Abstract
RAD52 is a member of the homologous recombination (HR) pathway that is important for maintenance of genome integrity. While single RAD52 mutations show no significant phenotype in mammals, their combination with mutations in genes that cause hereditary breast cancer and ovarian cancer like BRCA1, BRCA2, PALB2 and RAD51C are lethal. Consequently, RAD52 may represent an important target for cancer therapy. In vitro, RAD52 has ssDNA annealing and DNA strand exchange activities. Here, to identify small molecule inhibitors of RAD52 we screened a 372,903-compound library using a fluorescence-quenching assay for ssDNA annealing activity of RAD52. The obtained 70 putative inhibitors were further characterized using biochemical and cell-based assays. As a result, we identified compounds that specifically inhibit the biochemical activities of RAD52, suppress growth of BRCA1- and BRCA2-deficient cells and inhibit RAD52-dependent single-strand annealing (SSA) in human cells. We will use these compounds for development of novel cancer therapy and as a probe to study mechanisms of DNA repair.
INTRODUCTION
Mechanisms of DNA repair are essential for maintenance of genome integrity in all organisms. Multiple DNA repair systems evolved to eliminate a broad variety of DNA lesions caused by different exogenous agents or genotoxic products of metabolism. In normal cells, the specificities of different DNA repair mechanisms overlap to assure efficient genome protection (1–3). However, cancer cells often lose some DNA repair pathways due to intrinsic genome instability. In this case, cancer cell viability depends on the remaining alternative DNA repair mechanisms (4–6). Thus, it was demonstrated that PARP1, a protein involved in DNA damage signaling and repair of DNA single-stranded breaks (SSB), is essential for viability of cancer cells that are deficient in the homologous recombination (HR) pathway (7,8). Furthermore, hereditary breast cancer and ovarian cancer cells, which often carry mutations in HR proteins BRCA1 or BRCA2 can be eliminated using PARP1 inhibitors with a minimal harm to normal cells with at least one copy of functional BRCA1/2 genes (4,6).
Recently, S. Powell group discovered that BRCA1/2-deficient cancer cells are not viable when RAD52 protein is inactivated (9). In addition, RAD52 knockdown also causes lethality to human cells deficient in PALB2 (partner and localizer of BRCA2) and five RAD51 paralogs, including RAD51C (10,11). Mutations in PALB2 and RAD51C also contribute to hereditary breast and ovarian cancer (12,13). Previously, inviability of double mutations in RAD52 and RAD51C genes was reported in chicken DT-40 cells (14). Importantly, inactivation of PARP1 and RAD52 causes lethality of BRCA1/2-deficient and PALB2-deficient cells through different mechanisms. Inactivation of PARP1 causes disruption of repair of DNA SSBs. During DNA replication, unrepaired SSBs may cause formation of DNA double-stranded breaks (DSBs) or stalled replication forks which are repaired by the HR pathway. BRCA1/2/PALB2, constitute the major sub-pathway of HR; mutations in these proteins incapacitates HR making hereditary breast and ovarian cancer cells vulnerable to PARP1 inhibitors. However, recent data indicate that in addition to the BRCA1/2/PALB2 sub-pathway, the secondary HR sub-pathway operates in mammalian cell that depends on RAD52 protein (9,10). In normal mammalian cells, this pathway plays a minor role, as RAD52−/− mice are viable and fertile and do not display DNA damage sensitivity, abnormalities, or significant cancer predisposition (15). However, this sub-pathway becomes essential for viability in cells that lack the BRCA1/2/PALB2 sub-pathway. Thus, these findings identify RAD52 as a potential therapeutic target against familial breast cancer, familial ovarian cancer and other types of cancer with defective BRCA1/2/PALB2 genes. In accord with this view, one of the co-authors (T.S.) demonstrated that inhibition of RAD52 DNA binding activity by small peptide aptamer exerted synthetic lethality in BRCA1- and BRCA2-mutated cancer cells without any detectable side effects in normal cells and in mice (16).
Here, in order to develop small molecule inhibitors of the RAD52 ssDNA annealing activity we performed a high throughput screening of a 372,903-compound library using a fluorescence-quenching assay. We identified 70 putative inhibitors of RAD52, were further characterized using biochemical and cell-based assays. We found several compounds that specifically inhibit the biochemical activities of RAD52 and suppress growth of BRCA1- and BRCA2-deficient cells. One of these compounds, D-I03, also specifically inhibits RAD52-dependent single-strand annealing (SSA) in human cells. We will use these compounds for development of novel cancer therapy and also as a probe to study the mechanisms of DNA repair in human cells.
MATERIALS AND METHODS
Chemicals, proteins and DNA
Cisplatin, was purchased from Sigma-Aldrich. Human RAD52 and RAD51 (Supplementary Figure S1) were purified as described (17). The oligonucleotides (Supplementary Table S1) were purchased from IDT, Inc and further purified by electrophoresis (18). Supercoiled pUC19, pCBASce and pMX-GFP plasmid DNAs were purified using Qiagen kits. All DNA concentrations are expressed as moles of nucleotide.
Compound libraries, compounds
We used a 93672-compound Broad's diversity-oriented synthesis (DOS) library and a 279,231-compound Molecular Libraries Probe Center Network (MPLCN) library. All the compounds were dissolved in DMSO (Sigma, Cat # D8418). In the working solutions the DMSO concentration added with the stock of compounds was 2% (v/v), unless indicated otherwise. The compounds for confirmation analysis were purchased from Asinex Ltd., ChemBridge Co., ChemDiv Inc, Enamine, FCH Group, Frontier Scientific Services Inc., InterBioScreen Ltd., Life Chemicals Inc., Scientific Exchange Inc., Sigma-Aldrich Co. and Vitas-M Laboratory Ltd.
The fluorescence-quenching assay for RAD52 DNA annealing
Tailed dsDNA substrate was prepared by thermal annealing of ssDNA oligonucleotides 337-F and 1337-BHQ1 containing Fluorescein and Black Hole Quencher 1 residues at the 5′ and 3′ end, respectively. DNA annealing was initiated by adding RAD52 (20 nM) to the mixture of ssDNA oligonucleotide 265–55 (5 nM, molecules) and tailed dsDNA 337-F/1337-BHQ1 (5 nM, molecules) in buffer containing 25 mM Tris-acetate pH 7.5, 100 μg.ml−1 BSA and 1 mM DTT. The fluorescence intensity was measured in a 3-mm quartz cuvette (Starna Cells) using a FluoroMax-3 (HORIBA) fluorimeter with 492 nm excitation wavelength and 520 nm emission wavelength at 30°C.
HTS for RAD52 inhibitors
The fluorescence-quenching assay for RAD52-promoted DNA annealing was optimized to a 4 μl 1536 well protocol using 25 nM RAD52 and 8 nM (molecules) DNA in buffer containing 25 mM Tris-acetate pH 7.5, 100 μg.ml−1 BSA, 1 mM DTT and 0.01% Pluronic F-68. Wells containing no RAD52 were used as a positive control to estimate the activity of fully inhibited protein; wells in which the compounds were replaced with only the vehicle (DMSO) were used as neutral control. The HTS was performed using the 8 channel BioRAPTR 1536 (Beckman) for reagent dispensing. The reactions were carried out for 30 min followed by measurement of an endpoint fluorescence (485 nm excitation, 535 nm emission) using an EnVision multimode plate reader (Perkin Elmer). Wells containing no RAD52 enzyme were used to as positive control, and data were analyzed using Genedata. The compounds with an inhibitory effect of 30% or greater were tested further by measuring the concentration dependence (in a range from 1 nM to 100 μM) of their inhibition of RAD51. The most potent inhibitory compounds were analyzed further using non-fluorescent assays. Detailed methods for RAD52 screening are in PubMed: http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?aid=651660.
The D-loop formation by RAD52 or RAD51
To form RAD52 nucleoprotein complexes, RAD52 (0.45 μM) was incubated with a 32P-labeled ssDNA (oligo 90) (3 μM, nt) in buffer containing 25 mM Tris-Acetate, pH 7.5, 100 μg.ml−1 BSA, 0.3 mM magnesium acetate and 2 mM DTT at 37°C for 15 min. To form RAD51 nucleoprotein filament, RAD51 (0.45 μM) was incubated with 32P-labeled ssDNA (1.35 μM, nucleotides) in buffer containing 25 mM Tris-Acetate, pH 7.5, 100 μg.ml−1 BSA, 2 mM calcium chloride, 1 mM ATP and 2 mM DTT for 15 min at 37°C. Then inhibitors were added to both reactions and incubation continued for 15 min at 37°C. D-loop formation was initiated by addition of supercoiled pUC19 DNA (50 μM or 22.5 μM, nucleotides, for RAD52 and RAD51-promoted reactions, respectively) and was carried out 15 min at 37°C. The reactions were stopped and deproteinized by the addition of 1.5% SDS and proteinase K (0.8 mg/ml) for 15 min at 37°C, mixed with a 0.10 volume of loading buffer (70% glycerol, 0.1% bromphenol blue) and analyzed by electrophoresis in 1% agarose gels in TAE buffer (40 mM Tris acetate, pH 8.3 and 1 mM EDTA) at 5 V/cm for 3 h. The gels were dried on DEAE-81 paper (Whatman) and the yield of D-loops quantified using a Storm 840 PhosphorImager and ImageQuant 5.2 (GE Healthcare). The D-loop yield was expressed as a percentage of plasmid DNA carrying D-loops relative to the total plasmid DNA.
Calculation of the IC50 value for RAD52 inhibitors
IC50 values were calculated using GraphPad Prism V5.0 software. The data were obtained from three independent repeats of experiments.
Acridine orange displacement assay for DNA binding
To rule out DNA binding as an undesired mechanism of action, we tested the ability of the selected compounds to bind DNA. The compounds in varied concentrations were added into 30 μl reaction mixtures containing 50 nM acridine orange and 6 μg/ml salmon sperm DNA,10 mM HEPES, pH 7.5; 1 mM EDTA pH 7.5; 100 mM NaCl in 384-well plates, and the reactions were incubated at room temperature for 20 min followed by fluorescence polarization measurement using an EnVision (Perkin Elmer) equipped with a 480 nm excitation filter and 535 nm S and P emission filters with a D505 FP/D535 dichroic mirror. Mitoxantrone (10 μM) was used as a positive control. The S and P values are processed with the standard fluorescence polarization calculation formula (mP = 1000*(S-G*P)/(S+G*P) where G is the G-factor and is ≈1.
Luminescent cell viability assay
BxPC3 cells were kept in RPMI 1640 (ATCC) media supplemented with 10% FBS (Gibco); Capan-1 cells were kept in IMDM (ATCC) media containing 20% FBS (GIBCO); UWB1.298 and UWB1.298 (BRCA1+) cells were kept in 48.5% RPMI1640 (ATCC), 48.5% MEGM (Clonetics/Lonza, MEGM kit, CC-3150) and 3% FBS (GIBCO) respectively. Cells in log-phase were harvested and 100 μl cell suspensions were replated in a 96-well plate with a final density of 4000 cells/well. After overnight growth, cells were treated with indicated concentrations of compounds. Media containing the invariant concentration of compounds were refreshed every 3 days until cells were finally lysed by 30 μl/well of Promega CellTiter-Glo reagents and read on a Promega GloMax 96 reader on day 10 (9 days exposure). Promega CellTiter-Glo protocol is available on the web: http://www.promega.com/resources/protocols/technical-bulletins/0/celltiter-glo-luminescent-cell-viability-assay-protocol/.
The clonogenic survival assay
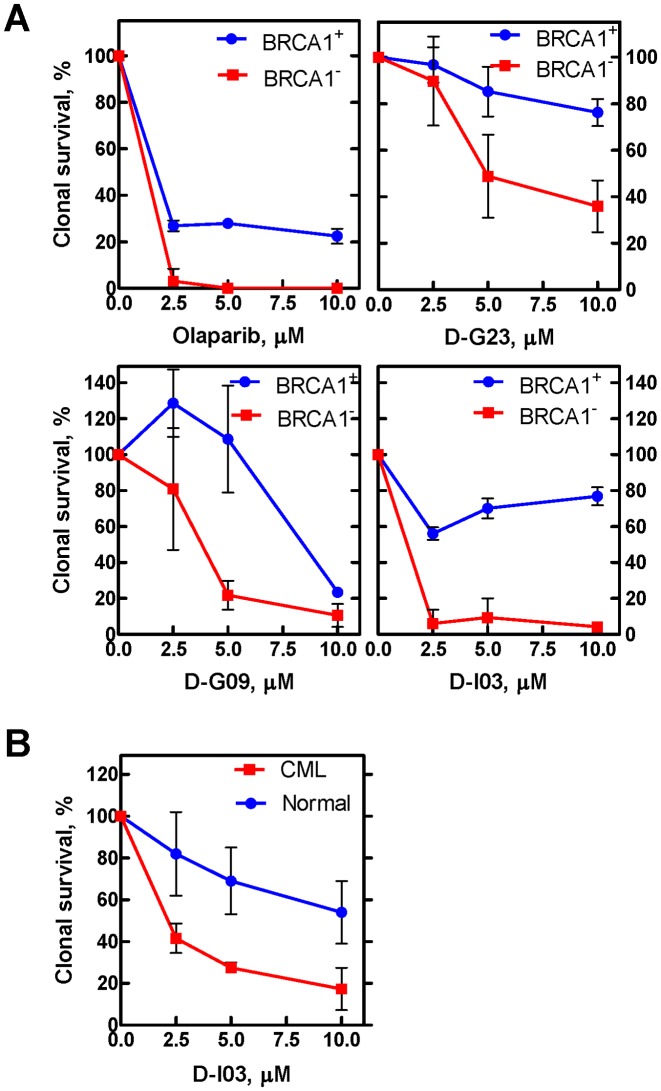
MDA-MB-436 cells were cultured in RPMI + 10% FBS. BRCA-proficient and BRCA-deficient cells were plated on day 0 in triplicate at 5000 cells/well. On days 1 and 3, the cells were treated with 0, 2.5 μM, 5 μM, or 10 μM D-I03, D-G23 or D-G09. Cells were counted on day 4 on a hemocytometer, using Trypan Blue exclusion, and immediately were plated in a clonogenic assay at a density of 500 cells/well in a 6 well plate, in RPMI + 10% FBS. After two weeks, the colonies were fixed/stained with 0.05% of 10 mg/ml ethidium bromide in 50% ethanol and visualized with Alphaimager gel imager (Alpha Innotech).
CML viability assay
Lin-CD34+ primary CML and normal cells were obtained by magnetic sorting using the EasySep negative selection human progenitor cell enrichment cocktail followed by treatment with human CD34 positive selection cocktail (StemCell Technologies), and were subsequently cultured in StemSpan H3000 media (StemCell Technologies) supplemented with a cocktail of growth factors (100 ng/ml stem cell factor, 20 ng/ml interleukin3 [IL-3], 100 ng/ml fms-related tyrosine kinase 3 ligand, 20 ng/ml granulocyte colony-stimulating factor, 20 ng/ml IL-6). For the viability assay, CD34+ CML (n = 3) and normal (n = 5) cells were plated at 1 × 104 cells/well in 96 well plates on day 0, and treated with 0 μM, 2 μM, 5 μM, or 10 μM D-I03 on days 0 and 2. Viable cells were counted on day 4 using Trypan Blue staining.
Measurement of compound binding to RAD52 by SPR
Experiments were performed using the ProteOn XPR36 SPR array system (Bio-Rad). ProteOn GLH sensor chips were preconditioned with two short pulses each (10 s) of 50 mM NaOH, 100 mM HCl and 0.5% SDS. Then the system was equilibrated with PBS-T buffer (20 mM Na-phosphate, 150 mM NaCl, and 0.1% polysorbate 20, pH 7.4). Individual ligand flow channels were activated for 5 min at 25°C with a mixture of 1-ethyl-3-[3-dimethylaminopropyl carbodiimide hydrochloride) (0.2 M) and sulfo-N-hydroxysuccinimide (0.05 M). Immediately after chip activation, either RAD52 (100 μg.ml−1 in 25 mM Tris-Acetate, 20mM KCl, 0.3 mM magnesium acetate, pH 7.5) or the anti-HIV mAb 2F5 (100 μg.ml−1 in 10 mM sodium acetate, pH 5.0) was injected across ligand flow channels for 5 min at a flow rate of 30 μl.min−1.
Excess active ester groups on the sensor surface were capped by a 5-min injection of 1 M ethanolamine HCl (pH 8.5). This resulted in the coupling of RAD52 and 2F5 at a density of 9000 RUs (response unit, which is an arbitrary unit that corresponds to 1 pg/mm2). The standard deviation in the immobilization level from the six spots within each channel was less than 4%. Compounds in indicated concentrations in 25 mM Tris-Acetate, 20 mM KCl, 0.3 mM magnesium acetate, pH 7.5, supplemented with 0.005% polysorbate 20 and 2% DMSO were injected over the control and RAD52 surfaces at a flow rate of 200 μl min−1, for either a 30s (D-I09) or 1-min association phase (D-I03, D-G23), followed by a variable dissociation phase at 25°C using the ‘one-shot’ functionality of the ProteOn (19).
Specific regeneration of the surfaces between injections was not needed owing to the nature of the interaction. Data were analyzed using the ProteOn Manager Software version 3.0 (Bio-Rad). The responses of a buffer injection and responses from the reference flow cell with the anti-HIV mAb 2F5 were subtracted to account for nonspecific binding. Experimental data were fitted globally to a simple 1:1 binding model. The average kinetic parameters (association [ka] and dissociation [kd] rates) generated from three data sets were used to define the equilibrium dissociation constant (KD). Data that could not be adequately fit to a binding model were analyzed using equilibrium analysis, plotting the response at equilibrium versus concentration and fitting to a steady state model.
Measuring the effect of inhibitors on GFP-RAD52 and RAD51 foci formation
GFP-RAD52 foci formation was measured in BCR-ABL1-positive BRCA1-deficient 32Dcl3 murine hematopoietic cell line that expresses GFP-RAD52 (16). RAD51 foci formation was measured in parental 32Dcl3. Both cell lines were cultured in IMDM plus 10% FBS. The cells were plated at 500 000 cells/ml and pretreated for 4 h with either D-G23 or D-I03 (2.5 μM) for GFP-RAD52 foci or with D-I03 (2.5 μM) for RAD51 foci (or no pretreatment for the control and cisplatin-treated cells). After 4 h of incubation, the cells were treated with 3 μg/ml cisplatin for 16 h. Following cisplatin treatment, cytospins were prepared using polylysine coated slides (Thermo Scientific). DNA was counterstained with DAPI. To detect RAD51 foci, cells were stained with an anti-RAD51 antibody (Thermo Scientific), followed by a secondary antibody conjugated with AlexaFluor 594. RAD51 and GFP-RAD52 foci were visualized with an inverted Olympus IX70 fluorescence microscope equipped with a Cooke Sensicam QE camera (The Cooke Co., Auburn Hills,MI, USA). Images from 25–60 cells/group were processed using SlideBook 3.0 (Intelligent Imaging Innovation).
Measuring the effect of D-I03 on single-strand annealing (SSA) and gene conversion (HDR) in U2OS cells
U2OS cells with chromosomally integrated SSA(U2OS-SSA) or gene conversion HDR(U20S-HDR)-reporter were cultured in DMEM (Sigma D-6429) containing 10% FBS (Gibco) supplemented with antibiotics (penicillin 100 U/ml, streptomycin 100 μg/ml, and plasmocin (2.5 μg/ml) (20,21). At 80% confluence, cells were trypsinized and plated in triplicate at a density of 2 × 105 cells/well in 6 well plates. After 22 h cells were washed with 1X PBS and further incubated for 2 h in antibiotic-free DMEM-10% FBS. Cells were transfected with pCBASce (0.8 μg) expressing I-SceI endonuclease or, in controls, with pUC19 (0.8 μg) or pMX-GFP (0.8 μg) plasmids using Lipofectamine 2000. After 3 h of transfection, cells were washed with antibiotic-free DMEM-10% FBS. Then, cells were incubated in DMEM-10% FBS supplemented with antibiotics and containing D-I03 at indicated concentrations followed by additional incubation for 48 h. In each well, cells were washed with 1X PBS, trypsinized and fixed with 3.3% formaldehyde. Fixed cells were kept on ice. The yield of GFP+ positive cells was measured by flow cytometry using Guava EasyCyte PRO (EMD Millipore).
RESULTS
HTS for RAD52 inhibitors
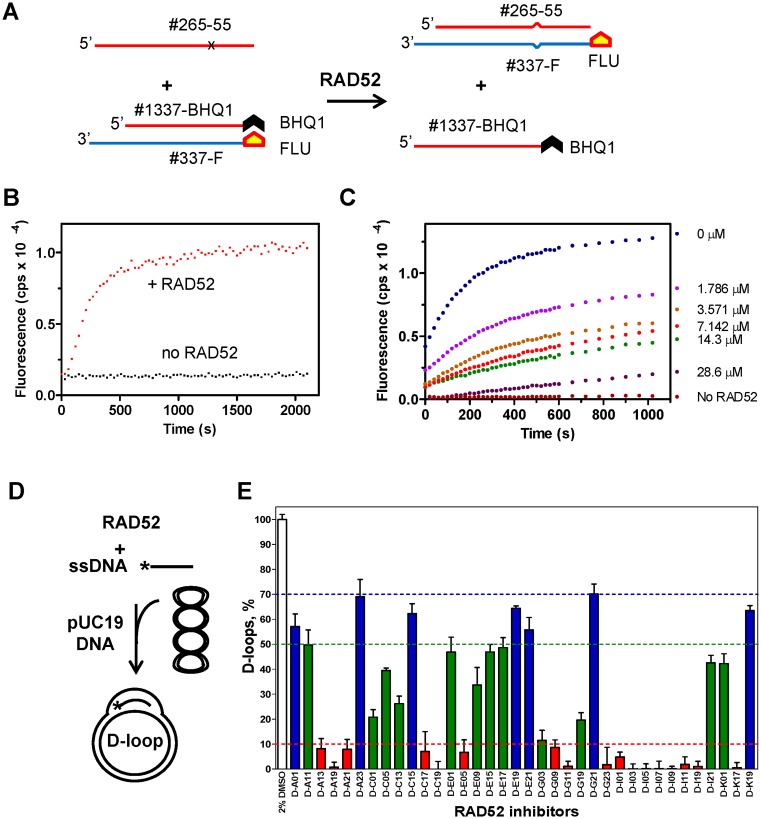
In order to target hereditary breast cancer and ovarian cancer cells we wanted to develop small molecule inhibitors of RAD52 using high throughput screening (HTS) of compound libraries. In vitro, RAD52 carries out annealing of complementary ssDNA molecules and an invasion of ssDNA into homologous duplex DNA. To screen for inhibitors of RAD52 ssDNA annealing activity, we developed a fluorescence-quenching assay (Figure 1). In this assay, RAD52 promotes DNA annealing between synthetic ssDNA (Oligo 265–55; 55 nt) and tailed dsDNA (tdsDNA) composed of a ssDNA 60-mer (Oligo 337-FLU) carrying fluorescein (FLU), a fluorescence donor group, and an ssDNA 39-mer (1337-BHQ1) carrying black hole quencher 1 (BHQ1), a non-fluorescent acceptor group (Figure 1A). The RAD52-promoted reaction included two steps: (i) annealing between Oligo 265–55 and a 15-nt ssDNA region of the tdsDNA and (ii) displacement of the ssDNA 1337-BHQ1 strand from the tdsDNA resulting in an increase in fluorescence due to secession of fluorescence quenching by the BHQ1 group (Figure 1B; an example of the effect of D-I03 inhibitor on RAD52-promoted ssDNA annealing is shown in Figure 1C and Supplementary Figure S2). Of these two steps, annealing was the limited step of the reaction in our assay, since RAD52 promotes DNA strand exchange between ssDNA and blunt end dsDNA poorly (22). In order to decrease RAD52-independent (uncatalyzed) reaction during branch migration step we incorporated a single mismatch in Oligo 265–55 ssDNA (Figure 1A).
Figure 1.
Identification and characterization of RAD52 small molecule inhibitors. (A) Fluorescence-quenching assay for RAD52 ssDNA annealing (plus DNA strand displacement). Experimental scheme. FLU-fluorescein; BHQ 1–black hole quencher 1. DNA substrates contain a mismatch (denoted by x) to block spontaneous reaction. (B) The kinetics of ssDNA annealing measured on a FluoroMax3 fluorimeter. The fluorescence intensity was expressed in arbitrary units (AU). Shown is representative result; the reactions were repeated at least three times. (C) RAD52 annealing activity was measured in the presence of D-I03 in indicated concentrations. (D) The scheme of the D-loop assay: RAD52 forms a complex with ssDNA and promotes its homologous pairing with pUC19 plasmid DNA. Asterisk denotes 32P label on ssDNA. (E) Inhibition of the RAD52 DNA pairing activity by tested compounds (30 μM). Error bars indicate standard deviation (SD).
The fluorescence-quenching assay was optimized for 1536 well plates (Z’ Avg = 0.64). The primary HTS of the 372,903-compound library yielded 1687 positive hits that caused more than 30% inhibition of the RAD52-dependent fluorescence increase (0.5% activities), including 628 hits in a 93672-compound Broad's diversity-oriented synthesis (DOS) library (0.7% activities) and 1115 hits in a 279,231-compound Molecular Libraries Probe Center Network (MPLCN) library (0.4% Activities). The hits were further analyzed for a concentration dependence in inhibiting of RAD52 and by testing their DNA binding affinity using the acridine orange assay. As a result, 187 compounds were identified that inhibited RAD52 with the IC50 lower than 10 μM and displayed no DNA binding. These remaining compounds were assessed for their potential chemical tractability by removing compounds with highly reactive or unstable functional groups and focusing on chemotypes that were synthetically accessible and attractive. The selected compounds as well as some closely related new analogs of certain hits were next purchased as dry powders from commercial sources. After executing this selection process, 70 compounds were obtained for further analyses.
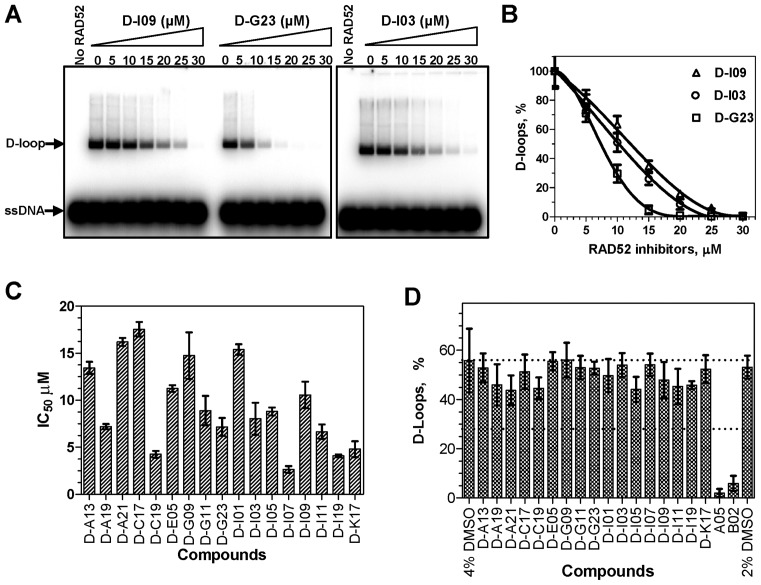
We tested the inhibitory effect of the 70 selected compounds using the D-loop assay, in which RAD52 promotes pairing between 32P-labeled ssDNA and homologous supercoiled plasmid pUC19 DNA (Figure 1D); the product of the reaction, D-loops, were analyzed by electrophoresis in 1% agarose gels. First, by testing the effect of the selected compounds at fixed (30 μM) concentration we identified 17 compounds that inhibited D-loop formation by more than 90% (Figure 1E). Then, we measured the effect of each of the 17 compounds on RAD52-dpendent D-loop formation in a concentration dependent manner. The IC50 of these compounds varied in a range between 2.7 μM and 17.5 μM (Figure 2A-C; Supplementary Table S2). In the D-loop assay the IC50 values were generally higher than in the fluorescence-quenching assay likely due to the higher RAD52 concentration employed by the former assay, 450 nM versus 25 nM, and due to the different nature of the assays.
Figure 2.
The effect of inhibitors on DNA pairing activity of RAD52. (A) The products of DNA pairing (D-loops) were analyzed by electrophoresis in 1% agarose gels. (B) Graphical representation of the data from A. (C) The IC50 values for 17 tested compounds. (D) The effect of the RAD52 inhibitors on DNA pairing activity of RAD51. The effect was measured using the D-loop assay at the inhibitors concentrations that correspond to their 10 x IC50 values (27–176 μM) for RAD52 pairing activity; concentrations of A05 and B02 were 100 μM. Error bars indicate SD.
Then, we examined the selectivity of the RAD52 inhibitors using RAD51 as a non-specific target. RAD51 is structurally unrelated to RAD52, but shares DNA pairing activity with RAD52. Using the D-loop assay, we found that at concentrations 10-fold higher than the IC50 for RAD52 DNA pairing none of the 17 tested compounds (Supplementary Figure S3) showed a significant inhibition of D-loop formation (Figure 2D). However, under suboptimal conditions for the D-loop assay at reduced Ca2+ concentration (1 mM) three tested N-Methyl thieno pyrazoles compounds, D-I07, D-I11 and D-I19 (see structures in Supplementary Figure S3), showed non-specific inhibition of RAD51 greater than 2-fold relative to the DMSO control (Supplementary Figure S4; Supplementary Table S2). Overall, as a result of the HTS and several confirmatory and selectivity assays, we identified 14 specific inhibitors of ssDNA annealing and ssDNA pairing activity of RAD52 in vitro.
Analysis of RAD52 inhibitors in human BRCA1- and BRCA2-deficient cells
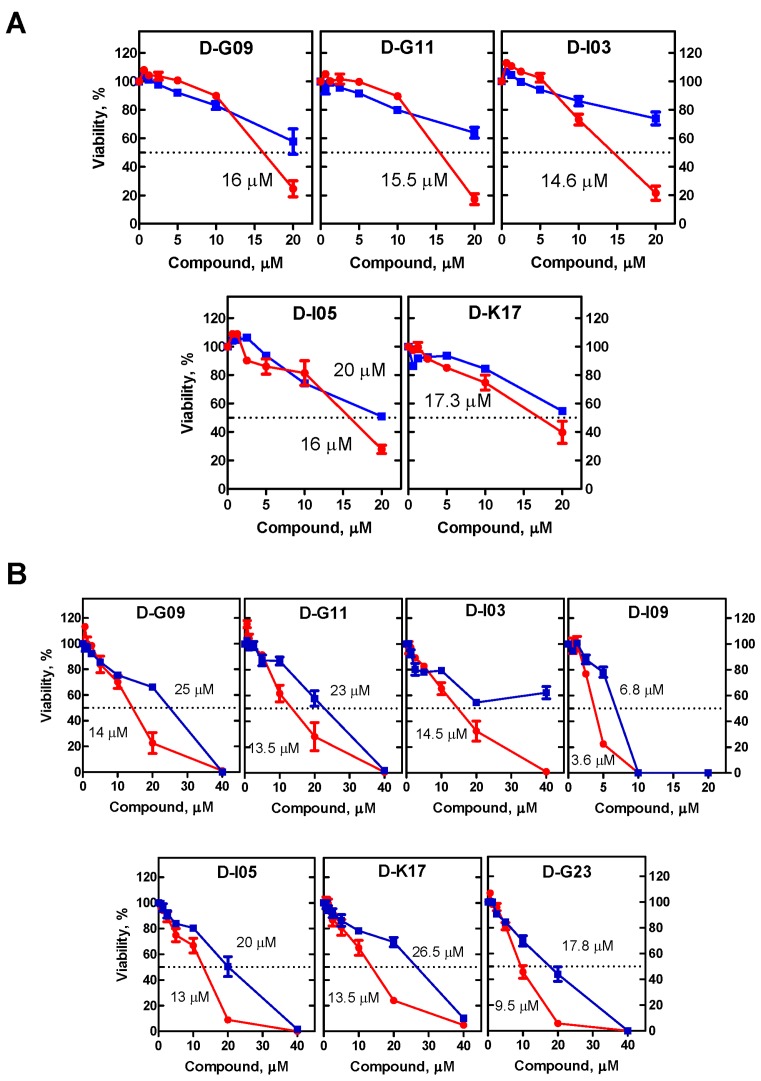
Since it was previously shown that inactivation of RAD52 by siRNA and peptide aptamer causes lethality of BRCA1- and BRCA2-deficient cells (10,16), we suggested that the RAD52 inhibitors can also inhibit growth of BRCA1/2-deficient cells. First, we tested the effect of 14 RAD52 inhibitors identified in the biochemical assays on the growth of BRCA2-deficient Capan-1 cells and BRCA2-proficient BxPC3 cells. BxPC3 cells are commonly used as a control for Capan-1 cells; these two well-differentiated pancreatic adenocarcinoma cell lines show significant similarity and share the mutation status of a number of tumor suppressor genes, including p53, p16, Rb, have strong expression of COX-2 and MMP-9 (23–26). We found that five of the tested inhibitors preferentially suppressed the growth of Capan-1 cells (Figure 3A).
Figure 3.
Effect of the RAD52 inhibitors on survival (A) Capan-1 (BRCA2−) and BxPC3 (BRCA2+) cells and (B) on UWB1.289 (BRCA1−) and [UWB1.289 (+BRCA1)] (BRCA1+) cells. Capan-1 and UWB1.289 are denoted by red line; BxPC3 and UWB1.289 (+BRCA1) indicated by blue line. The experiments were repeated at least three times, error bars indicate SD.
We then tested the effect of RAD52 inhibitors on the growth of BRCA1-deficient (UWB1.289 BRCA1-) and BRCA1-proficient (UWB1.289 BRCA+) cells. We found that 7 out 14 tested RAD52 inhibitors preferentially suppressed the growth of BRCA1-deficient cells; importantly, these 7 compounds included the 5 compounds that inhibited the growth of BRCA2-deficient cells (Figure 3B). We also tested the effect of the 14 RAD52 inhibitors on the survival of another BRCA1-deficient cells, MDA-MB-436. Three out 14 compounds, which also inhibited growth of UWB1.289 cells, showed an inhibitory effect on these cells compared with the isogenic MDA-MB-436 (BRCA1+) cells (Figure 4A). Finally, we tested the effect of D-I03, the strongest inhibitor on BRCA1- and BRCA2-deficient cells, on BCR-ABL1 –positive BRCA1-deficient chronic myeloid leukemia (CML) patient cells (27). BRCA1-deficiency in these cells is due to constitutive downregulation of this protein. We found that D-I03 selectively diminished the growth potential of BRCA1-deficient CML patient cells in comparison to BRCA1-proficient normal counterparts (Figure 4B).
Figure 4.
Effect of RAD52 inhibitors on survival of MDA-MB-436 (BRCA1−) and [MDA-MB-436 (BRCA1+)] (BRCA1+) cells (A) and on survival of on BCR-ABL1 –positive BRCA1-deficient CML cells and their BRCA1-proficient normal counterparts (B). The experiments were repeated at least three times, error bars indicate SD.
Overall, two compounds, D-G09 and D-I03, showed an inhibitory effect on all three tested BRCA1- and BRCA2-deficient cell lines. Importantly, these compounds showed significant structural similarity sharing the quinoline core (Supplementary Figure S3). Another member of this structural group, D-G11, showed activity on two of the tested cell lines (UWB1.289 and Capan-1). Remarkably, three other compounds, D-G23, D-I05 and D-K17, that preferentially inhibited at least two BRCA1/2-defficient cell lines also share similarity, but belong to another structural type with the quinazoline core. From all tested compounds, D-I03 showed the strongest and most consistent inhibitory effect on all tested BRCA1−/− and BRCA2−/− cell lines; moreover, it selectively inhibited growth of BRCA1-deficient CML cells from patients.
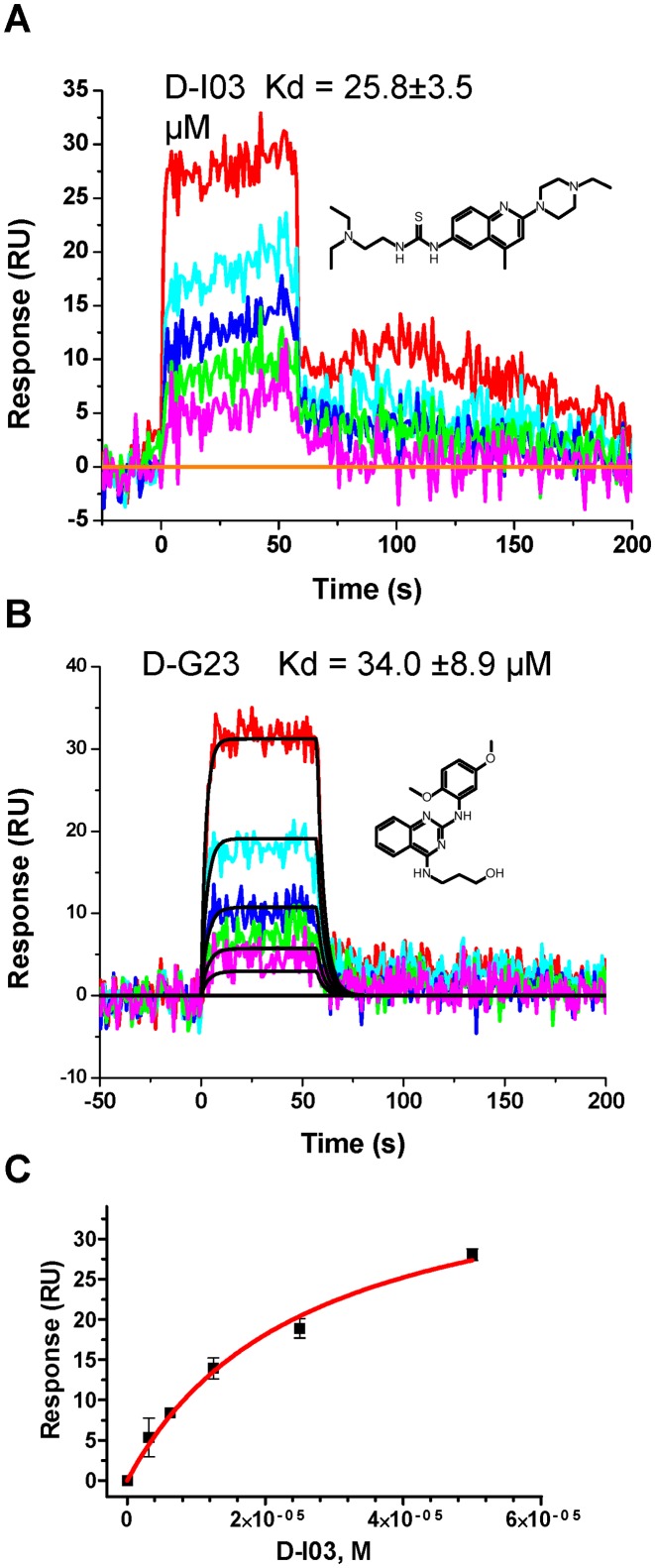
Measurement of inhibitors binding to RAD52 by SPR
We then tested whether RAD52 inhibitors that showed activity in the biochemical and cell-based assays physically interact with RAD52. Using the surface plasmon resonance (SPR) method, we demonstrated that D-I03 (3.12−50 μM) and D-G23 (3.125−25 μM) bind directly to RAD52 (Figure 5). The anti-HIV mAb 2F5 structurally unrelated to RAD52 was used in control and non-specific binding signal was subtracted from the RAD52 binding signal. The Kd values for D-I03 and D-G23 are 26.1 ± 4.5 μM and 34.0 ± 8.9 μM, respectively. These values are somewhat higher than one might expect from the IC50 values in the biochemical and cell-based assays with these inhibitors. Because the active form of RAD52 is a hexamer, the apparent differences in the activities of inhibitors may suggest that inhibition of RAD52 requires only partial saturation of the RAD52 hexamer with inhibitors. Using the acridine orange displacement assay we showed that D-I03 and D-G23 did not bind DNA. Thus, these compounds inhibit DNA annealing and pairing activities of RAD52 through direct binding, not through interaction with DNA substrates.
Figure 5.
Measurement of D-I03 (A) and D-G23 (B) binding to RAD52. Compound D-I03 at concentrations of 3.125, 6.25, 12.5, 25 and 50 μM or compound D-G23 at concentrations of 1.56, 3.125, 6.25, 12.5 and 25 were injected to the chip with immobilized RAD52. Colored lines indicate experimental data, whereas black lines indicate fitting to the simple 1:1 binding model using the ProteOn Manager Software version 3.0 (Bio-Rad). For D-G23 binding to RAD52, kinetic values are as follows: ka = 1.15 (± 0.44) × 104 M−1s−1; kd = 3.62 (± 0.7) × 10−1 s−1; Kd = 34.0 ± 8.9 μM. (C) For D-I03, the data did not fit to any available binding models and the Kd was determined using equilibrium analysis by plotting the response at equilibrium (Req) versus concentration of compound. Experiments were repeated at least three times; error bars indicate S.D.
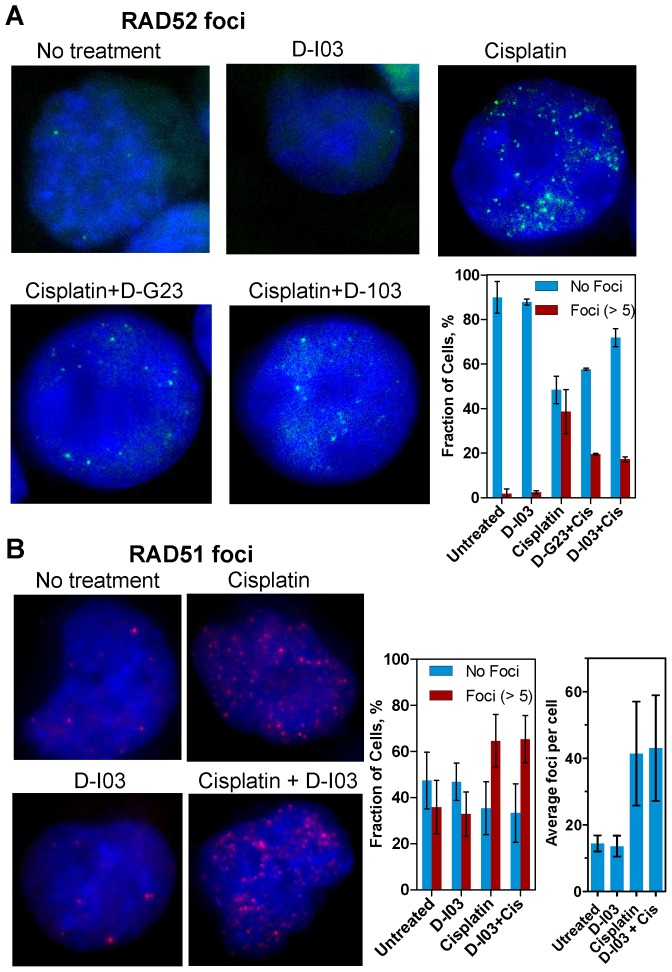
Inhibitors disrupt RAD52, but not RAD51, foci formation
We wished to test whether D-I03 and D-G23 can inhibit RAD52 activities in the cell. In response to DNA damage, RAD52 accumulates in the nucleus forming distinct structures known as foci (28,29). It is thought that the foci represent RAD52 complexes with DNA repair intermediates. Inhibitors of RAD52 may decrease RAD52 foci formation by disrupting its interaction with DNA substrates. Indeed, we found that both D-I03 and D-G23 inhibited RAD52 foci formation induced by cisplatin in BCR-ABL1-positive BRCA1-deficient 32Dcl3 murine hematopoietic cell line that expresses GFP-RAD52 (16) (Figure 6A). In the presence of D-I03 (2.5 μM) and D-G23 (2.5 μM), the fraction of cells with RAD52 foci (≥5 foci) was decreased ≈2.0–2.5-fold, from 38.7 ± 10% to 17 ± 1% and 19 ± 0.4%, respectively; at the same time, the fraction of cisplatin-treated cells without foci was increased from 48.4 ± 6.2% to 71.9 ± 4.1% and 57.6 ± 0.5% (Figure 6A, bottom right panel). Thus, compounds of two chemotypes, represented by D-I03 and G-23, were identified that inhibited the biochemical activities of RAD52 in vitro, showed preference in suppressing survival of BRCA1- and BRCA2-deficient cells and inhibited formation of damage-induced RAD52 foci formation. We also tested a non-specific effect of D-I03, a strongest of theRAD52 inhibitors, on RAD51 foci formation. Using parental 32Dcl3 (BRCA1-proficient) cells, we found that D-I03 does not have a significant effect on RAD51 foci induced by cisplatin (Figure 6B). Also, D-I03 alone induce neither RAD51 foci nor RAD52 foci (in BRCA1-deficient cells; Figure 6A) indicating low genotoxicity of this compound.
Figure 6.
(A) Effect of RAD52 inhibitors D-G23 and D-I03 (2.5 μM) on GFP-RAD52 foci formation in response to cisplatin (10 μM) treatment. GFP-RAD52 was constitutively expressed in p210BCR-ABL1-positive 32Dcl3 murine hematopoietic (BRCA1-deficient) cells. In control, cells were treated with D-I03 (2.5 μM) alone. On the bottom right, the data are shown as a graph. Cells with a small foci number (1–5) were excluded from calculations. Error bars represent SD. (B) Effect of D-I03 (2.5 μM) on RAD51 foci formation with and without cisplatin (10 μM) treatment in parental 32Dcl3 (BRCA-proficient) cells. On right, the data are shown as a graph. Average numbers of foci per cell on right panel were determined in cells with more than 10 foci. Error bars represent SD.
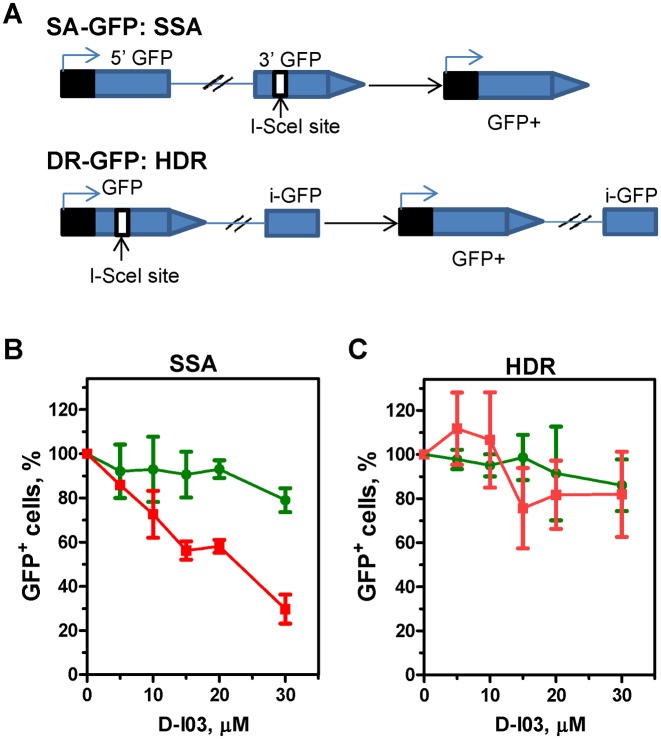
D-I03 inhibits single-strand DNA annealing, but not gene conversion in U2OS cells
In both yeast and mammalian cells, RAD52 promotes single-strand DNA annealing (SSA) (30,31). SSA is a type of HR that is initiated at DSBs and mediated by annealing of fortuitous direct repeats flanking DSB ends after their exonucleolitic resection. This mechanism leads to end re-joining with concomitant deletion of sequences between direct repeats. Here, using the SA-GFP construct integrated into the chromosomal DNA (20,21) we wished to examine the effect of RAD52 inhibitor D-I03 on SSA in U2OS cells. The SA-GFP reporter system contains a 5′ fragment of the GFP (5′-GFP) gene, and a 3′ fragment of the GFP (3′-GFP) that contains an 18-bp I-SceI site (Figure 7A). The GFP fragments are separated by 2.7 kb region and share a 266 bp region of homology. Transfection of cells with I-SceI expressing vector pCBASce induces DSB in the 3′-GFP. Repair of the DSB by SSA leads to formation of GFP+ cells. Thus, each SSA event can be scored by the appearance of a green fluorescent cell.
Figure 7.
D-I03 inhibits single-strand annealing (SSA), but not homology dependent recombination (HDR) (gene conversion) in U2OS cells. (A) The scheme of the reporter systems. The SSA-GFP reporter contains a 5′ fragment of the GFP (5′-GFP) gene, and a 3′ fragment of the GFP (3′-GFP) with an I-SceI site. The HDR-GFP reporter system contains the GFP gene interrupted by a Sce-I site, and a fragment of the GFP with truncated 3′- and 5′-rerminus. Repair of the I-SceI-induced DSB by either SSA or gene conversion leads to formation of GFP+ cells (21). Effect of D-I03 on the repair of the I-SceI-induced DSBs in U2OS cells carrying the chromosomally located SSA-GFP (B) or HDR-GFP (C) reporter is shown by red line. In a control, the effect of D-I03 on formation GFP+ cells was measured after transfection of U2OS cells with pMX-GFP plasmid expressing GFP protein (green line). Formation of GFP+ cells in the absence of D-I03 was expressed as 100%. Experiments were repeated at least three times; error bars indicate S.D.
We found that D-I03 reduces formation of GFP+ cells in a concentration dependent manner; at 30 μM D-I03 the yield of GFP+ cells was reduced ≈3.4-fold (Figure 7B, red line; Supplementary Figure S5). In control, we measured the effect of D-I03 on formation of GFP+ cells when U2OS cells were transfected with pMX-GFP plasmid encoding GFP (Figure 7B; green line). We found that up to 30 μM, D-I03 had no significant effect on the recovery of GFP+ cells.
We then tested the effect of D-I03 on DSB repair via the canonical homology dependent recombination (HDR) (also known as gene conversion) mechanism using the chromosomally integrated DR-GFP construct in U2OS cells (20,21). Previously, it was shown that RAD52 has no significant effect on the HDR in mammalian cells (30). The DR-GFP construct consists of two inactive copies of the GFP gene, one that is disabled by insertion of I-SceI recognition site and another (iGFP) is truncated at both ends (Figure 7A). A unique DSB is generated in this construct after the cells are transfected with pCBASce plasmid. The repair of this DSB by gene conversion using iGFP as a template gives rise to the functional GFP gene. We found that D-I03 does not have a significant effect on formation of GFP-positive cells (Figure 7C, red line; Supplementary Figure S6). D-I03 also has no effect on GFP expression or on recovery of GFP-positive cells when U2OS cells were transfected with pMX-GFP plasmid encoding GFP (Figure 7C, green line). Taken together, our results indicate that consistent with inhibition of RAD52 in human cells D-I03 reduces the level of DSB-induced SSA, but not HDR.
DISCUSSION
RAD52 is an evolutionarily conserved eukaryotic protein, a member of the HR pathway that is responsible for repair of DSBs, recovery of stalled replication forks and faithful chromosome segregation during meiosis (1,32–34). In yeast, Rad52 plays a key role in HR; RAD52 null mutation obliterates nearly all types of recombination events and renders cells extremely sensitive to DSB-inducing agents. Surprisingly, in mice RAD52 knockouts show virtually no DNA repair phenotype. Recent discovery that RAD52 mutations are synthetically lethal with mutations in BRCA1/2, PALB2 and RAD51 paralogs indicated that in mammalian cells RAD52 constitutes an alternative HR sub-pathway that may play a back-up role during DSB repair (9,10). This independent/redundant role of RAD52 in mammalian HR is also supported by cytological data. In response to DNA damage both RAD51 and RAD52 form nuclear foci, which are thought to play a role of the DNA repair centers. However, foci formation by these proteins show only partially overlap (28) and their formation is differently affected by mutations in other HR genes; whereas RAD51 foci formation depends on the functional BRCA1, BRCA2 and RAD51 paralogs, RAD52 foci formation is independent of these proteins (35).
The mechanistic basis for the RAD52 function in HR in mammalian cells remains to be elucidated. Biochemical studies indicate that in S. cerevisiae Rad52 may play a role of a mediator that helps to load Rad51 recombinase on ssDNA at the site of DSBs overcoming an inhibitory effect of Replicative Protein A (RPA), an abundant protein that has high affinity for ssDNA (36,37). The RAD51 nucleoprotein filament formed on ssDNA then searches for homologous dsDNA and invades it to form joint molecules (D-loops) that provide a template and a primer to recover the DNA lost at the site of DSB. However, the mediator activity was not demonstrated for human RAD52 in vitro (38).
RAD52 possesses ssDNA annealing activity both in vitro and in vivo which can contribute to HR in several different ways (30,39,40). This activity was implicated in the second DNA end capture during RAD51-dependent DSB repair resulted in formation of double D-loops and then of Holliday junctions (41–43). It was also proposed that the ssDNA annealing activity of RAD52 may be responsible for a step that follows DNA repair synthesis and D-loop dissociation: re-annealing of the displaced ssDNA with the second DNA end of the DSB (42). In addition, RAD52 ssDNA annealing activity may also contribute to HR in a RAD51-independent manner. Genetic data indicate that this activity may be responsible for the error-prone DNA single-strand annealing (SSA) sub-pathway of HR, which is independent of RAD51 (44). Furthermore, biochemical data show that RAD52, similar to RAD51, is able to promote D-loop formation, albeit with lower efficiency (45), suggesting that RAD52 may potentially substitute RAD51 in some HR events.
Importantly, the RAD52-dependent mechanism of DSB repair is essential for viability in mammalian cells that are defective in BRCA1, BRCA2, PALB2, or in five RAD51 paralogs (9,10). Synthetic lethality provides a conceptual framework for discovering drugs that selectively kill cancer cells while sparing normal tissues (7,8,46). In the current paper we exploited synthetic lethality between the RAD52 and BRCA1&2 genes (9,10). We proposed that targeting of RAD52 with small molecule inhibitors will disrupt the RAD52-dependent HR sub-pathway in BRCA1- and BRCA2-deficient cells causing their lethality. The data with peptide aptamer that disrupts RAD52 binding to ssDNA supported this hypothesis (16). RAD52 represents an attractive potential therapeutic target also because no RAD52 mutations or inactivation has been documented in human tumors (10). Using HTS, we identified several small molecule compounds that specifically inhibit RAD52 ssDNA annealing and DNA pairing activities. Importantly, the selected inhibitors of two different chemotypes showed inhibitory effect on tested BRCA1- and BRCA2-deficient cells. The compound, D-I03, with the strongest inhibitory effect in human cells also selectively inhibited the growth of CML patient cells, in which the expression of BRCA1 was constitutively reduced (27). Finally, RAD52 inhibitors D-I03 and, to smaller extent, D-G23 also disrupted formation of RAD52 foci induced by cisplatin. At the same time, D-I03 had no significant effect on formation of cisplatin-induced RAD51 foci indicating specific targeting of RAD52 in cells. In addition, D-I03 seems to have a low genotoxicity, as it did not induce RAD52 or RAD51 foci in BRCA1-defecient or BRCA1-proficient cells, respectively. In accord with specific targeting RAD52 in human cells, D-I03 inhibited the SSA in USO2 cells, without significantly affecting canonical HDR. Previously, genetic experiments demonstrated that in mammalian cells RAD52 plays an important role in SSA, but not in HDR (30).
Structurally six RAD52 inhibitors that show the biological effect in human cells belong to two scaffolds, the quinoline and quinazoline (Supplementary Figure S3). These scaffolds represent attractive starting points for medicinal chemistry optimization as at least three distinct regions of these chemotypes can be readily modified. Another important aspect to evaluate in prospective chemotypes is their calculated property profile. Gratifyingly, both chemotypes, represented by D-I03 and D-G23, have calculated properties within ranges generally considered ‘drug-like’ and favorable for important parameters such as oral bioavailability (Supplementary Table S2).
Previously, it was demonstrated that the N’-terminal domain is responsible for ssDNA annealing and DNA pairing activities of RAD52 (47). Therefore, we suggest that the selected compounds may specifically target this domain. However, more work is needed to investigate the mechanism of action of the inhibitors. We propose that RAD52 inhibitors can be used as prototypes for development of novel therapies against hereditary breast cancer and ovarian cancers with defective BRCA1 or BRCA2 proteins. This therapy can also be potentially applied against sporadic cancers in which BRCA1 or BRCA2 are either mutated or downregulated, e.g. due to constitutive promoter methylation (48,49). Specific RAD52 inhibitors can also be used as a research tool to study the RAD52 biochemical activities and cellular functions.
Supplementary Material
Acknowledgments
We are grateful to Dr. Jeremy Stark for providing the SA-GFP and DR-GFP constructs, Dr. Peter Wade for help with analysis of compound structures and purities and Dr. Jason Lamontagne for help with flow cytometry. We thank Drs. Jacqueline Emrich, Roger Greenberg, Lydia Komarnicky-Kocher, Patrick Lam, Olimpia Meucci, Richard Pomerantz, Simon Powell, Mauricio Reginato and Fiona Simpkins for discussion and advice.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH [1R01 CA100839, 1R01 CA188347, MH0957512 to A.V.M.; 1R01CA186238 to T.S.; 1R21AI087388 to S.C.]; Basser Innovation Award and Drexel Clinical and Translational Research Institute (CTRI) award [to A.V.M.]; Vanderbilt Specialized Chemistry Center for Accelerated Probe Development [1U54MH084659]; The Broad Institute Probe Development Center [1U54HG005032]. Funding for open access charge: NIH [CA188347]; Basser Innovation Award.
Conflict of interest statement. None declared.
REFERENCES
- 1.Moynahan M.E., Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marteijn J.A., Lans H., Vermeulen W., Hoeijmakers J.H. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell Biol. 2014;15:465–481. doi: 10.1038/nrm3822. [DOI] [PubMed] [Google Scholar]
- 3.Iyama T., Wilson D.M. 3rd. DNA repair mechanisms in dividing and non-dividing cells. DNA Repair (Amst.) 2013;12:620–636. doi: 10.1016/j.dnarep.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helleday T. DNA repair as treatment target. Eur. J. Cancer. 2011;47(Suppl. 3):S333–S335. doi: 10.1016/S0959-8049(11)70192-7. [DOI] [PubMed] [Google Scholar]
- 5.Lok B.H., Powell S.N. Molecular pathways: understanding the role of Rad52 in homologous recombination for therapeutic advancement. Clin. Cancer Res. 2012;18:6400–6406. doi: 10.1158/1078-0432.CCR-11-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lord C.J., Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481:287–294. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- 7.Farmer H., McCabe N., Lord C.J., Tutt A.N., Johnson D.A., Richardson T.B., Santarosa M., Dillon K.J., Hickson I., Knights C., et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 8.Bryant H.E., Schultz N., Thomas H.D., Parker K.M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N.J., Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 9.Feng Z., Scott S.P., Bussen W., Sharma G.G., Guo G., Pandita T.K., Powell S.N. Rad52 inactivation is synthetically lethal with BRCA2 deficiency. Proc. Natl. Acad. Sci. U.S.A. 2011;108:686–691. doi: 10.1073/pnas.1010959107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lok B.H., Carley A.C., Tchang B., Powell S.N. RAD52 inactivation is synthetically lethal with deficiencies in BRCA1 and PALB2 in addition to BRCA2 through RAD51-mediated homologous recombination. Oncogene. 2013;32:3552–3558. doi: 10.1038/onc.2012.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chun J., Buechelmaier E.S., Powell S.N. Rad51 paralog complexes BCDX2 and CX3 act at different stages in the BRCA1-BRCA2-dependent homologous recombination pathway. Mol. Cell. Biol. 2013;33:387–395. doi: 10.1128/MCB.00465-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antoniou A.C., Casadei S., Heikkinen T., Barrowdale D., Pylkas K., Roberts J., Lee A., Subramanian D., De Leeneer K., Fostira F., et al. Breast-cancer risk in families with mutations in PALB2. N. Engl. J. Med. 2014;371:497–506. doi: 10.1056/NEJMoa1400382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meindl A., Hellebrand H., Wiek C., Erven V., Wappenschmidt B., Niederacher D., Freund M., Lichtner P., Hartmann L., Schaal H., et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat. Genet. 2010;42:410–414. doi: 10.1038/ng.569. [DOI] [PubMed] [Google Scholar]
- 14.Fujimori A., Tachiiri S., Sonoda E., Thompson L.H., Dhar P.K., Hiraoka M., Takeda S., Zhang Y., Reth M., Takata M. Rad52 partially substitutes for the Rad51 paralog XRCC3 in maintaining chromosomal integrity in vertebrate cells. EMBO J. 2001;20:5513–5520. doi: 10.1093/emboj/20.19.5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rijkers T., Van Den Ouweland J., Morolli B., Rolink A.G., Baarends W.M., Van Sloun P.P., Lohman P.H., Pastink A. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol. Cell. Biol. 1998;18:6423–6429. doi: 10.1128/mcb.18.11.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cramer-Morales K., Nieborowska-Skorska M., Scheibner K., Padget M., Irvine D.A., Sliwinski T., Haas K., Lee J., Geng H., Roy D., et al. Personalized synthetic lethality induced by targeting RAD52 in leukemias identified by gene mutation and expression profile. Blood. 2013;122:1293–1304. doi: 10.1182/blood-2013-05-501072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bugreev D.V., Golub E.I., Stasiak A.Z., Stasiak A., Mazin A.V. Activation of human meiosis-specific recombinase Dmc1 by Ca2+ J. Biol. Chem. 2005;280:26886–26895. doi: 10.1074/jbc.M502248200. [DOI] [PubMed] [Google Scholar]
- 18.Rossi M.J., Mazina O.M., Bugreev D.V., Mazin A.V. Analyzing the branch migration activities of eukaryotic proteins. Methods. 2010;51:336–346. doi: 10.1016/j.ymeth.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bravman T., Bronner V., Lavie K., Notcovich A., Papalia G.A., Myszka D.G. Exploring ‘one-shot’ kinetics and small molecule analysis using the ProteOn XPR36 array biosensor. Anal. Biochem. 2006;358:281–288. doi: 10.1016/j.ab.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 20.Gunn A., Bennardo N., Cheng A., Stark J.M. Correct end use during end joining of multiple chromosomal double strand breaks is influenced by repair protein RAD50, DNA-dependent protein kinase DNA-PKcs, and transcription context. J. Biol. Chem. 2011;286:42470–42482. doi: 10.1074/jbc.M111.309252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gunn A., Stark J.M. I-SceI-based assays to examine distinct repair outcomes of mammalian chromosomal double strand breaks. Methods Mol. Biol. 2012;920:379–391. doi: 10.1007/978-1-61779-998-3_27. [DOI] [PubMed] [Google Scholar]
- 22.Bi B., Rybalchenko N., Golub E.I., Radding C.M. Human and yeast Rad52 proteins promote DNA strand exchange. Proc. Natl. Acad. Sci. U.S.A. 2004;101:9568–9572. doi: 10.1073/pnas.0403205101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bogliolo M., Taylor R.M., Caldecott K.W., Frosina G. Reduced ligation during DNA base excision repair supported by BRCA2 mutant cells. Oncogene. 2000;19:5781–5787. doi: 10.1038/sj.onc.1203951. [DOI] [PubMed] [Google Scholar]
- 24.Abbott D.W., Freeman M.L., Holt J.T. Double-strand break repair deficiency and radiation sensitivity in BRCA2 mutant cancer cells. J. Natl. Cancer Inst. 1998;90:978–985. doi: 10.1093/jnci/90.13.978. [DOI] [PubMed] [Google Scholar]
- 25.Su L.K., Wang S.C., Qi Y., Luo W., Hung M.C., Lin S.H. Characterization of BRCA2: temperature sensitivity of detection and cell-cycle regulated expression. Oncogene. 1998;17:2377–2381. doi: 10.1038/sj.onc.1202162. [DOI] [PubMed] [Google Scholar]
- 26.Yuan S.S., Lee S.Y., Chen G., Song M., Tomlinson G.E., Lee E.Y. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 1999;59:3547–3551. [PubMed] [Google Scholar]
- 27.Podszywalow-Bartnicka P., Wolczyk M., Kusio-Kobialka M., Wolanin K., Skowronek K., Nieborowska-Skorska M., Dasgupta Y., Skorski T., Piwocka K. Downregulation of BRCA1 protein in BCR-ABL1 leukemia cells depends on stress-triggered TIAR-mediated suppression of translation. Cell Cycle. 2014;13:3727–3741. doi: 10.4161/15384101.2014.965013. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y., Maizels N. Coordinated response of mammalian Rad51 and Rad52 to DNA damage. EMBO Rep. 2000;1:85–90. doi: 10.1093/embo-reports/kvd002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Essers J., Houtsmuller A.B., van Veelen L., Paulusma C., Nigg A.L., Pastink A., Vermeulen W., Hoeijmakers J.H., Kanaar R. Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO J. 2002;21:2030–2037. doi: 10.1093/emboj/21.8.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stark J.M., Pierce A.J., Oh J., Pastink A., Jasin M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol. Cell. Biol. 2004;24:9305–9316. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krogh B.O., Symington L.S. Recombination proteins in yeast. Annu. Rev. Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 32.Symington L.S., Gautier J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 33.van Gent D.C., Hoeijmakers J.H., Kanaar R. Chromosomal stability and the DNA double-stranded break connection. Nat. Rev. Genet. 2001;2:196–206. doi: 10.1038/35056049. [DOI] [PubMed] [Google Scholar]
- 34.Kowalczykowski S.C. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- 35.van Veelen L.R., Essers J., van de Rakt M.W., Odijk H., Pastink A., Zdzienicka M.Z., Paulusma C.C., Kanaar R. Ionizing radiation-induced foci formation of mammalian Rad51 and Rad54 depends on the Rad51 paralogs, but not on Rad52. Mutat. Res. 2005;574:34–49. doi: 10.1016/j.mrfmmm.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 36.Sung P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem. 1997;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- 37.Sugiyama T., Kowalczykowski S.C. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J. Biol. Chem. 2002;277:31663–31672. doi: 10.1074/jbc.M203494200. [DOI] [PubMed] [Google Scholar]
- 38.Jensen R.B., Carreira A., Kowalczykowski S.C. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mortensen U.H., Bendixen C., Sunjevaric I., Rothstein R. DNA strand annealing is promoted by the yeast Rad52 protein. Proc. Natl. Acad. Sci. U.S.A. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sugiyama T., New J.H., Kowalczykowski S.C. DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc. Natl. Acad. Sci. U.S.A. 1998;95:6049–6054. doi: 10.1073/pnas.95.11.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sugiyama T., Kantake N., Wu Y., Kowalczykowski S.C. Rad52-mediated DNA annealing after Rad51-mediated DNA strand exchange promotes second ssDNA capture. EMBO J. 2006;25:5539–5548. doi: 10.1038/sj.emboj.7601412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bugreev D.V., Hanaoka F., Mazin A.V. Rad54 dissociates homologous recombination intermediates by branch migration. Nat. Struct. Mol. Biol. 2007;14:746–753. doi: 10.1038/nsmb1268. [DOI] [PubMed] [Google Scholar]
- 43.McIlwraith M.J., West S.C. DNA repair synthesis facilitates RAD52-mediated second-end capture during DSB repair. Mol. Cell. 2008;29:510–516. doi: 10.1016/j.molcel.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 44.Pâques F., Haber J.E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kagawa W., Kagawa A., Saito K., Ikawa S., Shibata T., Kurumizaka H., Yokoyama S. Identification of a second DNA binding site in the human Rad52 protein. J. Biol. Chem. 2008;283:24264–24273. doi: 10.1074/jbc.M802204200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Helleday T., Lo J., van Gent D.C., Engelward B.P. DNA double-strand break repair: From mechanistic understanding to cancer treatment. DNA Repair (Amst). 2007;6:923–935. doi: 10.1016/j.dnarep.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 47.Kagawa W., Kurumizaka H., Ishitani R., Fukai S., Nureki O., Shibata T., Yokoyama S. Crystal structure of the homologous-pairing domain from the human Rad52 recombinase in the undecameric form. Mol. Cell. 2002;10:359–371. doi: 10.1016/s1097-2765(02)00587-7. [DOI] [PubMed] [Google Scholar]
- 48.Hansmann T., Pliushch G., Leubner M., Kroll P., Endt D., Gehrig A., Preisler-Adams S., Wieacker P., Haaf T. Constitutive promoter methylation of BRCA1 and RAD51C in patients with familial ovarian cancer and early-onset sporadic breast cancer. Hum. Mol. Genet. 2012;21:4669–4679. doi: 10.1093/hmg/dds308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Esteller M. Epigenetics in cancer. N. Engl. J. Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.