Abstract
The Sulfolobales host a unique family of crenarchaeal conjugative plasmids some of which undergo complex rearrangements intracellularly. Here we examined the conjugation cycle of pKEF9 in the recipient strain Sulfolobus islandicus REY15A. The plasmid conjugated and replicated rapidly generating high average copy numbers which led to strong growth retardation that was coincident with activation of CRISPR-Cas adaptation. Simultaneously, intracellular DNA was extensively degraded and this also occurred in a conjugated Δcas6 mutant lacking a CRISPR-Cas immune response. Furthermore, the integrated forms of pKEF9 in the donor Sulfolobus solfataricus P1 and recipient host were specifically corrupted by transposable orfB elements, indicative of a dual mechanism for inactivating free and integrated forms of the plasmid. In addition, the CRISPR locus of pKEF9 was progressively deleted when conjugated into the recipient strain. Factors influencing activation of CRISPR-Cas adaptation in the recipient strain are considered, including the first evidence for a possible priming effect in Sulfolobus. The 3-Mbp genome sequence of the donor P1 strain is presented.
INTRODUCTION
Members of the acidothermophilic Sulfolobales carry an exceptional class of crenarchaea-specific conjugative plasmids (1–4). Each plasmid carries a small cluster of conserved genes encoding proteins implicated in conjugation that encompass up to half of the plasmid. Most of these proteins carry transmembrane helical motifs and two of the larger conserved proteins show sequence motifs and domain structures characteristic of TraG and TrbE proteins that are involved in the bacterial DNA transfer and the Type IV secretion apparatus (2,5–7). The conjugative plasmids spread rapidly in cultures of different Sulfolobus species but they are generally unstable and tend to undergo rearrangements and deletions by recombination at direct repeat motifs and, moreover, they are often lost during continuous growth (2,5,6). Apart from a requirement for cell–cell contact, little is known about the conjugation mechanism (1), and the inherent instability of the plasmids has undermined attempts to investigate mechanisms of conjugation, DNA replication and copy number control.
CRISPR-Cas immune systems have been shown to target conjugative plasmids of both archaea and bacteria (8–10) and, given the complexity of Sulfolobus CRISPR-Cas systems (11), they are likely to contribute to plasmid instability. This inference is reinforced by the demonstration that CRISPR loci of different Sulfolobus species carry multiple spacers with significant sequence matches to conjugative plasmids (12–14). However, the potential interaction between host and plasmid is complicated by the finding that some conjugative plasmids, including pNOB8 and pKEF9, also carry small CRISPR loci (2,6). Moreover, for pKEF9, two of the spacers show significant sequence matches to Sulfolobus viruses (12) and, given that the CRISPR locus is transcribed, and likely processed by the host Cas6 endoribonuclease, it can yield mature crRNAs that can potentially assemble into host CRISPR-Cas interference modules and cleave at matching sequences of coinvading genetic elements, or their integrated forms in the host chromosome (13).
The first experimental studies on archaeal CRISPR-Cas adaptation from genetic elements of Sulfolobus species demonstrated that activation required the presence of two genetic elements, either a virus (SMV1) and conjugative plasmid (pMGB1) or two viruses (SMV1 + STSV2) (9,15,16). Subsequently, CRISPR-Cas adaptation was achieved with a single virus (STSV2) and it was then proposed that a threshold level of viral infection was critical for the activation (17). Possibly this leads to overexpression of Csa3a which regulates expression of the adaptation CRISPR-Cas module (18). It was also proposed for Sulfolobus species that CRISPR-Cas adaptation was stimulated by viral DNA replication because whereas non-replicating SMV1 was resistant to spacer acquisition, coinfecting and replicating genetic elements were susceptible (15,16). Moreover, this hypothesis received experimental support from the demonstration that formation of DNA breaks at replication forks stimulates spacer acquisition in a subtype I-E CRISPR-Cas system of Escherichia coli (19).
The present study was undertaken to examine factors affecting the stability of pKEF9, a conjugative plasmid originally isolated from a Sulfolobus islandicus KEF strain collected at Kerlingarfjöll, Iceland (3). S. islandicus REY15A was selected as the recipient host and growth properties were examined together with the CRISPR-Cas immune response and intracellular DNA profiles during pKEF9 conjugation. The recipient host encodes a single CRISPR-Cas adaptation module and three functionally diverse CRISPR-Cas interference modules of subtype I-A, III-B (Cmr-α) and III-B (Cmr-β) which target DNA, transcribing DNA and transcripts, respectively (20–22). The results demonstrate that post conjugation the plasmid replicates rapidly, resulting in strong retardation of cell growth, extensive degradation of cellular DNA and cell death. Moreover, the host CRISPR-Cas adaptation module is activated and the CRISPR locus of pKEF9 is progressively deleted.
MATERIALS AND METHODS
Sequencing of pKEF9-conjugated S. solfataricus P1
DNA from pKEF9-conjugated Sulfolobus solfataricus P1 was isolated using the Qiagen genomic DNA extraction kit (Qiagen, Westberg, Germany) and subjected to high-throughput DNA sequencing using the PacBio technology (GATC Biotech AG, Konstanz, Germany). Genome sequencing yielded 40 046 reads with a average length 4890 bp and after precleaning, 31 129 reads with a mean read length of 5010 bp, were assembled into the donor and plasmid genomes. Sequences were analysed using the CLC Workbench (Qiagen) and Artemis (23). Dot-plot comparison with the S. solfataricus P2 genome was visualized using the MUMmer package NUCmer 2.1 (24).
pKEF9 conjugation
Most experiments were performed with the genetic host S. islandicus E233S lacking the pyrEF genes (25), and with the Δcas6 mutant (26). They were cultured in Sulfolobus medium supplemented with 0.1% vitamin, 0.1% CAA, 0.2% sucrose and 0.1% uracil (SCVU medium) (27). Cells were diluted to A600 = 0.05 and grown to A600 = 0.1–0.2 before mixing pKEF9-conjugated S. solfataricus P1 cells at a donor:recipient ratio of 1:10 000. The mixtures were then incubated under moderate shaking at 75°C. Samples were removed at the indicated times and were either plated on Gelrite or used for optical density measurements and total DNA preparation. A600 values were measured twice daily. A total of 10 ml of cells were harvested by centrifugation (6000 g, 10 min), and plasmids were extracted using either alkaline lysis (28) or the Omega BAC/PAC DNA Kit (Omega, Georgia, USA).
qPCR determination of pKEF9 copy number
The copy number changes of pKEF9 were determined by semi-quantitative polymerase chain reaction (PCR). Real-time qPCR amplifications were performed using a BioRad CFX96 qPCR System. The SYBR Green PCR Master Mix (Thermo Fisher) was used for preparing real-time PCR reactions. The thermal cycling protocol was as follows: an initial denaturation step of 10 min at 95°C, followed by 40 cycles of 10 s at 95°C, 10 s at 55°C and 20 s at 72°C. A final step at 95°C was performed for 10 s after the 40th cycle. A melt curve was performed from 65 to 95°C with 0.5°C increases in 5 s increments. The fluorescence signal was measured at the end of each extension step. Two primer couples were employed to amplify: (i) a fragment of the pKEF9 trbE gene and (ii) a region of a cdc gene S. islandicus REY15A (Supplementary Table S1). The copy number of pKEF9 was calculated by dividing the yield of the pKEF9 gene by that of host gene.
Detection of CRISPR spacer acquisition
Cells were harvested by centrifugation (6000 g, 10 min) and DNA was isolated using the DNeasy® Blood &Tissue Kit (Qiagen). Leader end regions of S. islandicus CRISPR loci 1 and 2 were amplified by PCR. The products were separated in 1% agarose gels and bands larger than those produced from the unconjugated strain were excised and purified with QIAquick Gel Extraction Kit (Qiagen). The DNA was then cloned using InsTAcloneTM PCR Cloning Kit (Thermo Fisher Scientific, MA, USA) following the manufacturer's protocol. Plasmid purification and sequencing were performed by Eurofins MWG-Biotech (Ebersberg, Germany). PCR sequences were analysed using Artemis (23) and the CLC Workbench (Qiagen).
Location of pKEF9 integration sites
Southern hybridization was employed to determine the pKEF9 integration sites in S. islandicus and also to establish the presence of the pKEF9-matching spacer 44, CRISPR locus 2, in the recipient host. A standard procedure was used (29) in which about 4 µg total DNA from each sample was digested with EcoRI, fractionated in 1.0% agarose gels and transferred to IMMOBILON-NY+ membranes (Millipore, MA, USA) by capillary transfer. Membrane-bound DNA was then auto-crosslinked using a UV Cross-linker (Stratagene, CA, USA). Hybridization probes were amplified by PCR (Supplementary Table S1), purified and labelled with Digoxigenin Labelling kit (Roche, Basel, Switzerland). Hybridization was performed overnight at 42°C and hybridization signals were detected using a DIG detection kit with the CDP-star (Roche) and recorded on CP-BUl X-ray films (AGFA, Mortsel, Belgium). Integration of pKEF9 at two tRNAGlu (CTC and CTT) genes was established by PCR amplification using listed primers (Supplementary Table S1).
Flow cytometry
Sulfolobus cells were taken from the culture at different time points, fixed with 75% ethanol and stored at 4°C for at least 12 h. When all the samples were collected, fixed cells were centrifuged at 3000 rpm for 20 min. The pellet was resuspended in 950 μl 10 mM Tris-Cl, pH 7.5, 10 mM MgCl2. Samples were recentrifuged under the same conditions and pellets were resupended in 145 μl in the same buffer containing mithramycin (100 μg/ml) and ethidium bromide (20 μg/ml) and maintained in the dark for 1 h. At each step samples were kept on ice and they were analysed with an ApogeeFlow A-40 flow cytometer (Apogee Flow Systems, Hertfordshire, UK).
Transmission electron microscopy
Virus-like particles were adsorbed onto carbon-coated copper grids for 5 min and stained with 2% uranyl acetate. Images were recorded using a Tecnai G2 transmission electron microscope (FEI, Eindhoven, Holland), with a CCD camera, at an acceleration voltage of 120 kV.
RESULTS
Genome of the pKEF9 donor strain S. solfataricus P1
The stable pKEF9 donor strain S. solfataricus P1 was established, and provided, by Wolfram Zillig (30). DNA from the strain was subjected to high-throughput DNA sequencing using PacBio technology, and the plasmid and host genomes were assembled. The pKEF9 sequence was consistent with the published version (6). A low level of single nucleotide heterogeneity was observed, especially after mononucleotide runs, and these were likely to have arisen primarily from sequencing errors inherent in the PacBio sequencing technique. One major sequence difference was observed; the plasmid CRISPR locus was present in two forms. One form exhibited seven repeats and six spacers including a degenerate first repeat, as described earlier (6,13), while a minor component, estimated from sequence coverage at a molar ratio of about 1:7, carried only the degenerate repeat 1-spacer-repeat 2.
The sequence and gene synteny of the S. solfataricus P1 chromosome (ENA Accession Number: http://www.ebi.ac.uk/ena/data/view/LT549890) was similar to that of S. solfataricus P2 (31) but with several significant differences, as illustrated in the gene dot-plot (Supplementary Figure S1). Strain P1 carried an additional fuselloviral SSV2 genome integrated in the tRNAGly gene (positions 594 867–609 661) which, as for pKEF9, exhibited minor single nucleotide heterogeneities relative to the published sequence (32). Furthermore, strain P1 carried an additional region of about 63 kbp (1 133 458–1 196 503) encompassing genes involved in toluene metabolism (todA and tmoA) and it lacked an 18.6-kbp region of strain P2 (2 553 890–2 572 446) carrying genes implicated in energy and lipid metabolism (Supplementary Figure S1). Two further regions of about 22.5 kbp (460 293–482 781) and 18.7 kbp (1 645 876–1 664 610) were inverted with respect to strain P2, although the former region was also inverted in another sequenced P2 strain, denoted P2A (33). Remarkably, the six CRISPR loci of strain P1, carrying in total 430 spacer-repeat units, were identical to those sequenced earlier (12,13) and none of the spacers showed significant sequence matches (<7 mismatches) to either pKEF9 or SSV2.
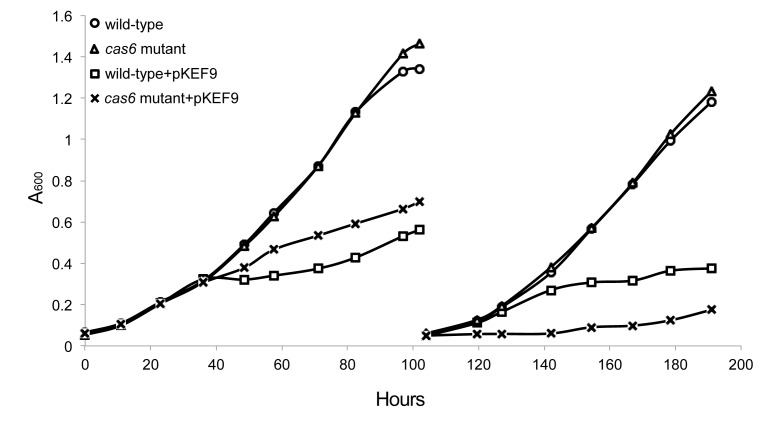
pKEF9 conjugation results in cell growth retardation
A recipient culture of wild-type S. islandicus was prepared and the donor strain P1 was added at a donor:recipient ratio of 1:10 000 and the culture was diluted successively every 4 days to A600 = 0.05, on reaching stationary growth. Growth retardation was first observed at about 37 hpc (Figure 1). Plasmid copy numbers were estimated for the recipient culture over time by qPCR, and the results showed that the plasmid replicated rapidly reaching on average about 16 copies per cell at 24 h post conjugation (hpc), 150 copies at 38 hpc and 48 copies at 85 hpc. (1.2) Lower values were observed in the donor P1 strain when pKEF9 peaked at about 60 copies per cell. However, it should be emphasized that both chromosomal and plasmid DNA are subject to degradation and therefore the copy number estimates are necessarily approximate.
Figure 1.
Growth curves of wild-type and Δcas6 strains of Sulfolobus islandicus unconjugated, and conjugated with Sulfolobus solfataricus P1-pKEF9 in the ratio 1:10 000. Similar growth curves were obtained in triplicate experiments.
The conjugation experiment was repeated for a mutant of S. islandicus lacking the single gene encoding the Cas6 endoribonuclease essential for processing CRISPR transcripts (26). The inability to produce crRNAs in this mutant renders all three CRISPR-Cas interference modules inactive (26). From 40 to 100 hpc the conjugated mutant grew slightly faster than the conjugated wild-type, and over this period the mutant pKEF9 copy numbers were 10–30% lower, but after 100 hpc, the wild-type recipient outgrew the mutant (Figure 1).
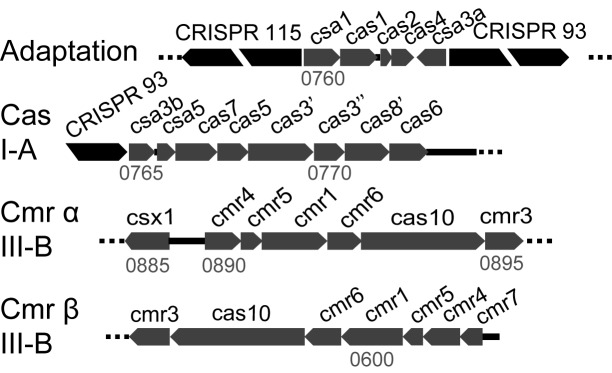
Host CRISPR-Cas adaptation is activated
The CRISPR loci and Cas/Cmr gene cassettes of S. islandicus (34,35) are illustrated in Figure 2. In order to test for active spacer acquisition, the leader ends of CRISPR loci 1 and 2 of the recipient host were amplified by PCR. The formation of additional larger PCR products in agarose gels indicated the presence of newly acquired spacers in both CRISPR loci at 37 hpc and the yields of the larger PCR products increased progressively with time (Figure 3A and B). At 37 hpc the plasmid copy number was also very high and strong growth retardation was initiated.
Figure 2.
Overview of the gene cassettes of the single adaptation and three interference CRISPR-Cas modules of Sulfolobus islandicus REY15A. CRISPR loci 1 (115 repeat-spacer units) and 2 (93 units) are indicated and genomic locations are given.
Figure 3.
Agarose gel showing PCR products from leader ends of Sulfolobus islandicus CRISPR loci before and after conjugation. (A) Locus 1: (1) no pKEF9, (2) 22 hpc. (3) 37 hpc. Locus 2: (4) no pKEF9, (5) 22 hpc. (6) 37 hpc. (B) Locus 1: (1) no pKEF9, (2) 85 hpc. Locus 2: (3) no pKEF9 (4) 85 hpc. Arrows denote bands carrying de novo spacers. M—DNA size markers.
Larger PCR products obtained after 12 days of continuous growth were cloned and sequenced. Of 107 sequenced clones, 53 yielded 73 de novo spacers from CRISPR locus 1 and 54 clones generated 81 de novo spacers from CRISPR locus 2. There was no major bias of protospacer distributions around the pKEF9 plasmid or between the DNA strands (Figure 4). For example, 12.3% of matching protospacers fell within intergenic regions and 12.9% of the genome is non-protein-coding. However, there was a relative increase in the frequency of protospacers in one plasmid region extending from about position 14 000, neighbouring the putative DNA replication origin, to position 20 000 beyond the CRISPR locus (Figure 4).
Figure 4.
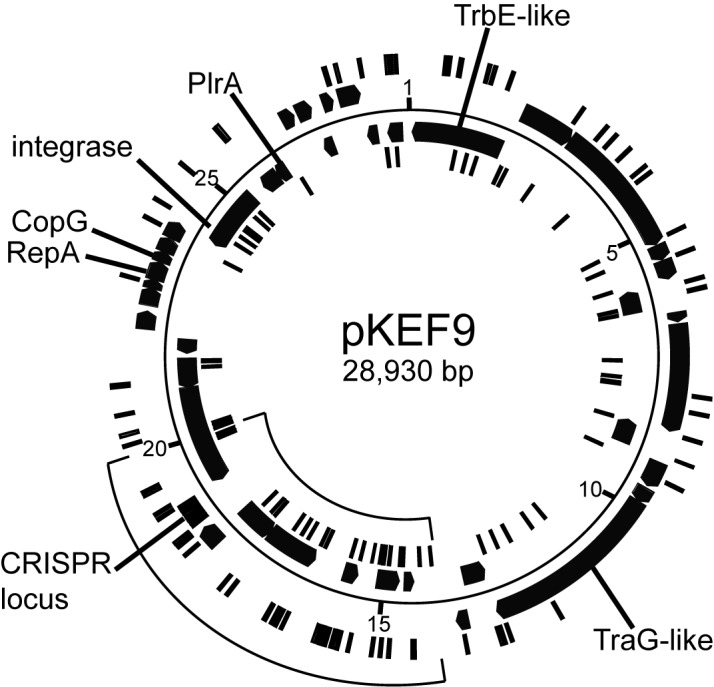
Circular genome map of pKEF9. Protospacers on each DNA strand are indicated on inner and outer concentric circles. Gene products with predicted functions are labelled. The CRISPR array is marked.
An additional eight de novo CRISPR spacers matched to SSV2 DNA, seven with perfect matches and one with a single mismatch and each protospacer carried a cognate CCN PAM sequence. This indicated that the integrated SSV2 had been excised from donor P1 strain DNA and had infected S. islandicus, albeit with a lower copy number than pKEF9. The presence of a low level of SSV2-like fusellovirus particles in the S. islandicus culture, post conjugation, was confirmed by low virus extraction yields and electron microscopy (Supplementary Figure S2).
Of the nine remaining de novo spacers, three yielded perfect matches to the single mobile orfB element of the donor and six spacers matched recipient host genes: isoleucine–tRNA synthetase (SiRe_1286), formate dehydrogenase α subunit (SiRe_2464), preprotein translocase subunit SecY (SiRe_1311), NAD(P)-dependent glycerol-1-phosphate dehydrogenase (SiRe_1249), FAD-dependent oxidoreductase (SiRe_1973) and an uncharacterized protein (SiRe_1317).
A similar PCR analysis was performed on CRISPR loci 1 and 2 of the Δcas6 mutant but no additional larger PCR products were produced at time points after 38 hpc and it was inferred that CRISPR-Cas adaptation was inactive in the mutant (data not shown).
The host carries a single spacer 44, CRISPR locus 2, with a single mismatch to pKEF9 and we confirmed its presence in the wild-type culture undergoing CRISPR-Cas adaptation by Southern blotting of the spacer (Supplementary Figure S3).
pKEF9 loses CRISPR spacers in the recipient host
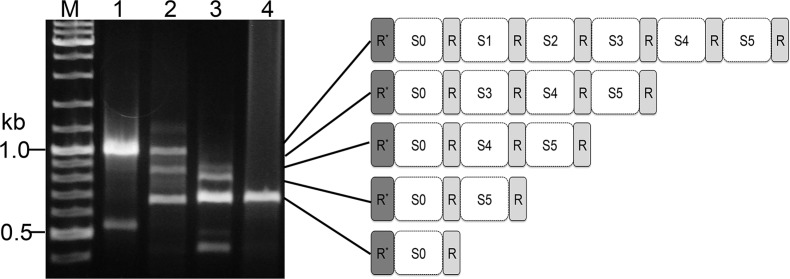
The CRISPR locus of pKEF9 carries an exceptional repeat structure and is transcribed and processed into small mature RNAs that could effect CRISPR-Cas interference when complexed with host Cas proteins (13). However, it is unlikely to undergo CRISPR-Cas adaptation because it lacks a leader region and carries a degenerate first repeat (9). Nevertheless, we tested for spacer acquisition by PCR amplifying the whole CRISPR locus before, and at increasing times after, conjugation but no larger PCR products were detected. Instead a series of smaller DNA fragments were resolved in agarose gels, suggestive of a gradual loss of spacer-repeat units over time (Figure 5). Sequencing of the PCR bands confirmed the absence of de novo spacers and revealed the presence of a heterogeneous mixture of five variant CRISPR structures (Figure 5). These results suggested that spacers 2 and 3 were preferentially lost over the first 200 hpc, followed by spacer 4 and then spacer 5 by 300 hpc. The resistant core CRISPR structure consisted of the degenerate repeat-spacer-repeat 1 that was also detected at a low level in the plasmid isolated from donor P1 strain.
Figure 5.
PCR products showing the heterogeneous CRISPR spacer contents of pKEF9 (S1–5). Lane 1—pKEF9 from conjugated Sulfolobus islandicus; lane 2—200 hpc; lane 3—280 hpc and lane 4—300 hpc. M—DNA size marker. R*—corrupted repeat.
A mechanism for selective inactivation of integrated pKEF9
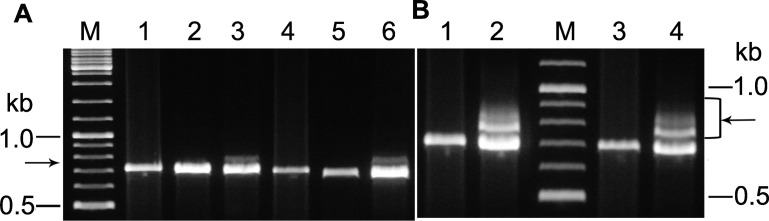
The genome sequence of S. solfataricus P1 revealed that pKEF9 was integrated exclusively at tRNAGlu (CTC). pKEF9 encodes an integrase and the attP site complements chromosomal attB sites at two tRNAGlu genes of donor and recipient strains, with a perfect match (CTC) and a single mismatch (CTT) to the anticodons (Supplementary Table S2). Therefore, we tested for plasmid integration at both genes in S. islandicus by Southern blotting at 24, 38 and 85 hpc and by PCR amplification at 85 hpc. The Southern blotting results showed that pKEF9 was integrated in the tRNAGlu (CTT) gene at 24 hpc and subsequently in the tRNAGlu (CTC) gene at 38 hpc (Figure 6A). PCR amplification of the 85 hpc sample confirmed the presence of integrated pKEF9 in both genes (Figure 6B). This raised the question as to whether CRISPR-Cas interference of free pKEF9 would be effective if the integrated form could be reversibly released into the cell. Therefore we investigated to what extent the integrated plasmid remained viable during the CRISPR-Cas response and found that that it was susceptible to transposition events.
Figure 6.
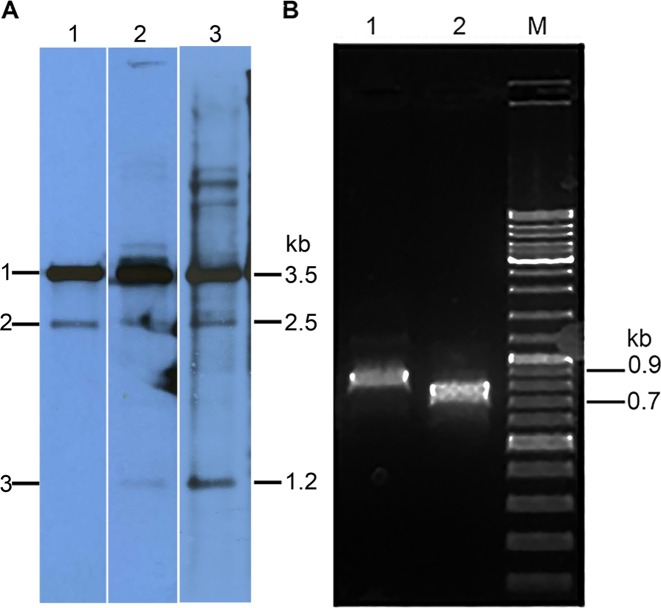
Analysis of pKEF9 integrated in the Sulfolobus islandicus genome. (A) Southern blot analysis of pKEF9 integration in S. islandicus. Lane 1—24 hpc; lane 2—38 hpc and lane 3 85 hpc. Bands 1, 2 and 3 derive from free pKEF9 and pKEF9 integrated at the tRNAGlu (CTT) and tRNAGlu (CTC) match sites, respectively. (B) PCR amplification products obtained at 85 hpc from pKEF9 integrated at lane 1—tRNAGlu (CTC) and lane 2—tRNAGlu (CTT). M—DNA size markers.
Strain P1 carries a single intact mobile orfB element of the IS605 family (36) that contains an orfB gene (positions 1 999 421–2 000 931) and encodes a 100-nt RNA Sso-109 (AJ786211) (positions 1 999 584–1 999 485) (37). Moreover, the genome sequencing revealed that conjugated strain P1 yielded several sequence reads and contigs in which the orfB element had transposed into pKEF9. Sequence analyses identified five transposition sites, each containing a GGC sequence (complementary to the terminal sequence of the orfB element), with insertions at positions 14 097/8 (44 sequences), 17 040/1 (21 sequences), 22 423/4 (23 sequences), 26 493/4 (11 sequences) and 28 710/1 (7 sequences). Furthermore, some of the plasmid copies had undergone rearrangements, and deletions, probably as a result of recombination between neighbouring orfB elements. In order to test whether orfB elements had transposed into free and/or integrated plasmid forms, PCR products were generated across the five potential transposition sites on pKEF9 isolated from strain P1, and cloned and sequenced. All sequences matched the plasmid and no transposed orfB elements were detected which suggested that orfB elements had selectively transposed into integrated pKEF9.
To test this further, the five transposition sites of pKEF9 were PCR amplified on: (i) free plasmid carrying the intact CRISPR locus (6,13), (ii) plasmid isolated from the donor S. solfataricus strain P1, (iii) chromosomal DNA from donor strain P1 and (iv) chromosomal DNA from the recipient S. islandicus strain. The PCR products from each sample were then resolved on agarose gels and larger bands, indicative of orfB transpositions, were only produced from chromosomal DNA of donor strain P1 and, to a lesser degree, from the chromosome of the recipient strain (Figure 7A and B). Moreover, the strongest PCR bands coincided with transposition at positions 14 097/8, 17 040/1 and 22 423/4, correlating well with the genome sequencing data (Figure 7A and B). Thus the results reinforce that the integrated form of the plasmid was selectively subjected to transposition but we cannot eliminate the possibility that some free plasmids underwent transposition and were then lost from the culture.
Figure 7.
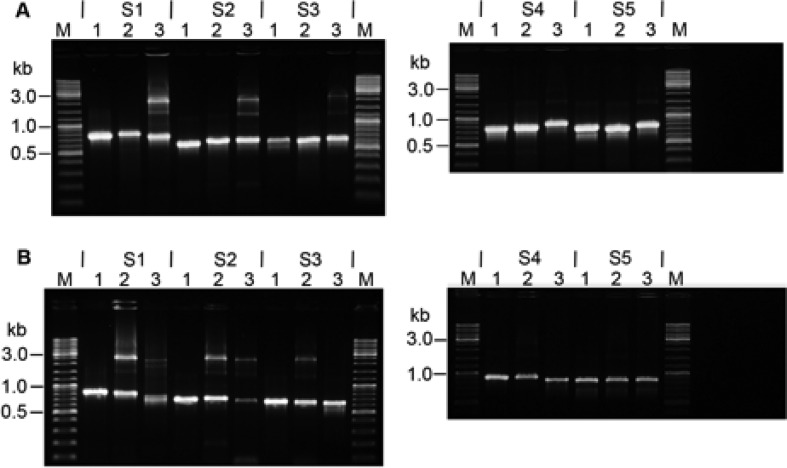
Agaose gels showing PCR products amplified from the five transposition sites of orfB elements in pKEF9: S1—14097/8; S2—17040/1; S3—22423/4; S4—26493/4 and S5—28710/1. (A) Donor strain. For each site: 1—free pKEF9 with full CRISPR locus; 2—pKEF9 from donor strain P1; 3—donor strain P1 DNA. (B) Recipient strain. For each site: 1—pKEF9 from donor strain P1; 2—donor strain P1 DNA and 3 recipient Sulfolobus islandicus DNA.
Changes in the cellular DNA profile
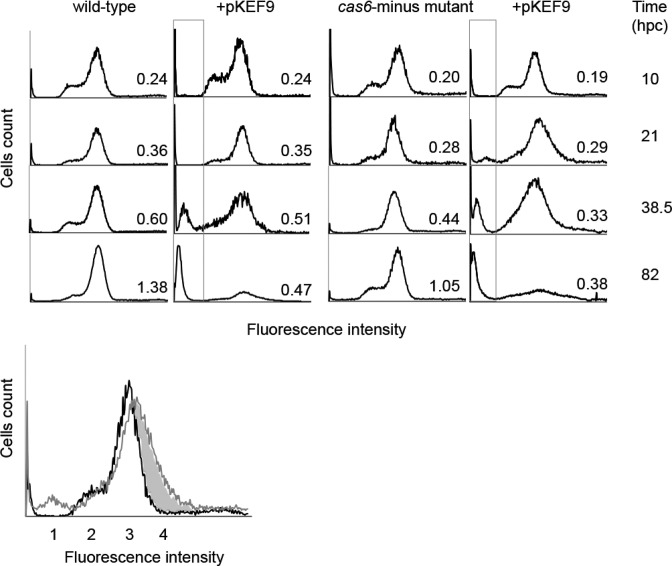
Next cellular DNA content distributions were monitored over time by flow cytometry in unconjugated and conjugated wild-type S. islandicus cultures, in order to determine differences in the levels, and integrity, of chromosomal and viral DNA post conjugation. The conjugated Δcas6 mutant was also examined to establish whether inactivation of CRISPR-Cas interference significantly affected the DNA profiles. In the exponentially growing unconjugated wild-type and Δcas6 mutant cultures, most cells carried two chromosome copies (peak 3) with fewer cells containing single copies (peak 2) (Figure 8), consistent with earlier Sulfolobus studies (38). For the conjugated wild-type and Δcas6 mutant cultures similar DNA content distributions were observed up to 21 hpc (Figure 8), except that the mutant showed a small proportion of DNA-deficient cells (peak 1) resulting from chromosomal DNA degradation and/or cellular fragmentation (39). Peak 1 was more prominent in both conjugated cultures at 38.5 hp and it increased dramatically at 82 hpc with a strong decrease in cells carrying one or two chromosomes and plasmid DNA (peaks 2, 3 and 4) (Figure 8). The results demonstrate that substantial degradation of chromosomal and plasmid DNA occurred in both conjugated cultures between the 21 and 82 hpc time points and the degradation effects were qualitatively similar in wild-type cells and in the Δcas6 mutant lacking a CRISPR-Cas response (Figure 8). Moreover, the extensive DNA degradation coincided approximately with the onset of growth retardation in both cultures (Figure 1), and with activation of CRISPR-Cas adaptation in the wild-type culture (Figure 3A).
Figure 8.
Flow cytometry analysis of DNA content distributions in the wild-type and Δcas6 mutant of Sulfolobus islandicus at increasing times post conjugation. Fluorescence intensity measurements were made at 10, 21, 38.5 and 85 hpc on unconjugated and conjugated wild-type and mutant strains. The upper panel shows the peak contents. Peak 1—degraded DNA and/or cellular fragmentation; peak 2—cells with single chromosomes; peak 3—cells with two chromosomes and peak 4—shaded area—mainly plasmid DNA present in conjugated cells.
pKEF9 did not activate CRISPR adaptation in S. solfataricus P2
pKEF9 conjugation induced a strong CRISPR-Cas adaptation response in S. islandicus whereas no spacer acquisition had occurred in the donor S. solfataricus P1 strain. It was inferred that CRISPR-Cas adaptation in strain P1 could have been inactivated by mutation. Therefore, we examined the effect of pKEF9 conjugation on the closely related P2 strain (Supplementary Figure S1). The growth curve of the pKEF9-conjugated P2 strain showed strong retardation after about 30 hpc and growth gradually recovered reaching near wild-type levels at 190 hpc (Supplementary Figure S4A), when pKEF9 was shown by PCR amplification to have integrated into both tRNAGlu genes (Supplementary Figure S4B). Plasmid extractions demonstrated that free pKEF9 was present in the culture until 100 hpc (data not shown) but none was detected at 190 hpc when growth had completely recovered. However, flow cytometry data obtained at 24, 33 and 47 hpc showed that strong degradation of intracellular DNA was initiated between 24 and 33 hpc, approximately coincident with the onset of growth retardation, while at 47 hpc most cells carried degraded DNA (Supplementary Figure S4B). In contrast, pKEF9-conjugated strain P1 showed a fairly stable DNA content and integrity over the same time period (Supplementary Figure S4C). Finally, we tested for CRISPR spacer acquisition from pKEF9 by PCR amplifying the leader ends of the six CRISPR loci (A to F) of conjugated strain P2. No larger bands were generated indicative of inactive CRISPR-Cas adaptation (Supplementary Figure S4D).
DISCUSSION
DNA degradation in conjugated cells and cell death
Strong growth retardation occurred in the conjugated wild-type S. islandicus culture at about 38 hpc that coincided with a very high pKEF9 copy number and the onset of CRISPR-Cas adaptation. Less dramatic retarded growth was observed after 38 hpc for the Δcas6 mutant that exhibited a lower pKEF9 copy number and lacked the capacity for CRISPR-Cas interference although the wild-type culture recovered more strongly after 100 hpc. Flow cytometry profiles revealed qualitatively similar levels of intracellular DNA fragmentation in the conjugated wild-type strain and Δcas6 mutant (Figure 8) which suggested that the CRISPR-Cas response had a minimal effect on the overall intracellular DNA integrity of the cell culture.
The copy number of pKEF9 increased strongly from about 16 to 150 copies per cell between 24 and 38 hpc in the wild-type strain suggesting that the observed increase in cellular DNA content (peak 4, Figure 8) resulted primarily from replicating pKEF9. Moreover, it is likely that the high plasmid level triggered cellular mechanisms that led to inhibition of cell division and, in turn, to chromosomal and viral DNA degradation and cell death. Rapid replication of pKEF9, with no effective copy number control, would result in a rapid increase in the number of replication forks due to the lack of some replication proteins. Moreover, the replication forks would tend to collapse yielding DNA breaks that would, in turn, lead to cell death owing to programmed cell death that is induced by extensive DNA damage (Han,W., Feng,X., Sun,M., Liang,Y.X., Shen,Y. and She,Q., unpublished data). This effect differs from the population-wide dormancy observed when S. islandicus was treated with low levels of active, or inactivated, fusellovirus SSV9, that could also lead to cell death (40).
The occurrence of conjugation-induced cell death receives support from another experiment with the conjugative plasmid pNOB8 (2), employing Sulfolobus NOB8H2 as donor and S. solfataricus P2 as recipient strain. Attempts to isolate pNOB8 conjugants from the conjugated cultures failed, and only donor colonies were obtained (Shen,B. and She,Q., unpublished data).
The rapid increase in DNA breaks could also stimulate CRISPR-Cas adaptation activity in the recipient wild-type strain, as demonstrated for the type I-E system of E. coli (19). Furthermore, consistent with the flow cytometry results on the wild-type and Δcas6 mutant (Figure 8), the overall effects of CRISPR-Cas activity on DNA integrity could be neutral; on one hand increasing viral DNA cleavage during adaptation and interference but on the other hand reducing DNA fragmentation in cells where the plasmid is eliminated at an early stage of conjugation. The latter process would also enable cells to recover, and explain the higher rate of growth recovery of the wild-type after 100 hpc relative to the Δcas6 mutant which still carries pKEF9 (Figure 1).
Specific targeting of pKEF9
Evidence was obtained for two cellular mechanisms, additional to the CRISPR-Cas response, that specifically targeted the plasmid. Firstly, the single orfB element of the donor S. solfaraticus P1 transposed into five GGC sites in some copies of integrated plasmid. Moreover, this led to plasmid degeneration as a result of recombination between adjacent orfB elements and prevented excision of viable plasmids from the host chromosome. The observation receives support from genome sequencing studies which demonstrated that Sulfolobus species frequently use transposable elements to gradually eliminate genetic elements integrated at tRNA genes (34,41). The results are also consistent with some Sulfolobus transposases undergoing enhanced expression on viral infection (17,39) and being activated during CRISPR-Cas adaptation (15).
The second effect was seen in the stepwise deletion of spacer-repeat units from the CRISPR locus of pKEF9, presumably as a result of recombination between repeat sequences (20). This locus can be transcribed and processed into crRNA-like RNAs that are likely to assemble into host-encoded Cas protein interference complexes (13). An inference that is supported by the CRISPR spacers showing potentially significant sequence matches to rudivirus SIRV2 (spacer 3–4 mismatches), fuselloviruses SSV4/5 (spacer 5–13 mismatches) and spacer 73 CRISPR locus F of S. solfataricus P1 and P2 (spacer 6–10 mismatches) (12,42). A possible explanation for the spacer deletions is that pKEF9 self-targeting occurs in S. islandicus whereby pKEF9 crRNAs are loaded into CRISPR-Cas effector complexes but fail to recognize pKEF9 as their source, and then induce plasmid self-interference until the spacer is removed. Given this hypothesis, the sequential loss of spacers from one end could reflect that crRNA spacer yields were higher at that end, as was observed in pKEF9-conjugated S. acidocaldarius (13).
Factors affecting activation of host CRISPR-Cas adaptation
Various factors appear to influence the activation of spacer acquisition in Sulfolobus species where the adaptation cas gene cassette is regulated by both a Csa3a protein and antisense RNA (17,18). Adaptation was first detected in Sulfolobus with mixtures of genetic elements at about 12 days post infection (9,16) and subsequently it was induced within 1–2 days after cold shock treatment (16). Recently, spacer acquisition was activated 5 or more days after infection depending on the culture medium (17).
Spacer acquisition is also dependent on active viral DNA replication in Sulfolobus (16). and experimental studies on the type I-E CRISPR-Cas system of E.coli have indicated that formation of DNA breaks at replication forks stimulates the process (19). Thus, it is likely that the rapid replication of pKEF9 observed between 24- and 38-hpc enhanced adaptation. It remains unclear whether coinfection by SSV2 DNA, excised from the donor genome and present at low levels in recipient cells during the first days post conjugation, also had a stimulatory effect, as was observed earlier for coinfecting genetic elements (15,16), but it did undergo a low level of spacer acquisition. In summary, multiple factors can influence CRISPR-Cas adaptation activation in Sulfolobus.
No evidence has yet been been found for type I-A CRISPR-Cas adaptation in Sulfolobus being activated by priming from matching host spacers, in contrast to bacterial type I-E and I-F systems (43–46) and the type I-B system of a haloarchaeon (47). Three main lines of evidence favour the absence of priming. (i) The virus SMV1 did not undergo adaptation in S. solfataricus P2 despite the host carrying multiple perfectly matching spacers and cognate PAM sequences (9,15,16). (ii) The STSV2 virus underwent spacer acquisition in S. islandicus despite the absence of host CRISPR spacers with significant sequence matches to the virus (6); and (iii) no biased distribution of viral or plasmid protospacers was observed during adaptation (15,16) that would be expected to result from priming effects (43–46). The latter inference carries the reservation that simultaneous multiple priming effects could mask any biases (11).
Nevertheless, it was surprising that no spacer acquisition was observed in either the donor S. solfataricus P1 strain or in the recipient P2 strain, in contrast to earlier studies of strain P2 conjugated with pMGB1 (9,15). One major difference between the three similar strains is that whereas no close sequence matches (<7 mismatches) were found between pKEF9 and spacers of S. solfataricus P1 and P2, CRISPR locus 2, spacer 44 of the S. islandicus recipient strain exhibited a single mismatch, at position 12, to pKEF9 (positions 17 869–17 831) and spacer 114 of locus 1 showed five mismatches (positions 6207–6248). Moreover, the former spacer match fell within the plasmid region 14 000–20 000 carrying the highest concentration of protospacers (Figure 4). Thus, the operation of a priming effect from the first of these spacers could explain the exclusive onset of CRISPR-Cas adaptation in the recipient S. islandicus strain and would also be compatible with the lack of adaptation in the Δcas6 strain.
Supplementary Material
Acknowledgments
Members of the Danish Archaea Centre are thanked for their support and insightful discussions. An anonymous referee is thanked for suggesting self-interference of pKEF9.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Danish Natural Science Research Council, Grant Number 34029. Funding for open access charge: Copenhagen University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Schleper C., Holz I., Janekovic D., Murphy J., Zillig W. A multicopy plasmid of the extremely thermophilic archaeon Sulfolobus effects its transfer to recipients by mating. J. Bacteriol. 1995;177:4417–4426. doi: 10.1128/jb.177.15.4417-4426.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.She Q., Phan H., Garrett R.A., Albers S.V., Stedman K.M., Zillig W. Genetic profile of pNOB8 from Sulfolobus: the first conjugative plasmid from an archaeon. Extremophiles. 1998;2:417–425. doi: 10.1007/s007920050087. [DOI] [PubMed] [Google Scholar]
- 3.Prangishvili D., Albers S-V., Holz I., Arnold H.P., Stedman K., Klein T., Singh H., Hiort J., Schweier A., Kristjansson J.K., et al. Conjugation in archaea: frequent occurrence of conjugative plasmids in Sulfolobus. Plasmid. 1998;40:190–202. doi: 10.1006/plas.1998.1363. [DOI] [PubMed] [Google Scholar]
- 4.Wang H., Peng N., Shah S.A., Huang L., She Q. Archaeal extrachromosomal elements. Microbiol. Mol. Biol. Rev. 2015;79:117–152. doi: 10.1128/MMBR.00042-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stedman K.M., She Q., Phan H., Holz I., Singh H., Prangishvili D., Garrett R., Zillig W. pING family of conjugative plasmids from the extremely thermophilic archaeon Sulfolobus islandicus: insights into recombination and conjugation in Crenarchaeota. J. Bacteriol. 2000;182:7014–7020. doi: 10.1128/jb.182.24.7014-7020.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greve B., Jensen S., Brügger K., Zillig W., Garrett R.A. Genomic comparison of archaeal conjugative plasmids from Sulfolobus. Archaea. 2004;1:231–239. doi: 10.1155/2004/151926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erauso G., Stedman K.M., van de Werken H.J., Zillig W., van der Oost J. Two novel conjugative plasmids from a single strain of Sulfolobus. Microbiology. 2006;152:1951–1968. doi: 10.1099/mic.0.28861-0. [DOI] [PubMed] [Google Scholar]
- 8.Marraffini L.A., Sontheimer E.J. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erdmann S., Garrett R.A. Selective and hyperactive uptake of foreign DNA by adaptive immune systems of an archaeon via two distinct mechanisms. Mol. Microbiol. 2012;85:1044–1056. doi: 10.1111/j.1365-2958.2012.08171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Westra E.R., Staals R.J.H., Gort G., Høgh S., Neumann S., de la Cruz F., Fineran P.C., Brouns S.J.J. CRISPR-Cas systems preferentially target the leading regions of MOBF conjugative plasmids. RNA Biol. 2013;10:749–761. doi: 10.4161/rna.24202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garrett R.A., Shah S.A., Erdmann S., Liu G., Mousaei M., León-Sobrino C., Peng W., Gudbergsdottir S., Deng L., Vestergaard G., et al. CRISPR-Cas adaptive immune systems of the Sulfolobales: unravelling the complexity and diversity of the adaptation and interference mechanisms. Life. 2015;5:783–817. doi: 10.3390/life5010783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lillestøl R.K., Redder P., Garrett R.A., Brügger K. A putative viral defence mechanism in archaeal cells. Archaea. 2006;2:59–72. doi: 10.1155/2006/542818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lillestøl R.K., Shah S.A., Brügger K., Redder P., Phan H., Christiansen J., Garrett R.A. CRISPR families of the crenarchaeal genus Sulfolobus: bidirectional transcription and dynamic properties. Mol. Microbiol. 2009;72:259–272. doi: 10.1111/j.1365-2958.2009.06641.x. [DOI] [PubMed] [Google Scholar]
- 14.Shah S.A., Hansen N.R., Garrett R.A. Distributions of CRISPR spacer matches in viruses and plasmids of crenarchaeal acidothermophiles and implications for their inhibitory mechanism. Biochem. Soc. Trans. 2009;37:23–28. doi: 10.1042/BST0370023. [DOI] [PubMed] [Google Scholar]
- 15.Erdmann S., Shah S.A., Garrett R.A. SMV1 virus-induced CRISPR spacer acquisition from the conjugative plasmid pMGB1 in Sulfolobus solfataricus P2. Biochem. Soc. Trans. 2013;41:1449–1458. doi: 10.1042/BST20130196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erdmann S., Le Moine Bauer S., Garrett R.A. Inter-viral conflicts that exploit host CRISPR immune systems of Sulfolobus. Mol. Microbiol. 2014;91:900–917. doi: 10.1111/mmi.12503. [DOI] [PubMed] [Google Scholar]
- 17.Leon-Sobrino C., Kot W.P., Garrett R.A. Transcriptome changes in STSV2-infected Sulfolobus islandicus REY15A undergoing continuous CRISPR spacer acquisition. Mol. Microbiol. 2016;99:719–728. doi: 10.1111/mmi.13263. [DOI] [PubMed] [Google Scholar]
- 18.Liu T., Li Y., Ye Q., Li H., Liang Y., She Q., Peng N. Transcriptional regulator-mediated activation of adaptation genes triggers CRISPR de novo spacer acquisition. Nucleic Acids Res. 2015;43:1044–1055. doi: 10.1093/nar/gku1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levy A., Goren M.G., Yosef I., Auster O., Manor M., Amitai G., Edgar R., Qimron U., Sorek R. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature. 2015;520:505–510. doi: 10.1038/nature14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gudbergsdottir S., Deng L., Chen Z., Jensen J.V., Jensen L.R., She Q., Garrett R.A. Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol. Microbiol. 2011;79:35–49. doi: 10.1111/j.1365-2958.2010.07452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng L., Garrett R.A., Shah S.A., Peng X., She Q. A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol. Microbiol. 2013;87:1088–1099. doi: 10.1111/mmi.12152. [DOI] [PubMed] [Google Scholar]
- 22.Peng W., Feng M., Feng X., Liang Y.X., She Q. An archaeal CRISPR type III-B system exhibiting distinctive RNA targeting features and mediating dual RNA and DNA interference. Nucleic Acids Res. 2015;43:406–417. doi: 10.1093/nar/gku1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rutherford K., Parkhill J., Crook J., Horsnell T., Rice P., Rajandream M.A., Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 24.Delcher A.L., Phillippy A., Carlton J., Salzber S.L. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002;30:2478–2483. doi: 10.1093/nar/30.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng L., Zhu H., Chen Z., Liang Y.X., She Q. Unmarked gene deletion and host-vector system for the hyperthermophilic crenarchaeon Sulfolobus islandicus. Extremophiles. 2009;13:735–746. doi: 10.1007/s00792-009-0254-2. [DOI] [PubMed] [Google Scholar]
- 26.Peng W., Li H., Hallstrom S., Peng N., Liang Y.X., She Q. Genetic determinants of PAM-dependent DNA targeting and pre-crRNA processing in Sulfolobus islandicus. RNA Biol. 2013;10:738–748. doi: 10.4161/rna.23798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zillig W., Kletzin A., Schleper C., Holz I., Janekovic D., Hain J., Lanzendörfer M., Kristjansson J. Screening for Sulfolobales, their plasmids and their viruses in Icelandic solfataras. Syst. Appl. Microbiol. 1993;16:609–628. [Google Scholar]
- 28.Birnboim H.C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J., Russell D. Molecular Cloning: a Laboratory Manual. 3rd edn. NY: Cold Spring Harbor Laboratory; 2001. [Google Scholar]
- 30.Zillig W., Arnold H.P., Holz I., Prangishvili D., Schweier A., Stedman K., She Q., Phan H., Garrett R., Kristjansson J.K. Genetic elements in the extremely thermophilic archaeon Sulfolobus. Extremophiles. 1998;2:131–140. doi: 10.1007/s007920050052. [DOI] [PubMed] [Google Scholar]
- 31.She Q., Singh R.K., Confalonieri F., Zivanovic Y., Gordon P., Allard G., Awayez M.J., Chan-Weiher C-Y., Clausen I.G., Curtis B., et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. U.S.A. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stedman K.M., She Q., Phan H., Arnold H.P., Holz I., Garrett R.A., Zillig W. Relationships between fuselloviruses infecting the extremely thermophilic archaeon Sulfolobus: SSV1 and SSV2. Res. Microbiol. 2003;154:295–302. doi: 10.1016/S0923-2508(03)00074-3. [DOI] [PubMed] [Google Scholar]
- 33.Redder P., Garrett R.A. Mutations and rearrangements in the genome of Sulfolobus solfataricus P2. J. Bacteriol. 2006;188:4198–4206. doi: 10.1128/JB.00061-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo L., Brügger K., Liu C., Shah S.A., Zheng H., Zhu Y., Wang S., Lillestøl R.K., Chen L., Frank J., et al. Genome analyses of Icelandic strains of Sulfolobus islandicus: Model organisms for genetic and virus-host interaction studies. J. Bacteriol. 2011;193:1672–1680. doi: 10.1128/JB.01487-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vestergaard G., Garrett R.A., Shah S.A. CRISPR adaptive immune systems of Archaea. RNA Biol. 2014;11:157–168. doi: 10.4161/rna.27990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brügger K., Redder P., She Q., Confalonieri F., Zivanovic Y., Garrett R.A. Mobile elements in archaeal genomes. FEMS Microbiol. Lett. 2002;206:131–141. doi: 10.1111/j.1574-6968.2002.tb10999.x. [DOI] [PubMed] [Google Scholar]
- 37.Tang T.H., Polacek N., Zywicki M., Huber H., Brügger K., Garrett R., Bachellerie J.P., Huttenhofer A. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol. Microbiol. 2005;55:469–481. doi: 10.1111/j.1365-2958.2004.04428.x. [DOI] [PubMed] [Google Scholar]
- 38.Bernander R., Poplawski A. Cell cycle characteristics of thermophilic archaea. J. Bacteriol. 2007;179:4963–4969. doi: 10.1128/jb.179.16.4963-4969.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Götz D., Paytubi S., Munro S., Lundgren M., Bernander R., White M.F. Responses of hyperthermophilic crenarchaea to UV irradiation. Genome Biol. 2007;8:R220. doi: 10.1186/gb-2007-8-10-r220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bautista M.A., Zhang C., Whitaker R.J. Virus-induced dormancy in the archaeon Sulfolobus islandicus. MBio. 2015;6 doi: 10.1128/mBio.02565-14. doi:10.1128/mBio.02565-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.You X-Y., Liu C., Wang S-Y., Jiang C.-Y., Shah S.A., Prangishvili D., She Q., Liu S-J., Garrett R.A. Genomic studies of Acidianus hospitalis W1 a host for studying crenarchaeal virus and plasmid life cycles. Extremophiles. 2011;15:487–497. doi: 10.1007/s00792-011-0379-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shah S.A., Garrett R.A. CRISPR/Cas and Cmr modules, mobility and evolution of adaptive immune systems. Res. Microbiol. 2011;162:27–38. doi: 10.1016/j.resmic.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 43.Datsenko K.A., Pougach K., Tikhonov A., Wanner B.L., Severinov K., Semenova E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat. Commun. 2012;3:945–951. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- 44.Xue C., Seetharam A.S., Musharova O., Severinov K., Brouns S.J.J., Severin A.J., Sashital D.G. CRISPR interference and priming varies with individual spacer sequences. Nucleic Acids Res. 2015;43:10831–10847. doi: 10.1093/nar/gkv1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richter C., Dy R.L., McKenzie R.E., Watson B.N., Taylor C., Chang J.T., McNeil M.B., Staals R.H., Fineran P.C. Priming in the type I-F CRISPR-Cas system triggers strand-independent spacer acquisition, bi-directionally from the primed protospacer. Nucleic Acids Res. 2014;42:8516–8526. doi: 10.1093/nar/gku527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vorontsova D., Datsenko K.A., Medvedeva S., Bondy-Denomy J., Savitskaya E.E., Pougach K., Logacheva M., Wiedenheft B., Davidson A.R., Severinov K., et al. Foreign DNA acquisition by the I-F CRISPR–Cas system requires all components of the interference machinery. Nucleic Acids Res. 2015;43:10848–10860. doi: 10.1093/nar/gkv1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li M., Wang R., Zhao D., Xiang H. Adaptation of the Haloarcula hispanica CRISPR–Cas system to a purified virus strictly requires a priming process. Nucleic Acids Res. 2014;42:2483–2492. doi: 10.1093/nar/gkt1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.