Abstract
A crucial component of the human mitochondrial DNA replisome is the ring-shaped helicase TWINKLE—a phage T7-gene 4-like protein expressed in the nucleus and localized in the human mitochondria. Our previous studies showed that despite being a helicase, TWINKLE has unique DNA annealing activity. At the time, the implications of DNA annealing by TWINKLE were unclear. Herein, we report that TWINKLE uses DNA annealing function to actively catalyze strand-exchange reaction between the unwinding substrate and a homologous single-stranded DNA. Using various biochemical experiments, we demonstrate that the mechanism of strand-exchange involves active coupling of unwinding and annealing reactions by the TWINKLE. Unlike strand-annealing, the strand-exchange reaction requires nucleotide hydrolysis and greatly stimulated by short region of homology between the recombining DNA strands that promote joint molecule formation to initiate strand-exchange. Furthermore, we show that TWINKLE catalyzes branch migration by resolving homologous four-way junction DNA. These four DNA modifying activities of TWINKLE: strand-separation, strand-annealing, strand-exchange and branch migration suggest a dual role of TWINKLE in mitochondrial DNA maintenance. In addition to playing a major role in fork progression during leading strand DNA synthesis, we propose that TWINKLE is involved in recombinational repair of the human mitochondrial DNA.
INTRODUCTION
Human mitochondrial (mt) DNA is replicated by a minimal T7-like replisome that consists of the nuclear-encoded DNA polymerase γ, DNA helicase TWINKLE and mtSSB (single strand DNA binding protein) (1–5). TWINKLE is a ring-shaped hexameric/heptameric helicase with structural and amino acid sequence homology to bacteriophage T7 gp4 helicase/primase (6–9,10,23). Unlike T7 gp4, however, TWINKLE has lost its primase function due to amino acid changes in the N-terminal domains, but the N-terminal domains appear to still bind to ssDNA (11). The linker region between the N-terminal and C-terminal domains is involved in intra-subunit interactions in T7 gp4 and is conserved between T7 gp4 and TWINKLE (9,12). Point mutations in the linker region of TWINKLE are associated with numerous mt diseases, including progressive external opthalmoplegia, infantile-onset spinocerebellar ataxia, premature ageing, dementia and certain types of cancer (13–15). In the mitochondria, TWINKLE mutations result in replication stalling and mtDNA depletion and deletions (16–18). The most common deletions are flanked by direct repeats (19), which indicates that DNA deletions may be caused by DNA recombination and slipped mispairing (20).
Previous biochemical studies have shown that TWINKLE and DNA polymerase γ along with mtSSB catalyze processive rolling circle DNA synthesis (21). On its own, however, TWINKLE has a poor DNA unwinding activity limited to short forked-duplex DNA, but this activity is enhanced in the presence of SSB (22). The poor unwinding activity of the isolated TWINKLE might be a way to prevent extensive DNA unwinding under situations where the helicase becomes uncoupled from the DNA polymerase. Alternatively, the poor unwinding activity observed in vitro could be due to DNA strand-annealing activity of TWINKLE that we have identified earlier (23), although the role of the annealing activity was not understood. Unlike the helicase activity that requires nucleotide triphosphate (NTP) hydrolysis, the DNA annealing activity is independent of NTP. A possible role of the DNA annealing activity could be in recombination where recombinase-mediated annealing of two ssDNA strands occurs during the strand exchange step. DNA recombination occurs in the yeast mitochondria and is reported in humans in specific organs such as the heart and brain (24,25). However, the proteins catalyzing recombination in the human mitochondria are largely unknown (26). A recent report showed stress-induced recruitment of Rad51 recombinase into mitochondria (27,28).
In this study, we show that TWINKLE on its own can catalyze DNA recombination reactions, including DNA strand-exchange and branch migration of four-way junction substrates. We demonstrate that the annealing activity of TWINKLE aids the strand-exchange reaction by catalyzing joint molecule formation between the recombining strands. The strand-exchange reaction is catalyzed by TWINKLE translocating along the 5′-tail strand of the fork DNA in an NTPase dependent manner while coupling dsDNA unwinding to DNA annealing between the recombining strands. Our findings suggest that TWINKLE may serve a dual role in mtDNA maintenance, both as a replicative helicase during leading strand synthesis and a recombinase, perhaps to aid the repair of double-stranded DNA breaks during replication.
MATERIALS AND METHODS
Proteins
The C-His6-TWINKLE lacking the first 42 amino acids was purified as described (23). The untagged TWINKLE was purified using N-His6-SUMO-fused TWINKLE construct made using Champion pET SUMO expression system from Invitrogen and Escherichia coli Rosetta (DE3) cells. Briefly, cells were lysed using lysozyme (0.2 mg/ml) and repeated freeze-thaw cycles in liquid nitrogen after pellet resuspension in Buffer A (50 mM Tris Cl pH 7.1, 600 mM KCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 10% Glycerol, 0.1% Tween, 0.2 mM 1.4-Dithiothreitol (DTT), 1 mM Phenylmethylsulfonyl fluoride (PMSF) and Roche protease inhibitor tablets). The cell lysate was clarified by centrifugation and precipitated with 65% ammonium sulfate. The ammonium sulfate pellet in Buffer A with 10 mM imidazole was loaded on the Ni-NTA column (GE Healthcare Life Sciences), which was washed with Buffer A with 60 mM imidazole and eluted with 60–350 mM imidazole gradient. Pure fractions were pooled and treated with Sumo-protease (1/100 mg/mg) for ∼16 h at 4°C in Buffer B (50 mM Tris Cl pH 7.9, 150 mM KCl, 5 mM EDTA, 10% Glycerol, 0.1% Tween, 5 mM DTT). The cleaved proteins were loaded on HiTrap Q-HP column (GE) in Buffer B with 200 mM KCl and the flow-through was concentrated and loaded on gel-filtration column (Superdex 200) in Buffer B with 600 mM KCl. Pure fractions were pooled and concentrated. Unless stated, TWINKLE concentration signifies hexamer concentration.
Nucleic acid substrates
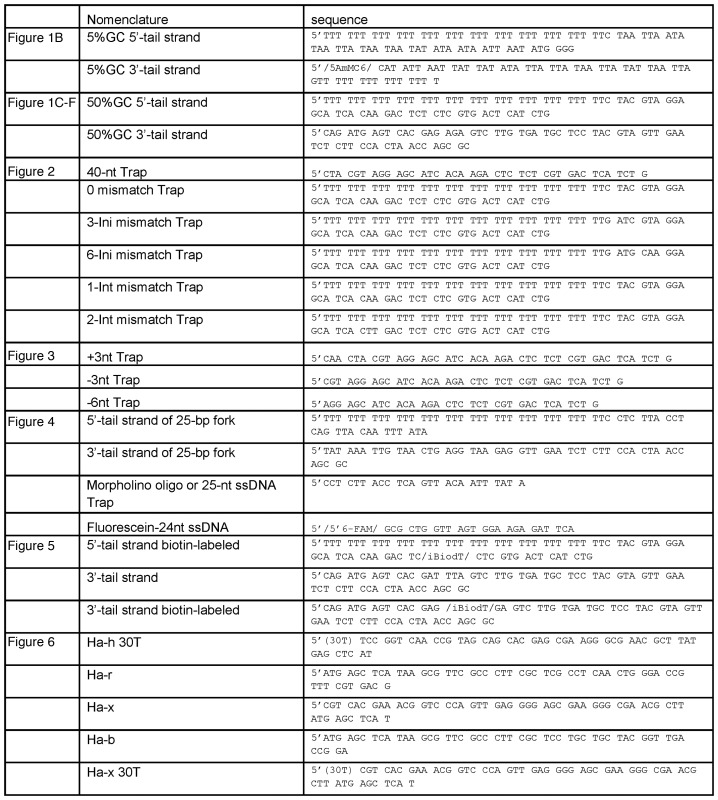
Oligodeoxynucleotides (Table 1) were purchased from Integrated DNA Technologies (Coralville, IA, USA) and polyacrylamide gelelectrophoresis purified in 7 M urea, electro-eluted and ethanol precipitated. The morpholino oligo was purchased from Gene Tools. The DNA concentrations were determined from absorbance at 260 nm and their calculated extinction coefficients. The DNA was 5′-labeled using T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (Perkin-Elmer) and purified by size exclusion chromatography (Bio-Gel P-30, BIORAD). Duplex DNA substrates were generated by heating complementary strands to 95°C followed by slow cooling. Holliday junction (HJ) substrates were prepared by separately annealing h-r in one vial and b-x in another and then mixing equimolar amounts of each of the annealed products at room temperature for 30 min (29).
Table 1. DNA oligonucleotides.
|
Radiometric DNA unwinding, strand-exchange and Holliday junction resolution assay
For unwinding and strand-exchange reactions 2 nM fork DNA and 10 nM TWINKLE hexamer was preincubated in reaction buffer (50 mM Tris-Acetate pH 7.5, 0.01% Tween 20, 5 mM dithiothreitol, 1 mM EDTA) with 4 mM Uridine triphosphate (UTP) for 15 min at 30°C and the reaction was started by adding 6 mM free Mg-acetate and ssDNA (or morpholino) trap (the particular trap used is indicated in the figure legends). The reactions were incubated at 30°C for indicated times and stopped with 12× the reaction volume of quenching solution (100 mM EDTA pH 8.0, 1% sodium dodecyl sulphate (SDS), 0.5% bromophenol blue). The unwinding and strand-exchange experiments with the biotin-streptavidin tagged fork DNAs were performed under the same conditions as above except 12 μM of biotin was added at the start of the reaction with Mg-acetate and 43-nt ssDNA trap. The amount of remaining dsDNA and unwound ssDNA was quantified from native polyacrylamide gel (1 × TBE) using PhosphorImager and ImageQuant and fitted using Sigma Plot software.
The branch migration reactions were carried out by mixing equal volumes of TWINKLE (100 nM), DNA substrate (2 nM), UTP (4 mM) and EDTA (1 mM) with 6 mM free Mg-acetate in unwinding buffer. After indicated times at 25°C, reactions were quenched with SDS-EDTA solution and loaded on a 12% native polyacrylamide gel in 1× TBE buffer. Since the HJ substrates are extremely sensitive to temperature, the gels were subjected to electrophoresis at 4°C at 60 volt to prevent spontaneous resolution. The C-His6-TWINKLE and untagged TWINKLE behaved similarly in unwinding assays (data not shown). The data in Figures 1, 2A–D, 6D–G were carried out with C-His6-TWINKLE and the rest with the untagged TWINKLE. All the experiments were carried out at least twice and representative gels and plots are shown.
Figure 1.
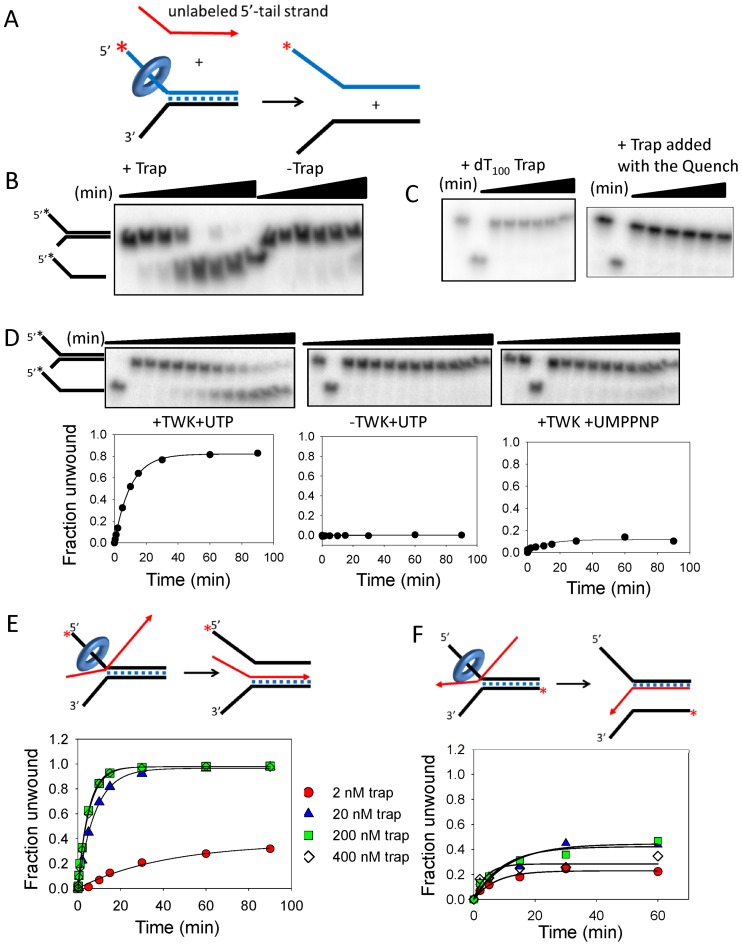
DNA unwinding activity of the TWINKLE depends on trapping of the unwound strand by complementary ssDNA. (A) Twinkle was incubated with the fork DNA with UTP minus Mg(II) and reactions were initiated with Mg(II) plus and minus trap DNA. (B) Gel image shows the time course of 40-bp fork DNA strand separation in the presence and absence of 20 nM ssDNA trap (unlabeled 5′-tail strand, displaced-strand trap). (C) Left, DNA unwinding with dT100 trap (0–60 min) and right, DNA unwinding with displaced-strand trap (200 nM) added with the quenching solution. (D) Top, gel images shows the time course of DNA unwinding in the presence of 20 nM trap with Twinkle plus UTP (left), minus Twinkle (middle) and Twinkle plus UMPPNP (right). Bottom, quantitation of the gel images shows 5′-tail strand generation with an initial rate (exponential rate × amplitude) of 0.08 strand/min. (E) The unwinding efficiency depends on the trap concentration. The initial rates of DNA unwinding with displaced-strand trap at 2 nM (0.009 ± 0.002 strand/min), 20 nM (0.1 ± 0.01 strand/min), 200 nM (0.2 ± 0.006 strand/min) and 400 nM (0.19 ± 0.004 strand/min) is obtained from the kinetics. (F) The initial rates of DNA unwinding with unlabeled 3′-tail strand at 2 nM (0.03 ± 0.006 strand/min), 20 nM (0.04 ± 0.013 strand/min), 200 nM (0.04 ± 0.013 strand/min) and 400 nM (0.07 ± 0.03 strand/min) is obtained from the kinetics. The cartoons show Twinkle (ring) tracking on the 5′-tail strand of the fork DNA to displace the 3′-tail strand and simultaneously annealing either the 3′-tail strand to unlabeled displaced-strand trap (left, 1E), or the 5′-tail strand to the unlabeled 3′-tail strand (right, 1F). The standard errors (SE) of the initial rates are calculated from the SE of the exponential rates and amplitudes using the error propagation formula.
Figure 2.
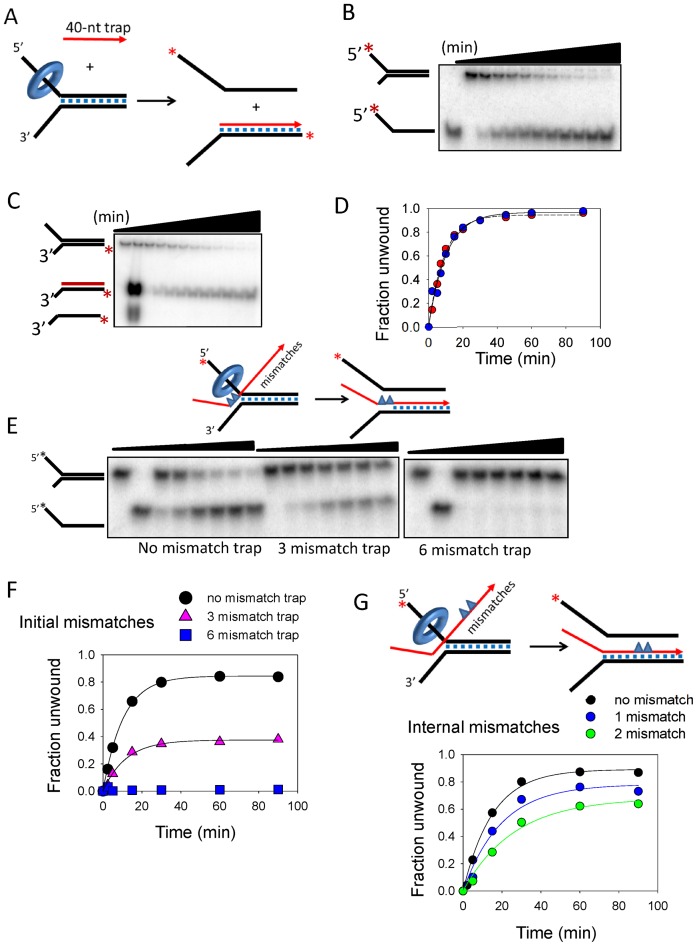
TWINKLE catalyzed strand-exchange reaction is affected by mismatches in the recombining strands. (A) Experimental design to observe the strand-exchanged product using a displaced strand trap (40-nt trap) that is complementary only to the duplex region of the 3′-tail strand. (B) Gel image shows the time course of 5′-tail strand release from the 50% GC fork by TWINKLE in the presence of 200 nM 40-nt trap. (C) Gel image shows the time-dependent generation of the radiolabeled 3′-tail strand annealed to the 40-nt trap. (D) Quantitation of the gel images shows close match between the rates (0.09 ± 0.02 strand/min) of 5′-tail strand release in (B), and accumulation of the strand-exchanged product in (C). (E) Gel images show the time-dependent 5′-tail strand release by TWINKLE in the presence of ‘0 mismatch Trap’ (left image), ‘3-Ini mismatch Trap’ with three consecutive mismatches (middle) or ‘6-Ini mismatch Trap’ with six consecutive mismatches (right) in the initially unwound region at the fork junction, as shown in the cartoon. (F) Quantitation of the gel images in (E) shows the effect of mismatched traps on the kinetics of DNA unwinding. The initial rate of unwinding with no mismatched trap is 0.08 ± 0.01 strand/min and with three mismatched trap is 0.04 ± 0.01 strand/min. (G) Kinetics of 5′-tail strand release in the presence of the displaced-strand trap that contains 1 (‘1-Int mismatch Trap’) or two mismatches in the middle of the 40-bp region (‘2-Int mismatch Trap’). The initial rate of unwinding with one mismatched trap is 0.04 ± 0.001 strand/min and with two mismatched trap is 0.02 ± 0.002 strand/min.
Figure 6.
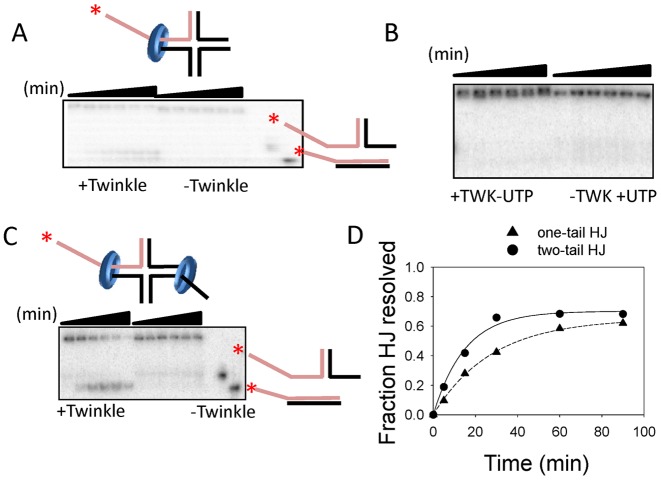
TWINKLE catalyzes homologous branch migration reaction. (A) Gel image shows the time course of HJ resolution of a substrate with one ss-DNA 5′-tail with and without TWINKLE in the presence of MgUTP. (B) Gel image shows control reactions where no HJ resolution was observed in the absence of UTP and absence of TWINKLE. (C) Gel image shows HJ resolution of substrate with two ssDNA 5′-tails in opposite arms. (D) The gels in (A and C) are quantified to obtain the rate of HJ resolution of a substrate with one ss-DNA tail (0.05 ± 0.002 strand/min) and two ssDNA tails (0.02 ± 0.001 strand/min).
Fluorescence anisotropy assays
Fluorescence anisotropy measurements were carried out using FluoroMax-4 spectrofluorimeter (Horiba JobinYvon) in the unwinding buffer at 30°C. Fluorescein labeled 24-nt ssDNA (5 nM) was titrated with Twinkle until there was no change in fluorescence anisotropy (excitation at 494 nm and emission at 516 nm) and then the complex was titrated with increasing concentration of either the non-fluorescent ssDNA or the morpholino oligo. The reported errors are standard deviations.
RESULTS
Trapping of the displaced strand forces TWINKLE to choose unwinding of fork dsDNA over re-annealing of unwound strands
The mtDNA helicase TWINKLE requires a fork DNA to unwind dsDNA (22), but cannot efficiently unwind greater than 20 bp of dsDNA (21). A possible reason for the poor unwinding activity of the TWINKLE is the strand annealing activity, reported previously (23). Here we show that addition of ssDNA that can base pair with one of the unwound strands of the fork DNA tips the balance between unwinding and reannealing toward unwinding in a dramatic way. These in vitro unwinding assays were carried out with the 40-bp fork DNA substrates with 5′- and 3′-non-complementary ssDNA tails (Figure 1A). TWINKLE was preassembled on the fork DNA (radiolabeled 5′-tail strand) in the presence of UTP without Mg(II) and the unwinding reactions were initiated with Mg(II) plus and minus a ssDNA trap. The ssDNA trap (referred to as the displaced-strand trap) was the unlabeled 5′-tail strand, which will base pair with the displaced 3′-tail strand. The time courses show that TWINKLE unwinds the fork DNA in the presence of the displaced-strand trap added at the start of the unwinding reaction with Mg(II) (Figure 1B, +Trap lanes), but not in the absence of the trap (Figure 1B, −Trap lanes). Similarly, no unwinding was observed when dT100 ssDNA was used as the trap (Figure 1C, left) or when the trap was added at the end of the reaction with the quenching solution consisting of EDTA (Figure 1C, right).
To investigate whether the displaced-strand trap DNA itself was causing spontaneous separation of the strands of the fork DNA, we left out TWINKLE or used the non-hydrolyzable analog of UTP, UMPPNP that cannot power processive unwinding. More than 80% DNA unwinding was observed with TWINKLE and UTP in the presence of the trap (Figure 1D, left panel), but not in the absence of TWINKLE (Figure 1D, middle panel), and only 10% unwinding was observed in the presence of UMPPNP, (Figure 1D, right panel). This indicates that the trap is not spontaneously unwinding the fork DNA, but this reaction is actively catalyzed by TWINKLE.
In the above experiments, we used 2 nM of fork DNA and 20 nM of the displaced-strand trap, but we found that even stoichiometric 2 nM concentration of trap supported unwinding (Figure 1E). With lower amount of trap (2 nM), the amplitude or the yield of the unwound product was ∼30% as opposed to almost 100% with 20 nM and higher concentrations of trap. The initial rate of unwinding with 200 and 400 nM trap was twice as fast as the rate with 20 nM trap (Figure 1E).
Next, we investigated whether unwinding was supported by trapping of the 5′-tail strand of the fork DNA with unlabeled 3′-tail strand as the trap. In these experiments, we radiolabeled the 3′-tail strand of the fork DNA to detect its release as ssDNA. Compared to the efficiency of unwinding observed with the displaced-strand trap, the amplitude of unwinding was significantly less when the 3′-tail strand was used as the trap; moreover, higher concentrations of the trap DNA did not increase the unwinding yield (Figure 1F). This is consistent with the model where TWINKLE binds to the 5′-tail strand (22); hence, its presence on that strand sterically hinders base-pairing of the 3′-tail strand trap with the unwound 5′-tail strand. Overall, the results are suggesting that trapping of the displaced-strand, which is favored in the absence of steric hindrance by protein binding, aids the unwinding reaction catalyzed by TWINKLE, as shown in the cartoons in Figure 1E and F. Accordingly, we have observed that TWINKLE can unwind a longer 70-bp fork DNA in the presence of the appropriate displaced strand trap (data not shown). Overall, the above results suggest that trapping of the displaced 3′-tail strand might be occurring concomitant with unwinding.
TWINKLE catalyzes DNA strand-exchange reaction
One way to confirm that TWINKLE is catalyzing concomitant unwinding and annealing is to detect the strand-exchanged DNA product. The expected products of the strand-exchange reaction with the displaced-strand trap are the free 5′-tail strand and the 3′-tail strand annealed to the displaced-strand trap (Figure 2A). We used a 40-nt ssDNA trap that lacks the 5′-tail region of 5'-strand of the fork DNA to distinguish the fork DNA substrate from the strand-exchanged product when resolved by electrophoresis. When the 5′-tail strand of the fork DNA was radiolabeled, the unwinding product was the expected free 5′-tail strand, which increased in amount with increasing reaction time (Figure 2B). When the 3′-tail strand of the fork DNA was radiolabeled, the unwinding product was duplexed 3′-tail strand annealed to the 40-nt trap DNA (Figure 2C). The kinetics of free 5′-tail strand and duplexed 3′-tail strand products overlay closely (Figure 2D). Previously, we have shown that under the conditions of the unwinding assay, spontaneous annealing of the complementary strands of the fork DNA is a slow process; e.g. in 30 min we had observed only ∼5% annealing in the absence of TWINKLE as compared to nearly 90% annealing in the presence of TWINKLE (23). Therefore, annealing of the displaced-strand trap with the unwound 3′-tail strand is unlikely to be a passive event. These results indicate that TWINKLE is catalyzing a homologous strand-exchange reaction between the ssDNA trap and the unwound 3'-tail strand by coupling DNA unwinding to annealing.
From here on, we refer to the trap-assisted DNA unwinding activity as the strand-exchange activity and the ssDNA trap as the invading strand.
Complementarity between the recombining strands is crucial for TWINKLE catalyzed strand-exchange reaction
To investigate the mechanism of strand-exchange catalyzed by TWINKLE, we introduced base changes in the invading strand to create mismatches with the complementary 3′-tail strand on the fork DNA. In one set, we introduced mismatches in the initially unwound region (Figure 2E, blue triangles in the trap strand in red). When the first 3-nt bases of the invading strand are mismatched, there is 50% reduction in the strand-exchange efficiency as compared to a fully matched invading strand (Figure 2F). When the first 6-nt bases of the invading strand are mismatched, there is no reaction (Figure 2F). Similarly, one or two mismatches in the middle of the invading strand decreased the efficiency of the strand-exchange reaction by 2–4-fold (Figure 2G).
When we deleted the first three nucleotides of the invading DNA that anneals to the initially unwound region of the fork DNA (−3 nt trap, Figure 3A), the unwinding yield reduced from 80% to 40% (with 20 nM trap) and the deletion of 6-nt reduced the unwinding yield to 10% (Figure 3B). Increasing trap DNA concentration recovered the unwinding yield with the −3 nt trap, but not with the −6-nt trap (Figure 3C). These results indicate that complementarity between the invading strand and first few nucleotide bases of the displaced strand of the fork DNA is important for efficient strand-exchange reaction.
Figure 3.
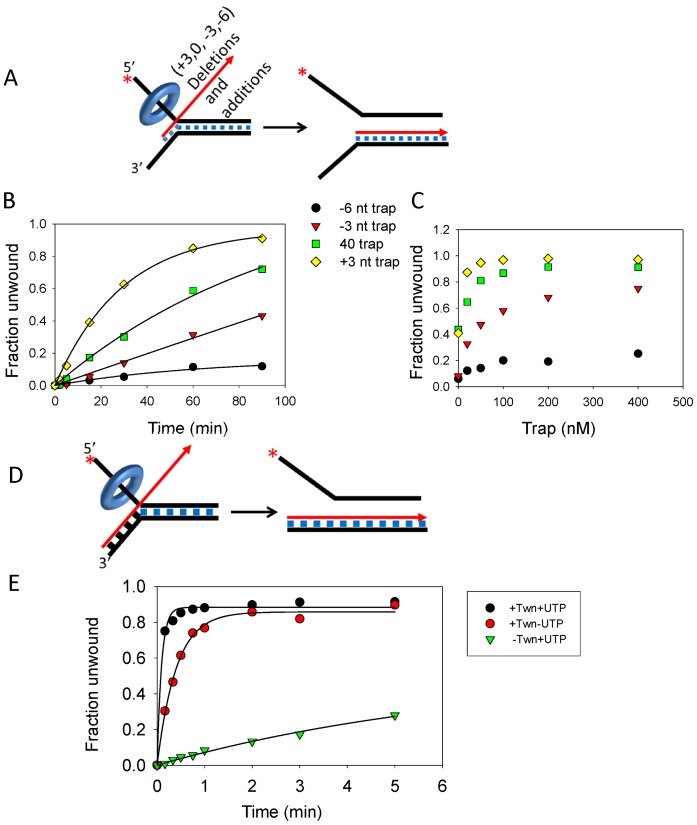
Effect of decreasing or increasing the homology region between the trap DNA and fork DNA on the strand-exchange reaction. (A) The 40-nt trap (designated ‘0’) was extended by three nucleotides at the 5′-end (‘+3 nt Trap’) to match the bases on the 3′-tail of the fork DNA or 3 and 6-nt were deleted (‘−3 nt Trap’ and ‘−6 nt Trap’, respectively) at the 5′-end so homology will initiate downstream from the unwinding region. (B) Kinetics of 5′-strand release by TWINKLE in the presence of 20 nM ‘40-nt Trap’ (0.012 ± 0.003 strand/min), ‘−3 nt Trap’ (0.005 ± 0.03 strand/min), ‘−6 nt Trap’ (0.002 ± 0.003 strand/min), and ‘+3 nt Trap’ (0.032 ± 0.002 strand/min). (C) The unwinding yield after 60 min of reaction with increasing trap DNA concentrations. (D) The 65-nt trap is completely complementary to the 3′-tail strand of the fork DNA. (E) Kinetics of DNA unwinding with 2 nM fully complementary trap with TWINKLE and UTP (9.3 ± 1 strand/min), with TWINKLE and without UTP (2 ± 0.12 strand/min) and without TWINKLE (0.08 ± 0.02 strand/min).
Interestingly, when we extended the region of homology between the invading strand and the 3′-tail strand of the fork DNA by adding three additional nucleotides that would base pair to the region immediately upstream of the unwinding duplex DNA (+3-nt trap), the efficiency of the reaction increased by ∼2-fold (Figure 3B, yellow diamonds). These effects were observed both in a time course of unwinding at fixed trap concentration (Figure 3B) and in a trap DNA concentration manner (Figure 3C).
To further test this idea, we increased the complementarity between the invading strand and the 3′-tail strand, such that the invading strand can form a 25-bp long joint molecule with the 3′-ssDNA tail of the fork DNA prior to the strand-exchange reaction (Figure 3D). The strand-exchange reaction in the presence of the 65-nt invading strand is very fast and complete within 10 s (Figure 3E). We estimate that the strand-exchange rate with the fully complementary 65-nt strand is ∼900 times faster than the rate with the trap where only 40-nt region of the duplex fork is complementary (see reaction in Figure 1E). Interestingly, the fully complementary 65-nt invading strand spontaneously strand-exchanges with the 3'-tail strand of the fork DNA in the absence of TWINKLE (Figure 3E). However, the spontaneous strand-exchange rate is 100 times slower as compared to the TWINKLE-catalyzed rate with UTP (Figure 3E). Interestingly, the TWINKLE-catalyzed strand-exchange rate without UTP is only five times slower as compared to with UTP (Figure 3E). This suggests that joint molecule formation is important and requires TWINKLE but not UTP, and once joint molecule is formed, the strand-exchange reaction can occur spontaneously, but the rate is much faster in the presence of TWINKLE. This is consistent with our previous studies that showed that TWINKLE catalyzes strand annealing in the absence of UTP (23). Taken together these results provide strong evidence that TWINKLE is catalyzing the reactions of strand-exchange and joint molecule formation by using its DNA unwinding and annealing activities.
The strand-exchange reaction catalyzed by TWINKLE requires interactions with the recombining strands
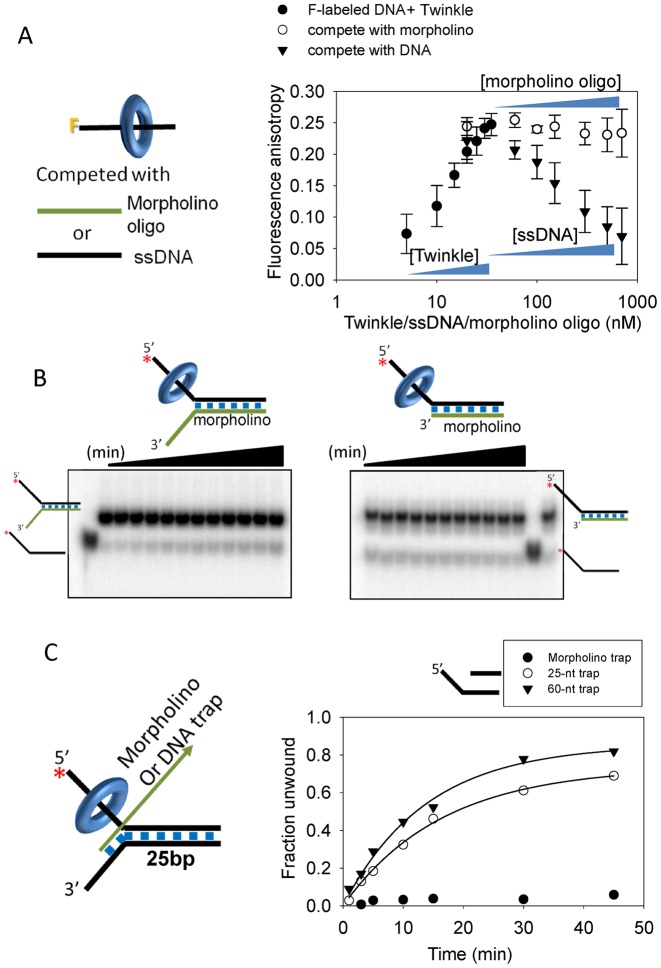
To catalyze joint molecule formation, TWINKLE must bring the invading strand and the homologous strand in the fork DNA in close proximity. To understand the mechanism of joint molecule formation, we replaced either the invading or the displaced-strand with a morpholino nucleic acid. A morpholino nucleic acid has standard bases that can base pair in the same manner as the DNA, but the bases are attached to morpholine rings instead of deoxyribose rings and linked through phosphorodiamidate groups that lack negative charges as in the phosphates of DNA. First, we established that TWINKLE does not interact with the morpholino oligo using a competition binding experiment. Fluorescence anisotropy measures binding of TWINKLE to fluorescein-labeled ssDNA, as described previously (23). A complex of TWINKLE with fluorescein-labeled ssDNA was titrated with increasing concentration of either the unlabeled ssDNA or a morpholino oligo. The decrease in fluorescence anisotropy upon addition of unlabeled ssDNA indicates that the fluorescein-labeled DNA strand is exchanged with the unlabeled DNA strand. When the complex was chased with morpholino oligo, a very slight decrease in fluorescence anisotropy was observed even at very high concentrations close to 1 μM. This indicates that the morpholino oligo is unable to displace the fluorescein-labeled ssDNA strand bound to TWINKLE. These DNA competition experiments show that TWINKLE does not have affinity for the morpholino oligo (Figure 4A).
Figure 4.
TWINKLE interacts with both the displaced-strand trap and the homologous 3′-tail strand to catalyze the strand-exchange reaction. (A) Fluorescence anisotropy competition assay to assess the binding of Twinkle to morpholino oligo. A complex of Twinkle (represented by blue ring) and ssDNA labeled with fluorescein (represented by black line with ‘F’ at one end in the cartoon) was formed first by titrating the labeled ssDNA with increasing TWINKLE, then the complex was titrated either with increasing concentrations of unlabeled ssDNA or the morpholino oligo and fluorescence anisotropy was measured in each case. A drop in fluorescence anisotropy indicating the ‘falling-off’ of TWINKLE from the pre-bound fluorescein-labeled ssDNA was observed only with ssDNA and not with the morpholino oligo. (B) Gel image shows the time course of unwinding the DNA/morpholino hybrid fork with 18-bp duplex region (left) or the DNA/morpholino 5'-overhang DNA with 25-bp duplex region (right). Quantitation shows about 3% unwinding in each case. (C) DNA unwinding in the presence of the morpholino oligo as trap. Kinetics of 5′-tail strand release from the 25-bp fork DNA in the presence of ssDNA 25-nt trap (0.045 ± 0.003 strand/min), ssDNA 60-nt trap (0.06 ± 0.006 strand/min) and morpholino oligo trap. The 60-nt trap is the unlabeled 5′ strand of the forked-substrate DNA.
When the 3′-tail strand of the unwinding substrate was replaced with a morpholino oligo and reactions were carried out in the presence of the displaced-strand DNA trap, TWINKLE did not unwind the fork DNA (or a 5′-overhang DNA) (Figure 4B and C). This is distinct from T7 gp4 helicase that unwinds the morpholino-DNA hybrid substrate (30). The results with TWINKLE indicate that the strand-exchange reaction requires interactions of TWINKLE with the 3'-strand of the unwinding substrate. Next, we used a DNA fork substrate and morpholino oligo as the trap added with the Mg(II) at the start of the reaction. Again, there was no strand-exchange reaction by TWINKLE in the presence of the morpholino oligo trap (Figure 4C). These experiments indicate that the strand-exchange reaction requires interactions of the TWINKLE with both the invading strand and the homologous strand of the fork DNA.
TWINKLE translocates on the 5′-strand of the fork DNA to catalyze the strand-exchange reaction
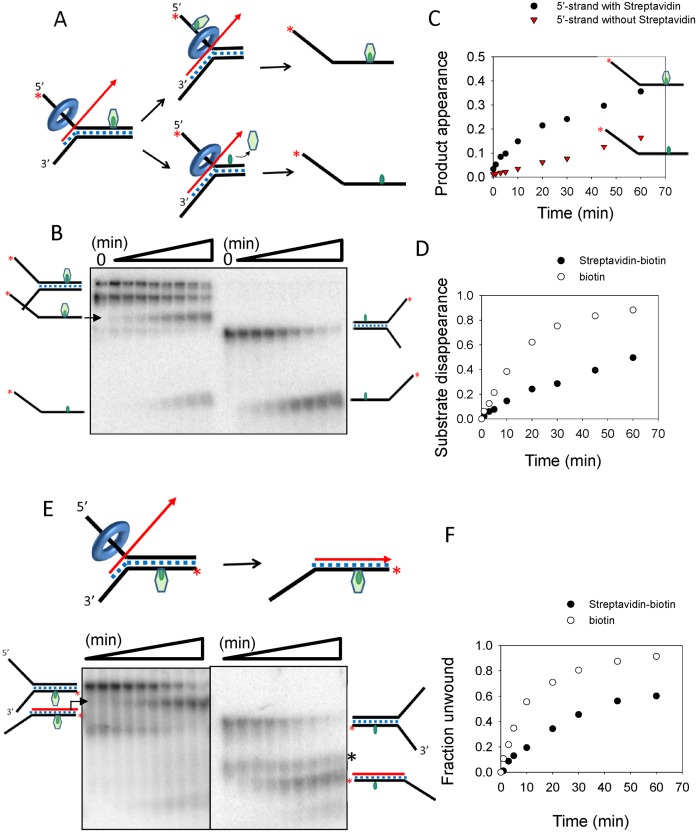
To further characterize the mechanism of the strand-exchange reaction, the fork DNA substrate was modified with a site-specific biotin-streptavidin, which is a bulky adduct that can determine which strand TWINKLE translocates along during the strand-exchange reaction and whether the bulky adduct inhibits the strand-exchange reaction (31). In one experiment, the biotin tag was introduced on the 5′-tail strand in the middle of the 40-bp duplex region and we expected the following outcomes (Figure 5A): (i) the bulky adduct blocks the strand-exchange reaction, in which case no 5′-tail strand product will be detected, (ii) strand-exchange occurs but TWINKLE bypasses the streptavidin, in which case the 5′-tail strand product still bound to the streptavidin will be detected, (iii) strand-exchange occurs and TWINKLE displaces the streptavidin, in which case the 5′-tail strand product without streptavidin will be detected. These outcomes were distinguished by monitoring the reactants and products on a native polyacrylamide gel.
Figure 5.
TWINKLE both bypasses and displaces the streptavidin attached to the fork DNA strands. (A) The 5′-tail strand of the 40-bp fork DNA was modified with biotin-streptavidin (green) and strand-exchange reaction was carried out in the presence of the displaced-strand trap (+3-nt trap, Table 1). (B) The gel image shows the time course of unwinding the 5′-tail strand biotin-streptavidin labeled fork DNA (left) and biotin-only fork DNA (right). (C) The kinetics of unwinding produces two types of 5′-tail strands, one with streptavidin bound (0.018 ± 0.005 strand/min) and one without streptavidin (0.003 ± 0.005 strand/min). (D) The kinetics of unwinding the 5′-tail strand streptavidin-biotin fork DNA (0.013 ± 0.003 strand/min) is compared to the biotin-only fork DNA (0.05 ± 0.002 strand/min). The fraction unwound represents total 5′-strand with and without streptavidin, which also corresponds to the substrate disappearance. (E) The gel image shows the time course of unwinding the fork DNA with biotin-streptavidin attached to the 3′-tail strand (left) and biotin-only fork DNA (right). The ‘*’ indicates a contaminant present in all the lanes. (F) The kinetics of unwinding the 3′-tail strand with biotin-streptavidin (0.02 ± 0.005 strand/min) is compared to biotin-only fork DNA (0.073 ± 0.005 strand/min).
The biotin-streptavidin labeled DNA fork was incubated with TWINKLE plus UTP and reactions were initiated with a mixture of Mg(II), displaced-strand trap and excess of biotin. Streptavidin can bind up to four biotin molecules, which explains the two species, monomer and dimer, of the streptavidin-complexed fork DNA as two bands on the gel at zero time (Figure 5B). Upon reaction, there was a time-dependent increase of both the streptavidin-free and streptavidin-bound 5′-tail strand products. These results indicate that TWINKLE can both displace and bypass the streptavidin block on the 5′-tail strand. The greater yield and six times faster rate of streptavidin-bound product indicates that the probability of bypass is higher than displacement (Figure 5C). This type of passage over a barrier has been observed for other ring-shaped helicases, such as the T7 gp4 helicase and MCM, where it was suggested that transient ring opening occurs during unwinding that allows these helicases to go over bulky moieties along the way (30,32). When we compare the strand-exchange rates of biotin-only fork DNA and streptavidin-biotin labeled fork DNA (Figure 5D), we find that the streptavidin adduct on the 5′-tail strand poses a barrier and slows the movement of the TWINKLE by about 4-fold.
On the other hand, when the biotin-streptavidin was introduced in the displaced 3′-tail strand, the major product is streptavidin-bound strand-exchanged product (Figure 5E). The small amount of streptavidin-free strand-exchanged product is produced from the small amount of fork DNA without streptavidin that was initially present in the reaction. This indicates that TWINKLE completely bypasses the bulky streptavidin on the 3′-tail strand during the strand-exchange reaction. Nevertheless, the 3-fold faster rate of unwinding the 3′-tail strand biotin-only fork DNA as compared to the 3′-tail strand streptavidin-biotin labeled fork DNA (Figure 5F) indicates that the bulky moiety on the displaced strand also inhibits the strand-exchange reaction. Overall, the experiments with the biotin-streptavidin modified fork DNAs indicate that TWINKLE catalyzes strand-exchange by translocating along the 5′-tail strand of the fork DNA while having some interactions with the 3′-tail strand, and it has the ability to both displace and bypass the streptavidin block on the translocating strand whereas it bypasses the block on the displaced strand.
TWINKLE catalyzes homologous branch migration reaction
To determine whether TWINKLE can resolve a four-way Holliday Junction (HJ) DNA substrate, we designed a homologous HJ that was used to demonstrate branch migration activity of T7 gp4 helicase (29). A single heterologous base was introduced at the center of the four-way junction to prevent spontaneous branch migration. Two opposite arms of the HJ substrate are 20 and 25 bp long and contain 5′-dT30 ssDNA tail on one or both the arms and the other arms are 30 bp long. It has been previously demonstrated that fork DNA with duplex 5′-tail cannot be unwound by TWINKLE suggesting that ssDNA 5′-tail is required for loading the hexamer (22). The requirement of ssDNA for loading TWINKLE enables efficient loading of the helicase on the ssDNA tails of the HJ and prevents loading of additional helicases on the other duplex arms. We observe branch migration in the presence of TWINKLE and UTP (Figure 6A), but not in the absence of TWINKLE or in the absence of UTP (Figure 6A and B). Similarly, we observe branch migration in HJ with two ssDNA tails on opposite arms (Figure 6C). The rate of HJ resolution is two times faster when the HJ has two dT30 tails as opposed to one dT30 tail (Figure 6D). These results indicate that TWINKLE like other hexameric helicases can catalyze the branch migration reaction.
DISCUSSION
In this study, we show that TWINKLE has four DNA modifying activities: strand separation, strand annealing, strand-exchange and branch migration. Only the DNA strand separation activity is needed for leading strand synthesis, which is catalyzed by the replisome complex of TWINKLE with polymerase γ and mtSSB (21). However, all four enzymatic activities of TWINKLE would be needed for recombinational repair of double strand breaks. The mtDNA undergoes extensive oxidative damage from reactive oxygen species continuously generated as a byproduct of the respiratory process in the mitochondria (33,34). The resulting base modifications and nicks in DNA cause replication fork arrest and collapse with deleterious consequences, if not repaired. In nuclear DNA, recombinational repair is a major pathway for repair of collapsed and arrested replication forks where sequence information from sister chromosome or the other parental DNA is used to restart the replication fork (35–37). Much is known about base excision repair and mismatch repair pathways in the human mitochondria (38), however little is known about recombinational repair.
Recombinational repair involves a series of steps, including strand separation and recession of the DNA break to create a single-stranded 3′-end for the strand invasion into a homologous dsDNA coupled to DNA unwinding and synthesis, and subsequently branch migration to resolve the products. The mt enzyme ExoG is a 5′-3′ nuclease associated with polymerase γ whose depletion causes persistent single strand breaks in mtDNA and this nuclease might be involved in the DNA recession reaction (39,40). The 3′-strand invasion is a recombination reaction and the RecA homolog Rad51 is a key protein that catalyzes this reaction in nuclear DNA. Interestingly, Rad51 was recently reported to localize into mitochondria under oxidative stress conditions (28). We show here that TWINKLE has strand-exchange activity of its own suggesting that TWINKLE may be involved in this reaction, either on its own or with Rad51.
To our knowledge, TWINKLE is the first replicative ring-shaped helicase with demonstrated strand-exchange activity of its own. The homologous T7 gp4 helicase catalyzes strand-exchange reaction, but only in the presence of the gp2.5 protein which also aids in joint molecule formation (41). Similarly, phage T4 helicase works with the UvsX recombinase (42) and herpes simplex virus type-1 helicase with ICP8 (43) to catalyze recombination. Joint molecule formation is critical for efficient strand-exchange reaction and TWINKLE is unique among these replicative helicases in that it contains a DNA strand annealing activity that aids in joint molecule formation. We show that a short region of homology between the invading ssDNA and the 3′-ssDNA tail of the fork DNA greatly increases the strand-exchange rates. When the homology region is 25 bp long, strand-exchange reaction rates are much faster. Increasing the homology length also promoted spontaneous strand-exchange reaction, but the TWINKLE-assisted reactions with UTP were ∼100-fold faster. The strand-exchange reaction by TWINKLE was sensitive to base pair mismatches, and both internal mismatches and mismatches and deletions near the initiation site affected the strand-exchange rates. One and two mismatches slowed the strand-exchange reaction and six mismatches drastically inhibited the reaction. The impact of internal mismatches on the rate of strand-exchange is not as dramatic as caused by initial mismatches. Lack of homology in the initially unwound region shows a drastic effect most likely because this homology is used by TWINKLE to recruit the displaced strand trap to base pair with the 3′-tail strand of the fork DNA substrate and initiate the strand-exchange reaction. If the invading strand was passively annealing to the displaced 3′-tail strand after it was completely unwound, we would not see such drastic effects from few mismatches over the 40-bp duplex region. It also appears that the unwinding and annealing are coupled at the base pair level, meaning that if annealing over one or two base pairs cannot occur, then the unwinding reaction is inhibited.
Although the detailed mechanism of how TWINKLE couples its unwinding activity to annealing during catalysis of strand-exchange remains to be determined, our studies have provided important new insights into the role of TWINKLE in mtDNA manipulations. We found that the strand-exchange reaction was sensitive to backbone modifications in the recombining strands. When the invading ssDNA was substituted with morpholino backbone DNA, the strand-exchange reaction was completely inhibited. Similarly, when the 3′-tail strand of the fork DNA was substituted with morpholino backbone DNA, the reaction was inhibited. This is in contrast to T7 gp4, which efficiently unwinds the 3′-morpholino backbone DNA (30). The results indicate that TWINKLE interacts with both the recombining strands during the strand-exchange reaction. Insights into how TWINKLE translocates on the fork DNA to unwind and strand-exchange were obtained from the streptavidin displacement assays. TWINKLE can displace streptavidin from biotin attached to ssDNA (data not shown). During the strand-exchange reaction, TWINKLE both displaces and bypasses the streptavidin on the 5′-tail strand, but only bypasses the streptavidin on the 3′-tail strand. This is consistent with 5′-3′ directionality of translocation and indicates that during strand-exchange TWINKLE threads the 5′-tail strand through its central channel and excludes the 3′-tail strand, similar to T7 gp4 unwinding by the strand-exclusion mechanism (44,45). Interestingly, like T7 gp4 (30), the TWINKLE ring can transiently open to bypass the streptavidin adduct on the translocating strand. We had proposed that the N-terminal domains of TWINKLE might be the secondary DNA binding sites involved in DNA annealing (23). Although the N-terminal domains of TWINKLE have lost the primase function, they may bind ssDNA (11). However, whether the N-teminal domains of TWINKLE have acquired a new function of DNA strand annealing to aid strand-exchange remains to be tested.
In addition to strand-exchange reaction, TWINKLE also catalyzes branch migration reaction on four-way HJ substrates. This activity is observed in many hexameric helicases, including T7 gp4, T4 gp41 and bacterial DnaB (29,46,47). However, the combination of strand-exchange and branch migration activities of TWINKLE is uniquely suitable for catalyzing recombinational repair of double-strand DNA breaks during replication. Evidence for TWINKLE being involved in recombination comes from studies of transgenic mouse model expressing TWINKLE mutants, which accumulate multiple mtDNA deletions and show elevated replication stalling (17,48). We speculate that the linker region mutations associated with mt-related diseases cause dysfunctions in the DNA modifying activities of the TWINKLE to increase the chances of DNA mispairing during recombination. The linker region is conserved between T7 gp4 and TWINKLE, and homologous mutations in ther linker region of T7 gp4 show helicase-deficiency, either from defects in DNA binding or uncoupled/defective NTP hydrolysis (49,50). One such mutant of T7 gp4, the A257T is analogous to the A359T mutant of TWINKLE, and its detailed characterization reported earlier revealed that the unwinding defect was due to its inability to load on the fork DNA (51). The DNA loading defect was partially rescued by T7 DNA polymerase, which interacts with T7 gp4. One can imagine that such defects in DNA loading would increase the chances of slipped mispairing by delaying replication fork restart. Detailed studies of recombinational repair by the mt replisome are necessary to understand this novel function of the TWINKLE helicase.
Acknowledgments
We thank members of the Patel Lab for constructive criticisms throughout these studies and Dr. Manjula Pandey for her guidance.
FUNDING
National Institutes of Health [GM55310 to S.S.P.]Funding for open access charge: NIH [GM55310].
Conflict of interest statement. None declared.
REFERENCES
- 1.Clayton D.A. Mitochondrial DNA replication: what we know. IUBMB Life. 2003;55:213–217. doi: 10.1080/1521654031000134824. [DOI] [PubMed] [Google Scholar]
- 2.Holt I.J., Reyes A. Human mitochondrial DNA replication. Cold Spring Harb. Perspect. Biol. 2012;4 doi: 10.1101/cshperspect.a012971. doi:10.1101/cshperspect.a012971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McKinney E.A., Oliveira M.T. Replicating animal mitochondrial DNA. Genet. Mol. Biol. 2013;36:308–315. doi: 10.1590/S1415-47572013000300002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miralles Fuste J., Shi Y., Wanrooij S., Zhu X., Jemt E., Persson O., Sabouri N., Gustafsson C.M., Falkenberg M. In vivo occupancy of mitochondrial single-stranded DNA binding protein supports the strand displacement mode of DNA replication. PLoS Genet. 2014;10:e1004832. doi: 10.1371/journal.pgen.1004832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szymanski M.R., Kuznetsov V.B., Shumate C., Meng Q., Lee Y.S., Patel G., Patel S., Yin Y.W. Structural basis for processivity and antiviral drug toxicity in human mitochondrial DNA replicase. EMBO J. 2015;34:1959–1970. doi: 10.15252/embj.201591520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spelbrink J.N., Li F.Y., Tiranti V., Nikali K., Yuan Q.P., Tariq M., Wanrooij S., Garrido N., Comi G., Morandi L., et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat. Genet. 2001;28:223–231. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- 7.Fernandez-Millan P., Lazaro M., Cansiz-Arda S., Gerhold J.M., Rajala N., Schmitz C.A., Silva-Espina C., Gil D., Bernado P., Valle M., et al. The hexameric structure of the human mitochondrial replicative helicase Twinkle. Nucleic Acids Res. 2015;43:4284–4295. doi: 10.1093/nar/gkv189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toth E.A., Li Y., Sawaya M.R., Cheng Y., Ellenberger T. The crystal structure of the bifunctional primase-helicase of bacteriophage T7. Mol. Cell. 2003;12:1113–1123. doi: 10.1016/s1097-2765(03)00442-8. [DOI] [PubMed] [Google Scholar]
- 9.Singleton M.R., Sawaya M.R., Ellenberger T., Wigley D.B. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- 10.Ziebarth T.D., Farr C.L., Kaguni L.S. Modular architecture of the hexameric human mitochondrial DNA helicase. J. Mol. Biol. 2007;367:1382–1391. doi: 10.1016/j.jmb.2007.01.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farge G., Holmlund T., Khvorostova J., Rofougaran R., Hofer A., Falkenberg M. The N-terminal domain of TWINKLE contributes to single-stranded DNA binding and DNA helicase activities. Nucleic Acids Res. 2008;36:393–403. doi: 10.1093/nar/gkm1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sawaya M.R., Guo S., Tabor S., Richardson C.C., Ellenberger T. Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7. Cell. 1999;99:167–177. doi: 10.1016/s0092-8674(00)81648-7. [DOI] [PubMed] [Google Scholar]
- 13.Copeland W.C. Defects in mitochondrial DNA replication and human disease. Crit. Rev. Biochem. Mol. Biol. 2012;47:64–74. doi: 10.3109/10409238.2011.632763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vandenberghe W., Van Laere K., Debruyne F., Van Broeckhoven C., Van Goethem G. Neurodegenerative Parkinsonism and progressive external ophthalmoplegia with a Twinkle mutation. Mov. Disord. 2009;24:308–309. doi: 10.1002/mds.22198. [DOI] [PubMed] [Google Scholar]
- 15.Wanrooij S., Falkenberg M. The human mitochondrial replication fork in health and disease. Biochim. Biophys. Acta. 2010;1797:1378–1388. doi: 10.1016/j.bbabio.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 16.Tyynismaa H., Mjosund K.P., Wanrooij S., Lappalainen I., Ylikallio E., Jalanko A., Spelbrink J.N., Paetau A., Suomalainen A. Mutant mitochondrial helicase Twinkle causes multiple mtDNA deletions and a late-onset mitochondrial disease in mice. Proc. Natl. Acad. Sci. U.S.A. 2005;102:17687–17692. doi: 10.1073/pnas.0505551102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wanrooij S., Goffart S., Pohjoismaki J.L., Yasukawa T., Spelbrink J.N. Expression of catalytic mutants of the mtDNA helicase Twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic Acids Res. 2007;35:3238–3251. doi: 10.1093/nar/gkm215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pohjoismaki J.L., Goffart S., Spelbrink J.N. Replication stalling by catalytically impaired Twinkle induces mitochondrial DNA rearrangements in cultured cells. Mitochondrion. 2011;11:630–634. doi: 10.1016/j.mito.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Mita S., Rizzuto R., Moraes C.T., Shanske S., Arnaudo E., Fabrizi G.M., Koga Y., DiMauro S., Schon E.A. Recombination via flanking direct repeats is a major cause of large-scale deletions of human mitochondrial DNA. Nucleic Acids Res. 1990;18:561–567. doi: 10.1093/nar/18.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krishnan K.J., Reeve A.K., Samuels D.C., Chinnery P.F., Blackwood J.K., Taylor R.W., Wanrooij S., Spelbrink J.N., Lightowlers R.N., Turnbull D.M. What causes mitochondrial DNA deletions in human cells. Nat. Genet. 2008;40:275–279. doi: 10.1038/ng.f.94. [DOI] [PubMed] [Google Scholar]
- 21.Korhonen J.A., Pham X.H., Pellegrini M., Falkenberg M. Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 2004;23:2423–2429. doi: 10.1038/sj.emboj.7600257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korhonen J.A., Gaspari M., Falkenberg M. TWINKLE Has 5′ -> 3′ DNA helicase activity and is specifically stimulated by mitochondrial single-stranded DNA-binding protein. J. Biol. Chem. 2003;278:48627–48632. doi: 10.1074/jbc.M306981200. [DOI] [PubMed] [Google Scholar]
- 23.Sen D., Nandakumar D., Tang G.Q., Patel S.S. Human mitochondrial DNA helicase TWINKLE is both an unwinding and annealing helicase. J. Biol. Chem. 2012;287:14545–14556. doi: 10.1074/jbc.M111.309468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Torregrosa-Munumer R., Goffart S., Haikonen J.A., Pohjoismaki J.L. Low doses of UV and oxidative damage induce dramatic accumulation of mitochondrial DNA replication intermediates, fork regression and replication initiation shift. Mol. Biol. Cell. 2015;26:4197–4208. doi: 10.1091/mbc.E15-06-0390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pohjoismaki J.L., Goffart S., Tyynismaa H., Willcox S., Ide T., Kang D., Suomalainen A., Karhunen P.J., Griffith J.D., Holt I.J., et al. Human heart mitochondrial DNA is organized in complex catenated networks containing abundant four-way junctions and replication forks. J. Biol. Chem. 2009;284:21446–21457. doi: 10.1074/jbc.M109.016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X.J. Mechanism of homologous recombination and implications for aging-related deletions in mitochondrial DNA. Microbiol. Mol. Biol. Rev. 2013;77:476–496. doi: 10.1128/MMBR.00007-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sage J.M., Gildemeister O.S., Knight K.L. Discovery of a novel function for human Rad51: maintenance of the mitochondrial genome. J. Biol. Chem. 2010;285:18984–18990. doi: 10.1074/jbc.M109.099846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sage J.M., Knight K.L. Human Rad51 promotes mitochondrial DNA synthesis under conditions of increased replication stress. Mitochondrion. 2013;13:350–356. doi: 10.1016/j.mito.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rasnik I., Jeong Y.J., McKinney S.A., Rajagopal V., Patel S.S., Ha T. Branch migration enzyme as a Brownian ratchet. EMBO J. 2008;27:1727–1735. doi: 10.1038/emboj.2008.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeong Y.J., Rajagopal V., Patel S.S. Switching from single-stranded to double-stranded DNA limits the unwinding processivity of ring-shaped T7 DNA helicase. Nucleic Acids Res. 2013;41:4219–4229. doi: 10.1093/nar/gkt133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morris P.D., Tackett A.J., Raney K.D. Biotin-streptavidin-labeled oligonucleotides as probes of helicase mechanisms. Methods. 2001;23:149–159. doi: 10.1006/meth.2000.1116. [DOI] [PubMed] [Google Scholar]
- 32.Yardimci H., Wang X., Loveland A.B., Tappin I., Rudner D.Z., Hurwitz J., van Oijen A.M., Walter J.C. Bypass of a protein barrier by a replicative DNA helicase. Nature. 2012;492:205–209. doi: 10.1038/nature11730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cadenas E., Davies K.J. Mitochondrial free radical generation, oxidative stress, and aging. Free Radic. Biol. Med. 2000;29:222–230. doi: 10.1016/s0891-5849(00)00317-8. [DOI] [PubMed] [Google Scholar]
- 34.Sykora P., Wilson D.M. 3rd, Bohr V.A. Repair of persistent strand breaks in the mitochondrial genome. Mech. Ageing Dev. 2012;133:169–175. doi: 10.1016/j.mad.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cox M.M. Recombinational DNA repair of damaged replication forks in Escherichia coli: questions. Annu. Rev. Genet. 2001;35:53–82. doi: 10.1146/annurev.genet.35.102401.090016. [DOI] [PubMed] [Google Scholar]
- 36.Michel B. Replication fork arrest and DNA recombination. Trends Biochem. Sci. 2000;25:173–178. doi: 10.1016/s0968-0004(00)01560-7. [DOI] [PubMed] [Google Scholar]
- 37.Kowalczykowski S.C. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- 38.Akhmedov A.T., Marin-Garcia J. Mitochondrial DNA maintenance: an appraisal. Mol. Cell. Biochem. 2015;409:283–305. doi: 10.1007/s11010-015-2532-x. [DOI] [PubMed] [Google Scholar]
- 39.Tann A.W., Boldogh I., Meiss G., Qian W., Van Houten B., Mitra S., Szczesny B. Apoptosis induced by persistent single-strand breaks in mitochondrial genome: critical role of EXOG (5′-EXO/endonuclease) in their repair. J. Biol. Chem. 2011;286:31975–31983. doi: 10.1074/jbc.M110.215715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cymerman I.A., Chung I., Beckmann B.M., Bujnicki J.M., Meiss G. EXOG, a novel paralog of Endonuclease G in higher eukaryotes. Nucleic Acids Res. 2008;36:1369–1379. doi: 10.1093/nar/gkm1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kong D., Richardson C.C. Single-stranded DNA binding protein and DNA helicase of bacteriophage T7 mediate homologous DNA strand exchange. EMBO J. 1996;15:2010–2019. [PMC free article] [PubMed] [Google Scholar]
- 42.Liu J., Morrical S.W. Assembly and dynamics of the bacteriophage T4 homologous recombination machinery. Virol. J. 2010;7:357. doi: 10.1186/1743-422X-7-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nimonkar A.V., Boehmer P.E. In vitro strand exchange promoted by the herpes simplex virus type-1 single strand DNA-binding protein (ICP8) and DNA helicase-primase. J. Biol. Chem. 2002;277:15182–15189. doi: 10.1074/jbc.M109988200. [DOI] [PubMed] [Google Scholar]
- 44.Ahnert P., Patel S.S. Asymmetric interactions of hexameric bacteriophage T7 DNA helicase with the 5′- and 3′-tails of the forked DNA substrate. J. Biol. Chem. 1997;272:32267–32273. doi: 10.1074/jbc.272.51.32267. [DOI] [PubMed] [Google Scholar]
- 45.Patel S.S., Picha K.M. Structure and function of hexameric helicases. Annu. Rev. Biochem. 2000;69:651–697. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- 46.Salinas F., Kodadek T. Phage T4 homologous strand exchange: a DNA helicase, not the strand transferase, drives polar branch migration. Cell. 1995;82:111–119. doi: 10.1016/0092-8674(95)90057-8. [DOI] [PubMed] [Google Scholar]
- 47.Kaplan D.L., O'Donnell M. DnaB drives DNA branch migration and dislodges proteins while encircling two DNA strands. Mol. Cell. 2002;10:647–657. doi: 10.1016/s1097-2765(02)00642-1. [DOI] [PubMed] [Google Scholar]
- 48.Goffart S., Cooper H.M., Tyynismaa H., Wanrooij S., Suomalainen A., Spelbrink J.N. Twinkle mutations associated with autosomal dominant progressive external ophthalmoplegia lead to impaired helicase function and in vivo mtDNA replication stalling. Hum. Mol. Genet. 2009;18:328–340. doi: 10.1093/hmg/ddn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Washington M.T., Rosenberg A.H., Griffin K., Studier F.W., Patel S.S. Biochemical analysis of mutant T7 primase/helicase proteins defective in DNA binding, nucleotide hydrolysis, and the coupling of hydrolysis with DNA unwinding. J. Biol. Chem. 1996;271:26825–26834. doi: 10.1074/jbc.271.43.26825. [DOI] [PubMed] [Google Scholar]
- 50.Rosenberg A.H., Griffin K., Washington M.T., Patel S.S., Studier F.W. Selection, identification, and genetic analysis of random mutants in the cloned primase/helicase gene of bacteriophage T7. J. Biol. Chem. 1996;271:26819–26824. [PubMed] [Google Scholar]
- 51.Patel G., Johnson D.S., Sun B., Pandey M., Yu X., Egelman E.H., Wang M.D., Patel S.S. A257T linker region mutant of T7 helicase-primase protein is defective in DNA loading and rescued by T7 DNA polymerase. J. Biol. Chem. 2011;286:20490–20499. doi: 10.1074/jbc.M110.201657. [DOI] [PMC free article] [PubMed] [Google Scholar]