Abstract
Genome-wide association studies (GWAS) have identified single nucleotide polymorphisms (SNPs) associated with increased risk for colorectal cancer (CRC). A molecular understanding of the functional consequences of this genetic variation is complicated because most GWAS SNPs are located in non-coding regions. We used epigenomic information to identify H3K27Ac peaks in HCT116 colon cancer cells that harbor SNPs associated with an increased risk for CRC. Employing CRISPR/Cas9 nucleases, we deleted a CRC risk-associated H3K27Ac peak from HCT116 cells and observed large-scale changes in gene expression, resulting in decreased expression of many nearby genes. As a comparison, we showed that deletion of a robust H3K27Ac peak not associated with CRC had minimal effects on the transcriptome. Interestingly, although there is no H3K27Ac peak in HEK293 cells in the E7 region, deletion of this region in HEK293 cells decreased expression of several of the same genes that were downregulated in HCT116 cells, including the MYC oncogene. Accordingly, deletion of E7 causes changes in cell culture assays in HCT116 and HEK293 cells. In summary, we show that effects on the transcriptome upon deletion of a distal regulatory element cannot be predicted by the size or presence of an H3K27Ac peak.
INTRODUCTION
In our previous studies, we identified a set of enhancers (defined as the presence of a H3K27Ac peak located farther than +/− 2 kb from a transcription start site) that harbor single nucleotide polymorphisms (SNPs) associated with an increased risk for colon cancer (1). Our working hypothesis is that the different nucleotide sequence between the ‘risk-associated’ vs. ‘non risk-associated’ SNPs affects activity of the enhancers, causing a change in expression in genes (coding or non-coding) that can influence the balance between normal tissue proliferation or differentiation versus tumor initiation or progression. Enhancers are composed of binding sites for many different site-specific DNA binding transcription factors (TFs) that are thought to work in concert to provide cell type-specific functionality. For example, one of the first characterized mammalian enhancers is the interferon β enhanceosome, which is bounded by eight different TFs (2,3). Recent studies from the ENCODE Project (4) and the Roadmap Epigenome Mapping Consortia (5) have identified hundreds of thousands of enhancers, most of which include motifs for a variety of different TFs. The overall function of a given enhancer is dependent upon several conditions, such as the number of motifs contained within it, the extent to which the nucleotides within the enhancer match consensus binding motifs, the expression level of the TFs that bind those motifs and the location of the enhancer with respect to chromatin boundaries. Because many TFs contribute to the overall function of an enhancer, it is likely that single nucleotide changes within an enhancer will have quite modest effects on the transcriptional output from a target promoter (6). Although modest effects in gene expression could have strong phenotypic outcomes over the course of a long time period, such as during tumor development, the consequences of a single nucleotide change in an enhancer may be difficult to observe in short term cell culture assays. Thus, rather than analyzing the effect of a single SNP, our approach is to determine the functional role of the enhancer as a whole by identifying genes that are responsive to loss of the enhancer in colon cancer cells. For comparison, we also analyzed an enhancer not associated with colorectal cancer (CRC) and a distal region that lacks the H3K27Ac mark. We show that deletion of distal regulatory elements associated with CRC can affect nearby genes and also have genome-wide effects on the transcriptome. Our results also suggest that effects on the transcriptome upon deletion of a distal regulatory element cannot be predicted by the size or presence of an H3K27Ac peak.
MATERIALS AND METHODS
Cell culture
The human cell lines (control and enhancer-deleted versions) HCT116 (ATCC #CCL-247) and HEK293 (ATCC #CRL-1573) were grown at 37°, in 5% CO2 in Dulbecco's Modified Eagle Medium with 10% fetal bovine serum and 1% penicillin and streptomycin.
CRISPR/Cas9-mediated genome editing
The guide RNAs (gRNAs) flanking the target enhancer regions were designed using a website tool (http://crispr.mit.edu), avoiding repeat regions in the hg19 genome. After identification of a potential guide RNA, the 16–17 nt region including the PAM sequence (NGG) was BLASTed against the hg19 genome to confirm that it was unique in the genome; all guide RNAs used in this study also did not have a 1 mismatch sequence in the human genome. The target DNA sequences of the gRNAs used in this study are listed in Additional File 1. The 100 bp oligonucleotides containing the gRNA sequences were inserted into the gRNA Empty Vector (Addgene, catalog#41824) according to the gRNA synthesis protocol (7). The sequences of gRNA expression plasmids were confirmed through Sanger sequencing. To delete an enhancer, two gRNA plasmids and a plasmid expressing Cas9-GFP (Addgene, catalog #44719) were transiently transfected into HCT116 or HEK293 cells in a 6-well plate with a Cas9: gRNAs molar ratio of 1:22 using Lipofectamine 3000 (Life Technologies, catalog #3000008). Genomic DNA was extracted from the transfected cells 48 h post-transfection using the QIAamp DNA mini kit (Qiagen, catalog# 51306). Subsequently, polymerase chain reaction (PCR) using primers flanking the target enhancer regions was performed to check the deletion efficiency. Once enhancer deletion was confirmed in a pool of transfected cells, cells with high Green Fluorescent Protein (GFP) expression were identified using fluorescence-activated cell sorting with the Aria II cell sorter (BD Biosciences). Sorted cells were plated into individual wells of a 24-well plate and then re-plated as single cells in 10 cm dishes and subsequently expanded for further analyses.
PCR detection of cells having enhancer deletions
Clonal colonies from the 10 cm dishes were transferred and passaged into 24-well plates. When cells were 80–90% confluent, genomic DNA was isolated from each clone using the QIAamp DNA mini kit (Qiagen, catalog# 51306) and tested for deletion by PCR using GoTaq Green Master Mix (Promega, catalog#M712) and primers flanking the target enhancer region. For those colonies that showed loss of the enhancer region, a second PCR was performed using primers that should detect the inner portion of the enhancer. In general, <10% of the clones tested after sorting for high GFP levels showed biallelic deletion. For the E7 CRC-associated enhancer, H3K27Ac ChIP-seq data was also used to confirm loss of the enhancer signal (Additional File 2).
RNA-seq
RNA samples were collected from cells using Trizol (Life Technologies, catalog #15596018) from clonal population. To remove batch effects, matched controls and deleted samples were plated with similar confluency and harvested at the same time, RNA was extracted at the same time, RNA libraries were prepared at the same time and barcoded libraries were pooled and sequenced together (see Additional File 3). All RNA libraries were prepared using the Illumina TruSeqV2 Sample Prep Kit (catalog #15596–026), starting with 1 μg total RNA. ERCC spike-in mix (Thermo Fisher, catalog # 4456740) was added when the libraries were made so that quality assessment could be performed on the RNA-seq data, which allowed the removal of outliers caused by technical variations. Libraries were sequenced on a Nextseq500 with 75 bp single reads. Raw reads were trimmed using the Quality Score method (minimal quality score 20, minimal read length 25, trimming from both ends) and mapped to hg19 (Ensembl 72) using Tophat2 (8) installed in the Partek Flow version 3 program (Partek Inc., St Louis, MO, USA). A matrix of raw fragments counts for each gene was generated from alignment files using HTSeq python package (9) with Genecode V19 annotation and these counts were used for differential gene expression analysis using an edgeR program (10). For the group of large datasets (Set A; see Additional File 3), where there are 12 controls versus 3 samples, genes with at least 1 counts per million (CPM) in at least 6 samples were kept for differential gene expression analysis. For groups of small datasets (Sets B and C; Additional File 3), consisting of 2 controls versus 2 samples, genes with at least 1 CPM in at least 2 samples were kept for differential gene expression analysis. In dataset A, after removing lowly counted genes, upper-quartile normalization was used for the GLM approach in edgeR which takes into account other covariates (e.g. date of RNA extraction; see Additional File 4) found in the PCA plots. For datasets B and C, filtered gene counts were normalized using the Trimmed Mean of M-value (TMM) method for the negative binomial model in edgeR. Among the differentially expressed genes with an FDR <0.05, lowly expressed genes were filtered out if the average CPM or average RPKM values (quantified using Partek software) was less than two in the controls for downregulated genes or two in the deleted samples for upregulated genes. Finally, genes with >1.5-fold change were defined as differentially expressed genes (i.e. the numbers shown in Table 1).
Table 1. Altered gene expression upon enhancer deletion.
| Enhancer | Total number of up regulated genes (>1.5 FC) | Upregulated genes +/− 1 Mb | Total number of downregulated genes (>1.5 FC) | Downregulated genes +/− 1 Mb | Putative direct targets | Average RPKM in Controls | Fold Change | Distance from enhancer to TSS |
|---|---|---|---|---|---|---|---|---|
| E7 | 590 | 0 | 565 | 5 | FAM84B | 7.45 | 1.50 | −843 000 |
| CCAT1 | 2.79 | 1.78 | −182 000 | |||||
| CASC8 | 2.33 | 1.55 | 81 000 | |||||
| MYC | 112.3 | 2.31 | 335 000 | |||||
| PVT1 | 7.96 | 1.59 | 395 000 | |||||
| E7 (HEK293) | 166 | 0 | 295 | 1 | MYC | 28.5 | 5.10 | 335 000 |
| 18qE | 13 | 0 | 3 | 0 | N/A | N/A | N/A | N/A |
| 18qNE | 14 | 0 | 3 | 0 | N/A | N/A | N/A | N/A |
Altered gene expression upon enhancer deletion. The number of genes having >1.5 fold increase or decrease in expression in the genome and within +/− 1 Mb after deletion of each distal region is indicated. Also shown are the names of the putative direct target genes, the average RPKM values for these genes in the control cells, the fold decrease in the enhancer-deleted cells and the distance of these genes from the distal regions.
Cell assays
Controls and E7-deleted HCT116 cells were counted by sorting single cells using a flow cytometer and then plated into a 96-well plate at a density of 1 × 104 or 2.5 × 103 cells per well. After 72 or 96 h, cells were rinsed with 1× PBS and then 100 µl of PBS mixed with 10 µl of WST-1 reagent (Roche, catalog # 05015944001) was added to each well. Cell proliferation was measured using a microplate reader (HIDEX Chameleon V4.43) after incubation for 15 min at 37°C. The wavelength for measuring the absorbance of formazan, the product of the WST-1 assay, was 450 nm.
Colony forming assays
HCT116 cells and HEK293 cells were counted by sorting single cells using a flow cytometer and were seeded in 6-well plates at a density of 1.5 × 103 cells per well and 6.5 × 103 cells per well, respectively. After a 2 week incubation, cells were fixed with 100% methanol for 10 min, stained with 0.5% crystal violet for 30 min, and colonies were assessed after rinsing the plates with water.
ChIP-seq and analysis
H3K27Ac ChIP-seq data from HCT116 cells (ENCODE accession number ENCSR000EUT) was downloaded from the UCSC genome browser and analyzed using the Sole-search ChIP-seq peak calling program (11,12) using the following parameters (Permutation:5; Fragment:250; αValue: 0.00010 = 1.0E-4; FDR: 0.00010 = 1.0E-4; PeakMergeDistance:0; HistoneBlurLength:1200). H3K27Ac ChIP-seq samples for control and enhancer-deleted HCT116 cells were prepared using an H3K27Ac antibody (Active motif catalog#39133, Lot#21311004), as previously described (13), with minor modifications (complete protocol available upon request). These ChIP-seq libraries were sequenced on a HiSeq2000 with 50 bp single end reads. All ChIP-seq FASTQ files were mapped to hg19 using BWA (default parameters). To identify decreased or increased H3K27Ac peaks in enhancer-deleted cells, two biological replicates for each group were used for differential peak analysis. A peak-calling prioritization pipleline (PePr) was used to identify significant differential binding sites of H3K27Ac caused by enhancer deletions (14); default parameters for histone marks were used for analysis and differential peaks having a p-value less than 1e−5 are reported.
RESULTS
Deletion of CRC risk-associated enhancers can cause widespread effects on the transcriptome
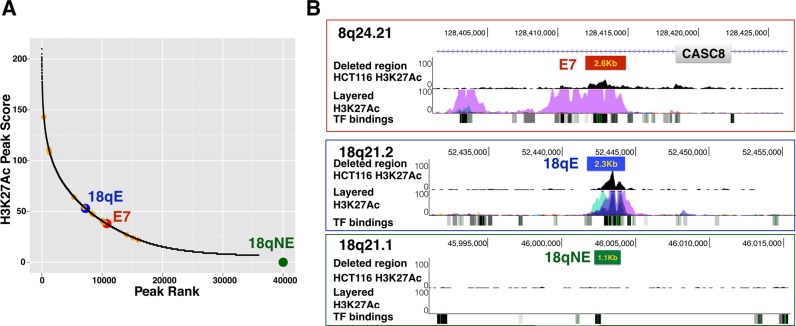
We have chosen to use HCT116 colon cancer cells for our analysis of CRC risk-associated enhancers because of the availability of histone modification data (15), whole genome DNA methylation data (16), and ChIP-seq TF binding sites (4) from these cells. Of the previously identified 28 CRC risk-associated enhancers, 14 are detected in HCT116 cells. However, they are not necessarily amongst the top ranked enhancers in HCT116 cells (Figure 1A). Interestingly, 12 of the 14 CRC risk-associated enhancers in HCT116 cells are located within the introns of 6 protein-coding genes and 1 non-coding RNA, whereas 2 of the enhancers are intergenic. In HCT116 cells, ∼50% of all H3K27Ac enhancers are intronic (16); it is not clear if the high preponderance of intronic enhancers in the set of CRC risk-associated enhancers is due to a bias involving the choice of index SNPs on the GWAS array or due to a biological or functional reason. However, others have also observed that GWAS SNPs (and SNPs in high linkage disequilibrium with GWAS index SNPs) are more enriched in introns than in intergenic regions (17). We selected to study a risk enhancer, which resides in an intron of the non-coding RNA CASC8 (enhancer E7, identified by rs6983267) for targeted deletion (Figure 1B). Although the function of CASC8 is not known, SNPs within the long non-coding RNA CASC8 have also been implicated in increased risk for gastric and breast cancer (18). However, enhancers can work at a long distance and in either orientation and thus it is not certain that the gene in which an enhancer resides is in fact regulated by that enhancer (19–22). For example, in a ChIA-PET study using an antibody for RNA polymerase II, Li et al. (23) identified ∼20 000–30 000 enhancer–promoter loops in MCF7 or K562 cells. Of these, more than 40% of the enhancers skipped over the nearest gene to loop to a farther one. Of course, it is not been proven that all loops represent bona fide regulatory interactions or that all enhancer-mediated regulation involves looping (24). However, these studies indicate that it is not a certainty that the genes in which the enhancers reside are the ones that should be linked to an increased risk for colon cancer. To gain further insight into the mechanisms by which the CRC risk-associated enhancers may influence tumor development, we analyzed gene expression changes in clonal populations of cells lacking E7.
Figure 1.
Genomic location and H3K27Ac profiles of E7, 18qE and 18qNE. (A) Shown is a graph representing HCT116 H3K27Ac peaks, as identified using Sole-search (11,12) to analyze the HCT116 H3K27Ac ChIP-seq data; all 35 932 peaks were plotted based on peak height (Y axis) versus peak rank (X axis). The locations of the 14 CRC risk-associated enhancers present in HCT116 cells are shown (orange circles), with the large red circle indicating the CRC risk-associated enhancer deleted in this study. Also shown is the location of the H3K27Ac peak called 18qE (blue circle); the green circle indicates that the 18qNE region was not called as a peak. (B) For the distal regions deleted in this study, the genomic location, the H3K27Ac pattern in HCT116 cells, the combined H3K27Ac pattern in multiple cell types (the ENCODE Regulation Layered H3K27Ac track) and TF binding data (the ENCODE Regulation Txn Factor ChIP track combines ENCODE ChIP-seq data from many different transcription factors (TFs) and cell lines in a relatively dense display) from the UCSC genome browser are shown; the boxes indicate the approximate region that was deleted.
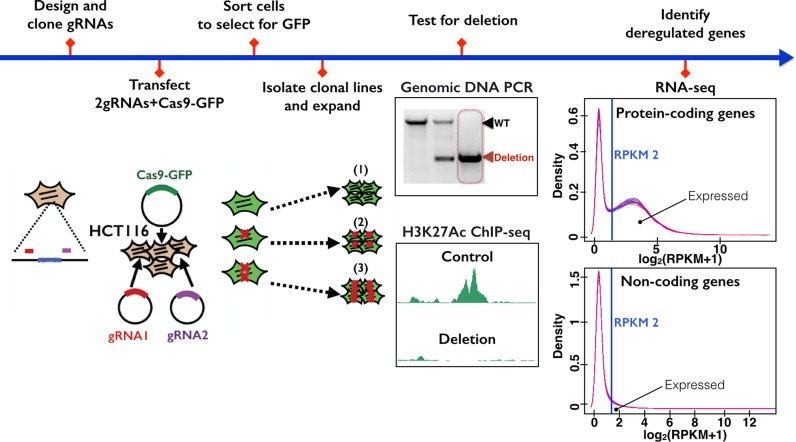
An overview of the strategy used to determine the function of enhancers is shown in Figure 2. Guide RNAs were designed that flank the H3K27Ac marks of an enhancer; see Additional File 1 for the genomic location and the sequences of all guide RNAs used in this study. Plasmids expressing the guide RNAs and Cas9-GFP were transfected into cells, 48 h later cells were sorted for high GFP expression and colonies were grown from single cells. PCR analysis using a set of primers that flank the targeted genomic region and a set of primers internal to the targeted region were used to identify clonal lines having deletion of the targeted region (Additional File 2). In general, <10% of the clones tested after sorting for high GFP levels showed deletion of all alleles. After confirming that the targeted region had been deleted, RNA-seq was performed to identify gene expression differences between the cells with enhancer deletion versus the control cells. We prepared control samples from four different clonal populations of FACS-selected HCT116 cells that had been transfected with Cas9-GFP plus the guide RNA vector lacking inserted guide RNAs. Each control clone was analyzed using RNA preps prepared from cell cultures grown on different days. For enhancer-deleted cells, we also prepared triplicate RNA samples from cells grown on the same days as the control clones, and sequenced matched control and deleted samples in the same lane of a sequencer to prevent batch effects. Information concerning all RNA-seq data sets analyzed in this study can be found in Additional File 3, quality control plots for the RNA-seq data can be found in Additional File 4 and gene expression data can be found in Additional File 5.
Figure 2.
Experimental schema. The overall approach used to analyze the function of an enhancer is shown, beginning with design of guide RNAs used to guide Cas9 to the targeted enhancers and ending with RNA-seq analysis of control and enhancer-deleted cells.
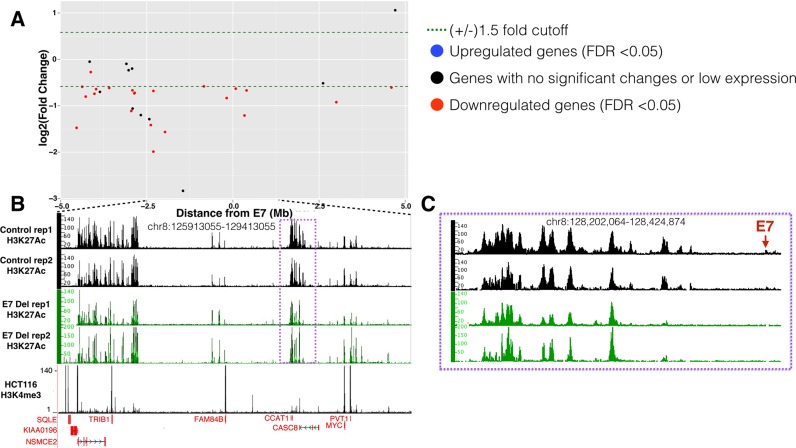
The E7 CRC risk-associated enhancer harbors a GWAS CRC index SNP and resides near, but not within, a region that contains a large number of super-enhancers (see Figure 3). Due to the small size of the H3K27Ac peak (Figure 3C), we initially thought that most genes in the region would be controlled by the super-enhancers and not by E7. Surprisingly, we found that many genes within +/− 5 Mb from E7 were downregulated upon deletion of the enhancer (Figure 3A). Specifically, five genes were downregulated within +/− 1 Mb of E7, including the non-coding CASC8 RNA (in which E7 resides), which was reduced 1.5-fold. Of note, deletion of E7 also had effects on genes that were not nearby, causing the up- or downregulation of more than 1000 genes in the genome, most of which were on other chromosomes (Table 1). In fact 25 genes were reproducibly downregulated more than 5-fold in the E7-deleted cells; a list of all downregulated genes in E7-deleted cells is found in Additional File 5. We also noted that deletion of the small E7 enhancer resulted in loss of H3K27Ac peaks at the edge of the nearby super-enhancer (Figure 3C) and at another super-enhancer located ∼2 Mb from E7.
Figure 3.
Gene expression and H3K27Ac changes upon deletion of E7 in HCT116 cells. (A) The Y-axis shows log2 fold changes in gene expression in the E7-deleted cells throughout a +/− 5 Mb region from E7, with genes showing decreased expression upon enhancer deletion having negative numbers and genes showing increased expression upon enhancer deletion having positive numbers. (B) Shown are the H3K27Ac patterns in independent replicates of control and enhancer-deleted cells within an ∼3 Mb region near the E7 enhancer (located within the purple box, which is shown in an expanded view in panel C). Also shown is the H3K4me3 pattern to identify promoter regions. The Y-axis indicates the peak height and the X-axis indicates the genomic location; only those genes significantly downregulated in the deleted cells are indicated in red below the H3K4me3 track. (C) The H3K27Ac patterns in control and deleted cells are shown for the region nearby E7 (red arrow), showing loss of the E7 H3K27Ac peak, as well as loss of H3K27Ac signal at the right side of the nearby large enhancer region.
The size or presence of an H3K27Ac peak does not always correlate with effects on the transcriptome
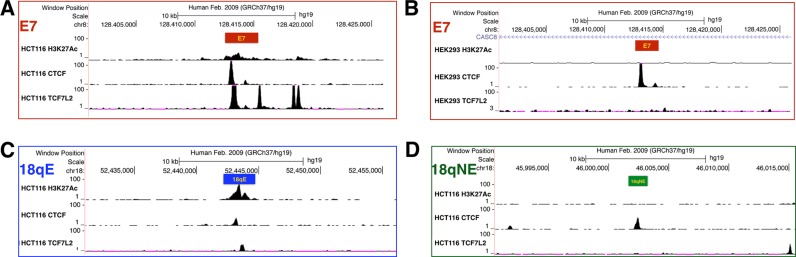
As an approach to understanding the genome-wide changes in gene expression seen upon deletion of E7, we also deleted a strong H3K27Ac peak on chromosome 18 (termed 18qE for ‘enhancer on chromosome 18q’). The 18qE region has a more robust H3K27Ac peak in HCT116 cells than does E7 (Figure 1A) and is covered by H3K27Ac in many cell types (see the layered H3K27Ac track in Figure 1B). Similar to E7 (Figure 4), 18qE is bound by TCF7L2, a site-specific TF that has been implicated as the downstream regulator of the WNT pathway that drives the development of colon cancer (25–29) and by CTCF, a multi-functional protein that is thought to be involved in gene regulation and chromosomal structure (30–34). However, we note that the binding of TCF7L2 and CTCF is not as strong at 18qE as at E7. For comparison, we also deleted another region on chromosome 18 (termed 18qNE for ‘no enhancer on chromosome 18q’) that does not have H3K27Ac or TCF7L2 binding but is bound by CTCF. Interestingly, we found that deletion of the large 18qE enhancer had little effect on gene expression, causing changes similar to those seen when the 18qNE region (which completely lacks H3K27Ac) was deleted (Table 1).
Figure 4.
H3K27Ac, TCF7L2, and CTCF binding profiles of E7, E24, 18qE and 18qNE. Shown are the H3K27Ac, TCF7L2 and CTCF binding patterns at (A) the E7 enhancer in HCT116 cells, (B) the E7 enhancer in HEK293 cells, (C) 18qE, an intergenic H3K27Ac peak on chromosome 18, in HCT116 cells and (D) 18qNE, an intergenic region on chromosome 18 that lacks the H3K27Ac mark, in HCT116 cells. The Y-axis indicates the peak height and the X-axis indicates the genomic location; the boxes indicate the region deleted by the CRISPR/Cas9 method.
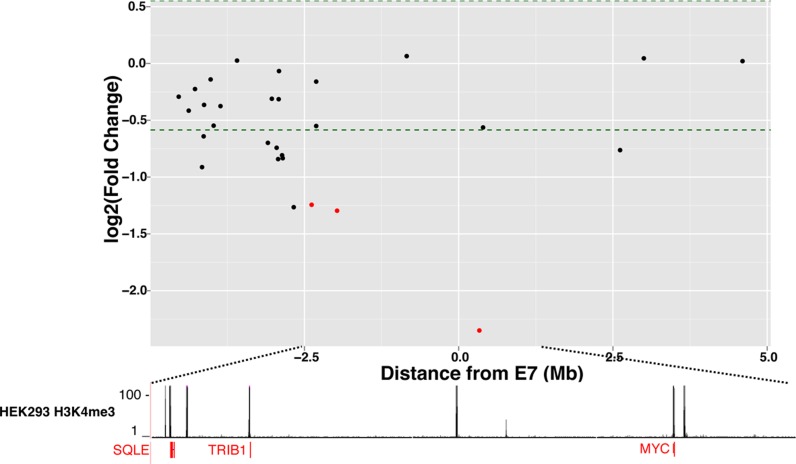
The differential effects on the transcriptome seen when E7 versus 18qE was deleted from HCT116 cells suggest that the presence of a robust H3K27Ac peak might not be strongly correlated with effects on the transcriptome, but that other, as of yet unknown characteristics of the region may be critical. Interestingly, although many cell types do show a strong H3K27Ac peak in the E7 region (see Figure 1B), there is no H3K27Ac peak in HEK293 cells (Figure 4). However, there is a very strong CTCF binding site at E7 in HEK293 cells. To compare the effects of deleting a genomic region that is marked by H3K27Ac in one cell type but not in another, we deleted E7 in HEK293 cells. We found that deletion of this region in HEK293 cells caused changes in hundreds of genes, including MYC, which showed a robust 5-fold decrease in expression (Figure 5 and Table 1); a list of all downregulated genes in HEK293 E7-deleted cells is found in Additional File 5. We note that HCT116 cells are derived from the colon whereas HEK293 cells are derived from the kidney. Therefore, downstream signaling pathways affected by MYC are likely to be quite different in the two cell types; as expected most of the genes (other than MYC) that were altered in the HCT116 and HEK293 cells upon deletion of E7 were different.
Figure 5.
Gene expression and H3K27Ac changes upon deletion of E7 in HEK293 cells. The Y-axis shows log2 fold changes in gene expression in the E7-deleted cells throughout a +/− 5 Mb region from E7, with genes showing decreased expression upon enhancer deletion having negative numbers and genes showing increased expression upon enhancer deletion having positive numbers; only those genes significantly downregulated in the deleted cells are indicated in red below the H3K4me3 track.
Characterization of the genome-wide changes in the epigenome upon enhancer deletion
As another approach to characterizing the effects of deletion of an specific enhancer, we performed a genome-wide analysis of H3K27Ac in the E7-deleted cells. We analyzed the H3K27Ac patterns within +/− 100 Kb of upregulated and downregulated genes in the E7-deleted cells. We found that many of the downregulated genes in E7-deleted cells (Additional File 6A) had decreased H3K27Ac peaks nearby. In contrast, many of the upregulated genes in E7-deleted cells (Additional File 6B) had increased H3K27Ac peaks nearby; see Additional File 7 for the differential H3K27Ac peak analysis. For example, in E7-deleted cells there was a large downregulation of DPEP1 and ANXA10 mRNA, with concomitant decrease in the H3K27Ac marks near those genes. In contrast, the H3K27Ac pattern was increased near upregulated genes such as FGF9 and VSNL1.
Cell growth is affected by deletion of enhancer E7
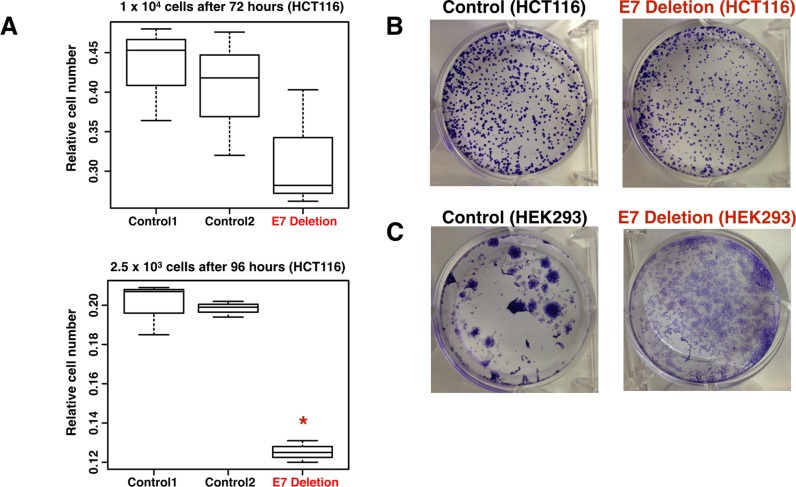
Deletion of E7 in both HCT116 cells and in HEK293 cells caused decreased expression of MYC, a well-characterized oncogene (35–37). Therefore, it is possible that deletion of the E7 enhancer affected cell proliferation via downregulation of MYC protein. To test this hypothesis, we performed cell number and colony forming assays in control and E7-deleted HCT116 and HEK293 cells. HCT116 control and E7-deleted cells were plated at two different densities and grown for 72 or 96 h. Cell number (Figure 6A) and colony formation (Figure 6B) is reduced in E7-deleted cells, as compared to control cells. As shown in Additional File 8, E7-deleted cells also have morphological changes and increased contact inhibition. The HEK293 cells in which the enhancer was deleted showed drastic changes in cell morphology, suggesting that the ability to form large colonies was affected upon enhancer deletion. Thus, deletion of the region containing rs6983267 caused downregulation of MYC, large changes in the transcriptome, and changes in responses in cell growth assays in both HCT116 and HEK293 cells, despite the fact that the epigenomic profile of E7 is quite different in the two cell types.
Figure 6.
Cell number is affected by deletion of enhancer 7. (A) Shown are the relative cell numbers 72 h after plating two different control clones or the E7 deleted clone at 1 × 10e4 cells (top panel) or 96 h after plating two different control clones or the E7 deleted clone at 2.5 × 10e3 cells (bottom panel). The Y-axis indicates relative cell numbers from an average of four replicate wells, as measured by absorbance of products of the WST-1 assay. Student's t-test was used for significance and * indicates a P-value < 0.005. (B) Shown are colony forming assays for control and E7-deleted HCT116 cells. A total of 1500 cells were seeded per well and after 2 weeks colonies were stained with crystal violet. At least three replicates were performed; additional replicates are shown in Additional File 8. (C) Shown are colony forming assays for control and E7-deleted HEK293 cells. A total of 6500 cells were seeded per well and after two weeks colonies were stained with crystal violet. At least three replicates were performed; additional replicates are shown in Additional File 8.
DISCUSSION
Employing CRISPR/Cas9 nucleases, we deleted a CRC risk-associated enhancer from the genome of HCT116 colon cancer cells and analyzed effects on the transcriptome and epigenome. We found widespread changes in expression and H3K27Ac patterning in the enhancer-deleted cells. However, deletion of a robust H3K27Ac peak (18qE) not associated with CRC did not cause similar changes in gene expression. Interestingly, deletion of the E7 region encompassing the CRC-associated risk SNP in HEK293 cells, at which there is no detectable H3K27Ac peak but there is a robust CTCF site, also caused widespread changes in gene expression. Finally, we show that deletion of the E7 region affects gene expression and cell culture assays in both HCT116 and HEK293 cells, likely due to downregulation of the MYC oncoprotein.
Large-scale GWAS efforts have identified sets of SNPs associated with a particular disease, such as colon cancer. If a SNP falls within an exon and changes the coding potential of that gene, investigators have suggested that the identified gene is likely to be associated with increased risk for that disease. However, most GWAS-identified SNPs do not fall within exons and thus the mechanism by which these SNPs might affect disease has not been clear (38,39). Recent studies have shown that many of the GWAS index SNPs and SNPs in high LD with the GWAS index SNPs fall within regulatory elements such as promoters and enhancers. In the case of the SNPs falling close to transcription start sites, it has been assumed that they cause changes in expression of the gene regulated by that promoter. In the case of SNPs falling within enhancer regions (as defined by the H3K27Ac mark), it is more difficult to understand how the SNP affects gene expression because enhancers can work at a great distance, in either orientation, and often skip over the nearest transcription start site to regulate genes farther away (21,23). We reasoned that precise deletion of an enhancer should identify genes regulated (directly and indirectly) by that enhancer, with nearby downregulated genes being possible direct target genes. As a test of this experimental approach, we deleted a CRC risk-associated enhancer from the human genome. We found that deletion of the CRC risk-associated enhancer, but not a non-risk-associated enhancer, caused reproducible changes in expression of at least one gene within +/− 1 MB of the enhancer. It is important to note that a promoter can be regulated by several different enhancers; if a gene is regulated equally by two enhancers, then a 50% drop in RNA levels is what would be expected upon deletion of one of the enhancers. Most of the nearby genes that were downregulated upon enhancer deletion were expressed at 20–65% of their expression level in control cells, suggesting that the enhancers contributed to expression of these genes, but were not the sole regulatory elements controlling the activity of the target promoters.
We note that deletion of the CRC risk-associated enhancer not only affected nearby genes but also affected the regulation of hundreds of other genes, some on the same chromosome but very far from the enhancer and still more located on other chromosomes. There have not yet been sufficient studies to know how far a gene can be from an enhancer (or even if it can be on a different chromosome) and still be a ‘direct’ target. One approach that is used to identify direct targets is to use 3-dimensional (3D) looping assays. A recent study showed that 57% of enhancer–promoter loops span more than 100 Kb (40). Importantly, our results are supported by a previous study in which the E7 enhancer region was shown to interact with the MYC and PVT1 promoters in colon cancer cells (41,42). Studies in other cell types have also provided evidence that E7 interacts with the MYC gene (43–46). We also observed a reduction in the lncRNA CCAT1 in the E7-deleted HCT116 cells. Others have suggested that reduction of CCAT1 can reduce long-range interactions between the MYC promoter and its enhancers (47). Thus, it is possible that direct downregulation of CCAT1 (which is located 182 Kb from E7) caused an indirect downregulation of MYC. However, looping assays should be interpreted with caution, as different assays give different results (48). Also, it is not yet known if all loops represent functional enhancer–promoter interactions or if some loops may play other roles, such as structural components of chromatin (24).
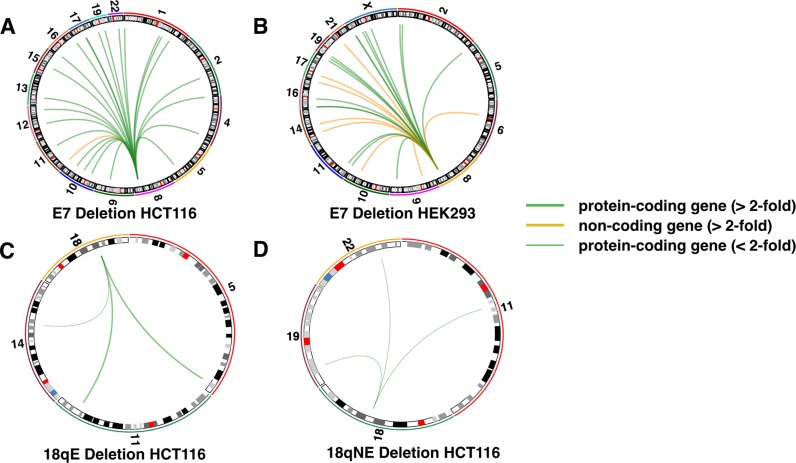
We have shown that deleting E7 not only affects expression of relatively ‘nearby’ genes but that it also causes major changes in the transcriptome. In fact, the genes that changed the most in expression are on other chromosomes (Figure 7). In our experiments, we have attempted to control for effects on the transcriptome not related to enhancer deletion. As described in detail in the ‘Materials and Methods’ section, all differential gene expression analysis was performed using a set of control clones that were selected in the same way as were the clones of enhancer-deleted cells (i.e. sorting via FACs for single colonies after transfection with Cas9-GFP and the guide RNA vector). Also, we show that deletion of other genomic regions does not lead to genome-wide changes in gene expression; the 18qE and 18qNE clones have essentially identical gene expression profiles as do the control clones, indicating that the robust changes in gene expression observed when E7 was deleted were not general effects due to the CRISPR/Cas9 technology. In addition, we note that we have used two independent sets of guide RNAs to delete E7. After showing that MYC was downregulated in HCT116 cells using a set of guide RNAs that deletes a 2.6 Kb region encompassing the E7 enhancer, we then shifted the guide RNAs slightly closer together such that they target a 2 Kb region within the original 2.6 Kb region (removing the same TF binding sites) and isolated another clone of HCT116 cells in which all alleles of E7 were deleted. Analysis of this clone by qRT/PCR showed that MYC was downregulated 2.9 fold (P < 0.002) using these independent guide RNAs. We used this second set of guide RNAs to delete E7 in HEK293 cells. Thus, in both HEK293 and HCT116 (using independent sets of guide RNAs), we identified MYC as a target gene. The use of two different sets of guide RNAs alleviates concerns that the effects on gene expression are due to specific off target effects of the guide RNAs targeting E7. We also note that the concomitant changes in expression and H3K27Ac patterns (two completely independent methodologies, performed on different days with different replicates of cells) support the conclusion that the genes identified as deregulated were not identified simply due to the method of RNA analysis.
Figure 7.
Circos plots for top downregulated genes. The circos plots show the chromosomal location of the top 25 downregulated genes (>2-fold) for each enhancer deletion (green for protein-coding genes and orange for non-coding genes, with the thickness of the line indicating the extent of the fold-change; for 18qE and 18qNE only three downregulated genes were identified and for 18qNE these downregulated genes were decreased <2-fold.
It is likely that most of the altered (indirect) gene regulation in the E7-deleted cells is initiated by reduction of MYC RNA, followed by changes in the MYC regulatory network. Using Ingenuity Pathway Analysis, we found that 61 of the downregulated genes are known components of the MYC network (Additional File 9); other genes showing altered expression may be downstream targets of the MYC-regulated genes. Deregulated expression of MYC expression is a hallmark feature of many types of cancer and frequently predicts a poor patient outcome (35,49). Accordingly, there is a strong push to develop therapeutic inhibitors of MYC function (36). Perhaps inactivation of cell type-specific enhancers that regulate MYC expression is an alternate to developing chemotherapeutic agents that target MYC itself. Interestingly, we have shown changes in cell culture responses upon deletion of E7 in both HCT116 and in HEK293 cells; the observed responses could be due to decreased cell proliferation, increased cell death or a combination of both. We note that Sur et al. deleted a region homologous to E7 in the mouse genome and observed slight changes in Myc expression in the colon, but a significant change in tumor formation in the colons of Apcmin mice (50). In addition, we have observed that E7-deleted cells are very difficult to trypsinize from the cell culture plates (data not shown). Others have previously reported that MYC is involved in suppressing cell adhesion molecules (51) and we find that many integrin molecules are significantly upregulated at least 2-fold in E7-deleted cells (e.g. ITGA3, ITGA4, ITGA5, and ITGA6 in HCT116 and ITGA3, ITGA7, and ITGAV in HEK293). These molecules are not upregulated in the other deleted cells. We note that both MYC and PVT1 RNAs are downregulated in HCT116 cells after deletion of E7. Tseng et al. (52) showed that PVT1 RNA levels and MYC protein expression correlate in primary human tumors and, in HCT116 cells, loss of PVT1 results in reduced MYC protein, reduced proliferation and impaired colony formation in soft agar. Therefore, the reduction in PVT1 RNA that occurs upon deletion of E7 may cause an even larger effect on MYC protein levels, and more effects on the MYC regulatory network, than would be expected by the changes in MYC RNA alone.
In conclusion, we have used the CRISPR/Cas9 technology to delete an enhancer that harbors a SNP associated with an increased risk for CRC. We show that loss of this enhancer causes changes in expression of hundreds of genes. However, we also show that the size of the H3K27Ac peak does not necessarily correlate with the effect of a distal element on gene regulation. Our results are supported by a recent study in which 2000 predicted enhancers were analyzed for activity in a reporter assay. The investigators found that enhancer fragments having ‘weaker’ H3K27Ac signals can drive expression as well as, if not better than, enhancer fragments having ‘stronger’ H3K27ac signals (53). Recent studies have shown that deleting or inverting a CTCF site can affect loop formation and cause gene expression changes (54,55). In the case of the E7 enhancer, it is possible that the effects on gene expression that we observed are due to changes in the 3D architecture of the chromosome due to removal of a CTCF site involved in looping; there is robust CTCF binding at the E7 enhancer in parental HCT116 and HEK293 cells. One could imagine that altering the 3D chromosomal domain in which a gene resides could have more dramatic effects on gene expression than removal of a single H3K27Ac-marked enhancer (such as 18qE) that is not robustly bound by CTCF. Clearly, further experiments are required before the activity of distal regulatory elements can be accurately predicted.
ACCESSION NUMBER
Supplementary Material
Acknowledgments
The H3K4me3 HCT116 ChIP-seq data (ENCODE accession number ENCSR000DTQ), the H3K4me3 HEK293 ChIP-seq data (ENCODE accession number ENCSR000DTU), and the CTCF HCT116 ChIP-seq data (ENCODE accession number ENCSR000DTW) were produced by the Stamatoyannopoulos laboratory; the H3K27Ac HCT116 ChIP-seq data (ENCODE accession number ENCSR000EUT), the H3K27Ac HEK293 ChIP-seq data (ENCODE accession number ENCSR000FCH), the TCF7L2 HCT116 ChIP-seq data (ENCODE accession number ENCSR000EUV), and the TCF7L2 HEK293 ChIP-seq data (ENCODE accession number ENCSR000EUY) were produced by the Farnham laboratory; and the CTCF HEK293 ChIP-seq data (ENCODE accession number ENCSR000BSE) was produced by the Myers laboratory and is available at https://www.encodeproject.org/. We thank the USC/Norris Cancer Center Next Generation Sequencing Facility for services rendered for production of the RNA-seq and ChIP-seq data, the USC Flow Cytometry Facility for assistance with sorting GFP positive cells, Vicky Yamamoto at USC for advice about cell proliferation assays, Kim Siegmund at USC for discussions about RNA-seq analysis, Meng Li and Yibu Chen from the USC Norris Medical Library Bioinformatics Service for assisting with sequencing data analysis (the bioinformatics software and computing resources used in the analysis are funded by the USC Office of Research and the Norris Medical Library) and members of the Segal and Farnham laboratory for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [R21HG006761, P30CA014089, R01CA136924, T32 HL086350 to M.S.B., in part]. Funding for open access charge: NIH [R01CA136924].
Conflict of interest statement. None declared.
REFERENCES
- 1.Yao L., Tak Y.G., Berman B.P., Farnham P.J. Functional annotation of colon cancer risk SNPs. Nat. Commun. 2014;5:5114. doi: 10.1038/ncomms6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Panne D. The enhanceosome. Curr. Opin. Struct. Biol. 2008;18:236–242. doi: 10.1016/j.sbi.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Maniatis T., Falvo J.V., Kim T.H., Lin C.H., Parekh B.S., Wathelet M.G. Structure and function of the interferon-B enhanceosome. Cold Spring Harb. Symp. Quant. Biol. 1998;63:609–620. doi: 10.1101/sqb.1998.63.609. [DOI] [PubMed] [Google Scholar]
- 4.ENCODE_Project_Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.RoadmapEpigenomicsConsortium. Integrative analysis of 111 reference human epigenomes. Nature. 2015;19:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corradin O., Scacheri P.C. Enhancer variants: evaluating functions in common disease. Genome Med. 2014;6:85. doi: 10.1186/s13073-014-0085-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blahnik K.R., Dou L., Echipare L., Iyengar S., O'Geen H., Sanchez E., Zhao Y., Marra M.A., Hirst M., Costello J.F., et al. Characterization of the contradictory chromatin signatures at the 3′ exons of zinc finger genes. PLoS One. 2011;6:e17121. doi: 10.1371/journal.pone.0017121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blahnik K.R., Dou L., O'Geen H., McPhillips T., Xu X., Cao A.R., Iyengar S., Nicolet C.M., Ludaescher B., Korf I., et al. Sole-search: an integrated analysis program for peak detection and functional annotation using ChIP-seq data. Nucleic Acids Res. 2010;38:e13. doi: 10.1093/nar/gkp1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Geen H., Echipare L., Farnham P.J. Using ChIP-seq technology to generate high-resolution profiles of histone modifications. Methods Mol. Biol. 2011;791:265–286. doi: 10.1007/978-1-61779-316-5_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y., Lin Y.H., Johnson T.D., Rozek L.S., Sartor M.A. PePr: a peak-calling prioritization pipeline to identify consistent or differential peaks from replicated ChIP-Seq data. Bioinformatics. 2014;30:2568–2575. doi: 10.1093/bioinformatics/btu372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frietze S., Wang R., Yao L., Tak Y.G., Ye Z., Gaddis M., Witt H., Farnham P.J., Jin V.X. Cell type-specific binding patterns reveal that TCF7L2 can be tethered to the genome by association with GATA3. Genome Biol. 2012;13:R52. doi: 10.1186/gb-2012-13-9-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blattler A., Yao L., Witt H., Guo Y., Nicolet C.M., Berman B.P., Farnham P.J. Global loss of DNA methylation uncovers intronic enhancers in genes showing expression changes. Genome Biol. 2014;15:469. doi: 10.1186/s13059-014-0469-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schork A.J., Thompson W.K., Pham P., Torkamani A., Roddey J.C., Sullivan P.F., Kelsoe J.R., O'Donovan M.C., Furberg H., Schork N.J., et al. All SNPs are not created equal: genome-wide association studies reveal a consistent pattern of enrichment among functionally annotated SNPs. PLoS Genet. 2013;9:e1003449. doi: 10.1371/journal.pgen.1003449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma G., Gu D., Lv C., Chu H., Xu Z., Tong N., Wang M., Tang C., Xu Y., Zhang Z., et al. Genetic variant in 8q24 is associated with prognosis for gastric cancer in a Chinese population. J. Gastroenterol. Hepatol. 2015;30:689–695. doi: 10.1111/jgh.12801. [DOI] [PubMed] [Google Scholar]
- 19.Sagai T., Hosoya M., Mizushina Y., Tamura M., Shiroishi T. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development. 2005;132:797–803. doi: 10.1242/dev.01613. [DOI] [PubMed] [Google Scholar]
- 20.Zhou H.Y., Katsman Y., Dhaliwal N.K., Davidson S., Macpherson N.N., Sakthidevi M., Collura F., Mitchell J.A. A Sox2 distal enhancer cluster regulates embryonic stem cell differentiation potential. Genes Dev. 2014;28:2699–2711. doi: 10.1101/gad.248526.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanyal A., Lajoie B.R., Jain G., Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farnham P.J. Insights from genomic profiling of transcription factors. Nat. Rev. Genet. 2009;10:605–616. doi: 10.1038/nrg2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li G., Ruan X., Auerbach R.K., Sandhu K.S., Zheng M., Wang P., Poh H.M., Goh Y., Lim J., Zhang J., et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yao L., Berman B.P., Farnham P.J. Demystifying the secret mission of enhancers: linking distal regulatory elements to target genes. Crit. Rev. Biochem. Mol. Biol. 2015;50:550–573. doi: 10.3109/10409238.2015.1087961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segditsas S., Tomlinson I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene. 2006;25:7531–7537. doi: 10.1038/sj.onc.1210059. [DOI] [PubMed] [Google Scholar]
- 26.Bright-Thomas R.M., Hargest R. APC, beta-Catenin and hTCF-4; an unholy trinity in the genesis of colorectal cancer. Eur. J. Surg. Oncol. 2003;29:107–117. doi: 10.1053/ejso.2002.1331. [DOI] [PubMed] [Google Scholar]
- 27.Gaddis M., Gerrard D., Frietze S., Farnham P.J. Altering cancer transcriptomes using epigenomic inhibitors. Epigenet. Chromatin. 2015;8:9. doi: 10.1186/1756-8935-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clevers H. Wnt breakers in colon cancer. Cancer Cell. 2004;5:5–6. doi: 10.1016/s1535-6108(03)00339-8. [DOI] [PubMed] [Google Scholar]
- 29.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 30.Holwerda S.J., de Laat W. CTCF: the protein, the binding partners, the binding sites and their chromatin loops. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2013;368:20120369. doi: 10.1098/rstb.2012.0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Narendra V., Rocha P.P., An D., Raviram R., Skok J.A., Mazzoni E.O., Reinberg D. Transcription. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science. 2015;347:1017–1021. doi: 10.1126/science.1262088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ong C.T., Corces V.G. CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet. 2014;15:234–246. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Splinter E., Heath H., Kooren J., Palstra R.-J., Klous P., Grosveld F., Galjart N., de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vietri Rudan M., Barrington C., Henderson S., Ernst C., Odom D.T., Tanay A., Hadjur S. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 2015;10:1297–1309. doi: 10.1016/j.celrep.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gabay M., Li Y., Felsher D.W. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb. Perspect. Med. 2014;4:a014241. doi: 10.1101/cshperspect.a014241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKeown M.R., Bradner J.E. Therapeutic strategies to inhibit MYC. Cold Spring Harb. Perspect. Med. 2014;4:a014266. doi: 10.1101/cshperspect.a014266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diolaiti D., McFerrin L., Carroll P.A., Eisenman R.N. Functional interactions among members of the MAX and MLX transcriptional network during oncogenesis. Biochim. Biophys. Acta. 2014:S1874–S9399. doi: 10.1016/j.bbagrm.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Freedman M.L., Monteiro A.N., Gayther S.A., Coetzee G.A., Risch A., Plass C., Casey G., De Biasi M., Carlson C., Duggan D., et al. Principles for the post-GWAS functional characterization of cancer risk loci. Nat. Genet. 2011;43:513–518. doi: 10.1038/ng.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coetzee G.A., Jia L., Frenkel B., Henderson B.E., Tanay A., Haiman C.A., Freedman M.L. A systematic approach to understand the functional consequences of non-protein coding risk regions. Cell Cycle. 2010;9:256–259. doi: 10.4161/cc.9.2.10419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin F., Li Y., Dixon J.R., Selvaraj S., Ye Z., Lee A.Y., Yen C.A., Schmitt A.D., Espinoza C.A., Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pomerantz M.M., Ahmadiyeh N., Jia L., Herman P., Verzi M.P., Doddapaneni H., Beckwith C.A., Chan J.A., Hills A., Davis M., et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat. Genet. 2009;41:882–884. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wright J.B., Brown S.J., Cole M.D. Upregulation of c-MYC in cis through a large chromatin loop linked to a cancer risk-associated single-nucleotide polymorphism in colorectal cancer cells. Mol. Cell. Biol. 2010;30:1411–1420. doi: 10.1128/MCB.01384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahmadiyeh N., Pomerantz M.M., Grisanzio C., Herman P., Jia L., Almendro V., He H.H., Brown M., Liu X.S., Davis M., et al. 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proc. Natl. Acad. Sci. U.S.A. 2010;107:9742–9746. doi: 10.1073/pnas.0910668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jager R., Migliorini G., Henrion M., Kandaswamy R., Speedy H.E., Heindl A., Whiffin N., Carnicer M.J., Broome L., Dryden N., et al. Capture Hi-C identifies the chromatin interactome of colorectal cancer risk loci. Nat. Commun. 2015;6:6178. doi: 10.1038/ncomms7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du M., Yuan T., Schilter K.F., Dittmar R.L., Mackinnon A., Huang X., Tschannen M., Worthey E., Jacob H., Xia S., et al. Prostate cancer risk locus at 8q24 as a regulatory hub by physical interactions with multiple genomic loci across the genome. Hum. Mol. Genet. 2015;24:154–166. doi: 10.1093/hmg/ddu426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dryden N.H., Broome L.R., Dudbridge F., Johnson N., Orr N., Schoenfelder S., Nagano T., Andrews S., Wingett S., Kozarewa I., et al. Unbiased analysis of potential targets of breast cancer susceptibility loci by Capture Hi-C. Genome Res. 2014;24:1854–1868. doi: 10.1101/gr.175034.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiang J.F., Yin Q.F., Chen T., Zhang Y., Zhang X.O., Wu Z., Zhang S., Wang H.B., Ge J., Lu X., et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014;24:513–531. doi: 10.1038/cr.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Williamson I., Berlivet S., Eskeland R., Boyle S., Illingworth R.S., Paquette D., Dostie J., Bickmore W.A. Spatial genome organization: contrasting views from chromosome conformation capture and fluorescence in situ hybridization. Genes Dev. 2014;28:2778–2791. doi: 10.1101/gad.251694.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Conacci-Sorrell M., McFerrin L., Eisenman R.N. An overview of MYC and its interactome. Cold Spring Harb. Perspect. Med. 2014;4:a014357. doi: 10.1101/cshperspect.a014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sur I.K., Hallikas O., Vaharautio A., Yan J., Turunen M., Enge M., Taipale M., Karhu A., Aaltonen L.A., Taipale J. Mice lacking a Myc enhancer that includes human SNP rs6983267 are resistant to intestinal tumors. Science. 2012;338:1360–1363. doi: 10.1126/science.1228606. [DOI] [PubMed] [Google Scholar]
- 51.Grandori C., Cowley S.M., James L.P., Eisenman R.N. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 52.Tseng Y.Y., Moriarity B.S., Gong W., Akiyama R., Tiwari A., Kawakami H., Ronning P., Reuland B., Guenther K., Beadnell T.C., et al. PVT1 dependence in cancer with MYC copy-number increase. Nature. 2014;512:82–86. doi: 10.1038/nature13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kwasnieski J.C., Fiore C., Chaudhari H.G., Cohen B.A. High-throughput functional testing of ENCODE segmentation predictions. Genome Res. 2014;24:1595–1602. doi: 10.1101/gr.173518.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanborn A.L., Rao S.S., Huang S.C., Durand N.C., Huntley M.H., Jewett A.I., Bochkov I.D., Chinnappan D., Cutkosky A., Li J., et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E6456–E6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Wit E., Vos E., Holwerda S.J., Valdes-Quezada C., Verstegen M.J., Teunissen H., Splinter E., Wijchers P.J., Kriger P.H., de Laat W. CTCF binding polarity determines chromatin looping. Mol. Cell. 2015;60:676–684. doi: 10.1016/j.molcel.2015.09.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.