Abstract
Uracil-DNA glycosylases (UDGs) are highly conserved proteins that can be found in a wide range of organisms, and are involved in the DNA repair and host defense systems. UDG activity is controlled by various cellular factors, including the uracil-DNA glycosylase inhibitors, which are DNA mimic proteins that prevent the DNA binding sites of UDGs from interacting with their DNA substrate. To date, only three uracil-DNA glycosylase inhibitors, phage UGI, p56, and Staphylococcus aureus SAUGI, have been determined. We show here that SAUGI has differential inhibitory effects on UDGs from human, bacteria, Herpes simplex virus (HSV; human herpesvirus 1) and Epstein-Barr virus (EBV; human herpesvirus 4). Newly determined crystal structures of SAUGI/human UDG and a SAUGI/HSVUDG complex were used to explain the differential binding activities of SAUGI on these two UDGs. Structural-based protein engineering was further used to modulate the inhibitory ability of SAUGI on human UDG and HSVUDG. The results of this work extend our understanding of DNA mimics as well as potentially opening the way for novel therapeutic applications for this kind of protein.
INTRODUCTION
Base excision repair (BER) is an important DNA repair system that deals with the most ubiquitous lesions in DNA such as alkylation, deamination, oxidative base damage, base loss and single-strand breaks (1–3). In the first step of this system, DNA glycosylases recognize and remove specific damaged or inappropriate bases to generate apurinic/apyrimidinic (AP) sites. The AP sites are then cleaved by an AP endonuclease. The resulting single-strand break can then be processed by either short-patch or long-patch BER to put the ‘right’ nucleotide into the DNA (1–3). Uracil DNA glycosylase (UDG) was the first BER-related glycosylase to be discovered (4). This glycosylase removes the uracils in DNA that are due to spontaneous deamination of cytosine or the misincorporation of dUMP during replication (1–3).
The DNA repair activity of UDG can be enhanced by cellular factors. For example, interaction with proliferating cell nuclear antigen (PCNA) stimulates the UDG activity (5). On the other hand, the DNA repair activities of UDG can also be strongly inhibited by uracil DNA glycosylase inhibitors (6–16). These inhibitors act through a mechanism of DNA mimicry (17–19). The first two reported uracil-DNA glycosylase inhibitors, UGI and p56, were originally found in bacterial phages (6–15). Interestingly, both UGI and p56 can inhibit the activities of UDGs from a wide range of sources. UGI inhibits the activities of UDGs from Bacillus subtilis, Escherichia coli, Micrococcus luteus, Saccharomyces cerevisiae, rat liver, herpes simplex virus (HSV), and humans, while p56 targets the UDGs from B. subtilis and HSV (6–15). Recently, we identified a new uracil-DNA glycosylase inhibitor from Staphylococcus aureus, called SAUGI, that interacts not only with S. aureus UDG (SAUDG), but also with human UDG with a relatively low binding affinity (16). The SAUGI/SAUDG complex has been determined, and shows that SAUGI binds to the SAUDG DNA binding region via several strong interactions, such as using a hydrophobic pocket to hold SAUDG's protruding residue. By binding to SAUDG in this way, SAUGI thus prevents SAUDG from binding to its DNA substrate and performing DNA repair activity (16).
In present study, we compared the binding affinities and inhibitory effects of SAUGI on five UDGs from S. aureus (SA), Mycobacterium tuberculosis (TB), human, Epstein–Barr virus (EBV) and Herpes simplex virus (HSV). Our results show that SAUGI had the greatest inhibitory activity on the two viral UDGs, followed by SAUDG and human UDG, and had almost no effect on TBUDG. We then determined the crystal structures of the SAUGI/human UDG and SAUGI/HSVUDG complexes and used them to explain these differential binding activities. Lastly, based on this structural information, we designed several site-directed mutants of SAUGI in an attempt to further modulate the inhibitory activities of SAUGI on human UDG and HSVUDG. Our results show that these differential inhibitory effects can be successfully modulated, and suggest the possible application of SAUGI mutant proteins to HSV-related studies.
MATERIALS AND METHODS
Preparation and purification of recombinant SAUGI, UGI and the UDGs
The recombinant proteins were prepared as described previously (16). Briefly, the full-length genes of (1) SAUGI (NCBI sequence ID: AAL26663.1, amino-acid residues 1–112) with the stop codon, (2) phage UGI (NCBI sequence ID: P14739.1, amino-acid residues 1–84) with the stop codon, (3) SAUDG (NCBI sequence ID: YP_040034.1, amino-acid residues 1–218) without the stop codon, (4) human UDG (NCBI sequence ID and PDB: 1SSP_E, amino-acid residues 1–223) without the stop codon, (5) Mycobacterium tuberculosis UDG (TBUDG; NCBI sequence ID: WP_003899565.1, amino-acid residues 1–227) without the stop codon, (6) EBVUDG (NCBI sequence ID: YP_401679.1, amino-acid residues 1–255) without the stop codon, and (7) HSVUDG (NCBI sequence ID: NP_044603.1, amino acid residues 1–244) without the stop codon, were ligated into pET21b expression vector. All expression vectors were transformed into E. coli BL21 (DE3). After the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), the recombinant proteins were expressed at 16°C for 16 h.
Soluble SAUGI and UGI were both purified by Q anion exchange chromatography (GE Healthcare) with a gradient of 0–1 M NaCl in the 20 mM Tris pH 7.4 buffer. The soluble C-terminal His6 tagged UDGs were purified by immobilized metal-ion chromatography with a Ni-NTA column. The purities of these recombinant proteins were further improved by gel filtration on a Superdex 75 column using gel filtration buffer (30 mM Tris–HCl pH 7.4, 100 mM NaCl, 5% glycerol, 1 mM EDTA and 1 mM DTT).
Determining the binding affinities between the Uracil DNA glycosylase inhibitors and the UDGs
The binding affinities of SAUGI to TBUDG, HSVUDG and EBVUDG were determined by surface plasmon resonance using a BIAcore T200 (GE Healthcare) according to the protocols described in the previous report (16). Briefly, the C-terminal His6 tagged UDGs were immobilized on the chip surface by the anti-His antibody on the CM5 chip. Subsequently, SAUGI in HBSP plus buffer (10 mM HEPES pH7.4, 150 mM NaCl and 0.05% v/v Surfactant P20) was injected for 1 min (flow rate was 30 μl/min). In HSVUDG related assays, 3 mM EDTA was added in HBSP plus buffer to improve the stability of immobilized HSVUDG. The temperature used in all studies was 25°C. BIAevaluation 3.1 software was used to calculate the association (ka) and dissociation (kd) kinetic constants using a simple 1:1 Langmuir model for all of the results expect the SAUGI/TBUDG curves. For SAUGI/TBUDG reaction, we found that increasing the association time improved the stability of the complex (Supplementary Figure S1). This suggested that the binding involved a conformational change, and we therefore used a two-state binding model for the SAUGI/TBUDG curves. The equilibrium dissociation constant (KD) values were calculated from the equation KD = kd/ka for the 1:1 Langmuir model, and from the equation KD = kd1*kd2/ka1*ka2 for two-state binding model. The Chi square values of all assays were <0.8, which indicate that the curve fitting of these data was good and that the quantitation of the binding affinities was reliable.
Determining the inhibiting effect of Uracil DNA glycosylase inhibitors on the activities of different UDGs
The methods used here to measure the uracil removing activity of four UDGs were modified from procedures described in a previous report (16). A 40-mer Hex-labeled oligodeoxynucleotide containing a single uracil (ssDNA19U; 5'-Hex-GTAAAACGACGGCCAGTGUATTCGAGCTCGGTACCCGGGG-3') was used as the DNA substrate. For each reaction, different amounts of purified recombinant SAUGI or phage UGI were mixed with the C-terminal His6 tagged UDGs. The reaction mixtures were adjusted to a volume of 9 μl in reaction buffer (20 mM Tris–HCl pH 8.0, 1 mM DTT, 1 mM EDTA) and pre-incubated at 37°C for 15 min. Subsequently, 1 μl of reaction buffer containing the DNA substrate was added to a final concentration of 0.5 μM, and incubation continued for 15 min. Lastly, NaOH was added to a final concentration of 0.2 M and allowed to react at 90°C for 30 min to cleave the apyrimidinic sites in the DNA substrate. The reaction products were then mixed with 2× sample loading buffer (95% formamide, 20 mM EDTA, 1 mg/ml bromophenol blue) and run on a 20% polyacrylamide gel (0.5× Tris–borate–EDTA buffer, 8 M urea). The results were visualized using ultraviolet light and the software imageJ was used to quantify the bands (20). For each concentration of SAUGI, the UDG activity was calculated as 100% × [cleavage product at the indicated SAUGI concentration]/ [cleavage product with no SAUGI added]. Three independent replications were performed for each assay.
Crystallization, data collection and structure determination of the SAUGI/human UDG and SAUGI/HSVUDG complexes
For crystallization of these complexes, 1 mg SAUGI and 2.5 mg C-terminal His6 tagged human UDG or HSVUDG were mixed in 100 μl of Q column buffer (30 mM Tris–HCl pH 7.4, 5% glycerol, 1 mM EDTA and 1 mM DTT) at RT for 1 h. A Q ion exchange column was then used to separate the complex from the uncomplexed monomers with a gradient of 0–1 M NaCl. A Superdex 75 gel filtration colum was then used to further improve the purity of the complexes with a gel filtration buffer (30 mM Tris–HCl pH 7.4, 100 mM NaCl, 5% glycerol, 1 mM EDTA and 1 mM DTT). After protein concentration, 2 μl of the SAUGI/human UDG (20 mg/ml) was mixed with 2 μl of a reservoir containing 0.1 M Tris–HCl pH 8.5, 0.2 M MgCl2·6H2O and 30% PEG 4000, while 2 μl of the SAUGI/HSVUDG (8.2 mg/ml) complex solution was mixed with 2 μl of a reservoir containing 0.2 M ammonium acetate, 0.1 M sodium citrate tribasic dihydrate pH 5.6, 30% PEG 4000. Equilibration with the reservoir was achieved by the sitting drop method. Prior to flash-cooling, ethylene glycol at a final concentration of 15% was added as a cryoprotectant for the complex crystals.
Native X-ray diffraction data from the crystals were collected on beamlines BL13B, BL13C and BL15A at the National Synchrotron Radiation Research Center in Hsinchu, Taiwan. The data were processed using the program HKL2000 (21). The molecular replacements were performed by the program Molrep (22). The UDGs (for human UDG: 1AKZ; for HSVUDG: 1LAU) were used as the initial search model. After fixing the UDG, the SAUGI obtained from the SAUGI/SAUDG complex (3WDG) was used as the second search model. The initial maps were produced after rigid body refinement using Refmac (23) and the resulting electron density map was sufficiently clear for model building (Supplementary Figure S2). The programs Buccaneer (24), Coot (25) and Refmac (23) were used in the subsequent model building and refinement. Statistics for the data collection and refinement are shown in Table 1. The PyMOL program (26) was used for the structural analyses and also for figure production. To confirm the accuracy of the electron density map for residues in the interaction regions of SAUGI Asp24, Gln52 and His62 to two UDGs, simulated annealing omit maps wereproduced by the programs sfall (27) and FFT (28) with the corresponding residues removed models (Supplementary Figure S3).
Table 1. Data collection and refinement statistics of SAUGI/human UDG and SAUGI/HSVUDG crystals.
| Data collection | SAUGI/human UDG | SAUGI/HSVUDG |
|---|---|---|
| Wavelength (Å) | 1.00000 | 1.00000 |
| Space group | C2 | P1 |
| Unit cell a, b, c (Å) | 177.83, 52.85, 82.34 | 53.30, 63.01, 65.51 |
| α, β, γ (°) | 90.00, 112.36, 90.00 | 65.10, 89.57, 88.38 |
| Resolution (Å) | 50–2.4 (2.49-2.40) | 20–2.1 (2.17–2.10) |
| Unique reflections | 26 778 (2701) | 43427 (4363) |
| Redundancy | 3.1 (3.0) | 2.1 (2.1) |
| Completeness (%) | 96.1 (97.7) | 95.6 (96.0) |
| I/σ(I) | 14.0 (1.7) | 12.9 (3.2) |
| Rmerge (%) | 5.4 (41.8) | 7.6 (27.1) |
| Refinement | ||
| Rwork (%) | 16.5 | 19.6 |
| Rfree (%) | 19.8 | 22.3 |
| Bond r.m.s.d. (Å) | 0.009 | 0.010 |
| Angle r.m.s.d. (°) | 1.46 | 1.47 |
| Mean B value/no. of atoms | ||
| UDG (human, HSV; Å2) | 40.7/3632 | 25.5/3670 |
| SAUGI (Å2)a | 40.9/1852 | 26.2/1850 |
| Water molecules (Å2) | 49.5/518 | 35.8/712 |
| Ramachandran plotb | ||
| Most favored (%) | 95.14 | 96.87 |
| Allowed (%) | 4.71 | 3.13 |
| Outliers (%) | 0.15 | 0.00 |
aThe determined SAUGI lengths in both complexes are the same. The two additional atoms found in the SAUGI/human UDG complexes are magnesium.
bRamachandran plots of these two complexes were produced by the program MolProbity (42). Only one outlier (SAUGI/human UDG Glu 303, C chain) was found.
Site directed mutagenesis of SAUGI
The website primer X (http://www.bioinformatics.org/primerx/) was used to design the mutagenic PCR primers for SAUGI (Supplementary Table S1). The site-directed mutagenesis was performed by using a QuikChange II Site-Directed Mutagenesis Kit (Agilent technologies). Briefly, 25ng pET21b containing the full-length SAUGI with the stop codon was used as the template DNA in a 50 μl reaction mixture containing one of the primer pairs. After PCR, the reaction products were incubated with 1.5 μl (9 U) of DpnI (New England Biolabs) for 1 h to digest the methylated parent plasmids. The resulting plasmids were transformed into DH5 alpha E. coli cells and successful mutagenesis was confirmed by DNA sequencing.
RESULTS
The binding affinities between SAUGI and five UDGs
In our previous study, we used surface plasmon resonance (BIAcore) to show that SAUGI can interact with both SAUDG and the fully active, conserved region from human mitochondrial UDG1 and nuclear UDG2 (16). To investigate the binding affinities of SAUGI with three other UDGs, here we used the same technique with EBVUDG, HSVUDG and TBUDG. The results show that SAUGI has a high equilibrium dissociation constant with EBVUDG (KD = 0.475 nM) and HSVUDG (KD = 0.508 nM) (Figure 1A and B; Table 2), and a binding affinity with TBUDG that is significantly lower (KD = 198 nM) than with the other UDGs (Figure 1C; Table 2). Compared to the equilibrium dissociation constant (KD) of SAUGI/SAUDG (∼1 nM; 16) and SAUGI/human UDG (∼2–2.5 nM; (16) and this study) (Table 2), we therefore conclude that SAUGI has the highest binding affinity to EBVUDG, followed by HSVUDG, SAUDG, and then human UDG and TBUDG.
Figure 1.
The binding affinities between SAUGI and (A) EBVUDG, (B) HSVUDG and (C) TBUDG. Increasing concentrations of SAUGI were interacted with immobilized HSVUDG, EBVUDG and TBUDG, and the sensograms were recorded on a BIAcore T200. The colored lines show experimentally recorded values at different concentrations, and the black lines are fitting curves of the data to a 1:1 Langmuir model in (A) and (B), and to a two-state model in (C).
Table 2. Differential binding of SAUGI to several UDGs.
| SAUGI to UDGs (1:1) | ka (1/Ms) | kd (1/s) | KD (nM) | Reference |
|---|---|---|---|---|
| SAUGI to SAUDG | 2472386 ± 16363 | 0.00296 ± 0.00010 | 1.198 ± 0.029 | (16) |
| SAUGI to human UDGa | 1846576 ± 130186 | 0.00361 ± 0.00084 | 1.959 ± 0.476 | This study |
| SAUGI to HSVUDG | 3400215 ± 153579 | 0.00173 ± 0.00008 | 0.508 ± 0.038 | This study |
| SAUGI to EBVUDG | 2535637 ± 343747 | 0.00120 ± 0.00019 | 0.475 ± 0.086 | This study |
| SAUGI E24AQ52R to human UDG | 1600443 ± 249179 | 0.00529 ± 0.00085 | 3.304 ± 0.102 | This study |
| SAUGI E24AQ52R to HSVUDG | 6917482 ± 897402 | 0.00111 ± 0.00007 | 0.163 ± 0.033 | This study |
| SAUGI to TBUDG (two-state) | ka1 and ka2 (1/Ms) | kd1 and kd2 (1/s) | KD (nM) | Reference |
| SAUGI to TBUDG | ka1: 4320235 ± 1244152 | kd1: 1.39320 ± 0.21470 | 198 ± 39 | This study |
| ka2: 20.00274 ± 0.00048 | kd2: 0.00400 ± 0.00056 |
aThe binding affinity of SAUGI to human UDG was measured in our previous report (KD = 2.5 nM, (16)). The same assay was also repeated here.
The inhibiting effects of SAUGI on five UDGs
We next used uracil removal assays to investigate the ability of SAUGI to inhibit the same five UDGs that were used in the surface plasmon resonance experiments. In the absence of SAUGI, all the UDGs removed the uracil from the uracil-containing DNA substrates effectively, and the resulting apyrimidinic site was cleaved by heat and alkali (Figure 2A-E, lane 2). The addition of SAUGI reduced the cleaved products in the SAUDG, EBVUDG, HSVUDG and human UDG reactions (Figure 2A-D, lanes 3–7; Supplementary Figure S4). SAUDG, EBVUDG and HSVUDG activity was initially inhibited when the molar ratio of SAUGI to UDG reached 1:1, but a ratio of 2:1 was needed for the inhibition of human UDG activity. Interestingly, although the structure of TBUDG is conserved relative to the other four UDGs, SAUGI had no significant effect on this UDG's activity (Figure 2E). These results correspond to those found in the BIAcore study. We therefore conclude that SAUGI has a relatively greater effect on the two viral UDGs than on human UDG. Since HSVUDG and EBVUDG have been shown to play an important role during virus replication (29), these results suggest a potential use of SAUGI as an anti-virus treatment.
Figure 2.
Differential inhibitory effects of SAUGI on the activities of (A) SAUDG, (B) EBVUDG, (C) HSVUDG, (D) human UDG and (E) TBUDG. Except for TBUDG, addition of SAUGI reduced the specific uracil-removing activity of SAUDG and the other three UDGs in a dosage-dependent manner.
Crystal structures of the SAUGI/human UDG and SAUGI/HSVUDG complexes
Since UDGs are highly conserved in terms of their amino acid sequences, structures and active sites (2), it is challenging to explain why SAUGI has a different binding affinity and inhibitory effect with different UDGs. To address this issue, we determined the structure of the SAUGI/human UDG and SAUGI/HSVUDG complexes (Figure 3A; resolutions for these two crystals are 2.4 and 2.1 Å, respectively). As with the binding between SAUGI and SAUDG, when SAUGI binds to human UDG or HSVUDG, it does not induce any large conformational changes (the RMSD fits of SAUGI-bound and SAUGI-unbound human UDG and HSVUDG are 0.390 and 0.459 Å; respectively). Further, a comparison of the SAUGI/SAUDG (16), SAUGI/human UDG and SAUGI/HSVUDG complexes revealed the conservation of several interactions that make a major contribution to the binding of SAUGI with these three UDGs (Figure 3B–E). The first interaction involves the side chain of the protruding residues SAUDG Leu184, human UDG Leu272 and HSVUDG Leu214. The side chains of these residues are held by the hydrophobic cavity of SAUGI that is formed by the side chains of Ile35, Phe69, Leu71, Ile83 and Met89 (Figure 3B). The second conserved interaction is mediated by van der Waals (VDW) forces between the Ile28 of SAUGI and His180 (SAUDG), His268 (human UDG) and His210 (HSVUDG) (Figure 3C). Thirdly, similar side chain orientations are found in the regions that contribute to the non-polar interactions between SAUGI and the three UDGs (Figure 3D). Lastly, the acidic side chain of Glu26 in SAUGI is directed towards the N-terminal end of a major helix, and makes hydrogen bonds with a serine residue (Ser83 in SAUDG, Ser169 in human and Ser112 in HSV) (Figure 3E).
Figure 3.
The conserved interactions between SAUGI and SAUDG, human UDG and HSVUDG. (A) A structural alignment of SAUGI/SAUDG, SAUGI/human UDG and SAUGI/HSVUDG complexes. (B–D) Conserved interactions found in all three complexes.
However, although the above major binding sites between SAUGI and three UDGs appear to be conserved, there are several other differential interactions in some of these complexes. Figure 4A shows that Glu24 of SAUGI may interact with Lys79 of SAUDG, but the corresponding residue in both human UDG and HSVUDG is a proline. In Figure 4B, while Gln52 of SAUGI is rotated away in the SAUDG and HSVUDG complexes, there is a very close proximity between the polar side chains of Gln52 of SAUGI and Asn215 of human UDG, and this may produce an unfavorable clash in the interacting region of these two proteins. Figure 4C shows His62 of SAUGI is stacked with Arg85 of SAUDG. An identical interaction is also observed in the SAUGI/HSVUDG complex with Arg114, but it is lost in the SAUGI/human UDG complex, where the arginine residue is replaced by Glu171 and the His62 side chain of SAUGI is rotated away. Lastly, although SAUDG and human UDG both employ a tyrosine to interact with the side chain of SAUGI His87, their different conformations suggest an alternative stacking mode (Figure 4D). Furthermore, when the tyrosine is replaced by Pro218 in HSVUDG, the His87 side chain of SAUGI is docked into an adjacent pocket. This interaction can readily stabilize the complex formation because it is stronger than a solvent-exposed salt bridge. It also explains the higher affinity of SAUGI to HSVUDG.
Figure 4.
Differential interactions between SAUGI and SAUDG, human UDG and HSVUDG. The panels show differential interactions with (A) SAUGI Glu24, (B) Gln52, (C) His62 and (D) His87.
Next, a structural superimposition of TBUDG and SAUDG in the complex was used to investigate the weak binding of SAUGI to TBUDG (Supplementary Figure S5). When these two structures were superimposed, a steric clash was seen found between Arg92 of TBUDG and Tyr67 of SAUGI. Since Tyr67 of SAUGI is in the center of the interacting region, it seems likely that this steric clash is disrupting the binding between SAUGI and TBUDG. However, we also found that increasing the association time improved the stability of the complex (Supplementary Figure S1). This suggests that the binding between SAUGI and TBUDG probably also involves a conformation change, ie, that the binding is consistent with a two-state model. In this model, although the steric clash is much more likely to occur, as indicated by the extremely high dissociation constant 1 (kd1) and the low association constant 2 (ka2; Table 2), the flexible Arg92 side chain might also be rotating to allow it to associate with the Tyr67 by van der Waals interaction.
The inhibitory effects of SAUGI mutants on the uracil-removing activities of human and HSV UDGs
In our previous study, we found phage UGI had an extremely high binding affinity to human UDG that was 12–13 times greater than that of SAUGI (16). We found here that phage UGI also inhibited both HSVUDG and human UDG more effectively than SAUGI (compare Supplementary Figure S6 with Figure 2C and D). We speculated that SAUGI's relatively low binding affinity to human UDG and its high binding affinity to viral UDG might potentially allow it to be used to prevent viral UDG repair activity without having too great an effect on human UDG. In our next experiment, we therefore used structural-based protein engineering to further modify SAUGI's activity on human UDG and HSVUDG.
Based on the differential interactions revealed by the structural features of the SAUGI complexes (Figure 4), we designed and made three SAUGI mutations. These were partly intended to verify the roles of some of the amino acid residues in the binding to HSV and human UDG, and also an attempt at modifying the strength of SAUGI's inhibition of these two UDGs. The SAUGI mutants were designed using the BeAtMuSiC server (http://babylone.ulb.ac.be/beatmusic/query.php). The predicted energy loss at the mutated interfaces in the SAUGI/human UDG and SAUGI/HSVUDG complexes suggested that the three mutations E24A (Glu24 to Ala), Q52R (Gln52 to Arg) and H62Q (His62 to Gln) would reduce the binding energy for HSVUDG by 0.65, 0.05 and 0.12 kcal/mol, but increase the binding energy for human UDG by 1.06, 0.31 and 1.44 kcal/mol, respectively. Details of the predicted energy changes for the mutations of these three residues are listed in Supplementary Table S2.
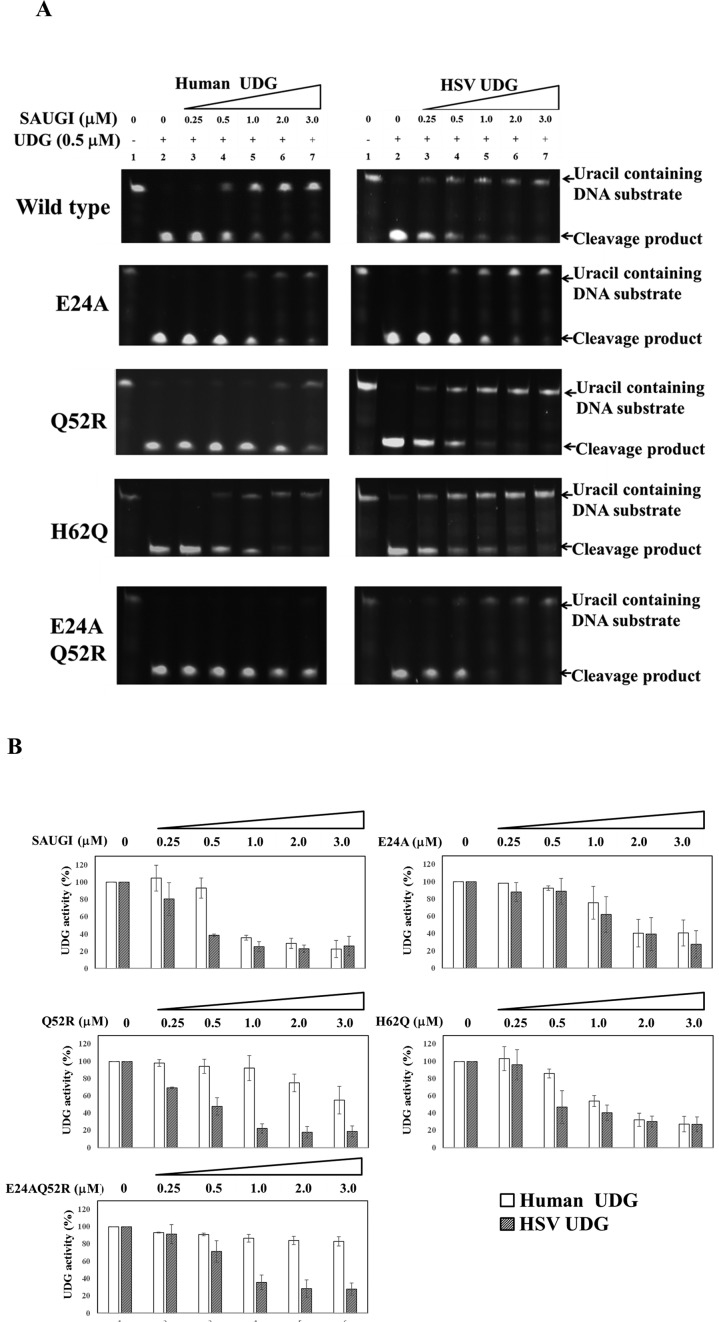
The inhibitory effects of the SAUGI mutants on human and HSV UDGs are shown in Figure 5. Compared to wild-type SAUGI, there was a slight decrease in the inhibitory activity of SAUGI E24A on both UDGs (Figure 5). Although the opposing residue to SAUGI Glu24 is a proline in both UDGs, its conformation showed some variation, directing the side chain towards Pro165 in human UDG but away from Pro108 in HSVUDG (Figure 4A). The reduced inhibition activity of the SAUGI E24A mutant is probably due to the conformational variations that contribute to shape complementarity and VDW interactions, both of which would be broken by removal of the Glu24 side chain because solvent molecules may intervene.
Figure 5.
Evaluation of the inhibitory effects of wild type and mutated SAUGI on human and HSV UDGs. (A) The reduction in change products with increasing SAUGI concentrations reflects the ability of the wild type and mutated SAUGIs (single mutants E24A, Q52R and H62Q, and the double mutant E24AQ52R.) to inhibit the uracil removal activity of human and HSV UDGs. (B) UDG activity was calculated from quantification of the cleavage products. Quantification data was obtained from three independent replications of each assay.
The second mutant, SAUGI Q52R, more closely matched our expectations. Compared to wild type SAUGI, this mutant showed reduced human UDG inhibition, but there was very little change in its activity toward HSVUDG (Figure 5). In the complex structure of SAUGI and human UDG, Gln52 is tightly packed against Asn215 (Figure 4B), and replacement by the larger arginine side chain should result in a more severe clash in the limited space. Conversely, in the other complexes with SAUDG or HSVUDG, the Gln52 side chain is directed away from the equivalent residue (Asn127 or Ala158) (Figure 4B). Consequently, the Q52R mutation should cause no problem in the binding, especially with the small partner side chain of Ala158 in HSVUDG.
The inhibitory effect of the third SAUGI mutant, H62Q, was very similar to that of the wild type SAUGI for both HSVUDG and human UDG (Figure 5). The result was unexpected because while the His62 side chain of SAUGI stacks with the Arg114 side chain of HSVUDG, no such interaction is observed in the complex with human UDG, where the corresponding residue is Glu171 (Figure 4C). Therefore, by mutating His62 into a glutamine, we hoped to obtain a more flexible side chain that could not only stack with Arg114 but also allow conformational changes that would result in additional hydrogen bonds. It was expected that H62Q would stabilize the complex formation of SAUGI and HSVUDG, but have little effect on human UDG. However, in light of the observed results, we now think that the flexible side chain of the glutamine acid may have rotated out of the interacting region.
Based on these results, we further tested the inhibitory activity of the double mutant SAUGI E24AQ52R on these two UDGs. The inhibitory activity of SAUGI E24AQ52R on human UDG was decreased strongly, while there was only a slight effect on HSVUDG. The BIAcore study further confirmed the binding affinity of this double mutant to HSVUDG is 20 times greater than that to human UDG (Supplementary Figure S7, Table 2). Additionally, SAUGI E24AQ52R also had very little effect on SAUGI's ability to inhibit EBVUDG (Supplementary Figure S8A). This was probably because the EBVUDG residue that corresponds to SAUGI Gln52 is Gly, which is even smaller than the corresponding Ala in HSVUDG. In a subsequent experiment using a higher concentration of SAUGI E24AQ52R, the human UDG activity was only found to be inhibited when the molar ratio of SAUGI E24AQ52R to UDG reached 10:1, and the inhibition was still far from complete even at the molar ratio of 30:1 (Supplementary Figure S8B). However, the binding affinity of this double mutant was only 1.6 times lower than that of wild type SAUGI (KD: 3.3 versus 2.0 nM). It is therefore hard to account for the decrease in inhibitory ability only in terms of a reduction in binding affinity. We speculate that the explanation may depend on the structure of the SAUGI E24AQ52R/human UDG complex. Unfortunately, we have not yet obtained crystals of this complex. To address this issue, we produced a binding model of SAUGI E24AQ52R and human UDG (Supplementary Figure S9). From this model, we hypothesize that although the SAUGI E24AQ52R double mutant still binds quite strongly to human UDG, it also fails to control two motifs of human UDG that both play important roles in DNA binding and damage recognition (i.e. the 4-Pro and Gly-Ser loops; (1)). The double mutant's failure to control these motifs therefore allows the human UDG to initiate the recognition and binding of damaged DNA. We further hypothesize that subsequent conformational changes in the human UDG (e.g. in the Leu272 loop) will cause the human UDG-bound SAUGI E24AQ52R to be replaced by DNA.
DISCUSSION
In our recent work, we have focused on identifying new DNA mimic proteins and exploring the potential applications of this novel kind of control factor (16,19,30–34). Among the five DNA mimic proteins that we have identified, SAUGI is of particular interest because it inhibits the activity of SAUDG and human UDG differentially (16). In this study, we found that SAUGI also shows a differential inhibitory effect on other UDGs, and in particular that it has a relatively higher affinity to two viral UDGs than to human UDG (0.5 nM versus 2 nM; Figures 1 and 2, Table 2). Compared to phage UGI, which has been found to show extremely high affinity to human UDG (∼0.2 nM; reference 16), SAUGI potentially provides a better starting point for designing a specific viral UDG inhibitor. In pursuit of this goal, we determined the complex structures of SAUGI/human UDG and SAUGI/HSVUDG, and produced a series of SAUGI mutants. While the inhibitory strength of the Q52R mutant to HSVUDG was at the same level as the wild type, the ability of this mutant to inhibit human UDG inhibition was decreased (Figure 5). The double mutant SAUGI E24AQ52R further reduced the inhibitory ability to human UDG while still having a slight effect on the HSVUDG inhibition (Figure 5). This SAUGI mutant may therefore have potential application in HSV-related studies.
Although the critical roles of UDGs and BER in virus replication are still unclear, there are already a number of studies that suggest their importance. For example, Human Immunodeficiency Virus (HIV) packages human UDG into its virus particles (35). Depletion of human UDG isoform 2 by shRNA in HIV-1-producing 293T cells inhibits viral infectivity and replication (36). Some large, complex dsDNA viruses such as herpesviruses and poxviruses also contain their own UDG genes in the genomes (29). Interestingly, the herpesvirus UDGs were shown to be dispensable for replication in cell culture, but they are important in non-dividing cells and animals (29). In human cytomegalovirus (CMV; human herpesvirus-5), the lack of CMV UDG expression will lead to a delay in replication in non-dividing cells (37). HSVUDG-minus mutants also replicate and spread poorly in mice (38). Furthermore, the association between the viral DNA polymerases (DNAP) and UDGs in HSV and EBV in their respective DNA replication complexes suggests that herpesvirus UDGs participate in viral DNA replication (39,40). Association with DNAP also increases the EBVUDG activity (40). Su et al's study showed evidences that the residues 164–255 of EBVUDG were necessary for the interaction with EBV DNAP (40). Using SAUGI/HSVUDG as a template, our model of the SAUGI/EBVUDG complex suggests that SAUGI interacts with residues 213–229 of EBVUDG (Supplementary Figure S10). This overlap implies that SAUGI may interfere with the binding between viral UDG and DNAP in the replication complex. Since herpesvirus DNA replication-associated enzymes have been considered good antiviral targets (40), SAUGI has potential application in the treatment of herpesvirus-related diseases by virtue of its ability to prevent viral UDG from binding to either DNA or to other proteins (e.g. viral DNAP). Meanwhile, the relatively low inhibitory effect of SAUGI E24AQ52R on human UDG suggests there should be only limited side effects on human cells. Nevertheless, all of these potentially favorable characteristics will need to be experimentally investigated.
Some other viral UDGs are encoded by poxviruses such as the smallpox and vaccinia viruses. The poxvirus UDGs have been found to be more closely associated with maintaining virus replication in cell culture (29). Like the herpesvirus UDGs, the vaccinia UDG is an essential component of the viral polymerase holoenzyme (41). Interestingly, another well-studied protein, phage UGI, acts on a wide range of UDGs but fails to inhibit vaccinia UDG (29). The reason for this may be related to UDG's protruding Leu residue (Leu184 in SAUDG; Leu272 in human UDG and Leu214 in HSVUDG; Figure 3B), which is surrounded by a hydrophobic pocket in the complexes found with phage UGI or SAUGI, but is replaced by an Arg in vaccinia UDG (Arg185; (29)). The longer, polar side chain of Arg185 in vaccinia UDG may produce unfavorable steric clashes in the hydrophobic pocket of the phage UGI. Since SAUGI uses a similar hydrophobic pocket to target the protruding Leu residue in the UDGs (Figure 3B), it is very likely that SAUGI will also fail to interact strongly with the poxviral UDGs. We will address this question experimentally in the near future.
In conclusion, this study provides evidence that SAUGI differentially inhibits a wide range of UDGs. Structural-based protein engineering further explored the potential applications of SAUGI, and the next step will be to investigate how SAUGI and its mutants affect viral and human UDG during virus infection. We believe this work will provide useful information in both the DNA mimic and BER research fields.
ACCESSION NUMBERS
The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org. PDB ID codes of SAUGI/human UDG and SAUGI/HSVUDG complexes are 5AYR and 5AYS, respectively.
Supplementary Material
Acknowledgments
We acknowledge the staffs of the beamlines BL13B, BL13C and BL15A at the Radiation Research Center (NSRRC) in Hsinchu, Taiwan for their help in X-ray crystal data collection. We also thank Dr. Shu-Chuan Jao of the Biophysics Core Facility, Department of Academic Affairs and Instrument Service at Academia Sinica for her help in BIAcore analyses.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Academia Sinica and National Science Council [NSC 102-2311-B-038-003 and MOST 103-2311-B-038-005]. Funding for open access charge: Academia Sinica and National Science Council [NSC 102-2311-B-038-003 and MOST 103-2311-B-038-005].
Conflict of interest statement. None declared.
REFERENCES
- 1.Parikh S.S., Mol C.D., Slupphaug G., Bharati S., Krokan H.E., Tainer J.A. Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J. 1998;17:5214–5226. doi: 10.1093/emboj/17.17.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parikh S.S., Putnam C.D., Tainer J.A. Lessons learned from structural results on uracil-DNA glycosylase. Mutat. Res. 2000;460:183–199. doi: 10.1016/s0921-8777(00)00026-4. [DOI] [PubMed] [Google Scholar]
- 3.Zharkov D.O. Base excision DNA repair. Cell Mol. Life Sci. 2008;65:1544–1565. doi: 10.1007/s00018-008-7543-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindahl T. An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc. Natl. Acad. Sci. U.S.A. 1974;71:3649–3653. doi: 10.1073/pnas.71.9.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ko R., Bennett S.E. Physical and functional interaction of human nuclear uracil-DNA glycosylase with proliferating cell nuclear antigen. DNA Repair. 2005;4:1421–1431. doi: 10.1016/j.dnarep.2005.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mol C.D., Arvai A.S., Sanderson R.J., Slupphaug G., Kavli B., Krokan H.E., Mosbaugh D.W., Tainer J.A. Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: protein mimicry of DNA. Cell. 1995;82:701–708. doi: 10.1016/0092-8674(95)90467-0. [DOI] [PubMed] [Google Scholar]
- 7.Putnam C.D., Shroyer M.J., Lundquist A.J., Mol C.D., Arvai A.S., Mosbaugh D.W., Tainer J.A. Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase. J. Mol. Biol. 1999;287:331–346. doi: 10.1006/jmbi.1999.2605. [DOI] [PubMed] [Google Scholar]
- 8.Kaushal P.S., Talawar R.K., Krishna P.D., Varshney U., Vijayan M. Unique features of the structure and interactions of mycobacterial uracil-DNA glycosylase: structure of a complex of the Mycobacterium tuberculosis enzyme in comparison with those from other sources. Acta Cryst. D. 2008;64:551–560. doi: 10.1107/S090744490800512X. [DOI] [PubMed] [Google Scholar]
- 9.Géoui T., Buisson M., Tarbouriech N., Burmeister W.P. New insights on the role of the gamma-herpesvirus uracil-DNA glycosylase leucine loop revealed by the structure of the Epstein-Barr virus enzyme in complex with an inhibitor protein. J. Mol. Biol. 2007;366:117–131. doi: 10.1016/j.jmb.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 10.Serrano-Heras G., Ruiz-Masó J.A., del Solar G., Espinosa M., Bravo A., Salas M. Protein p56 from the Bacillus subtilis phage phi29 inhibits DNA-binding ability of uracil-DNA glycosylase. Nucleic. Acids Res. 2007;35:5393–5401. doi: 10.1093/nar/gkm584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serrano-Heras G., Salas M., Bravo A. A uracil-DNA glycosylase inhibitor encoded by a non-uracil containing viral DNA. J. Biol. Chem. 2006;281:7068–7074. doi: 10.1074/jbc.M511152200. [DOI] [PubMed] [Google Scholar]
- 12.Serrano-Heras G., Bravo A., Salas M. Phage phi29 protein p56 prevents viral DNA replication impairment caused by uracil excision activity of uracil-DNA glycosylase. Proc. Natl. Acad. Sci. U.S.A. 2008;105:19044–19049. doi: 10.1073/pnas.0808797105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asensio J.L., Pérez-Lago L., Lázaro J.M., González C., Serrano-Heras G., Salas M. Novel dimeric structure of phage ϕ 29-encoded protein p56: insights into uracil-DNA glycosylase inhibition. Nucleic. Acids Res. 2011;39:9779–9788. doi: 10.1093/nar/gkr667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baños-Sanz J.I., Mojardín L., Sanz-Aparicio J., Lázaro J.M., Villar L., Serrano-Heras G., González B., Salas M. Crystal structure and functional insights into uracil-DNA glycosylase inhibition by phage ϕ 29 DNA mimic protein p56. Nucleic. Acids Res. 2013;41:6761–6773. doi: 10.1093/nar/gkt395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cole A.R., Ofer S., Ryzhenkova K., Baltulionis G., Hornyak P., Savva R. Architecturally diverse proteins converge on an analogous mechanism to inactivate uracil-DNA glycosylase. Nucleic. Acids Res. 2013;41:8760–8775. doi: 10.1093/nar/gkt633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H.C., Hsu K.C., Yang J.M., Wu M.L., Ko T.P., Lin S.R., Wang AHJ. Staphylococcus aureus protein SAUGI acts as a uracil-DNA glycosylase inhibitor. Nucleic. Acids Res. 2014;42:1354–1364. doi: 10.1093/nar/gkt964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dryden D.T.F. DNA mimicry by proteins and the control of enzymatic activity on DNA. Trends Biotechnol. 2006;4:378–382. doi: 10.1016/j.tibtech.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Putnam C.D., Tainer J.A. Protein mimicry of DNA and pathway regulation. DNA Repair. 2005;4:1410–1420. doi: 10.1016/j.dnarep.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 19.Wang H.C., Ho C.H., Hsu K.C., Yang J.M., Wang A.H.J. DNA mimic proteins: functions, structures and bioinformatic analysis. Biochemistry. 2014;42:1354–1364. doi: 10.1021/bi5002689. [DOI] [PubMed] [Google Scholar]
- 20.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 22.Vagin A., Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- 23.Vagin A.A., Steiner R.S., Lebedev A.A., Potterton L., McNicholas S., Long F., Murshudov G.N. REFMAC5 dictionary: organisation of prior chemical knowledge and guidelines for its use. Acta Cryst. D. 2004;60:2284–2295. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 24.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Cryst. D. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 25.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Cryst. D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.The PyMOL Molecular Graphics System. Version 1.2r3pre. Schrödinger, LLC; [Google Scholar]
- 27.Agarwal R. C. A new least-squares refinement technique based on the fast Fourier transform algorithm. Acta Cryst. A. 1978;34:791–809. [Google Scholar]
- 28.Read R. J., Schierbeek A. J. A phased translation function. J. Appl. Cryst. 1988;21:490–495. [Google Scholar]
- 29.Chen R., Wang H., Mansky L.M. Roles of uracil-DNA glycosylase and dUTPase in virus replication. J. Gen. Virol. 2002;83:2339–2345. doi: 10.1099/0022-1317-83-10-2339. [DOI] [PubMed] [Google Scholar]
- 30.Wang H.C., Wang H.C., Ko T.P., Lee Y.M., Leu J.H., Ho C.H., Huang W.P., Lo C.F., Wang A.H.J. White spot syndrome virus protein ICP11: A histone-binding DNA mimic that disrupts nucleosome assembly. Proc. Natl. Acad. Sci. U.S.A. 2008;105:20758–20763. doi: 10.1073/pnas.0811233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H.C., Ko T.P., Wu M.L., Ku S.C., Wu H.J., Wang A.H.J. Neisseria conserved protein DMP19 is a DNA mimic protein that prevents DNA binding to a hypothetical nitrogen-response transcription factor. Nucleic. Acids Res. 2012;40:5718–5730. doi: 10.1093/nar/gks177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H.C., Wu M.L., Ko T.P., Wang A.H.J. Neisseria conserved hypothetical protein DMP12 is a DNA mimic that binds to histone-like HU protein. Nucleic. Acids Res. 2013;41:5127–5138. doi: 10.1093/nar/gkt201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho C.H., Wang H.C., Ko T.P., Wang A.H.J. T4 phage DNA mimic protein Arn inhibits the DNA-binding activity of the bacterial histone-like protein H-NS. J. Biol. Chem. 2014;289:27046–27054. doi: 10.1074/jbc.M114.590851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chou C.C., Wang A.H.J. Structural D/E-rich repeats play multiple roles especially in gene regulation through DNA/RNA mimicry. Mol. Biosyst. 2015;11:2144–2151. doi: 10.1039/c5mb00206k. [DOI] [PubMed] [Google Scholar]
- 35.Guenzel C.A., Hérate C., Le Rouzic E., Maidou-Peindara P., Sadler H.A., Rouyez M.C., Mansky L.M., Benichou S. Recruitment of the nuclear form of uracil DNA glycosylase into virus particles participates in the full infectivity of HIV-1. J. Virol. 2012;86:2533–2544. doi: 10.1128/JVI.05163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weil A.F., Ghosh D., Zhou Y., Seiple L., McMahon M.A., Spivak A.M., Siliciano R.F., Stivers J.T. Uracil DNA glycosylase initiates degradation of HIV-1 cDNA containing misincorporated dUTP and prevents viral integration. Proc. Natl. Acad. Sci. U.S.A. 2013;110:448–457. doi: 10.1073/pnas.1219702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Courcelle C.T., Courcelle J., Prichard M.N., Mocarski E. S. Requirement for uracil-DNA glycosylase during the transition to latephase cytomegalovirus DNA replication. J. Gen. Virol. 2001;75:7592–7601. doi: 10.1128/JVI.75.16.7592-7601.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pyles R.B., Thompson R.L. Evidence that the herpes simplex virus type 1 uracil DNA glycosylase is required for efficient viral replication and latency in the murine nervous system. J. Virol. 1994;68:4963–4972. doi: 10.1128/jvi.68.8.4963-4972.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bogani F., Corredeira I., Fernandez V., Sattler U., Rutvisuttinunt W., Defais M., Boehmer P.E. Association between the herpes simplex virus-1 DNA polymerase and uracil DNA glycosylase. J. Biol. Chem. 2010;285:27664–27672. doi: 10.1074/jbc.M110.131235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su M.T., Liu I.H., Wu C.W., Chang S.M., Tsai C.H., Yang P.W., Chuang Y.C., Lee C.P., Chen M.R. Uracil DNA glycosylase BKRF3 contributes to EBV DNA replication through physical interactions with proteins in viral DNA replication complex. J. Virol. 2014;88:8883–8899. doi: 10.1128/JVI.00950-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boyle K.A., Stanitsa E.S., Greseth M.D., Lindgren J.K., Traktman P. Evaluation of the role of the vaccinia virus uracil DNA glycosylase and A20 proteins as intrinsic components of the DNA polymerase holoenzyme. J. Biol. Chem. 2011;286:24702–24713. doi: 10.1074/jbc.M111.222216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen V.B., Arendall W.B., Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Cryst. D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.