Abstract
In budding yeast, Set2 catalyzes di- and trimethylation of H3K36 (H3K36me2 and H3K36me3) via an interaction between its Set2–Rpb1 interaction (SRI) domain and C-terminal repeats of RNA polymerase II (Pol2) phosphorylated at Ser2 and Ser5 (CTD-S2,5-P). H3K36me2 is sufficient for recruitment of the Rpd3S histone deacetylase complex to repress cryptic transcription from transcribed regions. In fission yeast, Set2 is also responsible for H3K36 methylation, which represses a subset of RNAs including heterochromatic and subtelomeric RNAs, at least in part via recruitment of Clr6 complex II, a homolog of Rpd3S. Here, we show that CTD-S2P-dependent interaction of fission yeast Set2 with Pol2 via the SRI domain is required for formation of H3K36me3, but not H3K36me2. H3K36me3 silenced heterochromatic and subtelomeric transcripts mainly through post-transcriptional and transcriptional mechanisms, respectively, whereas H3K36me2 was not enough for silencing. Clr6 complex II appeared not to be responsible for heterochromatic silencing by H3K36me3. Our results demonstrate that H3K36 methylation has multiple outputs in fission yeast; these findings provide insights into the distinct roles of H3K36 methylation in metazoans, which have different enzymes for synthesis of H3K36me1/2 and H3K36me3.
INTRODUCTION
Chromatin structure is dynamically regulated and plays important roles in various genome functions including transcription, DNA replication, DNA repair and genome integrity. Several layers of mechanisms regulate chromatin structure; among them, histone modifications determine the basic landscape of chromatin structure by recruiting specific ‘readers,’ which recognize specific patterns of histone modifications and recruit other regulators such as chromatin-remodeling factors and other histone modifiers (1). Some specific histone methylations determine basic features of chromatin: for example, methylation of histone H3 at Lys9 (H3K9) is a mark of inactive chromatin structure. Sometimes the number of methyl groups attached to a lysine residue defines a histone modification's distinctive roles. In mammals, trimethylation of H3K9 (H3K9me3) is primarily observed in constitutive heterochromatin, such as pericentromeric heterochromatin, whereas dimethylation of H3K9 (H3K9me2) is primarily observed at silent regions in euchromatin. Importantly, different methyltransferases are responsible for generation of H3K9me3 and H3K9me2, which are synthesized by Suv39 H1/2 and G9a, respectively; this is consistent with the distinct roles of H3K9me2 and H3K9me3 (2,3).
In many organisms, methylation of H3K36 is observed in euchromatin; this modification primarily accumulates at transcribed regions, where it plays a role in transcriptional elongation (4). In addition, the function of H3K36 methylation was implicated in repression of transcription, alternative splicing, dosage compensation, DNA replication and repair (5).
In budding yeast, all states of H3K36 methylation (mono-, di- and trimethylation) accumulate in transcribed regions, and the level of the H3K36me3 is well-correlated with the level of transcription, suggesting that a co-transcriptional mechanism underlies H3K36 methylation (6–8). Indeed, Set2, the sole enzyme responsible for generation of all forms of H3K36me in budding yeast, associates with the C-terminal domain (CTD) of the largest subunit (Rpb1) of RNA polymerase II (Pol2) through its C-terminal Set2–Rpb1 interaction (SRI) domain (9). Loss of the SRI domain causes loss of both H3K36me2 and H3K36me3 (10), consistent with co-transcriptional H3K36 methylation. The Pol2 CTD consists of repeats of heptapeptides (Tyr1-Ser2-Pro3-Th4r-Ser5-Pro6-Ser7), which are well-conserved from yeast to mammals. The CTD functions as a platform with which various factors associate during transcription. These associations are primarily regulated by the co-transcriptional and differential phosphorylation of Ser, Thr and Tyr, and they regulate transcriptional elongation as well as RNA processing (11–13). Importantly, the SRI domain of Set2 preferentially associates with CTD phosphorylated at Ser2 and Ser5 (CTD-S2,5-P) (9,14). CTD-S5 is phosphorylated at the initial step of transcription and gradually decreases toward the 3′ ends of genes, whereas CTD-S2 is phosphorylated during the elongation step, and the phosphorylated state is maintained until the termination stage (15,16). As a result, the various H3K36 methylation states are distributed over the transcribed region in distinct patterns, with H3K36me1 primarily at the 5′ end, H3K36me2 ranging from the 5′ end to the early coding region and H3K36me3 primarily near the 3′ end (6). In addition to CTD phosphorylation, factors involved in transcription elongation, including Paf1 and histone chaperones, such as FACT, Asf1 and Spt6/Iws1, are also involved in H3K36 methylation (10,17–19).
Detailed analyses of the function of H3K36 methylation in budding yeast revealed that it plays a role in repressing cryptic transcription initiation in coding regions. Rpd3S histone deacetylase complex is first recruited to the coding region through its interaction with CTD-S2,5P of Pol2 (20,21). Next, Eaf3, a component of Rpd3S that contains a chromodomain that binds to H3K36me2 and H3K36me3, recruits the Rpd3S complex onto chromatin to promote deacetylation of histones (22–24). Because of the distribution of H3K36 methylation within genes, deacetylation occurs mainly in coding regions, thereby repressing initiation of both sense and antisense cryptic transcription in these regions (22–24). Importantly, H3K36me2 is sufficient for recruitment of Rpd3S and repression of cryptic transcription (25). However, other factors, such as FACT, Asf1 and Spt6, specifically contribute to H3K36me3; loss of these factors results in the loss of H3K36me3 but not H3K36me2 (10,17,18). Mammals possess multiple H3K36-specific methyltransferases; some of these enzymes exhibit mono/dimethylation activity, whereas others have trimethylation activity (5). These facts suggest that mono/dimethylation and trimethylation play distinct roles, although the exact output of each methylation state remains unclear.
The fission yeast Schizosaccharomyces pombe provides a useful model system for investigating chromatin regulation, especially heterochromatin, because in this species, heterochromatin is not essential for cell viability, and its basic features closely resemble that of metazoans. Heterochromatin is localized at centromeres, telomeres and the mating-type locus, and plays important roles at these loci (26). Centromeric heterochromatin is formed at repeated sequences known as dg and dh elements, and its formation largely depends on the RNA interference (RNAi) machinery and siRNA produced from non-coding RNAs transcribed from these repeats (27). In fission yeast, Set2 is the sole enzyme responsible for H3K36 methylation, and both Set2 and H3K36 methylation associate with transcribed regions (28,29). Moreover, deletion of set2 causes accumulation of heterochromatic non-coding RNAs, suggesting the involvement of H3K36 methylation in heterochromatic silencing (30,31). Clr6 complex II is a counterpart of budding yeast Rpd3S that contains a chromodomain protein, Alp13, which is a homolog of Eaf3 (30,32). Phenotypes of alp13 deletion resemble those of set2 deletion (30). Furthermore, H3K36me3 as well as Alp13 accumulates at heterochromatin during G1/S and early S phase, when heterochromatic transcripts are transcribed (31). These results suggest that the mechanism of silencing by Set2 is similar to that observed in repression of cryptic transcription in budding yeast; co-transcriptional methylation of H3K36me by Set2 recruits Clr6 complex II containing Alp13 (30,31). At euchromatin, deletion of set2 or alp13, causes accumulation of antisense RNAs (29,30,33), suggesting that co-transcriptional recruitment of HDAC complex by Set2-generated H3K36 methylation is also responsible for antisense repression. However, the cumulative effect of deletion of set2 and alp13 on de-repression of antisense transcript (30) and the smaller increase in histone acetylation in set2-deficient cells relative to alp13-deficient cells (33) suggest that mechanisms other than the Set2–H3K36me–HDAC pathway are responsible for the observed repression of antisense transcripts, although the details of such mechanisms remain unknown.
Set2 and H3K36 methylation have not been thoroughly analyzed in fission yeast. For example, a previous report pointed out that fission yeast Set2 has homology to the budding yeast SRI domain in its C-terminal region (9), but those authors did not investigate the interaction of Set2 with Pol2 or the function of the SRI domain. To address this issue, we generated Set2 mutants in which the SRI domain was deleted, and used these mutants to analyze the domain's role. We found that fission yeast Set2 interacted with Pol2 via the SRI domain, and that this interaction required CTD-S2P. Surprisingly, loss of SRI or CTD-S2P caused loss of H3K36me3, but H3K36me2 was retained, indicating that H3K36me2 is methylated via a different mechanism. The genomic distributions of Set2, Set2ΔSRI, H3K36me2 and H3K36me3 suggested that H3K36me2 is also methylated via a co-transcriptional mechanism. Transcriptome analysis revealed that Set2-generated H3K36me3 repressed heterochromatic non-coding RNAs, antisense RNAs and subsets of genes located at subtelomeric regions. Strikingly, analyses using set2ΔSRI revealed that H3K36me3, but not H3K36me2, was responsible for this repression. Further analysis indicated that heterochromatic non-coding RNA might be silenced mainly via a post-transcriptional mechanism involving the nuclear exosome, an RNA-degradation complex, whereas subtelomeric genes were primarily silenced via a transcriptional mechanism in which Clr6/Alp13 plays a minor role. These results reveal the mechanisms of Set2-generated co-transcriptional H3K36 methylation and the resultant gene silencing in fission yeast.
MATERIALS AND METHODS
Strains and media
The S. pombe strains used in this study are listed in Supplementary Table S1. Standard procedures (34) were used for the culture of yeast cells. Deletion and tagging of target genes were performed using the polymerase chain reaction (PCR)-based module method (35).
Antibodies
Mouse monoclonal antibodies against the following histone modifications were used: H3K9me2 (m5.1.1) (36), H3K36me2, H3K36me3, H3K9Ac and H3K14Ac (37,38). Mouse monoclonal antibodies against epitope tags, i.e. Myc (4A6, Millipore) and FLAG (M2, Sigma), were used in the ChIP assay and immunoblotting. Mouse monoclonal antibody against Pol2 CTD phosphorylated at Ser5 (4H8, Abcam), rabbit polyclonal antibody against the N-terminal half of Rpb1 (39) and rabbit polyclonal antibody against histone H3 C-terminal (Millipore) were used for immunoblotting.
Co-immunoprecipitation and western blotting
Cells (5 × 108) in YES were harvested and resuspended in 0.3-Buffer T (25 mM HEPES-KOH (pH 7.5), 0.1 mM ethylenediaminetetraacetic acid (EDTA), 10% glycerol, 0.3 M KCl and 0.1% Tween 20) containing 1 mM phenylmethylsulfonyl fluoride and protease inhibitor cocktail (0.35 μg/ml benzamidine, 0.7 μg/ml pepstatin, and 0.5 μg/ml leupeptin). The cells were homogenized with glass beads in a bead shocker (Yasui Kikai) seven times for 60 s at 4°C. The cell extract was centrifuged for 10 min at 15,000 rpm at 4°C. The resultant supernatant was incubated with Dynabeads Pan Mouse IgG (Invitrogen) for 2 h. After incubation, the beads were washed six times with 0.3-Buffer T. The beads were resuspended in loading buffer. Before immunoprecipitation, 50 μl of Dynabeads Pan Mouse IgG (Invitrogen) were equilibrated overnight in 5 ml of phosphate-buffered saline (PBS) containing 1 mg/ml of bovine serum albumin (BSA). The beads were incubated with an antibody for several hours at 4°C.
The immunoprecipitated and input samples were separated by polyacrylamide gel electrophoresis, and the proteins were blotted onto nitrocellulose membranes. The membranes were first probed with primary antibodies, and then incubated with horseradish peroxidase-conjugated anti-mouse IgG or anti-rabbit IgG (GE Healthcare Life Science).
ChIP assay
Cells (5 × 108) in 50 ml of YES were fixed with 1% formaldehyde (Nacalai Tesque) for 30 min at 30°C. Quenching of the fixative was performed with 150 mM glycine. The cells were harvested and resuspended in Buffer 1 (50 mM HEPES [pH 7.5], 140 mM NaCl, 1 mM EDTA, 1% Triton X-100 and 0.1% Na-deoxycholate) containing a protease inhibitor cocktail, and then homogenized with a bead shocker (Yasui Kikai) 30× for 60 s at 4°C. The cell extracts were centrifuged for 60 min at 15 000 rpm at 4°C. After discarding the supernatant, the pellets were resuspended in Buffer 1 containing a protease inhibitor cocktail and sonicated for 600 s with a Bioruptor (CosmoBio) set at level ‘H’. After sonication, the cell extract was centrifuged for 10 min at 15 000 rpm at 4°C, and the resultant supernatant was used for the input fraction. Before use, 50 μl of Dynabeads Pan Mouse IgG (Invitrogen) were equilibrated overnight in 5 ml of PBS containing 1 mg/ml of BSA. The beads were incubated with antibodies for several hours at 4°C. After washing with Buffer 1, the beads were incubated for 2 h with cell extract at 4°C. After immunoprecipitation, the beads were washed twice each with Buffer 1, Buffer 1′ (50 mM HEPES [pH 7.5], 500 mM NaCl, 1 mM EDTA, 1% TritonX-100 and 0.1% Na-deoxycholate) and Buffer 2 (10 mM Tris (pH 8.0), 250 mM LiCl, 0.5% NP-40, and 0.5% Na-deoxycholate). The beads were resuspended in TE buffer containing RNase A and incubated for 10 min at 37°C. Next, Proteinase K (0.5 μg/ml) was added, and the mixture was incubated for a further 1 h at 37°C. After reverse crosslinking at 65°C, DNA was purified using a PCR purification kit (Nippon Gene) and used for quantitative PCR. The primers used for quantitative PCR are listed in Supplementary Table S2.
RNA extraction and RT-PCR
Each strain was cultured in 25 ml of liquid YES to 5 × 106 cells/ml. The cells were harvested by centrifugation and washed with 10 ml of PBS. The cell pellet was suspended in buffer containing 1% sodium dodecyl sulfate (SDS) and acid phenol. Total RNA was extracted using a freeze-thaw treatment: rapid freezing in liquid N2 followed by an hour incubation in a water bath at 65°C. Next, 1 μg of total RNA was treated with 25 U recombinant DNase I (Takara Bio) at 37°C. The cDNAs were reverse transcribed using 200 U PrimeScript Reverse Transcriptase (Takara Bio) at 42°C with 3 pmol of target-specific reverse primers. The primer sets used in this study are listed in Supplementary Table S2.
siRNA detection
Small RNA was purified using the mirVana miRNA Isolation Kit (Ambion) from 5 × 108 cells. After separation in poly-acrylamide gel containing 15% urea, samples were blotted onto a Hybond-N membrane (Amersham Biosciences) using a Trans-Blot SD semi-dry electrophoretic transfer cell (Bio-Rad) and UV-crosslinked to the membrane. Oligonucleotide probes corresponding to siRNAs derived from dg and dh repeats (40) or tRNAAsn were labeled with [γ32P]dATP using T4 Polynucleotide Kinase (TOYOBO). After hybridization for 24 h, membranes were washed two times with 2× SSC/0.1% SDS for 10 min at 42°C. An imaging plate was exposed for 1–2 days.
Quantitative PCR
Quantitative PCR was performed with PCR cocktails containing SYBR Dye on a Thermal Cycler Dice Real Time System (Takara Bio). The primer sets used for quantitative PCR are listed in Supplementary Table S2. In each graph showing the results of qRT-PCR, error bars represent the standard error of the mean of biologically independent experiments (n = 3).
Microarray analysis
The microarray analysis was performed as described previously (41). Geometric means and expression ratios were calculated. The sequences of the probes and the original data from the microarray experiments were deposited in GEO (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE66821. Based on the expression ratio of each mutant relative to the wild-type strain, genes with a log2 fold change >1.5 (upregulated) or <1.5 (downregulated) were extracted. The hypergeometric distribution was calculated to evaluate the significant upregulation of RNAs in each mutant. Venn diagrams were produced in the R statistical environment (http://www.R-project.org) as previously described (42). The P-values of overlap between gene sets were calculated using Fisher's exact test.
ChIP-sequencing
Cells were cultured in 600 ml of YES to a density of 1 × 107 cells/ml. The following procedure is identical to the ChIP assay. Samples destined for ChIP-seq by Illumina platform were prepared according to the manufacturer's instructions. The sequencing data were deposited in DDBJ (http://www.ddbj.nig.ac.jp) under accession number DRA003220.
Sequence reads were processed using BWA (43), SAMtools (44) and MACS (45), and further analyzed in the R statistical environment (http://www.R-project.org) as previously described (42).
Purification of Set2-FLAG and analysis by LC-MS/MS mass spectrometry
Whole-cell extract preparation and immunopurification of Set2-FLAG was performed as described (46). Mass spectrometry was performed as previously described (47). Briefly, purified Set2 complex was separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis. After trypsinization of the sliced gel, the protein was analyzed by liquid chromatography/tandem mass spectrometry. The raw data files were analyzed using Mascot (Matrix Science) and queried against the S. pombe protein database.
RESULTS
The fission yeast Set2 SRI domain is necessary for the interaction with Pol2 phosphorylated at Ser2 of the CTD
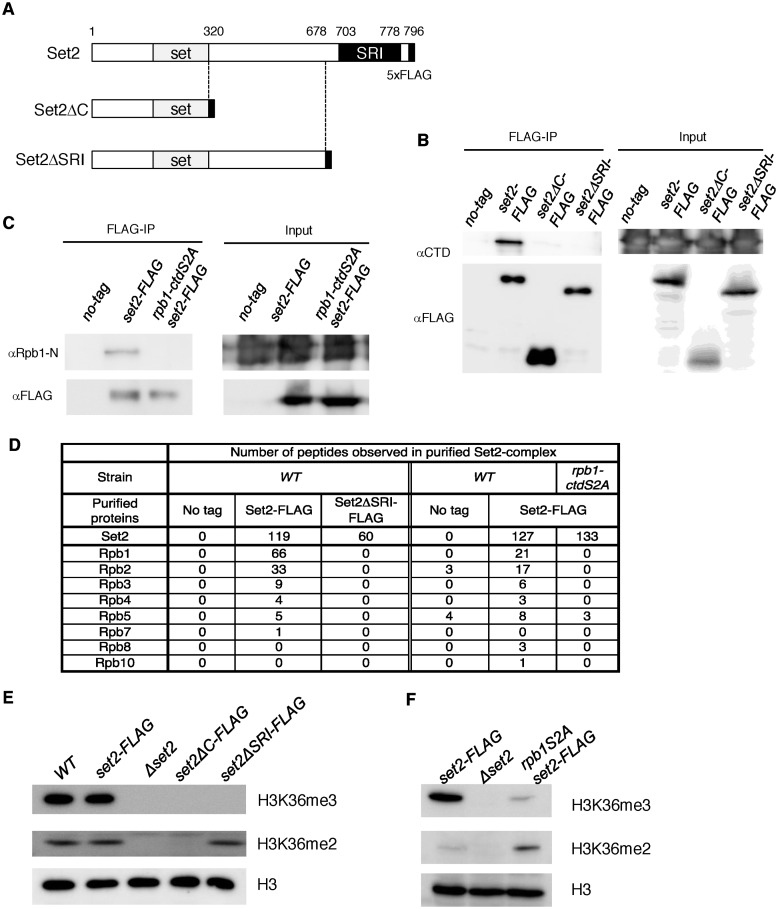
A previous report showed that the SRI (Set2–Rpb1-interacting) domain in the C-terminal region of budding yeast Set2 interacts with the Pol2 CTD, which is required for H3K36me2 and H3K36me3 (9,10). The fission yeast Set2 has a domain whose amino acids sequence is conserved with the SRI domain (17% identity, 42% similarity, (9)), but it remains unclear whether the domain of Set2 in fission yeast is required for interaction with the CTD and H3K36 methylation. To investigate this, we generated two types of C-terminal deletion mutants of set2 and introduced them into the endogenous set2 gene locus. One of the deletion mutants (Set2ΔC-FLAG) contained only the SET domain, which is the catalytic domain involved in H3K36 methylation, whereas the other mutant (set2ΔSRI-FLAG) lacked the SRI domain; both mutants had a C-terminal FLAG tag (Figure 1A). We confirmed that each mutant protein was expressed at levels comparable to that of the C-terminally tagged wild-type protein (Set2-FLAG) (Figure 1B, input). In a co-immunoprecipitation experiment, we investigated whether these mutants interact with Rpb1 (Figure 1B). As expected, Rpb1 co-precipitated with Set2-FLAG, but not with Set2ΔC-FLAG or Set2ΔSRI-FLAG. This result showed that the SRI domain of Set2 in fission yeast is necessary for the interaction with Pol2.
Figure 1.
Set2 interacts with Pol2 through its C-terminal SRI domain in a CTD-S2P–dependent manner, and this interaction is required for the generation of H3K36me3 but not H3K36me2. (A) Schematic diagrams of the C-terminal truncation mutants of Set2 used in this study. The SET and SRI domains are indicated by gray and black boxes, respectively. The black box at the C-terminal end of each mutant indicates the 5×FLAG tag. (B) Set2 interacts with Pol2 through its C-terminal SRI domain. The interaction between Set2 and Pol2 was examined by co-immunoprecipitation. Extracts prepared from the indicated strains were incubated with an antibody against the FLAG tag. Immunoprecipitated fractions were analyzed by immunoblotting using an antibody against CTD of Pol2. Input represents 15% of extracts used for immunoprecipitation. (C) Set2–Pol2 interaction depends on Pol2 CTD-S2P. Set2–Pol2 was examined using extracts prepared from the indicated strains by the co-immunoprecipitation scheme described in (B), except that an antibody against the N-terminal region of Rpb1 (39) was used to detect Rpb1 to avoid possible influences of the rpb1-ctdS2A mutation on antibody recognition. (D) Subunits of RNA polymerase II observed in Set2 complexes analyzed by mass-spectrometry analysis. (E) The SRI domain is essential for H3K36me3 but not for H3K36me2. Whole-cell extracts from the indicated strains were analyzed by immunoblotting using antibodies against H3K36me2, H3K36me3 (37) and the C-terminal region of histone H3. (F) CTD-S2P is required for efficient H3K36me3 but not for H3K36me2. H3K36me2, H3K36me3 and histone H3 in whole cell extracts were analyzed as (E).
In budding yeast, Set2 binds to the CTD phosphorylated at Ser2 and Ser5 (CTD-S2,5-P) (9,14). In fission yeast, the alanine substitution mutant of all Ser2 residues of the CTD (rpb1-ctdS2A) is viable, but rpb1-ctdS5A is lethal (48); therefore, we examined the requirement of CTD-S2 phosphorylation (CTD-S2P) for the Set2–Pol2 association using the rpb1-ctdS2A mutant. This mutation caused a significant reduction in Rpb1 co-immunoprecipitated with Set2 (Figure 1C), revealing that like the budding yeast SRI domain, the fission yeast SRI domain requires CTDS2-P in order to associate with Rpb1.
Analysis of Set2 complex by LC-MS/MS mass spectrometry further validated the interaction between Set2 and CTD-S2P, mediated by the Set2 SRI domain. Specifically, we purified Set2-FLAG and Set2ΔSRI-FLAG from wild-type and rpb1-ctdS2A cells and analyzed the interacting proteins by mass spectrometry. Six of twelve Pol2 subunits could be detected in the wild-type Set2 complex, but no Pol2 subunits were observed in the Set2 complex harboring deletion of the SRI. Similarly, Set2-FLAG purified from the rpb1-ctdS2A mutant did not contain Pol2 subunits except Rpb5, which could be detected in no-tag control (Figure 1D).
Set2 SRI domain is essential for H3K36 trimethylation, but not for dimethylation
We next examined the effects of set2 deletion mutants on histone H3K36 methylation by immunoblotting whole-cell extract with antibodies against H3K36me2 or H3K36me3 (Figure 1E). In wild-type cells, H3K36me3 was the predominant form of H3K36 methylation, and addition of the FLAG tag to C-terminus of Set2 did not affect the level of H3K36 di- or trimethylation. Deletion of set2 caused complete loss of H3K36me2 and H3K36me3, revealing that Set2 is the enzyme responsible for both methylated states of histone H3. In set2ΔC cells, neither H3K36me2 nor H3K36me3 was detectable. Notably, in set2ΔSRI cells, only H3K36 dimethylation was detected; the level was comparable to those in wild-type and set2-FLAG cells. These results revealed that in addition to the catalytic SET domain, the C-terminal region is essential for H3K36me2 and H3K36me3, and that the SRI domain is specifically required for H3K36me3 but not for H3K36me2. By contrast, in budding yeast, both H3K36 di- and trimethylation are lost upon deletion of the SRI domain, but dimethylation can be detected in set2ΔC cells at levels comparable to those in wild-type cells (10).
Because the CTD-S2P is necessary for efficient Rpb1–Set2 interaction (Figure 1C), we next analyzed the effect of rpb1-ctdS2A mutation on H3K36 methylation. In rpb1-ctdS2A cells, the amount of H3K36me3 was significantly reduced, as in the set2ΔSRI mutant, whereas the level of H3K36me2 was slightly elevated (Figure 1F). This result, together with the results obtained with the set2ΔSRI mutant, strongly suggests that Set2 associated with Pol2 CTD-S2P is responsible for trimethylation of histone H3K36 from the dimethylated state. Because the distribution of CTDS5-P is almost the same as that of Pol2 (48), and the budding yeast SRI domain binds to CTD-S2,5-P but not CTD-S2P or CTD-S5P (9,14), the fission yeast SRI domain may bind to CTD-S2,5P. We speculate that the weak or transient interaction between Set2 and Pol2 in the rpb1-ctdS2A mutant was responsible for the residual level of H3K36me3 and the increase in H3K36me2, although the Set1–Pol2 interaction revealed by co-immunoprecipitation experiments was abolished in rpb1-ctdS2A cells (Figure 1C). Alternatively, Set2 might has residual H3K36 tri-methylation activity in this mutant without its recruitment by Pol2.
Genome-wide analysis suggests that both H3K36me2 and H3K36me3 were methylated co-transcriptionally, but via different mechanisms
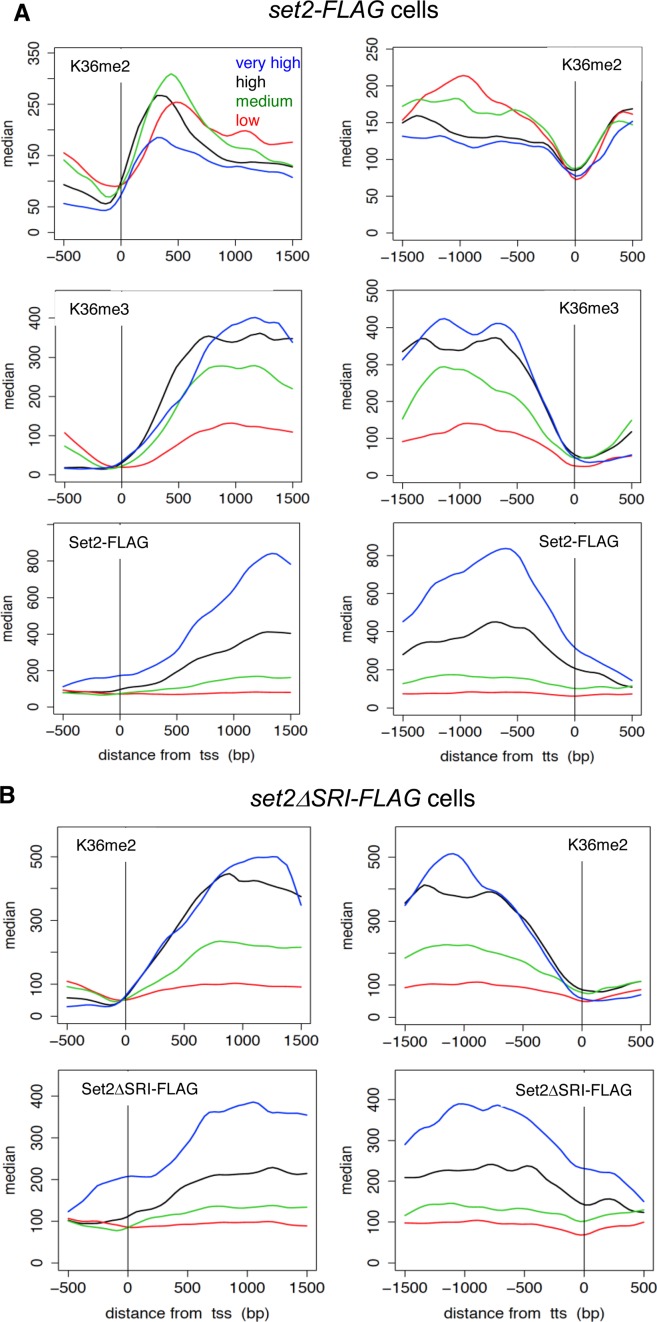
To gain further insight into functions of Set2 and H3K36 methylation in fission yeast, we analyzed genome-wide occupancies of H3K36me2 and H3K36me3, in wild-type and set2ΔSRI cells using chromatin immunoprecipitation-sequencing (ChIP-Seq). In addition, we analyzed the localization of Set2-FLAG and Set2ΔSRI-FLAG. H3K36me2, H3K36me3 and Set2-FLAG primarily localized in regions transcribed by Pol2, consistent with the interaction between Set2 and Pol2 (Figure 2A and B). To obtain clearer insights into the correlation between transcription and Set2-dependent H3K36 methylation, we classified coding genes into four classes (very high, high, middle and low) according to the level of Pol2 as previously reported (42).
Figure 2.
Distributions of methylation of H3K36 and Set2 proteins indicate co-transcriptional methylation by Set2. (A) ChIP-sequencing analysis of H3K36me2, H3K36me3 and Set2-FLAG in wild-type (set2-FLAG) cells. Positions (bp) relative to the transcription start site (TSS) (left panels) and termination site (TTS) (right panels) are shown on the x-axis. The gene sets (very high, high, medium, low) used were the same as those used in the previous study (42), which were classified according to the level of Pol2. The median tag counts of the indicated gene sets, which were transcribed at different levels, are shown on the y-axis in each upper panel. (B) ChIP-sequencing analysis of H3K36me2 and Set2ΔSRI-FLAG in set2ΔSRI-FLAG cells. Positions (bp) relative to the transcription start site (TSS) (left panels) and termination site (TTS) (right panels) are shown on the x-axis. The median tag counts of the indicated gene sets, which were the same as used in (A), are shown on the y-axis in each upper panel.
In wild-type cells, the level of H3K36me2 sharply increased at the 5′ ends of the genes (300–400 bp from transcription start site (TSS)) and gradually decreased through the gene bodies, whereas H3K36me3 was low at the beginning of transcription and accumulated toward the 3′ ends of the gene (Figure 2A and B). Both H3K36me2 and H3K36me3 sharply decreased at the 5′ end of the genes (400–300 to transcription termination site (TTS)). This distribution resembled the occupancy of Set2-FLAG, which gradually increased in the 3′ direction (Figure 2A)(29). Importantly, the distribution of Set2-FLAG was similar to that of Pol2, which was analyzed using antibody recognizing Rpb1-CTD-S5P (42) (Supplementary Figure S1A), at the 3′ region of the genes. In the 5′ region, there was a delay in increase in Set2-FLAG. The delay might represent the preferential binding of Set2 with CTD-S2P that is phosphorylated during elongation. The distribution pattern of H3K36me3 on the genes classified into ‘high’ was also similar to that of Set2-FLAG, while on the ‘very high’ genes, the level of H3K36me3 is low relative to the amount of Set2-FLAG (Supplementary Figure S1A). This may be due to the high turnover rate of histone H3/H4 at the highly transcribed genes and/or H3K36 level was saturated. In contrast, on the ‘medium’ and ‘low’ genes, H3K36me3 highly accumulated relative to Pol2 and Set2-FLAG (Supplementary Figure S1A). This may indicate that H3K36me3 was stably maintained after co-transcriptional methylation at the genes with relatively low expression level because of low turnover rate of histone H3/H4. Together with the Pol2–Set2 interaction, these results indicate that Set2 catalyzes H3K36 trimethylation co-transcriptionally.
In the case of set2ΔSRI cells, the occupancy of H3K36me2 was very similar to that of H3K36me3 in wild-type cells (Figure 2). Similarly, like Set2-FLAG, the occupancy of Set2ΔSRI-FLAG gradually increased in the 3′ direction (Figure 2B) but the level was low compared with that of Set2-FLAG. The decrease of Set2ΔSRI-FLAG was confirmed by ChIP-qPCR experiments (Supplementary Figure S2); the signals of Set2ΔSRI-FLAG were lower than those of Set2-FLAG at each gene chosen from each classes. The distribution pattern of H3K36me2 resembled that of Pol2 rather than Set2ΔSRI-FLAG (Supplementary Figure S1B). These results suggest that Set2ΔSRI-FLAG is recruited to transcribed regions in a transcription-dependent manner to catalyze H3K36 dimethylation. The recruitment might depend on direct or indirect interaction between Pol2 and Set2ΔSRI but the interaction is not strong or stable enough to give a strong signal in ChIP assay and co-immunoprecipitation experiments (Figure 1B). Other mechanisms such as recruitment of Set2ΔSRI to transcribed genes by co-transcriptionally modified histones are also possible.
Taken together, these data suggest that Set2 associates with Pol2 at the beginning of transcription via an as yet unknown binding partner, and promotes formation of H3K36me2; as CTD-S2,S5P accumulates, Set2 changes its binding partner to the CTD and catalyzes formation of H3K36me3.
H3K36me3 but not H3K36me2 repressed certain sets of genes
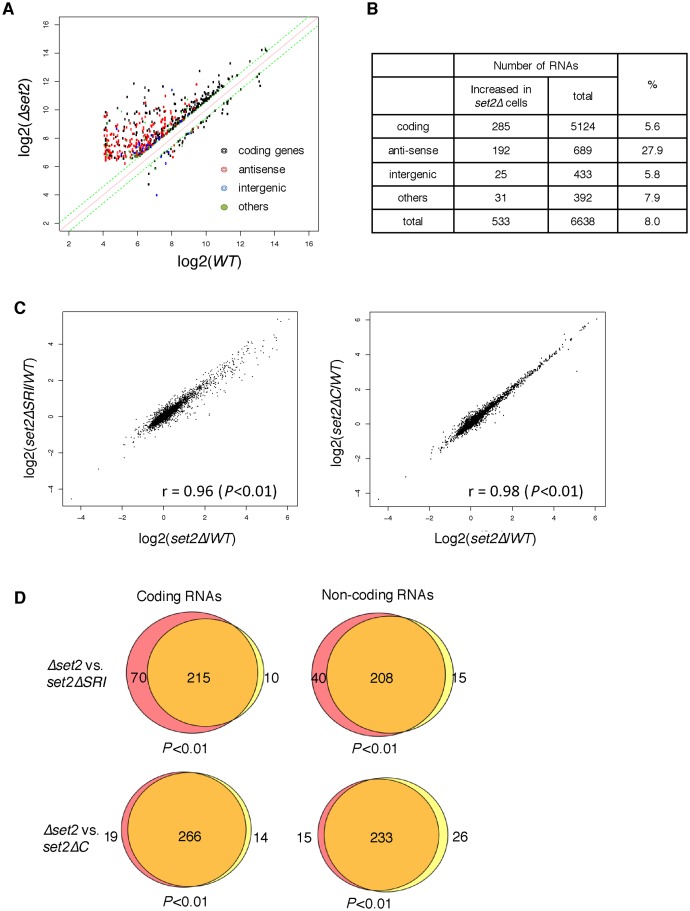
Because the specific absence of H3K36me3 in set2ΔSRI cells enabled us to examine the functional difference between H3K36me2 and H3K36me3, we performed transcriptome analysis using microarrays to compare the influence of set2Δ, set2ΔC and set2ΔSRI on RNA level. Complete loss of Set2 caused upregulation (>1.5-fold) of 533 RNAs and downregulation (<1.5-fold) of 61 RNAs, suggesting that Set2-generated H3K36 methylation primarily functions in downregulation of certain RNAs (Figure 3A). Of the RNAs downregulated by Set2, 248 out of 533 were non-coding, and 192 of those 248 were antisense RNAs (Figure 3B), consistent with previous reports that Set2 suppresses antisense RNAs (30,33).
Figure 3.
Comparison of change of RNA levels in set2 mutants. (A) RNAs whose levels changed in set2Δ cells more than 1.5-fold are plotted. The x-axis represents the log2 of the array signals in wild-type cells, and the y-axis represents the log2 of the array signals in set2Δ cells. RNAs are classified as coding, antisense, intergenic, and others, and shown by colored dots, as indicated in the inset. (B) A breakdown of RNAs that were elevated in set2Δ cells. (C) Scatter plots for comparisons of RNA levels in the indicated set2 mutants. The y-axis represents the log2 of the array signals in set2ΔSRI cells (left panel) or set2ΔC cells (right panel) over the array signals in wild-type, whereas the x-axis represents the log2 of the array signals in Δset2 over the array signals in wild-type. The correlation coefficients (r) are shown in the graph; P-values were calculated using Fisher's exact test. (D) Venn diagram showing the number of transcripts whose expression levels were more than 1.5-fold higher in the indicated mutants than in the wild-type. The transcripts were classified as coding and non-coding RNAs. Red color indicates transcripts up-regulated only in set2Δ, while yellow color shows transcripts up regulated only in set2ΔSRI cells (upper panels) or set2ΔC cells (lower panels). P-values were calculated using Fisher's exact test.
Next, we analyzed the correlations between the changes in RNA levels in set2Δ cells and those of set2ΔSRI or set2ΔC cells using scatter plots (Figure 3C). In both set2ΔSRI and set2ΔC cells, the expression levels of many genes were elevated, whereas the levels of a small number of genes were decreased. The correlation coefficients were close to 1 (0.98 for set2ΔSRI versus wild-type: 0.96 for set2ΔC versus wild-type), indicating that the effects of set2ΔSRI and set2ΔC on the expression levels of are almost the same as those of Δset2. We detected the RNAs that exhibited a >1.5-fold increase in expression in either of Δset2, set2ΔSRI, or set2ΔC cells, classified these genes into them as coding genes and non-coding genes and compared these sets of genes each other using Venn diagrams (Figure 3D). This analysis revealed that almost all upregulated genes in set2ΔSRI cells and set2ΔC cells overlapped with those in Δset2 cells, irrespective of their status as coding or non-coding genes. To confirm the genome wide specific decrease of H3K36me3 in set2ΔSRI cells, we chose a gene that showed the average expression level in each gene set (very high, high, medium and low) used in Figure 2 and measured the H3K36me2 and H3K36me3 in WT and set2ΔSRI cells by ChIP-qPCR (Supplementary Figure S2). In set2ΔSRI cells, H3K36me3 significantly decreased at all genes tested (<0.2-fold), while H3K36me2 did not. Together with the other results that also showed H3K36me2 was retained in set2ΔSRI cells (Figures 1E and 2B), these results indicate that H3K36me3 is important for the repression of certain genes, including both coding and non-coding genes but H3k36me2 is not sufficient for the silencing.
Loss of H3K36me3 caused accumulation of pericentromeric non-coding RNAs without affecting heterochromatin structure
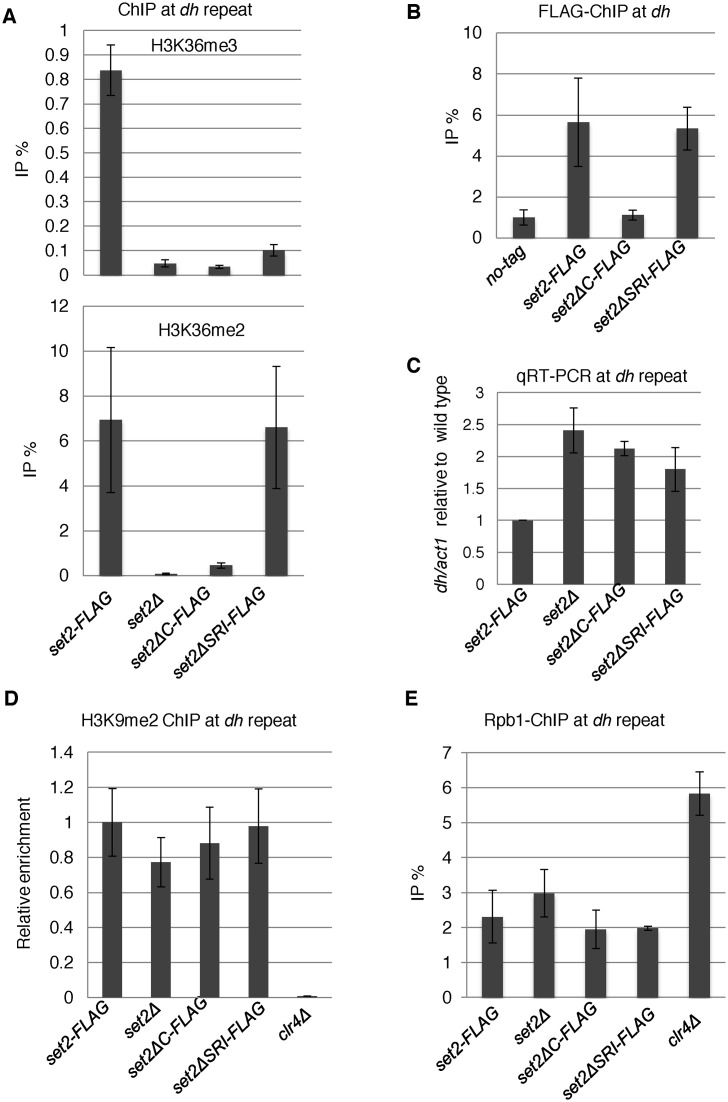
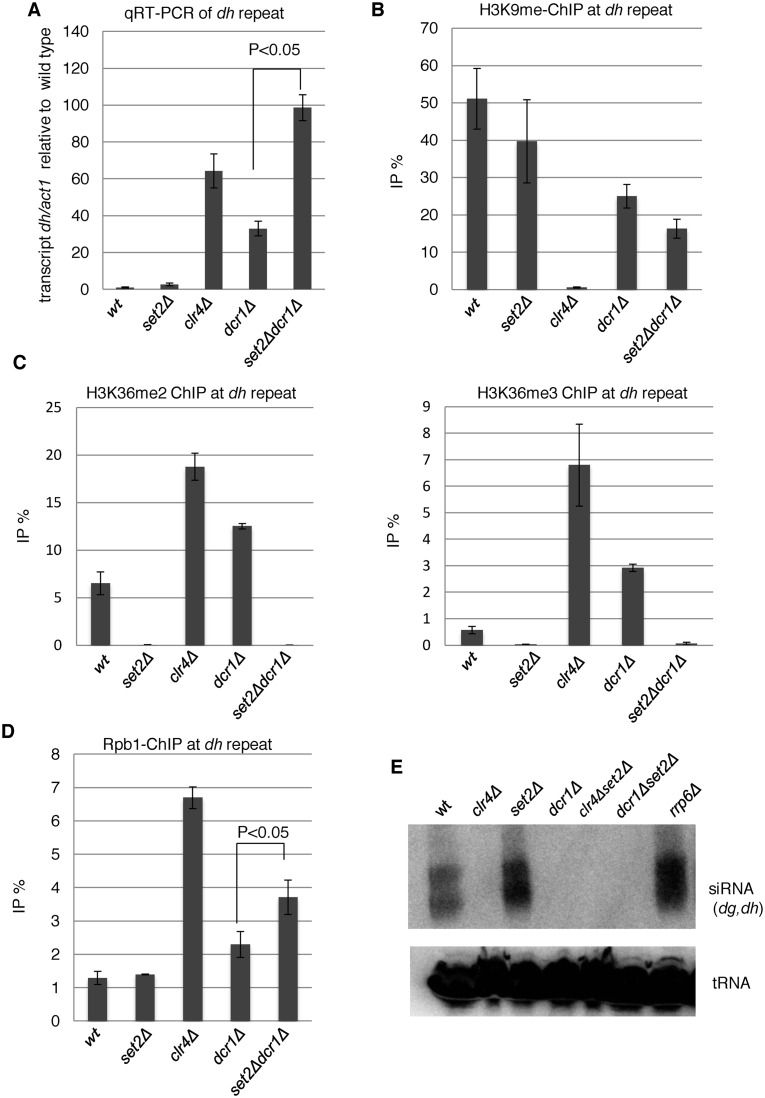
We noticed that the levels of some ncRNAs transcribed from pericentromeric heterochromatin were elevated in all set2 mutants (Supplementary Table S3), consistent with a previous report showing that deletion of set2 results in increased transcription of non-coding RNAs from pericentromeres in fission yeast (31). Note that the expression levels of heterochromatin-related factors such as clr4,dcr1, alp13, rrp6, did not significantly change in set2Δ cells (−0.19 < log2(set2Δ/wt) < 0.68). Because H3K36me2 was retained in setΔSRI cells, these data suggest that H3K36me3, but not H3K36me2, is required for silencing of heterochromatic transcripts, as in the case of euchromatic transcripts. To precisely examine the role of Set2-generated H3K36 methylation in heterochromatic silencing, we first used ChIP-qPCR to measure the methylation level of H3K36 at pericentromeric repeats in set2 mutants. set2Δ and set2ΔC cells lost both H3K36me2 and H3K36me3 from pericentromeric repeats, whereas set2ΔSRI decreased H3K36me3 without affecting the level of H3K36me2 (Figure 4A). These distributions were reminiscent of those at euchromatin. Consistent with these observations, ChIP analysis revealed that Set2-FLAG as well as Set2ΔSRI-FLAG localized at heterochromatin, whereas Set2ΔC-FLAG did not (Figure 4B). Next, we analyzed the accumulation of heterochromatic transcripts from dh repeats in set2 mutants using strand-specific qRT-PCR (Figure 4C). We observed ∼2-fold accumulation of dh-forward transcripts in all set2 mutants, demonstrating that H3K36me3, but not H3K36me2, played a role in heterochromatic silencing. The level of H3K9me2, a heterochromatic mark that is catalyzed by histone methyltransferase, Clr4, was not affected by set2 mutations (Figure 4D). In addition, level of total histone H3 was not affected by set2 mutations (Supplementary Figure S3). These data indicate that heterochromatin structure was maintained in the absence of H3K36 methylation. Next, we measured Pol2 occupancy by ChIP analysis using anti-Pol2 antibody 4H8, which preferentially binds to CTD-S5P, a marker for elongating Pol2 (Figure 4E). None of the set2 deletion mutants affected the amount of Pol2 at dh repeats, indicating that the observed accumulation of pericentromeric transcripts was not due to an increase in transcription. Thus, Set2-generated H3K36me3 may play a role in post-transcriptional gene silencing.
Figure 4.
Loss of Set2 compromised heterochromatic silencing without affecting heterochromatin structure. (A, B and D) ChIP analyses of H3K36me2 and H3K36me3 (A), Set2-FLAG (B), H3K9me2 (D) and Rpb1 (E) at heterochromatic dh repeats were performed using the indicated strains. Error bars show the standard deviation of three independent experiments. (C) Quantitative RT-PCR (qRT-PCR) analysis of the forward transcript from dh repeats was performed using the indicated strains. Error bars show the standard deviation of three independent experiments.
Pericentromeric heterochromatin formation mainly depends on the RNAi machinery, which produces siRNAs from pericentromeric transcripts; the resultant siRNAs are essential for heterochromatin formation (27). To investigate the relationship between H3K36me3-dependent heterochromatic silencing and the RNAi pathway, we performed epistasis analyses by generating a set2Δdcr1Δ double mutant (Figure 5). The dcr1 gene encodes the exonuclease Dicer, which is essential for siRNA production. The set2Δdcr1Δ double mutant exhibited a >2-fold increase in the level of heterochromatic transcripts from dh repeats relative to the dcr1Δ single mutant (Figure 5A), indicating that Set2 contributes to silencing independently of the RNAi pathway. Disruption of the RNAi system by dcr1Δ caused a partial disruption in centromeric heterochromatin due to the existence of another heterochromatin maintenance system that depends on the histone deacetylase (HDAC) Sir2 and the heterochromatin protein Swi6 (HP1 homolog) (49,50). In set2Δdcr1Δ cells, a substantial amount of H3K9me2 was maintained, although a slight decrease was observed (Figure 5B), suggesting a marginal role for Set2 in the alternative heterochromatin maintenance system. We observed a large increase in H3K36 methylation in clr4Δ cells and dcr1Δ cells: 3- and 2-fold increases in H3K36me2, and 2- and 5-fold increases in H3K36me3, in clr4Δ and dcr1Δ cells, respectively (Figure 5C). This increase was correlated with elevation of transcripts and Pol2 occupancy at pericentromeric repeats (Figures 4C and 5D), suggesting that upregulation of transcription induced co-transcriptional H3K36 methylation. In addition, the introduction of set2Δ into dcr1Δ cells caused an increase in Pol2, suggesting that Set2-generated H3K36me3 contributes to suppression of transcription in partially disrupted heterochromatin. We also analyzed the effect of depletion of Set2 on the amount of pericentromeric siRNA by northern blotting (Figure 5E). In clr4Δ cells, siRNA was not detected, which confirmed the requirement of H3K9 methylation for RNAi-dependent siRNA generation (51). In set2Δ cells, siRNA derived from pericentromeric repeats did not decreased, showing that Set2 and H3K36 methylation are not required for RNAi-dependent siRNA formation.
Figure 5.
Set2 functions at heterochromatin independently of RNAi. (A) qRT-PCR analyses of the forward transcripts from dh repeats were performed using the indicated strains. Error bars show the standard deviation of three independent experiments. P-values were determined using a two-sided Student's t-test to compare dcr1Δset2Δ cells with dcr1Δ cells. (B and D) ChIP analyses of H3K9me2 (B) and Rpb1 (D) at heterochromatic dh repeats were performed using the indicated strains. Error bars show the standard deviation of three independent experiments. P-values were determined using a two-sided Student's t-test to compare dcr1Δset2Δ cells with dcr1Δ cells. (C) H3K36 methylation was increased in mutants that compromised heterochromatin. ChIP analyses of H3K36me2 (left panel) and H3K36me3 (right panel) at heterochromatic dh repeats were performed in the indicated strains. Error bars show the standard deviation of three independent experiments. (E) siRNA analysis by northern blotting using oligonucleotide probes specific for dg and dh centromeric repeats (40). Oligonucleotide probe specific for tRNAAsn was used as a loading control.
H3K36me3 was required for post-transcriptional degradation of pericentromeric ncRNA
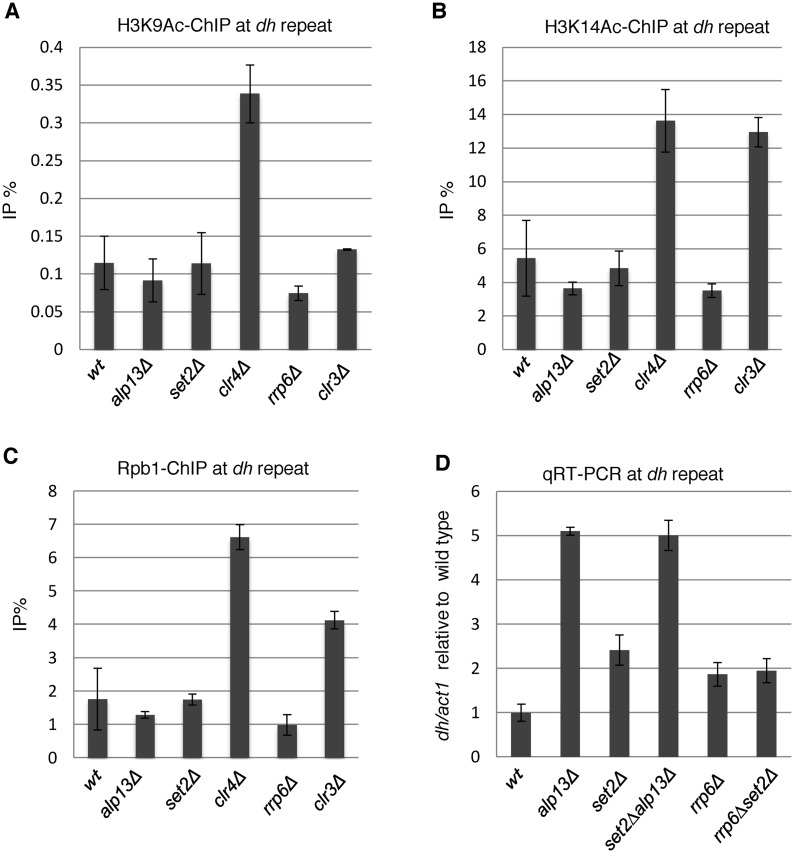
In both euchromatin and heterochromatin of fission yeast, H3K36 methylation is thought to recruit and/or activate an HDAC complex containing the catalytic subunit Clr6 via the chromodomain of the Alp13 subunit (30,31). The recruitment of the HDAC by H3K36me was suggested to contribute to heterochromatin formation via the same pathway (31). Failure of HDAC recruitment due to loss of H3K36 methylation in set2Δ cells, or to loss of Alp13, will cause an increase in histone acetylation. However, we did not detect any increase in acetylation of H3K9 or H3K14, which are targets of Clr6 (32,52) and the general mark for active transcription, in alp13Δ cells or set2Δ cells at heterochromatic repeats (Figure 6A and B). Furthermore, we did not detect any increase in the level of Pol2 in alp13Δ cells (Figure 6C). By contrast, clr6-1 mutation caused increase of heterochromatic transcripts accompanied by increases in the levels of H3K9Ac, H3K14Ac and Pol2 (Supplementary Figure S4). SHREC, another HDAC that functions at heterochromatin, contains Clr3 as a catalytic subunit and preferentially deacetylates H3K14 (52,53). Loss of Clr3 caused an increase in the acetylation of H3K14, accompanied by an increase in Pol2 (Figure 6B and C). These data suggest that Set2-generated H3K36me3 and Alp13 contribute post-transcriptionally to silencing of pericentromeric transcripts in a Clr6 complex II-independent manner. To further analyze the relationship between Alp13 and Set2/H3K36me3, we generated the alp13Δset2Δ double mutant and measured the amount of heterochromatic non-coding RNA at pericentromeres (Figure 6D). As indicated by a previous report (31), alp13Δ cells exhibited more accumulation of heterochromatic ncRNA than set2Δ cells. The double mutant exhibited the same level of accumulation as the alp13Δ single mutant. suggesting that Set2/H3K36me3 partly contributes to post-transcriptional silencing of pericentromeric transcripts by Alp13.
Figure 6.
Comparison of the states of heterochromatin among possible downstream factors of Set2. (A and C) ChIP analyses of H3K39Ac (A), H3K14Ac (B) and Rpb1 (C) at heterochromatic dh repeats in the indicated strains. Error bars show the standard deviation of three independent experiments. (D) Quantitative RT-PCR (qRT-PCR) analysis of the forward transcript from dh repeats was performed using the indicated strains. Error bars show the standard deviation of three independent experiments.
The nuclear exosome degrades pericentromeric ncRNAs (30,54) and also contributes to heterochromatin formation, in parallel with RNAi (55). A previous study observed a synergistic increase in pericentromeric transcripts in alp13Δrrp6Δ cells, showing that each gene product functions independently to suppress pericentromeric transcripts (30). Therefore, we started to analyze the possible involvement of Rrp6 in Set2/H3K36me3-dependent heterochromatic silencing. The absence of Rrp6, a catalytic subunit of the nuclear exosome, caused accumulation of pericentromeric transcripts, but did not increase the level of Pol2 (Figure 6C and D), indicating that the nuclear exosome contributes to heterochromatic silencing by degrading heterochromatic transcripts. To examine the genetic interaction between rrp6 and set2, we generated the set2Δrrp6Δ double mutant and measured the levels of pericentromeric transcripts (Figure 6D). The set2Δrrp6Δ double mutant exhibited the same level of accumulation of pericentromeric transcripts as those observed in each single mutant, suggesting Set2 and Rrp6 functions in the same pathway for heterochromatic silencing. We also observed that depletion of Rrp6 did not result in the decrease of H3K36me2/3 (Supplementary Figure S5). These results raise the possibility that the Rrp6/nuclear exosome is a downstream factor of Set2/H3K36me3 that post-transcriptionally silences pericentromeric transcripts.
One possible mechanism by which H3K36me3 could contribute to Rrp6/exosome-dependent heterochromatic RNA degradation is co-transcriptional recruitment of the exosome to heterochromatin. A ChIP assay using a strain expressing myc-tagged Rrp6 and an anti-myc antibody revealed the heterochromatic localization of Rrp6, as we demonstrated previously (56). However, the absence of H3K36 methylation due to deletion of set2 did not cause a decrease in the heterochromatic localization of Rrp6. Instead, it caused a modest increase (Supplementary Figure S5), suggesting that Set2/H3K36me3 functions in degradation of pericentromeric transcript after recruitment of Rrp6 to heterochromatin.
Set2-generated H3K36me3-dependent silencing of subtelomeric genes
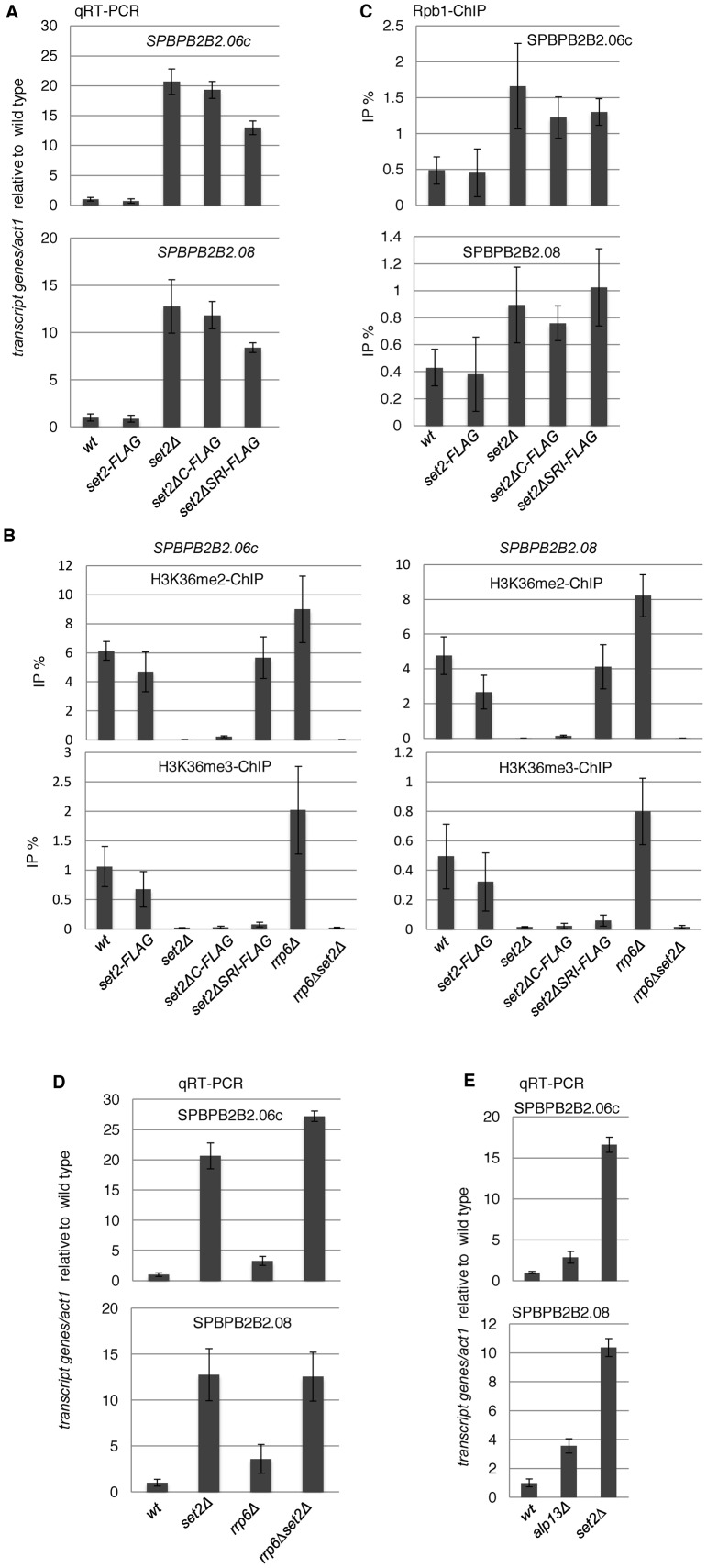
As described above, transcriptome analysis revealed that the absence of H3K36me3 caused induction of some euchromatic genes as well as antisense/non-coding transcripts (Figure 3A and B). Many of the highly upregulated genes were located close to telomeres: 18 of the 20 most upregulated genes in set2Δ cells are within 100 kb from the ends of chromosomes I and II (Supplementary Table S4 and Figure S6). In addition, many of these genes are upregulated during meiosis (9/20) or encode membrane proteins (6/20) (Supplementary Table S4). Hereafter, we focused on the two most upregulated genes (SPBPB2B2.06c and SPBPB2B2.08) in set2Δ cells, which are located close to the right telomere of chromosome II and whose transcripts are induced during meiosis (Supplementary Table S4). Analysis by qRT-PCR confirmed the significant increase in the RNA levels of these genes in set2ΔSRI, set2ΔC and set2Δ cells (Figure 7A). ChIP analysis of H3K36 methylation and histone H3 confirmed that the H3K36me3 level was reduced, but the H3K36me2 level was retained, at each gene in set2ΔSRI cells (Figure 7B and Supplementary Figure S3), showing that H3K36me3 was indeed important for the repression of these genes. To examine whether the observed repression occurs transcriptionally or post-transcriptionally, we checked the Pol2 occupancy at these genes by ChIP analysis (Figure 7C). Surprisingly, in all set2 mutants, the amount of Pol2 increased significantly, indicating that repression mainly occurs at the transcription step, which contrasted with the post-transcriptional silencing in heterochromatin.
Figure 7.
Set2 downregulates subtelomeric genes, SPBPB2B2.06c and SPBPB2B2.08 at the transcriptional level. (A, D and E) Quantitative RT-PCR (qRT-PCR) analyses of transcripts from both genes in the indicated strains were performed. Error bars show the standard deviation of three independent experiments. (B) Levels of H3K36 methylation at SPBBP2B2.06c and SPBPB2B2.08 in set2 mutants. ChIP analyses of H3K36me2 (upper panels) and H3K36me3 (lower panels) at SPBPB2B2.06c (left panels) and SPBPB2B2.08 (right panels) were performed in the indicated strains. Error bars show the standard deviation of three independent experiments. (C) ChIP analyses of Rpb1 at heterochromatic genes were performed using the indicated strains. Error bars show the standard deviation of three independent experiments.
In fission yeast, several meiotic genes are repressed by the Rrp6/exosome pathway (57), and the exosome appears to function with Set2/H3K36me3 at heterochromatic silencing as shown above. Therefore, we investigated whether Rrp6/exosome is involved in Set2/H3K36me3-dependent repression of these genes. Deletion of rrp6 caused induction of both SPBPB2B2.06c and SPBPB2B2.08 (Figure 7D), but the extent of induction was much lower than that observed in set2Δ cells, about 3–4-fold in rrp6Δ versus 12–20-fold in set2Δ cells, suggesting that Rrp6 functions in a different pathway than Set2/H3K36me3. The additive effects observed in the rrp6Δset2Δ double mutant at SPBPB2B2.06c suggested that Set2/H3K36me3 and Rrp6/exosome contribute independently to the repression of this gene. By contrast, the rrp6Δset2Δ double mutant exhibited the same level of induction of SPBPB2B2.08, suggesting that the exosome pathway and Set2/H3K36me3 pathway serve partially overlapping functions in the repression of this gene. We also observed that deletion of alp13 caused a smaller increase in both transcripts than deletion of set2 (Figure 7E), suggesting that Alp3 is not a primary downstream effector of Set2/H3K36me3.
DISCUSSION
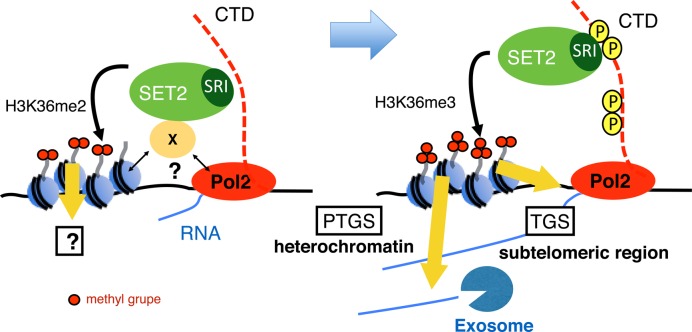
In this study, we characterized the roles of Set2-generated histone H3K36 methylation in the silencing of both heterochromatic and euchromatic transcription. Our data demonstrate that trimethylation, but not dimethylation, is important for the silencing of target transcripts; however, different mechanisms of silencing are employed at various loci. Our current model for Set2-generated H3K36 methylation is illustrated in Figure 8.
Figure 8.
Model of co-transcriptional methylation of H3K36 and its function in heterochromatic and subtelomeric regions in fission yeast. (Left panel) In the initial step of transcription, Set2 interacts with Pol2 or other factors with unknown mechanism and promotes H3K36me2 co-transcriptionally. The output of H3K36me2 is not yet known. (Right panel) When CTD-S2 (and probably CTD-S5) is phosphorylated, Set2 changes the binding mode to favor the interaction between the Set2 SRI domain and Pol2 CTD-S2,5P, thereby promoting formation of H3K36me3. H3K36me3 directs post-transcriptional gene silencing in heterochromatin, but assists in transcriptional gene silencing in the subtelomeric region.
Set2 differentially methylates H3K36 to the dimethylated and trimethylated states through differential interactions with Pol2
We showed that the SRI domain of Set2 is required for the interaction with Pol2 and trimethylation of H3K36, indicating that methylation of H3K36 by Set2 occurs co-transcriptionally, as in the case of budding yeast. However, in budding yeast, loss of the SRI domain causes loss of both H3K36me2 and H3K36me3 (10), whereas in fission yeast, it caused specific loss of H3K36me3. In addition, the budding yeast Set2 mutant corresponding to set2ΔC, which lacks the C-terminal region following the SET domain, exhibits specific loss of H3K36me3, as in the fission yeast set2ΔSRI strain, suggesting an inhibitory role for the region between the SET and SRI domains (10). Obviously, fission yeast Set2 does not have such an inhibitory domain. Alternatively, the difference between budding yeast and fission yeast could be due to a different truncation. In addition, we speculate that the region between the SET and SRI domains (amino acids 320–678) contains another domain that interacts with Pol2, either weakly or transiently, as discussed below.
Furthermore, we found that loss of the SRI domain, as well as the rpb1-ctdS2A mutation, diminished the interaction between Set2 and Pol2, as judged by the results of co-immunoprecipitation assays (Figure 1B and C), highlighting the specific requirement of CTD-S2P for the interaction between Set2 and Pol2. The budding yeast SRI domain preferentially binds to CTD-S2,5P (9,14). Considering the similarity between fission yeast SRI domain and budding yeast SRI, it is likely that CTD-S5P contributes to the interaction, although we did not test the requirement of CTD-S5P for the Set2–Pol2 interaction. Because the distribution of CTD-S5P is almost the same as that of Pol2 in fission yeast (48), the primary determinant of Set2 localization is likely to be CTD-S2 phosphorylation. Consistent with this, the distribution of Set2-FLAG accumulates slower than Pol2-CTD-S5P, which is the similar pattern as that of CTD-S2P: both accumulate in the latter half of transcription unit (48).
The distribution of the Set2ΔSRI-FLAG protein and the remaining H3K36me2 in the set2ΔSRI mutant are similar to those of Set2-FLAG and H3K36me3 in wild-type cells (Figure 2). The distribution of H3K36me2 in set2ΔSRI cells was correlated with that of Pol2 especially on the genes moderately transcribed (high and medium in Supplementary Figure S1B). However the levels of Set2ΔSRI on the gene were much lower than Set2-FLAG (Supplementary Figures S1A and S2). These data suggest that dimethylation also occurs co-transcriptionally by Set2ΔSRI associated with transcribing Pol2 through remaining domain. Since we could not detect association of Pol2 with set2ΔSRI in coimmunoprecipitation experiments, the interaction appears to be relatively weak or transient. Based on these findings, we propose that fission yeast Set2 differentially methylates H3K36 during transcription via two distinct modes of interaction with Pol2. In the first phase of transcription, Set2 weakly associates with Pol2 via amino acids 320–678 and promotes dimethylation of H3K36. When CTD-S2 is phosphorylated during elongation, Set2 binds to phosphorylated CTD with high affinity, and the strong binding mode promotes trimethylation. (Figure 8). However, our data did not exclude a possibility that Set2 is recruited by Pol2-independent mechanisms to promote H3K36 methylation in the first phase. Further experiments would clarify this point.
Mechanisms for silencing by Set2-generated H3K36me3
The specific loss of H3K36me3 in set2ΔSRI cells enabled us to analyze the differential roles of trimethylation and dimethylation of H3K36. Surprisingly, set2ΔSRI cells exhibited the same phenotype as set2Δ cells for genome-wide gene expression. This indicates that the ‘reader’ of H3K36 methylation for silencing in fission yeast specifically recognizes H3K36me3. The function of H3K36me3 at heterochromatin seems to be different from that of the subtelomeric region, as discussed below, suggesting that there are multiple H3K36me3-specific readers in fission yeast.
Heterochromatic non-coding RNAs are substrates of the RNAi machinery, and the resultant siRNAs are required for heterochromatin formation. These RNAs are repressed both transcriptionally and post-transcriptionally (27,54). In budding yeast, H3K36me2 and H3K36me3 transcriptionally represses cryptic transcription by recruiting Rpd3S HDAC complex via Eaf3, a subunit of Rpd3S and a ‘reader’ of H3K36me2 and H3K36me3 (22–24). A similar mechanism was proposed to function at fission yeast heterochromatin: Clr6 complex II, the fission yeast counterpart of Rpd3S that contains the Eaf3 homolog Alp13, is recruited to heterochromatin and promotes transcriptional silencing through deacetylation of histones (30,31). However, we found that Pol2 occupancy was not changed in the absence of H3K36me3 in set2 mutants, and that the level of H3K9me was not affected. Furthermore, acetylation of H3K9 and H3K14, which associate with active transcription, did not change in set2Δ cells, while both acetylation were elevated accompanied by elevated levels of Pol2 and transcription in clr6–1 mutant (Supplementary Figure S4). These results strongly suggest that Set2-generated H3K36me3 contributes to post-transcriptional silencing rather than transcriptional silencing by HDAC. Genetic analyses suggest that Set2 silenced heterochromatic transcripts via Rrp6/exosome and/or Alp13. Based on the additive effect of rrp6Δalp13Δ double mutant on de-silencing, a previous study suggested that Rrp6 and Alp13 function in different pathways in heterochromatic silencing (30). Since set2Δ did not show strong additive effect with rrp6Δ or alp13Δ on de-silencing, Alp13 and Rrp6 have overlapping function that requires Set2/H3K36me3. Loss of either Rrp6 or Alp13 caused a moderate decrease in H3K9me3, consistent with previous reports that both proteins contribute to H3K9 methylation (30,31); however, neither mutation affected the levels of Pol2 occupancy at heterochromatin, indicating that the remaining heterochromatin is sufficient for transcriptional silencing and that these gene products play roles in post-transcriptional silencing. Because loss of Alp13 did not affect the acetylation level of histone H3K9 and H3K14 while clr6 mutation caused the elevated levels of the acetylation accompanied by increase in transcription, the contribution of Alp13 in the heterochromatic silencing might not depend on the HDAC activity of Clr6 complex II. Because Eaf3 of budding yeast is a component of the Nu4A histone acetyltransferase complex (58), Alp13 might be a component of another complex that promotes post-transcriptional silencing.
Accumulation of heterochromatic nc RNA in set2Δ, rrp6Δ or alp13Δ cells was significantly lower than that observed in Clr4 or RNAi mutants (2–4-fold versus more than 10-fold). Previous analysis indicates that heterochromatic ncRNAs are degraded by two independent RNA degradation pathway, RNAi and exsosome (54).Thus, ncRNAs escaped from exosome-dependent degradation in rrp6Δ and set2Δ cells would be degraded by RNAi-dependent pathway. In other words, the degradation of heterochromatic transcripts by RNAi in set2 or rrp6 mutants may underestimate the contribution of post-transcriptional silencing by Set2 in wild-type cells.
Loss of H3K36me3 caused a marked increase in some transcripts, many of which localize close to telomeres. Subtelomeric regions are rich in pseudo-genes and non-coding RNA genes and are relatively transcriptionally inactive, although they do not contain the heterochromatic histone modification H3K9me (59,60). Therefore, subtelomeric regions appear to form distinct silent chromosomal domains, and H3K36me3 contributes to the formation of these domains. Indeed, a recent report showed that subtelomeric regions of chromosome I and II form special condensed chromatin structure, whose formation partly depends on Set2-directed H3K36 methylation (41). In contrast to the situation in heterochromatin, analysis of the two most upregulated genes in subtelomeric regions revealed that Pol2 occupancy was significantly increased by loss of H3K36me3, indicating that transcriptional gene silencing is the predominant mechanism for repression of these genes; however, involvement of post-transcriptional silencing cannot be entirely ruled out. Because relatively weak induction of these genes was observed in rrp6Δ or alp13Δ cells, we concluded that neither Rrp6/Exosome nor an Alp13-containing complex was a major downstream effector of H3K36me3. In dcr1Δ cells, in which heterochromatin is partially compromised, loss of H3K36 methylation caused an increase in Pol2 occupancy at heterochromatic repeats (Figure 5D), raising the possibility that H3K36me3 can repress transcription in heterochromatin. The predominant heterochromatin-dependent silencing might mask the role of H3K36me3-dependent transcriptional silencing in normal heterochromatin.
H3K36me2 versus H3K36me3
In budding yeast, a well-established role of H3K36 methylation by Set2 is suppression of aberrant transcription from cryptic promoters in coding sequences; this effect is mediated by recruitment of the Rpd3S HDAC complex (22–24). H3K36me2 is sufficient for this function (10,25), consistent with the fact that Eaf3, a reader of H3K36me in Rpd3S recognizes both H3K36me2 and H3K36me3 (24). However, several factors, such as FACT and Spt6, are specifically required for generation of H3K36me3 in budding yeast, suggesting the existence of H3K36me3-specific function. Mammals possess redundant H3K36 methyltransferases, some of which are specific for mono- and dimethylation, whereas others promote trimethylation (5), suggesting that H3K36me1/2 and H3K36me3 serve distinct functions. Indeed, mutations in different H3K36 methyltransferases are associated with different diseases (5).
Previously, no distinct roles had been established for H3K36me1/2 and H3K36me3. Our results clearly indicate that H3K36me3, but not H3K36me2, plays a specific role in repression of euchromatic and heterochromatic gene expression in fission yeast. Further analyses, e.g., searching for a H3K36me3-specific reader in fission yeast, will provide insight into the differential functions of H3K36 methylation states.
Supplementary Material
Acknowledgments
We thank R. Allshire and D. Hermand for the strains, and T. Urano, M. Kimura and A. Ishihama for antibodies. We also thank our laboratory members, especially A. Shimada and T. Kajitani for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grant-in-Aid for Scientific Research (A) from the Japan Society for the Promotion of Science [25250022]; Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan [25118502] (to Y.M.); JST PREST program (to H.K.); ChIP-sequencing was supported by a Grant-in-Aid for Scientific Research on Innovative Areas ‘Genome Science’ from the Ministry of Education, Culture, Sports, Science, and Technology of Japan [221S0002]. Funding for open access charge: Grant-in-Aid for Scientific Research (A) from the Japan Society for the Promotion of Science [25250022] (to Y.M.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Jenuwein T., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 2.Tachibana M., Sugimoto K., Nozaki M., Ueda J., Ohta T., Ohki M., Fukuda M., Takeda N., Niida H., Kato H., et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peters A.H.F.M., Mermoud J.E., O'Carroll D., Pagani M., Schweizer D., Brockdorff N., Jenuwein T. Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat. Genet. 2002;30:77–80. doi: 10.1038/ng789. [DOI] [PubMed] [Google Scholar]
- 4.Venkatesh S., Smolle M., Li H., Gogol M.M., Saint M., Kumar S., Natarajan K., Workman J.L. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature. 2012;489:452–455. doi: 10.1038/nature11326. [DOI] [PubMed] [Google Scholar]
- 5.Wagner E.J., Carpenter P.B. Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Biol. 2012;13:115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pokholok D.K., Harbison C.T., Levine S., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 7.Rao B., Shibata Y., Strahl B.D., Lieb J.D. Dimethylation of histone H3 at lysine 36 demarcates regulatory and nonregulatory chromatin genome-wide. Mol. Cell. Biol. 2005;25:9447–9459. doi: 10.1128/MCB.25.21.9447-9459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Venkatesh S., Workman J.L. Set2 mediated H3 lysine 36 methylation: regulation of transcription elongation and implications in organismal development. WIREs Dev. Biol. 2013;2:685–700. doi: 10.1002/wdev.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kizer K.O., Phatnani H.P., Shibata Y., Hall H., Greenleaf A.L., Strahl B.D. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol. Cell. Biol. 2005;25:3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Youdell M.L., Kizer K.O., Kisseleva-Romanova E., Fuchs S.M., Duro E., Strahl B.D., Mellor J. Roles for Ctk1 and Spt6 in regulating the different methylation states of histone H3 lysine 36. Mol. Cell. Biol. 2008;28:4915–4926. doi: 10.1128/MCB.00001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phatnani H.P., Greenleaf A.L. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 12.Medlin J., Scurry A., Taylor A., Zhang F., Peterlin B.M., Murphy S. P-TEFb is not an essential elongation factor for the intronless human U2 snRNA and histone H2b genes. EMBO J. 2005;24:4154–4165. doi: 10.1038/sj.emboj.7600876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egloff S., O'Reilly D., Chapman R.D., Taylor A., Tanzhaus K., Pitts L., Eick D., Murphy S. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science. 2007;318:1777–1779. doi: 10.1126/science.1145989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vojnic E., Simon B., Strahl B.D., Sattler M., Cramer P. Structure and carboxyl-terminal domain (CTD) binding of the Set2 SRI domain that couples histone H3 Lys36 methylation to transcription. J. Biol. Chem. 2006;281:13–15. doi: 10.1074/jbc.C500423200. [DOI] [PubMed] [Google Scholar]
- 15.Mosley A.L., Pattenden S.G., Carey M., Venkatesh S., Gilmore J.M., Florens L., Workman J.L., Washburn M.P. Rtr1 is a CTD phosphatase that regulates RNA polymerase II during the transition from serine 5 to serine 2 phosphorylation. Mol. Cell. 2009;34:168–178. doi: 10.1016/j.molcel.2009.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bataille A.R., Jeronimo C., Jacques P.-É., Laramée L., Fortin M.-È., Forest A., Bergeron M., Hanes S.D., Robert F. A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase, and isomerase enzymes along genes. Mol. Cell. 2012;45:158–170. doi: 10.1016/j.molcel.2011.11.024. [DOI] [PubMed] [Google Scholar]
- 17.Chu Y., Sutton A., Sternglanz R., Prelich G. The BUR1 cyclin-dependent protein kinase is required for the normal pattern of histone methylation by SET2. Mol. Cell. Biol. 2006;26:3029–3038. doi: 10.1128/MCB.26.8.3029-3038.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin L.-J., Minard L.V., Johnston G.C., Singer R.A., Schultz M.C. Asf1 can promote trimethylation of H3 K36 by Set2. Mol. Cell. Biol. 2010;30:1116–1129. doi: 10.1128/MCB.01229-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoh S.M., Lucas J.S., Jones K.A. The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes Dev. 2008;22:3422–3434. doi: 10.1101/gad.1720008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drouin S., Laramée L., Jacques P.-É., Forest A., Bergeron M., Robert F. DSIF and RNA polymerase II CTD phosphorylation coordinate the recruitment of Rpd3S to actively transcribed genes. PLoS Genet. 2010;6:e1001173. doi: 10.1371/journal.pgen.1001173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Govind C.K., Qiu H., Ginsburg D.S., Ruan C., Hofmeyer K., Hu C., Swaminathan V., Workman J.L., Li B., Hinnebusch A.G. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol. Cell. 2010;39:234–246. doi: 10.1016/j.molcel.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carrozza M.J., Li B., Florens L., Suganuma T., Swanson S.K., Lee K.K., Shia W.-J., Anderson S., Yates J., Washburn M.P., et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 23.Keogh M.-C., Kurdistani S.K., Morris S.A., Ahn S.H., Podolny V., Collins S.R., Schuldiner M., Chin K., Punna T., Thompson N.J., et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 24.Joshi A.A., Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol. Cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 25.Li B., Jackson J., Simon M.D., Fleharty B., Gogol M., Seidel C., Workman J.L., Shilatifard A. Histone H3 lysine 36 dimethylation (H3K36me2) is sufficient to recruit the Rpd3s histone deacetylase complex and to repress spurious transcription. J. Biol. Chem. 2009;284:7970–7976. doi: 10.1074/jbc.M808220200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grewal S.I. RNAi-dependent formation of heterochromatin and its diverse functions. Curr. Opin. Genet. Dev. 2010;20:134–141. doi: 10.1016/j.gde.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Volpe T.A., Kidner C., Hall I.M., Teng G., Grewal S.I.S., Martienssen R.A. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 28.Morris S.A., Shibata Y., Noma K.-I., Tsukamoto Y., Warren E., Temple B., Grewal S.I.S., Strahl B.D. Histone H3 K36 methylation is associated with transcription elongation in Schizosaccharomyces pombe. Eukaryot. Cell. 2005;4:1446–1454. doi: 10.1128/EC.4.8.1446-1454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeGennaro C.M., Alver B.H., Marguerat S., Stepanova E., Davis C.P., Bähler J., Park P.J., Winston F. Spt6 regulates intragenic and antisense transcription, nucleosome positioning, and histone modifications genome-wide in fission yeast. Mol. Cell. Biol. 2013;33:4779–4792. doi: 10.1128/MCB.01068-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicolas E., Yamada T., Cam H.P., FitzGerald P.C., Kobayashi R., Grewal S.I.S. Distinct roles of HDAC complexes in promoter silencing, antisense suppression and DNA damage protection. Nat. Struct. Mol. Biol. 2007;14:372–380. doi: 10.1038/nsmb1239. [DOI] [PubMed] [Google Scholar]
- 31.Chen E.S., Zhang K., Nicolas E., Cam H.P., Zofall M., Grewal S.I.S. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- 32.Nakayama J.-I., Xiao G., Noma K.-I., Malikzay A., Bjerling P., Ekwall K., Kobayashi R., Grewal S.I.S. Alp13, an MRG family protein, is a component of fission yeast Clr6 histone deacetylase required for genomic integrity. EMBO J. 2003;22:2776–2787. doi: 10.1093/emboj/cdg248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hennig B.P., Bendrin K., Zhou Y., Fischer T. Chd1 chromatin remodelers maintain nucleosome organization and repress cryptic transcription. EMBO Rep. 2012;13:997–1003. doi: 10.1038/embor.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moreno S., Klar A., Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Meth. Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 35.Bähler J., Wu J.Q., Longtine M.S., Shah N.G., McKenzie A., Steever A.B., Wach A., Philippsen P., Pringle J.R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 36.Sadaie M., Iida T., Urano T., Nakayama J.-I. A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. EMBO J. 2004;23:3825–3835. doi: 10.1038/sj.emboj.7600401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimura H., Hayashi-Takanaka Y., Goto Y., Takizawa N., Nozaki N. The organization of histone H3 modifications as revealed by a panel of specific monoclonal antibodies. Cell Struct. Funct. 2008;33:61–73. doi: 10.1247/csf.07035. [DOI] [PubMed] [Google Scholar]
- 38.Rechtsteiner A., Ercan S., Takasaki T., Phippen T.M., Egelhofer T.A., Wang W., Kimura H., Lieb J.D., Strome S. The histone H3K36 methyltransferase MES-4 acts epigenetically to transmit the memory of germline gene expression to progeny. PLoS Genet. 2010;6:e1001091. doi: 10.1371/journal.pgen.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kimura M., Sakurai H., Ishihama A. Intracellular contents and assembly states of all 12 subunits of the RNA polymerase II in the fission yeast Schizosaccharomyces pombe. Eur. J. Biochem. 2001;268:612–619. doi: 10.1046/j.1432-1327.2001.01911.x. [DOI] [PubMed] [Google Scholar]
- 40.Reinhart B.J., Bartel D.P. Small RNAs correspond to centromere heterochromatic repeats. Science. 2002;297:1831–1831. doi: 10.1126/science.1077183. [DOI] [PubMed] [Google Scholar]
- 41.Matsuda A., Chikashige Y., Ding D.-Q., Ohtsuki C., Mori C., Asakawa H., Kimura H., Haraguchi T., Hiraoka Y. Highly condensed chromatins are formed adjacent to subtelomeric and decondensed silent chromatin in fission yeast. Nat. Commun. 2015;6:7753–7764. doi: 10.1038/ncomms8753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kato H., Okazaki K., Iida T., Nakayama J.-I., Murakami Y., Urano T. Spt6 prevents transcription-coupled loss of posttranslationally modified histone H3. Sci. Rep. 2013;3:2186–2191. doi: 10.1038/srep02186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. 1000 Genome Project Data Processing Subgroup (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137.1–137.9. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki S., Nagao K., Obuse C., Murakami Y., Takahata S. A novel method for purification of the endogenously expressed fission yeast Set2 complex. Protein Expr. Purif. 2014;97:44–49. doi: 10.1016/j.pep.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 47.Nozawa R.-S., Nagao K., Masuda H.-T., Iwasaki O., Hirota T., Nozaki N., Kimura H., Obuse C. Human POGZ modulates dissociation of HP1alpha from mitotic chromosome arms through Aurora B activation. Nat. Cell Biol. 2010;12:719–727. doi: 10.1038/ncb2075. [DOI] [PubMed] [Google Scholar]
- 48.Coudreuse D., van Bakel H., Dewez M., Soutourina J., Parnell T., Vandenhaute J., Cairns B., Werner M., Hermand D. A gene-specific requirement of RNA polymerase II CTD phosphorylation for sexual differentiation in S. pombe. Curr. Biol. 2010;20:1053–1064. doi: 10.1016/j.cub.2010.04.054. [DOI] [PubMed] [Google Scholar]
- 49.Alper B.J., Job G., Yadav R.K., Shanker S., Lowe B.R., Partridge J.F. Sir2 is required for Clr4 to initiate centromeric heterochromatin assembly in fission yeast. EMBO J. 2013;32:2321–2335. doi: 10.1038/emboj.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buscaino A., Lejeune E., Audergon P., Hamilton G., Pidoux A., Allshire R.C. Distinct roles for Sir2 and RNAi in centromeric heterochromatin nucleation, spreading and maintenance. EMBO J. 2013;32:1250–1264. doi: 10.1038/emboj.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Noma K.-I., Sugiyama T., Cam H., Verdel A., Zofall M., Jia S., Moazed D., Grewal S.I.S. RITS acts in cis to promote RNA interference–mediated transcriptional and post-transcriptional silencing. Nat. Genet. 2004;36:1174–1180. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- 52.Bjerling P., Silverstein R.A., Thon G., Caudy A., Grewal S., Ekwall K. Functional divergence between histone deacetylases in fission yeast by distinct cellular localization and in vivo specificity. Mol. Cell. Biol. 2002;22:2170–2181. doi: 10.1128/MCB.22.7.2170-2181.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sugiyama T., Cam H.P., Sugiyama R., Noma K.-I., Zofall M., Kobayashi R., Grewal S.I.S. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell. 2007;128:491–504. doi: 10.1016/j.cell.2006.12.035. [DOI] [PubMed] [Google Scholar]
- 54.Bühler M., Haas W., Gygi S.P., Moazed D. RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell. 2007;129:707–721. doi: 10.1016/j.cell.2007.03.038. [DOI] [PubMed] [Google Scholar]
- 55.Reyes-Turcu F.E., Zhang K., Zofall M., Chen E., Grewal S.I.S. Defects in RNA quality control factors reveal RNAi-independent nucleation of heterochromatin. Nat. Struct. Mol. Biol. 2011;18:1132–1138. doi: 10.1038/nsmb.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oya E., Kato H., Chikashige Y., Tsutsumi C., Hiraoka Y., Murakami Y. Mediator directs co-transcriptional heterochromatin assembly by RNA interference-dependent and -independent pathways. PLoS Genet. 2013;9:e1003677. doi: 10.1371/journal.pgen.1003677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harigaya Y., Tanaka H., Yamanaka S., Tanaka K., Watanabe Y., Tsutsumi C., Chikashige Y., Hiraoka Y., Yamashita A., Yamamoto M. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature. 2006;442:45–50. doi: 10.1038/nature04881. [DOI] [PubMed] [Google Scholar]
- 58.Reid J.L., Moqtaderi Z., Struhl K. Eaf3 regulates the global pattern of histone acetylation in Saccharomyces cerevisiae. Mol. Cell. Biol. 2004;24:757–764. doi: 10.1128/MCB.24.2.757-764.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cam H.P., Sugiyama T., Chen E.S., Chen X., FitzGerald P.C., Grewal S.I.S. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 2005;37:809–819. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- 60.Kanoh J., Urano T., Ishikawa F. Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Curr. Biol. 2005;15:1808–1819. doi: 10.1016/j.cub.2005.09.041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.