Abstract
Long non-coding RNAs (lncRNAs) have emerged as regulators of gene expression across metazoa. Interestingly, some lncRNAs function independently of their transcripts – the transcription of the lncRNA locus itself affects target genes. However, current methods of loss-of-function analysis are insufficient to address the role of lncRNA transcription from the transcript which has impeded analysis of their function. Using the minimal CRISPR interference (CRISPRi) system, we show that coexpression of the catalytically inactive Cas9 (dCas9) and guide RNAs targeting the endogenous roX locus in the Drosophila cells results in a robust and specific knockdown of roX1 and roX2 RNAs, thus eliminating the need for recruiting chromatin modifying proteins for effective gene silencing. Additionally, we find that the human and Drosophila codon optimized dCas9 genes are functional and show similar transcription repressive activity. Finally, we demonstrate that the minimal CRISPRi system suppresses roX transcription efficiently in vivo resulting in loss-of-function phenotype, thus validating the method for the first time in a multicelluar organism. Our analysis expands the genetic toolkit available for interrogating lncRNA function in situ and is adaptable for targeting multiple genes across model organisms.
INTRODUCTION
Recent transcriptome analyses have revealed numerous transcripts that originate from the non-protein-coding part of the genome in metazoa. Long non-coding RNAs (lncRNAs) constitute a subset of these transcripts; they are >200 nt in length, produced by RNA pol II, and undergo RNA processing events and regulate gene expression, in transcript-dependent and -independent manner (1,2). Although linked to several disease conditions including cancers (3), only a fraction has been analyzed in detail and the mechanism of action elucidated. Current strategies to interrogate lncRNA functions include over-expression and knockdown analyses that either targets the endogenous locus (deletion or insertion of genetic elements) or the transcript (RNAi-mediated knockdown and antisense oligonucleotides (ASO)). While the former approach involves alteration of the DNA sequence and/or topology, the latter is dependent on the presence of host machinery; in addition, targeting nuclear RNAs by RNAi or ASO is inefficient. Furthermore, it remains challenging to separate the role of lncRNA transcription from the transcript which makes the interpretation of mutant phenotypes difficult (4).
The CRISPR/Cas9 technique had rapidly emerged as a simple, efficient and precise method for DNA modification in the recent years; it is independent of host factors and has been widely used for functional analyses of genomes across the animal kingdom (5,6). It is a two-component system consisting of the Cas9 nuclease which, in complex with a single guide RNA (sgRNA), generates double-stranded breaks. The cleavage specificity is determined by a 20 nt targeting sequence at the 5′ end of the sgRNA and 3 nt protospacer adjacent motif (PAM) sequence abutting the DNA-binding site. In addition to gene editing, modifications of the Cas9 protein and sgRNA have enabled the CRISPR system to be used as a platform for modulating gene expression, thus extending its usefulness. For example, a mutant Cas9 protein that lacks the endonuclease activity (dead Cas9, dCas9) has been used as a RNA-guided DNA-binding complex to program transcription of genes in the endogenous context; fusion with appropriate effector domains results in robust activation (CRISPR activator, CRISPRa) or repression (CRISPR interference, CRISPRi, Figure 1A) of transcription (7–11). The effectiveness of gene silencing by CRISPRi, however, has been variable. While targeting dCas9 to promoter regions of the endogenous genes resulted in substantial reduction in gene function in bacteria (∼ 5-fold, (9)) and yeast (up to 18-fold, (8)), it was inefficient in human cells (∼2.5-fold, (8)). Rather fusion of the transcription repressor KRAB domain to dCas9 protein increased the knockdown efficiency (up to 5-fold in human cells, (8)) presumably due to recruitment of chromatin-modifying factors on the DNA at the targeting site (8). In these studies, however, the knockdown efficiency was determined by protein function and RNA levels were not directly measured. Recently, dCas9-KRAB:sgRNA complex was used to knockdown lncRNAs (upto 7-fold, (7)) in human cells. However, tagging of dCas9 could affect its DNA-binding activity while ectopic assembly of chromatin-modifying complexes could have unintended consequences, especially in regions where genes are located close to each other. Therefore, it is desirable to develop new experimental strategies that would circumvent these limitations and enable functional characterization of lncRNAs and other genes.
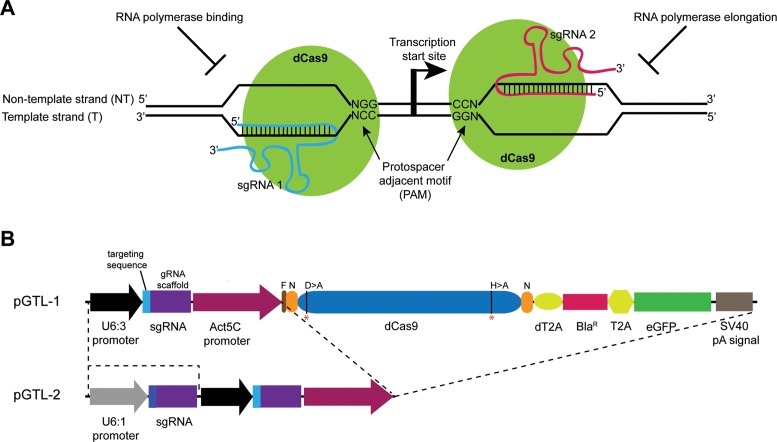
Figure 1.
CRISPRi in Drosophila. (A) Diagram showing the CRISPR interference (CRISPRi) system. To repress transcription, the catalytically inactive Cas9 protein (dCas9, green) is targeted either to the template or non-template DNA strand based on the targeting sequence of the sgRNA and an adjacent PAM sequence. Binding of the dCas9:sgRNA complex upstream of the transcription start site interferes with transcription initiation by preventing recruitment of the RNA polymerase while its assembly at a downstream site prevents transcription elongation. (B) Schematic representation of the transfection vectors pGTL-1 and pGTL-2. In pGTL-1, a single guide RNA with a 20 nt targeting sequence and the dCas9 protein are coexpressed under Drosophila constitutive promoters U6:3 and Actin5C, respectively. The dCas9 is separated from the blasticidin resistance gene (BlaR) and eGFP by self-cleaving T2A peptides (dT2A and T2A). The guide RNA (gRNA) scaffold contains the U6 transcription terminator sequence. The pGTL-2 vector contains an additional sgRNA scaffold under the U6:1 promoter, thus allowing production of two sgRNAs simultaneously with the dCas9 protein. The mutated amino acid residues (D10>A and H841>A) in dCas9 are marked with red asterisks (*). N = NLS sequence, F = FLAG epitope, pA = polyadenylation.
In this study, we show that both human and Drosophila codon-optimized dCas9 proteins are effective in silencing the lncRNA roX in Drosophila cells. Furthermore, we observe a robust knock down of the roX transcripts in vivo resulting in loss-of-function phenotypes, thus validating the effectiveness of the minimal CRISPRi system in Drosophila.
MATERIALS AND METHODS
Generation of transfection vectors
pGTL-1
The BlaR gene was PCR amplified from pCoBLAST (Life Technologies) using primers SG1 and SG2 and ligated between the XbaI and HindIII digested pAc5-STABLE2–neo (12) to replace the eGFP gene, thus generating pSG-Bla-neo. To insert the eGFP gene in place of neoR, the pSG-Bla-neo vector was digested with NheI and XhoI and used for ligation with PCR amplified eGFP using primers SG3 and SG4, resulting in pSG-Bla-GFP. The U6:3 cassette containing the promoter and sgRNA sequence was amplified from pCFD3 (13) using SG5 and SG6 primers while the dual promoter U6:1–6:3 cassette was derived from pCFD4 (13) using primers SG7 and SG6. Both PCR products were digested with BglII and ligated into the BglII site of pSG-Bla-GFP vector giving rise to plasmids pSG-Bla-GFP-U6:3 and pSG-Bla-GFP-U6:1–6:3, respectively. The human codon-optimized Cas9 gene together with 3xFLAG and NLS sequences was amplified from pAc5-sgRNA-Cas9-puro (14) and cloned into pUC19 vector. The human Cas9 was mutated by Quikchange II Site-Directed Mutagenesis kit (Stratagene) using primer pairs SG8/SG9 (for D10 > A) and SG10/SG11 (for H841 > A). Next, the human dCas9 cassette (FLAG-NLS-dCas9-NLS) was amplified from the pUC19 clone using primers SG12 and SG13, and used to replace the mCherry gene in pSG-Bla-GFP-U6:3 by KpnI and NotI digestion, resulting in pGTL-1 plasmid.
pGTL-2
To construct pGTL-2, the pSG-Bla-GFP-U6:1–6:3 vector backbone and the human dCas9 cassette were amplified using primer pairs SG14/SG15 and SG16/SG17, and used for two-fragment Gibson assembly reaction (New England Biolabs #E2611). The pGTL-2 vector containing the Drosophila dCas9 was generated as follows. The Drosophila codon-optimized Cas9 containing plasmid was a kind gift of Fillip Port (University of Cambridge) which was used as template to PCR amplify the gene using primers SG18 and SG19, subcloned into pUC19 vector and used for site-directed mutagenesis as described above using the primer pairs SG20/SG21 (for D10 > A) and SG22/SG23 (for H841 > A). The mutated Drosophila dCas9 was next amplified using SG24 and SG25 primers and used for Gibson assembly reaction with the pGTL-2 vector backbone amplified using primers SG26/SG27. This replaced the human dCas9 with the Drosophila dCas9 which is in-frame with the NLS sequences in the pGTL-2 vector.
sgRNA constructs
To minimize potential off-target activity, the targeting sequence predicted to have one target in the Drosophila genome was selected using the CRISPRdirect tool (15). The vectors expressing single guide RNA targeting roX1 were generated using pGTL-1 following the protocol described in the CRISPR fly design website (http://www.crisprflydesign.org/grna-expression-vectors/). Briefly, complementary oligonucleotides containing the 20 nt targeting sequence were phosphorylated, annealed, diluted and ligated to BbsI digested pGTL-1 vector. The oligonucleotide and guide RNA sequences are listed in Supplementary Tables S1 and S2, respectively.
To generate expression vectors containing two sgRNAs, the pGTL-2 plasmids with the human dCas9 and Drosophila dCas9 genes were PCR amplified with primers SG28 and SG29. Additionally, oligonucleotides containing the roX1 and roX2 targeting sequences were designed (listed in Supplementary Table S1) and used for PCR reaction with pCFD4 plasmid as template (as described for pCFD4 cloning at http://www.crisprflydesign.org/grna-expression-vectors/). The vector PCR fragment was then combined with the sgRNA fragments using the Gibson assembly reaction.
pGTL-3
The vector for fly transgenesis was constructed using 2-step Gibson assembly reaction. Four separate PCR reactions were performed, (i) using the pAct5c-Cas9 plasmid ((13), gift of Fillip Port), primers SG42/SG43 were used to generate the vector backbone that lacked the Actin5c:Cas9 cassette, (ii) the Actin5c promoter was amplified from pAct5c-Cas9 plasmid using SG44/SG45, (iii) the U6:1-rT8-U6:3-rT9 and U6:1-rTb-U6:3-rTc cassettes were generated from the corresponding pGTL-2 vectors using primers SG46/SG47 and (iv) the Drosophila dCas9 gene from pGTL-2 vector by SG48/SG49 primers. Following DpnI digestion, the vector backbone was assembled with the guide RNA cassettes while Actin5c with the Drosophila dCas9, and subsequently mixed together to generate the transgenesis plasmids. In addition to the Drosophila dCas9, the pGTL-3-roX1 and pGTL-3-roX2 plasmids support expression of rT8+rT9 and rTb+rTc guide RNAs, respectively.
All plasmid sequences were verified by sequencing.
Cell culture and transfection
Clone 8 cells (CME W1 cl.8+) were obtained from Drosophila Genomics Resource Center (DGRC) and maintained in M3 media containing 2% Fetal Bovine Serum (FBS) (Sigma #F4135), 5 μg/ml insulin (Sigma #I9278), 2.5% fly extract and penicillin-streptomycin (Life Technologies) in 25°C incubator. Transfection was carried out using Effectene Transfection Reagent (Qiagen) as per manufacturers’ instructions. Cells were seeded in 6-well plates (106cells/well) and incubated with 0.4 μg of plasmid DNA. After 3 days, blasticidin (Life Technologies #R210–01) was added at 25 μg/ml and cells were maintained in the antibiotic thereafter. Surviving cells were inspected for GFP expression and stable cell lines obtained in ∼3 weeks which was used for genomic DNA and RNA preparation.
RNA preparation and qRT PCR analysis
Total RNA was prepared from cells using the miRNeasy kit (Qiagen) as per manufacturers’ instructions. One microgram of RNA was used for reverse transcription using the QuantiTect Reverse Transcription Kit (Qiagen). For qPCR, the cDNA was diluted 1:10, mixed with primers and SYBRGreen Jumpstart Taq readymix (Sigma) and amplified in Applied Biosystems Fast Real-Time PCR system using primers SG30/SG31 (roX1), SG38/SG39 (roX1 RA), SG32/SG33 (roX2), SG34/SG35 (rp49).
The Ct values for roX were normalized with rp49 as the housekeeping gene and ΔΔCt values were calculated. The abundance values were normalized to control cells transfected with pGTL-1 or pGTL-2 producing sgRNAs with a non-targeting sequence (shown as 100%). The data are shown for two independent biological replicates with each replicate containing three technical replicates. The error bar shows the S.E.M. value.
For RT-PCR, total RNA was extracted from clone 8 cells and cDNA prepared as described above. One microliter of 1:10 diluted cDNA was used for PCR amplification using DreamTaq Green PCR master mix (Life Technologies) in a 20 μl volume as per instructions. The primer pairs used for the roX1 intergenic region (SG36/SG37), roX1 RA (SG38/SG39), roX1 RB (SG30/SG31) and rp49 (SG34/SG35) are shown in Supplementary Table S1.
DNA analysis
For genotyping and analysis of the roX loci, the cell lines expressing Drosophila dCas9 and sgRNAs (rNT7/rT8 and rTb/rTc) were pelleted and used for genomic DNA preparation using Gel Extraction kit (Qiagen) as described on the webpage of Klaus Forsterman lab (http://www.foerstemann.genzentrum.lmu.de/protocols/). A total of 20 ng of genomic DNA was used for PCR with primers SG31/SG40 (roX1) and SG33/SG41 (roX2) and the PCR product sequenced.
Western analysis
One microliter culture of the stable cell lines (∼107cells) were pelleted at 1000 g for 5 mins, suspended in 200 μl of SDS-sample buffer and boiled for 10 min. The proteins in the extract were separated on NuPAGE 4–12% Bis-Tris gels (Life Technologies) according to manufacturers’ instructions, blotted onto PVDF membrane (Amersham Hybond) using Mini Trans-Blot transfer cell (BioRad). The primary antibodies were mouse anti-FLAG antibody (Sigma #F3165) and mouse anti-Tubulin antibody (Sigma #T6199) used at 1:1000 dilution while HRP-conjugated donkey anti-mouse IgG (Jackson Immunoresearch, 1:1000 dilution) was used as the secondary antibody.
Fly transgenesis
The Drosophila transgenesis vector pGTL-3-roX1 and -roX2 were injected into nos-int; attP2 embryos for integration into the attP landing site on chromosome 3 (68A4) at the Microinjection Service of Department of Genetics, University of Cambridge. For coexpression of roX1 and roX2 guide RNAs, the GTL-3-roX1/TM6cSb and GTL-3-roX2/TM6cSb flies were crossed at 29°C and the progeny lacking Sb were scored. The male survival is based on the total number of progeny of the same genotype from the same cross. For comparison, y,w parents were kept at 29°C and the progeny counted.
RESULTS AND DISCUSSION
The Drosophila genome encodes numerous lncRNAs which shows tissue- and sex-specific expression (16,17). However, the functions of the majority of these transcripts remain unknown. We reasoned that repression of transcription of the lncRNAs by CRISPRi would overcome the limitations of current methods applied for their analysis and decided to re-evaluate the effectiveness of dCas9 in repressing transcription from endogenous lncRNA loci in Drosophila cells. The CRISPR/Cas9 system has been used extensively for gene editing in Drosophila (11,13–14,18–26). However, CRISPRi has not been applied till date in Drosophila.
To implement CRISPRi in Drosophila cells, we first constructed a single transfection vector that allows coexpression of the mutant Cas9 protein and sgRNA, similar to the one we previously described (14). This strategy eliminates the variability between experiments that arise from cotransfection of two plasmids. The transfection vector pGTL-1 (Figure 1B), derived from the multicistronic vector pAc5-STABLE2-neo (12), contains the BlaR gene which allows for efficient selection of the transfected cells and reduces the time to establish stable cell lines (27). In addition, presence of GFP enables easy monitoring of the transformed cells. The human codon-optimized Cas9 gene lacking nuclease activity (dCas9) flanked with NLS sequences and tagged with FLAG epitope at the N-terminal, is expressed under the Actin5C promoter. For optimal expression of the guide RNAs, the U6:3–sgRNA cassette was inserted into the vector; this promoter has a higher CRISPR/Cas9-mediated gene editing activity in Drosophila, as compared with the commonly used U6:2 promoter (13).
As candidate gene, we chose roX (RNA on X chromosome), which encodes the lncRNAs involved in dosage compensation in Drosophila males (28). The male-specific roX RNAs, roX1 and roX2, form complex with the MSL (male-specific lethal) proteins and facilitate targeting of the complex on the X chromosome, necessary for 2-fold hyperactivation of the X-linked genes in males. Genetic analysis shows that roX1 roX2 double mutants are male lethal which is rescued by either roX1 or roX2 cDNA, suggesting functional redundancy in their activity (29,30). Presently, the alleles used for loss-of-function analysis are genomic deletions of the loci or complex chromosomal arrangements. Furthermore, the transcription initiation sites and identity of the regulatory elements remain unclear. We designed multiple sgRNAs targeting the roX1 and roX2 locus (Figures 2A and 3A). To determine if effective silencing of roX RNAs is dependent on strand-specificity, we chose sequences complementary to both template (rT) and non-template (rNT) strand and in proximity of the predicted transcription start sites. For the assay, we generated stable cell lines using Drosophila clone 8 cells which expresses both roX RNAs (31).
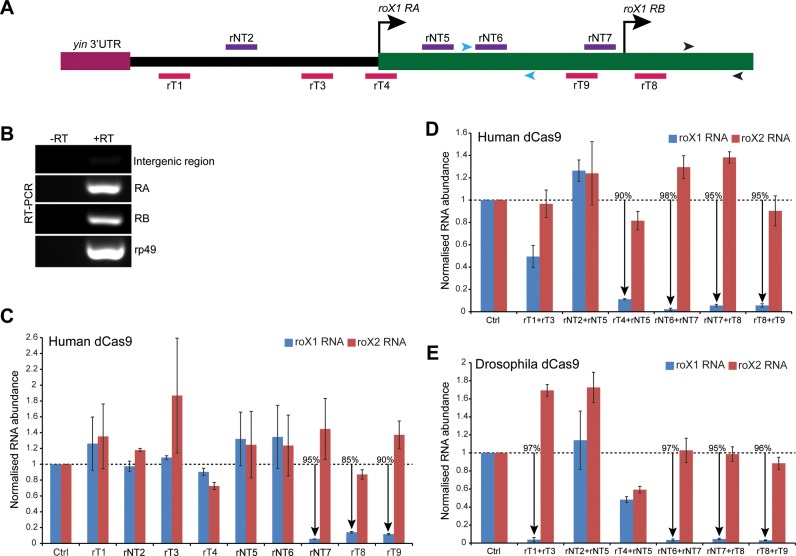
Figure 2.
The roX1 expression is efficiently silenced by CRISPRi. (A) Schematic representation of the Drosophila roX1 locus and the relative positions of the roX1-targeting sgRNAs. There are two transcription start sites of roX1 gene (270 nt apart) which produce isoforms RA and RB (FlyBase). The sgRNAs targeting the RNA polymerase template (rT, red) and non-template (rNT, violet) strands are shown. The 5′ intergenic region between roX1 RA and the upstream gene yin is shown in solid black line. The arrowheads mark the position of the primers used to detect roX1 RNA (black) and roX1 RA isoform (blue) for RT-qPCR analysis. (B) RT-PCR analysis using clone 8 cells shows the presence of roX1 RA and RB transcripts. The primers specific to sequences in the intergenic region, roX1 RA and roX1 RB isoform were used for PCR amplification. (C, D, E) RT-qPCR measurement of abundance of the roX1 and roX2 transcripts in cell lines coexpressing human (C, D) or Drosophila (E) dCas9 protein and sgRNAs (as shown on the x-axis) complementary to the rT or rNT DNA strand at the endogenous roX1 locus. The cell lines expressing single sgRNA (C), or two guide RNAs simultaneously (D, E) are shown. The gRNAs rNT7, rT8 and rT9 mediate efficient knockdown of the roX1 transcript, either alone or jointly with others. However, the combinations of rT1+rT3 and rT4+rNT5 which target the roX1 RA transcription initiation site, are effective only in pairs in the presence of human dCas9 and Drosophila dCas9, respectively. The y-axis shows the enrichment of RNAs relative to rp49 transcript and normalized to the cells transfected with pGTL-1 containing a non-targeting sequence (Ctrl). The data are shown from two biological replicates, each performed in triplicate. The error bars indicate SEM. The % shows knockdown in percentage as compared with the Ctrl.
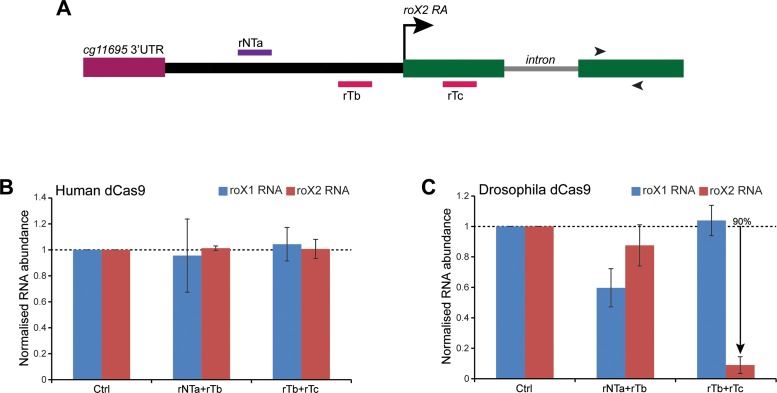
Figure 3.
The roX2 transcript is efficiently down-regulated by CRISPRi. (A) Schematic representation of the Drosophila roX2 locus showing relative positions of the roX2 targeting sgRNAs. The transcription start site is marked by an arrow while the intron within the roX2 gene is shown by a solid grey line. The intergenic region between the roX2 transcription start site and the adjacent gene (cg11695) is shown in solid black line. The sgRNAs targeting the rT and rNT strands are shown in red and violet, respectively. The arrowheads mark the position of the primers used to detect roX2 RNA for RT-qPCR analysis. (B and C) RT-qPCR measurement of abundance of the roX1 and roX2 transcripts in cell lines coexpressing human (B) or Drosophila (C) dCas9 protein and two sgRNAs (as shown on the x-axis). While sgRNAs rTb+rTc do not affect roX1 or roX2 levels in presence of human dCas9, they specifically knockdown roX2 transcript levels in cells coexpressing Drosophila dCas9. The y-axis shows the enrichment of RNAs relative to rp49 transcript and normalized to the cells transfected with pGTL-1 containing a non-targeting sequence (Ctrl). The data are shown from two biological replicates, each performed in triplicate. The error bars indicate SEM. The % shows knockdown in percentage as compared with the Ctrl.
First, we attempted to suppress transcription of the roX1 RNA by coexpressing a single guide RNA with the dCas9 protein. The FlyBase (http://flybase.org/) gene model for roX1 predicts two distinct transcription start sites resulting in a longer roX1 RA and a shorter roX1 RB isoform. Our data show that both transcripts are present in clone 8 cells (Figure 2B) validating the two initiation sites. Notably, the roX1 RB gene region alone was used in the transgenic constructs for rescue analysis (30). Our results show that recruitment of dCas9 adjacent to the roX1 RB transcription start resulted in efficient silencing of both RA and RB isoforms (Figure 2C, Supplementary Figure S1A). Interestingly, sgRNA targeting the non-template (rNT7) and template (rT8) strand showed similar efficacies (95% and 85%, respectively), indicating absence of DNA template bias for silencing as observed in human cells (7). Consistently, rT9, which targets the template strand and partially overlaps with rNT7, shows similar repressive activity. The knockdown is dependent on the roX1 targeting sequence and is specific as cells with significantly reduced roX1 levels did not show a concomitant reduction of roX2 RNA levels. Targeting of dCas9 to regions upstream of the roX1 transcription start sites (TSS) failed to affect RNA levels. Western analysis showed efficient expression of the dCas9 protein in these cell lines (Supplementary Figure S2A) underlining the critical role of guide RNA sequence in repression. The variability in dCas9 protein expression could be due to chromosomal position effects and/or chromosome copy number changes in this cell line (31). Taken together our results reveal potent sgRNAs for roX1 silencing and show that assembly of dCas9-sgRNA complex on either strand of DNA can effectively silence lncRNA expression, possibly by preventing the binding and/or elongation of RNA polymerase.
A previous study has shown that the efficiency of CRISPRi could be enhanced by using two sgRNAs targeting the same gene in bacteria (9). To test this in Drosophila cells, we inserted the U6:1-sgRNA cassette in the pGTL-1 vector immediately upstream of the U6:3 cassette, thus generating the pGTL-2 vector (Figure 1B); this allows expression of a second sgRNA under the control of U6:1 promoter which also shows a robust activity in Drosophila (13). For expression, we chose sgRNA combinations which are located >30 bp apart to allow for efficient binding of the dCas9 protein. As shown in Figure 2D, sgRNAs complementary to the sequences upstream of (rNT6+rNT7) and flanking the roX1 RB isoform (rNT7+rT8, rT8+rT9) showed a robust reduction of roX1 transcript. This is expected and can be attributed to the effectiveness of rNT7, rT8 and rT9 sgRNA as observed previously (Figure 2C), although rNT7 and rT8 are now expressed under the control of U6:1 promoter. Remarkably, we observe an effective silencing of roX1 RNA by sgRNAs targeting the roX1 RA transcription region. Although ineffective individually, the sgRNA combinations targeting the template strand (rT1+rT3) or both the template and non-template strand (rT4+rNT5) show striking loss in roX1 RNA levels (50% and 90%, respectively). We did not observe a strong correlation between the dCas9 levels and transcription inhibition (Supplementary Figure S2B). Collectively, these results suggest that binding of multiple dCas9 protein at the transcriptional start sites can augment the effectiveness of transcriptional silencing.
We next determined whether codon optimization of dCas9 protein could enhance the combinatorial activity of CRISPRi. The Drosophila codon-optimized Cas9 has been used recently for in vivo gene editing; however, expression of the protein with strong, ubiquitous Gal4 drivers led to substantial lethality for yet unknown reasons (13). We generated catalytically inactive Drosophila Cas9 by mutating the amino acids essential for nuclease activity (D10A and H841A) and substituted the human dCas9 in pGTL-2 vector. As shown in Figure 2E, coexpression of sgRNAs and Drosophila dCas9 recapitulates silencing activity of the sgRNAs to a similar extent as observed with the human dCas9 protein (rNT6+rNT7, rNT7+rT8 and rT8+rT9). Western analysis showed efficient translation of Drosophila dCas9 protein (Supplementary Figure S2B). The cell lines did not show any overt phenotype suggesting that Drosophila dCas9 expression is not toxic and the protein is active in silencing gene expression. However, as compared with human dCas9, we observed a striking reduction (97% for Drosophila dCas9 versus 50% for human dCas9) of roX1 RA and RB RNA with sgRNAs that target the 5′ region of roX1 RA TSS (rT1+rT3) (Figure 2E, Supplementary Figure S1B). The enhanced effects of this guide RNA combination, although ineffective individually, could be due to targeting of redundant regulatory elements in the roX1 intergenic region. We also noted that cells expressing rT4+rNT5 show ∼40% reduction in roX2 RNA levels (Figure 2E) – this is unlikely due to an off-target effect as Gapdh and CTPsyn RNA levels are not affected similarly (Supplementary Figure S3A). Importantly, DNA sequencing revealed that the silencing effect of the Drosophila dCas9-sgRNA complex at the endogenous loci does not involve changes in the DNA sequence (Supplementary Figure S4).
Since roX1 targeting sgRNAs showed a combinatorial effect in gene silencing and were potent in presence of Drosophila dCas9, we tested whether a similar strategy could be used to abrogate roX2 transcription. For analysis, we chose three targeting sequences around the roX2 RA TSS, complementary to both the non-template (rNTa) and template (rTb and rTc) strands (Figure 3A). Coexpression of the sgRNAs with human dCas9 did not affect the roX2 transcript level significantly (Figure 3B). However, when the same combinations of sgRNAs were expressed together with the Drosophila dCas9, we observed a significant reduction (90% knockdown) of roX2 transcript in the sgRNAs flanking the transcription start site (rTb+rTc, Figure 3C). Importantly, the roX1 transcript was unaffected in the same cell line, demonstrating the specificity of the knockdown. Consistently, Gapdh and CTPsyn RNA levels also remain unaltered (Supplementary Figure S3B). Notably, the dCas9 protein levels did not correspond to the degree of suppression (Supplementary Figure S2C). These data, together with our previous observation with roX1, supports the enhanced activity of Drosophila dCas9 in mediating transcription repression via CRISPRi.
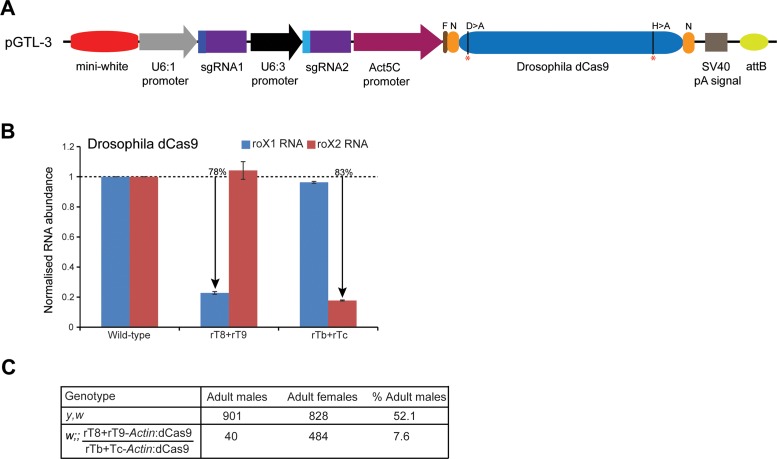
Till date, CRISPRi has not been applied in any multicellular organism. To determine the effectiveness of CRISPRi in repressing transcription of roX RNAs in vivo, we generated two transgenic flies expressing Drosophila dCas9 and sgRNAs under constitutively active promoters. For analysis, we chose the guide RNAs that showed potent activity in downregulating the corresponding roX genes in the Drosophila cell line – rT8+rT9 to target roX1 and rTb+rTc for roX2. For transgenesis, we modified the pAct5c-Cas9 vector (13) to generate pGTL-3 vector – the Cas9 gene was replaced by Drosophila dCas9 and the U6:1-sgRNA-U6:3-sgRNA cassettes were inserted between the mini-white gene and the Act5c promoter (Figure 4A). This vector allows for stable site-specific integration into the Drosophila genome, thus eliminating position-effect and variability in transgene expression. Indeed, the dCas9-roX1 and dCas9-roX2 fly lines efficiently express similar levels of the Drosophila protein (Supplementary Figure S2D) and were viable indicating that expression of these transgenes was not toxic. RT-qPCR showed a robust and specific reduction in the roX1 and roX2 RNA levels (Figure 4B) which is consistent with our analyses in the cell line and, thus, validates the repressive activity of Drosophila dCas9 in vivo. The knock down is likely to be an underestimation as Actin5c is not expressed in some adult tissues (32) and is excluded from the germline. Remarkably, as compared with females, we observed a drastic reduction in the viability of males coexpressing roX1 and roX2 guide RNAs (7.6% dCas9-roX1/dCas9-roX2 males versus 52.1% wild-type males, Figure 4C). The escaper males are fertile and show no obvious phenotypic defects. This is consistent with a strong loss-of-function of roX (33). Thus, we conclude that recruitment of the Drosophila dCas9 at roX1 and roX2 locus disrupts their expression by suppressing transcription, resulting in male lethality.
Figure 4.
Knock down of roX1 and roX2 RNAs in vivo. (A) Schematic representation of the Drosophila transgenesis vectors pGTL-3. The dominant selection marker mini-white is followed by the dual guide RNA expressing cassette under the control of U6:1 and U6:3 promoters while the expression of Drosophila dCas9 is controlled by the constitutively active promoter Actin5c. The mutated amino acid residues (D10 > A and H841 > A) in dCas9 are marked with red asterisks (*). N = NLS sequence, F = FLAG epitope, pA = polyadenylation. (B) RT-qPCR analysis showing robust depletion of roX1 and roX2 RNAs in the males of fly lines expressing the corresponding guide RNAs (shown on the x-axis). The y-axis shows the enrichment of RNAs relative to rp49 transcript and normalized to the wild-type (y,w) males. The data are shown from two biological replicates, each performed in triplicate. The error bars indicate SEM. The % shows knockdown in percentage as compared with the wild-type. (C) Table showing a severe reduction in the male progeny simultaneously expressing roX1 and roX2 guide RNAs (rT8+rT9-Act:dCas9/rTb+rTc-Act:dCas9). The percentage male survival is calculated based on the total number of progeny of the same genotype recovered in the same cross. The y,w strain was used as wild-type.
In this report, we describe an effective loss-of-function approach for studying lncRNA genes in Drosophila by a minimal CRISPRi system that does not require fusion of effector domains to dCas9. Using this straightforward method, we achieved high levels of knock down of the roX RNAs (up to 50-fold) by targeting the endogenous loci in Drosophila cells. Our strategy overcomes the previous limitations of genetic approaches used for assessing lncRNA function – it does not introduce changes in the DNA and eliminates ectopic recruitment of chromatin silencing complexes. Our results demonstrate that targeting multiple sgRNAs that flank the transcription start site of genes improves the knock down potency of CRISPRi. In this regard, recent development of engineered Cas9 proteins with flexible PAM recognition properties and enhanced specificity should increase the choice of targetable sequences while minimizing off-target effects (34,35). Since the guide RNA activity remains difficult to predict and characterization of promoter and transcription regulatory elements of lncRNAs remain incomplete, we recommend testing multiple guide RNAs to select potent guides for analysis. The design of our transfection vector and cell-based assay allows quick evaluation of the effectiveness of several sgRNAs before their use in in vivo studies, ranging from scaling to silencing of gene expression as well as promoter and/or enhancer mapping experiments. This strategy, in combination with traditional knock down approaches, should allow functional and mechanistic exploration of lncRNAs across metazoa, thus shedding light on these enigmatic transcripts that constitute the ‘dark matter’ of the genome.
Supplementary Material
Acknowledgments
We thank Fillip Port and Simon Bullock for the gift of plasmids, Drosophila Genomics Resource Center supported by NIH grant 2P40OD010949–10A1 for the cell line, Yong Huang for help with data analysis, Stuart Grice for critical reading of the manuscript and members of the Liu lab for helpful discussions. We also thank the University Of Cambridge Department Of Genetics Fly Facility for generation of the transgenic fly lines.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
UK Medical Research Council and the European Research Council (DARGENs, Project number 249869). Funding for open access charge: UK Medical Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wilusz J.E., Sunwoo H., Spector D.L. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petruk S., Sedkov Y., Riley K.M., Hodgson J., Schweisguth F., Hirose S., Jaynes J.B., Brock H.W., Mazo A. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell. 2006;127:1209–1221. doi: 10.1016/j.cell.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheetham S.W., Gruhl F., Mattick J.S., Dinger M.E. Long noncoding RNAs and the genetics of cancer. Br. J. Cancer. 2013;108:2419–2425. doi: 10.1038/bjc.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassett A.R., Akhtar A., Barlow D.P., Bird A.P., Brockdorff N., Duboule D., Ephrussi A., Ferguson-Smith A.C., Gingeras T.R., Haerty W., et al. Considerations when investigating lncRNA function in vivo. eLife. 2014;3:e03058. doi: 10.7554/eLife.03058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harrison M.M., Jenkins B.V., O'Connor-Giles K.M., Wildonger J. A CRISPR view of development. Genes Dev. 2014;28:1859–1872. doi: 10.1101/gad.248252.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilbert L.A., Horlbeck M.A., Adamson B., Villalta J.E., Chen Y., Whitehead E.H., Guimaraes C., Panning B., Ploegh H.L., Bassik M.C., et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilbert L.A., Larson M.H., Morsut L., Liu Z., Brar G.A., Torres S.E., Stern-Ginossar N., Brandman O., Whitehead E.H., Doudna J.A., et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choudhary E., Thakur P., Pareek M., Agarwal N. Gene silencing by CRISPR interference in mycobacteria. Nat. Commun. 2015;6:6267. doi: 10.1038/ncomms7267. [DOI] [PubMed] [Google Scholar]
- 11.Lin S., Ewen-Campen B., Ni X., Housden B.E., Perrimon N. In vivo transcriptional activation using CRISPR/Cas9 in Drosophila. Genetics. 2015;201:433–442. doi: 10.1534/genetics.115.181065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalez M., Martin-Ruiz I., Jimenez S., Pirone L., Barrio R., Sutherland J.D. Generation of stable Drosophila cell lines using multicistronic vectors. Sci. Rep. 2011;1:75. doi: 10.1038/srep00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Port F., Chen H.-M., Lee T., Bullock S.L. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E2967–E2976. doi: 10.1073/pnas.1405500111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bassett A.R., Tibbit C., Ponting C.P., Liu J.L. Mutagenesis and homologous recombination in Drosophila cell lines using CRISPR/Cas9. Biol. Open. 2014;3:42–49. doi: 10.1242/bio.20137120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naito Y., Hino K., Bono H., Ui-Tei K. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics. 2015;31:1120–1123. doi: 10.1093/bioinformatics/btu743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown J.B., Boley N., Eisman R., May G.E., Stoiber M.H., Duff M.O., Booth B.W., Wen J., Park S., Suzuki A.M., et al. Diversity and dynamics of the Drosophila transcriptome. Nature. 2014;512:393–399. doi: 10.1038/nature12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young R.S., Marques A.C., Tibbit C., Haerty W., Bassett A.R., Liu J.L., Ponting C.P. Identification and properties of 1,119 candidate lincRNA loci in the Drosophila melanogaster genome. Genome Biol. Evol. 2012;4:427–442. doi: 10.1093/gbe/evs020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bassett A.R., Kong L., Liu J.L. A genome-wide CRISPR library for high-throughput genetic screening in Drosophila cells. J. Genet. Genomics. 2015;42:301–309. doi: 10.1016/j.jgg.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kondo S., Ueda R. Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics. 2013;195:715–721. doi: 10.1534/genetics.113.156737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu Z., Ren M., Wang Z., Zhang B., Rong Y.S., Jiao R., Gao G. 2013. Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila. Genetics. 2013;195:289–291. doi: 10.1534/genetics.113.153825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bassett A.R., Tibbit C., Ponting C.P., Liu J.L. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 2013;4:220–228. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gratz S.J., Cummings A.M., Nguyen J.N., Hamm D.C., Donohue L.K., Harrison M.M., Wildonger J., O'Connor-Giles K.M. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013;194:1029–1035. doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xue Z., Wu M., Wen K., Ren M., Long L., Zhang X., Gao G. CRISPR/Cas9 mediates efficient conditional mutagenesis in Drosophila. G3 (Bethesda) 2014;4:2167–2173. doi: 10.1534/g3.114.014159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ren X., Yang Z., Xu J., Sun J., Mao D., Hu Y., Yang S.-J., Qiao H.-H., Wang X., Hu Q., et al. Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in Drosophila. Cell Rep. 9:1151–1162. doi: 10.1016/j.celrep.2014.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren X., Sun J., Housden B.E., Hu Y., Roesel C., Lin S., Liu L.-P., Yang Z., Mao D., Sun L., et al. Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc. Natl. Acad. Sci. U.S.A. 2013;110:19012–19017. doi: 10.1073/pnas.1318481110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chavez A., Scheiman J., Vora S., Pruitt B.W., Tuttle M., P R Iyer E., Lin S., Kiani S., Guzman C.D., Wiegand D.J., et al. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods. 2015;12:326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimura M., Takatsuki A., Yamaguchi I. Blasticidin S deaminase gene from Aspergillus terreus (BSD): a new drug resistance gene for transfection of mammalian cells. Biochim. Biophys. Acta. 1994;1219:653–659. doi: 10.1016/0167-4781(94)90224-0. [DOI] [PubMed] [Google Scholar]
- 28.Ilik I., Akhtar A. roX RNAs: non-coding regulators of the male X chromosome in flies. RNA Biol. 2009;6:113–121. doi: 10.4161/rna.6.2.8060. [DOI] [PubMed] [Google Scholar]
- 29.Park S.-W., Kuroda M.I., Park Y. Regulation of histone H4 Lys16 acetylation by predicted alternative secondary structures in roX noncoding RNAs. Mol. Cell. Biol. 2008;28:4952–4962. doi: 10.1128/MCB.00219-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stuckenholz C., Meller V.H., Kuroda M.I. Functional redundancy within roX1, a noncoding RNA involved in dosage compensation in Drosophila melanogaster. Genetics. 2003;164:1003–1014. doi: 10.1093/genetics/164.3.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee H., McManus C., Cho D.-Y., Eaton M., Renda F., Somma M., Cherbas L., May G., Powell S., Zhang D., et al. DNA copy number evolution in Drosophila cell lines. Genome Biol. 2014;15:R70. doi: 10.1186/gb-2014-15-8-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fyrberg E.A., Mahaffey J.W., Bond B.J., Davidson N. Transcripts of the six Drosophila actin genes accumulate in a stage- and tissue-specific manner. Cell. 1983;33:115–123. doi: 10.1016/0092-8674(83)90340-9. [DOI] [PubMed] [Google Scholar]
- 33.Deng X., Rattner B.P., Souter S., Meller V.H. The severity of roX1 mutations is predicted by MSL localization on the X chromosome. Mech. Dev. 2005;122:1094–1105. doi: 10.1016/j.mod.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 34.Kleinstiver B.P., Prew M.S., Tsai S.Q., Topkar V.V., Nguyen N.T., Zheng Z., Gonzales A.P.W., Li Z., Peterson R.T., Yeh J.-R.J., et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.