Abstract
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs) acquire new spacers to generate adaptive immunity in prokaryotes. During spacer integration, the leader-preceded repeat is always accurately duplicated, leading to speculations of a repeat-length ruler. Here in Haloarcula hispanica, we demonstrate that the accurate duplication of its 30-bp repeat requires two conserved mid-repeat motifs, AACCC and GTGGG. The AACCC motif was essential and needed to be ∼10 bp downstream from the leader-repeat junction site, where duplication consistently started. Interestingly, repeat duplication terminated sequence-independently and usually with a specific distance from the GTGGG motif, which seemingly served as an anchor site for a molecular ruler. Accordingly, altering the spacing between the two motifs led to an aberrant duplication size (29, 31, 32 or 33 bp). We propose the adaptation complex may recognize these mid-repeat elements to enable measuring the repeat DNA for spacer integration.
INTRODUCTION
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs) and their associated Cas (CRISPR-associated) proteins widely exist in archaeal (∼90%) and bacterial (∼50%) genomes (1,2), and they confer adaptive immunity against potentially harmful invaders (such as viruses/phages and plasmids) (3–8). These highly diversified CRISPR-Cas systems have been classified into 2 classes, 6 types and 17 subtypes (1,2).
CRISPRs are arrays of repetitive sequences (repeats) that are intervened by invader-derived fragments (spacers), and these structures are usually preceded by an adenine(A)/thymine(T)-rich leader sequence (9–12). Adjacent to CRISPR arrays, there is often (but not exclusively) a cluster of co-functional Cas protein-encoding genes (13,14). The repeat sequence on CRISPR RNA transcripts is recognized and cleaved by specific Cas endonuclease(s), which gives rise to small individual molecules termed CRISPR RNAs (crRNAs) (4,15–17). These small RNAs guide interfering Cas protein(s) to destroy foreign nucleic acids (DNA/RNA) based on complementary base pairing (5,18,19). Apparently, the adaptation process, i.e. acquisition of new spacers into the CRISPR memory, enables subsequent immunity and CRISPR adaptability (3). However, previous CRISPR studies have mainly focused on crRNA biogenesis and target interference processes, with the adaptation pathway still poorly understood.
CRISPR adaptation was first reported for the Streptococcus thermophilus Type II-A system in 2007 (3). In several S. thermophilus survivors of lytic phage infection, new spacers were added into their CRISPR arrays. Interestingly, this process was rarely reported for other systems in the subsequent five years, until the observation that the over-expression of Cas1 and Cas2 promoted infrequent adaptation events to be detectable in Escherichia coli (20). This Cas1–Cas2-mediated inefficient process was termed ‘naïve adaptation’ (21), to distinguish it from the priming pathway which has been reported for at least three different Type I systems, i.e. I-E of E. coli (22,23), I-B of Haloarcula hispanica (24) and I-F of Pectobacterium atrosepticum (25). During priming adaptation, the interfering effectors including Cas3, the Cascade (CRISPR-associated complex for antiviral defense) complex, and a pre-existing spacer with a full or partial match to the foreign DNA are further required (23,24,26).
The naïve and priming pathways appear to both involve two mechanistic steps. The first step is to recognize and process spacer substrates (called protospacers). Protospacer recognition depends in part on the presence of the conserved flanking sequence (2–5 bp) termed protospacer adjacent motif (PAM) (27,28). This motif is also critical for subsequent interference to discriminate the protospacer of invaders from the spacer in the CRISPR DNA (29). Other factors also influence protospacer recognition because new spacers are preferentially derived from the non-self DNA, even without the interference-mediated selection against self-derived spacers (20,30). It was recently reported that protospacer selection in E. coli is replication-dependent (i.e., impeded by the enriched Chi sites on the self chromosome while facilitated by the higher number of replication forks on the foreign DNA), which possibly underlies its intrinsic preference for non-self DNA (31). However, in H. hispanica, where naïve adaptation appears to be inactivated, our previous studies suggested that the additional priming step (which involves priming crRNA-guidance and PAM-authentication) guarantees self versus non-self discrimination at the adaptation level (24,32).
The second step of CRISPR adaptation is to integrate the selected protospacer into the CRISPR DNA, usually at its leader end (3,20,24). Note that the repeat at the integration site must get accurately duplicated, into two identical copies flanking the incoming spacer, to maintain the fixed repeat-spacer periodicity (22,23). Sequential and staggered cleavage of the repeat was then proposed : one strand of the CRISPR repeat is first cut at one end, and the other strand is cut subsequently at the other end by a ruler mechanism (30). Supporting this proposal, intermediates of this cleavage (which seemed to be concurrent with the end-joining between the protospacer and the ‘opened’ repeat DNA) was detected in E. coli (33). Doudna and colleagues recently demonstrated this process to be similar to DNA transposition and retroviral integration (34), and proposed a two-step integration model: (i) one of the two 3′-OH ends of the protospacer first attacks the minus strand of the CRISPR repeat at the leader-distal end, producing a half-site intermediate; (ii) then the other 3′-OH end attacks the plus strand by a ruler mechanism. However, there is a different view on the order of the two integration steps: the attack on the plus strand at the relatively conserved leader-repeat junction should be the initial step. Supporting this view, the disintegration activity of Cas1 from both E. coli and Sulfolobus solfataricus showed a clear preference for the nucleotides flanking the leader-repeat junction (35), and sequences in the vicinity of this junction were also reported to be important for adaptation in E. coli (20,30,33) and in S. thermophilus (36). Hence, it is intriguing to explore how the two termini of the first repeat are accurately recognized as integration sites, in a sequence-specific manner or by a molecular ruler.
The H. hispanica genome carries only one CRISPR array and an associated cas operon encoding the Cascade proteins (Cas5-8), the interfering Cas3 nuclease, and three putative adaptation-Cas proteins: Cas1, Cas2, and Cas4 (24). Its CRISPR adapts efficiently to the non-lytic virus HHPV-2 (H. hispanica pleomorphic virus-2), but strictly through the priming pathway where an invader-targeting spacer is required (24). Therefore, we engineered a strain with two separate CRISPR variants, namely priming-CRISPR (p-CRISPR) and adaptation-CRISPR (a-CRISPR), to produce the priming crRNAs and to accept new spacers, respectively. Using this modified system, we tested a series of a-CRISPR constructs with the 30-bp repeat differently mutated. Interestingly, two conserved DNA motifs in the middle of the repeat played critical roles during this process, and surprisingly, the position of one motif seemingly determined the repeat duplication size. In contrast, when nucleotides surrounding either of the two repeat ends were substituted, the mutated repeat was still accurately duplicated (though the adaptation process was often impaired by nucleotide substitutions adjacent to the leader-repeat junction). We propose a novel molecular ruler anchored in the middle of the repeat, and show the possibility that the repeat size or the periodicity of a CRISPR could be well manipulated.
MATERIALS AND METHODS
Strains and culturing conditions
The H. hispanica strains used in this study are listed in Supplementary Table S1. The strain DF60 (ΔpyrF strain of H. hispanica ATCC 33960) (37) and its derivatives DF60P and ΔCR (see below) were cultured at 37°C in AS-168 medium (per liter, 200 g of NaCl, 20 g of MgSO4·7H2O, 2 g of KCl, 3 g of trisodium citrate, 1 g of sodium glutamate, 50 mg of FeSO4·7H2O, 0.36 mg of MnCl2·4H2O, 5 g of Bacto casamino acids, 5 g of yeast extract, pH 7.2) with uracil added to a final concentration of 50 mg/l. Their transformant strains of pHAR-derived plasmids (Supplementary Table S1) were cultured in yeast extract-subtracted AS-168 medium.
E. coli JM109 used for molecular cloning was cultured in LB (lysogeny broth) medium. Ampicillin was added to a final concentration of 100 mg/l when needed.
Construction of DF60P variant and integrative plasmids
The plasmids that were used in this study are listed in Supplementary Table S1. For plasmid construction, DNA fragments were amplified using the high-fidelity KOD-Plus DNA polymerase (TOYOBO, Osaka, Japan), and validated by DNA sequencing. Restriction enzymes and T4 DNA ligase purchased from New England Biolabs (Beverly, MA, USA) were used for cloning. Transformation of H. hispanica cells was conducted according to the online Halohandbook (http://www.haloarchaea.com/resources/halohandbook/Halohandbook_2009_v7.2mds.pdf). Primers are listed in Supplementary Table S2.
To generate the H. hispanica strain DF60P, the wild-type CRISPR of DF60 was replaced by the p-CRISPR variant, which consists of a short-version (constitutive) promoter of a PHA synthesis-related gene (phaR) (38) and two repeat units flanking the original spacer13. The ∼500-bp chromosomal sequence immediately upstream of the DF60 CRISPR and that immediately downstream of spacer12 (i.e. the repeat-spacer13-repeat structure with its downstream ∼400-bp sequence) were amplified using P-UF/UR (upstream forward/ upstream reverse) and P-DF/DR (downstream forward/downstream reverse) primer pairs, respectively. The DNA sequence of the constitutive phaR promoter was designed on primers phaR-bridge1 and phaR-bridge2 and engineered onto the downstream fragment by two rounds of bridge PCR. These two fragments were cloned into the non-replicative vector pHAR and then transformed into DF60 cells to replace the wild-type CRISPR using the previously described pop-in-pop-out strategy (37). The ΔCR strain was similarly constructed by knocking out the only CRISPR locus in DF60.
To introduce a-CRISPR into the DF60P or ΔCR chromosome, an integrative plasmid (namely pHAR-in) was constructed by engineering a 460-bp chromosomal sequence downstream of the DF60 wild-type CRISPR (amplified using primers ChrSeq-F/R) into the non-replicative vector pHAR (37). The wild-type a-CRISPR (105-bp CRISPR leader and the first repeat) was amplified using primers A-F/R (forward/reverse), and then cloned into pHAR-in between the BamHI and KpnI restriction sites, generating the pCR-A plasmid. When pCR-A was transformed into DF60P or ΔCR, the a-CRISPR was knocked into the chromosome through homologous recombination. For construction of various a-CRISPR mutants, nucleotide substitutions were introduced into the forward or reverse primer to generate different mutations. If needed, bridge PCR with complementary primers containing mutations was performed.
Spacer integration analysis
Spacer acquisition was monitored by PCR as previously described (24) with a few modifications. For each transformant of the pHAR derivatives, three individual colonies were separately picked and cultured in yeast extract-subtracted AS-168 liquid medium to the exponential phase, and then diluted 1:15 with fresh medium containing HHPV-2 viruses at a multiplicity of infection of 10. Sub-inoculation was performed whenever the culture reached the stationary stage (after 7-day culturing). At different time points, these cultures were sampled as follows: 100 μl were centrifuged at 10 000 rpm for 2 min to collect the haloarchaeal cells, then the cells were lysed by 200 μl distilled water, and 0.3 μl were used as the template for each PCR reaction. The Exp-Fp/Rp and Exp-Fa/Ra primer pairs were used to detect spacer acquisition for p-CRISPR and a-CRISPR, respectively. The PCR program consists of the following steps: (i) 95°C for 5min; (ii) 35 cycles of 95°C for 30 s, 54°C for 30 s, and 72°C for 30 s; (iii) 72°C for 5 min. The PCR products were separated by 1.2% agarose gel electrophoresis, stained by ethidium bromide, and imaged using Bio-Rad's ChemiDoc™ MP System. Expanded PCR products from a-CRISPR were extracted from the agarose gel using the AxyPrep™ DNA Gel Extraction Kit (Corning, NY, USA), and then sequenced using primer Exp-Fa. The sequencing results were visualized using Vector NTI advance 10 (Thermo Fisher Scientific, MA, USA).
Relative quantification of the expanded PCR products was performed using the Quantity One software (Bio-Rad, CA, USA). For each lane on a gel, the parental and expanded bands were detected with an appropriate sensitivity after lane background subtraction, and then Gaussian modeling was performed. Under the Gaussian-fitted profile, the quantity of the parental or expanded band(s) was recorded to calculate the percentage of expanded PCR products (the expanded quantity divided by the total quantity) for each lane. For each mutant, three replicates (individual clones) were examined to get an average percentage with the standard deviation.
Repeat duplication analysis
To analyze repeat duplication during spacer integration, the infected culture was serially diluted and spread onto agar plates to obtain individual colonies. Colony PCR was then performed and the expanded products were sequenced using primer Exp-Fa. By a Basic Local Alignment Search Tool (BLAST) search against the HHPV-2 genome, the protospacers and their PAM were identified. Then the identical sequence flanking each spacer in the expanded a-CRISPR was generally regarded as the duplicated ‘repeat’ sequence (which not necessarily equals to the original designed repeat).
RESULTS
Construction of a system of two CRISPRs respectively for priming and adaptation
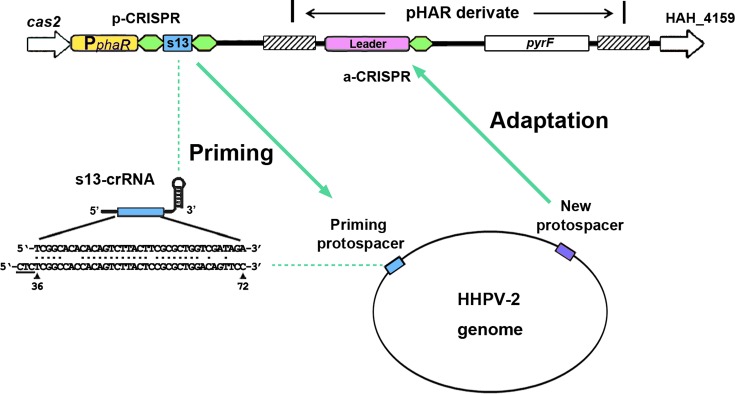
The only CRISPR array of H. hispanica is able both to prime adaptation to HHPV-2 and to acquire new spacers from this virus (24). To facilitate investigating CRISPR elements involved in the latter process, we designed a system of two separate CRISPRs respectively for priming and for spacer acquisition (Figure 1). We first replaced the wild-type CRISPR of DF60 (an auxotrophic H. hispanica strain with the pyrF gene deleted (37)) with a variant structure named priming-CRISPR (p-CRISPR) to generate the DF60P strain (Supplementary Figure S1A). Under the control of a strong constitutive promoter of phaR (a PHA synthesis-related gene) (38), p-CRISPR was designed to produce the s13-crRNA (crRNA of the original spacer13) molecules, which were shown able to prime efficient adaptation to HHPV-2 (24). However, this leaderless structure was unable to incorporate new spacers (see below). Therefore, we modified pHAR (a non-replicative vector carrying the selective marker pyrF (37)) to introduce another CRISPR for adaptation, named adaptation-CRISPR or a-CRISPR (Supplementary Figure S1B). Transformed into DF60P and cultured under the selection pressure (in yeast extract-subtracted AS-168 medium), a 460-bp chromosomal sequence designed in the pHAR derivate would facilitate it integrating (through homologous recombination) into the chromosome adjacent to the p-CRISPR (as depicted in Supplementary Figure S1). Thereby, when host cells containing these two CRISPRs are subjected to HHPV-2 infection, the priming activity of p-CRISPR would be independent of the a-CRISPR variations, which enables us to investigate the spacer integration process for an extensive series of a-CRISPR constructs.
Figure 1.
Design of two functionally separated CRISPRs to analyze the cis-elements during the primed spacer acquisition. The priming and adaptation steps are mediated by priming-CRISPR (p-CRISPR) and adaptation-CRISPR (a-CRISPR), respectively. First, the wild-type CRISPR of the H. hispanica auxotrophic strain DF60 was replaced by the p-CRISPR, and then into its downstream sequences, the a-CRISPR was integrated using a modified pHAR vector. The p-CRISPR consists of the constitutive phaR promoter and only two repeats (in green) flanking the wild-type spacer13. The s13-crRNA molecules from p-CRISPR have a partial match (indicated with a line of dots) to the HHPV-2 viral genome (genomic positions are indicated under the viral sequence, and the PAM sequence is underlined), which primes adaptation to HHPV-2. The a-CRISPR is designed containing the complete CRISPR leader to incorporate new spacers. A ∼460-bp homologous region (shadowed) facilitates recombination between the chromosome and the vector.
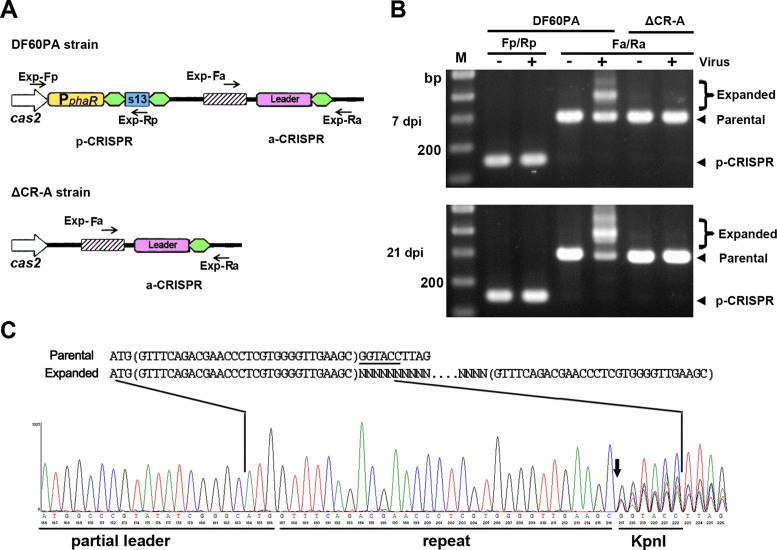
For naïve adaptation in E. coli and S. thermophilus, it was demonstrated or suggested that the leader and a single repeat together provide sufficient cis-elements (20,36). Hence, the initial a-CRISPR was constructed containing the leader and a single repeat. Note that the leader here is defined as the ∼105-bp CRISPR-flanking sequence that is relatively conserved among haloarchaeal I-B CRISPRs with similar repeat sequences (Supplementary Figure S2). This a-CRISPR was introduced into DF60P and ΔCR, respectively, giving rise to strains DF60PA and ΔCR-A (Figure 2A). In DF60PA, larger-sized or expanded PCR products were readily detected for a-CRISPR, but not for p-CRISPR, upon virus infection (Figure 2B). This result indicates that, as designed in Figure 1, the priming molecules (s13-crRNAs) had been functionally produced from p-CRISPR while new spacers were incorporated specifically into the a-CRISPR. Over a long-period monitoring, more and more PCR products from the a-CRISPR became expanded, but expansion was never observed for the leader-less p-CRISPR (Figure 2B). Therefore, the leader preceding a single repeat provides sufficient and essential elements also for the primed adaptation. Note that the a-CRISPR in ΔCR-A never expanded over the long-period monitoring (Figure 2B), which confirms the strict requirement for a priming step in this system (24).
Figure 2.
The leader preceding a single repeat provides sufficient cis-elements for the primed adaptation process. (A) Depiction of the CRISPRs in DF60PA and ΔCR-A. The a-CRISPR carries the 105-bp leader and a single repeat. Exp-Fp/Rp and Exp-Fa/Ra primer pairs (black arrows) were designed against sequences that surround p-CRISPR and a-CRISPR, respectively. (B) PCR assay detecting the expansion of p-CRISPR and a-CRISPR using their corresponding primers. Total DNA from virus-infected (+) or uninfected (-) H. hispanica cells was separately used as the PCR template. For the infected samples, cells were collected 7 or 21 days post infection (dpi). The parental and the expanded a-CRISPRs, respectively, gave rise to ∼300-bp and larger-sized PCR products. Lane Ms, dsDNA size markers. (C) Chromatograph map that shows the sequencing result of the expanded DNA mixture in panel B. The Exp-Fa primer (depicted in panel A) was used for DNA sequencing. The proposed spacer-integration process, in which new spacers are inserted between duplicated repeats (in brackets), is depicted above the chromatograph. Overlap of multiple signals (resulting from the insertion of various new spacers) was observed for positions downstream of the vertical arrow.
Spacer acquisition from HHPV-2 and its independence on interference
Expanded PCR products of the DF60PA a-CRISPR were extracted and the DNA mixture was sequenced using primer Exp-Fa. On the sequencing chromatograph (Figure 2C), an overlap of multiple signals started to appear at the first guanine (G) of the KpnI restriction site (designed immediately downstream of the repeat). As illustrated in Figure 2C, this result is consistent with the previously proposed spacer-integration model that the first repeat is duplicated into two identical copies flanking the incoming new spacer (22,23). We also sequenced the expanded a-CRISPR of 8 individual DF60PA colonies, which revealed that the 30-bp repeat was correctly duplicated during 15 spacer integration events (Supplementary Data S1). The new spacers were derived from viral sequences (protospacers) that are preceded by a conserved 5′-TTC PAM (i.e. 5′-TTC-protospacer-3′, reading along the spacer-sense or non-target strand) (Supplementary Table S3).
A colony (DF60PA_clone1 in Supplementary Data S1) with a new spacer was selected for the HHPV-2 re-infection assay, and a significant PFU (plaque-forming unit) reduction was observed relative to its parental strain DF60PA (Supplementary Figure S3). It was demonstrated that the H. hispanica leader and a single repeat together provide sufficient cis-elements to acquire immunity (or to adapt) to HHPV-2. However, when we disrupted the transcription of the a-CRISPR by mutating its core promoter element TATA box (so that it could not provide immunity after acquiring a new spacer), we observed expanded PCR products of a similar amount to DF60PA at different time points after HHPV-2 infection (Supplementary Figure S3). It was suggested that the a-CRISPR expansion observed in this assay was generally independent of interference (the interfering effects of new spacers), which may be attributed to the non-lytic feature of the virus HHPV-2.
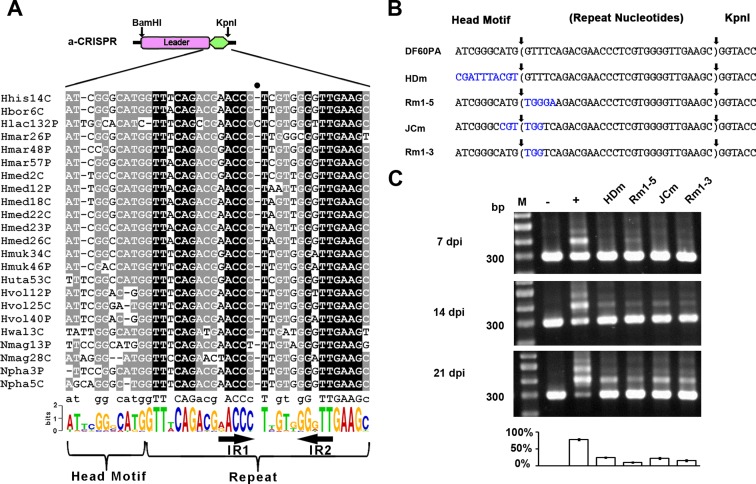
The leader-repeat junction provides a fixed integration site in spacer acquisition
The leader-repeat junction was supposed to be recognized to initiate spacer integration, because sequences spanning this junction are relatively conserved and have been shown important for this process (20,30,36). Correspondingly, the 10-bp leader sequence immediately upstream of the first repeat is also conserved among the haloarchaeal Type I-B CRISPRs (Figure 3A and also Supplementary Figure S2). Actually in our previous study on the Haloferax mediterranei CRISPRs, this conserved sequence was designated ‘head motif’ (we followed this name hereinafter) and shown to be not required for crRNA biogenesis (16). We mutated this motif of the DF60PA a-CRISPR, giving rise to the mutant HDm (Figure 3B). In DF60PA, expansion of the wild type a-CRISPR was detected as early as 4 days post-infection (dpi) (Supplementary Figure S4), and >70% of its 21-dpi PCR products were the expanded ones (Figure 3C). In contrast, expansion of the HDm a-CRISPR was not detected until 14 dpi, and of its 21-dpi PCR products, the larger-sized made up fewer than 30%. As exemplified in Supplementary Figure S5, the expanded percentage of the PCR products almost positively correlated with that of the PCR templates in our assay. Therefore, it is apparent that the adaptation (or expansion) process was significantly impaired in HDm. As depicted in Figure 3B, the first three or five repeat nucleotides of the DF60PA a-CRISPR were mutated to generate Rm1–3 or Rm1–5. In addition, we also mutated the six nucleotides surrounding the leader-repeat junction, giving rise to the mutant JCm (Figure 3B). Similarly for these mutants, a-CRISPR expansion was not detected until 14 dpi, and about 15%, 10% or 22% of the Rm1–3, Rm1–5 or JCm PCR products were the expanded ones on the 21st day post infection (Figure 3C). Taken together, these data suggest sequences spanning the leader-repeat junction should play important roles during adaptation in H. hispanica. It is worth mentioning that we have constructed more mutants with various mutations proximal to this junction, and interestingly observed very different influences on adaptation (Supplementary Figure S4). For example, a-CRISPR expansion was significantly impaired in the mutants HDmG, Rm1-5G and JCmG (‘G’ stands for the transition of A to G), but appeared to be less influenced in HDmT, Rm1–5T or JCmT (‘T’ stands for the transversion of A to T). It could be inferred that these mutations may differently influence, but not block, the recognition of the leader-repeat junction for spacer integration.
Figure 3.
Sequences spanning the leader-repeat junction are important for adaptation. (A) Sequences spanning the leader-repeat junction are conserved among haloarchaeal CRISPRs with similar repeats. Note that, for each CRISPR, the first repeat and its upstream ∼110-bp sequence were together retrieved and aligned (see Supplementary Figure S2 for details), and only a part of this multi-alignment is shown here. Using the WebLogo server (http://weblogo.berkeley.edu/logo.cgi), a sequence logo was generated from this multi-alignment. The inverted repeats (IR1 and IR2) within the CRISPR repeat are indicated. The full circle (•) indicates the position where an extra nucleotide from the Hlac132P repeat resulted in a gap in all the other repeat sequences during alignment. (B) Illustration of a series of a-CRISPR constructs with mutations (in blue) preceding, following, or flanking the leader-repeat junction. The repeat sequence is shown in brackets. Sequences between vertical arrows were duplicated during spacer integration. (C) Expansion of the a-CRISPRs shown in panel B at 7, 14 or 21 days post HHPV-2 infection (dpi). DNA from the infected (+) or uninfected (–) DF60PA cells was used as the positive or negative control. Three replicates were tested for each mutant, and each gel shows a representative result. The ∼300-bp (parental) and larger-sized (expanded) bands were relatively quantified for the 21-dpi (days post infection) samples, and the percentage of expanded PCR products in each lane is shown in the histogram. Lane Ms, dsDNA size markers.
Interestingly, when we sequenced the DNA mixture of the expanded a-CRISPRs using primer Exp-Fa, none of the above mutations influenced the appearance of multiple signals at the ‘G’ immediately downstream of the repeat sequence on the sequencing chromatogram (Figure 2C and Supplementary Figure S6). By sequencing the larger-sized PCR products from dozens of individual colonies (see Materials and Methods), we confirmed that the 30-bp repeat of each a-CRISPR mutant was accurately duplicated during spacer acquisition (indicated in Figure 3B and Supplementary Figure S4, and see Supplementary Data S1 for more information), which illustrates that recognition of the two repeat termini was not disturbed by these mutations. Apparently, the leader-repeat junction is important as a fixed integration site, although this site may be recognized not simply in a sequence-specific manner.
Critical adaptation elements that locate in the middle of the repeat
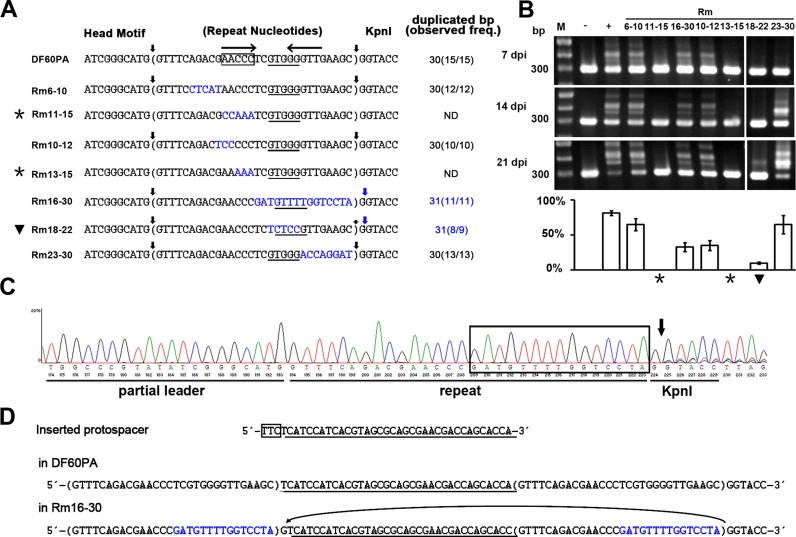
The H. hispanica repeat contains a pair of inverted repeats (IR1 and IR2 in Figure 3A), which implies its propensity to form a cruciform. To explore potential sequence and/or structural elements within the H. hispanica repeat, more mutational analyses were performed (Figure 4A). When nucleotides 6–10 were substituted, expansion of the mutated a-CRISPR was readily detected (Rm6–10 in Figure 4), suggesting conservation of these nucleotides may be dispensable. In contrast, mutation of the 11–15 nucleotides leaded to failure in spacer acquisition (Rm11–15 in Figure 4). For more information, another two mutants, Rm10–12 and Rm13–15, were further constructed (Figure 4A). As expected, expansion of the Rm13–15 a-CRISPR was not detected, but in Rm10–12, ∼35% of the 21-dpi PCR products were from the expanded a-CRISPR (Figure 4B). These data indicate that the conserved AACCC motif (corresponding to IR1) should be critical for adaptation in this system.
Figure 4.
Critical adaptation elements in the middle of the repeat. (A) Illustration of the a-CRISPR constructs with various repeat mutations (in blue). Sequences between vertical arrows were duplicated during spacer integration. The duplication size and frequency (duplication events/integration events) are summarized. The AACCC motif is framed for DF60PA, and for each a-CRISPR, the GTGGG motif or sequences that might be promiscuously recognized as this motif are underlined. The inverted repeats (IR1 and IR2) in the wild-type CRISPR repeat are indicated with convergent arrows. (B) Expansion of the a-CRISPRs shown in panel A at 7, 14 or 21 days post HHPV-2 infection (dpi). The percentage of expanded PCR products in each lane is shown in the histogram. Three replicates were tested for each mutant, and each gel shows a representative result. Lane Ms, dsDNA size markers. (C) DNA sequencing (using primer Exp-Fa in Figure 2A) result of the larger-sized PCR products from Rm16–30. Overlap of multiple signals was observed for positions downstream of the vertical arrow. The mutated sequence is framed. (D) A protospacer (underlined) was separately integrated into the wild-type a-CRISPR of DF60PA and the mutant a-CRISPR of Rm16–30. The PAM sequence is boxed. In Rm16–30, the mutated nucleotides are in blue, and the origination of the extra ‘G’ between the new spacer and its upstream repeat is indicated by an arrowed curve. Asterisks (*) or black triangles (▾) indicate CRISPR expansion was abrogated or seriously impaired.
But note that the cruciform-forming propensity of the repeat seemed to be dispensable, because when the palindrome was destroyed by mutating nucleotides 16–30 (including IR2, see Rm16-30 in Figure 4), a-CRISPR expansion was not abrogated, albeit moderately impaired. From the repeat alignment in Figure 3A, we noticed the conserved GTGGG (or GTTGG) sequence at positions 18–22. Notably, when we mutated this sequence into TCTCC (generating the mutant Rm18-22), a-CRISPR expansion was very seriously impaired: it was not observed until 21 dpi, and only ∼10% of the 21-dpi PCR products were the expanded ones (Figure 4B). In contrast, expansion was readily detected when nucleotides 23–30 were mutated (Figure 4B). These data suggest the conserved GTGGG motif should as well play an important role during adaptation.
The GTGGG motif involves in directing the leader-distal integration reaction
Interestingly, we found that the DNA sequencing result of the expanded PCR product mixture was different in Rm16–30: overlapping signals started to appear at the second ‘G’, instead of at the first ‘G’, of the KpnI restriction site (Figure 4C). By sequencing the expanded a-CRISPR of nine individual colonies, we confirmed that the ‘G’ immediately downstream of the repeat was always together duplicated during 11 spacer integration events (indicated in Figure 4A). Figure 4D shows an example of the same protospacer that has been separately integrated into the wild-type a-CRISPR of DF60PA and the mutated a-CRISPR of Rm16–30. The critical difference is the extra ‘G’ between the new spacer and its upstream new repeat in Rm16–30, which most likely have originated from the duplication of the ‘G’ immediately downstream of the original repeat (indicated in Figure 4D). Explicitly, the leader-distal terminus of the repeat was misrecognized when the leader-distal nucleotides 16–30 was mutated. This aberrant repeat duplication also revealed that the protospacers were acquired solely (without any PAM portions) in our experiments, which is different from the E. coli acquisition process (where the last PAM nucleotide is acquired in accompany with the protospacer) (39).
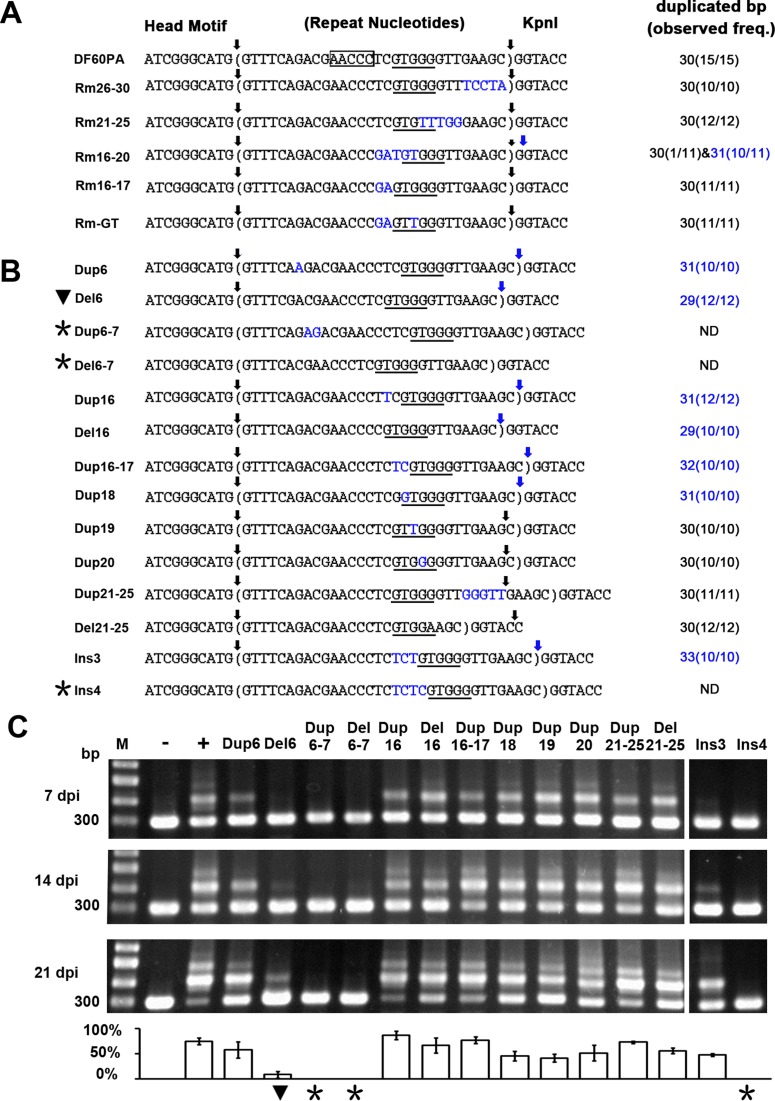
Interestingly, though the adaptation process was seriously impaired in Rm18–22 (Figure 4B), this mutated repeat was also duplicated together with its following ‘G’ during eight out of nine spacer integration events (Figure 4A). Therefore, we predicted a mid-repeat signal (possibly the GTGGG motif) may be involved in directing the leader-distal integration reaction. To confirm this prediction, we dissected the Rm16-30 mutation by constructing Rm16–20, Rm21–25 and Rm26–30 (Figure 5A). As expected, the repeat duplication size was consistently 30 bp in Rm21–25 and Rm26–30, but the Rm16–20 repeat was duplicated together with its following ‘G’ at a high frequency (10 out of 11 spacer insertion events). In addition, mutating nucleotides 16–17 did not alter the wild-type duplication size of 30 bp (Rm16–17 in Figure 5A). From the 30-bp and 31-bp duplicated ‘repeat’ sequences listed in Figures 4A and 5A, we noticed that the leader-distal terminus exhibited a constant distance (∼10 bp) from the centre of the GTGGG motif (positions 18–22). The most significant example may be the Rm16–20 mutant, in which the GTGGG moved 1 bp to positions 19–23, and thus the 31-bp duplication occurred (Figure 5A). Interestingly, when we exchanged its nucleotides T18 and G19 to generate a GTTGG sequence at positions 18–22, i.e. the canonical positions of the GTGGG (or GTTGG) motif (Figure 3A), the wild-type repeat duplication size (30 bp) was restored during 11 spacer integration events (Rm-GT in Figure 5A). These results support that the position of the conserved GTGGG (or GTTGG) motif (Figure 3A) is highly related to the repeat duplication size. It is noteworthy that some sequences at positions 18–22 or 19–23 (e.g. CTCCG, GTTTT and GTGTT from the mutants Rm18–22, Rm16–30 and Rm21–25, respectively) may be promiscuously recognized as this motif, thus generating the 30 or 31-bp ‘repeat’ duplication, though the adaptation process may be significantly impaired, for example, in Rm18–22 (Figure 4).
Figure 5.
The position of the GTGGG motif determines repeat duplication size. A series of a-CRISPR mutants were constructed with different repeat nucleotides substituted (A), deleted or duplicated (B). Nucleotide substitution and duplication are indicated in blue. The AACCC motif and the (promiscuously recognized) GTGGG motif are framed and underlined, respectively. Sequences between vertical arrows were duplicated during spacer integration, and for each a-CRISPR, the duplication size and corresponding frequency (duplication events/integration events) are summarized. (C) Expansion of the a-CRISPRs shown in panel B at 7, 14 or 21 days post HHPV-2 infection (dpi). The percentage of expanded PCR products in each lane is shown in the histogram. Three replicates were tested for each mutant, and each gel shows a representative result. Asterisks (*) or black triangles (▾) indicate CRISPR expansion was abrogated or seriously impaired.
To confirm the above observations and hypothesis, we altered the position of the GTGGG motif relative to the two repeat termini by duplicating or deleting different repeat-nucleotides (Figure 5B). As expected, duplicating (Dup6, Dup16 and Dup18) or deleting (Del6 and Del16) one nucleotide upstream of the GTGGG motif, which moved this motif downstream or upstream by 1 bp, resulted in 31-bp or 29-bp aberrant duplication. Furthermore, duplicating the nucleotides 16–17 (Dup16-17) or inserting three nucleotides (Ins3) between the AACCC and GTGGG motifs even resulted in 32- or 33-bp duplication (Figure 5B). In contrast, duplicating nucleotide 19 or 20 and duplicating/deleting nucleotides 21–25, which did not alter the position of the GTGGG motif relative to the leader-repeat junction, consistently resulted in the wild-type repeat duplication of 30 bp. In summary, repeat duplication consistently started from the leader-repeat junction, but terminated at a fixed distance away from the GTGGG motif rather than from the start site, which suggests a molecular ruler anchored at this motif.
It should be noted that duplicating or deleting two nucleotides upstream of the AACCC motif, like nucleotides 6–7 (Dup6–7 and Del6–7 in Figure 5B) or 8–9 (Dup8–9 and Del8–9 in Supplementary Figure S7), abrogated a-CRISPR expansion. Similarly, insertion of four (Ins4 in Figure 5) or more (Ins5 and Ins6 in Supplementary Figure S7) nucleotides between the AACCC and GTGGG motifs also abrogated adaptation. These data suggest the importance of suitable distances among the leader-repeat junction and the two mid-repeat motifs for spacer integration. Please refer to Supplementary Data S1 for more information about the duplicated sequences during spacer integration into different a-CRISPR mutants.
DISCUSSION
To generate adaptive immunity, the CRISPR-Cas system needs to integrate new spacers into the CRISPR locus, but this fundamental process remains poorly understood. An in vitro study on the E. coli spacer integration process observed products similar to those from retroviral integration and DNA transposition (34), suggesting their mechanistic similarities. Integration or transposition events usually cause target site duplication (TSD), which seemingly corresponds to repeat duplication during the spacer integration process. But notably, the TSD size is very short during retroviral integration (usually 4–6 bp) and DNA transposition. The largest one may be the 9–12 bp TSD of the Mutator/MuDR superfamily transposons (40), while the CRISPR repeat has an average size of 32 bp (41). Therefore in this view, the CRISPR spacer integration system is quite different. Interestingly, during preparation of our manuscript, a casposon-encoded Cas1 was reported to integrate specific sequences into non-specific target site and generate a 14–15 bp TSD (42), which is also much shorter than the CRISPR repeat.
During retroviral integration and DNA transposition, TSD derives from the staggered cleavage of their target site. Correspondingly, staggered cleavage of the CRISPR repeat during adaptation was also proposed (22,23), which was subsequently supported by the in vivo detection of the cleavage intermediates (33). In an interesting study using E. coli K-12 Cas proteins and a non-K12 CRISPR, spacer integration resulted in an aberrant new repeat which included two nucleotides from the upstream leader but lost the last two nucleotides of the original repeat (30). This observation led to the proposal that the non-cognate Cas proteins generated an initial cut at an incorrect position (possibly due to faulty sequence recognition), and then a second cut by a repeat-length molecular ruler. This proposal naturally raised the question about the order of the two cuts. The authors and many other studies proposed that the leader-repeat junction should be recognized for the initial cut (in a sequence-specific manner), because the sequences in there are more conserved and were shown important for adaptation (20,30,33,36). In contrast, Doudna and colleagues preferred an opposite model that the minus strand should be initially cut (by nucleophilic attack) at the leader-distal end of the first repeat, and then the plus strand be cut by a ruler mechanism (34). But as argued by Rollie et al., this preference may be true only for spacers with a C 3′-OH terminal (35).
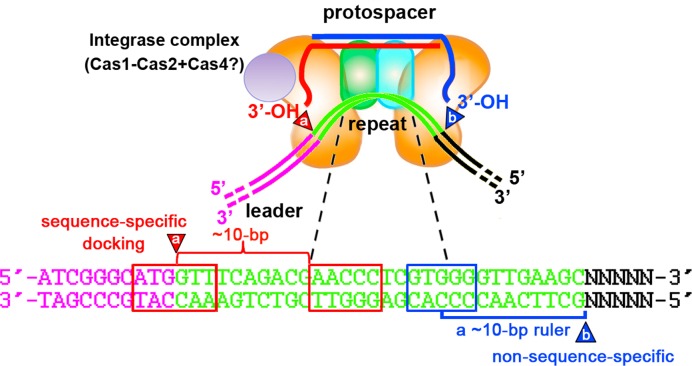
However, we wonder whether these two cuts occur sequentially with one depending on the other in all CRISPR systems. In this study on the H. hispanica I-B system, we fortunately observed repeat duplication with altered sizes (bigger or smaller than the repeat length) when spacers were integrated into a mutated repeat. Apparently, this observation indicates the repeat-length ruler should be not applicable for this system. In our study, the leader-proximal cut occurred consistently at the leader-repeat junction, supporting the critical role of the leader sequence during spacer integration. In contrast, the distal cut site was variable, but interestingly exhibited a constant distance to the conserved GTGGG motif (at positions 18–22 of the wild-type repeat), and this distance was not disturbed by any mutations (nucleotide substitutions, insertions or deletions) intervening them (Figure 5). However, mutations upstream of this motif, which altered its position relative to the leader-proximal cut site (the leader-repeat junction), caused aberrant duplication sizes. Therefore, we speculate the GTGGG motif may be recognized as an anchor of a shorter molecular ruler to generate the leader-distal cut. We also identified another mid-repeat motif, the conserved AACCC (at positions 11–15), which is essentially required for spacer integration (Figure 4). It's noteworthy that the ∼10-bp distance between the AACCC motif and the leader-repeat junction seemed to be important, because deletions and duplications, rather than substitutions, of the nucleotides between them seriously impaired or even abrogated spacer integration (Figure 5B). Based on our data and previous knowledge, we propose a possible model for the spacer integration process in H. hispanica (Figure 6). The mid-repeat AACCC and GTGGG motifs (or their complementary sequences on the minus strand), as well as the upstream leader-repeat junction, are probably recognized by the integrase complex. Together with the sequences spanning the leader-repeat junction, the AACCC motif may provide a docking site for the complex to direct the plus-strand cut precisely at the junction site (∼10 bp upstream of the AACCC motif). On the other side, a molecular ruler (∼10-bp) starting from the GTGGG recognition site may direct a downstream minus-strand cut in a non-sequence-specific manner. It should be noted that these two cuts occur not necessarily in order in this model. Interestingly, the two cut sites are both about 10 bp away from the corresponding mid-repeat recognition site, which reminds us of the length of one helical turn (10.4 bp) on the double helix DNA. We noticed that the CRISPR locus Hlac132P carried by the Halorubrum lacusprofundi plasmid pHLAC01 (GenBank identifier: CP001367.1) has a repeat sequence of 31 bp, rather than of the conserved repeat length (30 bp) for the haloarchaeal I-B CRISPRs. On the multi-alignment in Figure 3A, there is an extra nucleotide in the middle of this repeat, which caused an alignment gap in other repeat sequences. This extra nucleotide has expanded the conserved 2-bp interval between the AACCC and GTGGG motifs by 1 bp, which may explain its different repeat length and provide a natural evidence for our model.
Figure 6.
A possible model for the spacer integration process in H. hispanica. DNA motifs in the middle of the first repeat are recognized by the integrase complex (consisting of the protospacer, Cas1, Cas2 and perhaps Cas4) during spacer integration. The critical AACCC motif and the sequences spanning the leader-repeat junction require a specific distance (∼10 bp) between them, possibly to together provide a docking site for the integrase complex and to precisely direct the nucleophilic attack (indicated by a) at the junction site. In contrast, the leader-distal attack (indicated by b) is non-sequence-specific, and depends on a ∼10-bp molecular ruler starting from the GTGGG motif. Note that putative important interactions between the integrase complex (or other facilitative proteins) and upstream leader sections are not depicted.
The important roles of the mid-repeat motifs may help to explain their sequence conservation. Conserved mid-repeat motifs could be similarly observed for other repeat clusters defined by Kunin and colleagues (43), implying some of them may play similar roles during spacer integration. Though not required for adaptation, the 8 nucleotides downstream of the GTGGG motif are also conserved among the haloarchaeal I-B repeats, probably due to the interference requirement. Consistently, no virus immunity was observed for Rm21–25 and Rm26–30 clones that have acquired a new spacer from the virus (data not shown). These data also support the results in Supplementary Figure S3, which shows our adaptation assay doesn't rely on the interfering effects of new spacers.
In summary, our data demonstrated the significant roles of mid-repeat motifs during CRISPR adaptation of type I-B system. Though the generality of our spacer-integration model needs to be further verified for other subtypes, this study provides new insights into this fundamental but poorly characterized process. Besides, for the first time, we show that the repeat size or the periodicity of a CRISPR could be rationally modified to some extent.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31271334, 31571283]; CAS/SAFEA International Partnership Program for Creative Research Teams. Funding for open access charge: National Natural Science Foundation of China [31271334].
Conflict of interest statement. None declared.
REFERENCES
- 1.Makarova K.S., Wolf Y.I., Alkhnbashi O.S., Costa F., Shah S.A., Saunders S.J., Barrangou R., Brouns S.J., Charpentier E., Haft D.H., et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wright A.V., Nuñez J.K., Doudna J.A. Biology and applications of CRISPR systems: harnessing nature's toolbox for genome engineering. Cell. 2016;164:29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- 3.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 4.Brouns S.J.J., Jore M.M., Lundgren M., Westra E.R., Slijkhuis R.J.H., Snijders A.P.L., Dickman M.J., Makarova K.S., Koonin E.V., van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marraffini L.A., Sontheimer E.J. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiedenheft B., Sternberg S.H., Doudna J.A. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 7.Barrangou R., Marraffini L.A. CRISPR-Cas systems: prokaryotes upgrade to adaptive immunity. Mol. Cell. 2014;54:234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H., Peng N., Shah S.A., Huang L., She Q. Archaeal extrachromosomal genetic elements. Microbiol. Mol. Biol. Rev. 2015;79:117–152. doi: 10.1128/MMBR.00042-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sorek R., Kunin V., Hugenholtz P. CRISPR - a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 2008;6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 10.Bolotin A., Ouinquis B., Sorokin A., Ehrlich S.D. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 11.Mojica F.J.M., Díez-Villaseñor C., García-Martínez J., Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 12.Pourcel C., Salvignol G., Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 13.Haft D.H., Selengut J., Mongodin E.F., Nelson K.E. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 2005;1:474–483. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jansen R., van Embden J.D.A., Gaastra W., Schouls L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 15.Haurwitz R.E., Jinek M., Wiedenheft B., Zhou K.H., Doudna J.A. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li M., Liu H.L., Han J., Liu J.F., Wang R., Zhao D.H., Zhou J., Xiang H. Characterization of CRISPR RNA biogenesis and Cas6 cleavage-mediated inhibition of a provirus in the haloarchaeon Haloferax mediterranei. J. Bacteriol. 2013;195:867–875. doi: 10.1128/JB.01688-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carte J., Pfister N.T., Compton M.M., Terns R.M., Terns M.P. Binding and cleavage of CRISPR RNA by Cas6. RNA. 2010;16:2181–2188. doi: 10.1261/rna.2230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Semenova E., Jore M.M., Datsenko K.A., Semenova A., Westra E.R., Wanner B., van der Oost J., Brouns S.J.J., Severinov K. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. U.S.A. 2011;108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hale C.R., Zhao P., Olson S., Duff M.O., Graveley B.R., Wells L., Terns R.M., Terns M.P. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yosef I., Goren M.G., Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fineran P.C., Charpentier E. Memory of viral infections by CRISPR-Cas adaptive immune systems: acquisition of new information. Virology. 2012;434:202–209. doi: 10.1016/j.virol.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 22.Swarts D.C., Mosterd C., van Passel M.W.J., Brouns S.J.J. CRISPR interference directs strand specific spacer acquisition. PLoS One. 2012;7:e35888. doi: 10.1371/journal.pone.0035888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Datsenko K.A., Pougach K., Tikhonov A., Wanner B.L., Severinov K., Semenova E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat. Commun. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- 24.Li M., Wang R., Zhao D.H., Xiang H. Adaptation of the Haloarcula hispanica CRISPR-Cas system to a purified virus strictly requires a priming process. Nucleic Acids Res. 2014;42:2483–2492. doi: 10.1093/nar/gkt1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richter C., Dy R.L., McKenzie R.E., Watson B.N.J., Taylor C., Chang J.T., McNeil M.B., Staals R.H.J., Fineran P.C. Priming in the Type I-F CRISPR-Cas system triggers strand-independent spacer acquisition, bi-directionally from the primed protospacer. Nucleic Acids Res. 2014;42:8516–8526. doi: 10.1093/nar/gku527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fineran P.C., Gerritzen M.J.H., Suárez-Diez M., Künne T., Boekhorst J., van Hijum S.A.F.T., Staals R.H.J., Brouns S.J.J. Degenerate target sites mediate rapid primed CRISPR adaptation. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E1629–E1638. doi: 10.1073/pnas.1400071111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mojica F.J.M., Díez-Villaseñor C., García-Martínez J., Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 28.Shah S.A., Erdmann S., Mojica F.J.M., Garrett R.A. Protospacer recognition motifs: mixed identities and functional diversity. RNA Biol. 2013;10:891–899. doi: 10.4161/rna.23764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Westra E.R., Semenova E., Datsenko K.A., Jackson R.N., Wiedenheft B., Severinov K., Brouns S.J.J. Type I-E CRISPR-Cas systems discriminate target from non-target DNA through base pairing-independent PAM recognition. PLoS Genet. 2013;9:e1003742. doi: 10.1371/journal.pgen.1003742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Díez-Villaseñor C., Guzmán N.M., Almendros C., García-Martínez J., Mojica F.J.M. CRISPR-spacer integration reporter plasmids reveal distinct genuine acquisition specificities among CRISPR-Cas I-E variants of Escherichia coli. RNA Biol. 2013;10:792–802. doi: 10.4161/rna.24023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levy A., Goren M.G., Yosef I., Auster O., Manor M., Amitai G., Edgar R., Qimron U., Sorek R. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature. 2015;520:505–510. doi: 10.1038/nature14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li M., Wang R., Xiang H. Haloarcula hispanica CRISPR authenticates PAM of a target sequence to prime discriminative adaptation. Nucleic Acids Res. 2014;42:7226–7235. doi: 10.1093/nar/gku389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arslan Z., Hermanns V., Wurm R., Wagner R., Pul Ü. Detection and characterization of spacer integration intermediates in type I-E CRISPR-Cas system. Nucleic Acids Res. 2014;42:7884–7893. doi: 10.1093/nar/gku510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nuñez J.K., Lee A.S.Y., Engelman A., Doudna J.A. Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature. 2015;519:193–198. doi: 10.1038/nature14237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rollie C., Schneider S., Brinkmann A.S., Bolt E.L., White M.F. Intrinsic sequence specificity of the Cas1 integrase directs new spacer acquisition. eLife. 2015;4:e08716. doi: 10.7554/eLife.08716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei Y.Z., Chesne M.T., Terns R.M., Terns M.P. Sequences spanning the leader-repeat junction mediate CRISPR adaptation to phage in Streptococcus thermophilus. Nucleic Acids Res. 2015;43:1749–1758. doi: 10.1093/nar/gku1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu H.L., Han J., Liu X.Q., Zhou J., Xiang H. Development of pyrF-based gene knockout systems for genome-wide manipulation of the archaea Haloferax mediterranei and Haloarcula hispanica. J. Genet. Genomics. 2011;38:261–269. doi: 10.1016/j.jgg.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 38.Cai S., Cai L., Zhao D., Liu G., Han J., Zhou J., Xiang H. A novel DNA-binding protein, PhaR, plays a central role in the regulation of polyhydroxyalkanoate accumulation and granule formation in the haloarchaeon Haloferax mediterranei. Appl. Environ. Microbiol. 2015;81:373–385. doi: 10.1128/AEM.02878-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goren M.G., Yosef I., Auster O., Qimron U. Experimental definition of a clustered regularly interspaced short palindromic duplicon in Escherichia coli. J. Mol. Biol. 2012;423:14–16. doi: 10.1016/j.jmb.2012.06.037. [DOI] [PubMed] [Google Scholar]
- 40.Marquez C.P., Pritham E.J. Phantom, a new subclass of mutator DNA transposons found in insect viruses and widely distributed in animals. Genetics. 2010;185:1507–1582. doi: 10.1534/genetics.110.116673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bland C., Ramsey T.L., Sabree F., Lowe M., Brown K., Kyrpides N.C., Hugenholtz P. CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics. 2007;8:209. doi: 10.1186/1471-2105-8-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hickman A.B., Dyda F. The casposon-encoded Cas1 protein from Aciduliprofundum boonei is a DNA integrase that generates target site duplications. Nucleic Acids Res. 2015;43:10576–10587. doi: 10.1093/nar/gkv1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kunin V., Sorek R., Hugenholtz P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007;8:R61. doi: 10.1186/gb-2007-8-4-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.