Abstract
Chemically engineered small molecules targeting specific genomic sequences play an important role in drug development research. Pyrrole-imidazole polyamides (PIPs) are a group of molecules that can bind to the DNA minor-groove and can be engineered to target specific sequences. Their biological effects rely primarily on their selective DNA binding. However, the binding mechanism of PIPs at the chromatinized genome level is poorly understood. Herein, we report a method using high-throughput sequencing to identify the DNA-alkylating sites of PIP-indole-seco-CBI conjugates. High-throughput sequencing analysis of conjugate 2 showed highly similar DNA-alkylating sites on synthetic oligos (histone-free DNA) and on human genomes (chromatinized DNA context). To our knowledge, this is the first report identifying alkylation sites across genomic DNA by alkylating PIP conjugates using high-throughput sequencing.
INTRODUCTION
N-Methylpyrrole (P)—N-methylimidazole (I) polyamides (PIPs) are a class of programmable minor-groove binders that follow a canonical DNA recognition rule. The recognition rules are that an antiparallel arrangement of P opposite I (P-I) recognizes a C-G base pair; I-P recognizes a G-C base pair and P-P recognizes T-A or A-T base pairs (1). Several studies have investigated the binding specificity of these programmable PIPs (2–7). However, there is a bias toward PIPs binding chromatinized DNA. PIPs have the ability to penetrate the cell membrane and show an excellent DNA-binding efficiency, even in nanomolar concentrations (8), thus hindering the binding of transcription factors to their respective DNA sequences.
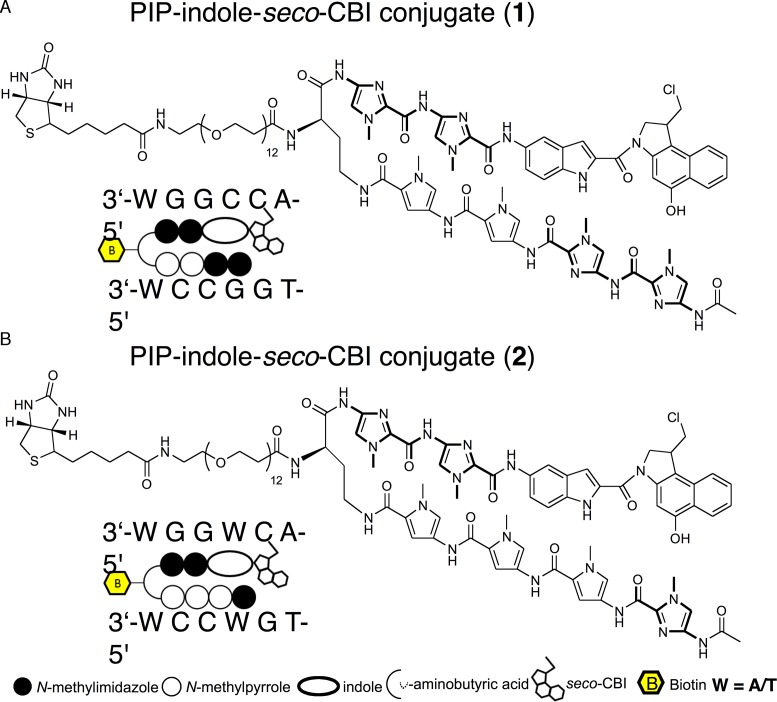
The normal transcriptional machinery sometimes becomes dysfunctional because of alteration of DNA bases causing variation in gene expression and development of disease. Such fluctuations in gene regulation, largely dictated by modifications such as DNA alkylation and methylation, are caused by various factors in day-to-day life (9,10). DNA damage induced by alkylating agents can modify the genetic code, resulting in faulty protein synthesis (9,11) that can cause abrupt cell-cycle arrest or apoptosis (12). This makes alkylating agents attractive as antitumor drugs. To date, many DNA-alkylating agents have been reported to exhibit anticancer activity toward a variety of leukemias and solid tumors (13). One of the major disadvantages of these agents is their non-selective DNA alkylation. Driving the alkylating agents toward tumor-specific target sequences in the human genome is a promising approach to advancing their efficacy as anticancer agents. Collectively, we developed various sequence-specific alkylating agents by coupling sequence-specific PIPs with alkylating moieties (14). Among the coupling linkers, the indole linker extends to two bases and its N-terminal sequence selectivity is achieved by the hydrogen bond of the amide group of indole with O2 of C or T, or with N3 of adenine (A) (15,16). Conjugating the alkylating moiety seco-CBI to a PIP can produce a covalent adduct with N3 of A within a predetermined sequence (Figure 1A and C). A PIP-indole-seco-CBI conjugate with unique sequence recognition was tested for antitumor activity by selective silencing of tumor-inducing genes (17). In this study, we have synthesized two PIP-indole-seco-CBI conjugates (1 with a symmetrical binding site of 5′-WGGCCA-3′ and 2 with asymmetrical binding site 5′-WGGWCA-3′ (Scheme 1)) to investigate their DNA-alkylating sites.
Figure 1.
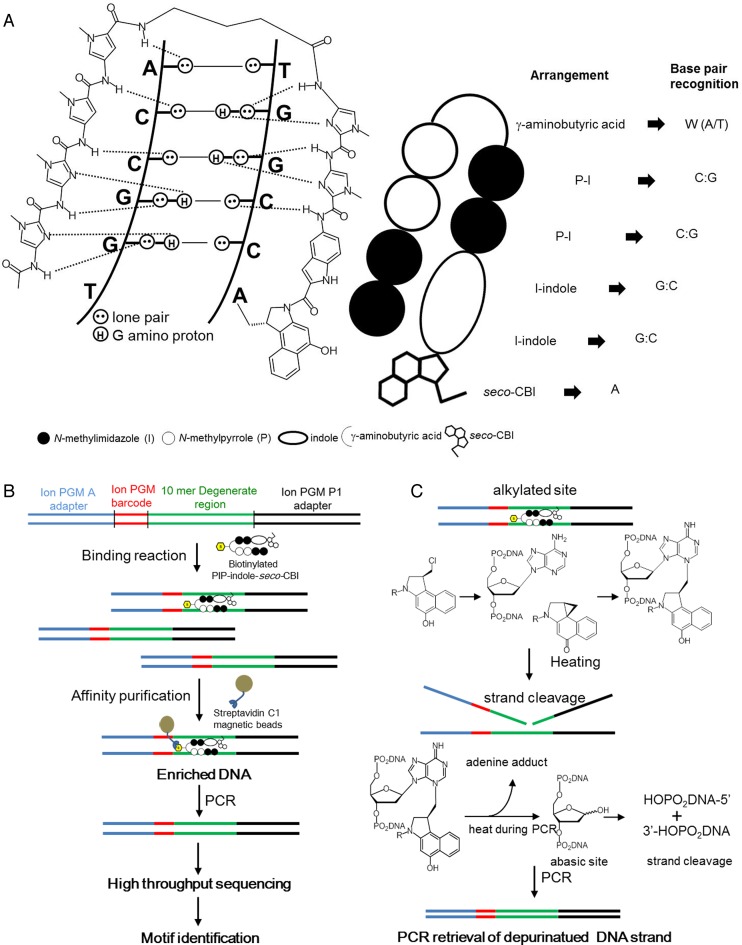
PIP conjugate-binding mode and Bind-n-Seq. (A) Recognition of DNA minor groove by PIP-indole-seco-CBI conjugates. (B) Workflow of Bind-n-Seq analysis with PIP-indole-seco-CBI conjugate. (C) PCR amplification-based depurinated strand retrieval and chemical reaction of DNA N3 alkylation by seco-CBI through the formation of CBI and heat treatment.
Scheme 1.
Chemical structures and representation of PIP-indole-seco-CBI conjugate. (A) 1 (Ac-I-I-P-P-(R)NH-PEG12-Biotinγ-I-I-Indole-seco-CBI). (B) 2 (Ac-I-P-P-P-(R)NH-PEG12-Biotinγ-I-I-Indole-seco-CBI). P = N-methylpyrrole and I = N-methylimidazole.
Previous studies have employed high-resolution denaturing polyacrylamide gel electrophoresis to test the sequence-specific DNA-alkylating efficiency of PIP-seco-CBI conjugates on pre-determined 200–300 bp template sequences. Under high-temperature conditions, alkylated sequences at the PIP-binding sites are cleaved, generating patterns of DNA fragments, which can be analyzed further (14–16). However, the selective DNA-alkylating ability of PIP-seco-CBI conjugates in a broad genome space using high-throughput sequencing has not yet been explored. Here, we describe the experimental design and data analysis methods to address this investigation.
Although PIP conjugates have many predicted binding sites across the human genome, only a small number of these binding sites play a significant role in gene regulation. This phenomenon is clearly observed in our previous report, where the whole genome expression analysis of a library of SAHA-PIP conjugates showed that individual compounds can trigger a distinctive set of genes in human dermal fibroblast (HDF) cells (18). The complex organization of chromatin packaging in the nucleus may be a critical factor for the PIP-binding preferences along the genome (19–21). A recent report by the Ansari group (22) has initiated the preliminary effort to map the PIP-binding sites in the human genome by developing an approach called ‘crosslinking of small molecules for isolation of chromatin’ or COSMIC, thereby directing the evolution in PIP design strategy for effectively targeted gene regulation. Data provided by ChIP-seq regarding the genome-wide mapping of key transcription factors and regulatory element-binding sites are helpful in the derivation of transcriptional regulation models that govern normal and diseased cell states (23,24). At this juncture, we have employed cost-efficient semiconductor-sequencing technology to study the affinity purification-based high-throughput sequencing of PIP-indole-seco-CBI conjugate-enriched human genomic regions. The present study will enable us to map the DNA-binding of small molecule DNA-alkylating sites all along the chromatin-packed genome utilizing high-throughput sequencing, which may provide a more detailed understanding of the mechanism of gene regulation by PIP conjugates.
MATERIALS AND METHODS
Synthesis of biotin-conjugated alkylating polyamide conjugates
Reagents and solvents were purchased from standard providers and used without further refinement. The EZ-Link NHS-PEG12-Biotin was obtained from Thermo Scientific, USA (No. 21312). Analytical High Performance Liquid Chromatography (HPLC) was performed using a COSMOSIL 5C18-MS-II reversed phase column (4.6 × 150 mm, Nacalai Tesque) in 0.1% Trifluoroacetic acid (TFA) in water with CH3CN as eluent at 1.0 ml/min, and a linear gradient elution of 0−100% CH3CN over 20 or 40 min with detection at 254 nm. The HPLC purification was performed with a COSMOSIL 5C18-MS-II reversed phase column (10 × 150 mm, Nacalai Tesque) in 0.1% TFA in water with CH3CN as the eluent. The final products were analyzed by ESI-TOF-MS (Bruker). The complete PIP synthesis procedure is provided in Supplementary Data.
Bind-n-Seq experiment and high-throughput sequencing
Bind-n-Seq experiments and high-throughput sequencing were performed based on our previous report (25). Broadly,
Synthesis of biotinylated alkylating PIPs and PIP-Conjugate 5 (a PIP conjugate where the alkylating CBI moiety was substituted with 3-dimethylaminopropylamine (Dp), synthesis details are given in Supplementary Data) (26), and a separate set of oligonucleotides with a 10- and 21-mer randomized region and Ion torrent adapters. (Oligomer designs and details are given in Supplementary Data) Oligonucleotides were duplexed by primer extension. Biotin conjugated alkylating PIPs and PIP-Conjugate 5 were allowed to bind and alkylate with their specific binding region of duplex randomized oligonucleotides separately at room temperature. Control experiments were performed without PIP-indole-seco-CBI conjugates/PIP-Conjugate 5, the data obtained were used for the normalization to acquire enrichment data. Biotin–streptavidin affinity-based purification was used to enrich the alkylating PIP attached DNA (washing steps were doubled for the PIP-indole-seco-CBI conjugates compared with the previously reported Bind-n-Seq to remove PIP simple binding).
Enriched DNA was subjected to polymerase chain reaction (PCR) to recover the alkylated DNA strand using sequencing library adapter-specific primers. The purified sequencing libraries were quantified using a BioAnalyzer with an Agilent DNA High Sensitivity BioAnalyzer kit, Agilent technologies, USA. Sufficient sequencing libraries with various barcodes were pooled for template preparation (Ion Personal Genome Machine (PGM) template OT2 200 kit) in an Ion OneTouch 2 system. The emulsion PCR amplified libraries were further enriched with Ion OneTouch ES. The enriched libraries were sequenced following the manufacturer's instructions with Ion PGM sequencer (Ion PGM sequencing 200 kit v2 and 318 chip V2 (Life Technologies, USA).
The sequenced reads were analyzed for a primary motif calling based on our previous reports (25–29).
Affinity purification-based high-throughput sequencing of human genomic regions enriched with PIP-indole-seco-CBI conjugate 2
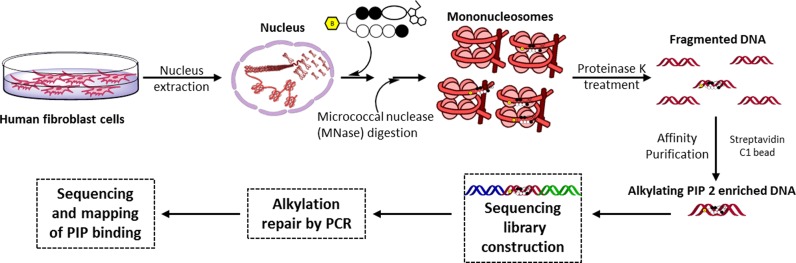
Human fibroblast BJ from neonatal foreskin (ATCC, USA), were maintained in 10% fetal bovine serum (FBS) (FBS, Japan Serum) supplemented with Dulbecco's modified eagle medium (DMEM, Nacalai Tesque, Japan), 10% HyClone FBS, non-essential amino acids, 100 U/ml penicillin, 100 μg/ml streptomycin and grown to 75–80% confluency in a humidified atmosphere of 5% CO2 at 37°C. Nuclei were isolated for alkylating PIP treatment (30–33). In brief, 2 × 106 P6 cells were washed with phosphate buffered saline (PBS) and isolated by 3 min trypsinization. The isolated cells were again washed 2× with ice-cold PBS. The cell pellet was suspended in 5 ml of ice-cold NP-40 lysis buffer (10 mM Tris–HCl (pH 7.4), 10 mM NaCl, 3 mM MgCl2, 0.5% Nonidet P-40, 0.15 mM spermine, 0.5 mM spermidine and 0.1× protease inhibitor cocktail) and incubated on ice for 5 min. The nuclei were pelleted by centrifugation at 300 g for 10 min. The pellet of nuclei was carefully resuspended in modified binding buffer (22) (10 mM Tris–Cl (pH 8.0), 5 mM MgCl2, 1 mM DTT, 0.3 M KCl, 0.3× protease inhibitor cocktail and 10% glycerol). The nuclei were incubated with 400 nM of 2 (dissolved in Dimethyl sulfoxide (DMSO), 0.1% final concentration) at 4°C for 16 h. Control experiments were performed without PIP-indole-seco-CBI conjugates and with a 0.1% final concentration of DMSO. We used the PIP concentrations from the previous report (22) that were consistent with the PIP quantity measured in the nuclei of treated cells (34). PIP-containing nuclei were washed with micrococcal nuclease (MNase) buffer (10 mM Tris–HCl (pH 7.4), 15 mM NaCl, 60 mM KCl, 0.15 mM spermine, 0.5 mM spermidine and 0.1× protease inhibitor cocktail) (30). Cell nuclei suspension was digested with MNase (TaKaRa, Japan) for 30 min at 37°C to obtain mononucleosomes in optimized reaction conditions. The reactions contained MNase buffer, MNase (0.2 μl of MNase (20 Units/μl) for nucleus extracted from 2 × 106 cells), RNase A and protease inhibitor cocktail. After digestion, the histone protein was removed by proteinase K treatment. After MNase digestion and proteinase K treatment the suspension was mixed with an equal volume of modified COSMIC buffer (20 mM Tris–Cl (pH 8.1), 2 mM ethylenediaminetetraacetic acid (EDTA), 150 mM NaCl, 0.1× protease inhibitor cocktail, 1% Triton-X100 and 0.1% sodium dodecyl sulphate (SDS)) (22). Ten percent of the sample was saved as input DNA. The assessment of the size distribution of DNA samples showed about 100–180 bp fragment distribution.
Preparation of magnetic beads. After removing the suspension solution from streptavidin-coated magnetic beads (Dynabeads MyOne C1, Life Technologies, USA). They were washed with 2× modified COSMIC buffers and resuspended in the same buffer. Resuspended streptavidin-coated magnetic beads (0.5 mg) were incubated with samples for 16 h at 4°C. After the incubation period, bound and unbound DNA samples were separated using affinity purification. Briefly, samples were washed 5 min once with 0.5 ml of washing buffer 1 (10 mM Tris–Cl (pH 8.0), 1 mM EDTA, 3% SDS), once with altered washing buffer 2 (10 mM Tris–Cl (pH 8.0), 250 mM LiCl, 1 mM EDTA, 0.5% NP40), 2× with altered washing buffer 3 (10 mM Tris–Cl (pH 7.5), 1 mM EDTA, 0.1% NP-40) and 3× with TE. The samples were then resuspended in elution buffer (10 mM Tris–HCl (pH 7.6), 0.4 mM EDTA and 100 mM KOH) (22) and DNA was eluted from magnetic beads after heating at 90°C for 30 min. The remaining DNA with the beads were eluted using elution buffer 2 (2% SDS, 100 mM NaHCO3 and 3 mM biotin) with heating to 65°C for 8–12 h. The detached samples were purified with a QIAquick PCR purification Kit (Qiagen, CA, USA) and quantified.
Polyamide-based affinity purification sequencing libraries were prepared using standard Ion Xpress Plus gDNA Fragment Library Preparation reagents and protocols (Life technologies, USA). Sequencing adapter ligated enriched DNA was subjected to a PCR to recover the alkylated DNA strand using sequencing library adapter specific primers and purified. The purified libraries were subjected to quality and quantity checks with an Agilent DNA High sensitivity BioAnalyzer kit (Agilent technologies, USA). The qualified libraries were used for high-throughput sequencing. The sequencing was performed, starting with template preparation using Ion PGM template OT2 200 v2 kit and an Ion PI template OT2 200 kit using an Ion OneTouch 2 system. The templates were then enriched using Ion OneTouch ES. The enriched libraries were sequenced with 260–300 flow of a single read performed with an Ion PGM sequencer using an Ion PGM sequencing 200 kit v2/318 v2 chip and an Ion Proton Sequencer using an Ion PI Sequencing 200 kit v3/Ion PI chip following the manufacturer's guidelines and we produced 25–30 million post filtered reads per library. The data were handled by employing standard program packages in the Ion torrent suite. A Torrent Mapping Alignment Program version 4.4.2 (TMAP) was used for aligning reads (mean read length 121 bp), ChIP-seq peaks were called using MACS version 1.4.2 (35) with the default parameters and a P-value < 0.001 (cutoff <10–2) (In total, 721 617 peaks were called). Enriched peak and PIP-indole-seco-CBI conjugate binding site annotations were made using Homer (36). To determine the high-affinity DNA-alkylating sites (motif) of 2 over the control sequence reads, a randomly sampled 10–15% of the uniquely mapped reads for each setting were used. The random sampling was performed using a Perl script (http://meme-suite.org/doc/fasta-subsample.html). We followed our previous analysis pipeline for motif calling (25–29).
We evaluated the genome-wide enrichment signature of 2 by calculating the cross-correlation of MACS peaks with the identified binding sites (DNA-alkylating motif). The peaks containing identified binding sites were considered as significant enrichment regions (total of 355 882 peaks). Analysis pipelines agplus (37), Position Weight Matrix model generation and evaluation-PWMScan (http://ccg.vital-it.ch/pwmscan) (38,39), ChIP-Cor Analysis Module (http://ccg.vital-it.ch/chipseq/chip_cor.php) and various platforms of the Signal Search Analysis Server (http://ccg.vital-it.ch/ssa) were used to evaluate the spatial precision of 2 enrichment data.
Validating PIP-indole-seco-CBI conjugate 2 bound and enriched region in the human genome
Human fibroblast BJ from neonatal foreskin and SKBR3 (breast adenocarcinoma, human) cell lines were purchased from ATCC. Fibroblast cells were grown in DMEM supplemented with 10% HyClone FBS, non essential amino acids, 100 U/ml penicillin, 100 μg/ml streptomycin, at 37°C in 5% CO2. SKBR3 cells were grown in ATCC-formulated McCoy's 5a modified medium complemented with 10% FBS and were maintained under an atmosphere of 5% CO2 at 37°C.
The effect of alkylating PIP 2 on the expression of ERBB2 mRNA was determined in both fibroblast and SKBR3 cell lines using real-time PCR. BJ skin fibroblast cells were seeded at a density of 5 × 104 cells/well of a 6-well plate and SKBR3 cells were plated into the 6-well plate at 4 × 105 cells/well. The cells were then treated with 50 and 100 nM of alkylating PIP 2 for 48 h with DMSO as a corresponding control sample. After 48 h, total RNA was isolated using RNEasy Kit (Qiagen) and cDNA was synthesized by ReverTra Ace qPCR RT Master mix with genomic DNA remover (Toyobo, Japan) following the manufacturer's instructions. The expression level of ERBB2 was normalized using β-actin, as an internal control.
The primers used in this study includes, β-actin sense, 5′-CAATGTGGCCGAGGACTTTG-3′ and antisense, 5′-CATTC TCCTTAGAGAGAAGTGG-3′. The sense primer of ERBB2 is 5′-AGCCGCGAGCA CCCAAGT-3′ and antisense, 5′-TTGGTGGGCAGGTAGGTGAGTT-3′.
RESULTS
Bind-n-Seq with PIP-indole-seco-CBI conjugate 1
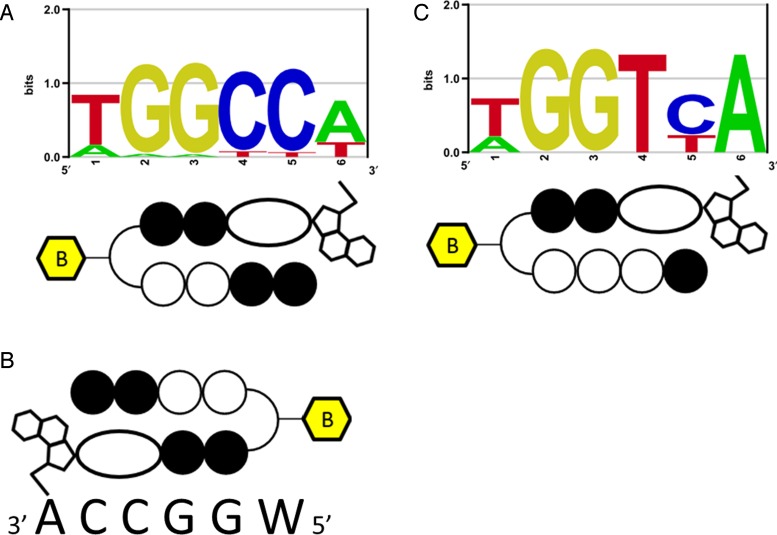
High-resolution denaturing polyacrylamide gel electrophoresis from our previous report showed multiple DNA-alkylating sites for the PIP-indole-CBI conjugate (B) (16). These results are biased toward identifying primary DNA-alkylating sites. To address this kind of issue, we synthesized a biotin-conjugated PIP-indole-seco-CBI conjugate 1 (synthetic procedure is given in Supplementary Data) and examined 1 for Bind-n-Seq (Figure 1B and C) (25,40–41) (experimental procedure is given in the ‘Materials and Methods’ section). The method is high-throughput sequencing-based, which may allow unbiased primary binding motif identification. Seco-CBI is readily converted to its cyclopropyl form CBI, and reacts with its DNA-reactive site (42). The CBI could possibly depurinate the DNA alkylation site during heat elution of enriched DNA in Bind-n-Seq. Therefore, we performed a PCR with sequencing adapter-specific primers to retrieve the damaged strand (Figure 1C). The purified enriched DNA was subjected to high-throughput sequencing. High quality uniquely randomized sequence reads were analyzed using the Bind-n-Seq analysis method (25) to obtain the high-affinity DNA-alkylating site of 1. The DNA showed a highly enriched ‘k-mer’ (k = 6) binding site. The enriched motif with 24.11-fold enrichment defined the DNA-alkylating sites of 1 and matches the PIP canonical binding rule. A graphical representation of the identified high-affinity motif for 1 is shown in Figure 2A; and its corresponding highly enriched sequences are given in Supplementary Table S1. Supplementary Table S1 also shows the other potential binding sites of 1 at ranks 3 and 6 (highlighted in green). Although the recognition site of 1 followed the PIP canonical binding rule, the alkylation site of 1 (sixth position from the 5′ end of the motif) was found to be W (A or T) instead of the expected base, A, because of the symmetrical nature of its binding. The results of the Bind-n-Seq analysis of 1 are vital because they show that this type of symmetrical PIP-indole-seco-CBI conjugate has the capability to bind with both strands of DNA in a forward-binding orientation (N-terminal to C-terminal of PIP binding with 5′ to 3′ of DNA) as shown in Figure 2A and B. When the binding of 1 is largely on symmetrical sequences, the acquired motif is acceptable with respect to the PIP-binding rule, but it is difficult to identify the precise alkylation site of CBI in 1 because of its symmetrical nature.
Figure 2.
PIP-indole-seco-CBI conjugate DNA-alkylating motifs. (A) Bind-n-Seq analysis identified high-affinity DNA-alkylating motif for 1. (B) One possible binding in its complementary recognition sequence. (C) Bind-n-Seq analysis identified a high-affinity DNA-alkylating motif for 2.
Bind-n-Seq with PIP-indole-seco-CBI conjugate 2
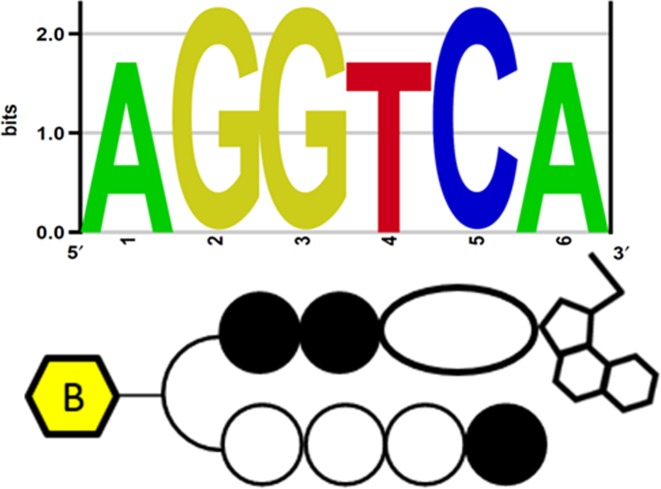
The investigation of Bind-n-Seq results for 1 showed the significance of PIP binding to its target site, but could not elucidate the CBI alkylation site. Taken together, our results for compound 1 motivated us to inspect the exact CBI alkylation site within a PIP conjugate. Therefore, we synthesized another asymmetrical PIP-indole-seco-CBI conjugate 2. We designed 2 by replacing I with P in 1, to obtain asymmetrical binding (synthetic procedure is given in the Supplementary Data). Accordingly, 2 was subjected to Bind-n-Seq analysis. Analysis of the sequence reads obtained from 2 enriched DNA is shown in Supplementary Table S2. The high-affinity DNA-alkylating motif of 2 (Figure 2C) was derived from 7.99- and 7.36-fold enrichment (corresponding highly enriched sequences are given in Supplementary Table S2). The motif obtained follows the binding rule and specific A alkylation site for CBI in 2. These findings support our earlier report of specific A alkylation by PIP-CBI conjugates within its recognition sequence (43).
Bind-n-Seq with PIP-conjugate 5
To confirm the significance of the enriched DNA-alkylating motif of PIP-indole-seco-CBI conjugates and PCR repossession of the alkylated DNA strand, we performed a Bind-n-Seq experiment (21-mer randomized region) with our previously reported (26) biotinylated PIP-Conjugate 5 (designed to target 8 bp). Bind-n-Seq data was analyzed for the 8 and 9 bp motif windows (k = 8 and k = 9) to obtain the fold enrichment based on the control experiment (experiment without PIP-conjugate 5). High-scoring motif hits (Supplementary Figure S2) clearly demonstrated that there is no sequence selectivity at the ninth position of the motif (from the 5′ end of the motif). By contrast, PIP conjugate 2 showed a distinct (A) adenine-specific alkylation at the targeted sixth position from the 5′ end of the motif. This base specific alkylation site identification could be possible only when there is recovery of previously damaged alkylated DNA. These results demonstrated that successful retrieval of a damaged DNA strand could be possible using PCR reaction.
Identification of PIP-indole-seco-CBI conjugate 2 high affinity DNA-alkylating site in chromatinized human genome
We then sought to extend the high-throughput sequencing approach to examine how chromatinized genomic architecture can impact the binding of 2 across the human genome in nuclei isolated from live cells. Consistent with this approach, we developed a method (Figure 3) based on the COSMIC approach (22) that includes affinity purification-based high-throughput sequencing of human genomic regions enriched with 2 without any photo-crosslinking procedure. We employed this method to elucidate the binding preferences of 2 in the biologically dynamic, histone-packed chromatinized surroundings of the nuclei (experimental procedure is described in the ‘Materials and Methods’ section). The qualified sequencing libraries were subjected to sequencing. The processed sequence data was mapped along the human genome. To comprehensively determine the DNA-alkylating site (primary motif), we performed motif detection using a Bind-n-Seq data analysis pipeline. Among the enriched sequences (normalized with the control experimental data), the high-scoring motif hit (rank 1 in Supplementary Table S3) was unique DNA-alkylation site of 2 (Figure 4) (enriched sequence details are given in Supplementary Table S3). Interestingly, the identified primary motifs of 2 propose that its sequence-specific DNA-alkylation of base A remains highly similar in both complex chromatinized human genomic DNA and randomized oligomer-based Bind-n-Seq analyses.
Figure 3.
Work flow for affinity purification-based high-throughput sequencing using PIP-indole-seco-CBI conjugates in the nucleus of human cells.
Figure 4.
Identified high-affinity DNA-alkylating motif of 2 in human genomic enriched sequence.
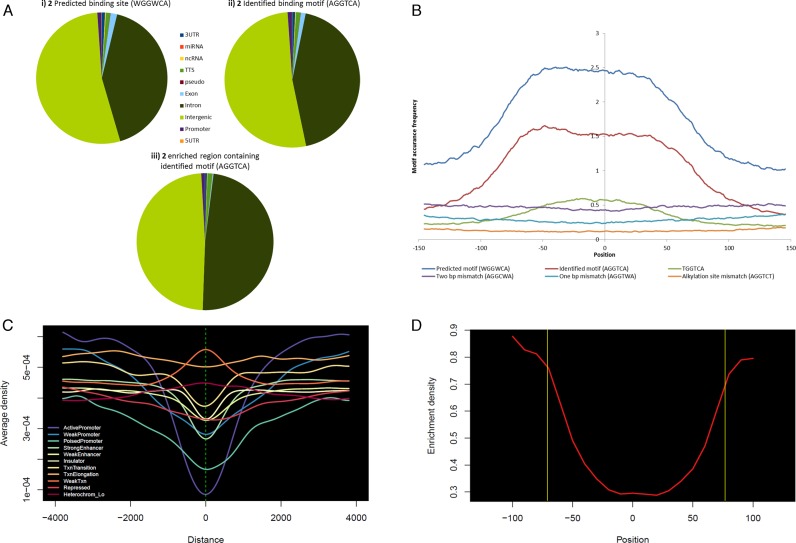
To compare the sensitivity of 2 enrichment, we have conducted MACS peak calling and the cross-correlation of identified DNA-alkylating motif with the obtained post-filtered genomic sequence data (25–30 million). Then we plotted (Figure 5B) the identified motif (5′-AGGTCA-3′) on the genome-wide enriched peaks with the window of 300 bp (–150 to 150 bp from the center (0) of the peak). This showed the greatest precision of distribution frequency with a 2 predicted recognition site (5′-WGGWCA-3′). Whereas, 1 bp mismatch (5′-AGGTWA-3′) and 2 bp mismatch (5′-AGGCWA-3′) sequence of 2 showed poor distribution in the enriched regions. In Figure 5B, we have also illustrated the mismatch DNA-alkylating site (5′-AGGTCT-3′) that displayed very low frequency. These results confirm the efficiency of the genomic pull-down enrichment by 2. Additionally, the γ-turn in 2 uniquely recognized the base A (adenine), which may be the result of chromatinized DNA binding (Figure 4). This exclusive detection was further verified with the results of poorly dispersed possible recognition of T (5′-TGGTCA-3′) by the γ-turn in the conjugate 2 (green line in Figure 5B). Reproducibility of the experiment is confirmed with R2 = 0.92 for two separate experimental enriched motifs (Supplementary Figure S1) and MACS peak annotation distribution (Supplementary Table S4).
Figure 5.
Genomic enrichment and DNA-alkylating site distribution of PIP-indole-seco-CBI conjugate 2: (A) (i) Predicted binding sites based on the canonical binding rule, (ii) experimentally identified high-affinity DNA-alkylating motif in human genome, (iii) affinity purification based sequencing enriched regions containing experimentally identified high-affinity DNA-alkylating motif. (B) 2 related possible binding sites (predicted, experimentally identified, possible recognition of T by the γ-turn in the conjugate, 1 bp mismatch, 2 bp mismatch and alkylation site mismatch) genome-wide distribution frequencies in the peak enriched region with the MACS peak window of 300 bp (−150 to 150 bp from the center (0) of the enriched peak). (C) 2 enriched region distribution on broad classes of chromatin states (six classes of chromatin states such as promoter (active, weak and poised), enhancer (strong and weak), insulator, transcribed (strongly (Txn transition and Txn elongation) and weakly transcribed regions), repressed and inactive states (heterochromatin)) (chromatin stated were organized based on the ENCODE-ChromHMM data). (D) Nucleosomal occupancy of the genome-wide 2 enriched region containing identified binding site. The region inside the yellow box corresponding to the nucleosomal region of ∼147 bps (nucleosomal positioning was measured based on the ENCODE nucleosomal positioning data).
Identification of PIP-indole-seco-CBI conjugate 2 representative enriched sites in chromatinized human genome
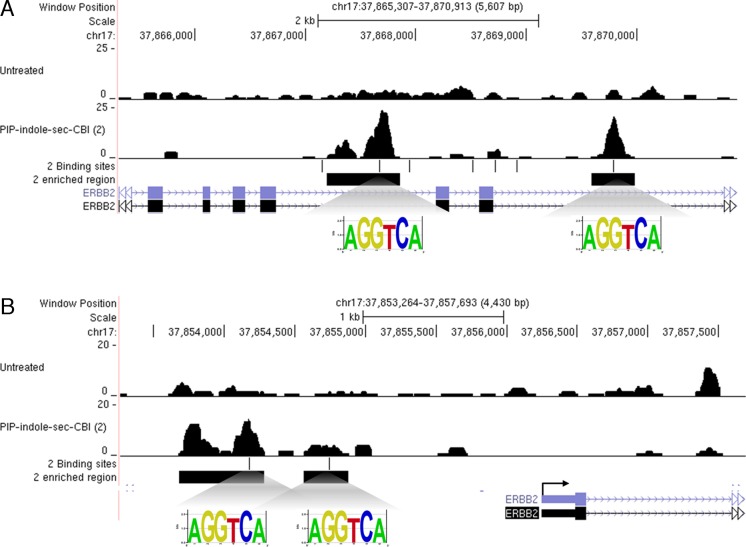
Targeted gene silencing can be achieved by PIPs, either by designing the PIPs to the transcription factor recognition site on the gene promoter (44) or by alkylating PIPs targeted to the coding region of the respective gene producing nonfunctional truncated mRNA (45). To obtain deeper insight into the gene of interest targeted by PIP-indole-seco-CBI conjugate 2, the enriched peak regions were mapped and correlated with the DNA-alkylating site of 2 on the human genome. One of the significantly enriched genomic regions ERBB2 is shown in Figure 6 (enrichment details are given in Supplementary Table S5) with the DNA-alkylating site. Several oncogenes have been studied in human cancers, but only a few have been reported to play a critical role in the progression of breast cancer. ERBB2 is one such oncogene overexpressed in many epithelial cancers and in about 20% of early-stage breast cancer patients with low survival rates, and confers chemo-resistance (46). By contrast, the universally expressed reference genes, such as ACTB and GAPDH did not show any enriched regions (Supplementary Figure S5). However, the quantification of ERBB2 mRNA (using real-time PCR) in conjugate 2 treated human BJ skin fibroblast cells and SKBR3 breast adenocarcinoma cells showed inhibition of ERBB2 mRNA expression with respect to reference genes in both cell types (Supplementary Figure S6).
Figure 6.
Genome-wide mapping of PIP-indole-seco-CBI (2) (A) Two binding and enriched genomic area in the ERBB2 gene coding region. (B) Two binding and enriched genomic area of the ERBB2 promoter region.
Genome-wide analysis of PIP-indole-seco-CBI conjugate 2 enriched sites distribution
To analyze the influence of complex eukaryotic chromatin conformation on the DNA-alkylation preferences of PIP conjugate, we performed a computational genome-wide distribution survey of 2 binding sites (predicted binding site 5′-WGGWCA-3′ based on the binding rule and genome-wide experimentally derived binding site 5′-AGGTCA-3′) in various annotated regions of the human genome (Supplementary Table S6 and Figure 5A i and ii). The total numbers of experimentally identified binding sites are significantly less than the predicted binding sites. This clearly shows that chromatin conformation plays a critical role in polyamide binding. We have also annotated the 2 enriched genomic regions across the human genome (Supplementary Table S6 and Figure 5A iii). The result showed high correlation with the genome-wide distribution pattern of DNA-alkylating sites.
We next sought to assess the annotated position of the predominant 2 enriched regions across the genome. We first compared the distribution of 2 identified motifs (DNA-alkylating site) with predicted binding sites (Supplementary Table S7 and Figure S3a) that retained comparable distribution of genomic positions (3′-UTR, miRNA, ncRNA, TTS, pseudo genes, exon, intron, intergenic, promoter and 5′-UTR). We again compared 2 enriched regions with identified and predicted binding regions that showed the rate of enrichment to be high in TTS, intron, intergenic and promoter regions when compared with the other genomic regions (Supplementary Table S7 and Figure S3b and c). To test this predominance, we generated an aggregation plot (37) with 2 enriched reads (Supplementary Figure S4). The average enrichment read density in the upstream (–4 kb) of TSSs (proximal and distal promoter) possess convincing read density and gene body detects crimped average enrichment read density because of the existence of 5′-UTR, 3′-UTR, coding exons and introns in the gene body (Supplementary Figure S4). These distribution marks are consistent with the enrichment distribution we determined earlier.
We next asked whether 2 enrichment profiling with various chromatin states could be used to infer a systematic means of perceiving PIP accessible regions; because the chromatin framework of a genome plays a central role in controlling DNA access. We examined the 2 enriched sites distribution on ChromHMM segments (47) and we present in Figure 5C, on a broad scale; (i) 2 differs in accessing various promoter states and its low average enrichment density on active promoters may support the target specific gene suppression by PIP conjugates. (ii) The positional distribution along enhancer, insulator and transcribed regions contain an almost equivalent form of enrichment, so designing PIPs to target such genomic positions may provide a correlative genome-wide effect. (iii) 2 showed characteristic patterns of chromatin accessibility that have been observed at repressed and inactive states. By contrast, the access of nucleosomal DNA by PIP is limited (20). In line with this, we sought to inspect 2 enrichment sites on nucleosomes at a fine scale of an approximately 147 bps window, we investigated the positioning of nucleosomes with the enriched regions using the MNase-Seq data (33). In this model, we were able to estimate the 2 binding density, which appears to be higher at the ends of nucleosome than in the core middle region (Figure 5D). Our data with greater precision at the genomic scale confirm the previously reported (studied at the defined nucleosomal core particle context) limitation of PIP accessibility; in addition, we report that this limitation might be as a result of the central core of well-positioned nucleosomes. Overall, our genome-wide enrichment assessment results provide a deeper understanding of the PIP accessibility toward chromatinized DNA.
DISCUSSION
New methods in chemical biology with deep sequencing applications and data analysis are constantly being developed (24). In this study, we have made use of high-throughput sequencing technology to show the significant sequence-specific DNA-alkylation of PIP-indole-seco-CBI conjugates corresponding to the proposed DNA-binding rule. The binding specificity of our small molecule remains similar in a broad sequence context of free DNA and in complex chromatinized human genome. However, our DNA-alkylating site mapping on the nucleosome resulting in this sequence-specific binding have limitations on the central core of chromatinized DNA. Progress in the field of sequencing technologies has allowed us to conduct this study cost-efficiently, and it may be a useful method for other DNA-binding small molecule design and redesign in the context of the complex genome space. Our results also indicate that the structural composition of PIP-indole-seco-CBI conjugates favorably alters the sequence specificity of PIPs. In future, this method may be an efficient tool for studying sequence-specific alkylation and in designing small molecules for targeted gene silencing compared with conventional polyacrylamide gel electrophoresis analysis.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan; JSPS KAKENHI [24225005]; ‘Basic Science and Platform Technology Program for Innovative Biological Medicine’; ‘JSPS-NSF International Collaborations in Chemistry (ICC)’ (to H.S.); JSPS KAKENHI [24310155 to T.B.]; JEES Mitsubishi Corporation Scholarship (to A.C., J.S.). Funding for open access charge: JSPS KAKENHI (Grant Numbers 24225005).
Conflict of interest statement. None declared.
REFERENCES
- 1.Dervan P.B., Edelson B.S. Recognition of the DNA minor groove by pyrrole-imidazole polyamides. Curr. Opin. Struct. Biol. 2003;13:284–299. doi: 10.1016/s0959-440x(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 2.Warren C.L., Kratochvil N.C.S., Hauschild K.E., Foister S., Brezinski M.L., Dervan P.B., Phillips G.N., Ansari A.Z. Defining the sequence-recognition profile of DNA-binding molecules. Proc. Natl. Acad. Sci. U.S.A. 2006;103:867–872. doi: 10.1073/pnas.0509843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Puckett J.W., Muzikar K., Tietjen J., Warren C.L., Ansari A.Z., Dervan P.B. Quantitative microarray profiling of DNA-binding molecules. J. Am. Chem. Soc. 2007;129:12310–12319. doi: 10.1021/ja0744899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keleş S., Warren C.L., Carlson C.D., Ansari A.Z. CSI-Tree: a regression tree approach for modeling binding properties of DNA-binding molecules based on cognate site identification (CSI) data. Nucleic Acids Res. 2008;36:3171–3184. doi: 10.1093/nar/gkn057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moretti R., Donato L.J., Brezinski M.L., Stafford R.L., Hoff H., Thorson J.S., Dervan P.B., Ansari A.Z. Targeted chemical wedges reveal the role of allosteric DNA modulation in protein-DNA assembly. ACS Chem. Biol. 2008;3:220–229. doi: 10.1021/cb700258r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ozers M.S., Warren C.L., Ansari A.Z. Determining DNA sequence specificity of natural and artificial transcription factors by cognate site identifier analysis. Methods Mol. Biol. 2009;544:637–653. doi: 10.1007/978-1-59745-483-4_41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tietjen J.R., Donato L.J., Bhimisaria D., Ansari A.Z. Sequence-specificity and energy landscapes of DNA-binding molecules. Methods Enzymol. 2011;497:3–30. doi: 10.1016/B978-0-12-385075-1.00001-9. [DOI] [PubMed] [Google Scholar]
- 8.Carlson C.D., Warren C.L., Hauschild K.E., Ozers M.S., Qadir N., Bhimsaria D., Lee Y., Cerrina F., Ansari A.Z. Specificity landscapes of DNA binding molecules elucidate biological function. Proc. Natl. Acad. Sci. U.S.A. 2010;107:4544–4549. doi: 10.1073/pnas.0914023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kondo N., Takahashi A., Ono K., Ohnishi T. DNA damage induced by alkylating agents and repair pathways. J. Nucleic Acids. 2010;2010:543531. doi: 10.4061/2010/543531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 11.Rouse J., Jackson S.P. Interfaces between the detection, signaling, and repair of DNA damage. Science. 2002;297:547–551. doi: 10.1126/science.1074740. [DOI] [PubMed] [Google Scholar]
- 12.Zhou B.B., Elledge S.J. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 13.Hurley L.H. DNA and its associated processes as targets for cancer therapy. Nat. Rev. Cancer. 2002;2:188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- 14.Bando T., Sugiyama H. Synthesis and biological properties of sequence-specific DNA-alkylating pyrrole-imidazole polyamides. Acc. Chem. Res. 2006;39:935–944. doi: 10.1021/ar030287f. [DOI] [PubMed] [Google Scholar]
- 15.Shinohara K.I., Bando T., Sasaki S., Sakakibara Y., Minoshima M., Sugiyama H. Antitumor activity of sequence-specific alkylating agents: pyrolle-imidazole CBI conjugates with indole linker. Cancer Sci. 2006;97:219–225. doi: 10.1111/j.1349-7006.2006.00158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shinohara K.I., Sasaki S., Minoshima M., Bando T., Sugiyama H. Alkylation of template strand of coding region causes effective gene silencing. Nucleic Acids Res. 2006;34:1189–1195. doi: 10.1093/nar/gkl005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiraoka K., Inoue T., Taylor R.D., Watanabe T., Koshikawa N., Yoda H., Shinohara K., Takatori A., Sugimoto H., Maru Y., et al. Inhibition of KRAS codon 12 mutants using a novel DNA-alkylating pyrrole–imidazole polyamide conjugate. Nat. Commun. 2015;6:6706. doi: 10.1038/ncomms7706. [DOI] [PubMed] [Google Scholar]
- 18.Pandian G.N., Taniguchi J., Junetha S., Sato S., Han L., Saha A., AnandhaKumar C., Bando T., Nagase H., Vaijayanthi T., et al. Distinct DNA-based epigenetic switches trigger transcriptional activation of silent genes in human dermal fibroblasts. Sci. Rep. 2014;4:3843. doi: 10.1038/srep03843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jespersen C., Soragni E., James Chou C., Arora P.S., Dervan P.B., Gottesfeld J.M. Chromatin structure determines accessibility of a hairpin polyamide-chlorambucil conjugate at histone H4 genes in pancreatic cancer cells. Bioorg. Med. Chem. Lett. 2012;22:4068–4071. doi: 10.1016/j.bmcl.2012.04.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gottesfeld J.M., Melander C., Suto R.K., Raviol H., Luger K., Dervan P.B. Sequence-specific recognition of DNA in the nucleosome by pyrrole-imidazole polyamides. J. Mol. Biol. 2001;309:615–629. doi: 10.1006/jmbi.2001.4694. [DOI] [PubMed] [Google Scholar]
- 21.Dudouet B., Burnett R., Dickinson L.A., Wood M.R., Melander C., Belitsky J.M., Edelson B., Wurtz N., Briehn C., Dervan P.B., et al. Accessibility of nuclear chromatin by DNA binding polyamides. Chem. Biol. 2003;10:859–867. doi: 10.1016/j.chembiol.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 22.Erwin G.S., Bhimsaria D., Eguchi A., Ansari A.Z. Mapping polyamide-DNA interactions in human cells reveals a new design strategy for effective targeting of genomic sites. Angew. Chem. Int. Ed. Engl. 2014;53:10124–10128. doi: 10.1002/anie.201405497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Northrup D.L., Zhao K. Application of ChIP-Seq and related techniques to the study of immune function. Immunity. 2011;34:830–842. doi: 10.1016/j.immuni.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anandhakumar C., Kizaki S., Bando T., Pandian G.N., Sugiyama H. Advancing small-molecule-based chemical biology with next-generation sequencing technologies. Chembiochem. 2014;16:20–38. doi: 10.1002/cbic.201402556. [DOI] [PubMed] [Google Scholar]
- 25.Anandhakumar C., Li Y., Kizaki S., Pandian G.N., Hashiya K., Bando T., Sugiyama H. Next-generation sequencing studies guide the design of pyrrole-imidazole polyamides with improved binding specificity by the addition of β-alanine. Chembiochem. 2014;8501:1–6. doi: 10.1002/cbic.201402497. [DOI] [PubMed] [Google Scholar]
- 26.Taylor R.D., Chandran A., Kashiwazaki G., Hashiya K., Bando T., Nagase H., Sugiyama H. Selective targeting of the KRAS Codon 12 mutation sequence by pyrrole-imidazole polyamide seco -CBI conjugates. Chemistry. 2015;21:14996–15003. doi: 10.1002/chem.201501870. [DOI] [PubMed] [Google Scholar]
- 27.Zykovich A., Korf I., Segal D.J. Bind-n-Seq: high-throughput analysis of in vitro protein-DNA interactions using massively parallel sequencing. Nucleic Acids Res. 2009;37:e151. doi: 10.1093/nar/gkp802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bailey T.L. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics. 2011;27:1653–1659. doi: 10.1093/bioinformatics/btr261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Workman C.T., Yin Y., Corcoran D.L., Ideker T., Stormo G.D., Benos P. V. enoLOGOS: a versatile web tool for energy normalized sequence logos. Nucleic Acids Res. 2005;33:389–392. doi: 10.1093/nar/gki439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Published in association with Cold Spring Harbor Laboratory Press. Micrococcal nuclease–Southern blot assay. Nat. Meth. 2005;2:719–720. [Google Scholar]
- 31.Richard-Foy H., Hager G.L. Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J. 1987;6:2321–2328. doi: 10.1002/j.1460-2075.1987.tb02507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Enver T., Brewer A.C., Patient R.K. Simian virus 40-mediated cis induction of the Xenopus beta-globin DNase I hypersensitive site. Nature. 1985;318:680–683. doi: 10.1038/318680a0. [DOI] [PubMed] [Google Scholar]
- 33.Gaffney D.J., McVicker G., Pai A.A., Fondufe-Mittendorf Y.N., Lewellen N., Michelini K., Widom J., Gilad Y., Pritchard J.K. Controls of Nucleosome Positioning in the Human Genome. PLoS Genet. 2012;8:e1003036. doi: 10.1371/journal.pgen.1003036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsu C.F., Dervan P.B. Quantitating the concentration of Py-Im polyamide-fluorescein conjugates in live cells. Bioorg. Med. Chem. Lett. 2008;18:5851–5855. doi: 10.1016/j.bmcl.2008.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng J., Liu T., Qin B., Zhang Y., Liu X.S. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 2012;7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maehara K., Ohkawa Y. agplus: a rapid and flexible tool for aggregation plots. Bioinformatics. 2015;31:3046–3047. doi: 10.1093/bioinformatics/btv322. [DOI] [PubMed] [Google Scholar]
- 38.Iseli C., Ambrosini G., Bucher P., Jongeneel C.V. Indexing strategies for rapid searches of short words in genome sequences. PLoS One. 2007;2:e579. doi: 10.1371/journal.pone.0000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meier J.L., Yu A.S., Korf I., Segal D.J., Dervan P.B. Guiding the design of synthetic DNA-binding molecules with massively parallel sequencing. J. Am. Chem. Soc. 2012;134:17814–17822. doi: 10.1021/ja308888c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang J.S., Meier J.L., Dervan P.B. Design of sequence-specific DNA binding molecules for DNA methyltransferase inhibition. J. Am. Chem. Soc. 2014;136:3687–3694. doi: 10.1021/ja500211z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bando T., Sasaki S., Minoshima M., Dohno C., Shinohara K.I., Narita A., Sugiyama H. Efficient DNA alkylation by a pyrrole-imidazole cbi conjugate with an indole linker: sequence-specific alkylation with nine-base-pair recognition. Bioconjug. Chem. 2006;17:715–720. doi: 10.1021/bc060022w. [DOI] [PubMed] [Google Scholar]
- 43.Bando T., Narita A., Sasaki S., Sugiyama H. Specific adenine alkylation by pyrrole-imidazole CBI conjugates. J. Am. Chem. Soc. 2005;127:13890–13895. doi: 10.1021/ja052412j. [DOI] [PubMed] [Google Scholar]
- 44.Syed J., Pandian G.N.N., Sato S., Taniguchi J., Chandran A., Hashiya K., Bando T., Sugiyama H. Targeted suppression of EVI1 oncogene expression by sequence-specific pyrrole-imidazole polyamide. Chem. Biol. 2014;21:1–11. doi: 10.1016/j.chembiol.2014.07.019. [DOI] [PubMed] [Google Scholar]
- 45.Shinohara K.I., Narita A., Oyoshi T., Bando T., Teraoka H., Sugiyama H. Sequence-specific gene silencing in mammalian cells by alkylating pyrrole-imidazole polyamides. J. Am. Chem. Soc. 2004;126:5113–5118. doi: 10.1021/ja031673v. [DOI] [PubMed] [Google Scholar]
- 46.Arteaga C.L., Sliwkowski M.X., Osborne C.K.a, Perez E., Puglisi F., Gianni L. Treatment of HER2-positive breast cancer: current status and future perspectives. Nat. Rev. Clin. Oncol. 2011;9:16–32. doi: 10.1038/nrclinonc.2011.177. [DOI] [PubMed] [Google Scholar]
- 47.Ernst J., Kheradpour P., Mikkelsen T.S., Shoresh N., Ward L.D., Epstein C.B., Zhang X., Wang L., Issner R., Coyne M., et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.