Abstract
Epigenetic modifications are highlighted for their great importance in regulating plant development, but their function associated with single-cell differentiation remains undetermined. Here, we used the cotton fibre, which is the epidermal hair on the cotton ovule, as a model to investigate the regulatory role of DNA methylation in cell differentiation. The level of CHH (H = A, T, or C) DNA methylation level was found to increase during fibre development, accompanied by a decrease in RNA-directed DNA methylation (RdDM). Examination of nucleosome positioning revealed a gradual transition from euchromatin to heterochromatin for chromatin dynamics in developing fibres, which could shape the DNA methylation landscape. The observed increase in DNA methylation in fibres, compared with other ovule tissue, was demonstrated to be mediated predominantly by an active H3K9me2-dependent pathway rather than the RdDM pathway, which was inactive. Furthermore, integrated multi-omics analyses revealed that dynamic DNA methylation played a role in the regulation of lipid biosynthesis and spatio-temporal modulation of reactive oxygen species during fibre differentiation. Our study illustrates two divergent pathways mediating a continuous increase of DNA methylation and also sheds further light on the epigenetic basis for single-cell differentiation in plants. These data and analyses are made available to the wider research community through a comprehensive web portal.
INTRODUCTION
DNA methylation is the most extensively studied heritable epigenetic modification in eukaryotes. In plants, DNA methylation occurs in the symmetric CG and CHG (H = A, T or C) contexts and also in the asymmetric CHH context (1). Classically, DNA methylation is crucial for transposable element (TE) silencing, genome imprinting and heterosis (2–4). Recent studies have also demonstrated that DNA methylation plays a vital role in plant development, including gametogenesis (5), flower and seed development (6–7), fruit ripening (8), and stress responses (9).
In Arabidopsis, genome-wide mapping and functional studies have uncovered pathways mediating DNA methylation (10). The de novo establishment of DNA methylation in all sequence contexts is catalysed by the DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2) protein and maintained by different pathways. CG methylation is maintained by METHYLTRANSFERASE 1 (MET1) (1). Non-CG DNA methylation (CHG and CHH) patterns are mediated by DRM2 through the RNA-directed DNA methylation (RdDM) pathway (11) and also by CHROMOMETHYLASE 2/3 (CMT2/3) proteins through linker histone H1 and DECREASE-IN-DNA-METHYLATION 1 (DDM1) activity in a self-reinforcing loop (12). The main roles of CMT2 and CMT3 are to methylate CHH and CHG sites respectively, but CMT2 can also methylate CHG sites in association with CMT3 in a partially redundant manner (13). Both DRM2 and CMT2/3-mediated methylation patterns are cooperatively regulated by dimethylated H3K9 (H3K9me2) (14). RdDM involves 24-nt small interfering RNAs (siRNAs) and preferentially functions in gene-rich regions, short TEs and ends of long TEs, while CMT2/3-mediated methylation predominantly occurs in heterochromatic regions, especially in long TEs (12).
Epidermal hairs, differentiated from epidermal cells, are widely present in flowering plants. They are of great importance in plant adaptation, and aid in the resistance to insect herbivory (represented by trichomes), the absorption of nutrients and water (represented by root hairs), and in seed dispersal (such as for cotton fibres) (15). Cotton (Gossypium spp.) is cultivated in more than 80 countries for the utilization of its single-cell fibres as the most important natural renewable sources for textiles. The most widely cultivated allotetraploid cotton species are Gossypium hirsutum and Gossypium barbadense, among which G. hirsutum contributes to approximately 95% of world cotton production, whereas G. barbadense is also exploited somewhere for its superior length, strength, and fineness of fibres. Generally, cotton fibre undergoes four overlapping stages of development to form a mature lint fibre: initiation, elongation, secondary cell wall biosynthesis and maturation (16). A recent study divided this developmental process into five stages by introducing a transition stage between the rapid elongation and secondary cell wall biosynthesis stages (17). Due to progressive features of its development, the cotton fibre is regarded as an excellent experimental system for studying cell differentiation, including cell fate determination, polarized cell elongation and cell wall synthesis in plants (17).
Many studies have achieved a theoretical understanding of the molecular mechanisms controlling epidermal patterning (18–20). In cotton, systematic functional studies also have identified several important genes that contribute to fibre initiation and elongation (21–24). However, limited studies have focused on the epigenetic regulation of cotton fibre development, or epidermal patterning in Arabidopsis (25,26). Therefore, systematic maps of the epigenome are necessary to explore the intriguing biological question of how the architecture of the allotetraploid genome of cotton is organized at each stage of fibre development.
Here, we present integrated maps of DNA methylation and chromatin dynamics of developing cotton fibres. With these dynamic maps, we analysed the regulatory roles of asymmetric DNA methylation in contributing to homoeologous gene expression bias, and also examined a possible role for dynamic DNA methylation in the spatiotemporal modulation of reactive oxygen species during fibre development. This study used Gossypium barbadense, given its recently published genome and superior quality fibre (27). Our study opens up the way for understanding the role of DNA methylation during cotton fibre development and provides novel insights into the epigenetic regulation of single-cell differentiation in plants.
MATERIALS AND METHODS
Plant material
Cotton (Gossypium barbadense L. cv 3–79) plants were cultivated in a field in Wuhan, China, under normal farming conditions. To collect cotton fibre samples, flowers were tagged on the day of blooming (0 day post anthesis (DPA)). Ovules (0 DPA) and fibres (10 DPA, 20 DPA and 30 DPA) were carefully excised from developing cotton bolls. Samples from different plants were pooled and immediately stored at –70°C until further use.
In vitro ovule culture and treatment
Cotton bolls at 2 DPA were harvested from the field, disinfected with 0.1% (w/v) HgCl2 for 15 min and washed three times in sterilized distilled water. Ovules were carefully excised from the bolls under sterile condition and immediately floated in liquid BT medium (50 ml) with optimized phytohormone concentrations (0.5 μM GA3, 5 μM IAA) (28). Zebularine (Selleck, S7113), a DNA methylation inhibitor, was added to the liquid BT medium for treatment (29). The ovules were cultured at 30°C in the dark without agitation (28). The cultured ovules (5 DPA) and fibres (10 DPA and 20 DPA) were sampled for RNA sequencing.
Bisulfite-treated DNA library construction and sequencing
Genomic DNA was extracted from cotton ovules and fibres using the CTAB method (30). For each sample, a total of 2 μg DNA was fragmented to 200–500 bp by sonication. DNA libraries were constructed using the Illumina TruSeq DNA Sample Prep Kit following the manufacturer's instructions with minor modifications necessary for the bisulfite treatment. Briefly, the fragmented DNA was processed by repairing ends, adenylating 3′ ends and ligating adapters. Adapter-ligated DNA fragments were then treated with bisulfite, followed by 10 cycles of PCR amplification. Finally, the bisulfite-treated PCR products were purified using AMPure XP Beads. Bisulfite conversion of DNA samples was conducted using the EZ DNA Methylation-Gold™ Kit (Catalog No. D5005). The unmethylated lambda DNA (Promega) was converted simultaneously as a control. The paired-end sequencing of bisulfite-treated DNA libraries was performed using the Illumina HiSeq 2000 system (paired-end 100-bp reads).
Bisulfite-treated DNA sequencing data analysis
The raw bisulfite-treated DNA sequencing data were filtered by clipping adapters and low-quality reads were trimmed using Trimmomatic (version 0.32) (31). The clean reads were aligned to the G. barbadense genome using Bismark (version 0.13.0) software under the following parameters (-N 1, -L 30) (32). The uniquely mapped reads were retained to extract potentially methylated cytosines using the Bismark methylation extractor under standard parameters. The bisulfite non-conversion rate (0.004) was calculated by sequencing the bisulfite-converted lambda DNA (Promega). Methylated cytosines covered by at least three reads were identified using binomial distribution (P-value cutoff 1e−5). The weighted methylation levels in CG, CHG and CHH contexts per gene were calculated by programming customized Perl scripts. Differentially methylated regions (DMRs) were identified using Fisher's exact test (false discovery rate (FDR) < 0.01) in 200 bp windows of the G. barbadense genome. Windows containing less than 10 methylated cytosines were removed.
Stranded RNA-Seq and data analysis
Total RNA was isolated from ovule and fibre samples as previously described (33). RNA-Seq libraries were constructed using the Illumina TruSeq Stranded RNA Kit (Illumina, San Diego, CA, USA) following the manufacturer's recommendations and were sequenced with the Illumina HiSeq 2000 system (paired-end 100-bp reads) with two biological replicates. The clean RNA-Seq reads were mapped to the G. barbadense genome using Tophat (with -G parameter) (34). The uniquely mapped reads were extracted for estimating the expression levels of protein-coding genes and long non-coding RNAs using Cufflinks (35). The Cuffdiff procedure was followed to identify differentially expressed genes (FDR < 0.01) (35).
Small RNA-Seq and data analysis
The small RNA-Seq libraries were constructed using the Illumina TruSeq Small RNA Library Preparation Kit (Illumina, San Diego, CA, USA) and were sequenced with two biological replicates. After adapter clipping, the cleaned reads (18–26 bp) were filtered for tRNA, rRNA, snRNA, snoRNA and putative miRNAs. The remaining 24-nt siRNA reads were mapped to the G. barbadense genome using Bowtie (version 1.1.1) allowing for no mismatches with a maximum of mapping 200 times (36). The uniquely mapped reads were retained for further analysis. A 24-nt siRNA cluster was defined as a region containing at least ten distinct siRNA reads, and each was separated from the nearest neighbor by a maximum of 200 nucleotides (37).
MNase digestion of chromatin
The ovule and fibre samples were ground into powder with liquid nitrogen. A total of 1 g ovule at 0 DPA and 50, 100 and 150 g fibres at 10 DPA, 20 DPA and 30 DPA, respectively, were used for chromatin extraction. The MNase digestion of chromatin was conducted according to Gent et al. (2014) with minor modifications (38). Briefly, samples were resuspended in nucleus isolation buffer (containing 0.4% triton X-100 instead of 0.5% Tween 40) and gently shaken for 20 min. Then, samples were filtered twice with two layers of Miracloth (Calbiochem, 475855-1R) and centrifuged at 1500 x g for 6 min at 4°C. The pelleted nuclei were washed with isolation buffer without triton X-100. The collected nuclei were resuspended with 600 μl 1× MNase buffer before MNase (NEB; M2047S) digestion. All the steps above were conducted on ice or at 4°C. Half of the nuclei of each sample were used as a control. The remaining half nuclei were added to MNase at a final concentration of ∼4 gel units/μl. Both the control and MNase-treated nuclei were incubated at 37°C for 5 min. Histone removal and DNA purification were performed immediately on the incubated samples. Finally, the precipitated DNA was dissolved in 30 μL TE buffer. All purified DNA samples were treated with 40 μg/ml RNase A. A portion of each sample was kept for gel electrophoresis (2% agarose). The digested samples were then purified with AMPure XP Beads (Beckman Coulter; A63881), and 100–200 bp fragments were isolated with a Pippin HT (Sage Science, Beverly, MA, USA).
Chromatin immunoprecipitation (ChIP)
Chromatin was extracted as described (39), with the same amount of sample for each as in the MNase digestion experiment. To obtain 200–500 bp fragments, the extracted chromatin was sonicated for a total time of 35 min with a 30 s on and 30 s off cycle on ice, except for the 0 DPA ovule sample, which had a total of 5 min of sonication. ChIP was performed using the antibody H3K9me2 (Abcam; ab1220). The antibody was cross-linked with Dynabeads® protein A (Life Technologies; Lot#165116310) and was added into the sonicated samples for immunoprecipitation. For each sample, 20 μl supernatant was kept as an input control. The input and immunoprecipitated DNA were then de-cross-linked with sodium chloride at 65°C overnight and were further treated with protease K and RNase A. Finally, the DNA was precipitated with absolute ethanol, sodium acetate and glycogen. For each sample, two biological replicates were prepared.
MNase-Seq, ChIP-Seq and data analysis
The isolated fragments in MNase digestion and precipitated DNA in ChIP were used for library construction using the Illumina TruSeq Sample Prep Kit following the manufacturer's recommendations. For each sample, a total of 3 ng DNA and Input control DNA were used for library construction. MNase-Seq and ChIP-Seq were performed with the Illumina HiSeq 3000 system (paired-end 150-bp reads). After clipping adapters and trimming low-quality reads, the clean sequencing reads were mapped to the G. barbadense genome using Bowtie2 (version 2.2.4) (40). After potential PCR duplicates were removed, MACS software (version 2.1.0) was used to call histone modification peaks with the default parameters (FDR < 0.05) (41). The genes (including upstream 2 kb and gene body regions) overlapping with identified peaks were considered to have epigenetic modifications.
Quantitative RT-PCR
Quantitative RT-PCR was performed as previously described (42). The expression levels were normalized to Ubiquitin 7. Three biological replicates were performed for each gene.
Data deposition
The raw bisulfite-treated DNA sequencing data, stranded mRNA-Seq data, small RNA-Seq data, MNase-Seq data and ChIP-Seq data have been submitted to the NCBI Sequence Read Archive under the Bioproject ID PRJNA266265.
RESULTS
Global disruption of DNA methylation depresses cotton fibre development
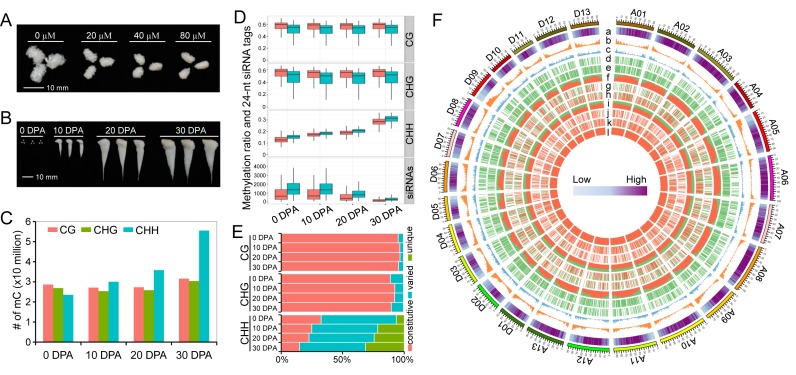
The relationship between DNA methylation status and cotton fibre development has not been well studied. To investigate this phenomenon, we treated ovule cultures with zebularine, a long half-life inhibitor of DNA (cytosine-5) methyltransferases. We observed an increasingly inhibitory effect on fibre development with increasing concentration of zebularine (Figure 1A), which supports the hypothesis that DNA methylation contributes to the regulation of cotton fibre development (25,26).
Figure 1.
Identification of methylated cytosines and DNA methylation dynamics during cotton fibre development. (A) Zebularine treatment suppresses fibre development in vitro. This figure shows that 0 day post anthesis (DPA) ovules cultured for 10 days exhibit different fibre lengths. Different concentrations of zebularine (20, 40 and 80 μM) were applied. (B) Samples used for genome-wide methylation profiling. The samples collected from field include ovules at 0 DPA and fibres at 10 DPA, 20 DPA and 30 DPA. (C) Identification of methylated cytosines in each of the four samples. (D) Comparisons of methylation levels and 24-nt siRNAs between At (shown in red boxplots) and Dt (shown in green boxplots) subgenomes. For the calculation of methylation and siRNA levels, chromosomes from the At and Dt subgenomes were divided into 1 Mb windows that slide 200 kb. (E) Comparison of differentially methylated cytosines in CG, CHG and CHH contexts during fibre development. Methylated cytosines that are detected in all the four stages are termed constitutive methylcytosines. Methylated cytosines detected in two or three stages are termed varied cytosines. Methylated cytosines detected in only one stage are termed as unique cytosines. (F) A circos plot shows the methylation dynamics between two consecutive stages. The outer track represents the 26 chromosomes in the G. barbadense genome (A01-A13 of At subgenome and D01-D13 of Dt subgenome). The other histogram tracks represent the TE content (a); the number of protein-coding genes (b); the number of lincRNAs (c); differentially methylated regions (DMRs) between fibres at 10 DPA and ovules at 0 DPA in CG (d), CHG (e) and CHH (f) contexts; DMRs between fibres at 20 DPA and fibres at 10 DPA in CG (g), CHG (h) and CHH (i) contexts; and DMRs between fibres at 30 DPA and fibres at 20 DPA in CG (j), CHG (k) and CHH (l) contexts. The data for each chromosome were analysed in 1 Mb windows sliding 200 kb. For tracks d-l, DMR in each windows was determined as either hypermethylated DMR or hypomethylated DMR in consecutive developmental stages. The ratio of hypermethylated DMRs against hypomethylated DMRs is indicated by the ratio of red against green.
To further explore the effect of zebularine treatment on cotton fibre development, we sequenced the small RNAs and transcriptomes of cultured ovules at 5 days post anthesis (DPA) and fibres at 10 and 20 DPA (Supplementary Tables S1 and S2). An overall decrease in the levels of 24-nt siRNAs was observed in each treated sample (Supplementary Figure S1A), and we identified 24-nt siRNA clusters that were less abundant than those in controls (Supplementary Figure S1B). The expression patterns and levels of genes and transposable elements (TEs) that overlapped with the differentially enriched siRNA clusters were analysed to inverstigate their possible direct regulation by the RdDM pathway. The results showed that the overlapping protein-coding genes were generally expressed at higher levels at 5 DPA, while the expression of long intergenic non-coding RNAs (lincRNAs) and TEs was dramatically increased after zebularine treatment in all the three stages (Supplementary Figure S1C). In addition, the numbers of up- and down-regulated protein-coding genes were comparable in the three developmental stages but the numbers of up-regulated lincRNAs and TEs were relatively high (Supplementary Figure S1D), consistent with our previous study (43). These findings showed that global disruption of DNA methylation re-activated a large number of genomic loci and suppressed cotton fibre development, suggesting a close relationship between DNA methylation and fibre development. This prompted us to analyse DNA methylation systematically.
Single-base resolution maps of DNA methylation
To investigate the role of DNA methylation, we carried out whole-genome bisulfite sequencing to construct single-base resolution maps. Samples at four developmental stages were selected (Figure 1B), representing cotton fibre initiation (ovule at 0 DPA), rapid elongation (fibre at 10 DPA), transition to secondary cell wall (SCW) synthesis (fibre at 20 DPA) and SCW synthesis (fibre at 30 DPA), respectively. We obtained a coverage of at least 20× paired-end sequencing reads for each sample and the clean data were uniquely aligned to the Gossypium barbadense genome to identify methylcytosines (mCs; Supplementary Table S3).
We identified ∼79–117 million mCs from the four samples (Figure 1C). Among these mCs, 28-31 million mCs were in a CG context, 26–30 million were in a CHG context and 23-56 million were in a CHH context. We observed that mCs in the CG and CHG contexts remained approximately constitutive, while the number of mCs in the CHH context increased from the cotton fibre initiation stage to the SCW synthesis stage. A chromosome-scale view of methylation levels in all three contexts showed that mCs were enriched predominantly in pericentromeric regions (Supplementary Figure S2) (44).
We then compared overall methylation levels between the At and Dt subgenomes. The CG and CHG methylation levels of the At subgenome were remarkably higher than in the Dt subgenome, whereas the opposite situation was observed for CHH methylation (Figure 1D). Consistent with CHH DNA methylation, the enrichment of 24-nt siRNAs in the At subgenome was also lower than in the Dt subgenome at all four developmental stages. Typically, the CHH methylation level increased during fibre development, but the enrichment of 24-nt siRNAs decreased (Figure 1D and Supplementary Figure S3). A similar decreasing pattern of 24-nt siRNAs was observed in ovule cultures described here (Supplementary Figure S1A) and in previous studies (45,46). A comprehensive publicly available web portal has been produced for data interrogation (http://cotton.cropdb.org/cgi-bin/gb2/gbrowse/Gbmethy/).
DNA methylation dynamics during cotton fibre development
Based on the DNA methylation maps, our data provide insights into the regulatory roles in consecutive developmental stages in the cotton fibre. We first investigated the differential methylation of cytosines (DMCs) at different stages. The results showed that more than 95% of cytosines in the CG context of all four stages were constitutively methylated, while 89% to 93% of the cytosines were methylated in the CHG context (Figure 1E). The number of mCs in the CHH context varied, with only 15% (30 DPA) to 33% (0 DPA) constitutive mCs at all stages and 6% (0 DPA) to 32% (30 DPA) stage-specific methylated CHH sites (Figure 1E).
We next identified differentially methylated regions (DMRs) between two consecutive developmental stages. Consistent with the DMC results, the numbers of the CHH DMRs (95 528–254 129) of the three comparisons were much larger than those of the CG DMRs (1480–1734) and CHG DMRs (2641–3451) (Supplementary Figure S4). An analysis of hypo- and hyper-DMRs showed that the majority of DMRs in CG and CHG methylation contexts were hypo-methylated in 10 DPA fibres when compared with 0 DPA ovules (Figure 1F and Supplementary Figure S4). The numbers of hypo- and hyper-DMRs in the CG and CHG contexts in 20 DPA and 10 DPA fibres were comparable, and hyper-DMRs represented the largest group when 30 DPA fibres were compared with 20 DPA fibres. However, the majority of DMRs in the CHH context across the three comparisons were hyper-methylated (Figure 1F and Supplementary Figure S4). Hyper-DMRs in the CHH context were preferentially distributed in TE-rich regions, peaking in pericentromeric heterochromatic regions in 10 DPA fibres and 0 DPA ovules (Supplementary Figure S5), suggesting the distinct methylation patterns between fibres and ovules at the chromosomal level (Figure 1F).
DMR overlapping genes may regulate differential gene expression during developmental stage transition in an epigenetic manner. In this study, only 163–182 and 208–226 protein-coding genes were obtained from CG and CHG context-specific DMRs, respectively (Supplementary Figure S6). Much larger numbers of genes (4144–6792) in CHH context-specific DMRs were identified, consistent with the remarkable methylation differences in the CHH context across consecutive developmental stages. Considering the pattern of increasing CHH DNA methylation levels and differences between euchromatic and heterochromatic regions, we next investigated whether DNA methylation increase related to chromatin dynamics and in turn to cotton fibre development.
DNA hypermethylation relates to a gradual transition from euchromatin to heterochromatin
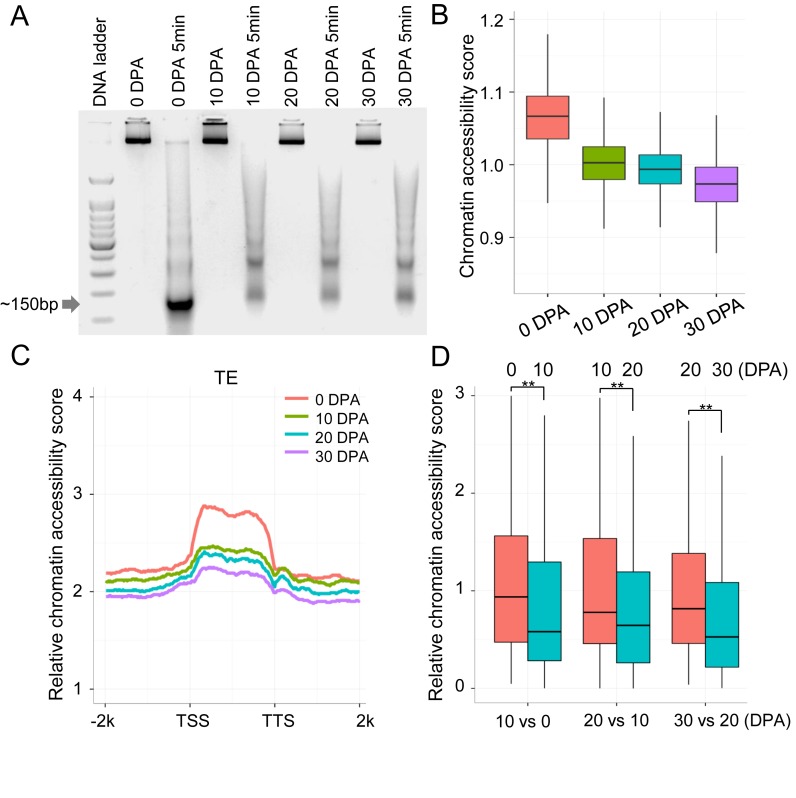
We were interested in determining whether DNA methylation related to chromatin dynamics. To explore the relationship between DNA methylation and heterochromatinization, we measured chromatin accessibility with micrococcal nuclease (MNase), which possesses the ability to digest linker DNA that is not bound to nucleosomes. Theoretically, euchromatic nucleosomes are rapidly released and heterochromatic nucleosomes generally show strong resistance to MNase digestion, indicative of a condensed chromatin state (47). We digested chromatin at the four developmental stages and isolated bound DNA fragments from single nucleosomes for MNase-Seq (Figure 2A).
Figure 2.
Chromatin accessibility revealed by MNase-Seq. (A) MNase digestion of chromatin and single nucleosome-bound DNA fragments (∼150 bp) were excised for sequencing. (B) The relative chromatin accessibility in pericentromeric regions of chromosomes. Each pericentromeric region was divided into 100 kb windows. The number of MNase-Seq reads for each window was compared with genome coverage of the control sample. (C) Relative chromatin accessibility of long TEs (>4 kb). (D) Relative chromatin accessibility of hypermethylated regions in CHH context between fibres at 10 DPA and ovules at 0 DPA, between fibres at 20 DPA and fibres at 10 DPA, and between fibres at 30 DPA and fibres at 20 DPA (**P-value < 0.001).
Investigation of nucleosome positioning provided evidence for chromatin dynamics in fibre development. Clean MNase-Seq reads were compared with the genome coverage of each control sample to obtain high-confidence, accessible nucleosome maps (Supplementary Table S4). Comparisons of pericentromeric chromatin accessibility between consecutive developmental stages showed a remarkable drop-down pattern, suggesting that heterochromatin increased during cotton fibre development (Figure 2B). Then, we compared the relative chromatin accessibility of TE regions among developmental stages, which was normalized using the chromatin accessibility of pericentromeric regions in each developmental stage. Consistent with the observation in pericentromeric regions, the chromatin of ovules at 0 DPA exhibited the highest accessibility and fibres at 30 DPA exhibited the lowest accessibility (Figure 2C).
The relationship between DNA methylation and chromatin accessibility was explored. As expected, chromatin accessibility was broadly negatively correlated with DNA methylation at the whole genome level (Figure 3A). We analysed the chromatin accessibility of CHH hyper-DMRs, which showed the vast majority of DNA methylation dynamics. The result showed that hypermethylated regions in consecutive developmental stages corresponded to lower chromatin accessibility (Wilcoxon rank sum test, P-value < 2.2e−16; Figure 2D). These observations suggest that a genome-wide increase of DNA methylation might be related to a gradual transition from euchromatin to heterochromatin during cotton fibre development.
Figure 3.
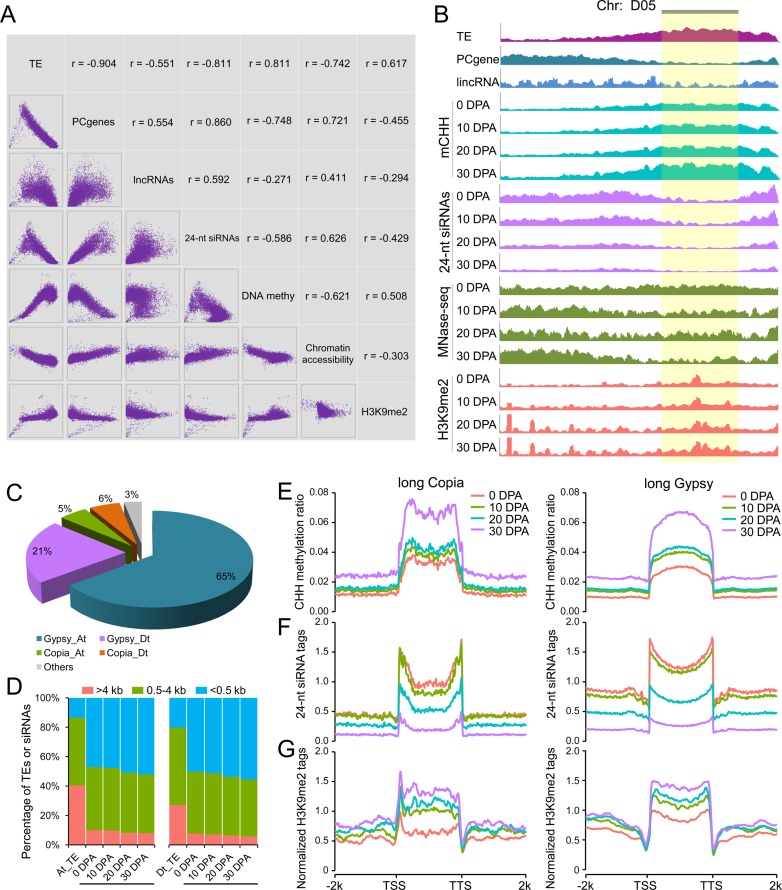
H3K9me2-dependent pathway contributes to CHH hypermethylation in long TEs. (A) Correlation matrix between H3K9me2 modification patterns with TE content, number of protein-coding genes (PCgenes), number of lincRNAs, number of 24-nt siRNAs, DNA methylation level and chromatin accessibility. Each type of data was measured in 1 Mb windows sliding 200 kb. x and y axes of scatter plots under the diagonal represent the normalized data (log2) in each window. The corresponding pearson correlation for each comparison is indicated above the diagonal. (B) Chromosome landscape (D05) showing several genomic features and H3K9me2 modification patterns. Each type of data was normalized to the same level for comparison between consecutive stages. The pericentromeric region is shown with a gold background. (C) Proportions of Copia and Gypsy long terminal repeat (LTR) retrotransposons occupying long TEs (>4 kb) in the At and Dt subgenomes. (D) Distribution of uniquely mapped 24-nt siRNAs in long TEs (>4 kb, red color), middle-sized TEs (0.5–4 kb, green color) and short TEs (<0.5 kb, blue color). (E) Patterns of CHH methylation level in long Copia and long Gypsy TEs from upstream 2 kb to downstream 2 kb. (F) Patterns of 24-nt siRNA enrichment in long Copia and long Gypsy TEs from upstream 2 kb to downstream 2 kb. (G) Patterns of H3K9me2 ChIP-Seq modification marks in long Copia and long Gypsy TEs from upstream 2 kb to downstream 2 kb.
H3K9me2-dependent pathway predominantly mediates heterochromatic DNA hypermethylation
To understand how DNA methylation increases, we explored the activity of pathways mediating DNA methylation, including RdDM and H3K9me2-dependent pathways. We first analysed the relationship between the RdDM pathway and DNA methylation dynamics. An analysis of the enrichment of 24-nt siRNAs in hyper-DMRs showed that the majority of DMRs were not correlated with the dynamics of 24-nt siRNAs (Supplementary Figure S7), which might relate to the negative correlation between genome-wide enrichment of 24-nt RNAs and levels of DNA methylation (Figure 3A). The overall content of 24-nt siRNAs decreased but the level of DNA methylation increased during cotton fibre development. These results imply that the RdDM pathway might perform a limited role in DNA hypermethylation in fibres, as observed by down-regulated expression of genes in this pathway (Supplementary Figure S8).
To investigate whether the H3K9me2-dependent pathway mediates DNA methylation independent of the RdDM pathway, we performed chromatin immunoprecipitation followed by massively parallel sequencing (ChIP-Seq) of H3K9me2 across the four developmental stages (Supplementary Table S5). Genome-wide correlation analysis showed that the enrichment of H3K9me2 modification marks was negatively correlated with gene density and 24-nt siRNAs, but was positively correlated with TE density, DNA methylation and inaccessibility of chromatin (Figure 3A). The chromosome-scale view of these genomic features showed a clear increase in the enrichment of H3K9me2 modification marks consistent with patterns of DNA methylation (Figure 3B), especially in pericentromeric heterochromatin regions as described in previous studies (8,12,13). Considering the genome-wide preference of H3K9me2 modification and the observed chromatin dynamics, we next examined the putative role of the H3K9me2-dependent pathway in mediating DNA methylation increase in heterochromatic regions.
Heterochromatic regions have a high abundance of TEs, and exploring DNA methylation and chromatin dynamics in these TE regions could reveal new information on genomic architecture. Our study first showed that long TEs (>4 kb) were predominantly enriched in pericentromeric regions, whereas short TEs (<0.5 kb) were preferentially enriched in chromosomal arms (Supplementary Figure S9). Therefore, we focused on long TEs which might reflect DNA methylation dynamics in heterochromatic regions. A detailed categorization of long TEs showed that the proportion of Gypsy long terminal repeat (LTR) retrotransposons in the At subgenome was three times that in the Dt subgenome, whereas the content of Copia LTR retrotransposons was similar in the At and Dt subgenomes (Figure 3C).
Before starting to analyse H3K9me2 modification marks in long TEs, we observed a significantly low proportion of 24-nt siRNAs in long TEs when comparing their genomic ratio with short TEs (Figure 3D). This observation primarily suggests that DNA hypermethylation in long TEs is not predominantly mediated by the RdDM pathway. Then, CHH DNA methylation levels, 24-nt siRNA enrichment and H3K9me2 modifications in long Copia and Gypsy TEs were explored. The CHH methylation levels in long TE regions were higher than in TE-flanking regions for both Copia and Gypsy (Figure 3E). Typically, H3K9me2 modification marks were highly enriched in long TEs, different from the distribution of 24-nt siRNAs (Figure 3F and G), which was consistent with previous studies in Arabidopsis (12,13). Comparisons of these features in consecutive stages showed a consistently increasingly common relationship between CHH DNA methylation and the enrichment of H3K9me2 modification marks, but a reverse pattern between CHH DNA methylation and 24-nt siRNA levels. Collectively, these findings suggest a prominent role for the H3K9me2-dependent pathway in mediating DNA hypermethylation in long TEs, as observed by up-regulated expression of CMT2 gene in this pathway (Supplementary Figure S8). This could explain the commonly observed down-regulated expression of long TEs (Supplementary Figure S10).
Contribution of RdDM may somehow decrease in euchromatic regions
We next explored the contribution of the RdDM pathway to dynamic DNA methylation in euchromatic regions. Average CHH methylation levels around siRNA mapping regions were determined and compared in consecutive developmental stages. Typically, CHH methylation levels peaked at siRNA loci and decreased at flanking regions for both overlapping and stage-specific siRNAs, suggesting a true RdDM effect (Figure 4A). For the three comparisons, we observed that CHH methylation levels in overlapping siRNA regions were higher than in stage-specific mapping regions. However, the stage-specific siRNAs in two consecutive developmental stages failed to correlate with higher methylation levels in the corresponding stages (Figure 4A). These observations imply that the 24-nt siRNA mapping regions might establish additional CHH methylation, independent of RdDM, in developing fibres.
Figure 4.
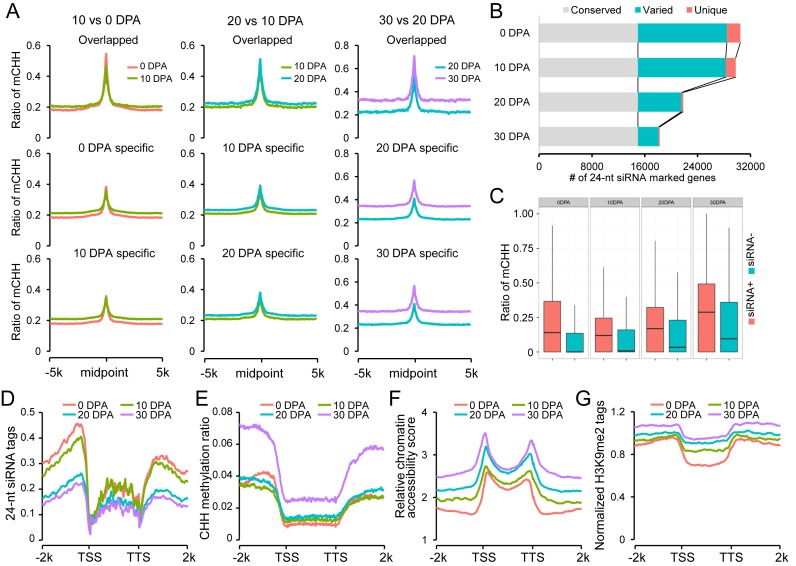
Examination of the effect of RdDM pathway on gene regions. (A) Patterns of CHH methylation levels in 24-nt siRNA mapping regions. For each pairwise comparison, CHH methylation levels of overlapped siRNA loci and stage-specific siRNA loci were investigated from upstream 5 kb to downstream 5 kb. (B) Comparisons of 24-nt siRNA marked genes between consecutive development stages. (C) Box-plots show the CHH methylation levels of 24-nt siRNA marked genes (+) and un-marked genes (-) in each developmental stage. (D) Patterns of 24-nt siRNA enrichment in gene regions. (E) Patterns of CHH methylation level in gene regions. (F) Patterns of relative chromatin accessibility in gene regions. For each sample, relative chromatin accessibility was calculated by comparison with pericentromeric regions. (G) Patterns of H3K9me2 ChIP-Seq tags in gene regions. The H3K9me2 ChIP-Seq tags were normalized with Input control DNA sequencing data.
We focused on genes in which DNA methylation could be mediated by the RdDM pathway. We defined an RdDM-marked gene with at least one siRNA product in the upstream 2 kb regions. A total of 33 036 genes were detected in all four developmental stages (30 467 in ovule and a total of 30 997 in the three fibre stages; Figure 4B). A detailed comparison showed that ∼45.3% (∼82.0% related to fibre at 30 DPA) of these genes could be detected with 24-nt siRNA production in all four stages. Interestingly, fibres at the three stages only showed ∼7.8% unique gene loci related to ovules. Indeed, these 24-nt siRNA marked genes showed higher methylation levels than the unmarked genes at each development stage (Figure 4C). However, no direct correlation was observed between the enrichment of stage-specific 24-nt siRNAs and gene methylation levels in consecutive stages. These results implied that RdDM was conserved for the vast majority of marked genes in ovules and fibres, but its contribution was largely limited in the differentiation of fibre cells from ovules.
To further dissect the effect of RdDM, we measured 24-nt siRNA enrichment in genic regions. We noticed a strong trend for siRNA enrichment in gene flanking regions near transcription start sites (TSS) and transcription termination sites (TTS) (Figure 4D). This trend could indicate a possible methylation island mediated by RdDM, so regulating gene transcription. However, a direct examination of CHH methylation did not reveal a pattern matching the enrichment of siRNAs except for ovule at 0 DPA (Figure 4E). The opposite patterns observed in 24-nt siRNAs and CHH methylation in fibres were compared with the patterns of chromatin accessibility and H3K9me2 modification marks. Strikingly, chromatin accessibility in gene flanking regions decreased in fibres but slightly increased in ovules (Figure 4F). However, a reverse trend was observed in the enrichment of H3K9me2 modification marks in gene flanking regions, which could be responsible for DNA hypermethylation (Figure 4G).
These findings imply that the observed pattern of CHH methylation in euchromatic regions was consistent with classical RdDM in cotton ovules. The ovule-bearing fibres established additional CHH methylation that might be mediated largely by the H3K9me2-dependent pathway, independent of RdDM.
Asymmetric DNA methylation contributes to homoeologous gene expression bias
Allopolyploid formation appears to be accompanied by widespread non-additive gene expression in allopolyploid plants (48,49). The dynamic DNA methylation maps of allopolyploid cotton fibres provide a new avenue to examine the potential relationship between them. Correlation of gene expression with the methylation levels of promoter regions showed that highly methylated genes tended to be expressed at lower levels in all four developmental stages (Supplementary Figure S11). Therefore, we analysed homoeologous gene pairs in the At and Dt subgenomes to detect its effects on fibre development.
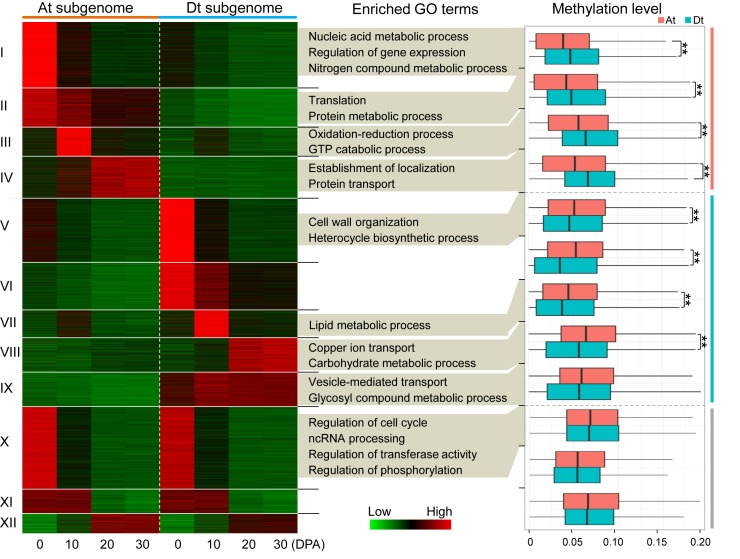
Using a combined approach of genome synteny and reciprocal best hits, we identified 16 077 homoeologous gene pairs, of which 12 304 were expressed in at least one developmental stage (FPKM > 0.5). A k-means clustering analysis showed that 4254 (34.6%) gene pairs in four clusters (I to IV) were expressed biased to the At-subgenome; 5017 (40.8%) gene pairs in five clusters (V to IX) were expressed biased to the Dt subgenome; and gene pairs in the other three clusters (X to XII) exhibited similar expression levels in both the At and Dt subgenomes (Figure 5). Gene Ontology (GO) analysis showed that genes in each cluster were enriched in specific biological pathways except for clusters XI and XII (false discovery rate (FDR) < 0.01); for instance, genes in cluster III were involved in the oxidation-reduction process and genes in cluster VII were involved in the lipid metabolic process.
Figure 5.
The relationship between homoeologous gene expression bias and DNA methylation levels. All expressed homoeologous gene pairs between the At and Dt subgenomes were categorized into 12 clusters (I to XII). Gene Ontology (GO) terms with significant enrichment levels were present for each cluster (FDR < 0.01). The methylation levels (upstream 2 kb) of gene pairs between the At and Dt subgenomes were compared and the differentially methylated clusters (I to VIII) were indicated (**P-value < 0.001).
The methylation levels of promoter regions for each gene pair were analysed. Strikingly, we observed generally higher methylation levels in the Dt subgenome in At-biased expression clusters and the converse in the At subgenome in Dt-biased expression clusters except for cluster IX (Wilcoxon rank sum test, P-value < 0.001; Figure 5). The overall methylation levels in the At and Dt subgenomes exhibited no significant differences for the other three clusters (X to XII). These findings imply that DNA methylation could lead to the asymmetric contribution of subgenomes to allotetraploid cotton fibre development.
Dynamic DNA methylation contributes to staged cotton fibre development
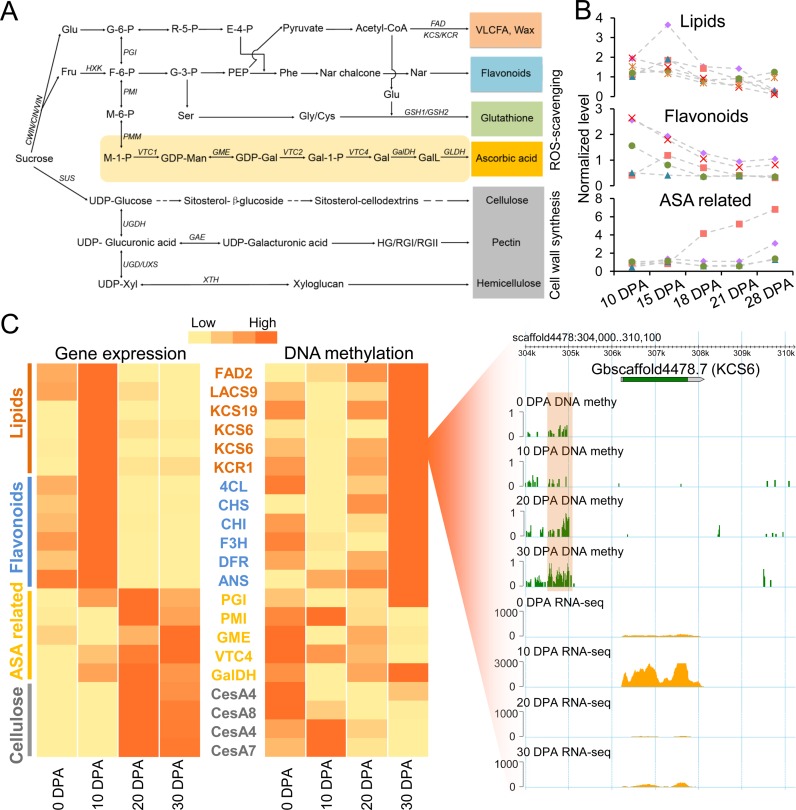
Each developmental stage of developing fibre has different biological features (50). Previous studies indicated that the lipid metabolic pathway, especially for very-long-chain fatty acids, plays a vital role in fibre elongation (Figure 6A and Supplementary Figure S12) (16,51). Flavonoids, which might be acting as antioxidants, function primarily in the elongation stage (Supplementary Figure S13) (42). A recent metabolomics analysis demonstrated that the ascorbate-glutathione cycle might be a major contributor to enzymatic reactive oxygen species (ROS) scavenging due to the observation of high concentrations of main metabolites in the SCW stage (coupled with prolonged fibre elongation) (52). Deep mining of the metabolomics data also revealed that representative lipids and flavonoids reached concentration peaks at the rapid elongation stage (represented by 10 and 15 DPA) and gradually decreased at the transition to the SCW stage (Figure 6B). Here, we explored the putative regulatory role of DNA methylation in staged fibre development by focusing on these pathways.
Figure 6.
DNA methylation regulates cotton fibre elongation and secondary cell wall synthesis. (A) The carbohydrate metabolism pathway involved in lipid, flavonoid, ascorbate-glutathione and cell wall biosynthesis. The full names of these genes are shown in Supplementary Table S6. (B) Concentrations of representative metabolites in lipid, flavonoid and ascorbate biosynthesis pathways during cotton fibre elongation and secondary cell wall synthesis. These data were explored in the publication of Tuttle et al. (52) and are also presented in Supplementary Table S7. (C) Heatmaps show the expression levels and promoter methylation levels of key functional genes. The genome browser snapshot in the right panel shows the methylation level and expression level of 3-ketoacyl-CoA synthase 6 (KCS6) in each developmental stage. The orange box shows a differentially methylated region.
We first confirmed that these biological processes were significantly enriched in each developmental stage with a comparative transcriptome analysis. GO enrichment analysis indicated that significantly enriched processes could represent relative biological features for each developmental stage (Supplementary Figure S14), consistent with the abundance of metabolites (Figure 6B). Notably, the oxidation-reduction process was enriched at 10 DPA and 30 DPA fibres. A detailed analysis of this process revealed that the genes in flavonoid metabolic pathway were highly expressed at 10 DPA and the genes in the ascorbate biosynthesis pathway had higher expression levels at 20 or 30 DPA (Figure 6C; Supplementary Figure S15). We next explored DNA methylation levels of those key genes involved in these pathways. Strikingly, genes associated with lipid and flavonoid biosynthesis had low methylation levels at 10 DPA and high levels at other stages. For example, a hypomethylated DMR was detected for 3-ketoacyl-CoA synthase 6 (KCS6) at 10 DPA, when it exhibited a higher expression level than in other stages (Figure 6C). However, a reverse pattern was observed for ascorbate biosynthesis related genes and cellulose synthases (CesAs), which exhibited low methylation levels at 20 or 30 DPA. Therefore, our integrated analysis of multi-omics data including the DNA methylome, transcriptome and metabolome indicate that dynamic DNA methylation is associated with the regulation of lipid biosynthesis and spatio-temporal modulation of ROS levels during cotton fibre development.
DISCUSSION
Extensive gene functional studies have uncovered the mysteries of differentiation and development of epidermal hairs in plants, but limited studies have focused on epigenetic regulatory mechanisms. Here, applying the single-celled cotton fibre as a model, we carried out epigenetic mapping through bisulfite-treated DNA sequencing, RNA-Seq, small RNA-Seq, MNase-Seq and ChIP-Seq of developing fibres. Integration of these multi-omics data is able to reveal DNA methylation and chromatin dynamics, and thus elucidates their roles in fibre development.
Chromatin dynamics has been studied in female reproductive tissues (53–54), but dissection of the relationship between DNA methylation and chromatin dynamics remains fragmentary in plant development as well as in single-cell differentiation. In this study, we carried out an incomplete MNase digestion of chromatin in developing fibres because this approach can reveal the differentially condensed chromatin state in consecutive stages (38). The analysis of nucleosome positioning revealed a gradual transition from euchromatin to heterochromatin, which was coupled with an continuing increase in the CHH methylation context. A previous study showed that nucleosome positioning could influence DNA methylation patterning (55), so we predict that heterochromatinization is responsible for shaping the DNA methylation landscape. Nucleosome formation usually inhibits the access of regulatory proteins to DNA sequences (56), so the increase of heterochromatin is likely to have a role in regulation of gene transcription during staged cotton fibre differentiation.
In plants, CHH methylation can be mediated through the RdDM pathway and a CMT2-pathway dependent on H3K9me2 (10). RdDM often occurs in euchromatic regions, whereas the H3K9me2-dependent pathway plays a major role in heterochromatic regions (12). In this study, a sharply decreased enrichment of 24-nt siRNAs contrasted with an increasing frequency of CHH methylation. This relationship was different from previous studies which compared the levels of 24-nt siRNAs and DNA methylation in mutant plants (12,13), suggesting that cotton fibre development uses different mechanisms to regulate gene expression. We observed an up-regulated expression of CMT2 and increased deposition of H3K9me2 modification marks. Genome-wide H3K9me2 deposition was positively correlated with TE density and DNA methylation level, but was negatively correlated with 24-nt siRNAs. These relationships were confirmed in heterochromatic long TEs and euchromatic gene regions. The observed increase of H3K9me2 can be partially explained by the euchromatin to heterochromatin transition, because heterochromatic regions are preferentially deposited with repressive chromatin modification marks (57). Therefore, it may be the H3K9me2-dependent pathway that plays a predominant role in mediating DNA hypermethylation during cotton fibre development. The increase of DNA methylation could further suppress the production of 24-nt siRNAs, acting as a feedback mechanism. This finding will enhance our understanding the role of H3K9me2-dependent CMT2-pathway in mediating DNA methylation in plant development.
This study provided new information for the regulatory role of dynamic DNA methylation in cotton fibre development. Since DNA methylation exhibits a general negative correlation with gene expression (Supplementary Figure S11), it seems likely that the increase of CHH methylation may be responsible for transcriptional silencing of some fibre-related genes, in part supported by the decreased number of transcribed genes during fibre development (Supplementary Figure S14). Previous studies show that the At and Dt subgenomes might exhibit partially divergent contributions to polyploid cotton fibre development (48). Here, we found that asymmetric DNA methylation between two subgenomes is associated with homoeologous gene expression bias in each developmental stage of fibre development, suggesting that this partitioned expression could be regulated in an epigenetic manner. In addition, we examined a regulatory role for DNA methylation in the rapid elongation and transition to secondary cell wall synthesis stages. Dynamic DNA methylation was associated with the modulation of ROS levels at different developmental stages.
In summary, this study is the first to build dynamic DNA methylation maps of a single plant cell type by integrating multi-omics data. It is also the first to demonstrate that a continuously increasing DNA methylation level is related to a gradual transition from euchromatin to heterochromatin. We demonstrates that the increasing level of DNA methylation is predominantly mediated by the H3K9me2-dependent pathway rather than the RdDM pathway. The established maps and identified divergent patterns between two pathways provide novel insights into the regulation of cotton fibre development by epigenetic mechanisms. With the rapid development of single-cell sequencing techniques, these findings can be excellent references for exploring epigenetic regulation of other important biology processes at the single-cell level, such as trichome differentiation, stomatal development or pollen development.
Supplementary Material
Acknowledgments
We are very grateful to the laboratory of Candace H. Haigler for releasing the metabolome data of cotton fibre development.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31230056, 31201251]; Huazhong Agricultural University Independent Scientific & Technological Innovation Foundation [2014bs03]. Funding for open access charge: National Natural Science Foundation of China [31230056, 31201251]; Huazhong Agricultural University Independent Scientific & Technological Innovation Foundation [2014bs03].
Conflict of interest statement. None declared.
REFERENCES
- 1.Law J.A., Jacobsen S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Slotkin R.K., et al. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136:461–472. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang M., et al. Genome-wide high resolution parental specific DNA and histone methylation maps uncover patterns of imprinting regulation in maize. Genome Res. 2013;24:167–176. doi: 10.1101/gr.155879.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Z.J. Genomic and epigenetic insights into the molecular bases of heterosis. Nat. Rev. Genet. 2013;14:471–482. doi: 10.1038/nrg3503. [DOI] [PubMed] [Google Scholar]
- 5.Le Trionnaire G., Twell D. Small RNAs in angiosperm gametophytes: from epigenetics to gamete development. Genes Dev. 2010;24:1081–1085. doi: 10.1101/gad.1936110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang H., et al. Whole-genome DNA methylation patterns and complex associations with gene structure and expression during flower development in Arabidopsis. Plant J. 2015;81:268–281. doi: 10.1111/tpj.12726. [DOI] [PubMed] [Google Scholar]
- 7.Xing M.Q., et al. Global analysis reveals the crucial roles of DNA methylation during rice seed development. Plant Physiol. 2015;168:1417–1432. doi: 10.1104/pp.15.00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhong S., et al. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotech. 2013;31:154–159. doi: 10.1038/nbt.2462. [DOI] [PubMed] [Google Scholar]
- 9.Dowen R.H., et al. Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl. Acad. Sci. U.S.A. 2012;109:2183–2191. doi: 10.1073/pnas.1209329109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du J., Johnson L.M., Jacobsen S.E., Patel D.J. DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 2015;16:519–532. doi: 10.1038/nrm4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matzke M.A., Mosher R.A. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014;15:394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- 12.Zemach A., et al. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153:193–205. doi: 10.1016/j.cell.2013.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stroud H., et al. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 2014;21:64–72. doi: 10.1038/nsmb.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du J., et al. Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell. 2012;151:167–180. doi: 10.1016/j.cell.2012.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wagner G.J., Wang E., Shepherd R.W. New approaches for studying and exploiting an old protuberance, the plant trichome. Ann. Bot. 2004;93:3–11. doi: 10.1093/aob/mch011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin Y.M., Zhu Y.X. How cotton fibers elongate: a tale of linear cell-growth mode. Curr. Opin. Plant Biol. 2011;14:106–111. doi: 10.1016/j.pbi.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 17.Haigler C.H., Betancur L., Stiff M.R., Tuttle J.R. Cotton fibre: a powerful single-cell model for cell wall and cellulose research. Front Plant Sci. 2012;3:104. doi: 10.3389/fpls.2012.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grebe M. The patterning of epidermal hairs in Arabidopsis-updated. Curr. Opin. Plant Biol. 2012;15:31–37. doi: 10.1016/j.pbi.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 19.Ishida T., Kurata T., Okada K., Wada T. A genetic regulatory network in the development of trichomes and root hairs. Annu. Rev. Plant Biol. 2008;59:365–386. doi: 10.1146/annurev.arplant.59.032607.092949. [DOI] [PubMed] [Google Scholar]
- 20.Pesch M., Hulskamp M. One, two, three...models for trichome patterning in Arabidopsis? Curr. Opin. Plant Biol. 2009;12:587–592. doi: 10.1016/j.pbi.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 21.Machado A., Wu Y., Yang Y., Llewellyn D.J., Dennis E.S. The MYB transcription factor GhMYB25 regulates early fibre and trichome development. Plant J. 2009;59:52–62. doi: 10.1111/j.1365-313X.2009.03847.x. [DOI] [PubMed] [Google Scholar]
- 22.Walford S.A., Wu Y., Llewellyn D.J., Dennis E.S. GhMYB25-like: a key factor in early cotton fibre development. Plant J. 2011;65:785–797. doi: 10.1111/j.1365-313X.2010.04464.x. [DOI] [PubMed] [Google Scholar]
- 23.Deng F., et al. GbPDF1 is involved in cotton fibre initiation via the core cis-element HDZIP2ATATHB2. Plant Physiol. 2012;158:890–904. doi: 10.1104/pp.111.186742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X.B., Fan X.P., Wang X.L., Cai L., Yang W.C. The cotton ACTIN1 gene is functionally expressed in fibres and participates in fibre elongation. Plant Cell. 2005;17:859–875. doi: 10.1105/tpc.104.029629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osabe K., et al. Genetic and DNA methylation changes in cotton (Gossypium) genotypes and tissues. PLoS One. 2014;9:e86049. doi: 10.1371/journal.pone.0086049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin X., et al. A potential role for CHH DNA methylation in cotton fibre growth patterns. PLoS One. 2013;8:e60547. doi: 10.1371/journal.pone.0060547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan D., et al. The genome sequence of Sea-Island cotton (Gossypium barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres. Sci. Rep. 2015;5:17662. doi: 10.1038/srep17662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beasley C.A., Ting I.P. The effects of plant growth substances on in vitro fibre development from fertilized cotton ovules. Am. J. Bot. 1973;60:130–139. [Google Scholar]
- 29.Baubec T., Pecinka A., Rozhon W., Scheid O.M. Effective, homogeneous and transient interference with cytosine methylation in plant genomic DNA by zebularine. Plant J. 2009;57:542–554. doi: 10.1111/j.1365-313X.2008.03699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paterson A., Brubaker C., Wendel J. A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP or PCR analysis. Plant Mol. Biol. Rep. 1993;11:122–127. [Google Scholar]
- 31.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krueger F., Andrews S.R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu D., Zhang X., Tu L., Zhu L., Guo X. Isolation by suppression-subtractive hybridization of genes preferentially expressed during early and late fibre development stages in cotton. Mol. Biol. 2006;40:741–749. [PubMed] [Google Scholar]
- 34.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trapnell C., et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li A., et al. mRNA and small RNA transcriptomes reveal insights into dynamic homoeolog regulation of allopolyploid heterosis in nascent hexaploid wheat. Plant Cell. 2014;26:1878–1900. doi: 10.1105/tpc.114.124388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gent J.I., et al. Accessible DNA and relative depletion of H3K9me2 at maize loci undergoing RNA-directed DNA methylation. Plant Cell. 2014;26:4903–4917. doi: 10.1105/tpc.114.130427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun Q.W., Zhou D.X. Rice jmjC domain-containing gene JMJ706 encodes H3K9 demethylase required for floral organ development. Proc. Natl. Acad. Sci. U.S.A. 2008;105:13679–13684. doi: 10.1073/pnas.0805901105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng J., Liu T., Qin B., Zhang Y., Liu X.S. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 2012;7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan J., et al. A genetic and metabolic analysis revealed that cotton fibre cell development was retarded by flavonoid naringenin. Plant Physiol. 2013;162:86–95. doi: 10.1104/pp.112.212142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang M., et al. Long noncoding RNAs and their proposed functions in fibre development of cotton (Gossypium spp.) New Phytol. 2015;207:1181–1197. doi: 10.1111/nph.13429. [DOI] [PubMed] [Google Scholar]
- 44.Wang S., et al. Sequence-based ultra-dense genetic and physical maps reveal structural variations of allopolyploid cotton genomes. Genome Biol. 2015;16:108. doi: 10.1186/s13059-015-0678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xue W., Wang Z.M., Du M.J., Liu Y.D., Liu J.Y. Genome-wide analysis of small RNAs reveals eight fibre elongation-related and 257 novel microRNAs in elongating cotton fibre cells. BMC Genomics. 2013;14:629. doi: 10.1186/1471-2164-14-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu N., et al. Small RNA and degradome profiling reveals a role for miRNAs and their targets in the developing fibres of Gossypium barbadense. Plant J. 2014;80:331–344. doi: 10.1111/tpj.12636. [DOI] [PubMed] [Google Scholar]
- 47.Telford D.J., Stewart B.W. Micrococcal nuclease: Its specificity and use for chromatin analysis. Int. J. Biochem. 1989;21:127–138. doi: 10.1016/0020-711x(89)90100-6. [DOI] [PubMed] [Google Scholar]
- 48.Hovav R., et al. Partitioned expression of duplicated genes during development and evolution of a single cell in a polyploid plant. Proc. Natl. Acad. Sci. U.S.A. 2008;105:6191–6195. doi: 10.1073/pnas.0711569105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu C., et al. Genome-wide disruption of gene expression in allopolyploids but not hybrids of rice subspecies. Mol. Biol. Evol. 2014;31:1066–1076. doi: 10.1093/molbev/msu085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gou J.Y., Wang L.J., Chen S.P., Hu W.L., Chen X.Y. Gene expression and metabolite profiles of cotton fibre during cell elongation and secondary cell wall synthesis. Cell Res. 2007;17:422–434. doi: 10.1038/sj.cr.7310150. [DOI] [PubMed] [Google Scholar]
- 51.Qin Y.M., et al. Saturated very-long-chain fatty acids promote cotton fibre and Arabidopsis cell elongation by activating ethylene biosynthesis. Plant Cell. 2007;19:3692–3704. doi: 10.1105/tpc.107.054437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tuttle J.R., et al. Metabolomic and transcriptomic insights into how cotton fibre transitions to secondary wall synthesis, represses lignification, and prolongs elongation. BMC Genomics. 2015;16:477. doi: 10.1186/s12864-015-1708-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.She W.J., et al. Chromatin reprogramming during the somatic-to-reproductive cell fate transition in plants. Development. 2013;140:4008–4019. doi: 10.1242/dev.095034. [DOI] [PubMed] [Google Scholar]
- 54.Baroux C., Autran D. Chromatin dynamics during cellular differentiation in the female reproductive lineage of flowering plants. Plant J. 2015;83:160–176. doi: 10.1111/tpj.12890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chodavarapu R.K., et al. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466:388–392. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang T., Zhang W., Jiang J. Genome-wide nucleosome occupancy and positioning and their impact on gene expression and evolution in plants. Plant Physiol. 2015;168:1406–1416. doi: 10.1104/pp.15.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu C., Lu F., Cui X., Cao X. Histone methylation in higher plants. Annu. Rev. Plant Biol. 2010;61:395–420. doi: 10.1146/annurev.arplant.043008.091939. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.