Abstract
It is well established that the correct identification of the messenger RNA targeted by a given microRNA (miRNA) is a difficult problem, and that available methods all suffer from low specificity. We hypothesize that the correct identification of the pairing should take into account the effect of the Argonaute protein (AGO), an essential catalyst of the recognition process. Therefore, we developed a strategy named MiREN for building and scoring three-dimensional models of the ternary complex formed by AGO, a miRNA and 22 nt of a target mRNA that putatively interacts with it. We show here that MiREN can be used to assess the likelihood that an RNA molecule is the target of a given miRNA and that this approach is more accurate than other existing methods, usually based on sequence or sequence-related features. Our results also suggest that AGO plays a relevant role in the selection of the miRNA targets. Our method can represent an additional step for refining predictions made by faster but less accurate classical methods for the identification of miRNA targets.
INTRODUCTION
MicroRNAs (miRNAs) are endogenous non-coding RNAs that control gene expression at the post-transcriptional level. Their mechanism of action involves the binding of mature miRNAs to the Argonaute (AGO) proteins to recruit other components and form the miRNA-induced silencing complex (miRISC) (1). The miRISC complex recognizes complementary sequences in the 3′untranslated regions (3UTR) or in coding regions (2) of target mRNAs, and inhibits protein synthesis, either indirectly, by promoting mRNA decay, or directly by inhibiting the initiation and elongation steps of translation.
The diffusion of powerful ‘omic’ analytical techniques, including microarray, HITS-CLIP/PAR-CLIP and MIR-seq, highlighted the variety of biological processes in which these small molecules are involved, and provided new insights into the mechanisms through which molecular targets are recognized by endogenous miRNAs.
The new miRNA targets discovered by ‘omic’ analyses led to a revision of the rules dictating the mode of recognition of their targets. For example, the requirement, previously considered essential, that a contiguous ‘seed’ (3) region should base pair with nucleotides located at the 5′ end of the miRNA (positions 2–8) has been shown not to be an indispensable requisite. In fact multiple mismatches (4,5) or even lack of complementarity to the seed sequence have been observed in some cases (6). This is true not only for animal organisms but also in plants where evidence of non-canonical miRNA binding modes has been reported (7,8). Similarly, many new experimentally verified miRNA target sites have been found in non-conserved genomic regions suggesting that evolutionary conservation of the sites might be less important than assumed so far (9).
Existing computational methods for the prediction of miRNA target sites generally base their predictions on the search for sequences complementary to a given miRNA and rank the candidate sites according to properties derived from primary and secondary structures and sometimes on evolutionary information.
Some methods, such as MIRANDA (10), estimate the thermodynamic stability of the secondary structure of the putative miRNA-target pair. Differently, PITA (9) predicts the local secondary structure of a region of 140 nt surrounding the hypothetical target site and estimates the energetic gain (if any) in opening this region to bind the miRNA. Other tools, such as TARGETSCAN (11) and PICTAR (12) filter the putative target sites on the basis of their sequence conservation in different organisms.
Regardless of their strategy, a limit of currently available methods resides in their low specificity (13) resulting from our still incomplete understanding of the rules governing target recognition.
Crystallographic studies(14)revealed the nature of the interaction between the AGO protein and a duplex formed by a miRNA (mimicked by a DNA molecule in the crystal) and its target, as well as the specific structural requirements for mRNA cleavage. The AGO protein structure can be divided into two lobes, one containing the PAZ domain and the other including the middle (MID) domain and the PIWI domain connected by a linker region. The crystal structure of the complex shows that the miRNA nucleotides from 18 to 20 inserted into the PAZ domain of AGO cannot be base-paired with the mRNA molecule. Moreover the presence of single nucleotide bulges opposite to positions 4–5 and 5–6 of the miRNA strand 5′-seed segment has minimal effect on cleavage (15–17), while cleavage is strongly reduced after insertion of dual bulges at the same positions.
In contrast to the miRNA strand, the sugar-phosphate backbone of the target strand complementary to the seed segment, is able to accommodate single nucleotide and dinucleotide bulges, without impairing the target RNA cleavage and reinforcing the evidence that helical imperfections that disrupt pairing interactions in the 3′ region of the guide strand have a minimal effect on cleavage activity. This suggests that AGO could exert an effect in the selection of targets sites that can more easily adopt specific three-dimensional (3D) conformations compatibles with its binding site.
Recently, Gan et al. (18) explored the possibility of using 3D RNA modeling to predict the structure of known miRNA–target complexes. With their approach, they were able to predict the conformation of two constructs of Caenorhabditis elegans let-7 miRNA target duplexes with an error of 3.8 Angstrom RMSD with respect to the experimental nuclear magnetic resonance structures. These authors also built 3D models of eight miRNA–target pairs bound to AGO with and without single nucleotides mismatches. The comparison of the estimated energies of models with experimental results from titration calorimetric measurements (19) highlighted a good agreement between the computed and measured data.
This notwithstanding, a method that estimates the likelihood of a target being recognized by a miRISC complex taking into account the structural properties of the AGO binding site still does not exists.
In this work, we assess whether a given miRNA–mRNA pair is likely to be functional taking advantage of a strategy, named MiREN, whereby we model the ternary complex of the AGO, the miRNA and the 22 nt of the putative target mRNA and use the estimated energy of the interaction as a scoring function.
The results obtained with our strategy compare favorably with those obtained by the most widely used prediction methods (20) when applied to a human dataset, constituted by experimentally validated positive and negative miRNA and mRNA pairs, and to a rather challenging dataset from Mus musculus (MM) including data extracted from a HITS-CLIP experiment on cells overexpressing a single miRNA.
MATERIALS AND METHODS
Datasets
Our method was applied on two independent and non-overlapping datasets of miRNA–target interactions for which experimental evidence in Homo sapiens (HS) and MM exists.
The HS dataset was built starting from experimentally validated miRNA target pairs annotated in mirtarbase release 3.5. We initially selected a total of 234 interactions (117 randomly extracted from those annotated as functional and 117 among those annotated as non-functional). Each example was further analyzed by reviewing the literature. We found and discarded 16 cases annotated as non functional in mirtarbase that were discovered to be functional according to the literature. The final dataset consists of 133 functional pairs and 101 non-functional pairs. For each target gene, we retrieved the mRNA RefSeq sequence with the longest 3′ UTR and identified all the sites the sequence of which was complementary to a given miRNA using MIRANDA with default parameters. Among the sites proposed by MIRANDA, the one with the maximum number of paired bases was selected.
The MM dataset was based on data provided in the study of Loeb et al. (21). We extracted the data from a HITS-CLIP experiment performed on mouse CD4+ activated T cells over-expressing mmu-miR-155 with respect to sites detected in T cells from mouse knockout for mmu-miR-155. In their work the authors provide the complete sequences of the 3′ UTR transcriptome used to map the HITS CLIP peaks together with 97 sequences that are differentially expressed between the two systems and harbor a mmu-miR-155 canonical seed binding sequence. The 22-nt long regions complementary to the seed region of miR-155 present in the transcripts formed our MMPos dataset. The negative counterpart (MMneg) dataset was built using 97 sequences (22 nt long) containing the miR-155 seed sequence in the 3′ UTR showing no difference between the wild-type and knock out mouse samples. A list of all the miRNA–target pairs included in analyzed datasets is provided in the Supplementary Information.
The secondary structures of the sites in the positive and negative datasets were predicted using MIRANDA (10). The PITA scores were computed by running PITA (11) with default parameters. The TARGETSCAN scores were retrieved from the TARGETSCAN 6.2 database (12). The score was set to 0 if the modeled site was not detected by TARGETSCAN in the corresponding RefSeq sequence.
Model building
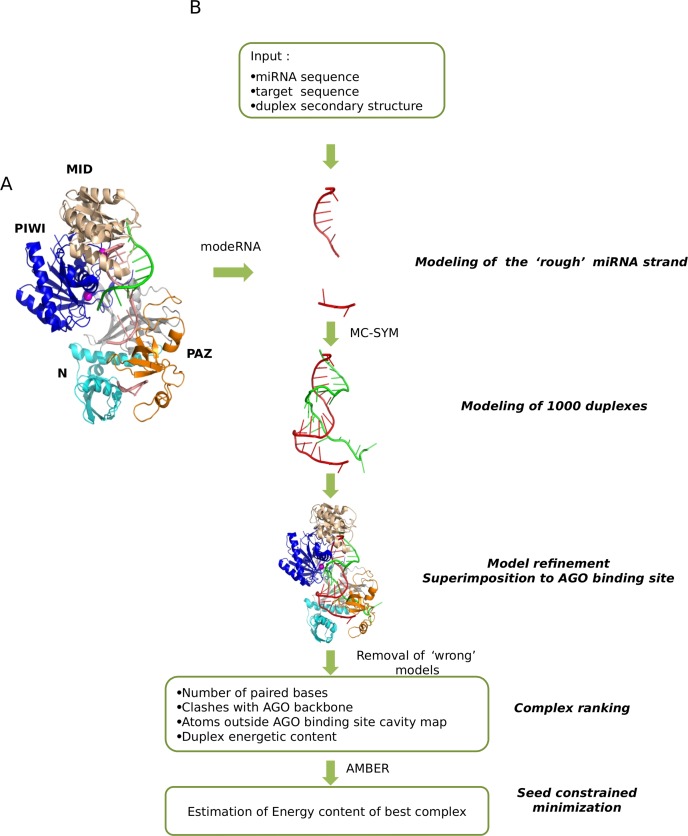
We used the Thermus thermophilus AGO protein structure (PDB code: 3F73) that is considered an appropriate model for the eukaryotic members of the AGO family given its structural and functional similarities with the latter (22,23). This structure corresponds to the active form of AGO complexed with a duplex of a guide DNA (meant to mimic the miRNA) bound to its target. In the crystal, the bases of the guide DNA can be traced for the 1–10 segment, in which the 5′-phosphate is inserted into the binding pocket of the Mid domain of AGO and for the 19–21 segment, in which nucleotides 20 and 21 at the 3′ end are inserted into the binding pocket of the AGO PAZ domain (Figure 1).
Figure 1.
(A) Computational strategy for building the three-dimensional (3D) models of the miRNA–target in complex with the AGO protein. The method consists of five steps. The initial input is the secondary structure and sequence of a miRNA paired with its putative target site (B). The seed nucleotides of the miRNA are modeled using as a template the DNA (that mimics the miRNA) bound to its target, present in the crystallographic structure of Thermus thermophilus AGO. The remaining part of the structure is modeled by 3D RNA fragments assembly. The best model is minimized and its estimated energy recorded.
RNA models were built and analyzed by a two-step procedure. Both steps are performed in a semi automated way through Perl scripts.
In the first step, the modeRNA (24) software is used to model nucleotides 1–9 and 19–21 of the miRNA using as template the guide DNA crystallized in the 3F73 structure. The remaining nucleotides are modeled using MC-SYM (25). This latter tool builds models by assembling RNA di- and tetra-nucleotides 3D structures (NCMs) specified as input. We generated an appropriate input using an ad hoc developed Perl scripts. Multiple canonical Watson–Crick paired bases were assembled according to their secondary structure by selecting 2:2 double strand tetra-nucleotide NCMs; bulges inserted in the RNA duplex were modeled by assembling double strand NCMs including a bulge; unpaired or bulged regions longer than three nucleotides were assembled combining single strand NCMs. All the MC-SYM parameters were kept to their default values except for relaxing the penalty for clashes between non bonded atoms during the progression of fragments assembly to 0.9 (clash threshold parameter), setting the maximum computing time for building 1000 miRNA–target duplex models to two hours and for requesting that all the models differ from each other by an all atom RMSD of at least 1 Angstrom. The time limit threshold was sufficient to obtain 1000 models in more than 90% of the cases.
In order to obtain the correct stereochemistry at the site of junction of consecutive backbone atoms deriving from different fragments we perform a short minimization with AMBER 11 (26) (400 steps in vacuo using the AMBER force field, the LBFGS algorithm and setting the non-bonded cut-off to 14 Å). The computed energies of the RNA duplexes after minimization are one of the components used for model ranking (see later).
The RNA models are superimposed to the RNA moiety in the AGO binding site and discarded if they intertwine with the AGO loops pointing into the binding site. This automatic check is performed using CHIMERA (27). We place 126 pseudo atoms inside the loop pointing into the AGO binding site. If the distance of the phosphorus atom of any nucleotide is shorter than 2.75 Å from one of the 128 pseudo atoms, the model is discarded. We also discard models where the 5′ tail of target is displaced from the cavity located between the PAZ and N-domain, opposite to the PIWI domain. We create a pseudo atom in the centroid of the aforementioned region and measure its distance from the last nucleotide of the target and discard models in which the phosphorous atom of the last target nucleotide is more than 23 Å away from the pseudo atom and less than 35 Angstrom from the Mg++ cleavage site. These distance thresholds are selected after inspection of the distances of the pseudo- and Mg++ atoms from the C alpha of the amino acids that form the AGO cavity.
Ranking of models
We used CHIMERA to estimate the number of duplex paired bases and the number of clashes between the RNA and the protein binding site residues.
To detect models in which a substantial fraction of the mRNA strand is outside the binding site, we computed a molecular map corresponding to the volume occupied by the solvent into the binding site using the 3VV server (28). Subsequently with CHIMERA we fit the models into the molecular map to estimate the number of atoms of the model falling outside the map.
Models are sorted according to the number of duplex paired bases, number of clashes between the RNA and protein moieties, number of atoms outside the AGO binding site and the estimated energy of the RNA duplex.
The best ranking model of the ternary complex is further minimized keeping the seed nucleotides fixed. This is performed in vacuo for 6000 steps with the AMBER software utilizing the AMBER force field, LBFGS algorithm and a non bonded cut-off of 14 Å. The number of steps was selected by analyzing the energy values obtained by running a long minimization of 20 000 steps on a test dataset of 20 models. In all cases, the energy values did not decrease after 6000 steps. The final total energy after minimization is collected and used for subsequent analyses.
RESULTS
Test of the procedure on experimentally validated miRNA target sites
We estimated the probability of a given pair to be functional on the human dataset described in ‘Materials and Methods’ section and composed by 133 human experimentally validated pairs of miRNA and their target sites (HSpos dataset) and 101 human potential miRNA target pairs showing exact complementarity in the seed region but known not to be functional (HSneg dataset). Some of the cases (8 in the positive and 15 in the negative sets, respectively) were discarded at this stage because none of their models were stereochemically plausible (either their atoms overlapped with the AGO backbone or were intertwined with the AGO loops. (See the ‘Model building’ section of ‘Material and Methods’ section for more details).
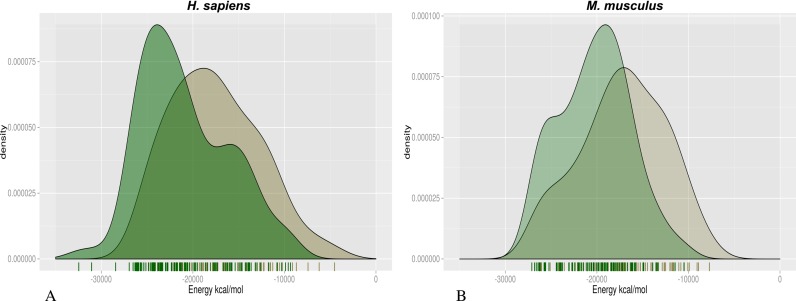
The density plot of the number of the pairs in the HSpos and HSneg datasets as a function of the estimated energy is shown in Figure 2A. It can be seen that models of HSpos pairs have significantly lower estimated energy than models of the HSneg dataset (Wilcoxon test = 3382, P-value = 4.82*10−6).
Figure 2.
Histogram of the estimated complex energy for positive and negative cases in the analyzed datasets. The plot shows a histogram of the number of cases as a function of their estimated energy after 6000 steps of minimization. Positive and negative cases are shown in green and gray, respectively. HS dataset (A) and MM dataset (B). The difference between the negative and positive distributions is statistically significant (Wilcoxon P-value: HS dataset = 4.82*10−6; MM dataset = 1. 2*10−5).
We also tested our procedure on a second, more challenging, dataset (MM) consisting of sites extracted from a HITS-CLIP experiment for mmu-miR-155 as described in ‘Materials and Methods’ section (MMPos) and appropriate controls (MMNeg). It is worth noticing that this latter dataset represents a more stringent test, since the MMPos and MMNeg pairs only differ for the nucleotides of the target downstream the seed region.
In this case, as for the human dataset, we discarded 11 pairs from both the positive and negative set because, none of their models showed plausible stereochemistry. In summary, we modeled 86 sites from the MMpos dataset and 86 from the MMneg dataset (Supplementary Table S1).
For the HS dataset, true positives (TPs) are defined as the miRNA–target pairs for which mirtarbase reports strong evidence of interaction (luciferase assay, western blot) while TPs in the MM dataset correspond to targets for which a HITS-CLIP peak is observed.
The results shown in Figure 2B indicate that, also in this case, the distribution of the estimated energy of the positive sites is lower, on average, than that of negative ones. The difference is statistically significant (Wilcoxon test = 2268, P-value = 1.2*10−5) although the estimated energy distributions overlap more in this case than in the HS dataset.
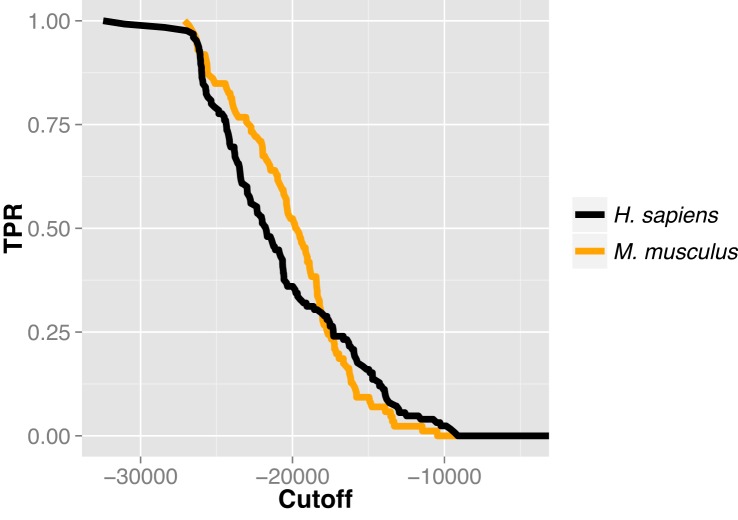
The cumulative distribution of the sensitivity as function of the computed energy in the tested datasets shows that models of miRNA/target duplexes bound into AGO binding site with an estimated complex energy below −25 000 kcal/mol have a 75% probability of being true miRNA targets (Figure 3).
Figure 3.
Distribution of the true positive (TP) rate in function of the estimated energy. The plot shows the distribution of the sensitivity defined as the number of TP cases divided by the total number of positive predicted cases (TPR) as a function of the estimated energy of models.
Comparison with other predictors
We compared the accuracy of our strategy for discriminating positive from negative target sites with that of three widely used methods, namely PITA (11), MIRANDA (10) and TARGETSCAN (12).
PITA computes the difference between the free energy gained from the formation of the miRNA–target duplex and the energetic cost of unpairing a region of 70 nt upstream and downstream the target to make it accessible to the miRNA. MIRANDA first scans candidate UTRs for a miRNA to find target sites by base complementarity and next estimates the free energy of optimal strand–strand interaction between the miRNA and the target site. TARGETSCAN predicts the biological targets of miRNAs by searching for the presence of conserved 8mer and 7mer sites that match the seed region of each miRNA.
All these methods provide a score, which in our case is the estimated energy of the complex, rather than a binary prediction. In order to be able to compare them, we ordered the predictions by the corresponding scores and considered the following partitions: the Top10, Top20 and Top30 lists include predictions with scores in the top 10, 20 and 30% of the ranked list, the Bottom10, Bottom20 and Bottom30 partitions contain the 10, 20 and 30% predictions with the lowest scores. Clearly, one expects most of the positives (negatives) to be found among the highest (lowest) scoring pairs.
For each of the partitions, we consider TP the positive sites in the Top partitions, false positives (FP) the negative sites incorrectly positioned in the Top partitions. True negatives (TN) are negative sites correctly ranked with the lowest scores (i.e. in the Bottom partitions) and false negatives (FN) the positive sites incorrectly located in the Bottom partitions.
For the HS dataset, MiREN has a better precision than all other methods regardless of the partition considered (Table 1A) and is more accurate in all cases.
Table 1. Comparison of the performance of MiREN with that of widely used methods.
| Partitions | TP | TN | FP | FN | Accuracy | Precision | AUC | |
|---|---|---|---|---|---|---|---|---|
| A HS dataset | ||||||||
| Top10 and Bottom10 | PITA | 14 | 14 | 7 | 7 | 0.67 | 0.67 | 0.72 |
| MIRANDA | 10 | 13 | 11 | 8 | 0.55 | 0.48 | 0.54 | |
| TARGETSCAN | 15 | 14 | 6 | 7 | 0.69 | 0.71 | 0.71 | |
| MiREN | 20 | 15 | 1 | 6 | 0.83 | 0.95 | 0.84 | |
| MiREN ∩ PITA | - | 2 | - | - | - | - | ||
| Top20 and Bottom20 | PITA | 29 | 25 | 13 | 17 | 0.64 | 0.69 | 0.67 |
| MIRANDA | 25 | 20 | 17 | 22 | 0.54 | 0.60 | 0.54 | |
| TARGETSCAN | 26 | 21 | 16 | 21 | 0.56 | 0.62 | 0.62 | |
| MiREN | 34 | 25 | 8 | 17 | 0.70 | 0.81 | 0.78 | |
| MiREN ∩ PITA | 8 | 9 | 3 | 2 | 0.77 | 0.73 | ||
| Top30 and Bottom30 | PITA | 42 | 40 | 21 | 24 | 0.65 | 0.67 | 0.66 |
| MIRANDA | 41 | 30 | 22 | 33 | 0.56 | 0.65 | 0.54 | |
| TARGETSCAN | 40 | 28 | 23 | 35 | 0.54 | 0.63 | 0.57 | |
| MiREN | 51 | 34 | 12 | 29 | 0.67 | 0.81 | 0.73 | |
| MiREN ∩ PITA | 13 | 17 | 7 | 6 | 0.70 | 0.65 | ||
| B MM dataset | ||||||||
| Top10 and Bottom10 | PITA | 13 | 15 | 4 | 2 | 0.82 | 0.76 | 0.81 |
| MIRANDA | 12 | 12 | 5 | 5 | 0.71 | 0.71 | 0.69 | |
| TARGETSCAN | 12 | 12 | 5 | 5 | 0.71 | 0.71 | 0.69 | |
| MiREN | 11 | 15 | 6 | 2 | 0.76 | 0.65 | 0.80 | |
| MiREN ∩ PITA | 1 | 3 | - | - | - | |||
| Top20 and Bottom20 | PITA | 23 | 26 | 11 | 8 | 0.72 | 0.68 | 0.77 |
| MIRANDA | 24 | 21 | 10 | 13 | 0.66 | 0.71 | 0.69 | |
| TARGETSCAN | 22 | 25 | 12 | 9 | 0.69 | 0.65 | 0.70 | |
| MiREN | 23 | 28 | 11 | 6 | 0.75 | 0.68 | 0.77 | |
| MiREN ∩ PITA | 11 | 8 | 1 | 1 | 0.90 | 0.91 | ||
| Top30 and Bottom30 | PITA | 35 | 35 | 17 | 17 | 0.67 | 0.67 | 0.74 |
| MIRANDA | 34 | 30 | 18 | 22 | 0.62 | 0.65 | 0.65 | |
| TARGETSCAN | 33 | 36 | 19 | 16 | 0.66 | 0.63 | 0.69 | |
| MiREN | 34 | 38 | 18 | 14 | 0.69 | 0.65 | 0.74 | |
| MiREN ∩ PITA | 16 | 14 | 3 | 2 | 0.86 | 0.84 | ||
Accuracy is defined as (TP + TN)/(TP + FP + FN + TN) and Precision as TP/(TP + FP). (A) Results on the HS dataset. (B) Results on the MM dataset. The MiREN ∩ PITA rows report targets predicted by both MiREN and PITA in the corresponding partitions. Best performances for each partition are highlighted in bold.
For the MM dataset, MiREN is more accurate in all the partitions excepted for the top 10% and lower 10% of scoring sites for which PITA performs better (Table 1B).
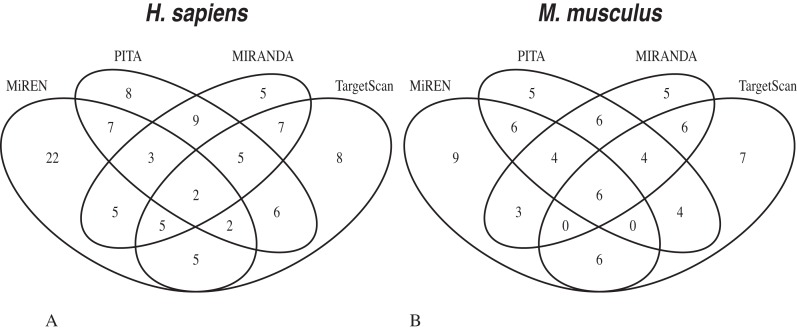
Interestingly, our method correctly predicts a substantial number of TPs that are not predicted by the other methods (Figure 4). For the 30% partition, our method correctly predicts 43 and 26% of the TPs in the HS and MM datasets, respectively. The corresponding figures for PITA, MIRANDA and TARGETSCAN are 19, 12 and 20% for the HS dataset and 14, 15 and 21% for the MM dataset.
Figure 4.
Venn diagram showing the number of true target sites correctly predicted in the Top30 and Bottom30 partitions by the different methods. The plot shows the number of true target sites correctly predicted by the different methods in the HS (A) and MM (B) datasets. MiREN predicts 39 and 26% of the TPs in the HS and MM datasets. The corresponding figures for PITA, MIRANDA and TARGETSCAN are 19, 12 and 20% for the HS dataset and 14, 15 and 21% for the MM dataset.
In Table 1 we also show the quality of the predictions resulting from the intersection of the PITA and miREN method (i.e. for the cases where the two strategies agree). In this case we obtain rather high accuracy and precision, obviously at the expense of a substantial loss in coverage. This suggests that miREN can be used to filter the predictions obtained by PITA.
DISCUSSION
Widely used methods for miRNA targets prediction take advantage of the sequence properties to identify potential miRNA target sites. Here we described a different approach that can complement the existing ones, especially since it is able to identify a larger fraction of TPs.
The overall procedure is rather computationally expensive. We performed a benchmark test by modeling 40 mirna-target pairs randomly selected from our datasets. The total computation time for the benchmark test was 46 h on a machine with eight threads and 16GB of RAM, the time required for individually predicting the best structure in each case is shown in Supplementary Figure S1.
The above implies that MiREN is not suitable for transcriptome-wide screenings, but, in our opinion, can be effectively used as a filter to assess the reliability of a sequence-based prediction of a miRNA site prior to perform time consuming and expensive experiments. Indeed, as mentioned before, the method can reach very respectable accuracy and precision when used on the subset of sites predicted by PITA and therefore be instrumental in prioritizing experiments.
Last but no least, our work highlights that the AGO protein plays a relevant role in the selection of the correct target site of a miRNA.
The method is available and can be downloaded together with the complete runs of some examples of the modeled target sites at the URL: http://circe.med.uniroma1.it/MiREN.
Supplementary Material
Acknowledgments
The authors are also grateful to Dr Marc Parisien who helped to setup MC-SYM, Dr Daniele Di Marino and all the other members of the Biocomputing Unit for useful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Epigenomics Flagship Project—EPIGEN; KAUST [KUK-I1–012–43]; PRIN Project [20108XYHJS]. Funding for open access charge: Epigenomics Flagship Project—EPIGEN.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fabian M.R., Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat. Struct. Mol. Biol. 2012;19:586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 2.Brummer A., Hausser J. MicroRNA binding sites in the coding region of mRNAs: extending the repertoire of post-transcriptional gene regulation. Bioessays. 2014;36:617–626. doi: 10.1002/bies.201300104. [DOI] [PubMed] [Google Scholar]
- 3.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 4.Chi S.W., Zang J.B., Mele A., Darnell R.B. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M., Jr, Jungkamp A.C., Munschauer M., et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin H.C., Wani S., Steptoe A.L., Krishnan K., Nones K., Nourbakhsh E., Vlassov A., Grimmond S.M., Cloonan N. Imperfect centered miRNA binding sites are common and can mediate repression of target mRNAs. Genome Biol. 2014;15:R51. doi: 10.1186/gb-2014-15-3-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brousse C., Liu Q., Beauclair L., Deremetz A., Axtell M.J., Bouché N., Betel D., Koppal A., Agius P., Sander C., et al. A non-canonical plant microRNA target site. Nucleic Acids Res. 2014;42:5270–52799. doi: 10.1093/nar/gku157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kakrana A., Hammond R., Patel P., Nakano M., Meyers BC. sPARTA: a parallelized pipeline for integrated analysis of plant miRNA and cleaved mRNA data sets, including new miRNA target-identification software. Nucleic Acids Res. 2014;42:e139. doi: 10.1093/nar/gku693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grosswendt S., Filipchyk A., Manzano M., Klironomos F., Schilling M., Herzog M., Gottwein E., Rajewsky N. Unambiguous identification of miRNA: target site interactions by different types of ligation reactions. Mol. Cell. 2014;54:1042–1054. doi: 10.1016/j.molcel.2014.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Betel D., Koppal A., Agius P., Sander C., Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kertesz M., Iovino N., Unnerstall U., Gaul U., Segal E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 12.Friedman R.C., Farh K.K., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krek A., Grun D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 14.Witkos T.M., Koscianska E., Krzyzosiak W.J. Practical aspects of microRNA target prediction. Curr. Mol. Med. 2011;11:93–109. doi: 10.2174/156652411794859250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y., Juranek S., Li H., Sheng G., Tuschl T., Patel D.J. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sheng G., Zhao H., Wang J., Rao Y., Tian W., Swarts D.C., van der Oost J., Patel D.J., Wang Y. Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage. Proc. Natl. Acad. Sci. U.S.A. 2014;111:652–657. doi: 10.1073/pnas.1321032111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y., Juranek S., Li H., Sheng G., Wardle G.S., Tuschl T., Patel D.J. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gan H.H., Gunsalus K.C. Tertiary structure-based analysis of microRNA-target interactions. RNA. 2013;19:539–551. doi: 10.1261/rna.035691.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parker J.S., Parizotto E.A., Wang M., Roe S.M., Barford D. Enhancement of the seed-target recognition step in RNA silencing by a PIWI/MID domain protein. Mol. Cell. 2009;33:204–214. doi: 10.1016/j.molcel.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peterson S.M., Thompson J.A., Ufkin M.L., Sathyanarayana P., Liaw L., Congdon C.B. Common features of microRNA target prediction tools. Front. Genet. 2014;5:23. doi: 10.3389/fgene.2014.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loeb G.B., Khan A.A., Canner D., Hiatt J.B., Shendure J., Darnell R.B., Leslie C.S., Rudensky A.Y. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol. Cell. 2012;48:760–770. doi: 10.1016/j.molcel.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y., Li Y., Ma Z., Yang W., Ai C. Mechanism of microRNA-target interaction: molecular dynamics simulations and thermodynamics analysis. PLoS Comput. Biol. 2010;6:e1000866. doi: 10.1371/journal.pcbi.1000866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia Z., Clark P., Huynh T., Loher P., Zhao Y., Chen H.W., Ren P., Rigoutsos I., Zhou R. Molecular dynamics simulations of Ago silencing complexes reveal a large repertoire of admissible ‘seed-less’ targets. Sci. Rep. 2012;2:569. doi: 10.1038/srep00569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rother M., Rother K., Puton T., Bujnicki J.M. ModeRNA: a tool for comparative modeling of RNA 3D structure. Nucleic Acids Res. 2011;39:4007–4022. doi: 10.1093/nar/gkq1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parisien M., Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008;452:51–55. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 26.Case D.A., Cheatham T.E. 3rd, Darden T., Gohlke H., Luo R., Merz K.M., Jr, Onufriev A., Simmerling C., Wang B., Woods R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 28.Voss N.R., Gerstein M. 3V: cavity, channel and cleft volume calculator and extractor. Nucleic Acids Res. 2010;38:W555–W562. doi: 10.1093/nar/gkq395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.