Abstract
Genome instability triggers cellular senescence and is a common cause of cancer. The ribosomal RNA genes (rDNA), due to their repetitive structure, form a fragile site with frequent rearrangements. To identify eukaryotic factors that connect reduced genome stability to senescence we screened 4,876 strains of a Saccharomyces cerevisiae deletion library for aberrant rDNA and found 708 genes that contribute to its upkeep. 28 mutants caused abnormalities in non-rDNA chromosomes and among them 12 mutants have abnormalities both in rDNA and in non-rDNA chromosomes. Many mutated genes have not previously been implicated with genome maintenance nor their homologues with tumorigenesis in mammals. The link between rDNA state and senescence was broken after deletion of factors related with DNA polymerase ϵ. These mutations also suppressed the short lifespan phenotype of a sir2 mutant, suggesting a model in which molecular events at the heart of the replication fork induce abnormal rDNA recombination and are responsible for the emergence of an aging signal.
INTRODUCTION
Repetitive sequences make chromosomes fragile. A small repeating unit can form a secondary DNA structure that inhibits progression of replication forks, potentially inducing the formation of DNA double-strand breaks (DSBs) (1,2). When damage of a large repeating unit, like in the huge clusters of tandemly repeated ribosomal RNA genes (rDNA), gets repaired by homologous recombination with a neighboring unit, the number of repeats decreases (Supplementary Figure S1b-2) (3). Due to the unstable nature of repetitive sequences fluctuations in rDNA copy number are common and specialized mechanisms have evolved to maintain in each species the number of rDNA repeats at a particular level, e.g. ∼150 in budding yeast Saccharomyces cerevisiae (4) and ∼300 in human (5). In budding yeast, rDNA copies on chromosome XII (chr.XII) lost by recombination are recovered by a gene amplification mechanism that relies on stalling of the replication fork by a complex formed by Fob1 and the replication fork barrier (RFB) sequence (4). This induces a DSB that is repaired by homologous recombination between sister-chromatids (Supplementary Figure S1). This repair is regulated by the histone deacetylase Sir2 and its target, the non-coding promoter E-pro (6,7). When the copy number falls below wild-type level and Sir2 becomes inactive, E-pro transcription is activated bi-directionally and removes cohesin from the surrounding regions. Then, recombination can occur ‘unequally’, with the broken end integrating at a different unit so that the rDNA copy number increases (Supplementary Figure S1b-1). In contrast, when the copy number reaches wild-type level, Sir2 represses transcription from E-pro which is then bound by cohesin, leading to ‘equal’ sister-chromatid recombination that does not change copy number (Supplementary Figure S1a). As a result, contraction and amplification of rDNA occurs continuously to counteract the changes incurred by its instability.
In budding yeast, rDNA stability affects replicative lifespan as measured by the number of cell divisions until death (8,9), while general genome instability is caused by defects in repair genes which also shorten lifespan in yeast and humans (10,11). The lifespan of a sir2 mutant, in which the rDNA is quite unstable, is about half of that of wild-type yeast (12). In contrast, fob1 mutants have very stable rDNA and an increased lifespan (13,14). Stability of the whole genome may be determined by that of the rDNA region (occupying ∼10% of the whole genome in yeast), from which a putative ‘aging signal’ may originate that induces cellular senescence (15). But what exactly constitutes such a signal?
rDNA stability seems to be affected by several factors. Fork arrest and DSB efficiencies change the recombination frequency. Replication initiation frequency and replication fork stability affect recombination at the RFB (and other) sites in the genome (16). Factors that affect E-pro transcription and the extent of cohesin association are expected to alter the ratio of equal and unequal sister-chromatid recombination. Although the mechanism how DSB is induced at the RFB remains unknown, repair and recombination enzymes are expected to be involved. Most of these rDNA maintenance factors may also affect stability of non-rDNA regions because of the general occurrence of fork arrest and non-coding transcription (17,18). Instability of non-rDNA regions leads to genome rearrangements, such as translocations, that may cause cancer in mammals (19). Translocations occur at non-specific sites and with low frequencies, so that these events are difficult to study. Rearrangements in rDNA, however, are easily detected by a change in the copy number as described below.
In order to identify genes that affect rDNA stability, we screened a yeast deletion library covering 4876 genes by pulsed field gel electrophoresis (PFGE) and isolated 708 strains with notable changes in the rDNA. We named these strains ribosomal DNA unstable mutants, or RiUMs. As could be expected, RiUMs carried deletions in genes related to genome stability, but many mutants did not. Overall, an unexpectedly large number of genes contribute to rDNA stability and maybe to that of non-rDNA regions, with implications for a better understanding of tumorigenesis in mammals.
Strains with unstable rDNA were found that carried deletions in genes coding DNA polymerase ϵ (Pol ϵ) subunits Dpb3, Dpb4 and its associating factor Mrc1. Intriguingly, the lifespan of these strains was normal, while the same mutations partially suppressed the short lifespan phenotype of a sir2 mutant. We propose a model how these genes are acting on the same pathway to senescence and how this relates to the generation of an aging signal.
MATERIALS AND METHODS
Strains, culture condition, medium and PFGE
The deletion library (in BY4741, MATa) and the wild-type strain were purchased from Open Biosystems (now at GE/Dharmacon). Strains used in the 1D, 2D and northern analyses were listed in Supplementary Table S9. The 5153 strains of the library were tested, but in some strains similar loci had been disrupted by the kanamycin marker, so that 4876 genes were analyzed. We opened each mutant stock (−80°C) on the yeast extract-peptone-dextrose (YPD) plates and cultured in 0.5 ml YPD medium with G418 in 96-wells plates at 30°C (>5.0 OD, 600 nm) without single colony isolation, harvested the cells and isolated the DNA in 1.0% agarose plugs for PFGE on a RioRad CHEF MAPPER, essentially as described previously (6), alongside a Hansenula wingei chromosomal size marker (Biorad). The gels were stained by Ethidium bromide (EtBr) and blotted for Southern analysis. Chr. XII was detected with an rDNA specific probe (IGS1) (4).
Ranking of mutants and GO analysis
The 4876 deletion mutants were ranked according to rDNA size and stability as described in the ‘Results’ section (Table 1). Broadness of chr. XII bands relative to that of the wild-type in the same EtBr-stained gel was assessed by eye. Two persons independently judged and the differences were discussed to determine the final h-Ranks and s-Classes.
Table 1. Categorization of mutants according to rDNA stability into h-Ranks 1–4, or according to rDNA size (copy number) into s-Classes 1–4.
| rDNA Stability | mutants | reference | ||
|---|---|---|---|---|
| h-Rank 4 | very unstable, like sir2 | 43 | Supplementary Table S1 | |
| h-Rank 3 | less stable than WT | 665 | Supplementary Table S2 | |
| h-Rank 2 | WT stability | 4410 | ||
| h-Rank 1 | more stable than WT | 22 | Supplementary Table S3 | |
| Chr.XII size (Mb) | copy no | mutants | reference | |
| s-Class 4 | >5 | >450 | 44 | Supplementary Table S4 |
| s-Class 3 | ∼3–5 | ∼200–450 | 185 | Supplementary Table S5 |
| s-Class 2 | ∼1.8–3 | ∼80–200 | 4643 | |
| s-Class 1 | <1.8 | <80 | 9 | Supplementary Table S6 |
For the detail, see the tables in the reference.
Gene ontology (GO) analysis of the mutants was performed with the GO Slim Mapper using the Saccharomyces Genome Data base (SGD, http://www.yeastgenome.org/).
Analysis of RFB- or DSB activity, E-pro transcription and lifespan.
For detection of DNA replication fork blocking (RFB) activity, two dimensional agarose gel electrophoresis (2D analysis) was performed as described previously (6) albeit that DNA from early log phase cells (∼3 × 106 cells/ml in YPD medium) were digested in agarose plugs (5 × 107 cells/plug) by BglII for ∼20 h at 37°C. DSB frequency in similarly prepared DNA was detected by one dimensional agarose gel electrophoresis (1D analysis) as previously described (6). Briefly, the plug was heated at 65°C for 10 min before the first BglII digestion (2 h, 37°C), and then heated at 70°C for 10 min to melt the DNA completely, followed by the second BglII digestion (2 h, 37°C) (20). Finally, the plug was melted again to load DNA into the agarose gel before electrophoresis.
Northern analysis was performed with RNA prepared from cells grown in 10 ml YPD (∼0.8 OD, 600 nm), washed with ice-cold water and suspended in 10 mM Tris–HCl (pH 7.5), 10 mM ethylenediaminetetraacetic acid, 0.5% sodium dodecyl sulphate (SDS) and extracted with Phenol/Chloroform (pH 5.2) for 1 h at 65°C, vortexing every 10 min. For electrophoresis, 10 μg RNA was separated on a 1% formaldehyde/1xMOPS gel in Northern Max Denaturing Gel Buffer (Life Technologies) at 50 V for 3 h) (T.Iida, unpublished). Non-coding transcript levels were plotted relative to wild-type after ACT1 normalization. Measurement of replicative lifespan, was done as described previously (21) on YPD medium. The Mann–Whitney non-parametric test was used to determine statistical significance between lifespans.
Quantification of instability of chr. XII
To quantify instability of rDNA in PFGE, signal intensity of chr. XII was measured by NIH image in the EtBr stained gel. A fixed square area of the peak of chr. XII band that fits to the size of chr. IV band was measured. The intensity of chr. XII was normalized by that of the internal control, chr. IV. As the signal intensity of band in the gel becomes weaker in proportion to the size of chromosome, for example, 2.7 Mb band of the size marker is broader (weaker) than 1.66 Mb band (see Hansenula wingei size marker in Figure 4, upper), the intensity was normalized by the standard curve drawn by the size marker.
Figure 4.
rDNA stability in mutants of Pol ϵ related factors. (A and B) rDNA stability in single and double mutants of dpb3 or dpb4 with sir2 (A) or fob1 (B) was detected by PFGE. Gels were stained with EtBr (upper panel), blotted and hybridized to an rDNA specific probe (middle panel). The wild-type control (WT) and the position of the Hansenula wingei size marker (M) are shown. Three independent clones were tested. (bottom) Quantification of broadness of chr. XII. To quantify instability of rDNA, the signal intensity of chr. XII was measured in the EtBr stained gel. A broader band has weaker signal in a fixed size area. The intensity was normalized by that of chr. IV. As the signal intensity in the gel become weaker in proportion to the size, they were normalized by the size marker. The values are relative to the wild-type and standard deviations are shown (thin lines on the bars). In the fob1,dpb3 andfob1,dpb4 double mutants (B, lanes 13 and 16), chr. XII and chr. IV are overlapped. Therefore, these lanes were not used for the calculation.
RESULTS
Huge variation in rDNA stability and copy number
The budding yeast S. cerevisiae is composed of ∼6000 genes, and among them, ∼4900 are not essential for viability and they can be deleted (22,23). To identify novel factors involved in the maintenance of rDNA stability, a library of 4876 mutants in which these non-essential genes were deleted (24) was screened for a change in rDNA copy number by PFGE. The rDNA occupies ∼60% of chr. XII and copy number variation changes the length of the chromosome. Compared to wild-type, chr. XII of rDNA unstable mutants (RiUMs) migrates aberrantly with respect to size or appearance [Figure 1; and the Yeast rDNA Stability DataBase (YRSD at http://lafula-com.info/kobayashiken/geldata/index.php)]. We categorized mutants according to increasing size-heterogeneity (heterogeneity) into h-Ranks 1–4 and, with respect to increasing average length (size) of chr. XII, into s-Classes 1–4 (Table 1). Mutants with wild-type chr. XII were classified as h-Rank 2, s-Class 2.
Figure 1.
Detection of rDNA stability by PFGE. Four example gels, as accessible in the database, after staining with EtBr (upper panels) or after blotting and hybridization to an rDNA specific probe (lower panels). The wild-type control (W) and the position of the Hansenula wingei size marker are shown.
We identified 43 mutants, falling in h-Rank 4, with very unstable rDNA, such as sir2, in which the chr. XII band is completely smeared out and almost invisible by Ethidium bromide (EtBr) staining (Figure 1A-a) (6). In s-Class 4 mutants like rtt109 migration of chr. XII is retarded due a large increase in rDNA copy number (Figure 1B-b) (25,26), whereas in s-Class 1 mutants like ssn8 the copy number is reduced (Figure 1C-c).
h-Rank 4 rDNA stability mutants
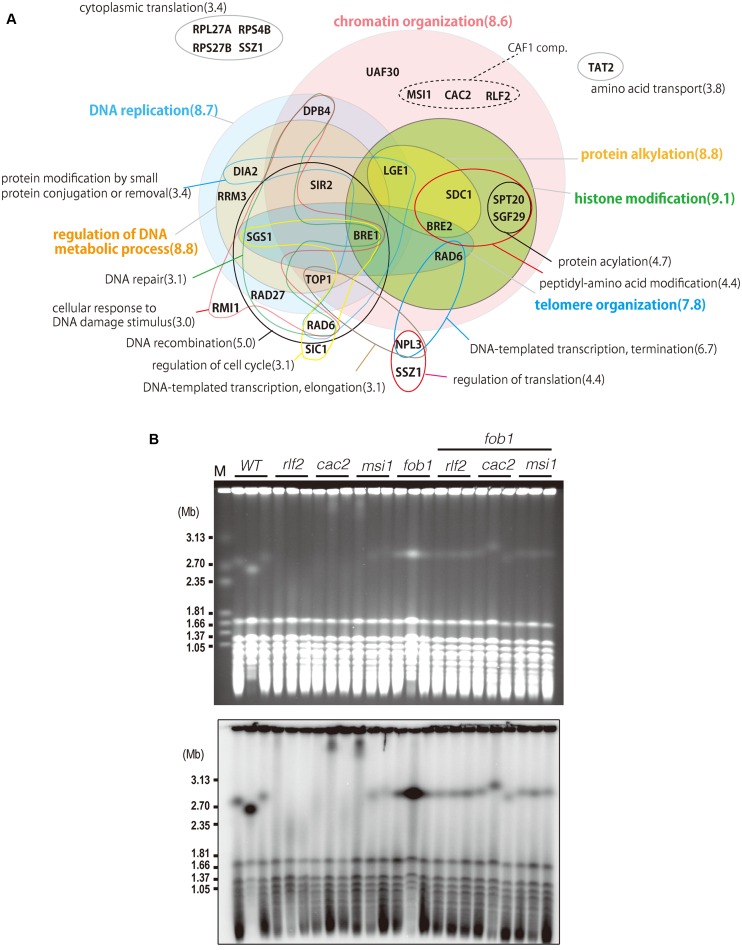
GO-term analysis of h-Rank 4 mutants showed enrichment for genes involved in DNA replication and damage repair (Figure 2A, Supplementary Table S1). Many of these genes, like SIR2, RLF2 or DPB4, are also important for chromatin organization and histone modification. Rlf2, together with Msi1 and Cac2, forms the CAF1 complex—a histone chaperone that assembles newly synthesized DNA into nucleosomes (27) and is required for post-replication repair (28). The rDNA instability observed in absence of either of these genes was rescued by deletion of FOB1 (Figure 2B) and is therefore directly linked to stalling of the replication fork.
Figure 2.
Analysis of RiUM genes (h-Rank4). (A) GO-term analysis of RiUM genes (h-Rank4). Venn Diagrams of biological processes related to genes deleted in strains with impaired rDNA stability of h-Rank 4 (>3.0, enrichment). The most highly enriched biological processes (>7.0) are colored. The CAF1 complex is shown in a dotted circle. The number in parentheses is the value of enrichment, i.e. the ratio of the frequency of that GO-annotation for genes in h-Rank 4 to the frequency of that annotation among all 6338 yeast ORFs. (B) rDNA instability in CAF1 mutants is FOB1 dependent. PFGE was performed to test rDNA stability in CAF1 single mutants and double mutants with fob1. M is the Hansenula wingei chromosomal size marker.
The stability of rDNA depends on silencing of the non-coding transcription from E-pro, which might require, apart from Sir2 (7), the RNA polymerase II repressors Srb8 and Spt21 (29,30) as their deletion gave a h-Rank 4 phenotype. Transcription of rDNA by RNA polymerase I depends on h-Rank 4 genes UAF30 (31) and TOP1, with topoisomerase I known to cleave at RFB sites in a Fob1-dependent manner (32). As replication of rDNA initiates downstream of transcriptionally active genes (33), release of torsional stress by topoisomerase I appears to stabilize rDNA when Fob1 is present to stall replication forks.
h-Rank 3 rDNA stability mutants
The band of chr. XII is in the 665 deletion mutants of h-Rank3 (Supplementary Table S2; e.g. slx5, Figure 1A-f) apparent but much broader than that of the wild-type. GO-term analysis revealed that many of the affected genes acted on DNA-templated transcription, or elongation (Supplementary Figure S2). Deletion of such genes has been associated with enhanced genome instability (34).
Mitochondrial and vacuolar functions are known to affect cellular senescence (35,36). Suggestive for the theory that cellular senescence depends on rDNA stability, many of the h-Rank 3 mutants are related to organelle organization, but how mechanistically this could influence rDNA stability remains unclear.
h-Rank 1 mutants with very stable rDNA
For 22 deletion strains, classified as h-Rank 1, chr. XII migrated as a sharper band, i.e. was more stable than wild-type (Supplementary Table S3, e.g. Figure 1D-g). It is possible that some genes disrupted in these strains act opposite to genes whose deletion destabilizes rDNA (i.e. the h-Rank 4 or 3 mutants).
Two h–Rank 1 strains, rim1 and tif4631, have a longer lifespan than the wild-type (37) while erg2, in which the gene for an isomerase required for ergosterol biosynthesis was disrupted, has a shorter lifespan (38). RIM1 encodes a single strand DNA-binding protein essential for mitochondrial genome maintenance and TIF4631 codes for a translation initiation factor (39,40). As for the other mutants in this rank, five of them (elp2, yta7, caf40, ynl296w, eaf3) are related to pol II transcription, suggesting that the mutations repress E-pro transcription.
s-Class 4 and s-Class 3 mutants with elevated rDNA copy numbers
Apart from rDNA stability, we also scored mutants for the length of their rDNA, i.e. rDNA copy number. In 44 s-Class 4 mutants the copy number exceeded 450, that is three times higher than that of the wild-type (Supplementary Table S4) (25,26). In them, 36 mutants are in h-Rank 3. In one of these 36 mutants the gene for histone acetyltransferase Rtt109, involved in recombination and repair (41,42), was deleted (Figure 1B-b). In absence of Rtt109, DNA synthesis continues resulting in a rolling circle type amplification of rDNA copies (Supplementary Figure S1b-3) (26). GO-term analysis revealed enrichment of RNA metabolic genes POP2, CCR4 and DHH1 that are components of the CCR4–NOT1 complex, implicated in the regulation of mRNA and pre-rRNA transcription (43,44).
s-Class 3 mutants have an elevated rDNA copy-number of ∼200–450. We identified 185 gene-deletions causing this characteristic, 111 of which with wild-type stability (h-Rank 2) and the others (74 mutants) are in h-Rank 3, but GO-term analysis did not reveal obvious functional roles for genes in this class (Supplementary Table S5), possibly indicating that rDNA maintenance relies on many different cellular signals.
s-Class 1 mutants with abnormally short rDNA clusters
We found nine mutants with very low rDNA copy numbers (<80, Supplementary Table S6), four of which with wild-type stability (h-Rank 2), suggestive of a role in a mechanism by which the rDNA copy number is monitored. A fairly broad band of chr. XII indicating unstable rDNA was observed for five of these s-Class 1 mutants, like the arp5 and vps72 strains that carry a deletion of a gene implicated in chromosome maintenance.
In terms of the relationship between size-heterogeneity and copy number, the ratio of unstable rDNA (h-Rank3) in s-Class 4 (82%) is higher than those in s-Class 3 (40%), s-Class 2 (10%) and s-Class 1 (47%). This suggests that for high copy number strains, it is more difficult to maintain rDNA stability.
Mutants with chromosome abnormalities
For 219 mutants southern hybridization revealed multiple bands for chr. XII, indicating a fault with chromosome segregation during mitosis (e.g. Figure 1C-d). GO-term analysis revealed an enrichment of genes related to changes in cell-morphology (Supplementary Table S7). As chr. XII is the largest chromosome, it may be the most vulnerable for aberrant cell-division processes (see ‘Discussion’ section for detail). In each of these mutants (219), the chromosomes belong to the same rDNA stability h-Rank (Supplementary Table S7), suggesting that rDNA clusters are maintained irrespective of the chromosome they are on. Compared to the frequency of RiUMs in the library (708 h-Rank 3 and 4 mutants over 4876 strains, i.e. 15%), a very high proportion of mutants with segregation-phenotype has unstable rDNA (99 h-Rank 3 mutants out of 219, i.e. 45%). Some of the multiple bands may also arise from divergent populations following unequal sister chromatid recombination.
Indicative that rDNA stability is related to chromosome separation is the finding that rDNA is unstable in 12 out of 28 mutants with aberrant numbers of the largest non-rDNA chromosome, chr. IV (Supplementary Table S8). In more than half of these mutants (15/28), this chromosome (chr. IV) was duplicated and changed in size (e.g. Figure 1A-e), maybe because of the presence of a recombinational hotspot (45). We speculate that the chromosome (IV) duplicated, reciprocal translocation occurred between them and the size changed. Any abnormalities in smaller chromosomes than chr. XII and IV would not have been detected under the electrophoresis conditions used. These data suggest a general link between rDNA stability and chromosome segregation during cell division.
Factors associated with Pol ϵ link rDNA stability to lifespan
Genome instability is known to affect cellular senescence (replicative lifespan) in yeast and mammalian cells (10,11) and a link between proper maintenance of rDNA, being the largest unstable region in the genome, and cell-growth has been established (9,46). Therefore, if rDNA damage produces a major aging signal (15), strains with unstable rDNA could be expected to have a shorter lifespan unless the deleted genes are involved in propagating such a signal. Five of the seven h-Rank 4 mutants for which the lifespan has been tested have shorter lifespan (sgs1 rad27 rad6 rlf2 sir2, Supplementary Table S1) (47–49). Life span is lengthened on inhibition of tryptophan import by depletion of the aromatic amino acid transporter Tat2 (49), but how this links to rDNA stability is unclear. Absence of Dpb4, a subunit of DNA Pol ϵ that synthesizes the leading strand during replication (50), and of Mrc1, a Pol ϵ associating factor, leads to reduced rDNA stability (h-Ranks 4 and 3) without lifespan interference (51). Mrc1 supports Pol ϵ in stabilizing an arrested fork in vivo (52). Therefore, we wondered whether loss of this fork stability is instrumental in the production of an aging signal to induce cellular senescence.
Previously, we constructed a series of mutants for testing RFB activity and lifespan (4,8), and in this genetic background, deletion-strains of Pol ϵ related factors Dpb4, Dpb3 (a subunit of DNA Pol ϵ, h-Rank 3) and Mrc1 were created (Supplementary Table S9). We found that the lifespans of the novel dpb4 and mrc1 strains were similar to that of the parental strain though that of the dpb3 mutant was reduced but not to the extent as seen for sir2 (Figure 3) (12). Their rDNA stabilities were also reduced (Figure 4A, lanes 7-15, see below). Interestingly, in the absence of especially Dpb4 or Mrc1, the reduced-lifespan phenotype of the sir2 mutant was suppressed (Figure 3). Furthermore, rDNA instability in sir2 was suppressed by deletion of either Dpb4, or Mrc1 (Figure 4A, lanes 16–27). In agreement with this, the abundance of extra-chromosomal rDNA circles (ERCs) that are produced by unequal sister chromatid recombination (Figure S1b-2) were reduced in sir2 after deletion of either of the three Pol ϵ relating factors (Figure 5A, Supplementary Figure S3). These findings suggest that increased E-pro transcription and unequal sister-chromatid recombination that occurs in absence of Sir2 activity is reduced or not effective when the stability of the Pol ϵ complex on the leading strand in the replication fork has been compromised.
Figure 3.
Replicative lifespan in mutants of Pol ϵ related factors. The number of buds that emerged from a mother was counted. The average lifespan is shown above each bar. Ninety-five percent confidential intervals are indicated (thin lines on the bars). Calculated P-values are shown above the graph.
Figure 5.
Detection of ERC and E-pro transcripts in mutants of Pol ϵ related factors. (A) Quantification of ERC-formation in mutants of Pol ϵ related factors. Non-digested DNA was separated, blotted and ERCs were detected with an rDNA specific probe (Supplementary Figure S3). The fold increase of ERC formation relative to the wild-type and standard deviations are shown (thin lines on the bars). URA3 was used as an internal standard for normalization. For accurate comparison among the different gels, we added control lanes (WT, fob1and sir2) every time and the graphs were drawn separately. (B and C) E-pro transcripts were measured by northern analysis (Supplementary Figure S6) using probes for the RFB (B) or 5S rRNA (C). Signals were normalized to ACT1 to calculate the average amount of E-pro transcription relative to the wild-type (shown above the bar). Standard deviations are indicated (thin lines on the bars). For accurate comparison among the different gels, we added control lanes (WT, sir2) every time and the graphs were drawn separately.
We note that in the single mutants mrc1 and dpb4 in Figure 4A, chr.XII is not so unstable (not so broad) even though they are ranked h-Rank 3 and 4 in the deletion library, respectively. This may be because of different strain background (W303). By the quantitation of the broadness (Figure 4, bottom), the chromosomes are still unstable. Moreover, in the wild-type and mrc1 mutant, the size of chr.XII varies in each lane (each colony). As the variation is not observed in the fob1 mutant, this is also due to rDNA instability.
The fob1 mutant has a longer lifespan than the wild-type (Figure 3) (13,14) which implies that blocking of replication forks at the RFB site directly relates to a reduced number of cell-divisions, possibly because unequal recombination events, induced by a DSB upon stalling of replication, become more frequent. Also, in the absence of Fob1, no ERCs were formed (Figure 5A, Supplementary Figure S3). Reduced stability of the replication fork around the RFB site, as could be expected in the case of deficient Pol ϵ, combined with collisions with rDNA transcription in the absence of Fob1 (17) would increase rDNA instability. Indeed, deletion of Pol ϵ subunits Dpb3 and Dpb4 in a fob1 mutant led to a decrease of rDNA stability and size (Figure 4B, lanes 13–18), and an increase of ERC-formation (Figure 5A). In the mrc1, fob1 double mutant, the rDNA is unstable to compare with the fob1 single mutant though the size is similar to the mutant (Figure 4B, lanes 10–12). In agreement with a direct link with lifespan, the double mutants of fob1 with mrc1, dpb3 or dpb4 had a shorter lifespan than the single fob1 mutant (Figure 3). These results show that, for the control of lifespan, Pol ϵ activity is connected to fork-blocking activity of Fob1 at RFB sites.
Pol ϵ activity stimulates non-coding transcription in the absence of Sir2
To gain a better understanding of the tight linkage between Pol ϵ related factors and RFB activity, we measured the rate of replication fork arrest in the various mutants by two dimensional gel electrophoresis (2D analysis) of replication intermediates which were visualized by Southern blotting (6). The number of stalled replication forks, as shown by the relative intensity of Y-spot signals normalized to a replication intermediate signal (Supplementary Figure S4) was not altered in the wild-type or in sir2 after deletion of MRC1, DPB3 or DPB4, indicating comparable replication fork blocking activities in these strains. Similarly, deletion of MRC1, DPB3 or DPB4 did not restore RFB activity, normally absent in the fob1 mutant (as indicated by lack of an Y-spot).
Similarly, between the wild-type and sir2 strains after deletion of Pol ϵ subunits no significant changes were observed in the frequency of DSB at the RFB site, which was measured by comparing—for identical samples—the amount of broken DNA (resolved on 1D gel electrophoresis and visualized by Southern blotting, Supplementary Figure S5) to the number of stalled replication forks (i.e. the corresponding Y-spot intensity in the 2D-analysis of Supplementary Figure S4).
Apart from RFB-activity and DSB frequencies, a major factor that controls rDNA stability is E-pro transcription, mainly through its effect on the rate of unequal sister-chromatid recombination (7,8). As determined by northern blot analysis of RNA extracted from logarithmically growing cells, bi-directional E-pro transcription (producing downstream transcripts over the 5S rRNA gene and upstream products containing RFB sequences) was hardly changed after deletion of Pol ϵ subunits in comparison to wild-type cells (Figure 5BC, Supplementary Figure S6). In strains lacking Sir2, however, deletion of either Dpb4 or Mrc1, in contrast to that of Dpb3, caused a marked reduction in the amount of non-coding RNA transcribed from E-pro, indicating that the mrc1 and dbp4 alleles suppress the rDNA instability of sir2 mutants by slowing down the rate of unequal sister-chromatid recombination, thereby increasing lifespan. Thus, elevated E-pro transcription observed in the absence of Sir2, appears to be dependent on Pol ϵ activity, i.e. the stabilization of the leading strand during replication.
DISCUSSION
The highly repetitive structure of rDNA makes it the most and largest fragile site in any genome. This instability is manifested in copy number variations that can be detected by PFGE and enabled us to identify at least 708 non-essential genes that contribute to rDNA maintenance in the eukaryote S. cerevisiae, as deposited in the Yeast rDNA Stability DataBase.
The number of rDNA unstable mutants (RiUMs) corresponded to ∼15% of the total number of genes tested (4876), and exceeded by far the 244 genes known to be related to DNA repair. We found 142 genes involved in genome maintenance and chromatin organization (including histone modification) mutated among the strains with the strongest RiUM phenotypes (of h-Rank 3 and 4), as well as those with a structurally altered rDNA copy number (of s-Class 1, 3 and 4). For many mutants not only missegregation of chr. XII with the rDNA locus was observed but also for the second largest chr. IV. Our database will supply valuable information not only on rDNA maintenance, but also on general genome stability. For more than half of the 708 h-Rank 3 and 4 genes no reasonable explanation is available yet that would help understand how their deletion affect rDNA stability, like, for example, a number of ribosomal protein genes or an aromatic amino acid transporter. The biological function for 172 genes (in h-Ranks 3 and 4) in the database still has to be elucidated. These insights will open a new direction for the study on genome stability that could be critical for the understanding of cancer, development and aging.
The maintenance of rDNA is associated with RFB-activity of Fob1 and the regulation of E-pro transcription by Sir2 (Supplementary Figure S1). The balance of these factors determines the extent of unequal sister-chromatid recombination that can induce loss of rDNA copies as well as to recover them. The resulting stability of the rDNA is linked to cellular lifespan and somehow the interplay around the RFB appears to result in an aging signal that triggers senescence. In our database we found a set of proteins, Dpb4, Dpb3 and Mrc1 that form part of or interact with Pol ϵ and whose deletion leads to unstable rDNA without the aging signal being emitted. Pol ϵ synthesizes the leading strand, which gets broken at a RFB site when this is occupied by Fob1 (53). This activity still occurs normally in the absence of Dpb4, Dpb3 or Mrc1 (Supplementary Figures S4 and S5) and seems required to break the link between unstable rDNA and reduced life-span in these mutants as in the absence of Sir2 and Fob1 lifespan changes were observed in these strains. In the absence of Sir2, E-pro transcription is high but gets reduced when Dpb4 and Mrc1 are not present either; concomitantly lifespan increases. In the absence of Fob1, when the replication fork can collide with rDNA transcription (14), the reverse was observed when these Pol ϵ related factors were lacking, i.e. the fob1-extended lifespan became shorter due to increased rDNA instability. The increase of ERC-formation in these fob1 double mutants suggest an increase of unequal sister-chromatid recombination in the rDNA not observed in the absence of Fob1 alone (Figure 5, Supplementary Figure S3). In these cases a change of E-pro transcription correlates with a change in rDNA stability and lifespan. This however, did not occur in single mutants of Dpb4, Dpb3 or Mrc1: their rDNA was unstable, but E-pro transcription was not increased accordingly, nor was their lifespan reduced. This suggests that the level of E-pro transcription is linked to the production of an aging signal. Therefore, a simple idea is that when the broken-end is repaired by equal sister-chromatid recombination less aging signal is produced from the RFB, but when repair occurs via unequal sister-chromatid recombination that changes copy number and increases rDNA instability, more aging signal is produced (Figure 6).
Figure 6.
Summary of aging signal production at RFB. In the wild-type (WT), the end broken at the RFB site is repaired by equal sister chromatid recombination that doesn't change rDNA copy number (1). In the sir2 mutant, E-pro is activated and repair of the broken end occurs via unequal sister chromatid recombination that changes rDNA copy number (2). The recombination produces an aging signal that shortens lifespan. In the mutants mrc1, dpb4 and to a lesser extent dpb3 (dpb3), E-pro is slightly activated with a lesser effect on rDNA copy number change. However, the aging signal in these mutants is repressed (3). Compared to sir2, in the mrc1, dpb4, (dpb3) and sir2 double mutants, E-pro transcription is reduced and with it the frequency of unequal sister chromatid recombination. As a result, the aging signal decreases and the lifespan is recovered (4). E-pro transcriptions, rDNA instabilities and aging signals (−, ++, +++) are estimated from the results in Figures 3–5.
Supplementary Material
Acknowledgments
We thank Drs. Tetsushi Iida (IMCB), Yufuko Akamatsu (IMCB), Satoru Ide (NIG) and Hiroyuki Araki (NIG) for technical advice and discussion.
Author contributions: K.S. screened the yeast deletion library, identified RiUMs and analyzed the relationship between rDNA stability and lifespan. A.T. analyzed the relationship between RiUM phenotypes and RFB activities. M.S. analyzed the data and wrote the paper. T.K. conceived and designed the experiments, analyzed the data and wrote the paper.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grants-in-aid for Scientific Research from the Ministry of Education [23114002, in part]; Culture, Sports, Science and Technology (MEXT), Japan (to T.K.). Funding for open access charge: Grants-in-aid for Scientific Research from the Ministry of Education [23114002, in part]; Culture, Sports, Science and Technology (MEXT), Japan (to T.K.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Samadashwily G.M., Raca G., Mirkin S.M. Trinucleotide repeats affect DNA replication in vivo. Nat. Genet. 1997;17:298–304. doi: 10.1038/ng1197-298. [DOI] [PubMed] [Google Scholar]
- 2.Freudenreich C.H., Kantrow S.M., Zakian V.A. Expansion and length-dependent fragility of CTG repeats in yeast. Science. 1998;279:853–856. doi: 10.1126/science.279.5352.853. [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi T. Regulation of ribosomal RNA gene copy number and its role in modulating genome integrity and evolutionary adaptability in yeast. Cell. Mol. Life Sci. 2011;68:1395–1403. doi: 10.1007/s00018-010-0613-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kobayashi T., Heck D.J., Nomura M., Horiuchi T. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 1998;12:3821–3830. doi: 10.1101/gad.12.24.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakai M., Ohta T., Minoshima S., Kudoh J., Wang Y, de Jong PJ, Shimizu N. Human ribosomal RNA gene cluster: identification of the proximal end containing a novel tandem repeat sequence. Genomics. 1995;26:521–526. doi: 10.1016/0888-7543(95)80170-q. [DOI] [PubMed] [Google Scholar]
- 6.Kobayashi T., Horiuchi T., Tongaonkar P., Vu L., Nomura M. SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell. 2004;117:441–453. doi: 10.1016/s0092-8674(04)00414-3. [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi T., Ganley A.R.G. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science. 2005;309:1581–1584. doi: 10.1126/science.1116102. [DOI] [PubMed] [Google Scholar]
- 8.Saka K., Ide S., Ganley A.R.D., Kobayashi T. Cellular senescence in yeast is regulated by rDNA noncoding transcription. Curr. Biol. 2013;23:1794–1798. doi: 10.1016/j.cub.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 9.Ganley A.R.D., Kobayashi T. Ribosomal DNA and cellular senescence: new evidence supporting the connection between rDNA and aging. FEMS Yeast Res. 2014;14:49–59. doi: 10.1111/1567-1364.12133. [DOI] [PubMed] [Google Scholar]
- 10.Ellis N.A., Groden J., Ye T.Z., Straughen J., Lennon D.J., Ciocci S., Proytcheva M., German J. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 11.Park P.U., Defossez P.A., Guarente L. Effects of mutations in DNA repair genes on formation of ribosomal DNA circles and life span in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:3848–3856. doi: 10.1128/mcb.19.5.3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaeberlein M., McVey M., Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Defossez P.A., Prusty R., Kaeberlein M., Lin S.J., Ferrigno P., Silver P.A., Keil R.L., Guarente L. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol. Cell. 1999;3:447–455. doi: 10.1016/s1097-2765(00)80472-4. [DOI] [PubMed] [Google Scholar]
- 14.Takeuchi Y., Horiuchi T., Kobayashi T. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev. 2003;17:1497–1506. doi: 10.1101/gad.1085403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi T. A new role of the rDNA and nucleolus in the nucleus- rDNA instability maintains genome integrity- Bioessays. 2008;30:267–272. doi: 10.1002/bies.20723. [DOI] [PubMed] [Google Scholar]
- 16.Ganley A.R.G., Ide S., Saka K., Kobayashi T. The effect of replication initiation on gene amplification in the rDNA and its relationship to aging. Mol. Cell. 2009;35:683–693. doi: 10.1016/j.molcel.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 17.Ivessa A.S., Lenzmeier B.A., Bessler J.B., Goudsouzian L.K., Schnakenberg S.L., Zakian V.A. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol. Cell. 2003;12:1525–1536. doi: 10.1016/s1097-2765(03)00456-8. [DOI] [PubMed] [Google Scholar]
- 18.Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 19.Stratton M.R., Campbell P.J., Futreal P.A. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murakami H., Borde V., Nicolas A., Keeney S. Gel electrophoresis assays for analyzing DNA double-strand breaks in Saccharomyces cerevisiae at various spatial resolutions. Methods Mol. Biol. 2009;557:117–142. doi: 10.1007/978-1-59745-527-5_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kennedy B.K., Austriaco N.R., Jr, Guarente L. Daughter cells of Saccharomyces cerevisiae from old mothers display a reduced life span. J. Cell. Biol. 1994;127:1985–1993. doi: 10.1083/jcb.127.6.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winzeler E.A., Shoemaker D.D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H., et al. Functional characterization of the Saccharomyces cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 23.Giaever G., Chu A.M., Ni L., Connelly C., Riles L., Véronneau S., Dow S., Lucau-Danila A., Anderson K., André B., et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 24.Wach A., Brachat A., Poehlmann R., Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 25.Houseley J., Tollervey D. Repeat expansion in the budding yeast ribosomal DNA can occur independently of the canonical homologous recombination machinery. Nucleic Acids Res. 2011;39:8778–8791. doi: 10.1093/nar/gkr589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ide S., Saka K., Kobayashi T. Rtt109 Prevents Hyper-Amplification of Ribosomal RNA Genes through Histone Modification in Budding Yeast. PLoS Genet. 2013;9:e1003410. doi: 10.1371/journal.pgen.1003410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaufman P.D. Nucleosome assembly: the CAF and the HAT. Curr. Opin. Cell. Biol. 1996;8:369–373. doi: 10.1016/s0955-0674(96)80012-3. [DOI] [PubMed] [Google Scholar]
- 28.Game J.C., Kaufman P.D. Role of Saccharomyces cerevisiae chromatin assembly factor-I in repair of ultraviolet radiation damage in vivo. Genetics. 1999;151:485–497. doi: 10.1093/genetics/151.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balciunas D., Ronne H. Three subunits of the RNA polymerase II mediator complex are involved in glucose repression. Nucleic Acids Res. 1995;23:4421–4425. doi: 10.1093/nar/23.21.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang J.S., Winston F. Spt10 and Spt21 are required for transcriptional silencing in Saccharomyces cerevisiae. Eukaryot. Cell. 2011;10:118–129. doi: 10.1128/EC.00246-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siddiqi I.N., Dodd J.A., Vu L., Eliason K., Oakes M.L., Keener J., Moore R., Young M.K., Nomura M. Transcription of chromosomal rRNA genes by both RNA polymerase I and II in yeast uaf30 mutants lacking the 30 kDa subunit of transcription factor UAF. EMBO J. 2001;20:4512–4521. doi: 10.1093/emboj/20.16.4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Di Felice F., Cioci F., Camilloni G. FOB1 affects DNA topoisomerase I in vivo cleavages in the enhancer region of the Saccharomyces cerevisiae ribosomal DNA locus. Nucleic Acids Res. 2005;33:6327–6337. doi: 10.1093/nar/gki950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muller M., Lucchini R., Sogo J.M. Replication of rDNA initiates downstream of transcriptionally active genes. Mol. Cell. 2000;5:767–777. doi: 10.1016/s1097-2765(00)80317-2. [DOI] [PubMed] [Google Scholar]
- 34.Aguilera A. The connection between transcription and genomic instability. EMBO J. 2002;21:195–201. doi: 10.1093/emboj/21.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jazwinski S.M. Yeast replicative life span—the mitochondrial connection. FEMS Yeast Res. 2004;5:119–125. doi: 10.1016/j.femsyr.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 36.Hughes A.L., Gottschling D.E. An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature. 2012;492:261–265. doi: 10.1038/nature11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delaney J.R., Sutphin G.L., Dulken B., Sim S., Kim J.R., Robison B., Schleit J., Murakami C.J., Carr D., An E.H., et al. Sir2 deletion prevents lifespan extension in 32 long-lived mutants. Aging Cell. 2011;10:1089–1091. doi: 10.1111/j.1474-9726.2011.00742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang F., Watkins J.W., Bermudez M., Gray R., Gaban A., Portie K., Grace S., Kleve M., Craciun G. A life-span extending form of autophagy employs the vacuole-vacuole fusion machinery. Autophagy. 2008;4:874–886. doi: 10.4161/auto.6556. [DOI] [PubMed] [Google Scholar]
- 39.van Dyck E., Foury F., Stillman B., Brill S.J. A single-stranded DNA binding protein required for mitochondrial DNA replication in S. cerevisiae is homologous to E. coli SSB. EMBO J. 1992;11:3421–3430. doi: 10.1002/j.1460-2075.1992.tb05421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goyer C., Altmann M., Lee H.S., Blanc A., Deshmukh M., Woolford J.L., Jr, Trachsel H., Sonenberg N. TIF4631 and TIF4632: two yeast genes encoding the high-molecular-weight subunits of the cap-binding protein complex (eukaryotic initiation factor 4F) contain an RNA recognition motif-like sequence and carry out an essential function. Mol. Cell. Biol. 1993;13:4860–4874. doi: 10.1128/mcb.13.8.4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Driscoll R., Hudson A., Jackson S.P. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science. 2007;315:649–652. doi: 10.1126/science.1135862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Endo H., Kawashima S., Sato L., Lai M.S., Enomoto T., Seki M., Horikoshi M. Chromatin dynamics mediated by histone modifiers and histone chaperones in postreplicative recombination. Genes Cells. 2010;15:945–958. doi: 10.1111/j.1365-2443.2010.01435.x. [DOI] [PubMed] [Google Scholar]
- 43.Kruk J.A., Dutta A., Fu J., Gilmour D.S., Reese J.C. The multifunctional Ccr4-Not complex directly promotes transcription elongation. Genes Dev. 2011;25:581–593. doi: 10.1101/gad.2020911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laribee R.N., Hosni-Ahmed A., Workman J.J., Chen H. Ccr4-Not regulates RNA polymerase I transcription and couples nutrient signaling to the control of ribosomal RNA biogenesis. PLoS Genet. 2015;11:e1005113. doi: 10.1371/journal.pgen.1005113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.St Charles J., Petes T.D. High-resolution mapping of spontaneous mitotic recombination hotspots on the 1.1 Mb arm of yeast chromosome IV. PLoS Genet. 2013;9:e1003434. doi: 10.1371/journal.pgen.1003434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flach J., Bakker S.T., Mohrin M., Conroy P.C., Pietras E.M, Reynaud D., Alvarez S., Diolaiti M.E., Ugarte F., Forsberg E.C., et al. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature. 2014;512:198–202. doi: 10.1038/nature13619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Managbanag J.R., Witten T.M., Bonchev D., Fox L.A., Tsuchiya M., Kennedy B.K., Kaeberlein M. Shortest-path network analysis is a useful approach toward identifying genetic determinants of longevity. PLoS One. 2008;3:e3802. doi: 10.1371/journal.pone.0003802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dang W., Sutphin G.L., Dorsey J.A., Otte G.L., Cao K., Perry R.M., Wanat J.J., Saviolaki D., Murakami C.J., Tsuchiyama S., et al. Inactivation of yeast Isw2 chromatin remodeling enzyme mimics longevity effect of calorie restriction via induction of genotoxic stress response. Cell Metab. 2014;19:952–966. doi: 10.1016/j.cmet.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He C., Tsuchiyama S.K., Nguyen Q.T., Plyusnina E.N., Terrill S.R., Sahibzada S., Patel B., Faulkner A.R., Shaposhnikov M.V., Tian R., et al. Enhanced longevity by ibuprofen, conserved in multiple species, occurs in yeast through inhibition of tryptophan import. PLoS Genet. 2014;10:e1004860. doi: 10.1371/journal.pgen.1004860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pursell Z. F., Kunkel T.A. DNA polymerase e: a polymerase of unusual size (and complexity) Prog. Nucleic Acid Mol. Biol. 2008;82:101–145. doi: 10.1016/S0079-6603(08)00004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feser J., Truong D., Das C., Carson J.J., Kieft J., Harkness T., Tyler J.K. Elevated histone expression promotes life span extension. Mol. Cell. 2010;39:724–735. doi: 10.1016/j.molcel.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lou H., Komata M., Katou Y., Guan Z., Reis C.C., Budd M., Shirahige K., Campbell J. L. Mrc1 and DNA polymerase ϵ function together in linking DNA replication and the S phase checkpoint. Mol. Cell. 2008;32:106–117. doi: 10.1016/j.molcel.2008.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burkhalter M.D., Sogo J.M. rDNA enhancer affects replication initiation and mitotic recombination: Fob1 mediates nucleolytic processing independently of replication. Mol. Cell. 2004;15:409–421. doi: 10.1016/j.molcel.2004.06.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.