Abstract
Horizontal gene transfer (HGT) is the non-inherited acquisition of novel DNA sequences. HGT is common and important in bacteria because it enables the rapid generation of new phenotypes such as antibiotic resistance. Here we show that in vivo and in vitro DNA methylation patterns can be horizontally transferred into bacterial chromosomes to program cell phenotypes. The experiments were performed using a synthetic system in Escherichia coli where different DNA methylation patterns within the cis-regulatory sequence of the agn43 gene turn on or off a fluorescent reporter (CFP). With this system we demonstrated that DNA methylation patterns not only accompany the horizontal transfer of genes into the bacterial cytoplasm but can be transferred into chromosomes by: (i) bacteriophage P1 transduction; and (ii) transformation of extracellular synthetic DNA. We also modified the experimental system by replacing CFP with the SgrS small RNA, which regulates glucose and methyl α-D-glucoside uptake, and showed that horizontally acquired DNA methylation patterns can increase or decrease cell fitness. That is, horizontally acquired DNA methylation patterns can result in the selection for and against cells that have HGT. Findings from these proof-of-concept experiments have applications in synthetic biology and potentially broad implications for bacterial adaptation and evolution.
INTRODUCTION
Horizontal gene transfer (HGT) is important in bacterial evolution (1,2). HGT refers to the non-inherited acquisition of DNA, which may be integrated into the chromosome of the recipient cell (3) or may be extra-chromosomal (e.g. plasmids or lytic bacteriophage). Horizontal transfer (HT) of DNA may occur via: (i) contact between bacteria (a process termed conjugation, which is often mediated by type IV secretion systems (4,5)); (ii) bacteriophage and gene transfer agents (a process termed transduction); (iii) uptake from the environment (a process termed transformation); (iv) nanotubes; and (v) membrane vesicles (1,3). The horizontally acquired DNA often must evade destruction by restriction-modification (RM) systems (6) and other surveillance mechanisms including the CRISPR-Cas system (7). In addition, the expression of acquired genes may be silenced by factors such as the histone-like nucleoid structuring protein (H-NS) (8). HGT is more common between closely related bacteria because of these surveillance and silencing mechanisms and because homologous recombination is more likely between bacteria with similar DNA sequences and proteins (e.g. receptors that bind bacteriophage), and shared ecological niches (9).
One important aspect of HGT that has not been investigated is whether the presence or absence of DNA methylation on acquired DNA can be incorporated into chromosomes. DNA methylation is the covalent attachment of methyl groups at specific recognition sequences in the genome (e.g. GATC), which are mediated by DNA methyltransferases (‘DNA methylases’). At these recognition sequences, methylation may occur on zero (‘unmethylated’), one (‘hemi-methylated’) or two (‘methylated’ or ‘fully methylated’) DNA strands. A region of DNA may have different methylation states at different sites and this combination of states is referred to as the ‘DNA methylation pattern’. A sequence that is fully methylated usually can only become unmethylated through DNA replication. DNA replication creates an unmethylated copy of the methylated template DNA strand resulting in hemi-methylated DNA. If potential sites for DNA methylation on the unmethylated strand are blocked by proteins or randomly missed by DNA methylases then a second round of DNA replication can generate double-stranded unmethylated DNA (10,11). DNA methylation may affect the thermostability, bending and steric hindrance of DNA, which can alter the binding of transcription factors and other proteins (10). Because DNA methylation encodes important regulatory information, just like DNA bases, it has been referred to as the ‘fifth base’ (12). DNA methylation patterns regulate the transcription of genes in diverse processes including biofilm formation, conjugation, bacteriophage replication, transposition, the timing of chromosome replication and mismatch-repair (10).
Our knowledge of DNA methylation in relation to HGT into the chromosome is very limited. Studies of bacteriophages and plasmids have shown that methylated and unmethylated DNA can enter cells (9,13). It is known that acquired DNA that does not have a specific methylation pattern may be cleaved or methylated by RM systems (9,13). It has also been shown that transfection of bacteriophage lambda and plasmids with mismatched hemi-methylated DNA results in the recognition and repair of the unmethylated DNA strand by DNA adenine methylase (Dam) directed mismatch repair systems (14,15). From all of these studies it is clear that methylation patterns on horizontally acquired DNA can at least temporarily persist in the bacterial cytoplasm but it has not been shown that these methylation patterns can then be transferred into the chromosome. Specifically, it is not known whether extra-chromosomal bacteriophage DNA and extracellular DNA that are acquired with unmethylated sites, which do not have proteins bound to them to ‘protect’ them from DNA methylases, can be transferred into the chromosome before they are methylated. In addition, for methylation patterns on horizontally acquired DNA to be transferred to the chromosome with high fidelity they need to remain unmodified during the processes of homologous recombination and subsequent chromosome replication; and it is not clear that this is the case given the involvement of DNA methylation and DNA replication in these processes. Lastly, for the HT of DNA methylation patterns into chromosomes to contribute to bacterial adaptation and evolution (16,17) they must not only be able to persist through the entire process of HGT but also modulate the expression of genes and alter bacterial fitness, and this has not been demonstrated.
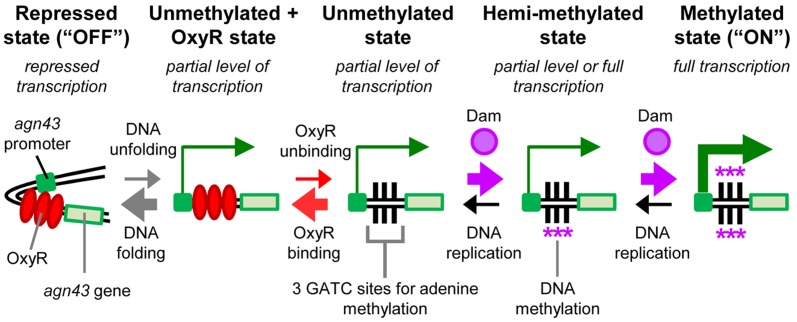
To investigate whether HT of DNA methylation patterns into the chromosome can occur we created a synthetic system. Synthetic systems complement and can provide advantages over theoretical and field studies (18) particularly for transient behaviors and mechanisms (including errors in transcription, translation and protein folding) (18–22). Our synthetic system was based on the agn43 system in Escherichia coli (Figure 1). In this system, Dam methylates N6 in adenine at three GATC sites in the cis-regulatory sequence of the agn43 gene (23,24). DNA methylation at the GATC sites results in full transcription (‘ON’). When the GATC sites are unmethylated they can be bound by the OxyR protein, which represses transcription (‘OFF’) (23,24). Competition between Dam and OxyR for the GATC sites and the kinetics of the system result in stable ON and OFF expression states that are heritable with random switching between the states occurring in ∼10−2–10−3 cells per generation (11,23–26). If neither Dam nor OxyR is present then agn43 is transcribed at a ‘partial’ level between ON and OFF expression (11,27). We previously found it difficult to distinguish the ON and OFF expression states by placing a fluorescent reporter directly under its control (11). Therefore, we used an amplification system where the agn43 cis-regulatory sequence, which is referred to as the ‘agn43 region’, regulates transcription of the T7 RNA polymerase gene (‘T7RNAP’), which in turn regulates transcription of the cyan fluorescent protein gene (cfp) from the T7 promoter (PT7) (11). We stress that the system is a convenient tool to determine the DNA methylation pattern at a specific site and that our experiments were not designed to investigate agn43 regulation (including switching rates) or determine whether the native agn43 system is horizontally transferred in natural populations.
Figure 1.
Regulation of the agn43 switch. Competition between OxyR and Dam for three GATC sites in the agn43 region results in five possible states, some of which have different levels of transcription (11). The relative magnitude of transition events between the states is indicated by arrow thickness. The kinetics of the system strongly favors the repressed, unmethylated (OFF) state and the fully methylated (ON) state. The other states are unstable in wild-type cells with OxyR and Dam. Notes: (i) hemi-methylation may occur on either DNA strand; and (ii) objects are not to scale.
We performed three sets of experiments with our system. In the first set of experiments we demonstrated HT of DNA methylation patterns at a region of the chromosome that was transferred from donor cells to the chromosome of recipient cells by bacteriophage P1 (‘phage’) transduction. In the second set of experiments we showed that methylation patterns on extracellular DNA generated by polymerase chain reaction (PCR) and in vitro DNA methylation can be transformed and integrated into chromosomes. To our knowledge these two sets of experiments show for the first time that methylation patterns on acquired DNA may not only be transferred into the cytoplasm of bacteria but they may also be integrated into the chromosome and maintained for many generations. In the third set of experiments we modified our system by replacing the gene for CFP with the gene for SgrS RNA and then demonstrated that HT of DNA methylation patterns can increase or decrease cell fitness on different growth media. This last set of experiments shows that methylation patterns on acquired DNA can result in the positive or negative selection of cells that have had HGT and thus they may potentially contribute to the evolution of the genome.
MATERIALS AND METHODS
Strains and plasmids
Details of strains, plasmids and oligonucleotides are in the Supplementary Methods, Tables S1 and S2, and Figure S1.
Bacteriophage P1 transduction
Bacteriophage P1 transduction was performed with donor phage prepared from Δdam (HL23) and ΔoxyR (HL24) strains and added to wild-type, Δdam, ΔoxyR and ΔdamΔoxyR recipient cells (HL5786, HL6712, HL6794 and HL6713 respectively). A standard transduction protocol was used (Supplementary Methods) except for the SgrS experiments where sodium citrate-1× LB media was substituted with sodium citrate-1× M9 minimal media + 2% v/v glycerol. Cells were plated on agar with LB media, 1× M9 minimal media + 0.2% w/v glucose or 1× M9 minimal media + 2% v/v glycerol + 0.5% w/v methyl α-D-glucoside (α-MG; CAS#97-30-3, Acros Organics from Fisher Scientific, Pittsburgh, PA, USA). All plates had 50 μg·ml−1 kanamycin, 34 μg·ml−1 chloramphenicol and 9 × 10−2 mg·ml−1 bovine liver catalase (Sigma Aldrich, St. Louis, MO, USA).
Transformations
Transformations were performed using the lambda Red method (28). PCR product was amplified with the yeepupf and t7seqr991 oligonucleotides. An aliquot of the PCR product was methylated for >12 h with Dam plus s-adenosyl methionine (additional methionine was added after 4 h) (New England Biolabs, MA, USA). The methylated PCR product was digested with MboI restriction endonuclease (New England Biolabs), which cuts unmethylated GATC sequences, and then the full length PCR product was gel extracted (Supplementary Figure S2). The number of colonies that grew on antibiotic selection divided by the number of colonies that grew without antibiotic selection (note: the latter is the number of viable cells after electroporation) yielded similar values for the transformation of unmethylated DNA and methylated DNA into wild-type recipient cells (∼1.44 × 10−5 and ∼1.36 × 10−5 integrations per viable cell respectively).
RESULTS
Bacteriophage P1 transduction of DNA methylation patterns
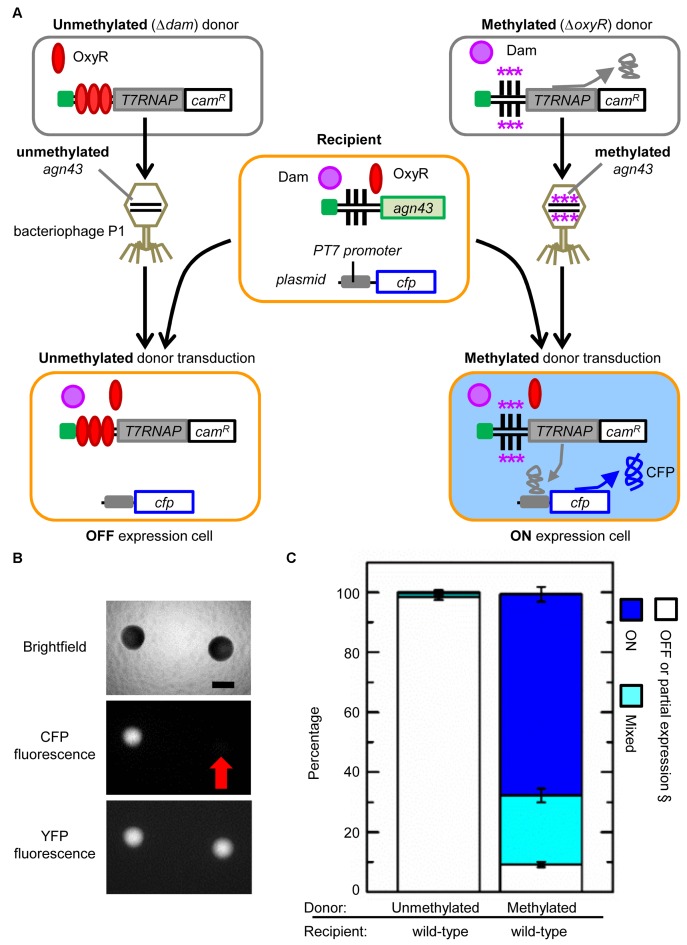
We first examined whether methylated and unmethylated agn43 regions can be transferred by bacteriophage P1 transduction from donor cell chromosomes to recipient cell chromosomes (Figure 2A). The basic steps of the experiment are: (i) a strain was created with the agn43 region regulating T7RNAP and with an adjacent chloramphenicol resistance gene (camR); (ii) from this strain we generated two donor strains that had dam or oxyR deleted and these were used to produce phage carrying unmethylated and methylated donor DNA respectively; (iii) these donor phages infected recipient cells, which have cfp under the control of PT7; (iv) cells that had successfully integrated the donor agn43 region were selected on media with chloramphenicol; and (v) the percentage of selected cells with OFF and ON expression was measured. Recipient cells contained chromosomal yfp, which was constitutively transcribed, so that colonies arising from successful transduction of donor DNA into recipient cells could be distinguished from any possible donor cell carry-through (i.e. in case some cells survived treatment of the lysate with chloroform) by measuring YFP fluorescence.
Figure 2.
Bacteriophage P1 transduction of methylated and unmethylated DNA sequences. Symbols are defined in Figure 1. (A) Schematic of experimental workflow. Asterisks indicate DNA adenine methylation. All genes are chromosomal except cfp, which is on a plasmid. Recipient cells also contained a chromosomal copy of yfp that was constitutively transcribed (not shown). Note: OxyR and Dam proteins shown in transduced cells are from the recipient cell and are not transmitted from the donor cell via phage. Donor strains are HL23 (Δdam) and HL24 (ΔoxyR). Recipient strains are HL5786 (wild-type), HL6712 (Δdam), HL6794 (ΔoxyR) and HL6713 (ΔdamΔoxyR). (B) Fluorescence and brightfield (with phase condenser annulus 3) microscopy of colonies. Scale bar is 500 μm. Red arrow indicates the location of the OFF colony. (C) Percentage of OFF, ON and mixed colonies. Error bars are the standard error of the mean (SEM) of 15 biological replicates. § OFF expression could not be distinguished from the partial level of expression expected in ΔdamΔoxyR strains (11) by fluorescence microscopy.
We predicted that the transduction of methylated and unmethylated donor DNA into wild-type recipient cells (which have Dam and OxyR) would lead to a greater percentage of ON and OFF cells respectively. Importantly, we did not expect the percentages to be 100% ON and 100% OFF respectively because cells can switch states. We compared the percentage of OFF and ON expression cells rather than the absolute number of colonies so the results did not depend on the donor phage titer or whether recipient cells preferentially uptake, cleave and/or integrate methylated or unmethylated DNA. We tested two methods for measuring the percentage of cells with OFF and ON expression: (i) counting colonies; and (ii) flow cytometry (Supplementary Methods and Figure S3).
The colony counting method has the advantage that cells are plated on selective media 4 h after transduction and each cell gives rise to a colony. Therefore, the percentage of colonies with predominantly OFF and ON expression provides a snap-shot of the percentage of cells with OFF and ON expression at the time of plating. Another advantage of this method is that large and small colonies are equivalent in calculating the percentage of OFF and ON cells therefore it is to some extent independent of their growth rates. ‘Mixed’ colonies with OFF and ON cells (defined as <70% OFF or ON cells) occur due to switching during the growth of the colony (Supplementary Figure S4). The founding cell of most mixed colonies is likely to be an ON cell (see transformation experiments below). In contrast to colony counting, flow cytometry was unable to measure the percentage of OFF and ON cells at 4 h post-transduction (or even after 7 or 8 h; Supplementary Figure S3C and D) but it could do so after 16 or more hours (Supplementary Figure S3E and F). However, the 16 h growth period increased the percentage of OFF cells (Supplementary Figure S3G) because of their faster growth (doubling times for OFF and ON cells are 25.42 ± 0.16 and 29.92 ± 0.36 min respectively; Supplementary Figure S5). Flow cytometry measurements also showed more variation among the replicate cultures in the percentage of OFF cells than colony counting measurements; this was probably due to the long growth period in the former amplifying small differences in the percentage of OFF and ON cells (Supplementary Figure S3F, G). Because colony counting can provide more accurate and precise measurements of the percentage of OFF and ON cells, and can measure these percentages after a shorter period post-transduction, it was used for subsequent experiments.
We found the percentages of OFF, ON and mixed colonies arising from the transduction of methylated and unmethylated donor DNA were significantly different (chi-square test; χ2 = 5240.6, n = 7581, P < 0.0001). Transduction of wild-type recipient cells with an unmethylated agn43 region (Δdam donor) resulted in 98.4 ± 1.0% OFF, 0.3 ± 0.1% ON and 1.3 ± 0.8% mixed colonies (15 biological replicates; n = 1856) (Figure 2B and C). In comparison, transduction of wild-type recipient cells with a methylated agn43 region (ΔoxyR donor) resulted in 9.2 ± 0.8% OFF, 67.1 ± 2.5% ON and 23.1 ± 2.3% mixed colonies (15 biological replicates; n = 5725). ON and OFF colonies had reversible switching, which is consistent with an epigenetically determined phenotype. When streaked out, OFF colonies gave rise to 5.0 ± 2.2% ON colonies (three biological replicates; n = 173) and ON colonies gave rise to 57.0 ± 0.9% OFF colonies (three biological replicates; n = 200).
In control transduction experiments with Δdam, ΔoxyR or ΔdamΔoxyR recipient cells, the methylation state of the donor DNA did not affect the percentages of OFF, ON or mixed colonies. This result was expected because no matter what methylation pattern is present on the HT DNA, only an unmethylated or methylated state can be maintained when dam or oxyR is respectively deleted. Specifically, we found that the transduction of methylated or unmethylated donor DNA into Δdam recipients resulted in only OFF colonies (n = 1309 and 747 respectively) [note: the transduction of methylated donor DNA into Δdam recipient cells resulted in one ON and one mixed colony but these were found to be due to the concurrent transduction of the dam gene from the donor strain]. Conversely, the transduction of unmethylated or methylated donor DNA into ΔoxyR recipients always resulted in ON colonies (n = 668 and 1522 respectively). The transduction of unmethylated or methylated donor DNA into ΔdamΔoxyR recipient cells always generated colonies that were OFF or had a partial level of expression (n = 1005 and 1739 respectively).
Our finding that the transduction of unmethylated and methylated donor DNA into wild-type cells generates primarily OFF and ON colonies respectively is consistent with DNA methylation patterns at the agn43 region in donor cell chromosomes being successfully transferred to recipient cell chromosomes. The results cannot be explained by preferential degradation or recombination of unmethylated or methylated donor DNA in recipient cells. These mechanisms would be expected to produce different numbers of colonies but they would not be expected by themselves to produce different percentages of OFF and ON colonies with transduction of unmethylated or methylated donor DNA. For example, if only methylated donor DNA could avoid degradation by RM systems and recombine with the recipient cell chromosome then only ON colonies should form with methylated and unmethylated donor DNA (assuming some of the latter was methylated soon after entering the cell). An additional point of note is that the very high percentage of OFF colonies arising from the transduction of unmethylated donor DNA indicates that very little donor DNA is methylated by bacteriophage P1 encoded Dam (29).
In vitro DNA methylation and transformation of the agn43 region
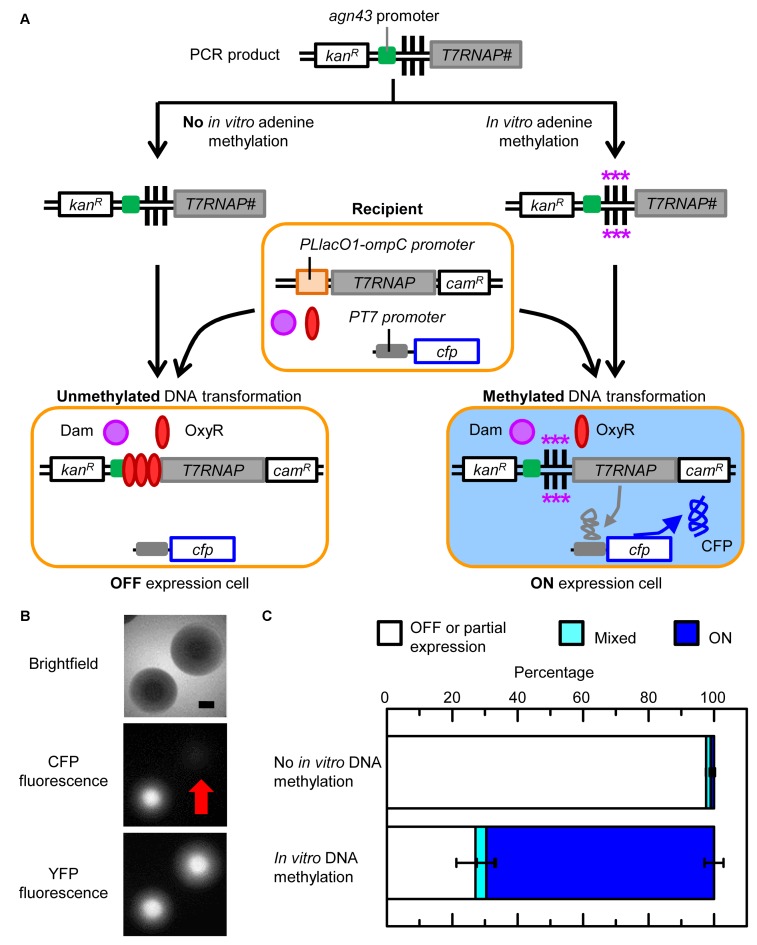
We next investigated whether the methylation state of ‘naked’ extracellular DNA (i.e. without any bound proteins) can be transferred via transformation to the chromosome of recipient cells (Figure 3A). The basic outline of this experiment is as follows: (i) PCR product was amplified containing 137 bp of the yeeP’ pseudogene sequence (for recombination), the kanamycin resistance gene (kanR), 525 bp of the agn43 region, and the first 48 bp of agn43 coding sequence fused in frame with 1014 bp of T7RNAP sequence (for recombination); (ii) the PCR product was either methylated in vitro with Dam or not methylated; (iii) the methylated and unmethylated PCR products were transformed into wild-type recipient cells, which did not have the agn43 region, but had dam, oxyR and cfp under the control of PT7 in the chromosome; (iv) successful chromosomal integrations were selected on media with kanamycin; and (v) the percentage of cells with OFF and ON expression was determined by colony counting. As previously mentioned, we compared the percentage of OFF and ON colonies rather than the absolute number of colonies so the results did not depend on whether recipient cells preferentially uptake, cleave and/or integrate methylated or unmethylated DNA. Note: complete in vitro methylation of the PCR product was confirmed using restriction endonucleases (DpnI and MboI) that are sensitive to methylation of the adenine in GATC sites (Supplementary Figure S2).
Figure 3.
In vitro DNA methylation and transformation of the agn43 region. Symbols are defined in Figure 1. (A) Schematic of experimental workflow. Asterisks indicate DNA adenine methylation. All genes are chromosomal except the extracellular PCR product before transformation. # indicates the PCR product only contained the first 1014 base pairs (bp) of T7RNAP. The recipient strain is HL6494 (wild-type). (B) Fluorescence and brightfield (with phase condenser annulus 3) microscopy of colonies. Recipient cells contained a chromosomal copy of yfp that was constitutively transcribed (not shown). Scale bar is 500 μm. Red arrow indicates the location of the OFF colony. (C) Percentage of OFF, ON and mixed colonies. Error bars are the SEM of two biological replicates.
The transformation experiments differ from the transduction experiments in several aspects: (i) there is no agn43 region in recipient cells; (ii) no donor cell factors (e.g. Dam or OxyR) can be horizontally transferred; (iii) the acquired DNA is completely synthesized and methylated in vitro; (iv) the length of the transformed DNA is much shorter than typically acquired by transduction; (v) cfp is on the chromosome at the location of the yfjV’ pseudogene instead of on a plasmid; (vi) recipient cells are in an electrocompetent state instead of exponentially growing; and (vii) cells were grown for 4 h at 30°C post-transformation (instead of 37°C post-transduction).
Transformation of wild-type recipient cells with unmethylated DNA resulted in 97.6 ± 0.0% OFF, 1.4 ± 0.3% ON and 1.0 ± 0.3% mixed colonies (two biological replicates; n = 205), whereas transformation of wild-type recipient cells with methylated DNA resulted in 27.1 ± 5.8% OFF, 69.6 ± 2.9% ON and 3.4 ± 2.9% mixed colonies (two biological replicates; n = 151) (Figure 3B and C). The percentages of OFF, ON and mixed colonies resulting from the transformation of the unmethylated and methylated agn43 region were significantly different (χ2 value = 198.9, n = 356, P < 0.0001). The OFF and ON colony phenotypes were shown to be reversible which is consistent with epigenetic regulation. When streaked out, OFF colonies gave rise to 6.3 ± 1.6% ON colonies (three biological replicates; n = 186) and ON colonies gave rise to 51.6 ± 0.2% OFF colonies (three biological replicates; n = 206).
The above experiments demonstrate that the methylation state of extracellular DNA can be horizontally transferred via transformation to recipient cell chromosomes. The percentage of OFF colonies arising from the HT of unmethylated DNA into wild-type recipient cells was similar in the transformation and transduction experiments (97.6 ± 0.0% and 98.4 ± 1.0% respectively). The percentage of ON cells arising from the HT of methylated DNA into wild-type recipient cells was also similar in the transformation and transduction experiments (69.6 ± 2.9% and 67.1 ± 2.5% respectively). As with the transduction experiments, the results cannot be explained by preferential transformation or survival of recipient cells with unmethylated or methylated extracellular DNA, which would be expected to produce different numbers of colonies but not different percentages of OFF and ON colonies. The transformation experiments show definitively that donor factors other than DNA are not required for the transfer and stability of acquired DNA methylation states.
The observation that transformation and transduction of unmethylated DNA result in almost 100% of recipient colonies being in the OFF state indicates the unmethylated agn43 region can be stably maintained through the entire HGT process. In contrast, transformation and transduction of methylated DNA results in ∼70% of the recipient colonies being in the methylated state. The latter indicates there must be at least one step in the HGT process and/or subsequent rounds of DNA replication where DNA methylation is not maintained and cells switch to the unmethylated OFF state. Furthermore, while it appears that switching from the methylated ON state to the unmethylated OFF state occurs in the same percentage of cells it tends to occur later in the transduction experiments than in the transformation experiments. In the transduction experiments, cells tend to switch to the OFF state at least one cell division after plating resulting in 2.5-fold as many mixed colonies as OFF colonies. In the transformation experiments, cells tend to switch to the OFF state before plating resulting in 8-fold as many OFF colonies as mixed colonies. There are many possible reasons for the difference in the delay in switching from ON to OFF following transformation and transduction including different conditions and media used in the experiments, phage factors, host responses to phage, and the effects of being in an electrocompetent state.
Horizontal transfer of DNA methylation patterns affects cell fitness
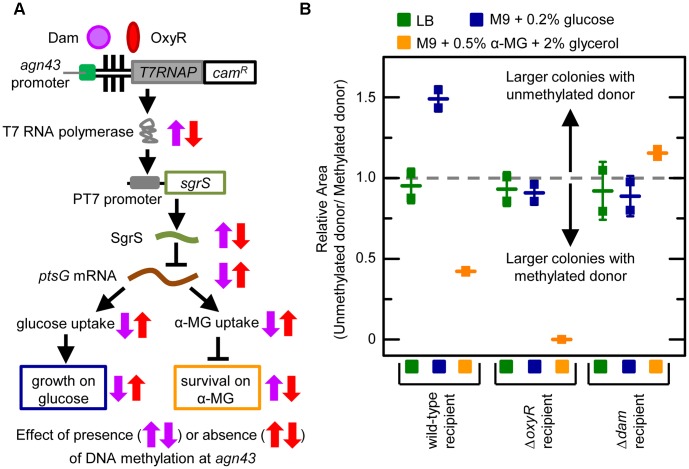
We sought to demonstrate that the acquisition of DNA methylation patterns at a locus can in principle alter cell fitness through its effect on gene expression (Figure 4A). To do this we modified our synthetic system so that DNA methylation patterns in the agn43 region control transcription of T7 RNA polymerase which in turn regulates transcription of the SgrS small RNA (in a strain with native sgrS deleted). SgrS can bind to the ptsG mRNA and decrease its translation and lifetime (30). The ptsG mRNA encodes the EIICBGlc protein that is part of the phosphoenolpyruvate phosphotransferase system (PTS). Because this system transports glucose into the cell (31) it is often referred to as ‘glucose permease’. EIICBGlc also transports other sugars including methyl α-D-glucoside (α-MG), which is a toxic non-metabolizable analogue of glucose that competes for enzymes in glycolytic pathways (32). Therefore, SgrS inhibits glucose and α-MG uptake via its actions on the ptsG mRNA. SgrS is also translated under some conditions resulting in a small protein (SgrT) that directly inhibits glucose and α-MG transport (33,34).
Figure 4.
Horizontal transfer of DNA methylation patterns affects cell fitness. Symbols are defined in Figure 1. (A) Schematic of synthetic circuit and the effect of DNA methylation patterns at the agn43 region on SgrS transcription, glucose and α–MG uptake, cell growth and cell survival. All genes are chromosomal except sgrS which is on a plasmid. Purple arrows indicate whether a chemical species or process in the system increases or decreases with methylation at the agn43 region. Red arrows indicate whether a chemical species or process in the system increases or decreases with no methylation at the agn43 region. (B) Relative area of colonies generated by transduction of unmethylated and methylated donor DNA into recipient cells on different media. Horizontal line symbols indicate the mean relative area (which is unitless) of colonies for each strain and error bars are the SEM. Horizontal dash line indicates a relative area of 1, which occurs when colonies transduced with methylated and unmethylated donor DNA have the same size (i.e. the methylation pattern on the horizontally transferred DNA has no effect on fitness). Transduction of unmethylated and methylated donor DNA into: (i) wild-type recipient cells (HL6866) generated HL6870 and HL6871; (ii) ΔoxyR recipient cells (HL6869) generated HL6876 and HL6877; and (iii) Δdam recipient cells (HL6868) generated HL6874 and HL6875.
We chose to regulate SgrS because its transcription can increase or decrease cell fitness depending on conditions. DNA methylation at the agn43 region was predicted to turn on SgrS transcription causing decreased cell growth and colony size on media containing glucose as the only carbon source (M9 minimal media + 0.2% w/v glucose), and increased cell growth and colony size on media containing α-MG (M9 minimal media + 0.5% w/v α-MG + 2% v/v glycerol) (note: glycerol is a carbon source that is not imported by glucose permease). An unmethylated agn43 region was predicted to not induce SgrS transcription thereby causing increased cell growth and colony size on M9 minimal media + 0.2% w/v glucose and decreased growth and colony size on M9 minimal media + 0.5% w/v α-MG + 2% v/v glycerol. We confirmed all of these predictions in a pilot study (Supplementary Methods and Figure S6).
Having established that methylation patterns at the agn43 region can positively and negatively affect cell fitness through our SgrS system we used it to demonstrate that HT of DNA methylation patterns via transduction into a wild-type recipient cell can affect the fitness of successful transductants. In this experiment, phage from methylated and unmethylated donor cells were transduced into wild-type, Δdam or ΔoxyR recipient cells (which did not contain cfp or yfp). The procedure was similar to that in Figure 2A except that following transduction the cells were plated onto M9 minimal media + 0.2% w/v glucose, M9 minimal media + 0.5% w/v α-MG + 2% v/v glycerol or lysogeny broth (LB) media. LB media was a control for differences in transduction efficiency and growth rate. To measure fitness we calculated the ‘relative colony area’, which is a unitless metric, by taking the mean area of colonies transduced with unmethylated donor DNA and dividing it by the mean area of colonies transduced with methylated donor DNA. This metric is preferable to the absolute number of colonies because: (i) it is independent of donor phage titer and transduction efficiency; (ii) we found it works well for weak selection; and (iii) it only compares fitness in the same background and under the same conditions.
We found that HT of methylated and unmethylated agn43 regions into wild-type recipient cells impacted cell fitness and selection as predicted (Figure 4B). The relative colony area was 1.49 ± 0.06 (two biological replicates, n = 837) when cells were plated on M9 minimal media + 0.2% w/v glucose indicating that cell fitness was greater when wild-type recipient cells were transduced with an unmethylated agn43 region (resulting in no SgrS transcription and no inhibition of glucose uptake). In contrast, the relative colony area was 0.42 ± 0.01 (two biological replicates, n = 409) when cells from the same experiment were plated on M9 minimal media + 0.5% w/v α-MG + 2% v/v glycerol indicating cells were fitter when transduced with a methylated agn43 region (resulting in SgrS transcription and inhibition of α-MG import). The relative colony area was ∼1 (0.95 ± 0.08; two biological replicates, n = 217) when the same cells were plated on LB media indicating there was no fitness advantage for the methylated or unmethylated donor DNA. In control experiments, methylated and unmethylated agn43 regions were transduced into recipient cells with dam or oxyR deleted. In these control experiments, the relative colony areas were ∼ 1, which was expected because recipient cells have the same phenotype regardless of whether the donor DNA is methylated or unmethylated (Figure 4B). The exception was with unmethylated donor DNA transduced into ΔoxyR recipient cells and grown on M9 minimal media + 0.5% w/v α-MG + 2% v/v glycerol; in this case cells were non-viable and the colony size and relative colony area were zero. Therefore the fitness advantages associated with transduction of methylated or unmethylated DNA are not only reversed under different conditions but they also disappear in the controls with alternative carbon sources or no differences in SgrS transcription. Together these results demonstrate that transduction of methylated or unmethylated donor DNA can affect cell fitness and selection due to the effect of HT DNA methylation patterns on the regulation of gene expression.
DISCUSSION
In this study, we showed that DNA methylation patterns at the agn43 cis-regulatory region can be horizontally transferred into bacterial chromosomes by phage and by the transformation of extracellular synthetic DNA. Phage transduction is not believed to transmit donor cell proteins to recipient cells unlike conjugation (35) and in our transformation experiments the DNA was synthesized in vitro and then purified so there were no proteins bound to the DNA. Therefore, our experiments show it is the DNA methylation pattern itself, and not OxyR or some other factors bound to unmethylated or methylated DNA that propagates the methylation state of the agn43 region from donor DNA or extracellular DNA into the chromosome of recipient cells. To our knowledge this is the first time that local DNA methylation patterns have been definitively shown to be horizontally transferred to chromosomes (note: prior studies had only shown the HT of DNA methylation patterns extending as far as the cytoplasm as described in the Introduction). Furthermore, our experiments show the HT of DNA methylation patterns at a cis-regulatory region can program different gene expression states in recipient cells to increase or decrease their fitness in different environments, and that this in turn can cause selection for and against cells that have had HGT. This suggests that in principle, DNA methylation patterns that accompany HGT could contribute to genome evolution (this is discussed in detail further below).
Our experimental system can be adapted to examine the HT of methylation patterns at other DNA sequences by substituting them for the agn43 region, and used to track the spread of HGT and DNA methylation patterns in communities of bacteria including biofilms (36). In addition, our demonstration that in vitro DNA methylation can specify the state of a genetic switch and that this state can then be integrated into the chromosome and then potentially transferred to other cells via inheritance or additional HT could be harnessed in synthetic biology. This process could be used to turn on or off the transcription of acquired or pre-existing genes in recipient cells to create subpopulations of differentiated cell types that are genetically identical. These differentiated cell types can be used to divide the labor of complex and metabolically burdensome processes thereby making individual cells fitter, create communities that are capable of more complex behaviors, decrease the emergence of cheaters and mutants in engineered populations and generate diversity so the population is better able to survive a wider variety of conditions (37–40). One of the advantages of using the agn43 system as a switch to program gene expression states is that the ON and OFF states are determined by DNA methylation, which is a stable covalent modification that can be horizontally transferred to cells by many mechanisms. Other advantages of using the agn43 switch are: (i) the number of regulatory components does not need to increase with the number of switches as with most systems because the state of each switch is independent; (ii) the stability of the expression states can be modulated independently of the transcription levels by altering OxyR and Dam concentrations and/or modifying the cis-regulatory sequence (11,27); and (iii) the capacity for in vitro programming to avoid the use of in vivo induction agents and their effects on cell physiology (41).
Our synthetic system was designed to only test whether it is mechanistically possible for DNA methylation patterns to be horizontally transferred to chromosomes, program gene expression and alter fitness. Care must be taken in attempting to directly extrapolate specific findings to specific natural systems because of differences. For example, we transformed double stranded DNA into the cytoplasm of electrocompetent cells whereas single stranded DNA is translocated into the cytoplasm with natural transformation in many bacteria (4,5). Having said this, there are features that are shared between our experiments and HGT in natural populations, and the general principles arising from our findings are likely to have broad relevance.
As mentioned in the introduction, HGT is most common among bacteria that have similar DNA sequences, proteins and RM systems, and shared ecological niches, which is the scenario examined by our experiments. HGT is less common between distantly related bacteria and organisms but it is certainly not rare. There has been substantial HGT between E. coli and Salmonella after they diverged 100–160 million years ago (42,43) resulting in some genes that are almost identical (44). There is also evidence for HGT between bacteria and archaea, and between bacteria and eukaryotes (2,45–47). In addition to transduction and transformation, HGT can occur via conjugation (48) and it may occur in multiple steps (e.g. novel DNA sequences and their associated methylation patterns could be acquired on plasmid DNA and then transferred to the chromosome via recombination (9)).
DNA methylation regulates the expression of a wide array of genes in many types of bacteria including those involved in the timing of chromosome replication (10,49,50). Some of these genes that are regulated by DNA methylation have discrete alternate expression states that display random switching and heritability; this pattern of expression, which occurs with agn43, is termed ‘phase variation’ and is important in pathogenesis. In some cases, such as the pyelonephritis-associated pili (pap) operon in E. coli, phase variation is directly regulated by Dam. In other cases, it is the production of DNA methylases, which are part of type I, II and III RM systems, that are phase variable. The latter results in phase variable DNA methylation patterns in the genome (and consequently phase variable transcription of some genes) and has been observed in important human pathogens including Neiserria meningitidis and Haemophilus influenzae (51). One explanation for the evolution of phase variable regulation of RM systems is that it enables the temporary turning off of the RM system in a subset of cells to remove it as a barrier to HGT and the acquisition of new traits (52,53). While there is no strong evidence to support this, it is noteworthy that DNA methylation also regulates the expression of genes in other processes that are important for HGT including phage replication and conjugation (10). DNA methylation can alter gene transcription through general effects on DNA by altering its thermostability, bending and steric hindrance (10), and by blocking sigma factor binding (50). Therefore the HT of DNA methylation patterns may affect gene transcription in recipient cells even if they do not have specific transcription factors such as OxyR whose binding is sensitive to DNA methylation.
HGT could possibly transfer DNA methylation patterns between bacteria that not only involve N6-methyladenine (which was examined in this study) but also N4-methylcytosine and 5-methylcytosine bases. Archaea and eukaryotes also have N6-methyladenine and 5-methylcytosine (10) therefore it is conceivable that DNA methylation patterns that are horizontally transferred among bacteria, archaea and eukaryotes could alter gene transcription, especially given the many mechanisms by which DNA methylation can alter transcription (10). However, HT of DNA methylation patterns between distantly related organisms will in most cases probably have only a transient effect because recipient cells are less likely to have DNA methylases and proteins with similar functions to those in donor cells, which are necessary to maintain the acquired patterns.
Any direct or indirect effects of horizontally transferred DNA methylation patterns on the expression of acquired or pre-existing genes can potentially alter cell fitness. Even a fitness advantage lasting less than one generation could under the right circumstances lead to selection as can occur with transient errors in transcription, translation and protein folding (19,20,54). Genotypes that increase the propensity of bacteria for HGT and HT of DNA methylation patterns could be selected for in the same way and for the same reasons as genotypes that increase DNA mutation rates; that is, these genotypes accelerate evolution and increase phenotypic diversity which is beneficial under some conditions (55). The advantages of HT of DNA methylation patterns to recipient cells include: (i) generating alternate transcription profiles from the same acquired DNA sequences; (ii) the ability to easily revert from a new phenotype back to the original phenotype by reversing the epigenetic mechanism (in contrast, it can be difficult to reverse a new phenotype that arises by random mutations) (54); (iii) providing pre-programmed epigenetic instructions so that transcription of acquired genes can begin immediately after transfer; and (iv) turning off the transcription of acquired genes so they do not immediately impact phenotype (but they can be turned on in the future when altering the phenotype is beneficial such as when the environment changes (54,56)). The HT of DNA methylation patterns may also benefit phage through the above effects on host cell fitness resulting in the selection of phage that help mediate HT of DNA methylation patterns and do not alter these patterns with their own methylases (as we observed).
It seems more likely than not that the HT of DNA methylation patterns has had a role in adaptation and evolution in natural systems during the last three billion years, at least under some conditions and in some organisms based on all of the factors mentioned above. That is, HGT is very common among bacteria, DNA methylation patterns readily accompany HGT (as we have shown), DNA methylation patterns can alter gene transcription by many mechanisms, and even transient changes in gene expression caused by altered DNA methylation could affect cell fitness. Because HGT often occurs between bacteria that are genetically similar it follows that acquired DNA sequences will often be the same or very similar to pre-existing sequences in recipient cells thereby making any co-acquired DNA methylation patterns potentially more important in generating alternate phenotypes. To be clear, under most circumstances the HT of DNA methylation patterns is not likely to be more important to adaption and evolution than the acquisition of novel DNA sequences which can have longer and more profound effects on phenotype. However, the HT of DNA methylation patterns and the HT of DNA sequences are not mutually exclusive and the acquisition of both DNA methylation patterns and DNA sequences will likely provide an organism with more information and greater flexibility in creating different phenotypes than acquisition of DNA sequences alone.
The contribution of HT of DNA methylation patterns to adaptation and evolution would be consistent with other mechanisms of storing epigenetic information that are horizontally transferred and can reprogram gene expression in recipient cells (including prions and other macromolecules) (35,57,58), which are also thought to play a role in evolution (59,60). As with these other epigenetic mechanisms, future studies seeking to demonstrate the contribution of HT of DNA methylation patterns to the evolution of specific natural populations will be challenging because DNA methylation patterns and their effects are often transient (59). However, these studies should nonetheless be pursued because they will greatly add to our general understanding of adaptation and evolution via HGT, and because HGT is important (or potentially so) in the spread of antibiotic resistance and pathogenesis, reprogramming the gut microbiome and the development of human diseases (61,62).
In conclusion, our findings provide direct evidence that the HT of DNA methylation patterns into bacterial chromosomes is mechanistically possible. This mechanism could contribute to bacterial adaptation and evolution, and it can be harnessed for novel strategies to program gene expression states in synthetic gene circuits.
Supplementary Material
Acknowledgments
We thank Ayelet Cohen, Dena Block and Katya Frazier for constructing some strains and/or performing preliminary experiments, and we appreciate Razika Hussein's comments on the manuscript. We thank Huanjie Sheng for assistance with the ImageJ software.
Author Contributions: J.-E.S. performed experiments and analyzed data. C.L. performed experiments. H.N.L. designed the project, performed experiments, analyzed data and wrote the paper.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
University of California, Berkeley. Funding for open access charge: Berkeley Research Impact Initiative (BRII) sponsored by the UC Berkeley Library.
Conflict of interest statement. None declared.
REFERENCES
- 1.Popa O., Dagan T. Trends and barriers to lateral gene transfer in prokaryotes. Curr. Opin. Microbiol. 2011;14:615–623. doi: 10.1016/j.mib.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 2.Boto L. Horizontal gene transfer in evolution: facts and challenges. Proc. Biol. Sci. 2009;277:819–827. doi: 10.1098/rspb.2009.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Darmon E., Leach D.R. Bacterial genome instability. Microbiol. Mol. Biol. Rev. 2014;78:1–39. doi: 10.1128/MMBR.00035-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cabezon E., Ripoll-Rozada J., Pena A., de la Cruz F., Arechaga I. Towards an integrated model of bacterial conjugation. FEMS Microbiol. Rev. 2015;39:81–95. doi: 10.1111/1574-6976.12085. [DOI] [PubMed] [Google Scholar]
- 5.Guglielmini J., de la Cruz F., Rocha E.P. Evolution of conjugation and type IV secretion systems. Mol. Biol. Evol. 2012;30:315–331. doi: 10.1093/molbev/mss221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Labrie S.J., Samson J.E., Moineau S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010;8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 7.Marraffini L.A., Sontheimer E.J. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 2010;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorman C.J. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2004;2:391–400. doi: 10.1038/nrmicro883. [DOI] [PubMed] [Google Scholar]
- 9.Thomas C.M., Nielsen K.M. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 2005;3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 10.Marinus M.G., Casadesus J. Roles of DNA adenine methylation in host-pathogen interactions: mismatch repair, transcriptional regulation, and more. FEMS Microbiol. Rev. 2009;33:488–503. doi: 10.1111/j.1574-6976.2008.00159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim H.N., van Oudenaarden A. A multistep epigenetic switch enables the stable inheritance of DNA methylation states. Nat. Genet. 2007;39:269–275. doi: 10.1038/ng1956. [DOI] [PubMed] [Google Scholar]
- 12.Lister R., Ecker J.R. Finding the fifth base: genome-wide sequencing of cytosine methylation. Genome Res. 2009;19:959–966. doi: 10.1101/gr.083451.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loenen W.A., Raleigh E.A. The other face of restriction: modification-dependent enzymes. Nucleic Acids Res. 2014;42:56–69. doi: 10.1093/nar/gkt747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parker B.O., Marinus M.G. Repair of DNA heteroduplexes containing small heterologous sequences in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1992;89:1730–1734. doi: 10.1073/pnas.89.5.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pukkila P.J., Peterson J., Herman G., Modrich P., Meselson M. Effects of high levels of DNA adenine methylation on methyl-directed mismatch repair in Escherichia coli. Genetics. 1983;104:571–582. doi: 10.1093/genetics/104.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beaber J.W., Hochhut B., Waldor M.K. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature. 2004;427:72–74. doi: 10.1038/nature02241. [DOI] [PubMed] [Google Scholar]
- 17.Wiedenbeck J., Cohan F.M. Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol. Rev. 2011;35:957–976. doi: 10.1111/j.1574-6976.2011.00292.x. [DOI] [PubMed] [Google Scholar]
- 18.Smith R., Tan C., Srimani J.K., Pai A., Riccione K.A., Song H., You L. Programmed Allee effect in bacteria causes a tradeoff between population spread and survival. Proc. Natl. Acad. Sci. U.S.A. 2014;111:1969–1974. doi: 10.1073/pnas.1315954111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gordon A.J., Satory D., Halliday J.A., Herman C. Lost in transcription: transient errors in information transfer. Curr. Opin. Microbiol. 2015;24:80–87. doi: 10.1016/j.mib.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drummond D.A., Wilke C.O. The evolutionary consequences of erroneous protein synthesis. Nat. Rev. Genet. 2009;10:715–724. doi: 10.1038/nrg2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.True H.L., Lindquist S.L. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- 22.Tanouchi Y., Pai A., Buchler N.E., You L. Programming stress-induced altruistic death in engineered bacteria. Mol. Syst. Biol. 2013;8:626. doi: 10.1038/msb.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waldron D.E., Owen P., Dorman C.J. Competitive interaction of the OxyR DNA-binding protein and the Dam methylase at the antigen 43 gene regulatory region in Escherichia coli. Mol. Microbiol. 2002;44:509–520. doi: 10.1046/j.1365-2958.2002.02905.x. [DOI] [PubMed] [Google Scholar]
- 24.Haagmans W., van der Woude M. Phase variation of Ag43 in Escherichia coli: Dam-dependent methylation abrogates OxyR binding and OxyR-mediated repression of transcription. Mol. Microbiol. 2000;35:877–887. doi: 10.1046/j.1365-2958.2000.01762.x. [DOI] [PubMed] [Google Scholar]
- 25.Hasman H., Schembri M.A., Klemm P. Antigen 43 and type 1 fimbriae determine colony morphology of Escherichia coli K-12. J. Bacteriol. 2000;182:1089–1095. doi: 10.1128/jb.182.4.1089-1095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Owen P., Meehan M., de Loughry-Doherty H., Henderson I. Phase-variable outer membrane proteins in Escherichia coli. FEMS Immunol. Med. Microbiol. 1996;16:63–76. doi: 10.1111/j.1574-695X.1996.tb00124.x. [DOI] [PubMed] [Google Scholar]
- 27.Wallecha A., Munster V., Correnti J., Chan T., van der Woude M. Dam- and OxyR-dependent phase variation of agn43: essential elements and evidence for a new role of DNA methylation. J. Bacteriol. 2002;184:3338–3347. doi: 10.1128/JB.184.12.3338-3347.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lobocka M.B., Rose D.J., Plunkett G. III, Rusin M., Samojedny A., Lehnherr H., Yarmolinsky M.B., Blattner F.R. Genome of bacteriophage P1. J. Bacteriol. 2004;186:7032–7068. doi: 10.1128/JB.186.21.7032-7068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morita T., Maki K., Yagi M., Aiba H. Analyses of mRNA destabilization and translational inhibition mediated by Hfq-binding small RNAs. Methods Enzymol. 2008;447:359–378. doi: 10.1016/S0076-6879(08)02218-0. [DOI] [PubMed] [Google Scholar]
- 31.Richards G.R., Vanderpool C.K. Induction of the Pho regulon suppresses the growth defect of an Escherichia coli sgrS mutant, connecting phosphate metabolism to the glucose-phosphate stress response. J. Bacteriol. 2012;194:2520–2530. doi: 10.1128/JB.00009-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richards G.R., Patel M.V., Lloyd C.R., Vanderpool C.K. Depletion of glycolytic intermediates plays a key role in glucose-phosphate stress in Escherichia coli. J. Bacteriol. 2013;195:4816–4825. doi: 10.1128/JB.00705-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balasubramanian D., Vanderpool C.K. Deciphering the interplay between two independent functions of the small RNA regulator SgrS in Salmonella. J. Bacteriol. 2013;195:4620–4630. doi: 10.1128/JB.00586-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kosfeld A., Jahreis K. Characterization of the interaction between the small regulatory peptide SgrT and the EIICBGlc of the glucose-phosphotransferase system of E. coli K-12. Metabolites. 2012;2:756–774. doi: 10.3390/metabo2040756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heinemann J.A. Genetic evidence of protein transfer during bacterial conjugation. Plasmid. 1999;41:240–247. doi: 10.1006/plas.1999.1392. [DOI] [PubMed] [Google Scholar]
- 36.Madsen J.S., Burmolle M., Hansen L.H., Sorensen S.J. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol. Med. Microbiol. 2012;65:183–195. doi: 10.1111/j.1574-695X.2012.00960.x. [DOI] [PubMed] [Google Scholar]
- 37.Wintermute E.H., Silver P.A. Emergent cooperation in microbial metabolism. Mol. Syst. Biol. 2010;6:407. doi: 10.1038/msb.2010.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rossetti V., Bagheri H.C. Advantages of the division of labour for the long-term population dynamics of cyanobacteria at different latitudes. Proc. Biol. Sci. 2012;279:3457–3466. doi: 10.1098/rspb.2012.0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mee M.T., Wang H.H. Engineering ecosystems and synthetic ecologies. Mol. Biosyst. 2012;8:2470–2483. doi: 10.1039/c2mb25133g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brenner K., You L., Arnold F.H. Engineering microbial consortia: a new frontier in synthetic biology. Trends Biotechnol. 2008;26:483–489. doi: 10.1016/j.tibtech.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 41.Kosinski M., Rinas U., Bailey J. Isoprpoyl-B-D-thiogalactopyranoside influences the metabolism of Escherichia coli. Appl. Microbiol. Biotechnol. 1992;36:782–784. [Google Scholar]
- 42.Karberg K.A., Olsen G.J., Davis J.J. Similarity of genes horizontally acquired by Escherichia coli and Salmonella enterica is evidence of a supraspecies pangenome. Proc. Natl. Acad. Sci. U.S.A. 2011;108:20154–20159. doi: 10.1073/pnas.1109451108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gordienko E.N., Kazanov M.D., Gelfand M.S. Evolution of pan-genomes of Escherichia coli, Shigella spp., and Salmonella enterica. J. Bacteriol. 2013;195:2786–2792. doi: 10.1128/JB.02285-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blattner F.R., Plunkett G. 3rd, Bloch C.A., Perna N.T., Burland V., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 45.Heinemann J.A., Sprague G.F., Jr Transmission of plasmid DNA to yeast by conjugation with bacteria. Methods Enzymol. 1991;194:187–195. doi: 10.1016/0076-6879(91)94016-6. [DOI] [PubMed] [Google Scholar]
- 46.Bock R. The give-and-take of DNA: horizontal gene transfer in plants. Trends Plant Sci. 2009;15:11–22. doi: 10.1016/j.tplants.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 47.Dunning Hotopp J.C. Horizontal gene transfer between bacteria and animals. Trends Genet. 2011;27:157–163. doi: 10.1016/j.tig.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adelberg E.A., Pittard J. Chromosome Transfer in bacterial conjugation. Bacteriol. Rev. 1965;29:161–172. doi: 10.1128/br.29.2.161-172.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robbins-Manke J.L., Zdraveski Z.Z., Marinus M., Essigmann J.M. Analysis of global gene expression and double-strand-break formation in DNA adenine methyltransferase- and mismatch repair-deficient Escherichia coli. J. Bacteriol. 2005;187:7027–7037. doi: 10.1128/JB.187.20.7027-7037.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shell S.S., Prestwich E.G., Baek S.H., Shah R.R., Sassetti C.M., Dedon P.C., Fortune S.M. DNA methylation impacts gene expression and ensures hypoxic survival of Mycobacterium tuberculosis. PLoS Pathog. 2013;9:e1003419. doi: 10.1371/journal.ppat.1003419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Srikhanta Y.N., Fox K.L., Jennings M.P. The phasevarion: phase variation of type III DNA methyltransferases controls coordinated switching in multiple genes. Nat. Rev. Microbiol. 2010;8:196–206. doi: 10.1038/nrmicro2283. [DOI] [PubMed] [Google Scholar]
- 52.Fox K.L., Dowideit S.J., Erwin A.L., Srikhanta Y.N., Smith A.L., Jennings M.P. Haemophilus influenzae phasevarions have evolved from type III DNA restriction systems into epigenetic regulators of gene expression. Nucleic Acids Res. 2007;35:5242–5252. doi: 10.1093/nar/gkm571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hallet B. Playing Dr Jekyll and Mr Hyde: combined mechanisms of phase variation in bacteria. Curr. Opin. Microbiol. 2001;4:570–581. doi: 10.1016/s1369-5274(00)00253-8. [DOI] [PubMed] [Google Scholar]
- 54.Herman J.J., Spencer H.G., Donohue K., Sultan S.E. How stable 'should' epigenetic modifications be? Insights from adaptive plasticity and bet hedging. Evolution. 2013;68:632–643. doi: 10.1111/evo.12324. [DOI] [PubMed] [Google Scholar]
- 55.Denamur E., Matic I. Evolution of mutation rates in bacteria. Mol. Microbiol. 2006;60:820–827. doi: 10.1111/j.1365-2958.2006.05150.x. [DOI] [PubMed] [Google Scholar]
- 56.Veening J.W., Stewart E.J., Berngruber T.W., Taddei F., Kuipers O.P., Hamoen L.W. Bet-hedging and epigenetic inheritance in bacterial cell development. Proc. Natl. Acad. Sci. U.S.A. 2008;105:4393–4398. doi: 10.1073/pnas.0700463105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Christie P.J. Type IV secretion: intercellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol. Microbiol. 2001;40:294–305. doi: 10.1046/j.1365-2958.2001.02302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wickner R.B., Edskes H.K., Roberts B.T., Baxa U., Pierce M.M., Ross E.D., Brachmann A. Prions: proteins as genes and infectious entities. Genes Dev. 2004;18:470–485. doi: 10.1101/gad.1177104. [DOI] [PubMed] [Google Scholar]
- 59.Heinemann J.A., Roughan P.D. New hypotheses on the material nature of horizontally mobile genes. Ann. N.Y. Acad. Sci. 2000;906:169–186. doi: 10.1111/j.1749-6632.2000.tb06609.x. [DOI] [PubMed] [Google Scholar]
- 60.Soto C. Transmissible proteins: expanding the prion heresy. Cell. 2012;149:968–977. doi: 10.1016/j.cell.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robinson K.M., Sieber K.B., Dunning Hotopp J.C. A review of bacteria-animal lateral gene transfer may inform our understanding of diseases like cancer. PLoS Genet. 2013;9:e1003877. doi: 10.1371/journal.pgen.1003877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brown J.M., Hazen S.L. The gut microbial endocrine organ: bacterially derived signals driving cardiometabolic diseases. Annu. Rev. Med. 2015;66:343–359. doi: 10.1146/annurev-med-060513-093205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.