Abstract
G-quadruplexes are dynamic structures folded in G-rich single-stranded DNA regions. These structures have been recognized as a potential nucleic acid based mechanism for regulating multiple cellular processes such as replication, transcription and genomic maintenance. So far, their transcriptional role in vivo during vertebrate embryonic development has not yet been addressed. Here, we performed an in silico search to find conserved putative G-quadruplex sequences (PQSs) within proximal promoter regions of human, mouse and zebrafish developmental genes. Among the PQSs able to fold in vitro as G-quadruplex, those present in nog3, col2a1 and fzd5 promoters were selected for further studies. In cellulo studies revealed that the selected G-quadruplexes affected the transcription of luciferase controlled by the SV40 nonrelated promoter. G-quadruplex disruption in vivo by microinjection in zebrafish embryos of either small ligands or DNA oligonucleotides complementary to the selected PQSs resulted in lower transcription of the targeted genes. Moreover, zebrafish embryos and larvae phenotypes caused by the presence of complementary oligonucleotides fully resembled those ones reported for nog3, col2a1 and fzd5 morphants. To our knowledge, this is the first work revealing in vivo the role of conserved G-quadruplexes in the embryonic development, one of the most regulated processes of the vertebrates biology.
INTRODUCTION
G-quadruplexes are dynamic structures folded in G-rich single-stranded DNA regions. These structures have been recognized as a potential nucleic acid based mechanism for regulating multiple cellular processes such as replication, transcription and genomic maintenance. The main unit of a quadruplex consists of G-quartets formed by four guanine residues arranged in a planar structure. The stacking of three or more planar G-quartets establishes the G-quadruplex, which is stabilized mainly by potassium cations. One of the widely used consensus putative G-quadruplex sequence (PQS) is G≥3 N1–7 G≥3 N1–7 G≥3 N1–7 G≥3, wherein G-tracts are connected by loops of varying length and nucleotide (N) composition (1). The relative orientation of G-tracts defines parallel or antiparallel G-quadruplex topologies (2,3). G-quadruplexes were initially found in telomeric DNA sequences; however, the discovery of the enriched presence of PQSs in gene promoters from bacteria to humans suggested a role of these structural motifs in transcriptional regulation, as well as a natural selection along evolution (4–6). G-quadruplexes are stable under physiological conditions in vitro (7) and their existence in the human genome has been recently demonstrated in cellulo (8). In addition, a number of studies using small G-quadruplex-targeting ligands support a transcriptional role of G-quadruplex in cellulo (9) and in vivo in early zebrafish (Danio rerio) embryos (10). However, nonspecific or pleiotropic effects of these ligands could not be ruled out, as animal genomes possess numerous genes containing at least one G-quadruplex near the transcription start site (TSS) (11). Reinforcing the notion of a fundamental role of G-quadruplexes in the biology of living organisms, numerous reports have indirectly demonstrated the involvement of G-quadruplexes in human disease (12). While in recent years the knowledge about the biological role of the G-quadruplex has made significant progress, the challenge still lies in proving the direct role of these structures on a specific biological process in a complex living organism.
During embryonic development, gene expression is orchestrated by specific and highly evolutionarily conserved mechanisms that take place accurately, both at spatial and temporal levels (13). An intricate array of cis-regulatory sequences controlling individual genes leads to a fine-tuning of gene expression in different developmental processes, which in turn may set up specific phenotypes both in health and disease. The last decades have provided compelling evidence that not only protein-mediated transcriptional control but also chromatin state play essential roles in orchestrating all stages of embryonic development (14,15). In this context, we wondered whether G-quadruplexes contribute to the transcriptional control of genes required for the proper vertebrate embryonic development.
In this work we present evidences gathered by using combined computational and experimental analyses showing that evolutionarily conserved G-quadruplexes found in the promoters of the developmentally related genes nog3, col2a1 and fdz5 regulate in vivo their transcriptional activity during zebrafish embryonic development. To our knowledge, this is the first work reporting the G-quadruplexes as cis-acting elements contributing to the complex regulatory network that orchestrates the success of vertebrate embryonic development.
MATERIALS AND METHODS
Bioinformatics
Gene promoter sequences were searched and retrieved by using Ensembl Biomart tool (http://www.ensembl.org/biomart/martview) (16) and genome versions Zv9 (Danio rerio); GRCh37.p10 (Homo sapiens), and GRCm38.p1 (Mus musculus). Proximal promoter regions (PPRs) were defined as the region spanning 1000 bp upstream from reported TSS. Unique orthologous gene lists were generated using Mus musculus gene information as reference nomenclature. PQSs were searched using Quadparser algorithm (1). Parameters were set to search for quadruplexes formed by the stacking of at least three guanine tetrads with loop sequence lengths spanning from 1 to 7 nucleotides. Gene lists intersection was made using BioVenn (17). Gene Ontology (GO) data and GO term enrichment was analyzed with Biological Networks Gene Ontology (BiNGO) plugin using Cytoscape v2.8 (http://www.cytoscape.org/) (18). GO terms statistical significance was analyzed using a hypergeometric test and a Benjamini & Hochberg false discovery rate correction (FDR).
Oligonucleotides and compounds
Synthetic desalted single-stranded oligodeoxyribonucleotides were purchased from InvitrogenTM, dissolved in bidistilled water and stored at −20°C. Concentrations of all oligonucleotides were determined by spectrometry using extinction coefficients provided by the manufacturer. Oligonucleotides used as primers for polymerase chain reactions (PCRs) were designed using Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) and their specificity checked using MFE primer 2.0 (http://biocompute.bmi.ac.cn/CZlab/MFEprimer-2.0/). Sequences are listed in Supplementary Table S1. Mutations in G-quadruplex forming oligonucleotides were rationalized based on G-tracts disruption and were tested in silico using the QGRS Mapper software (19) and Quadparser algorithm (1).
Thioflavin T (ThT or 3,6-Dimethyl-2-(4-dimethylaminophenyl) benzothiazolium cation) was obtained from Sigma-Aldrich (Ref. T3516) and used without further purification. The concentration was calculated using the molar extinction coefficient in water at 412 nm of 36000 M−1cm−1 (20). The cationic porphyrins meso-Tetra (N-methyl-4-pyridyl) porphine tetrachloride (TMPyP4) and meso-Tetra (N-methyl-2-pyridyl) porphine tetrachloride (TMPyP2) were purchased from Frontier Scientific (Logan, Utah, USA) and used without further purification. The concentration of each porphyrin drug was calculated using the molar extinction coefficients in water at 424 nm of 226000 M−1cm−1 for TMPyP4, and at 414 nm of 182000 × M−1cm−1 for TMPyP2 (21).
ThT fluorescence assays
ThT fluorescence assays were performed as described elsewhere (20) with slight modifications. Briefly, before analysis, oligonucleotides were heated at 95°C for 5 min at 2 μM concentration in 100 mM Tris-HCl, pH 7.5, 100 mM KCl and slowly cooled to room temperature over 2 h. Then, oligonucleotides and ThT were mixed at 1 and 0.5 μM final concentrations, respectively, in 50 mM Tris-HCl, pH 7.5 and 50mM KCl in a volume of 200 μl using 96-well microplates (Greiner). Fluorescence emission measurements were performed at room temperature using a microplate reader (Synergy 2 Multi-Mode Microplate Reader, BioTek) with excitation filter of 485 ± 20 nm and an emission filter of 528 ± 20 nm. Each oligonucleotide was tested by triplicate and fluorescence values were relativized to ThT fluorescence in the absence of oligonucleotides. Pu18 and Pu27 oligonucleotides representing G-quadruplex from human c-MYC promoter were used as positive controls, while mutated versions of Pu18 and Pu27 that disrupt G-quadruplex formation as negative controls (22).
Circular dichroism (CD) spectroscopy
Oligonucleotides (Supplementary Table S1) were heated at 95°C for 5 min at 2 μM concentration in 10 mM Tris-HCl, pH 7.5, with or without the addition of 100 mM KCl, and slowly cooled to room temperature over 2 h. CD spectra were recorded at room temperature over a wavelength range of 220–320 nm with a Jasco-810 spectropolarimeter (1 cm quartz cell, 100 nm/min scanning speed, 1 s response time, average of four scans). The spectral contribution of buffers, salts and drugs were appropriately subtracted by using the software supplied with the spectropolarimeter.
In cellulo luciferase reporter experiments
In cellulo experiments were performed as described elsewhere (23). Briefly, duplex DNAs were generated by annealing oligonucleotides representing PQSs (or their mutated versions impeding G-quadruplex formation) with their complementary strands (Supplementary Table S1) and then cloned upstream the basal SV40 promoter by blunt-end ligation in SmaI digested pGL3-promoter vector plasmid (Promega). Several clones were sequenced and those that contained the PQSs or their mutated versions in the same strand (coding or template) as they are found in genomes were used for transfections. Mouse neuroblastoma cell line Neuro-2a (ATCC CCL-131) was cultured and transfected as previously described (23) with 0.5 μg of unmodified pGL3-promoter vector plasmid or reporter constructs. In addition, all dishes were co-transfected with 0.5 μg of pCMV-β-galactosidase (Promega) as a control for transfection efficiency. Luciferase activity was measured as described (23), normalized to β-galactosidase activity and expressed as a ratio of luciferase/β-galactosidase. Finally, values determined for wild-type and mutated PQSs constructs were relativized to those for pGL3-promoter vector plasmid. Experiments were repeated three times.
Animal handling and microinjection of zebrafish embryos
Animal handling during this study was carried out in strict accordance with relevant local, national and international guidelines. Protocols were approved by the Committee on the Ethics of Animal Experiments of the Universidad Nacional de Rosario (Expedient No. 6060/132; Resolution No. 298/2012). Adult zebrafish were maintained at 28°C on a 14 h light/10 h dark cycle. One-cell embryos were injected and embryos and larvae were staged according to development at 28°C as described elsewhere (24). For TMPyP4/2, 5 nl of each drug were injected in several dilutions in water ranging from 0.5 to 10 μM to determine the highest sub-toxic dose. For assays, 5 nl of TMPyP4/2 in a concentration of 1 μM were injected, and embryos were raised up to 48 h post-fertilization (hpf) stage to perform RT-qPCR. For oligonucleotides, 5 nl of CTRL oligonucleotide (Supplementary Table S1) were injected in several dilutions in KCl 0.1 M ranging from 0.001 to 100 ng/μl in order to determine the highest sub-toxic dose. For assays, 5 nl of 5 ng/μl of each oligonucleotide (Supplementary Table S1) in KCl 0.1 M were injected, and embryos were raised up to the corresponding stage to perform RT-qPCR. For nog3 rescue experiments, zebrafish nog3 capped-mRNA was synthesized using zebrafish nog3 coding sequence cloned in pCS2+ plasmid (25) using SP6 mESSAGE mACHINE SP6 kit (Ambion) following the manufacturer's instructions. Five ng/μl of zebrafish nog3 capped-mRNA were co-injected with the nog3-ASO or CTRL solutions.
RT-qPCR assays
RNA extraction from zebrafish embryos at different stages was followed by reverse transcription and real-time quantitative PCR following previously reported procedures (24). Three biological and technical repeats were performed for each condition, resulting in similar trends. Data from one biological repeat are presented. The validity of the RT-qPCR data was assured by following the MIQE guidelines (26).
Cartilage staining
Four-days post-fertilization (dpf) staged larvae were fixed for 24 h in 4% (w/v) paraformaldehyde (PFA) in phosphate-buffered saline 1X (PBS) containing 0.1% (v/v) Tween-20 and cartilages stained with Alcian Blue as previously described (24).
Whole-mount in situ hybridization (WISH)
Embryos at different stages were fixed overnight in 4% (w/v) PFA in PBS at 4°C. After washing, embryos were stored in methanol at −20°C until use. The procedure for WISH was carried out as previously described (24). Digoxigenin-UTP-labeled riboprobes were synthesized by in vitro transcription using plasmid cloned template cDNAs for col2a1a, krox20 (erg2), myoD, fzd5 (27) and tbx2b (28) genes, or generated by PCR from genomic DNA for nog3 gene (29).
Observation and measurement of embryonic morphologies
Live and fixed/stained embryos and larvae were observed with a MVX10 Stereo Microscope (Olympus) and recorded with a MVXTV1XC digital camera (Olympus). Lengths and angles for morphological features were measured from registered images using ImageJ software (30).
RESULTS
Searching for conserved PQSs in PPRs of vertebrate genes
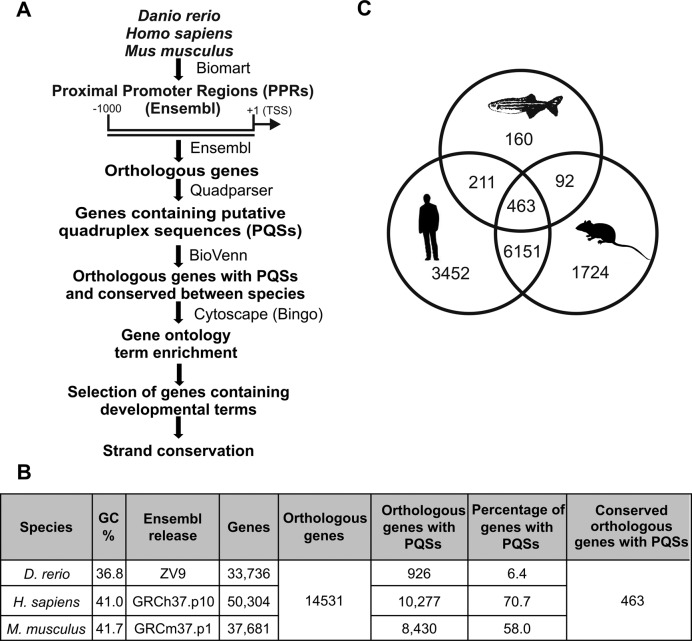
PPRs (herein defined as 1000 bp upstream from the TSS) from Homo sapiens, Mus musculus and Danio rerio genomes were retrieved from ENSEMBL by using the BioMart tool. Orthologous genes were collated to generate a unique list according to the mouse nomenclature (Supplementary Table S2). PPRs from orthologous genes from each species were analyzed searching for at least one PQS by using the Quadparser algorithm (Figure 1A,B; Supplementary Tables S3–S5). No asymmetric pattern of PQS distribution between coding and template DNA-strands was observed. The absolute number and the percentage of PPRs containing PQSs increased from fish to mammals consistently with the increase of GC-percentage observed in the respective genomes (Figure 1B) (31). Then, the three lists of genes were intersected through the web application BioVenn (Figure 1A,C; Supplementary Table S2). Although the number of PPRs containing PQSs in the zebrafish genome is much lower than those found in mouse and human genomes, 50% of them are shared among the three species, 73% are shared with human and 60% with mouse. Similarly, PPRs containing PQSs shared between human and mouse are 64% of the human sequences and 72% of the mouse sequences (Figure 1C). Percentages higher than expected by chance suggest a positive selection of PQSs in PPRs throughout vertebrate evolution as well as a potential role for G-quadruplexes to act as transcriptional control elements. Identified genes encode proteins associated to a variety of biological processes (Supplementary Table S6) according to the BiNGO tool (Figure 1A). GO-Biological Process terms related to development were significantly enriched (Supplementary Table S6), in agreement with previous report (6). Among the 120 genes related to development, 22 were selected since they are involved in different developmental processes (Supplementary Table S6). The 13 genes with conserved PQS-strand location (coding or template) were selected for further studies (Figure 1A and Supplementary Table S6). When more than one PQS was identified in the same PPR, the one presenting similar relative position respect to the TSS in the three species was selected (Supplementary Table S6).
Figure 1.
Selection strategy of conserved PQSs in silico. (A) Schematic representation of conserved PQS selection strategy. (B) Chart representing the number of genes retrieved in each step of the bioinformatic search for the analyzed species. GC contents (%) were obtained from Kai et al. (31). (C) The three lists of genes containing PQSs in their PPRs obtained for the three species were intersected using BioVenn software.
Testing the formation of G-quadruplex in selected PQSs in vitro
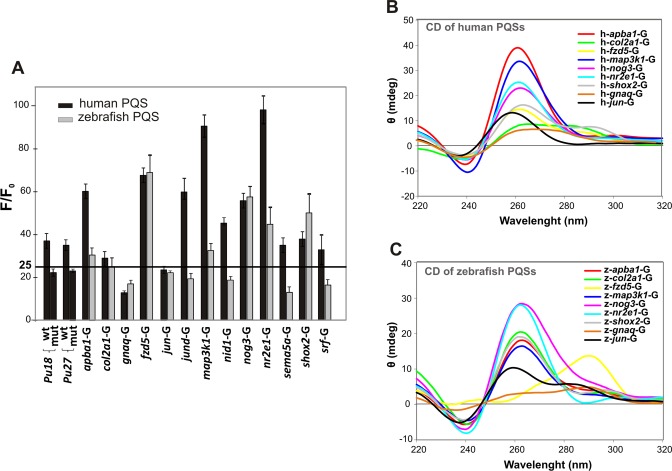
Synthetic single-stranded oligodeoxyribonucleotide sequences representing the human and zebrafish PQSs of the 13 selected genes (Supplementary Table S1) were used to assess in vitro whether they fold as G-quadruplexes. First, we tested G-quadruplexes folding in an assay using ThT. This benzothiazole dye becomes fluorescent in the presence of the G-quadruplex structure but not in presence of duplex or single-stranded DNA (20). Those PQSs from human and zebrafish that increased the fluorescence of ThT by at least 25-times (Figure 2A) were selected for further CD spectroscopy analyses (32). CDs performed on PQSs folded in the presence of 100 mM K+ showed a sharp positive peak around 262 nm and a negative peak around 240 nm for most PQSs (Figure 2B,C; and Supplementary Figure S1), indicative of parallel topologies of G-quadruplexes. The oligonucleotide sequence representing the PQS from zebrafish frizzled-5 (fzd5) displayed a CD spectrum exhibiting a maximum around 295 nm (Figure 2B,C; and Supplementary Figure S1), indicating an antiparallel topology. Results show that seven genes from human and zebrafish contain sequences capable of folding as G-quadruplex in their PPRs.
Figure 2.
In vitro analysis of G-quadruplex formation by ThT assays and circular dicroism. (A) Bar graph of fluorescence enhancement (F/F0) of ThT in the presence of the different oligonucleotides representing human and zebrafish selected PQSs. Each bar represents the mean of three technical repeats and error bars correspond to standard deviation (SD). (B) CD spectra of oligonucleotides representing human PQSs of the seven selected genes positive for ThT assay (apba1, col2a1, fzd5, map3k1, nog3, nr2e1 and shox2) and two control genes negative for ThT assay (jun and gnaq). (C) CD spectrum of oligonucleotides representing zebrafish PQSs of the seven selected genes positive for ThT assay (apba1, col2a1, fzd5, map3k1, nog3, nr2e1 and shox2) and two control genes negative for ThT assay (jun and gnaq).
Selection of genes for studying the role of G-quadruplex in cellulo and in vivo
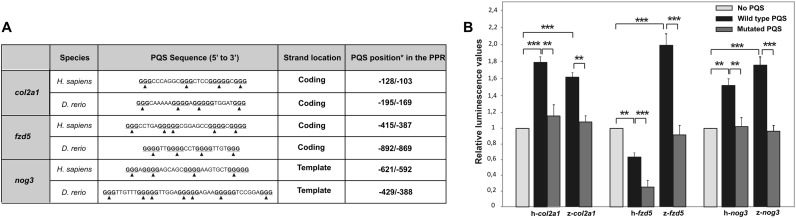
Among the seven potential key genes, we have selected noggin 3 (nog3), collagen type II alpha 1 (col2a1) and fzd5 for further studies because: (i) their PQSs increased by at least 25-times the fluorescence of ThT; (ii) in the case of zebrafish, PQSs present in col2a1 and nog3 PPRs fold as parallel while the PQS in the fzd5 PPR fold as antiparallel G-quadruplex (Figure 2B); (iii) PQSs in col2a1 and fzd5 PPRs locate to the coding DNA-strand whereas PQS in nog3 PPR locates to the template DNA-strand (Supplementary Table S6); and (iv) changes in the abundance of their transcripts during embryonic development lead to well-defined and non-fully overlapping phenotypes (25,33–35). Sequences, strand locations and relative positions of the human and zebrafish PQSs for the three selected genes are summarized in Figure 3A.
Figure 3.
Col2a1, fzd5 and nog3 PQSs and their role in transcriptional expression control in cellulo. (A) Chart presenting the sequences, strand locations and relative positions for the PQSs present in the PPRs of the three selected human and zebrafish genes. G-tracts are bold and underlined. Arrowheads point the G to A replacements in mutated PQSs. *Relative positions are considering TSS as +1. (B) Luciferase assay performed in Neuro-2a cells transfected with pGL3-promoter vector plasmid (no PQS) or pGL3-promoter vector plasmid containing the wild type (wild-type PQS) or mutated (mutated PQS) sequence of human or zebrafish PQSs of the three selected genes upstream the basal promoter SV40. Each bar represents the luciferase activity normalized to β-galactosidase activity and relativized to that for the unmodified pGL3-promoter vector plasmid. Bars represent the mean of three independent experiments and error bars correspond to standard deviation (SD). **P < 0.01, ***P < 0.001, t-Student test.
Role of identified PQSs on transcriptional expression control in cellulo
The in vitro studies presented so far have established that the conserved PQSs located to the PPRs of nog3, col2a1 and fzd5 are capable of forming G-quadruplexes. However, the key question is whether they are critical for regulating the transcriptional activity. To address this, luciferase reporter plasmids were built by cloning the six sequences representing each PQS (three from human PPRs and three from zebrafish PPRs) upstream the basal SV40 promoter in the pGL3 promoter vector. Each PQS was cloned in the same DNA-strand (coding or template) as they are found in genomes. Additional constructs were done replacing guanines with adenines in G-tracts thus impeding G-quadruplex formation (Figure 3A, Supplementary Table S1 and Supplementary Figure S2). Plasmids were transiently transfected in Neuro-2a cells and the luciferase activity was measured as a transcriptional reporter. With the exception of human fzd5-PQS, luciferase expression controlled by PQSs was significantly higher than that detected for the basal SV40 promoter. This result confirms that the analyzed PQSs are able to regulate the activity of the unrelated SV40 promoter. Reinforcing this finding, luciferase expression controlled by mutated PQSs was significantly lower than that controlled by the wild-type ones, and reaching similar levels than the expression controlled by the SV40 promoter. The human fzd5-PQS caused a repression in the transcription of the reporter gene. Yet, the mutated sequence led to luciferase-transcript abundance even lower than that detected for the wild-type sequence (Figure 3B). Results indicate that PQSs present in the PPRs of human and zebrafish nog3, col2a1 and fzd5 genes are able to fold as G-quadruplexes and regulate in cellulo the SV40-dependent transcription.
Role of G-quadruplex in transcriptional expression control in vivo
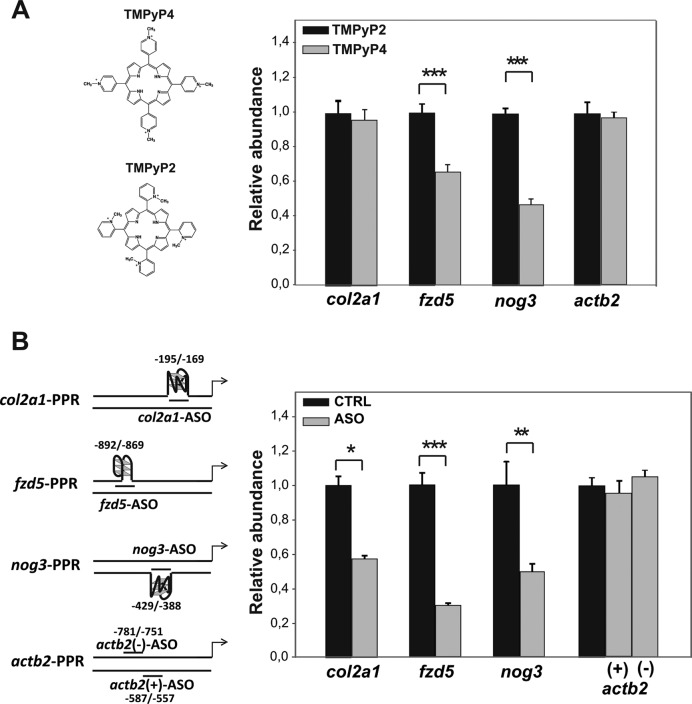
Data generated in cellulo confirmed that G-quadruplexes are able to regulate the activity of the SV40 promoter thus influencing the expression of a reporter gene. However, the real effect of these structures in a cellular and genomic natural context might be different. Thereby, the key question was if G-quadruplexes were really involved in transcriptional regulation in vivo during development. To answer this, we disrupted the G-quadruplex structures by microinjecting small G-quadruplex-targeting ligands in one-cell staged zebrafish embryos and then measuring the relative abundance of transcripts by RT-qPCR. We used the small cationic porphyrin TMPyP4 that has been reported to act as both a stabilizer and a destabilizer of G-quadruplexes depending upon the target (36–39). Although TMPyP4 can interact with both G-quadruplexes and duplex DNA, it has a higher selectivity for the G-quadruplex than for the duplex structure (40). We also used the cationic porphyrin TMPyP2, which is a positional isomer of TMPyP4 and has a lower affinity for G-quadruplexes (41). Compared to TMPyP2, the presence of TMPyP4 did not significantly modify the abundance of col2a1 transcripts at 48-hpf; however, it led to a significant decrease in the abundance of nog3 and fzd5 transcripts measured. The relative abundance of actin beta 2 (actb2) transcripts did not show significant changes, as expected because it is a housekeeping gene homogeneously expressed throughout embryonic development and that does not contain PQSs in its PPR (Figure 4A). The magnitude of CD signals revealed that TMPyP4 destabilized G-quadruplexes when comparing with TMPyP2 (Supplementary Figure S3), suggesting that the transcriptional effect of TMPyP4 was due to an in vivo destabilization of G-quadruplexes. Nevertheless, unspecific or pleiotropic effects of small ligands could not be ruled out because more than 6% of zebrafish genes contain at least one PQS motif near the TSS (Figure 1B). Thereby, a more specific experimental approach consisting of microinjecting ASOs complementary to the selected PQSs was performed. In previous report, ASOs had been used in zebrafish embryos for in vivo testing the role of the human c-MYC G-quadruplex in controlling the expression of luciferase (42). Worth noticing, these data were generated by microinjecting plasmid DNA containing the targeted G-quadruplex and might not reflect what is really happening on promoters of endogenous genes. In the present study, in addition to the ASOs complementary to the selected PQSs (Supplementary Figure S4A), ASOs complementary to either the template or the coding strands of the actb2 PPR were designed to be used as specificity controls. An oligonucleotide that does not anneal to the zebrafish genome was used as unspecific control (CTRL, Supplementary Table S1). The effect of ASOs on the G-quadruplexes - duplex DNA equilibrium was tested in vitro by PAGE and ThT assays. Duplex DNA was the predominant form observed in PAGE when ASOs were present during or posterior to the G-quadruplexes folding (Supplementary Figure S4B). Data from ThT assays confirmed these observations (Supplementary Figure S4C). In agreement with Triplexator software predictions (43), duplex DNA containing the PQSs incubated in the presence of ASOs did not form triplex DNA structures (Supplementary Figure S4D). Thereby, results show that ASOs interfere the G-quadruplex folding by forming duplex- but not triplex-DNA structures with PQSs.
Figure 4.
Disruption of G-quadruplexes by G-quadruplex ligands and ASO injection affects transcriptional expression in zebrafish embryos. (A) Relative abundance of transcripts of col2a1, fzd5, nog3 and the control actb2 genes measured by RT-qPCR in 48-hpf embryos injected with the drugs TMPyP2 and TMPyP4. (B) Relative abundance of transcripts of col2a1, fzd5, nog3 and the control actb2 genes measured by RT-qPCR in 90% epiboly, 30-hpf and 48-hpf staged embryos, respectively, injected with the ASO complementary to the corresponding PQS. In the case of actb2 gene, two ASOs were used, one complementary to the template (actb2(+)-ASO) and the other to the coding strand (actb2(-)-ASO). In all cases, three biological and technical repeats were performed for each condition, resulting in similar trends. Bars represent the mean of the three technical repeats for one representative biological repeat. Error bars correspond to standard deviation (SD) of the three technical repeats. *P < 0,05, **P < 0.01, ***P < 0.001, t-Student test.
Then, the toxicity of ASOs was defined in vivo by injecting different concentrations of CTRL in one-cell staged zebrafish embryos and assessing the survival of specimens until 48-hpf. The higher sub-toxic dose was defined as 25 pg-ASO/embryo (i.e. 5 nl of 5 ng/μl; Supplementary Figure S5). The relative abundance of nog3, col2a1 and fdz5 transcripts in ASO-injected embryos was measured by RT-qPCR at those developmental stages in which either a sharp increment of the transcription (Supplementary Figure S6) or well-defined knock-down phenotypes were detected (25,33). Compared to CTRL injected embryos, the presence of specific ASOs led to a significant decrease in the relative abundance of nog3, col2a1 and fdz5 but not of actb2 transcripts (Figure 4B), ruling out pleiotropic and unspecific effects due to the toxicity of oligonucleotides. In addition, the absence of transcriptional effect in the presence of actb2-ASOs indicated that changes in transcripts abundances were due to the G-quadruplexes disruption and not solely to the annealing of ASOs with PPRs. Overall, results indicate that G-quadruplexes interference reduces in vivo the transcription of the three genes under study.
G-quadruplex disruption mimics the effect of col2a1, fdz5 and nog3 knock-down during zebrafish embryonic development
Next, we wondered whether the impediment of G-quadruplex formation had any effect on the patterning of embryonic developmental structures. To answer this issue, we compared the phenotypes and the spatiotemporal expression patterns generated in the presence of ASOs with those ones reported when the expression of the three genes under study had been knocked-down by specific Morpholinos (MO).
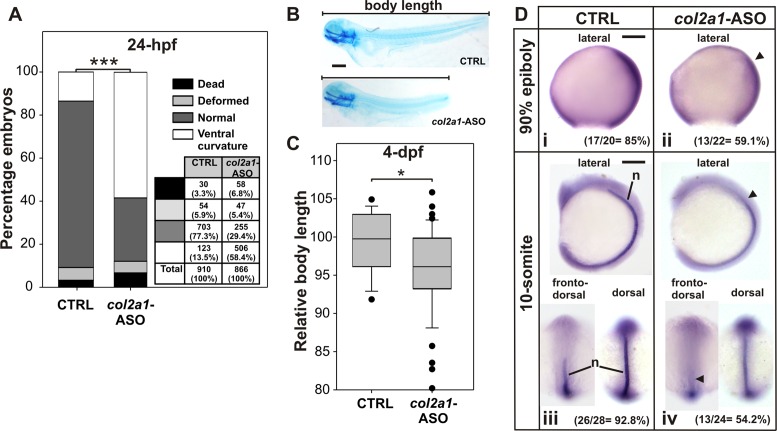
Collagen II, coded by the col2a1 gene, is the major fibril-forming collagen in cartilage. Complete absence of collagen II in mice is not compatible with life and mutations in the human's COL2A1 gene lead to osteochondrodysplasias with diverse phenotypes, from prenatally lethal and short stature to relatively mild defects that may be apparent only in adulthood (44). In zebrafish, col2a1-MO-knocking-down leads to ventrally curved organisms (34) displaying aberrant notochord morphology (35). Compared to CTRL injected embryos, the presence of col2a1-ASO did not cause a noticeable effect on the percentage of dead and deformed embryos but led to a significant increase in the amount of 24-hpf staged embryos showing a ventral curvature phenotype (Figure 5A) and high number of 4-dpf staged larvae displaying significant shortening in the body length (Figure 5B,C). The presence of col2a1-ASO did not significantly affect the craniofacial cartilage development (Supplementary Figure S7A). WISH revealed that col2a1-ASO injection produced slightly lower abundance of col2a1-mRNA at 90% epiboly and a thinner and anteriorly shorter notochord at 10-somite staged embryos (Figure 5D), which may explain the body shortening detected among larvae. No differences in the expression pattern of other developmental marker genes were observed (Supplementary Figure S7B), ruling out unspecific effects of the col2a1-ASO injection.
Figure 5.
Effect of col2a1 G-quadruplex disruption by col2a1-ASO injection in zebrafish embryos. (A) Numbers and percentages of embryos showing dead, deformed, normal or ventral curvature phenotypes are shown in a table and represented in a stacked bar graph for 24-hpf staged embryos injected with CTRL or col2a1-ASO. ***P < 0.001, chi-square test. (B) Representative picture of 4-dpf staged larvae injected with CTRL or col2a1-ASO and stained with Alcian Blue to determine the body length. Lateral views, anterior to the left. (C) Box-plot of the relative body length of 4-dpf staged larvae injected with CTRL or col2a1-ASO. *P < 0.05, t-Student test. (D) WISH assessing the expression of col2a1 mRNA in 90% epiboly (i and ii) and 10-somite (iii and iv) staged embryos injected with CTRL (i and iii) or col2a1-ASO (ii and iv). Arrowheads point regions of lower expression. In lateral views anterior is to the left, and in dorsal and fronto-dorsal views anterior is up. Numbers and percentages of embryos/larvae with the shown phenotype are indicated in each panel. n: notochord. Scale bars (200 μm) are represented in B, Di and Diii.
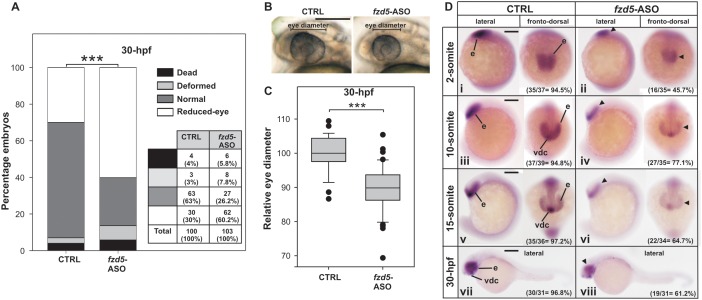
Fzd5 expression is highly restricted to eye, forebrain and gut in zebrafish developing embryos (27) and its knocking-down causes a reduction of the eye field (33). The microinjection of fzd5-ASO did not cause a noticeable effect on the percentage of dead and deformed embryos when comparing to controls but led to a significant increase in the number of embryos showing a reduced-eye phenotype (Figure 6A) and a statistically significant reduction in the eye diameter (Figure 6B,C). In addition, WISH revealed lower fzd5 expression in the eye territory mainly from 10-somite staged embryos onward (Figure 6D). No difference in expression pattern of a typical developmental marker gene was detected (Supplementary Figure S8), ruling out unspecific effects of the fzd5-ASO injection.
Figure 6.
Effect of fzd5 G-quadruplex disruption by fzd5-ASO injection in zebrafish embryos. (A) Numbers and percentages of embryos showing dead, deformed, normal or reduced-eye phenotypes are shown in a table and represented in a stacked bar graph for 30-hpf staged embryos injected with CTRL or fzd5-ASO. ***P < 0.001, chi-square test. (B) Representative picture of 30-hpf staged embryos injected with CTRL or fzd5-ASO used to determine eye diameter. Lateral views, anterior to the left. (C) Box-plot of the relative eye diameter of 30-hpf staged embryos injected with CTRL or fzd5-ASO. ***P < 0.001, t-Student test. (D) WISH assessing the expression of fzd5 mRNA in 2-somite (i and ii), 10-somite (iii and iv), 15-somite (v and vi) and 30-hpf (vii and viii) staged embryos injected with CTRL (i, iii, v and vii) or fzd5-ASO (ii, iv, vi and viii). Arrowheads point regions of lower expression. In lateral views anterior is to the left, and in fronto-dorsal views anterior is up. Numbers and percentages of embryos/larvae with the shown phenotype are indicated in each panel. e: eye; vdc: ventral diencephalon. Scale bars (200 μm) are represented in B, Di, Diii, Dv and Dvii.
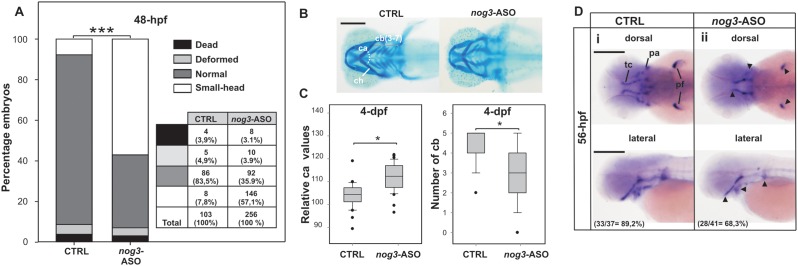
Nog3 seems to play an important role in chondrogenic progenitor survival during zebrafish pharyngeal development since its knock-down led to ventral pharyngeal cartilages severely reduced in size (25). The microinjection of nog3-ASO did not cause a noticeable effect on the percentage of dead and deformed embryos when comparing to controls, but led to a significant increase in the number of embryos showing a small-head phenotype (Figure 7A). Statistically significant changes in the angle formed by ceratohial cartilages and in the number of ceratobranchial cartilages were detected by Alcian Blue staining (Figure 7B,C). Defects were specifically caused by nog3-ASO since they were fully rescued by nog3-mRNA co-injection (Supplementary Figure S9). WISH assessing the expression of the marker of differentiated chondrocytes col2a1 in nog3-ASO injected embryos showed shorter trabeculae cranii development and a compression of pharyngeal arches while no significant changes were observed in the otic vesicle expression territory (Supplementary Figure S10). Besides, WISH revealed lower expression of nog3 in pharyngeal arches and pectoral fins and in a shortened trabeculae cranii (Figure 7D). No significant changes in the expression of col2a1 in the otic vesicle territory along with rescue data point out the specificity of nog3-ASO. In agreement with the results described above, embryos injected with both actb2-ASOs did not produced evident developmental phenotypes (Supplementary Figure S11).
Figure 7.
Effect of nog3 G-quadruplex disruption by nog3-ASO injection in zebrafish embryos. (A) Numbers and percentages of embryos showing dead, deformed, normal or small head phenotypes are shown in a table and represented in a stacked bar graph for 48-hpf staged embryos injected with CTRL or nog3-ASO. ***P < 0.001, chi-square test. (B) Representative picture of Alcian Blue staining of craniofacial cartilages in 4-dpf staged larvae injected with CTRL or nog3-ASO. Ventral views, anterior to the left. (C) Box-plots of the relative ceratohyal cartilages angle and ceratobranchial cartilages number of 4-dpf staged larvae injected with CTRL or nog3-ASO. *P < 0.05, t-Student test. (D) WISH assessing the expression of nog3 mRNA in 56-hpf staged larvae injected with CTRL (i) or nog3-ASO (ii). Arrowheads point regions of lower expression. In lateral and dorsal views anterior is to the left. Numbers and percentages of embryos/larvae with the shown phenotype are indicated in each panel. ca: ceratohyal cartilages angle; cb (3-7): ceratobranchial cartilages 3 to 7; ch: ceratohyal cartilage; pa: pharyngeal arches; pf: pectoral fin; tc: trabeculae cranii. Scale bars (200 μm) are represented in B and Di.
Collectively, data presented here show that conserved G-quadruplexes present in the PPRs of developmentally related genes play transcriptional regulatory roles and are required to achieve the proper development of embryonic structures.
DISCUSSION
In this work we presented results supporting G-quadruplexes as additional members of the intricate array of cis-regulatory sequences responsible for the transcriptional control during embryonic development. Enrichment of genes related to developmental processes among those containing conserved PQSs in their PPRs suggests a functional selection of G-quadruplexes likely participating in accurate transcriptional regulation throughout development. Although this analysis was performed in zebrafish, the vast majority of candidate sequences are conserved between fish and mammals. Therefore, it is likely that the transcriptional control by G-quadruplexes characterized in fish also exists in other vertebrate systems. As mammalian PPRs contain higher number of PQSs due to a genome GC-enrichment along evolution, it is tempting to speculate that the contribution of G-quadruplexes to the transcription of mammalian developmentally related genes is even more relevant than in zebrafish. In this work we have only assessed the 1000 bp upstream from TSS region of a set of developmentally related genes. Because G-quadruplexes may also regulate transcription at long distances from the TSS (45–47), the contribution of this non-canonical form of DNA to developmental transcriptional control may be even higher than that shown in this work. Although the zebrafish genome possesses a relatively scant number of PQSs within PPRs, most of them are conserved in mouse and human orthologous genes turning zebrafish into a good model for studying the role of G-quadruplexes in different developmental pathways, as well as other cellular processes.
Several studies performed in silico or in cellulo using reporter constructs suggest that G-quadruplexes act as transcriptional repressors by impeding transcription factor binding to duplex-DNA or stalling the progression of RNA polymerase, mostly when they are located downstream the TSS in the template strand (3,12,48,49). Conversely, it was also shown that G-quadruplexes may enhance the transcription of particular genes by favoring the binding of specific transcription factors (3,42,49,50) or by holding the DNA molecule open thus facilitating the re-initiation of transcription (3,49,51). With the exception of human fdz5-PQS, the experimental evidences gathered in this work indicate that G-quadruplexes located upstream the TSS of col2a1, fzd5 and nog3 enhance the transcription in cellulo. This enhancer effect would be due to the recruitment of specific activators on folded G-quadruplexes, the removal of a repressive process, and/or the fact that G-quadruplex holds the DNA molecule open thus facilitating the re-initiation of transcription. Regarding the human fzd5-PQS, we speculate that the inhibitory effect of the G-quadruplex on luciferase transcription might be due to the recruitment/binding of transcription suppressors or to an impediment in the correct transcriptional machinery assembly. In cellulo experiments also demonstrated that all of mutated sequences impairing G-quadruplex formation diminished the expression of luciferase. In the particular case of human fdz5-PQS, mutations might generate novel cis-repressor elements in the duplex-DNA that in turn recruit suppressors causing an even greater decrease in transcription. Collectively, the transcriptional effect was observed regardless of whether the tested sequences represented PQSs found in either human or zebrafish genomes, were located to the coding or template strand, or adopted parallel or antiparallel G-quadruplex topologies. Even more, as human and zebrafish PQSs are not conserved in primary sequence and luciferase was measured in Neuro2a cells (i.e. in a mouse cellular context), results strongly suggest that the transcriptional diminishing was actually due to deficiencies in G-quadruplexes folding and not to environmental or pleiotropic effects.
The ASO strategy, which had been successfully employed for assessing the transcriptional and translational role of G-quadruplex in vivo (42,52), led to reduced transcription of the three analyzed genes along with phenotypes similar to those ones reported when the knocking-down had been performed by other experimental strategies. The sole presence of an ASO complementary to the PQS within the PPR of a particular gene generated phenotypes greatly related with its developmental function. This fact minimizes the chances that the obtained results were due to nonspecific or pleiotropic effects. Our results suggest that G-quadruplexes favor the transcription of zebrafish nog3, col2a1 and fzd5 likely by recruiting activators or impeding the binding of suppressors to the duplex-DNA, as well as by facilitating transcription re-initiation. Altogether, data support a role of G-quadruplexes in the transcriptional regulation of developmentally related genes and highlight ASO-strategy as a tool to get insight into the role of G-quadruplex during the embryonic development of complex living organisms.
Adjustments in the fine-tuning of gene expression controlling developmental processes may lead to phenotypic variations among healthy individuals, as well as stronger alterations leading to disease. A recent work has reported the existence of G-quadruplex polymorphisms affecting G-tracts or even loop-regions, which may result in significant changes in gene expression among individuals (53). In this context, it is tempting to speculate that G-quadruplex polymorphisms might contribute to the fine-tuning of the transcriptional regulation of critical developmental genes, with potential biological or even pathological consequences. If so, our knowledge and ability to manipulate the folding of the G-quadruplexes could become a powerful tool for preventing and treating diseases derived from aberrant embryonic developmental processes.
To our knowledge, this is the first work showing in vivo the direct role of evolutionarily conserved G-quadruplexes in one of the most strictly regulated biological process of vertebrates; i.e. the embryonic development.
Supplementary Material
Acknowledgments
A.P.D. and P.D. are fellows of CONICET. E.M., C.B., P.A. and N.B.C. are staff members of CONICET and UNR. We are thankful to Dr Dario Krapf for critical reading of the manuscript and to Sebastian Graziati for excellent fish husbandry. We thank Dr Kelsh and Dr Ning for providing the plasmid containing fzd5 and nog3 cDNA, respectively. We also thank Stephanie Cariker for the language correction of this manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Agencia Nacional de Promoción Científica y Tecnológica [PICT 2014-0741 to P.A.]; Consejo Nacional de Investigaciones Científicas y Técnicas [PIP 00773 to N.B.C.]. Funding for open access charge: Agencia Nacional de Promoción Científica y Tecnológica [PICT 2014-0741 to P.A.]; Consejo Nacional de Investigaciones Científicas y Técnicas [PIP 00773 to N.B.C.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Huppert J.L., Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murat P., Balasubramanian S. Existence and consequences of G-quadruplex structures in DNA. Curr. Opin. Genet. Dev. 2014;25:22–29. doi: 10.1016/j.gde.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 3.Bochman M.L., Paeschke K., Zakian V.A. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012;13:770–780. doi: 10.1038/nrg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Capra J.A., Paeschke K., Singh M., Zakian V.A. G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae. PLoS Comput. Biol. 2010;6:e1000861. doi: 10.1371/journal.pcbi.1000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verma A., Halder K., Halder R., Yadav V.K., Rawal P., Thakur R.K., Mohd F., Sharma A., Chowdhury S. Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J. Med. Chem. 2008;51:5641–5649. doi: 10.1021/jm800448a. [DOI] [PubMed] [Google Scholar]
- 6.Huppert J.L., Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sen D., Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 8.Biffi G., Tannahill D., McCafferty J., Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balasubramanian S., Neidle S. G-quadruplex nucleic acids as therapeutic targets. Curr. Opin. Chem. Biol. 2009;13:345–353. doi: 10.1016/j.cbpa.2009.04.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agarwal T., Lalwani M.K., Kumar S., Roy S., Chakraborty T.K., Sivasubbu S., Maiti S. Morphological effects of G-quadruplex stabilization using a small molecule in zebrafish. Biochemistry. 2014;53:1117–1124. doi: 10.1021/bi4009352. [DOI] [PubMed] [Google Scholar]
- 11.Zhao Y., Du Z., Li N. Extensive selection for the enrichment of G4 DNA motifs in transcriptional regulatory regions of warm blooded animals. FEBS Lett. 2007;581:1951–1956. doi: 10.1016/j.febslet.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 12.Maizels N. G4-associated human diseases. EMBO Rep. 2015;16:910–922. doi: 10.15252/embr.201540607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolpert L. Do we understand development. Science. 1994;266:571–572. doi: 10.1126/science.7939707. [DOI] [PubMed] [Google Scholar]
- 14.Misteli T. Self-organization in the genome. Proc. Natl. Acad. Sci. U.S.A. 2009;106:6885–6886. doi: 10.1073/pnas.0902010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajapakse I., Perlman M.D., Scalzo D., Kooperberg C., Groudine M., Kosak S.T. The emergence of lineage-specific chromosomal topologies from coordinate gene regulation. Proc. Natl. Acad. Sci. U.S.A. 2009;106:6679–6684. doi: 10.1073/pnas.0900986106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinsella R.J., Kahari A., Haider S., Zamora J., Proctor G., Spudich G., Almeida-King J., Staines D., Derwent P., Kerhornou A., et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database (Oxford) 2011;2011:bar030. doi: 10.1093/database/bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hulsen T., de Vlieg J., Alkema W. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kikin O., D'Antonio L., Bagga P.S. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006;34:W676–W682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Renaud de la Faverie A., Guedin A., Bedrat A., Yatsunyk L.A., Mergny J.L. Thioflavin T as a fluorescence light-up probe for G4 formation. Nucleic Acids Res. 2014;42:e65. doi: 10.1093/nar/gku111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagesh N., Sharma V.K., Ganesh Kumar A., Lewis E.A. Effect of Ionic Strength on Porphyrin Drugs Interaction with Quadruplex DNA Formed by the Promoter Region of C-myc and Bcl2 Oncogenes. J. Nucleic Acids. 2010;2010 doi: 10.4061/2010/146418. 146418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seenisamy J., Rezler E.M., Powell T.J., Tye D., Gokhale V., Joshi C.S., Siddiqui-Jain A., Hurley L.H. The dynamic character of the G-quadruplex element in the c-MYC promoter and modification by TMPyP4. J. Am. Chem. Soc. 2004;126:8702–8709. doi: 10.1021/ja040022b. [DOI] [PubMed] [Google Scholar]
- 23.Domizi P., Aoyama C., Banchio C. Choline kinase alpha expression during RA-induced neuronal differentiation: role of C/EBPbeta. Biochim. Biophys. Acta. 2014;1841:544–551. doi: 10.1016/j.bbalip.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 24.Margarit E., Armas P., Garcia Siburu N., Calcaterra N.B. CNBP modulates the transcription of Wnt signaling pathway components. Biochim. Biophys. Acta. 2014;1839:1151–1160. doi: 10.1016/j.bbagrm.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 25.Ning G., Liu X., Dai M., Meng A., Wang Q. MicroRNA-92a upholds Bmp signaling by targeting noggin3 during pharyngeal cartilage formation. Dev. Cell. 2013;24:283–295. doi: 10.1016/j.devcel.2012.12.016. [DOI] [PubMed] [Google Scholar]
- 26.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 27.Nikaido M., Law E.W., Kelsh R.N. A systematic survey of expression and function of zebrafish frizzled genes. PLoS One. 2013;8:e54833. doi: 10.1371/journal.pone.0054833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dheen T., Sleptsova-Friedrich I., Xu Y., Clark M., Lehrach H., Gong Z., Korzh V. Zebrafish tbx-c functions during formation of midline structures. Development. 1999;126:2703–2713. doi: 10.1242/dev.126.12.2703. [DOI] [PubMed] [Google Scholar]
- 29.Thisse C., Thisse B. High Throughput Expression Analysis of ZF-Models Consortium Clones. http://zfin.org ZFIN Direct Data Submission. 2005 [Google Scholar]
- 30.Schindelin J., Rueden C.T., Hiner M.C., Eliceiri K.W. The ImageJ ecosystem: An open platform for biomedical image analysis. Mol. Reprod. Dev. 2015;82:518–529. doi: 10.1002/mrd.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kai W., Kikuchi K., Tohari S., Chew A.K., Tay A., Fujiwara A., Hosoya S., Suetake H., Naruse K., Brenner S., et al. Integration of the genetic map and genome assembly of fugu facilitates insights into distinct features of genome evolution in teleosts and mammals. Genome Biol. Evol. 2011;3:424–442. doi: 10.1093/gbe/evr041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Randazzo A., Spada G.P., da Silva M.W. Circular dichroism of quadruplex structures. Top. Curr. Chem. 2013;330:67–86. doi: 10.1007/128_2012_331. [DOI] [PubMed] [Google Scholar]
- 33.Cavodeassi F., Carreira-Barbosa F., Young R.M., Concha M.L., Allende M.L., Houart C., Tada M., Wilson S.W. Early stages of zebrafish eye formation require the coordinated activity of Wnt11, Fz5, and the Wnt/beta-catenin pathway. Neuron. 2005;47:43–56. doi: 10.1016/j.neuron.2005.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mangos S., Lam P.Y., Zhao A., Liu Y., Mudumana S., Vasilyev A., Liu A., Drummond I.A. The ADPKD genes pkd1a/b and pkd2 regulate extracellular matrix formation. Dis. Model. Mech. 2010;3:354–365. doi: 10.1242/dmm.003194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gansner J.M., Mendelsohn B.A., Hultman K.A., Johnson S.L., Gitlin J.D. Essential role of lysyl oxidases in notochord development. Dev. Biol. 2007;307:202–213. doi: 10.1016/j.ydbio.2007.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weitzmann M.N., Woodford K.J., Usdin K. The development and use of a DNA polymerase arrest assay for the evaluation of parameters affecting intrastrand tetraplex formation. J. Biol. Chem. 1996;271:20958–20964. doi: 10.1074/jbc.271.34.20958. [DOI] [PubMed] [Google Scholar]
- 38.Monchaud D., Granzhan A., Saettel N., Guedin A., Mergny J.L., Teulade-Fichou M.P. ‘One ring to bind them all’-part I: the efficiency of the macrocyclic scaffold for g-quadruplex DNA recognition. J. Nucleic Acids. 2010 doi: 10.4061/2010/525862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Del Toro M., Bucek P., Avino A., Jaumot J., Gonzalez C., Eritja R., Gargallo R. Targeting the G-quadruplex-forming region near the P1 promoter in the human BCL-2 gene with the cationic porphyrin TMPyP4 and with the complementary C-rich strand. Biochimie. 2009;91:894–902. doi: 10.1016/j.biochi.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 40.Martino L., Pagano B., Fotticchia I., Neidle S., Giancola C. Shedding light on the interaction between TMPyP4 and human telomeric quadruplexes. J. Phys. Chem. B. 2009;113:14779–14786. doi: 10.1021/jp9066394. [DOI] [PubMed] [Google Scholar]
- 41.Grand C.L., Han H., Munoz R.M., Weitman S., Von Hoff D.D., Hurley L.H., Bearss D.J. The cationic porphyrin TMPyP4 down-regulates c-MYC and human telomerase reverse transcriptase expression and inhibits tumor growth in vivo. Mol. Cancer Ther. 2002;1:565–573. [PubMed] [Google Scholar]
- 42.Kumar N., Patowary A., Sivasubbu S., Petersen M., Maiti S. Silencing c-MYC expression by targeting quadruplex in P1 promoter using locked nucleic acid trap. Biochemistry. 2008;47:13179–13188. doi: 10.1021/bi801064j. [DOI] [PubMed] [Google Scholar]
- 43.Buske F.A., Bauer D.C., Mattick J.S., Bailey T.L. Triplexator: detecting nucleic acid triple helices in genomic and transcriptomic data. Genome Res. 2012;22:1372–1381. doi: 10.1101/gr.130237.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kannu P., Bateman J., Savarirayan R. Clinical phenotypes associated with type II collagen mutations. J. Paediatr. Child Health. 2012;48:E38–E43. doi: 10.1111/j.1440-1754.2010.01979.x. [DOI] [PubMed] [Google Scholar]
- 45.Zhang C., Liu H.H., Zheng K.W., Hao Y.H., Tan Z. DNA G-quadruplex formation in response to remote downstream transcription activity: long-range sensing and signal transducing in DNA double helix. Nucleic Acids Res. 2013;41:7144–7152. doi: 10.1093/nar/gkt443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hegyi H. Enhancer-promoter interaction facilitated by transiently forming G-quadruplexes. Sci. Rep. 2015;5:9165. doi: 10.1038/srep09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Du Z., Zhao Y., Li N. Genome-wide colonization of gene regulatory elements by G4 DNA motifs. Nucleic Acids Res. 2009;37:6784–6798. doi: 10.1093/nar/gkp710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Agarwal T., Roy S., Kumar S., Chakraborty T.K., Maiti S. In the sense of transcription regulation by G-quadruplexes: asymmetric effects in sense and antisense strands. Biochemistry. 2014;53:3711–3718. doi: 10.1021/bi401451q. [DOI] [PubMed] [Google Scholar]
- 49.Smestad J.A., Maher L.J. 3rd. Relationships between putative G-quadruplex-forming sequences, RecQ helicases, and transcription. BMC Med. Genet. 2015;16:91. doi: 10.1186/s12881-015-0236-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uribe D.J., Guo K., Shin Y.J., Sun D. Heterogeneous nuclear ribonucleoprotein K and nucleolin as transcriptional activators of the vascular endothelial growth factor promoter through interaction with secondary DNA structures. Biochemistry. 2011;50:3796–3806. doi: 10.1021/bi101633b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Du Z., Zhao Y., Li N. Genome-wide analysis reveals regulatory role of G4 DNA in gene transcription. Genome Res. 2008;18:233–241. doi: 10.1101/gr.6905408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang K., Donnelly C.J., Haeusler A.R., Grima J.C., Machamer J.B., Steinwald P., Daley E.L., Miller S.J., Cunningham K.M., Vidensky S., et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525:56–61. doi: 10.1038/nature14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baral A., Kumar P., Halder R., Mani P., Yadav V.K., Singh A., Das S.K., Chowdhury S. Quadruplex-single nucleotide polymorphisms (Quad-SNP) influence gene expression difference among individuals. Nucleic Acids Res. 2012;40:3800–3811. doi: 10.1093/nar/gkr1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.