Abstract
CRISPR/Cas9-induced site-specific DNA double-strand breaks (DSBs) can be repaired by homology-directed repair (HDR) or non-homologous end joining (NHEJ) pathways. Extensive efforts have been made to knock-in exogenous DNA to a selected genomic locus in human cells; which, however, has focused on HDR-based strategies and was proven inefficient. Here, we report that NHEJ pathway mediates efficient rejoining of genome and plasmids following CRISPR/Cas9-induced DNA DSBs, and promotes high-efficiency DNA integration in various human cell types. With this homology-independent knock-in strategy, integration of a 4.6 kb promoterless ires-eGFP fragment into the GAPDH locus yielded up to 20% GFP+ cells in somatic LO2 cells, and 1.70% GFP+ cells in human embryonic stem cells (ESCs). Quantitative comparison further demonstrated that the NHEJ-based knock-in is more efficient than HDR-mediated gene targeting in all human cell types examined. These data support that CRISPR/Cas9-induced NHEJ provides a valuable new path for efficient genome editing in human ESCs and somatic cells.
INTRODUCTION
Zinc-finger nucleases (ZFNs) (1), transcription activator-like effector nucleases (TALENs) (2) and bacterial clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein 9 (Cas9) system (3) have achieved great success in introducing site-specific DNA double-strand breaks (DSBs) with high accuracy and efficiency. They have been developed into versatile tools to introduce a broad range of genomic modifications, such as targeted mutation, insertion, large deletion or gene knock-out, in various prokaryotic, eukaryotic cells and organisms (4). Among these tools, CRISPR/Cas9 has rapidly gained popularity due to its superior simplicity (5,6). In this system, a single guide RNA (sgRNA) complexes with Cas9 nuclease, which can recognize a variable 20-nucleotide target sequence adjacent to a 5′-NGG-3′ protospacer adjacent motif (PAM) and introduce a DSB in the target DNA (7,8). The induced DSB then triggers DNA repair process mainly via two distinct mechanisms, namely, the non-homologous end joining (NHEJ) and the homology-directed repair (HDR) pathways.
The NHEJ pathway repairs DNA DSBs by joining the broken ends through a homology-independent mechanistically flexible process, which often results in random small insertions or deletions (indels) (9). Thus, CRISPR/Cas9-introduced DNA cleavage followed by NHEJ repair has been exploited to generate loss-of-function alleles in protein-coding genes (10). In contrast, the HDR pathway mediates a strand-exchange process to repair DNA damage accurately based on existing homologous DNA sequences (11). Utility of this repair mechanism enables intentional replacement of endogenous genome segments with plasmid sequences, allowing targeted DNA insertion into genome and precise genetic modification in living cells. CRISPR/Cas9-introduced site-specific DNA cleavage greatly promotes HDR at nearby regions and enhances the efficiency of HDR-based gene targeting (12).
In human cells, efficient knock-in of foreign DNA into a selected genomic locus has been long awaited. It is anticipated to facilitate various applications, ranging from gene function study to therapeutic genome editing. Currently, most studies have focused on HDR-based strategies, and the rate of targeted integration was reported to be low (13). This is because HDR in human cells is intrinsically inefficient, whereas NHEJ-mediated DNA repair is prevalent (14). These properties result in generation of few target clones amid a large number of random integrations. Notably, in human embryonic stem cells (ESCs) (15) and induced pluripotent stem cells (iPSCs) (16), which are pluripotent and possess unprecedented potentials for basic research and cell-based therapies (17), gene targeting via HDR is found to be particularly difficult and has impeded the application of these cells (18,19). Even in the presence of ZFN, TALEN or CRISPR/Cas9, the efficiency of HDR-based gene targeting in human pluripotent stem cells is found to be consistently low (20,21). In a recent study by Merkle et al., the efficiency of CRISPR/Cas9-induced HDR-mediated knock-in was estimated to be around 1 × 10−5 without pre-selection (19). Hence, technical expertise for sophisticated selections and cumbersome screening of a large number of clones are required to obtain genetically modified cells (19,21–23).
To date, it still remains unclear whether the extremely low efficiency of HDR is a feature unique to human pluripotent stem cells. Furthermore, it has not been investigated whether the prevalent NHEJ repair can be employed to mediate high-efficiency knock-in in a wide range of human cells, especially in ESCs. In order to address these questions, we constructed a universal reporter system, by targeting the GAPDH locus in human genome with a promoterless fluorescent reporter. Through systematic investigation into the potentials of both HDR and NHEJ repair in mediating CRISPR/Cas9-induced reporter integration, we demonstrated that CRISPR/Cas9-induced NHEJ can mediate reporter knock-in more efficiently than HDR-based strategy, in various human cells types including human ESCs. This finding paves a new path for efficient genome editing in human ESCs and somatic cells, and it offers a great potential in their subsequent applications.
MATERIALS AND METHODS
Cas9 and sgRNA constructs
The human codon-optimized Cas9 (Addgene # 41815) and nickase Cas9D10A (Addgene # 41816) plasmids were obtained from Addgene (8). sgRNAs were designed and constructed as described previously (8,24). Briefly, target sequences (20 bp or 17 bp) starting with guanine and preceding the PAM motif (5′-NGG-3′) were selected from the target genomic regions (8,25). Potential off-target effects of sgRNA candidates were analyzed using the online tool CRISPR Design developed by Zhang's laboratory (http://crispr.mit.edu/), and the sgRNA sequences with fewer off-target sites in human genome were selected for further analysis. Target sequences of sgRNAs used in this study are shown in Supplementary Table S1, and the potential off-target sites for sg-1–4 were listed in Supplementary Table S2.
Donor constructs
Various donor plasmids were constructed. Details of the cloning work were provided in Supplementary Data.
LIG4 overexpression construct
Human LIG4 cDNA was amplified by RT-PCR from the RNA extracted from wild type LO2 cells, and cloned into pCAG-ires-Hyg vector at the BglII and XhoI sites (26). Primers used were listed in Supplementary Table S4.
Cell cultures
H1 human ESCs (WiCell Research Institute) were cultured as previously described (27), on mitomycin-C-inactivated MEF feeders. Prior to transfection, H1 human ESCs were cultured feeder-free in mTeSR1 medium (Stemcell Technologies), on Matrigel (BD Biosciences). Medium was changed daily and cells were subcultured with collagenase IV (Life technologies) every three days (27). Human somatic cell lines were obtained from ATCC. LO2 and HEK293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS); SMMC-7721, BEL-7402, BEL-7404 and H1299 cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 supplemented with 10% FBS; HK2 cells were cultured in 1:1 F-12/DMEM medium supplemented with 10% FBS; and HCT116 cells were cultured in McCoy 5A medium supplemented with 10% FBS. All media and sera were purchased from Life Technologies. All cells were incubated at 37°C and 5% CO2.
Generation of LIG4 null LO2 cells
Wild type LO2 cells were co-transfected twice with Cas9 together with combined sgLIG4-i–iv. The transfected cells were dissociated into single cells and seeded at low density (2000 cells/10cm dish) for clonal expansion. Individual clones were then isolated and analyzed by genome PCR and western blot (details were provided in Supplementary Data). Primers used are shown in Supplementary Table S4.
Transfection and gene targeting assays
H1 human ESCs were transfected using Amaxa nucleofection (Lonza) according to the manufacturer's instructions. Briefly, human ESCs were dissociated using TrypLE into single cells. For each transfection, 5 × 106 cells were mixed with 100 μl pre-warmed nucleofection reagents (82 μl solution-1 and 18 μl solution-B); the cell suspension was then mixed with 16 μg DNA (6 μg donor + 6 μg Cas9 + 4 μg sgRNA) and electroporated. Electroporated H1 human ESCs were cultured on inactivated MEF feeders (27). Medium was changed daily for 4–5 days and cells were dissociated to prepare single cells for FACS analysis. The estimated transfection efficiency was around 53.5% using 16 μg pEGFP-N1 plasmid.
LO2, HEK293T and HCT116 cells were transfected using Lipofectamine 2000 (Life Technologies). SMMC-7721, BEL-7402, BEL-7404, H1299 and HK2 cells were transfected using FuGENE HD (Promega). Cells were seeded into 12-well plates at a density of 5 × 105 cells/well. 1.6 μg DNA (0.6 μg donor + 0.6 μg Cas9 + 0.4 μg sgRNA) and 4 μl Lipofectamine 2000 or 6 μl FuGENE HD were used for transfection into each well, following the manufacturer's instruction. When more than one sgRNA were used, 0.4 μg of total sgRNAs, divided equally by the number of plasmids, was added. In regard of LIG4 rescue assays in Figure 3B, additional 0.6 μg LIG4 cDNA overexpression plasmid was combined with the 0.6 μg donor + 0.6 μg Cas9 + 0.4 μg sgRNA, and 5.5 μl Lipofectamine 2000 was used for the transfection. The transfected cells were passaged once or twice before FACS analysis (BD LSRFortessa Cell Analyzer). Transfection efficiency in these cell lines was estimated by transfection of 1.6 μg pEGFP-N1 plasmid followed by FACS analysis after 48 h.
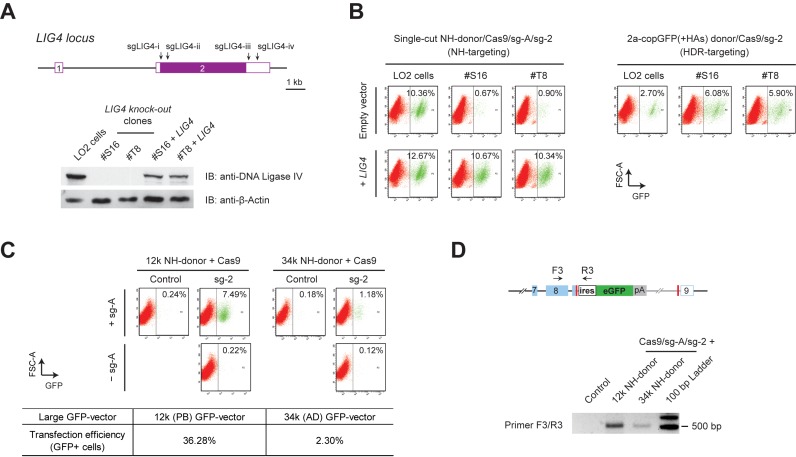
Figure 3.
Conventional NHEJ repair mediates efficient knock-in of large reporter genes. (A) Upper panel shows the schematics of the sgLIG4-i–iv target positions at the LIG4 locus. These sgRNAs were combined and co-transfected with Cas9 into LO2 cells to generate LIG4 knock-out clones. Lower panel is the western blot, showing the loss of DNA ligase IV in the obtained LIG4 null clones, and LIG4 expression introduced by transfection of LIG4 cDNA construct into these cells. (B) FACS analysis of the LIG4 knock-out LO2 cells. Homology-independent knock-in was induced by single-cut NH-donor/Cas9/sg-A/sg-2, and HDR-based knock-in was introduced using 2a-copGFP(+HAs) donor/Cas9/sg-2, in both wild type as well as LIG4 knock-out LO2 cells. Drastic decrease of NH-targeting and rescue by LIG4 overexpression were observed in both LIG4 null clone #S16 and #T8 (left panel). Significant increase of HDR knock-in was also observed in LIG4 null cells (right panel). (C) FACS results showing NHEJ-mediated knock-in with large size donors. 12k and 34k NH-donors were co-transfected with Cas9/sg-A/sg-2 into wild type LO2 cells. Controls were transfected without sg-2 or sg-A. GFP+ cells are gated to the right of the dashed line in each panel. At the same time, constant GFP-expressing 12k (PB) and 34k (AD) GFP-vectors were transfected in parallel; and the transfection efficiencies examined at day 2 by FACS are shown at the lower panel. (D) PCR detection of the reporter integration in the transfected cells (unsorted) in C. Primer pair F3/R3 detected the 5′-junctions of the12k and 34k NH-donors integrated at GAPDH 3′-UTR. PCR amplifications showed DNA fragments at expected sizes.
RESULTS
Quantification of HDR-mediated knock-in in various human cells
To directly quantify and compare the efficiency of CRISPR/Cas9-induced HDR-mediated DNA integration (HDR-targeting) across human ESCs and different somatic cell types, we constructed a reporter system targeting the GAPDH locus in human genome. Three sgRNAs (sg-1–3) were designed to target the GAPDH 3′-UTR in close proximity to the coding sequences (CDS); while a common donor plasmid was generated to carry a promoterless 2a-copGFP sequence flanked by two homology arms (HAs) at each end, thus named 2a-copGFP(+HAs) donor (Figure 1A and Supplementary Figure S1A). When the DSBs induced by Cas9/sg-1–3 are successfully repaired via homologous recombination between the genome and donor template, the 2a-copGFP fragment will be inserted in frame with the genomic GAPDH CDS and result in GFP expression (Figure 1A). This allows direct assessment of the knock-in efficiency by FACS analysis.
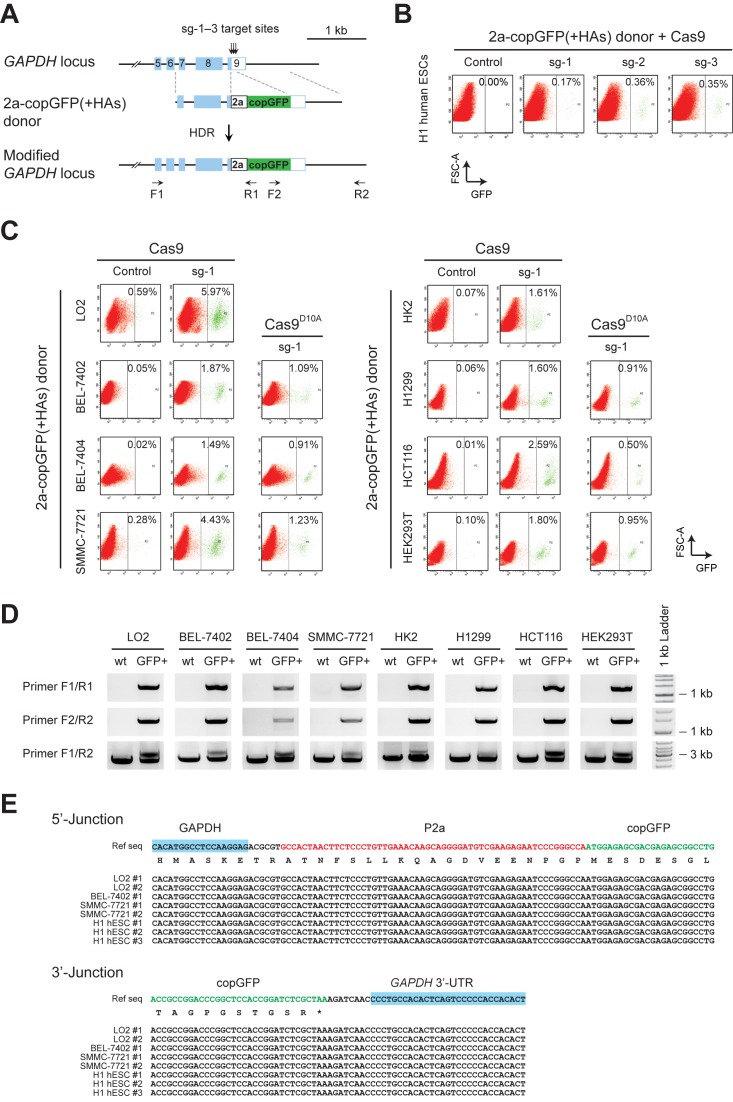
Figure 1.
Varied frequency of HDR-mediated gene targeting in different human cell lines. (A) Schematics of the donor plasmid and targeting strategy for HDR-mediated knock-in of the 2a-copGFP reporter at GAPDH 3′-UTR. Dashed lines indicate sections of homology between the genomic locus and plasmid DNA. Positions of PCR primers used for detection of reporter knock-in are shown. (B) FACS analysis of H1 human ESCs, showing HDR-mediated integration of 2a-copGFP in the presence of Cas9 and sg-1, 2 or 3. Human ESCs were co-transfected with donor/Cas9/sgRNAs by nucleofection, and analyzed four days after transfection. X and Y-axis denote levels of GFP signal and forward scatter area, respectively. (C) FACS results showing varied frequencies of HDR-mediated 2a-copGFP knock-in in different human cell lines. 2a-copGFP(+HAs) donor and Cas9 were co-transfected, with or without sg-1, into eight human cell lines. GFP+ cells from the Cas9/sg-1 targeted cells (green signals gated to the right of the dashed line in each panel) were sorted for further analysis. To compare with the fully functional Cas9, nickase Cas9D10A were co-transfected with 2a-copGFP(+HAs) donor and sg-1 in selected cell lines, and the FACS results were shown in the column 3 and 6. (D) Genome PCR analysis of GFP+ cells produced with Cas9/sg-1 in C. Primer binding sites are indicated in A. Primer pair F1/R1 and F2/R2 amplify the 5′-junction (1350 bp) and the 3′-junction (1473 bp) of the 2a-copGFP integration, respectively. Primers F1/R2 amplified two DNA fragments that represent the wild type (2480 bp) and modified alleles (3241 bp). All amplified DNA fragments exhibited the expected sizes, indicating correct integration of 2a-copGFP via HDR into the genome. (E) Sequencing results of the PCR fragments amplified from the junctions. Expected modifications were confirmed at both 5′- and 3′-junctions, indicating precise integration of 2a-copGFP through HDR-mediated repair.
Consistent with previous studies (19,23), we observed HDR-mediated reporter integration at a low frequency in H1 human ESCs. In the absence of either sgRNAs or Cas9, no GFP+ cells were detected within 105 cells. When the 2a-copGFP(+HAs) donor plasmid was co-transfected with Cas9 and sg-1, 2 or 3, GFP+ cells were observed at frequencies of 0.17–0.36% (Figure 1B) with a transfection efficiency at 53.5% (Supplementary Figure S1B). Variation in targeting efficiencies may reflect the intrinsic properties of different sgRNAs, as indicated by T7E1 assays (Supplementary Figure S1C).
On the other hand, the analysis using this reporter system revealed varied but generally higher efficiencies of CRISPR/Cas9-induced HDR-mediated knock-in in human somatic cell lines. In the presence of Cas9/sg-1, immortalized human cells LO2 showed a targeting efficiency at 5.97%, whereas HK2 and HEK293T cells produced 1.61% and 1.80% GFP+ cells (Figure 1C). Among the human cancer cell lines examined, BEL-7402, BEL-7404 and SMMC-7721 exhibited a targeting efficiency of 1.87%, 1.49% and 4.43% respectively; while H1299 and HCT116 produced 1.60% and 2.59% GFP+ cells, respectively (Figure 1C). Genome PCR and sequencing analysis of the sorted GFP+ cells showed that the 2a-copGFP indeed integrated precisely at the GAPDH 3′-UTR in the genome (Figure 1D,E), which supported that the targeting processes were mediated by the HDR pathway. Transfection efficiency in these somatic cell lines ranged from 50.0% to 73.5% (Supplementary Figure S1D); while frequency of indels induced by Cas9/sg-1, which indicates its genome-targeting activity, ranged from 6.8% to 50.1% in different cell lines (Supplementary Figure S1E). No apparent correlation was observed among the transfection efficiencies, Cas9/sg-1 targeting activities and HDR-mediated knock-in efficiencies. Compared to the fully functional Cas9, nickase Cas9D10A induced reporter knock-in at a lower efficiency (Figure 1C).
Together, these data showed that the HDR-mediated DNA integration occurred at varied frequencies in different human cell types; the knock-in frequency was indeed lower in human ESCs than that in somatic cells, by approximatively 10–20 fold.
Homology-independent knock-in via CRISPR/Cas9-induced NHEJ repair
To explore the potential of CRISPR/Cas9-induced NHEJ in mediating DNA integration (non-homology (NH)-targeting), we constructed two donor plasmids that carry promoterless ires-eGFP, but without homology sequences to the GAPDH locus (Figure 2A). In these NH-donors, we inserted a single sgRNA (sg-A) target site at 5′ of ires-eGFP (single-cut donor), or two sg-A sites at both sides of ires-eGFP (double-cut donor), to introduce cleavage for desired integration and to generate ires-eGFP fragments in different lengths (Figure 2A). The ires element was used to bypass any frameshift caused by NHEJ-introduced indels and to ensure GFP expression after reporter integration.
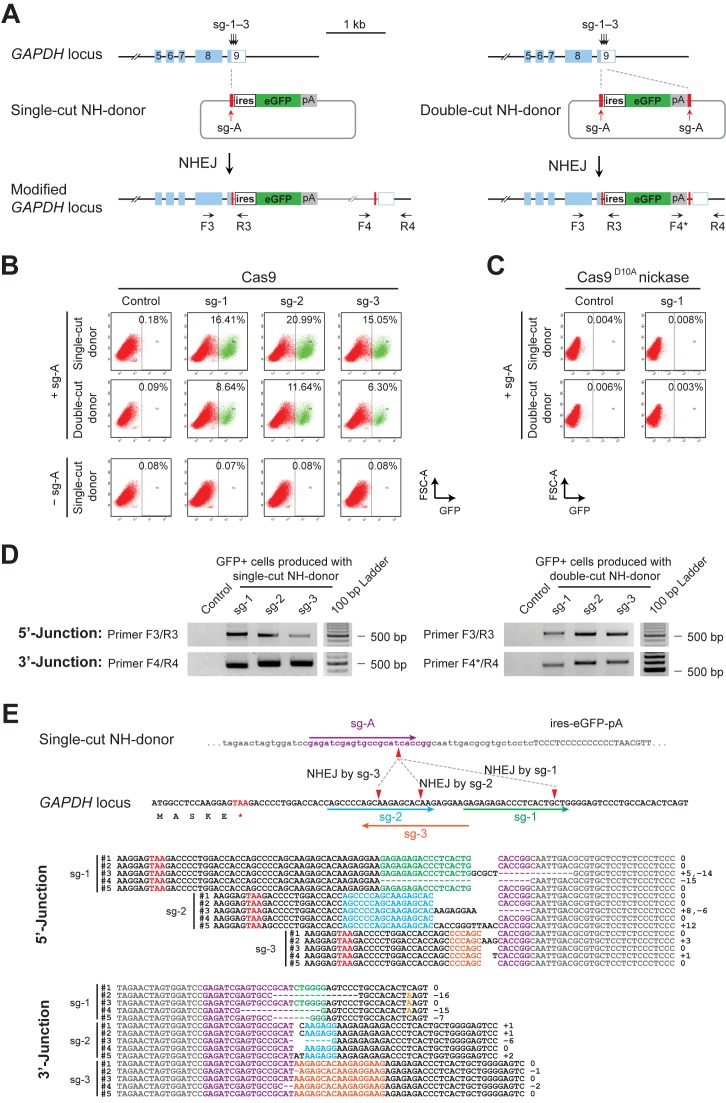
Figure 2.
Homology-independent knock-in of reporter genes into CRISPR/Cas9-induced DSBs in genome. (A) Schematics of the donor plasmids and targeting strategy for CRISPR/Cas9-induced homology-independent insertion of the ires-eGFP reporter at the GAPDH 3′-UTR. Two NH-donor plasmids were generated; one plasmid (left panel) carries a single sg-A target site at the 5′ of ires-eGFP reporter (single-cut donor), and the other (right panel) carries two sg-A target sites at both the 5′ and 3′ sides of ires-eGFP (double-cut donor). Dashed lines indicate positions of DSBs introduced and the cleaved genome and plasmids rejoined. (B) FACS analysis of LO2 cells showing homology-independent knock-in of the ires-eGFP reporter, induced by different combinations of donor plasmids and sgRNAs in the presence of fully functional Cas9. The top row shows the targeting results produced with single-cut NH-donor/Cas9/sg-A/sg-1, 2 or 3, while the middle row shows the results obtained when the double-cut donor plasmid was used. Controls without sg-A are shown in the bottom row. GFP+ cells are gated to the right of the dashed line in each panel. (C) FACS analysis showing no detectable reporter integration in LO2 cells when nickase Cas9D10A was used instead of fully functional Cas9, either with the single- or the double-cut NH-donor plasmid. (D) Genomic PCR of GFP+ cells sorted from samples in B. Positions of primers used for junction detection are indicated in A. PCR amplifications showed DNA fragments at expected sizes, indicating correct integration of the ires-eGFP donors at the GAPDH 3′-UTR. (E) The upper panel shows the schematics of sg-1, 2 and 3 target sites in the GAPDH genomic locus, sg-A target site in the single-cut NH-donor, and the positions of cleavage and rejoining between the genome and the donor DNAs. The lower panel shows sequences of the integration junctions amplified from GFP+ cells produced with the single-cut donor (in D, left panel). 5′- and 3′-junctions of sg-1, 2 or 3-induced integrations were analyzed separately. For each junction, multiple sequences are shown. Nucleotides of different sgRNA target sites and PAMs are color-coded. Sequences from donor templates are shown in gray and the genomic DNA sequences flanking the integration junction are shown in black.
We co-transfected these NH-donor plasmids with Cas9/sg-A/sg-1, 2 or 3 into LO2 cells. Intriguingly, we detected a high frequency of reporter insertion when the single-cut donor was used. GFP+ cells were detected in the presence of sg-1, 2 or 3 at a frequency of 16.41%, 20.99% and 15.05%, respectively (Figure 2B, top row). The targeting efficiencies decreased with all sg-1–3 when the double-cut donor vector was used to produce a shorter ires-eGFP fragment (Figure 2B, middle row). Importantly, no obvious reporter knock-in could be detected in the absence of either sg-1–3 or sg-A (Figure 2B, left column and bottom row), or when nickase Cas9D10A was used to introduce single strand breaks (SSBs; Figure 2C). This indicated that, unlike HDR-based knock-in, site-specific DSBs in both genome and donor DNAs are stringently required for reporter knock-in at a selected genomic locus via the NH-targeting.
PCR analysis of GFP+ cells produced with the single-cut donor verified the integration of ires-eGFP fragment together with vector backbone at the GAPDH 3′-UTR in the genome (Figure 2D, left panel). Similarly, in the GFP+ cells produced with double-cut donor, PCR analysis confirmed the genomic insertion of the short ires-eGFP fragment, which was located between the two sg-A target sites (Figure 2D, right panel). Sequencing analysis of integration junctions in both types of GFP+ cells confirmed the cleavage by specific sgRNAs as well as the rejoining between genome and donor templates at the cleavage sites (Figure 2E and Supplementary Figure S2), suggesting that the integrations indeed occurred at Cas9/sgRNA-induced DSB sites.
To explore whether this NH-targeting approach could produce stable knock-in clones at high efficiency, we expanded the cells transfected with single-cut NH-donor/Cas9/sg-A/sg-2 and 3 at a low density. Among the colonies raised from the unsorted cells, we observed pure GFP+ clones (Supplementary Figure S3A). Among 90 clones randomly isolated from the cells transfected with sg-2, 13 were found to be GFP+ (14.44%). PCR and sequencing analysis confirmed that these clones indeed carried the correct reporter knock-in in their genomes (Supplementary Figure S3B,C), suggesting a success in generating stable knock-in clones without any pre-selection.
To further uncover the molecular basis underlying these homology-independent reporter integrations, we generated DNA ligase IV (LIG4) knock-out LO2 cells by deleting large pieces of the LIG4 CDS using Cas9/sgRNAs (Figure 3A). In the two LIG4 knock-out clones examined, we observed drastic decrease of reporter knock-in after transfection with the single-cut NH-donor/Cas9/sg-A/sg-2, as compared to that in wild type LO2 cells (Figure 3B, left panel, top row). Moreover, the decrease of NH-targeting in these LIG4 null cells could be rescued by a plasmid carrying LIG4 overexpression cassette (Figure 3B, left panel, bottom row). Consistent with the recent studies by Maruyama et al. (28) and Chu et al. (29), we also observed a significant increase of the HDR-based knock-in of 2a-copGFP reporter in these LIG4 null cells (Figure 3B, right panel), which correlated with the loss of NHEJ activity. Collectively, these data showed that the homology-independent reporter integrations observed were indeed largely mediated by the conventional DNA ligase IV-dependent NHEJ pathway.
NHEJ-mediated knock-in is non-directional and it accommodates large DNA inserts
Next, we speculated that the linearized NH-donor or fragments might also integrate via NHEJ repair in the reverse direction and result in no GFP expression (Supplementary Figure S4A). Moreover, the cleavages at both sg-A target sites in the double-cut donor likely produced two fragments, which might compete for genomic integration and lower the efficiency of GFP+ integration (Supplementary Figure S4A, right panel). PCR analysis indeed confirmed the presence of these non-GFP expressing integrations (Supplementary Figure S4B). The detection of non-GFP expressing integrations in the sorted GFP+ cells, which carried the correct reporter knock-in in at least one allele, suggested that different integrations might occur at the two genomic alleles in a single cell. These data indicated that NHEJ-mediated knock-in is non-directional and non-selective, and GFP+ cells observed represented only a portion of cells that carried DNA integrations. This also explained why a lower rate of reporter knock-in was observed when the double-cut donor was used. Given that single-cut donor/Cas9/sg-A/sg-2 produced 20.99% GFP+ cells in LO2 cells (Figure 2B), together with non-GFP expressing events, the total frequency of NHEJ-mediated integration at the single target site deduced might reach up to 40%.
Off-target effect is a general concern to all CRISPR/Cas9-based technology (30). Because of the homology-independent and non-directional nature, the NHEJ-mediated knock-in approach faces a higher chance to introduce DNA insertion at an off-target site than the HDR approach does. To evaluate the off-target effect, we searched for the potential off-target sites that contain ≤ 2 mismatches to the used sgRNAs, throughout the entire human genome (hg19). We found no strong off-target site for sg-A. For sg-1, 2 and 3 targeting GAPDH, we identified 15, 14 and 6 potential off-target sites respectively, and none of these off-targets are located in an exon of a known transcript (Supplementary Table S2). We further selected the top 3 off-targets of sg-2, and performed PCR analysis on off-target integrations. Among the 90 single-cell clones that were expanded previously, none were found to carry reporter integration at the off-target site #1, while integration at off-target site #2 and #3 were found in two and three clones, respectively. Compared with the number of correct knock-in clones obtained (13 out of 90; Supplementary Figure S3), these results indicated that off-target integrations might occur during the NHEJ-mediated knock-in, but at a much lower frequency than the on-target insertion.
Furthermore, we examined whether the NHEJ-mediated knock-in could accommodate a larger insert. We constructed new plasmids named 12k and 34k NH-donors, by inserting the promoterless ires-eGFP reporter together with the 5′ sg-A target sequence into a large PiggyBac vector (12 kb) and an adenoviral vector (34 kb), respectively. These donors can be cleaved at the sg-A target sequence upon the co-transfection with Cas9/sg-A, thus providing linear donors that carry the ires-eGFP in a 12 kb or 34 kb backbone for NHEJ-based knock-in. After co-transfection with the Cas9/sg-A/sg-2, we detected 7.49% GFP+ cells with the 12k NH-donor, and 1.18% with the 34k NH-donor (Figure 3C, upper panel). Together with the 20.99% GFP+ cells observed using the single-cut NH-donor (4.6 kb; Figure 2B), it was apparent that the knock-in frequencies decreased when larger donors were used. This might be caused, at least partially, by the reduced transfection efficiencies of the larger plasmids (Figure 3C, lower panel). PCR analysis of the transfected cells further confirmed the correct knock-in of these large donors at the GAPDH locus (Figure 3D).
Comparison between the frequencies of HDR- and NHEJ-mediated knock-in
To further clarify whether NHEJ repair facilitates DNA integration at a higher efficiency than HDR does, we constructed another donor plasmid that carries an identical ires-eGFP reporter flanked by homology arms to the GAPDH locus. The 5′ homology arm in this plasmid is longer than that in 2a-copGFP(+HAs) donor, covering the GAPDH stop codon as well as sg-2 and sg-3 target sites (Figure 4A, upper panel). When this donor, namely ires-eGFP(+HAs) donor-1, was co-transfected with Cas9/sg-1 in LO2 cells, we detected HDR-mediated reporter knock-in at 7.11% (Figure 4B, Supplementary Figure S5A–C). This frequency was comparable to that produced by HDR-targeting using the 2a-copGFP(+HAs) donor with Cas9/sg-1 (Figure 1C), but lower than that produced by NH-targeting using either single- or double-cut donor and Cas9/sg-1 (Figure 2B).
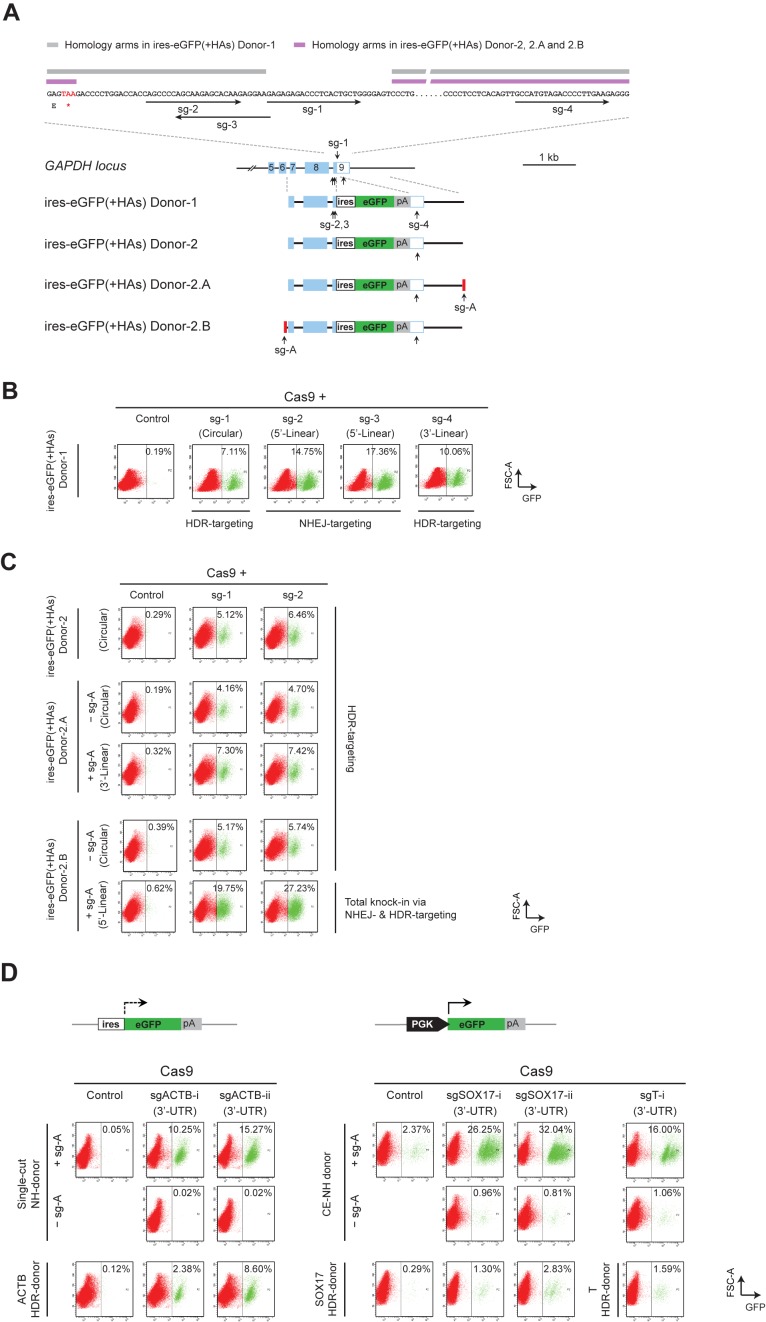
Figure 4.
Comparison between HDR- and NHEJ-mediated reporter knock-in. (A) Schematics showing a zoomed-in view of the sg-1–4 target sites and their positions on the genomic GAPDH locus, as well as the design of ires-eGFP(+HAs) Donor-1, 2, 2.A and 2.B plasmids. Homology arm regions used in the ires-eGFP(+HAs) donor-1 are highlighted in gray, and the HAs used in donor-2, 2.A and 2.B are highlighted in purple. Donor-2.A carries a single sg-A target site at the 3′, and Donor-2.B carries a sg-A target site at the 5′ of the ires-eGFP(+HAs) cassette. (B) FACS analysis of LO2 cells transfected with the ires-eGFP(+HAs) donor-1/Cas9 and sg-1, 2, 3 or 4. Due to the different target positions on the genome and the donor, sg-1 induced HDR-mediated knock-in; sg-2 and sg-3 induced NHEJ-based knock-in; and sg-4 mainly produced GFP+ cells via the HDR-based knock-in through the intact 5′ homology arm. (C) FACS analysis showing HDR-mediated knock-in with circular and linear donor templates. The ires-eGFP(+HAs) Donor-2, 2.A or 2.B were transfected together with Cas9/sg-1 or Cas9/sg-2. The Donor-2.A and 2.B were both examined in the presence of sg-A (linear) as well as in the absence of sg-A (circular). Cas9/sg-A cleaves the Donor-2.A at 3′ of the ires-eGFP(+HAs) cassette and the linearized Donor 2.A produced GFP+ cells via HDR-mediated knock-in. Distinctly, Cas9/sg-A cleaves the Donor-2.B at 5′ of the ires-eGFP(+HAs) cassette, and the linearized Donor 2.B produced high proportion of GFP+ cells via both NHEJ- and HDR-mediated knock-in. (D) FACS results showing NHEJ- and HDR-mediated reporter knock-in at ACTB, SOX17 and T gene loci. Upper panel shows the schematics of ires-eGFP and PGK-eGFP reporters used for knock-in at ACTB and SOX17 or T gene loci, respectively. Single-cut NH-donor was co-transfected with Cas9/sg-A/sgACTB-i or sgACTB-ii to target the ACTB locus (lower left panel, top two rows); while the CE NH-donor was co-transfected with Cas9/sg-A/sgSOX17-i, sgSOX17-ii or sgT-i to target the SOX17 or T gene loci (lower right panel, top two rows). ACTB HDR-donor carrying ires-eGFP, and SOX17 and T HDR-donors containing PGK-eGFP, were co-transfected with Cas9 and corresponding sgRNAs to examine the HDR-based knock-in (lower panel, bottom row). Control samples were transfected without gene-specific sgRNA or sg-A. FACS analysis for the tests at ACTB locus was performed at day 5 after transfection. Cells transfected with PGK-eGFP containing donors for the tests at SOX17 and T loci, were maintained for five passages before FACS analysis. GFP+ cells are gated to the right of the dashed line in each panel.
Interestingly, when we co-transfected the ires-eGFP(+HAs) donor-1 with Cas9/sg-2 or sg-3, which targets to the 5′ homology arm in both genome and donor plasmid, GFP+ cells increased to 14.75% and 17.36% respectively (Figure 4B). These knock-in efficiencies were comparable to NH-targeting with the single-cut donor (Figure 2B, top row). Genome PCR and sequencing confirmed end-joining between genome and donor plasmids beyond the 3′ homology arm (Supplementary Figure S5A,B,D), suggesting that Cas9/sg-2 or sg-3 cleaved both genomic and donor DNAs, and induced NHEJ-mediated integration of the reporters. On the other hand, when Cas9/sg-4 was used to target the 3′ homology arm, GFP+ cells decreased to 10.06% (Figure 4B). Sequencing analysis detected no indels at the 5′-junctions (Supplementary Figure S5A,B,E), suggesting that the intact 5′ homology arms mediated HDR-based integrations, which explained the knock-in observed at a lower frequency.
Next, we constructed the ires-eGFP(+HAs) donor-2 by using a shortened 5′ homology arm that does not contain the sg-2 and sg-3 target sites (Figure 4A, upper panel). This plasmid will not be cleaved by Cas9/sg-2 or sg-3 and can only serve as donor for HDR-based knock-in. Indeed, co-transfection of Cas9/sg-2 with this new donor yielded 6.46% GFP+ cells (Figure 4C, top row). This frequency was much lower than the NHEJ-based knock-in introduced with ires-eGFP(+HAs) donor-1/Cas9/sg-2 (Figure 4B), while it was comparable to the HDR-mediated reporter integrations produced using Cas9/sg-1 together with either type of the (+HAs) donors (Figures 1C and 4B,C).
To compare the NHEJ- and HDR-based knock-in in the identical conditions, we further examined HDR-mediated reporter insertion using a linearized donor. We constructed the ires-eGFP(+HAs) donor-2.A and donor-2.B, by inserting a sg-A target sequence at the 3′ or 5′ of the ires-eGFP(+HAs) cassette, respectively (Figure 4A). These donors thus can be cleaved at the sg-A target site by Cas9/sg-A to provide linear templates carrying homology arms. Using the ires-eGFP(+HAs) donor-2.A in presence of sg-A, we observed 7.30% GFP+ cells with sg-1, and 7.42% with sg-2 (Figure 4C, third row), which were indeed higher than the results obtained using circular donors (Donor-2, or Donor-2.A and 2.B without sg-A; Figure 4C, top, second and fourth rows). These frequencies, however, were still much lower than that produced through NHEJ-based reporter knock-in (Figure 2B, top row; and Figure 4B, with sg-2 and sg-3). Interestingly, using the ires-eGFP(+HAs) donor-2.B and Cas9/sg-A, we observed 19.75% GFP+ cells with sg-1, and 27.23% with sg-2 (Figure 4C, bottom row). It indicated that the linearized donor-2.B enabled NHEJ-based knock-in, and the high proportion of GFP+ cells likely represented a combinatory result of both NHEJ- and HDR-mediated GFP+ knock-in events.
Collectively, these data are consistent with the results observed using 2a-copGFP(+HAs) donor (Figure 1C) or single-cut NH-donor (Figure 2B); and they clearly showed that the simultaneous introduction of DSBs in genome and donors induced targeted DNA integration via NHEJ, at a higher efficiency compared with that mediated by an HDR-based approach.
CRISPR/Cas9-coupled NHEJ introduces efficient knock-in at both active and silenced gene loci
Next, we examined whether the chromatin architecture in a local genomic context influences the efficiency of NHEJ-mediated reporter knock-in, by targeting another actively transcribed locus ACTB and several silenced gene loci, including SOX17, T, OCT4, NANOG and PAX6.
We designed two sgRNAs targeting ACTB 3′-UTR to examine the HDR- and NHEJ-mediated knock-in at the ACTB locus. By co-transfecting the single-cut NH-donor/Cas9/sg-A together with sgACTB-i or sgACTB-ii, we observed GFP+ cells at 10.25% and 15.27%, respectively (Figure 4D, lower left panel, top row). Using the newly constructed ACTB HDR-donor, which carried the ires-eGFP flanked by homology arms to ACTB gene locus, we observed the HDR-based knock-in at 2.38% with sgACTB-i, and 8.60% with sgACTB-ii (Figure 4D, lower left panel, bottom row). Both the NHEJ- and HDR-based knock-in frequencies were comparable to that observed at the GAPDH locus.
In order to examine knock-in at a silenced gene locus directly by FACS analysis, we employed the PGK-eGFP reporter (Figure 4D, upper right panel), which will express GFP after integration regardless whether the target locus is actively transcribed or not. We constructed a constant expression (CE) NH-donor which carries the sg-A target sequence at 5′ of the PGK-eGFP cassette; meanwhile, we generated sgRNAs targeting the SOX17 and T 3′-UTRs. It is noteworthy that because the expression of PGK-eGFP reporter is independent from integration orientations, the GFP+ cells observed in these assays represented knock-in events in either orientation. After transfected with the CE NH-donor/Cas9/sg-A and one of the gene-specific sgRNAs, the LO2 cells were maintained for five passages to eliminate the transient GFP expression before FACS analysis. Indeed, we detected 26.25% and 32.04% GFP+ cells for sgSOX17-i and sgSOX17-ii respectively, and observed 16.00% GFP+ cells with sgT-i (Figure 4D, lower right panel, top row). In contrast, only around 2–3% GFP+ cells were observed in the absence of gene-specific sgRNA; and around 1% GFP+ cells were detected in the absence of sg-A. Using this CE NH-donor, we also examine the NHEJ-mediated knock-in at various positions of OCT4, NANOG, T and PAX6 gene loci, which are largely silenced in LO2 cells. Indeed, we observed varied knock-in frequencies, which correlated neither with the target positions in a gene, nor the transcriptional status of the target loci (Supplementary Figure S6A,B), suggesting that the actual targeting efficiency was largely determined by the intrinsic properties of a sgRNA.
Furthermore, we examined the HDR-based knock-in at the SOX17 and T genomic loci, using donor plasmids carrying PGK-eGFP flanked by homology arms to SOX17 or T genomic regions respectively. Similarly, the transfected cells were passaged for five times before FACS analysis. By transfecting the SOX17 HDR-donor together with Cas9/sgSOX17-i or sgSOX17-ii, we observed 1.30% and 2.83% GFP+ cells, which indicated the HDR-mediated knock-in at SOX17 locus; while usage of T HDR-donor together with Cas9/sgT-i produced 1.59% GFP+ cells (Figure 4D, lower right panel, bottom row). These frequencies were indeed much lower than that produced by the NHEJ-based knock-in at the same target sites (Figure 4D, lower right panel, top two rows). Moreover, they were also lower than the HDR-based knock-in observed in actively transcribed GAPDH and ACTB loci (Figure 4B,C and D, lower left panel, bottom row), which is consistent with previous studies showing that active transcription enhances homologous recombination (31,32).
Collectively, these results indicated that CRISPR/Cas9-coupled NHEJ could mediate efficient knock-in at both active and silenced gene loci, and the efficiencies were higher than that produced by an HDR-based approach.
Efficient knock-in via CRISPR/Cas9-coupled NHEJ in human ESCs and somatic cell lines
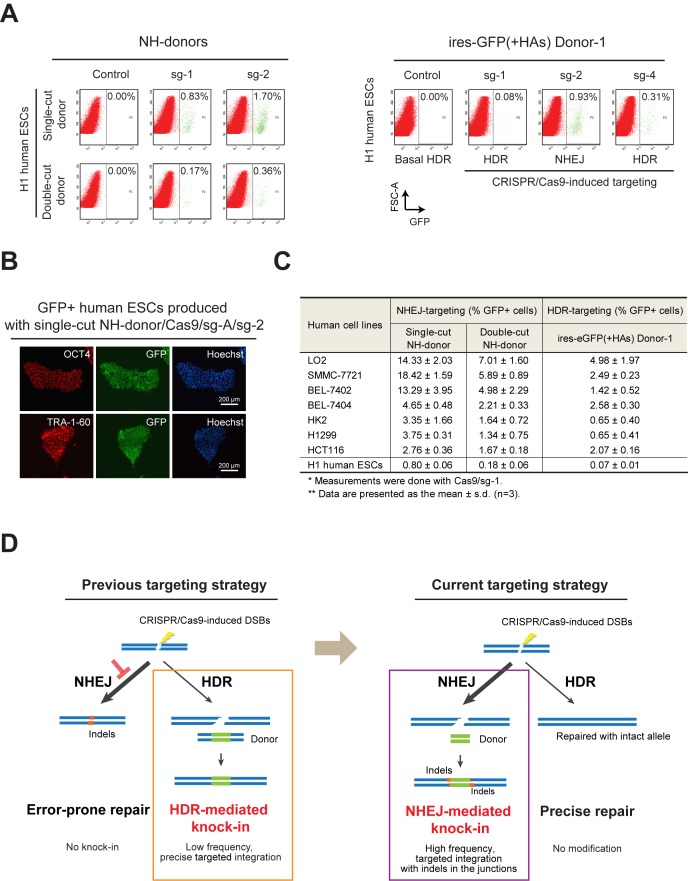
Using the ires-eGFP donors with or without HAs, we examined the efficiency of NHEJ-mediated reporter knock-in in human ESCs. Indeed, the reporter knock-in observed was more efficient compared to that introduced using the HDR-based approach. Co-transfection of single-cut NH-donor/Cas9/sg-A/sg-1 produced 0.83% GFP+ cells, and the proportion of GFP+ cells increased up to 1.70% when sg-2 was used (Figure 5A, left panel). These NHEJ-mediated integrations indeed occurred at a higher frequency than the HDR-based knock-in, induced either by Cas9/sg-1–3 with the 2a-copGFP(+HAs) donor (Figure 1B), or by Cas9/sg-1 with the ires-eGFP(+HAs) donor-1 (Figure 5A, right panel, sg-1). Consistently, when we co-transfected the ires-eGFP(+HAs) donor-1 with Cas9/sg-2, which can simultaneously cleave both genome and donor DNAs and induce NHEJ-mediated donor integration, the yield of GFP+ cells increased to 0.93% (Figure 5A, right panel, sg-2). This insertion rate is also higher than that in HDR-targeting (Figure 1B and Figure 5A, right panel, sg-1), and closer to that produced with the single-cut NH-donor/Cas9/sg-A/sg-1 or sg-2 (Figure 5A, left panel). We further sorted the GFP+ cells produced with single-cut donor/Cas9/sg-A/sg-2. These cells maintained the human ESC morphology in culture and expressed the pluripotency markers OCT4 and TRA-1–60 (Figure 5B), suggesting that the NHEJ knock-in process did not interfere with the maintenance of pluripotency state, and it allows the generation of stable knock-in cells simply by FACS sorting.
Figure 5.
CRISPR/Cas9-coupled NHEJ mediates more efficient reporter knock-in than HDR in human ESCs and somatic cell lines. (A) Knock-in of ires-eGFP reporter via CRISPR/Cas9-coupled NHEJ repair in human ESCs. Left panel shows the NHEJ-mediated knock-in of ires-eGFP reporter. Both single-cut and double-cut NH- donors were examined in combination with sg-1 or sg-2. Right panel shows the reporter integration introduced with the ires-eGFP(+HAs) donor-1 and Cas9/sg-1, 2 or 4. Sg-1 and sg-4 mainly produced GFP+ cells via the HDR-based knock-in; while sg-2 induced NHEJ-based knock-in, due to the presence of sg-2 target site at the 5′-HA in the donor. FACS analyses were performed four days after nucleofection. GFP+ cells are gated to the right of the dashed line in each panel. (B) Immunofluorescence detection of pluripotency markers OCT4 (upper panel) and TRA-1–60 (lower panel) in the GFP+ cells sorted from H1 human ESCs transfected with single-cut donor/Cas9/sg-A/sg-2. Nuclei were counterstained with Hoechst. (C) Summary of Cas9/sg-1-induced NHEJ- and HDR-targeting in human ESCs and somatic cell lines examined. Single-cut and double-cut NH-donor were used for NHEJ-based targeting. The ires-eGFP (+HAs) donor-1 plasmid was used for HDR-based targeting. Percentages of GFP+ cells, presented as the mean ± s.d., were derived from three independent experiments. (D) Schematic diagram illustrating the balance between NHEJ- and HDR-mediated DSB repair in human cells, as well as the strategies (boxes) used for gene targeting.
To verify whether the CRISPR/Cas9-coupled NHEJ-targeting strategy can knock-in the reporter into other genomic loci in human ESCs efficiently, we co-transfected the single-cut NH-donor/Cas9/sg-A together with sgRNAs targeting the OCT4 or ACTB genes at their 3′-UTRs. Both OCT4 and ACTB genes are actively transcribed in human ESCs; hence, knock-in of ires-eGFP reporter at their 3′-UTRs will produce GFP and allow direct analysis by FACS. Indeed, we observed 0.55% and 0.40% GFP+ cells in the H1 human ESCs transfected with sgOCT4-iv or sgACTB-ii, respectively (Supplementary Figure S7A). PCR and sequencing analysis on the OCT4 locus further confirmed the integration of single-cut donor at the target site (Supplementary Figure S7B,C). Collectively, these data showed that CRISPR/Cas9-coupled NHEJ repair can mediate efficient knock-in of reporter genes into a selected genomic locus in human ESCs.
To compare in a broader range of human cells, we further quantified NHEJ- and HDR-mediated reporter knock-in in other human somatic cell lines. We found that, indeed, the NHEJ-based reporter knock-in was more efficient than HDR-mediated integration in all human somatic cell lines examined (Figure 5C). The ratio of GFP+ cells ranged from 2.76% in HCT116 cells to 18.42% in SMMC-7721 cells, when single-cut donor/Cas9/sg-A/sg-1 were used (Figure 5C). Interestingly, LO2, BEL-7402, and SMMC-7721 cells exhibited relatively higher efficiency in both NHEJ and HDR-mediated targeting, whereas, HCT116, H1299 and HK2 were relatively inefficient in both targeting strategies (Figure 5C). Notably, among all the cell lines examined, human ESCs showed the lowest efficiency in both NHEJ- and HDR-targeting (Figure 5C). These results implied that there may be intrinsic restrictions hampering efficient gene targeting in human ESCs, via either HDR or NHEJ repair. This observation is consistent with previous literatures (19,33–34), suggesting that human ESCs possess unique properties in repairing DNA damage. Therefore, further investigation is needed to uncover the deliberate mechanisms and resolve existing discrepancy regarding DNA repair in human ESCs.
DISCUSSION
In summary, our results demonstrate that the NHEJ repair can enable efficient rejoining of genome and plasmids following CRISPR/Cas9-induced DNA DSBs, which permits knock-in of large DNAs at remarkably higher efficiency than HDR-mediated integrations, in all human cell lines examined. These data have established CRISPR/Cas9-coupled NHEJ repair as a valuable path for efficient knock-in in human ESCs and somatic cells (Figure 5D), providing great potential in biomedical research and therapeutic applications.
Efficient knock-in of exogenous DNA is a highly desirable technology for studies carried out in human cells. Following previous far-reaching success in generating genetically modified mice (35,36), tremendous effort has been made to exploit HDR-mediated approaches for precise DNA insertion or replacement at a selected genomic locus (18,37). Even after the emergence of ZFNs, TALENs and CRISPR/Cas9 technologies, the competence and potential of other DNA repair mechanisms were not well explored, and most gene targeting studies still focused on HDR-mediated approaches to introduce genomic knock-in (19–21,23,28–29).
In fact, NHEJ repair is predominant in mammalian cells. Random DNA integration via NHEJ has been widely used to generate transgenic animals and cell lines, and the frequency was estimated to be over-1000-fold higher than HDR-mediated DNA insertion (37). In the studies of HDR-based gene targeting using TALEN and CRISPR/Cas9, NHEJ-introduced indels were found to occur at a higher frequency than HDR-mediated reporter knock-in (38), and the prevalence of NHEJ-based knock-in was observed when single strand oligonucleotide donors were provided for HDR-mediated gene correction (39). However, very few studies have directly investigated the nature of these NHEJ-based integration processes, and their application potential as a biological technology has not attracted attention until recently. Following pioneer studies that showed the NHEJ-mediated capture of exogenous DNA at genomic DSBs (40), Orlando et al. (2010) first found that short oligonucleotides (<100 bp) could be inserted efficiently at ZFN-induced genomic DSBs via NHEJ repair (41). By introducing a ZFN or TALEN target sequence in donor plasmids, Cristea et al. and Maresca et al. (2013) further showed that simultaneous cleavage on both plasmid and genome DNAs by nucleases enabled targeted integration of large plasmid DNA at the genomic DSBs via NHEJ repair (42,43). This design was then coupled to CRISPR/Cas9 system for reporter knock-in in zebrafish (44–48) and Xenopus (49), in which, the HDR-mediated gene insertion was extremely inefficient. To date, the potential of CRISPR/Cas9-induced NHEJ in mediating large DNA insertion has not been systematically investigated in human cells, and the efficient knock-in in human ESCs still remains a challenge.
In this study, we constructed promoterless fluorescent reporters targeted to the GAPDH locus in human genome. The ubiquitously active nature of the GAPDH gene enables GFP expression upon correct knock-in, allowing rapid assessment by FACS and direct comparison among different human cell types without any cumbersome process of raising single-cell clones. In addition, we employed a sg-A target site taken from prokaryotic DNA sequence, which has no homology to human genome, thus it makes the Cas9/sg-A universal in providing linearized donors for NHEJ knock-in and suitable for the direct comparison across multiple human cell lines. Using these reporter systems, with or without homology arms, we quantified the homology-dependent and independent reporter knock-in directly in various human cell lines. We found that, indeed, CRISPR/Cas9-coupled NHEJ mediates efficient DNA insertion when DSBs are introduced to both genome and donor DNAs; and the knock-in efficiency is much higher than that mediated by HDR-based approach, in all human cell lines examined. Our analysis using LIG4 null LO2 cells showed that the high-frequency homology-independent knock-in events were indeed largely mediated by the conventional NHEJ (C-NHEJ) pathway. Clonal expansion further demonstrated that, with this NHEJ-based targeting approach, stable knock-in clones could be generated in a fluorescence-independent selection-free manner. Together, these data suggest that CRISPR/Cas9-coupled NHEJ repair can provide a valuable path for efficient knock-in in human cells; meanwhile, the results obtained have established our reporter systems as valuable tools for rapid quantification of the HDR and NHEJ activities across different cell lines/clones, which could be useful for dissecting a given molecular pathway, or for screening of therapeutic compounds to restore the impaired DNA repair responsible for human diseases.
In addition, we have unraveled distinct features of the NHEJ-mediated knock-in. Unlike the HDR-based DNA insertion, the NHEJ-based knock-in stringently relies on the presence of DSBs in both genome and donor DNAs; it allows integration at either orientation and it can accommodate insertion of large DNAs (up to 34 kb). Importantly, the homology-independent nature also rendered this NHEJ knock-in approach an advantage in targeting silenced genomic loci, which have been shown to be difficult to access through traditional HDR-based knock-in strategy (50,51). Currently, the CE-NH donor we employed still produced non-specific GFP signal due to transient expression and random integration; hence, further work is needed to improve the reporter design for knock-in at a silenced gene locus.
On the other hand, we showed that the high-efficiency NHEJ knock-in approach could potentially introduce undesired DNA integration at low frequency, due to off-target cleavage by Cas9/sgRNA (30). It is possible that off-target integrations may also produce GFP if the target locus is actively transcribed or the PGK-eGFP reporter is used. Therefore, it is important to follow high-stringency criteria in sgRNA design for minimizing the off-target effect. Usage of the new Cas9 that has been further optimized to reduce off-target effect is also likely beneficial (52). Moreover, as shown by our results as well as by other studies (38,39), high-frequency NHEJ-mediated repair events may occur in many types of genome editing without being detected in a particular assay; hence, unwanted genomic modification should be considered and controlled during data analysis and interpretation. Together, to beware and to confine these limitations are important for further improving the CRISPR/Cas9-based genome editing technology, via either HDR- or NHEJ-mediated DNA repair.
Interestingly, although the C-NHEJ machinery often introduces indels by repairing non-compatible or damaged DNA ends (9), the alternative NHEJ (A-NHEJ) pathway initiates a repair process by single-strand resection (9), and the 3′-5′ exonuclease activity of Cas9 can also introduce deletions by trimming the DNA ends (7), we observed substantial precise ligations between cleaved plasmid and genome DNAs during the NHEJ-mediated knock-in (Figure 2E and Supplementary Figure S4). This is consistent to the analysis results on ZFN- or TALEN-induced DNA integration by Cristea et al. and Maresca et al. (42,43), as well as on CRISPR/Cas9-induced chromosomal inversions by Li et al. (53). These data, together with previous evidence on the precise repair by C-NHEJ (54,55), support that the C-NHEJ pathway can largely mediate precise ligation of DNA ends generated by engineered nucleases, prompting a greater potential of the NHEJ-mediated gene targeting in a wider range of applications.
Supplementary Material
Acknowledgments
We thank Huck-Hui Ng, Wenyi Feng, Jia-Hui Ng, Wing Ki Wong and Tsz Yau Wong Gerald for critical comments on the manuscript.
Footnotes
Present address: Bo Feng, School of Biomedical Sciences, The Chinese University of Hong Kong, Room 105A, Lo Kwee-Seong Integrated Biomedical Sciences Building, Area 39, Shatin, N.T., Hong Kong.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Research Grants Council of Hong Kong [CUHK478812, CUHK14102214 and CUHK14104614 to B.F.; HKUST T13-607/12R to Y.I.]; National Natural Science Foundation of China [NSFC 31171433 to B.F., in part]; National Basic Research Program of China [973-Program 2015CB964700 to Y.L., in part]; Shenzhen SZSIA foundation [JCYJ20140425184428469 to J.R., in part]. X.J., C.T. and W.Y. are supported by the CUHK graduate school scholarship. Funding for open access charge: The Research Grants Council of Hong Kong; National Basic Research Program of China.
Conflict of interest statement. None declared.
REFERENCES
- 1.Maeder M.L., Thibodeau-Beganny S., Osiak A., Wright D.A., Anthony R.M., Eichtinger M., Jiang T., Foley J.E., Winfrey R.J., Townsend J.A., et al. Rapid ‘open-source’ engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol. Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reyon D., Tsai S.Q., Khayter C., Foden J.A., Sander J.D., Joung J.K. FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotech. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhaya D., Davison M., Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 2011;45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 4.Cox D.B., Platt R.J., Zhang F. Therapeutic genome editing: prospects and challenges. Nat. Med. 2015;21:121–131. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1077. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 6.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lieber M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F., Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heyer W.D., Ehmsen K.T., Liu J. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang H., Wang H., Shivalila C.S., Cheng A.W., Shi L., Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kan Y., Ruis B., Lin S., Hendrickson E.A. The mechanism of gene targeting in human somatic cells. PLoS Genet. 2014;10:e1004251. doi: 10.1371/journal.pgen.1004251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mao Z., Bozzella M., Seluanov A., Gorbunova V. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair (Amst.) 2008;7:1765–1771. doi: 10.1016/j.dnarep.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 17.Wu S.M., Hochedlinger K. Harnessing the potential of induced pluripotent stem cells for regenerative medicine. Nat. Cell Biol. 2011;13:497–505. doi: 10.1038/ncb0511-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song H., Chung S.K., Xu Y. Modeling disease in human ESCs using an efficient BAC-based homologous recombination system. Cell Stem Cell. 2010;6:80–89. doi: 10.1016/j.stem.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 19.Merkle F.T., Neuhausser W.M., Santos D., Valen E., Gagnon J.A., Maas K., Sandoe J., Schier A.F., Eggan K. Efficient CRISPR-Cas9-mediated generation of knockin human pluripotent stem cells lacking undesired mutations at the targeted locus. Cell Rep. 2015;11:875–883. doi: 10.1016/j.celrep.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lombardo A., Genovese P., Beausejour C.M., Colleoni S., Lee Y.L., Kim K.A., Ando D., Urnov F.D., Galli C., Gregory P.D., et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 21.Hockemeyer D., Soldner F., Beard C., Gao Q., Mitalipova M., DeKelver R.C., Katibah G.E., Amora R., Boydston E.A., Zeitler B., et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hockemeyer D., Wang H., Kiani S., Lai C.S., Gao Q., Cassady J.P., Cost G.J., Zhang L., Santiago Y., Miller J.C., et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rong Z., Zhu S., Xu Y., Fu X. Homologous recombination in human embryonic stem cells using CRISPR/Cas9 nickase and a long DNA donor template. Protein Cell. 2014;5:258–260. doi: 10.1007/s13238-014-0032-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu J., Lei Y., Wong W.K., Liu S., Lee K.C., He X., You W., Zhou R., Guo J.T., Chen X., et al. Direct activation of human and mouse Oct4 genes using engineered TALE and Cas9 transcription factors. Nucleic Acids Res. 2014;42:4375–4390. doi: 10.1093/nar/gku109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu Y., Sander J.D., Reyon D., Cascio V.M., Joung J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng B., Jiang J., Kraus P., Ng J.H., Heng J.C., Chan Y.S., Yaw L.P., Zhang W., Loh Y.H., Han J., et al. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat. Cell Biol. 2009;11:197–203. doi: 10.1038/ncb1827. [DOI] [PubMed] [Google Scholar]
- 27.Chia N.Y., Chan Y.S., Feng B., Lu X., Orlov Y.L., Moreau D., Kumar P., Yang L., Jiang J., Lau M.S., et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468:316–320. doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- 28.Maruyama T., Dougan S.K., Truttmann M.C., Bilate A.M., Ingram J.R., Ploegh H.L. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu V.T., Weber T., Wefers B., Wurst W., Sander S., Rajewsky K., Kuhn R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 30.Mali P., Aach J., Stranges P.B., Esvelt K.M., Moosburner M., Kosuri S., Yang L., Church G.M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nickoloff J.A. Transcription enhances intrachromosomal homologous recombination in mammalian cells. Mol. Cell. Biol. 1992;12:5311–5318. doi: 10.1128/mcb.12.12.5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gottipati P., Helleday T. Transcription-associated recombination in eukaryotes: link between transcription, replication and recombination. Mutagenesis. 2009;24:203–210. doi: 10.1093/mutage/gen072. [DOI] [PubMed] [Google Scholar]
- 33.Rocha C.R., Lerner L.K., Okamoto O.K., Marchetto M.C., Menck C.F. The role of DNA repair in the pluripotency and differentiation of human stem cells. Mutat. Res. 2013;752:25–35. doi: 10.1016/j.mrrev.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 34.Weissbein U., Benvenisty N., Ben-David U. Quality control: genome maintenance in pluripotent stem cells. J. Cell Biol. 2014;204:153–163. doi: 10.1083/jcb.201310135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koller B.H., Hagemann L.J., Doetschman T., Hagaman J.R., Huang S., Williams P.J., First N.L., Maeda N., Smithies O. Germ-line transmission of a planned alteration made in a hypoxanthine phosphoribosyltransferase gene by homologous recombination in embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 1989;86:8927–8931. doi: 10.1073/pnas.86.22.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Capecchi M.R. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat. Rev. Genet. 2005;6:507–512. doi: 10.1038/nrg1619. [DOI] [PubMed] [Google Scholar]
- 37.Vasquez K.M., Marburger K., Intody Z., Wilson J.H. Manipulating the mammalian genome by homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8403–8410. doi: 10.1073/pnas.111009698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hendel A., Bak R.O., Clark J.T., Kennedy A.B., Ryan D.E., Roy S., Steinfeld I., Lunstad B.D., Kaiser R.J., Wilkens A.B., et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 2015;33:985–989. doi: 10.1038/nbt.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang L., Guell M., Byrne S., Yang J.L., De Los Angeles A., Mali P., Aach J., Kim-Kiselak C., Briggs A.W., Rios X., et al. Optimization of scarless human stem cell genome editing. Nucleic Acids Res. 2013;41:9049–9061. doi: 10.1093/nar/gkt555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin Y., Waldman A.S. Capture of DNA sequences at double-strand breaks in mammalian chromosomes. Genetics. 2001;158:1665–1674. doi: 10.1093/genetics/158.4.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orlando S.J., Santiago Y., DeKelver R.C., Freyvert Y., Boydston E.A., Moehle E.A., Choi V.M., Gopalan S.M., Lou J.F., Li J., et al. Zinc-finger nuclease-driven targeted integration into mammalian genomes using donors with limited chromosomal homology. Nucleic Acids Res. 2010;38:e152. doi: 10.1093/nar/gkq512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cristea S., Freyvert Y., Santiago Y., Holmes M.C., Urnov F.D., Gregory P.D., Cost G.J. In vivo cleavage of transgene donors promotes nuclease-mediated targeted integration. Biotechnol. Bioeng. 2013;110:871–880. doi: 10.1002/bit.24733. [DOI] [PubMed] [Google Scholar]
- 43.Maresca M., Lin V.G., Guo N., Yang Y. Obligate ligation-gated recombination (ObLiGaRe): custom-designed nuclease-mediated targeted integration through nonhomologous end joining. Genome Res. 2013;23:539–546. doi: 10.1101/gr.145441.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Auer T.O., Duroure K., De Cian A., Concordet J.P., Del Bene F. Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res. 2014;24:142–153. doi: 10.1101/gr.161638.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Irion U., Krauss J., Nusslein-Volhard C. Precise and efficient genome editing in zebrafish using the CRISPR/Cas9 system. Development. 2014;141:4827–4830. doi: 10.1242/dev.115584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kimura Y., Hisano Y., Kawahara A., Higashijima S. Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Sci. Rep. 2014;4:6545. doi: 10.1038/srep06545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hisano Y., Sakuma T., Nakade S., Ohga R., Ota S., Okamoto H., Yamamoto T., Kawahara A. Precise in-frame integration of exogenous DNA mediated by CRISPR/Cas9 system in zebrafish. Sci. Rep. 2015;5:8841. doi: 10.1038/srep08841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li J., Zhang B.B., Ren Y.G., Gu S.Y., Xiang Y.H., Du J.L. Intron targeting-mediated and endogenous gene integrity-maintaining knockin in zebrafish using the CRISPR/Cas9 system. Cell Res. 2015;25:634–637. doi: 10.1038/cr.2015.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi Z., Wang F., Cui Y., Liu Z., Guo X., Zhang Y., Deng Y., Zhao H., Chen Y. Heritable CRISPR/Cas9-mediated targeted integration in Xenopus tropicalis. FASEB J. 2015;29:4914–4923. doi: 10.1096/fj.15-273425. [DOI] [PubMed] [Google Scholar]
- 50.Thomson A.J., Marques M.M., McWhir J. Gene targeting in livestock. Reprod. Suppl. 2003;61:495–508. [PubMed] [Google Scholar]
- 51.Marques M.M., Thomson A.J., McCreath K.J., McWhir J. Conventional gene targeting protocols lead to loss of targeted cells when applied to a silent gene locus in primary fibroblasts. J. Biotechnol. 2006;125:185–193. doi: 10.1016/j.jbiotec.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 52.Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2015;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y., Park A.I., Mou H., Colpan C., Bizhanova A., Akama-Garren E., Joshi N., Hendrickson E.A., Feldser D., Yin H., et al. A versatile reporter system for CRISPR-mediated chromosomal rearrangements. Genome Biol. 2015;16:111. doi: 10.1186/s13059-015-0680-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin W.Y., Wilson J.H., Lin Y. Repair of chromosomal double-strand breaks by precise ligation in human cells. DNA Repair (Amst.) 2013;12:480–487. doi: 10.1016/j.dnarep.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Betermier M., Bertrand P., Lopez B.S. Is non-homologous end-joining really an inherently error-prone process? PLoS Genet. 2014;10:e1004086. doi: 10.1371/journal.pgen.1004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.