we investigated the clinical correlations among hypoxia, miR-215 and KDM1B by ISH and IHC assays on the same set of GBM tissue microarrays and GBM xenografts. As displayed in Figure 7D, miR-215 expression was correlated positively with protein levels of HIF1α while negatively with KDM1B level in the GBM specimens. Consistently, an inverse correlation was observed between levels of HIF1α and KDM1B in serial sections from GBM xenografts (Figure S7I). Consequently, these results support the notion that the HIF-miR-215-KDM1B signaling pathway plays an important role in the GBM progression, likely through regulation of hypoxic adaptation of GICs.
Figure 7. Clinical correlation of HIF1α-miR-215-KDM1B expression profiles and prognosis of GBM patients.
(A) Expression of KDM1B in tumor tissues of GBM patients compared with normal tissues, as analyzed from multiple published datasets.
(B–C) Kaplan-Meier curves were drawn to show the correlation between expression profiles of KDM1B and clinical outcome, as analyzed from GBM tissue microarray (B) and two public datasets (C).
(D) Representative images of ISH of miR-215 and IHC of HIF1α, KDM1B in the GBM tissue microarray (left panel). Scale bar: 50 µm. Quantification of the correlation among the expression level of miR-215 and the immunoreactivity of HIF1α and KDM1B protein in the tissue microarray (right panel).
Data are represented as the mean ± SEM. (* p<0.05, **p<0.01, ***p<0.001)
DISSCUSSION
Several miRNAs have been functionally implicated during hypoxic responses in cancer cells. In the context of GBM, for instance, miR-210 has been reported to assist cell survival in glioma cell lines (Agrawal et al., 2014; Yang et al., 2014). As a pivotal niche for GICs, hypoxia maintains the distinct biological properties of this specific group of cells. In this report, we have combined miRNA qPCR array with an in vivo functional screen and identified miR-215 as an essential miRNA modulating the GICs’ responses to hypoxia. Since miR-210 was considered to be a predominant miRNA downstream of hypoxia, we have also examined its expression level and biological function in GICs under hypoxia. The level of miR-210 is not elevated consistently in GICs from multiple lines. Suppressing the function of miR-210 in GICs could not alter the intracranial GBM progression (data not shown), suggesting that miR-210 does not serve as a universal key miRNA during the hypoxic responses in GICs.
Instead, miR-215 has emerged as a critical mediator for the hypoxic adaptation of GICs. Notably, expression levels of miR-215 and KDM1B are altered by hypoxia only in GICs rather than paired non-GICs, indicating that the miR-215-KDM1B network functions specifically in the GICs to enhance their fitness in the hypoxic niche environment and there are differential regulations of miR-215 biogenesis by hypoxia in GICs and non-GICs. One potential mechanism is that unknown factors such as specific RNA binding proteins may be required and associated with HIF-Drosha complex under hypoxia to enhance miR-215 biogenesis specifically in GICs. In this case, either expression level of those factors or the interactions between HIF-Drosha and the unknown factors may be differentially regulated in GICs verses non-GICs. Notably, we performed the microRNA profiling in the non-GICs cultured under hypoxic and normoxic conditions and found that overall expression pattern of the 88 miRNAs in the non-GICs is strikingly different from that in the GICs under hypoxia (data not shown), although both HIF1α and HIF2α were induced in those cells, indicating that mechanisms regulating the general miRNA biogenesis pathway by hypoxia might be different in GICs versus non-GICs. The differential responses to hypoxia and the underlying regulatory mechanisms between GICs and non-GICs remain to be further investigated. Because of the initial observations on the potential biological impact and regulatory mode of biogenesis, we focused on miR-215 in the current study. In the meantime, several other miRNAs including let-7i and miR-1260a shared a similar regulatory mode of their biogenesis as miR-215 by hypoxia in GICs. As those miRNAs were identified subsequently from the RNA sequencing after the initial characterization of miR-215, their potential roles in the GICs’ responses to hypoxia still remain unknown and need to be examined in the future.
Genetic knockout of KDM1B in mice was embryonic lethal due to its critical roles in maintaining the maternal genomic imprints of specific genes (Ciccone et al., 2009). In somatic cells, KDM1B possesses a great potential to modulate the expression of a large number of genes as an epigenetic regulator, which may help to maintain or reprogram the cellular state in response to environmental changes. Indeed, this notion is supported by our observations that multiple genes and pathways are regulated by KDM1B in GICs, which work collectively to affect different aspects of GICs’ responses to hypoxia and consequently assist their survival within the stress microenvironment. Among these pathways, biological processes of glucose metabolism and angiogenesis have been well explored to mediate the hypoxic responses. In this study, we have uncovered a potential role of the modification process of CSPG in hypoxia. The chondroitin sulfate chains in the ECM have been reported to bind ECM proteins, chemokines, as well as certain growth factors such as FGFs so as to enhance cancer cell proliferation and migration (Hirose et al., 2001; Takada et al., 2013). Therefore, induction of the two enzymes could lead to accelerated biosynthesis of the chondroitin-4-sulfate (C4S) chain and enhanced pro-survival signal via interactions between C4S and ECM-associated growth factors. Indeed, this assumption is supported by a report demonstrating that the level of C4S is accumulated during hypoxia, which further mediates the effects of hypoxia through regulating its binding with galectin-3 in human bronchial epithelial cells (Bhattacharyya and Tobacman, 2012). Although we have observed that disruption of the chondroitin sulfate chain by chondroitinase impairs the GIC growth under hypoxic stress, the exact activities of the chondroitin sulfate chain in affecting GIC function at the mechanistic level remain to be further explored.
As key transcriptional factors, HIFs are well known to stimulate transcription of miRNA genes, such as miR-210 (Huang et al., 2009). In concordance with this previous report, we also observed a transcriptional induction of miR-210 in GICs derived from one of the xenografted lines. On the other hand, studies have shown that hypoxia could affect microRNA processing at the post-transcriptional level through altering the expression level or modification of microprocessors, which are mainly HIF-indirect regulations (Bandara et al., 2014; Rupaimoole et al., 2014; Shen et al., 2013). Here we reveal a direct role of HIFs in mediating miRNA processing post-transcriptionally during hypoxic response. Currently two transcription factors, Smads and p53, have been shown to promote miRNA biogenesis directly through modulating the processing activity of Drosha complex on a subset of pri-miRNAs. These findings indicate that transcription factors may in general play multi-faceted functions, not only stimulate gene transcription but are also involved in the biogenesis of specific sets of miRNAs. This level of post-transcriptional regulation in miRNA biogenesis may help cells respond quickly to a specific extracellular signal in the environment via immediate production of a group of miRNAs and suppression of their mRNA targets. One question that remains to be fully answered is the detailed mechanism underlying regulation of the interaction between HIFs and the specific pri-miRNAs. Unlike Smad proteins that bind to a subset of pri-miRNAs directly through association of their DNA binding domain and consensus binding sequences embedded in the pri-miRNAs (Davis et al., 2008; Davis et al., 2010), it appears that HIFs interact with the pri-miRNAs independently from their DNA binding domains, since a HIF1α mutant with mutations in its bHLH domain was still able to promote the incorporation of pri-miR-215 into HIF1α complex or affecting the processing activity of miR-215 by Drosha (data not shown). It is possible that other domains of HIFs and/or additional RNA-binding factors associated with HIFs could directly interact with pri-miRNAs, consequently conferring the specificity of the pri-miRNAs with enhanced processing activities under hypoxia.
EXPERIMENTAL PROCEDURES
GIC isolation
GBM xenografted lines (D456MG, 110040, 080695 and 080326) were isolated from patient specimen and passaged in nude mice as subcutaneous xenografts. Xenografted tumors were dissociated by Papain Dissociation System (Worthington Biochemical). Cells were incubated with the microbead-conjugated CD133 antibody and separated by magnetic columns (Miltenyi Biotec) or with anti-Human CD15 FITC (eBioscience) and separated by the flow cytometry to enrich GICs. GICs and non-GICs were cultured as described (Wang et al., 2014b). Within 7 days after CD133 separation, same number of CD133+ and CD133− cells were implanted intracranially to measure their abilities of tumor formation.
Patient specimens were dissociated by the Papain Dissociation System and cultured in Neurobasal media as described (Wang et al., 2014b) to enrich GICs. GSCs (GSC20, GSC23, GSC17 and GSC11) provided by Dr. Erik Sulman were enriched and maintained as described (Bhat et al., 2013).
Measurement of pri-, pre- and mature miRNAs
Total RNA was extracted using the mirVana™ miRNA Isolation Kit (Ambion) and small RNAs less than 200bp were enriched by the same kit according to the manufacturer’s instructions. Total and the small RNAs were treated with DNaseI (Ambion) and reverse transcribed by the Superscript III Reverse Transcriptase (Invitrogen) using gene specific RT primers (Jiang et al., 2005). Expression of the pri-miRNAs was measured from the total RNAs while expression of the pre-miRNAs was determined from the small RNAs. To measure miRNA, total RNA was reverse transcribed by the RT2 miRNA First Strand Kit (Qiagen). Specific miRNA levels were measured by RT2 qPCR primer assays (Qiagen) with the RT2 SYBR Green mastermix (Qiagen). ACTB was used as an internal control for pri-miRNAs; RNU6 was used as the control for pre- and mature miRNAs.
Statistical Analysis
Significance of Kaplan Meier curves was tested by Log-rank (Mantel-Cox) test. Assays of cell growth curve and microprocessor activity reporter were analyzed by two-way ANOVA with p values determined from test of interaction effects. Assays of miRNA expression among different time points and luciferase reporter of KDM1B 3’UTR were analyzed by one-way ANOVA to examine significance comparing three groups. Expression profiles of miRNAs from the qPCR array and from GICs isolated from multiple xenografted lines were analyzed by two-tailed paired t-test. All other p values were determined using two-tailed unpaired student t-test. For mice experiments, n means the number of mice. For cell culture assay, n means the number of biological replicates of a given experiment and the same experiment was repeated multiple times.
Study Approval
The GBM tissue microarray study was approved by the institutional review board and the ethics committee of Harbin Medical University. The GBM patient specimens were obtained from Duke University Brain Tumor Tissue Bank. The study was approved by the Institutional Review Board of Duke University. Informed consent was obtained from all patients. All the mouse studies including subcutaneous and intracranial tumor cell implantation were conducted in accordance with a protocol approved by the Duke University Institutional Animal Care and Use Committee.
Supplementary Material
HIGHLIGHTS.
Hypoxia-induced miR-215 is required for GIC survival and GBM progression
Biogenesis of miR-215 is enhanced post-transcriptionally by HIF-Drosha interaction
MiR-215 suppresses KDM1B expression and modulates activities of multiple pathways
MiR-215 level correlates with level of HIF1α, KDM1B and GBM progression in patients
SIGNIFICANCE.
Glioblastoma (GBM) is the most common and aggressive primary brain tumor in adults, with glioma-initiating cells (GICs) implicated to be critical for tumor occurrence and recurrence. The hypoxic tumor microenvironment serves as a niche for GICs. Multiple aspects of GICs need to be modulated to reprogram cells to a state favorable for residing in the hypoxic environment and maintaining their distinct biological features. Here we reveal the HIF-miR-215-KDM1B-mediated signaling required for GIC adaptation to hypoxia through modulating activities of multiple pathways. In the meantime, we uncover an unconventional role of HIF in regulating miRNA biogenesis post-transcriptionally. Therefore, our study has provided critical insights into molecular mechanisms underlying the miRNA biogenesis pathway and the essential activities of GICs under hypoxia.
Acknowledgments
We thank M. Cook and B. Li for assistance with flow cytometry, D. Satterfield, B. Wiener and L. Ehinger for assistance with collecting fresh GBM patient specimens, and Duke Pathology Facility for assistance with paraffin processing and embedding. We thank the laboratories of Didier Trono, Thomas Tuschl, William Kaelin, Garry Nolan and Feng Zhang for providing us with plasmids via Addgene. We thank Dr. Chuan-Yuan Li for the Flag-tagged HIF2α plasmid and Dr. Erik Sulman for the GSC lines isolated from GBM patients. This work was supported by NIH grants CA164791 to X-F.W and AI091878 to Q.-J.L; the National Natural Science Foundation of China 81201978 and 81200362, the Natural Science Foundation of Jiangsu Province BK2012483, the Program for Advanced Talents within Six Industries of Jiangsu Province 2012-WSN-019 to H-B.W. The Duke University Brain Tumor Tissue Bank is supported by the Duke University Brain Cancer SPORE.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have declared that no conflict of interest exists.
Accession Number
All original microarray and RNA-Seq data were deposited in the NCBI’s Gene Expression Omnibus (GEO GSE73556, GSE73573, GSE73625).
Please refer to the supplemental information for additional experimental procedures.
REFERENCE
- Agrawal R, Pandey P, Jha P, Dwivedi V, Sarkar C, Kulshreshtha R. Hypoxic signature of microRNAs in glioblastoma: insights from small RNA deep sequencing. BMC genomics. 2014;15:686. doi: 10.1186/1471-2164-15-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babar IA, Czochor J, Steinmetz A, Weidhaas JB, Glazer PM, Slack FJ. Inhibition of hypoxia-induced miR-155 radiosensitizes hypoxic lung cancer cells. Cancer Biol Ther. 2011;12:908–914. doi: 10.4161/cbt.12.10.17681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandara V, Michael MZ, Gleadle JM. Hypoxia represses microRNA biogenesis proteins in breast cancer cells. BMC cancer. 2014;14:533. doi: 10.1186/1471-2407-14-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Bhat KP, Balasubramaniyan V, Vaillant B, Ezhilarasan R, Hummelink K, Hollingsworth F, Wani K, Heathcock L, James JD, Goodman LD, et al. Mesenchymal differentiation mediated by NF-kappaB promotes radiation resistance in glioblastoma. Cancer cell. 2013;24:331–346. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S, Tobacman JK. Hypoxia reduces arylsulfatase B activity and silencing arylsulfatase B replicates and mediates the effects of hypoxia. PloS one. 2012;7:e33250. doi: 10.1371/journal.pone.0033250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruning U, Cerone L, Neufeld Z, Fitzpatrick SF, Cheong A, Scholz CC, Simpson DA, Leonard MO, Tambuwala MM, Cummins EP, Taylor CT. MicroRNA-155 promotes resolution of hypoxia-inducible factor 1alpha activity during prolonged hypoxia. Molecular and cellular biology. 2011;31:4087–4096. doi: 10.1128/MCB.01276-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, Parada LF. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RH, Nishimura MC, Bumbaca SM, Kharbanda S, Forrest WF, Kasman IM, Greve JM, Soriano RH, Gilmour LL, Rivers CS, et al. A Hierarchy of Self-Renewing Tumor-Initiating Cell Types in Glioblastoma. Cancer cell. 2010;17:362–375. doi: 10.1016/j.ccr.2009.12.049. [DOI] [PubMed] [Google Scholar]
- Ciccone DN, Su H, Hevi S, Gay F, Lei H, Bajko J, Xu GL, Li E, Chen TP. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature. 2009;461:415–U115. doi: 10.1038/nature08315. [DOI] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol Cell. 2010;39:373–384. doi: 10.1016/j.molcel.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng YJ, Huang ZX, Xu YJ, Jin J, Zhuo W, Zhang C, Zhang XT, Shen MH, Yan XY, Wang LJ, et al. MiR-215 modulates gastric cancer cell proliferation by targeting RB1. Cancer Lett. 2014;342:27–35. doi: 10.1016/j.canlet.2013.08.033. [DOI] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang R, Barbera AJ, Xu YF, Rutenberg M, Leonor T, Bi Q, Lan F, Mei PC, Yuan GC, Lian C, et al. Human LSD2/KDM1b/AOF1 Regulates Gene Transcription by Modulating Intragenic H3K4me2 Methylation. Mol Cell. 2010;39:222–233. doi: 10.1016/j.molcel.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation (vol 5, pg 522 2004) Nat Rev Genet. 2004;5:522. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Hirose J, Kawashima H, Yoshie O, Tashiro K, Miyasaka M. Versican interacts with chemokines and modulates cellular responses. J Biol Chem. 2001;276:5228–5234. doi: 10.1074/jbc.M007542200. [DOI] [PubMed] [Google Scholar]
- Huang X, Ding L, Bennewith KL, Tong RT, Welford SM, Ang KK, Story M, Le QT, Giaccia AJ. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol Cell. 2009;35:856–867. doi: 10.1016/j.molcel.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang JM, Lee EJ, Gusev Y, Schmittgen TD. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res. 2005;33:5394–5403. doi: 10.1093/nar/gki863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MF, Hara T, Francis P, Li XL, Bilke S, Zhu Y, Pineda M, Subramanian M, Bodmer WF, Lal A. The CDX1-microRNA-215 axis regulates colorectal cancer stem cell differentiation. Proc Natl Acad Sci U S A. 2015;112:E1550–E1558. doi: 10.1073/pnas.1503370112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur B, Khwaja FW, Severson EA, Matheny SL, Brat DJ, Van Meir EG. Hypoxia and the hypoxia-inducible-factor pathway in glioma growth and angiogenesis. Neuro-oncology. 2005;7:134–153. doi: 10.1215/S1152851704001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumenis C, Naczki C, Koritzinsky M, Rastani S, Diehl A, Sonenberg N, Koromilas A, Wouters BG. Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2alpha. Molecular and cellular biology. 2002;22:7405–7416. doi: 10.1128/MCB.22.21.7405-7416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulshreshtha R, Davuluri RV, Calin GA, Ivan M. A microRNA component of the hypoxic response. Cell Death Differ. 2008;15:667–671. doi: 10.1038/sj.cdd.4402310. [DOI] [PubMed] [Google Scholar]
- Li Z, Bao S, Wu Q, Wang H, Eyler C, Sathornsumetee S, Shi Q, Cao Y, Lathia J, McLendon RE, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer cell. 2009;15:501–513. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling N, Gu J, Lei Z, Li M, Zhao J, Zhang HT, Li X. microRNA-155 regulates cell proliferation and invasion by targeting FOXO3a in glioma. Oncology reports. 2013;30:2111–2118. doi: 10.3892/or.2013.2685. [DOI] [PubMed] [Google Scholar]
- Liu FB, You XN, Chi XM, Wang T, Ye LH, Niu JQ, Zhang XD. Hepatitis B virus X protein mutant HBx Delta 127 promotes proliferation of hepatoma cells through up-regulating miR-215 targeting PTPRT. Biochem Bioph Res Co. 2014;444:128–134. doi: 10.1016/j.bbrc.2014.01.004. [DOI] [PubMed] [Google Scholar]
- Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew LK, Skuli N, Mucaj V, Lee SS, Zinn PO, Sathyan P, Imtiyaz HZ, Zhang ZF, Davuluri RV, Rao S, et al. miR-218 opposes a critical RTK-HIF pathway in mesenchymal glioblastoma. P Natl Acad Sci USA. 2014;111:291–296. doi: 10.1073/pnas.1314341111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Triboulet R, Mohseni M, Schlegelmilch K, Shrestha K, Camargo FD, Gregory RI. Hippo Signaling Regulates Microprocessor and Links Cell-Density-Dependent miRNA Biogenesis to Cancer. Cell. 2014;156:893–906. doi: 10.1016/j.cell.2013.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallamshetty S, Chan SY, Loscalzo J. Hypoxia: A master regulator of microRNA biogenesis and activity. Free Radical Bio Med. 2013;64:20–30. doi: 10.1016/j.freeradbiomed.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrom QT, Gittleman H, Liao P, Rouse C, Chen Y, Dowling J, Wolinsky Y, Kruchko C, Barnholtz-Sloan J. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2007–2011. Neurooncology. 2014;16(Suppl 4):iv1–iv63. doi: 10.1093/neuonc/nou223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupaimoole R, Wu SY, Pradeep S, Ivan C, Pecot CV, Gharpure KM, Nagaraja AS, Armaiz-Pena GN, McGuire M, Zand B, et al. Hypoxia-mediated downregulation of miRNA biogenesis promotes tumour progression. Nature communications. 2014;5:5202. doi: 10.1038/ncomms6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senanayake U, Das S, Vesely P, Alzoughbi W, Frohlich LF, Chowdhury P, Leuschner I, Hoefler G, Guertl B. miR-192, miR-194, miR-215, miR-200c and miR-141 are downregulated and their common target ACVR2B is strongly expressed in renal childhood neoplasms. Carcinogenesis. 2012;33:1014–1021. doi: 10.1093/carcin/bgs126. [DOI] [PubMed] [Google Scholar]
- Shen J, Xia W, Khotskaya YB, Huo L, Nakanishi K, Lim SO, Du Y, Wang Y, Chang WC, Chen CH, et al. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature. 2013;497:383–387. doi: 10.1038/nature12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeda A, Park M, Lee D, Mintz A, Androutsellis-Theotokis A, McKay RD, Engh J, Iwama T, Kunisada T, Kassam AB, et al. Hypoxia promotes expansion of the CD133-positive glioma stem cells through activation of HIF-1 alpha. Oncogene. 2009;28:3949–3959. doi: 10.1038/onc.2009.252. [DOI] [PubMed] [Google Scholar]
- Son MJ, Woolard K, Nam DH, Lee J, Fine HA. SSEA-1 Is an Enrichment Marker for Tumor-Initiating Cells in Human Glioblastoma. Cell Stem Cell. 2009;4:440–452. doi: 10.1016/j.stem.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm D, Bender S, Jones DT, Lichter P, Grill J, Becher O, Hawkins C, Majewski J, Jones C, Costello JF, et al. Paediatric and adult glioblastoma: multiform (epi)genomic culprits emerge. Nature reviews Cancer. 2014;14:92–107. doi: 10.1038/nrc3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–U111. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- Takada W, Fukushima M, Pothacharoen P, Kongtawelert P, Sugahara K. A sulfated glycosaminoglycan array for molecular interactions between glycosaminoglycans and growth factors or anti-glycosaminoglycan antibodies. Anal Biochem. 2013;435:123–130. doi: 10.1016/j.ab.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Tong YQ, Liu B, Zheng HY, Gu J, Liu H, Li F, Tan BH, Hartman M, Song C, Li Y. MiR-215, an activator of the CTNNBIP1/beta-catenin pathway, is a marker of poor prognosis in human glioma. Oncotarget. 2015 doi: 10.18632/oncotarget.4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 similar to 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Zhao Y, Zheng Y. MiR-122/Wnt/beta-catenin regulatory circuitry sustains glioma progression. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014a;35:8565–8572. doi: 10.1007/s13277-014-2089-4. [DOI] [PubMed] [Google Scholar]
- Wang H, Sun T, Hu J, Zhang R, Rao YH, Wang S, Chen R, McLendon RE, Friedman AH, Keir ST, et al. miR-33a promotes glioma-initiating cell self-renewal via PKA and NOTCH pathways. J Clin Invest. 2014b;124:4489–4502. doi: 10.1172/JCI75284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- Yan Z, Wang J, Wang C, Jiao Y, Qi W, Che S. miR-96/HBP1/Wnt/beta-catenin regulatory circuitry promotes glioma growth. FEBS letters. 2014;588:3038–3046. doi: 10.1016/j.febslet.2014.06.017. [DOI] [PubMed] [Google Scholar]
- Yang W, Wei J, Guo T, Shen Y, Liu F. Knockdown of miR-210 decreases hypoxic glioma stem cells stemness and radioresistance. Experimental cell research. 2014;326:22–35. doi: 10.1016/j.yexcr.2014.05.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.