Abstract
Human respiratory syncytial virus (hRSV) is a leading cause of acute lower respiratory tract infection in infants, elderly and immunocompromised individuals. To date, no specific antiviral drug is available to treat or prevent this disease. Here, we report that the Smoothened receptor (Smo) antagonist cyclopamine acts as a potent and selective inhibitor of in vitro and in vivo hRSV replication. Cyclopamine inhibits hRSV through a novel, Smo-independent mechanism. It specifically impairs the function of the hRSV RNA-dependent RNA polymerase complex notably by reducing expression levels of the viral anti-termination factor M2-1. The relevance of these findings is corroborated by the demonstration that a single R151K mutation in M2-1 is sufficient to confer virus resistance to cyclopamine in vitro and that cyclopamine is able to reduce virus titers in a mouse model of hRSV infection. The results of our study open a novel avenue for the development of future therapies against hRSV infection.
Bronchiolitis is a severe lower-respiratory tract infectious disease primarily caused by members of the Paramyxoviridae family. Human respiratory syncytial virus (hRSV) is the principal cause of morbidity in children less than 2 years of age1,2 as well as the elderly, immunocompromised and transplant patients3,4,5,6,7. To date, there are neither vaccines nor approved small molecule drugs available to prevent or treat hRSV infection. The immuno-prophylactic antibody palivizumab8 is approved for high-risk patients only such as premature babies and infants suffering from underlying diseases8,9. The broad-spectrum small molecule antiviral ribavirin is available to treat infection, but it has considerable side-effects and limited efficacy10,11. During the past decade, a number of drug candidates targeting hRSV entry12,13,14,15,16 or replication steps17,18,19 have been advanced to pre-clinical or clinical development.
The hRSV genomic RNA (vRNA) is packaged by the viral nucleoprotein (N) at all times, forming a N:RNA complex, called nucleocapsid. This ribonucleoprotein complex is used as a template for mRNA transcription and genomic or antigenomic RNA replication by the RNA-dependent RNA polymerase (RdRp), which is composed of 2 major viral proteins: the phosphoprotein P and the large polymerase L20. In this complex, the phosphoprotein is an essential co-factor of the L polymerase by binding to L and N and targeting the polymerase L to vRNA21,22,23. Two co-factors, M2-1 and M2-2, are required for the RdRp to process RNA efficiently during the viral cycle. M2-1 is a tetrameric transcription processivity factor that binds in a competitive manner to RNA and P via its core domain21,22,24. M2-1 functions as an anti-terminator of transcription that prevents premature termination of transcription both intra- and inter-genetically20,25. Although in vitro experiments have shown that M2-1 binds preferentially to positive-sense viral gene end (GE) and poly-A sequences21,26, the exact mechanisms by which M2-1 improves transcription efficiency is not fully understood.
By screening libraries of known bioactive compounds, we identified cyclopamine (CPM) and jervine as highly potent and selective inhibitors of hRSV replication in vitro. CPM is a well-known antagonist of the smoothened protein (Smo), a 7-transmembrane receptor of the Sonic hedgehog signaling pathway (Shhp)27,28 involved in embryonic development, cell differentiation and tumorigenesis29. The discovery of the CPM anti-proliferative activity has led to the development of novel, CPM-competitive Smo antagonists for cancer therapy including GDC-0449 (vismodegib), approved for the treatment of basal-cell carcinoma30, and LY294068031.
The present report shows that CPM is a potent inhibitor of hRSV infection. Inhibition is not mediated by the cognate Smo receptor, but through the hRSV viral processivity factor M2-1. The CPM-mediated suppression of hRSV infection could also be demonstrated in vivo, where hRSV lung titers could be significantly reduced in CPM-treated mice. This novel off-target effect of CPM paves the way to the future development of therapies against hRSV infection, using CPM analogues that target hRSV replication complex, in particular M2-1.
Results
Identification of Veratrum steroidal alkaloids as inhibitors of hRSV infection in vitro
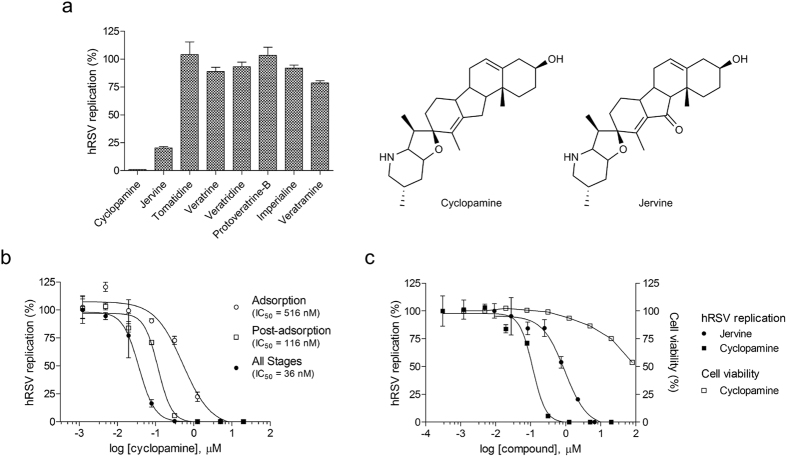
We performed phenotypic, cell-viability-based screens designed to measure the antiviral effect of compounds on hRSV replication in HEp-2 cells. We screened a 502 compound natural product library and a 3,244 compound approved drug and probe-compound library (hit rates 1.85% and 2.39%, respectively). They allowed us to identify jervine and CPM, two Veratrum steroidal alkaloids, as potent in vitro anti-hRSV molecules. Other compounds of this class such as veratrine, portoveratrine-B, imperialine or veratramine were inactive against hRSV, indicating a specificity of action of jervine and CPM (Fig. 1a).
Figure 1. Inhibition of hRSV infection by CPM and jervine in vitro.
(a) Single dose inhibition of hRSV infection of HEp-2 cells by analogues of CPM tested at 3.5 μg/mL (cyclopamine: 8.5 μM, jervine: 8.2 μM, tomatidine: 7.7 μM, veratrine: 5.9 μM, veratridine: 5.2 μM, protoveratrine B: 4.3 μM, imperialine: 8.1 μM, veratramine: 8.5 μM). (b) Dose-dependent inhibition of hRSV replication in HEp-2 cells by CPM when present during viral adsorption stage, post-adsorption or at all stages of infection. Antiviral activities were measured by focus reduction assay. Foci numbers (adsorption stage) and sizes (post-adsorption and all stages) were measured automatically using the software FiJi57, and were normalized as percentage of the untreated control. (c) Dose-dependent inhibition of hRSV replication in HEp-2 cells by CPM and jervine at post-adsorption stage, and cytotoxicity of CPM towards HEp-2 cells. All infections were carried out for 72 h. The cytotoxicity of CPM was measured over 72 h of incubation with cells, by cell viability assay. Each data point represents the mean of duplicate values, and the SEM is represented by the error bars. The graphs and the IC50 and IC95 values were created and calculated using the software GraphPad Prism v.5 (GraphPad Software, La Jolla California, USA), and are representative of 3 independent experiments.
A focus reduction assay was used to further characterize the 50%, 90%, 95% or 99% inhibitory concentrations of the compounds (IC50, IC90, IC95 and IC99, respectively). CPM (11-deoxojervine) was identified as the most potent inhibitor of hRSV infection with an IC50 of 36 nM, IC90 of 151 nM, IC99 of 336 nM and a selectivity index >2000 when present throughout infection (Fig. 1b). The concentration of CPM resulting in a 50% decrease of cell viability (CC50), was found to be >80 μM (Fig. 1c). CPM remained the most active against hRSV when added exclusively post-adsorption as demonstrated by an IC50 of 116 nM and IC90 of 404 nM (Fig. 1b). In contrast, the potency of CPM was decreased when present exclusively during virus adsorption (IC50 516 nM, IC90 5.4 μM). Jervine also inhibited hRSV replication at post-adsorption stage with higher IC50 and IC95 values of 994 nM and 6.9 μM, respectively (Fig. 1c). These results suggest that CPM and jervine likely act on a post-entry mechanism of hRSV replication. Due to its greater potency and to the wealth of available literature data on its biological activity, CPM was then used for all further studies.
CPM inhibits hRSV infection independently of the Smo receptor
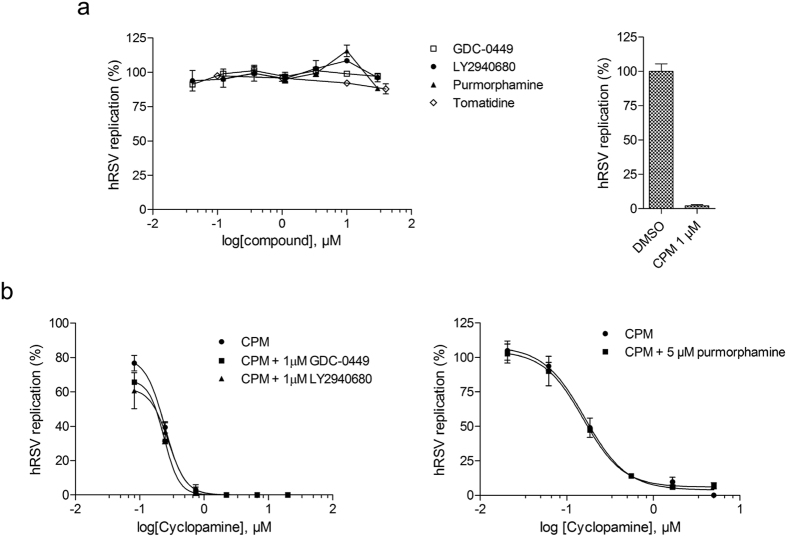
To assess whether Smo is involved in CPM-mediated inhibition of hRSV replication, two Shhp antagonists (GDC-0449, LY2940680) and one agonist (purmorphamine), as well as one non Smo-binding structural analogue of CPM (tomatidine) were evaluated on hRSV infection in vitro. As shown in Fig. 2a, all compounds except CPM failed to inhibit hRSV replication at concentrations up to 30 μM. Since GDC-0449, LY2940680 and purmorphamine are known competitors of CPM for binding to Smo32,33,34, we performed a competition assay with increasing concentrations of CPM in presence of the compounds. None of the competitors induced a shift in IC50 for CPM-mediated hRSV inhibition (Fig. 2b), leading us to the conclusion that the inhibition of hRSV by CPM is not mediated by its binding to Smo but through a novel, Shhp-independent mechanism.
Figure 2. Effect of Smo modulators and a CPM analog on hRSV infection in vitro.
(a) Dose-dependent inhibition of hRSV replication in HEp-2 cells of Smo inhibitors (GDC-0449, LY2940680), an Smo activator (Purmorphamine) and a CPM structural analog (Tomatidine) (left), versus the effect of 1 μM of CPM (right). (b) Comparative effect of increasing concentrations of CPM in combination with 1 μM of GDC-0449, LY294068 (left), or 5 μM of purmorphamine (right) on hRSV replication in HEp-2 cells. Antiviral activities in (a,b) were measured by focus reduction assay, with compounds applied post-adsorption for 72 h. The average size of foci was measured using the software Fiji57, and normalized as percentage of the untreated control. Each data point represents the mean of duplicate values, and the SEM is represented by error bars. The graphs were created using the software GraphPad Prism v.5 (GraphPad Software, La Jolla California USA), and are representative of at least 2 independent experiments.
The R151K mutation in M2-1 confers resistance to CPM
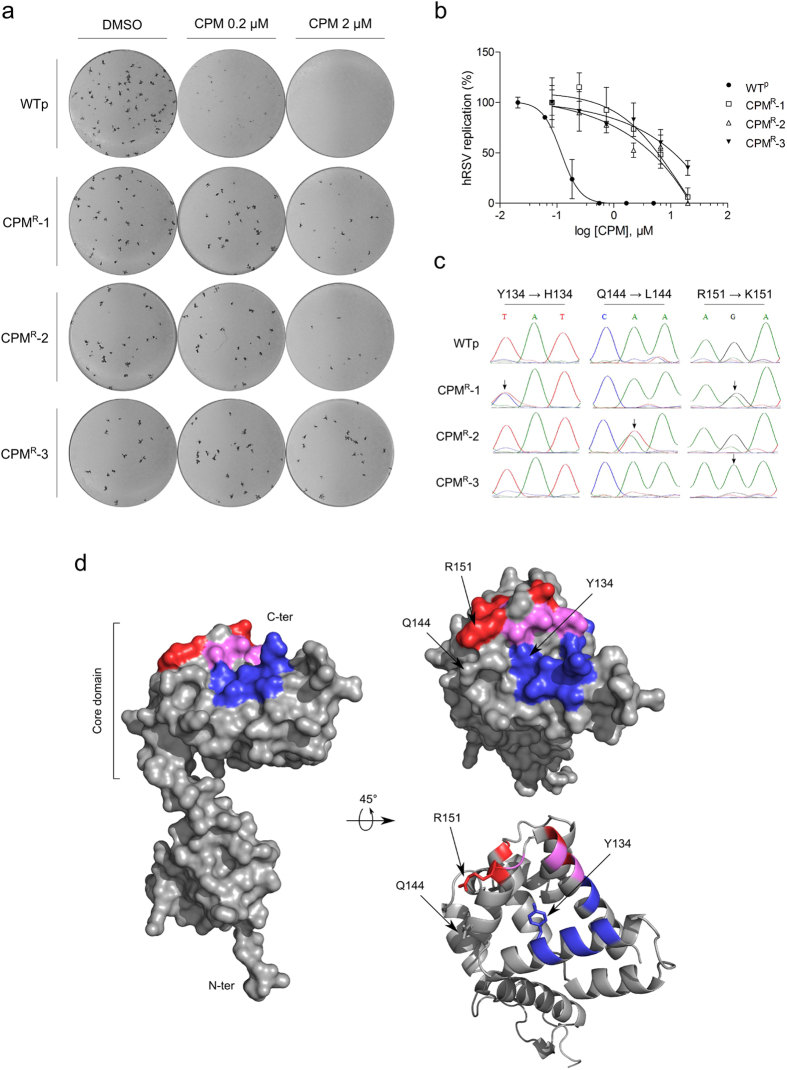
To determine whether a particular viral protein was implicated in the inhibition of hRSV by CPM, we selected CPM-resistant mutants of hRSV by 8 sequential passages in presence of increasing CPM concentrations, from 100 nM to 1 μM. Three CPM-resistant hRSV culture supernatants were generated independently, CPMR-1, CPMR-2 and CPMR-3. They possessed growth characteristics comparable to a wild-type virus passaged in parallel (WTp) and displayed similar focus morphology in the presence and absence of CPM (Fig. 3a). Interestingly, the virus was unable to regain fitness and induce CPE if the concentration of cyclopamine was increased too quickly (i.e. from 400 nM to 800 nM).
Figure 3. hRSV mutants resistant to CPM.
(a) Comparison of the focus formation by CPM-resistant viruses CPMR-1, CPMR-2, CPMR-3, and a passaged-wild-type virus (WTp) on HEp-2 cells in the presence or absence of CPM. (b) Dose-response of CPM on the 3 resistant mutants and WTp, measured by focus reduction assay on HEp-2 cells. The compound in (a,b) was applied post-adsorption, for 72 h. The average size of foci, measured using the software Fiji57, was normalized as percentage of the untreated control. The mean of duplicated values are plotted, along with the SEM represented by the error bars. The results are representative of 3 independent experiments. (c) Sequencing chromatograms of WTp and the 3 resistant virus supernatants, showing the base changes and amino-acid substitutions. The black arrows indicate the sites of clear mutation. (d) Mapping of the three mutation sites on a monomeric M2-1. Left: Surface representation of a full-length M2-1. Right: Surface (top) and cartoon (bottom) representations of the C-terminal side of M2-1 core domain. The RNA and P binding sites described by Blondot et al.21 are colored in red and blue respectively. The amino-acids that belong to both binding sites are colored in magenta. The three sites of mutation are indicated with black arrows. The models (PDB accession code 4C3B) were rendered using the PyMOL Molecular Graphics System, Version 1.7.1.0, Schrödinger, LLC.
The potency of CPM, added post-adsorption, was evaluated for each of the CPM-resistant supernatants. As shown in Fig. 3b, the WTp supernatant was CPM-sensitive with an IC50 value of 119 nM, comparable to the original WT hRSV. However, the potency of CPM in resistant virus supernatants was decreased 38- to 96-fold with IC50 values of 6 μM, 4.5 μM and 11.22 μM for CPMR-1, CPMR-2 and CPMR-3, respectively, confirming the presence of resistant virus in the culture supernatants.
Genome sequencing and alignment of the supernatants revealed three coding mutations located in the hRSV M2-1 gene (Fig. 3c). While WTp was found to be identical to the original hRSV sequence, CPMR-1 and CPMR-2 consisted of a mixed population of wild-type and M2-1 variants containing Y134H/R151K and Q144L/R151K mutations, respectively. In contrast, CPMR-3 was a homogeneous population displaying only the M2-1 R151K mutation. Interestingly, CPMR-3 showed the highest resistance to CPM as measured by a 96-fold increase of the IC50. Although the three mutated residues are located within the core domain of M2-1, R151 is located in the α-helix α8, a region previously described for its role in RNA and P binding21,22 (Fig. 3d). Together, these data demonstrate that the R151K mutation is sufficient to confer resistance to CPM, suggesting that the function of M2-1 as part of the RdRp complex is affected by the compound.
CPM inhibits M2-1-dependent viral transcription
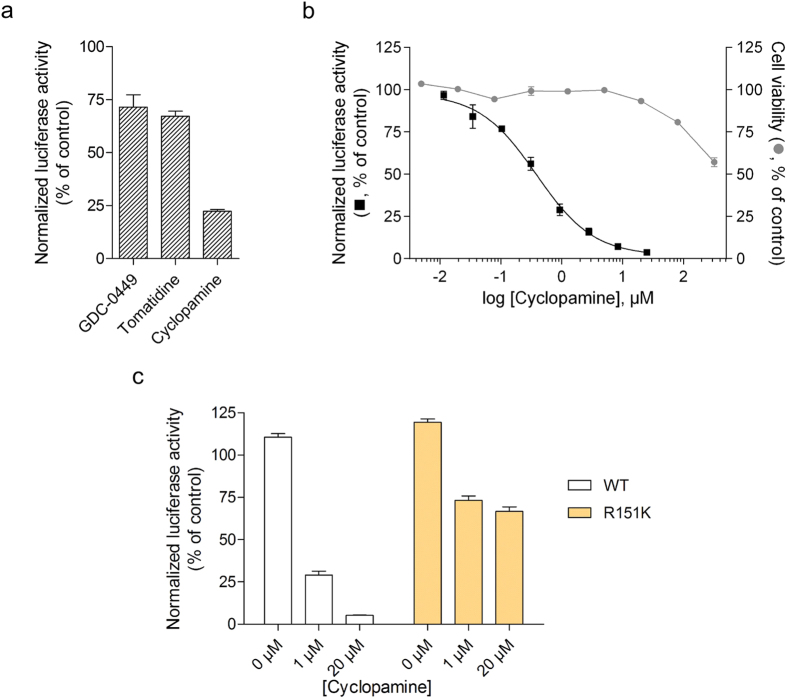
To confirm that the M2-1 mutations were sufficient to confer hRSV resistance to CPM, we used a well-established hRSV-specific minigenome assay system22,26. It contains the authentic M/SH gene junction, and the Luc reporter gene downstream of the gene start sequence present in this gene junction. The expression of the Luc gene in this system is absolutely dependent on the presence of a functional M2-122. The pM/Luc plasmid was co-transfected in BHK-21 BSR-T7/5 cells expressing T7 RNA polymerase together with p-β-gal, pL, pP, PN, and pM2-1 (WT or mutant). Luciferase activities were determined and normalized based on β-galactosidase expression. The effect of CPM on viral transcription was analyzed using either M2-1WT or M2-1R151K. Figure 4a,b shows that CPM, unlike GDC-0449 and tomatidine, inhibits the function of the hRSV RdRp complex in a dose-dependent manner with an IC50 of 380 nM while showing low toxicity towards BSR-T7/5 cells at working concentrations (CC50 > 320 μM, Fig. 4b). However, in cells expressing M2-1R151K the RdRp activity was highly resistant to CPM with a maximum inhibition of 26% at 1 μM, which could not be significantly increased using higher CPM concentrations (Fig. 4c). This result further validates that CPM blocks the RdRp complex by specifically inhibiting M2-1-mediated hRSV transcription.
Figure 4. CPM inhibition of hRSV M2-1-dependent transcription.
Inhibition of hRSV RdRp by 8.3 μM of CPM, GDC-0449, tomatidine (a) and dilutions of CPM (b), measured by RdRp-specific minigenome assay in BSR-T7/5 cells. The cytotoxicity of CPM towards BSR-T7/5 cells is shown in (b). (c) Inhibition of hRSV replication complex by CPM measured by minigenome assay in BSR-T7/5 cells transfected with plasmids coding for a WT or a R151K variant M2-1. Viral RNA synthesis in (a–c) was quantified by measuring the luciferase activity after cell lysis 24 h after transfection. The error bars represent the standard error of the mean of luciferase activity measured in triplicates, normalized against β-galactosidase activity and expressed as percentage of a control treated with PBS. For the data points at 0 μM, DMSO was used in place of CPM. The results are representative of 2 independent experiments performed in triplicate.
CPM reduces M2-1 expression levels
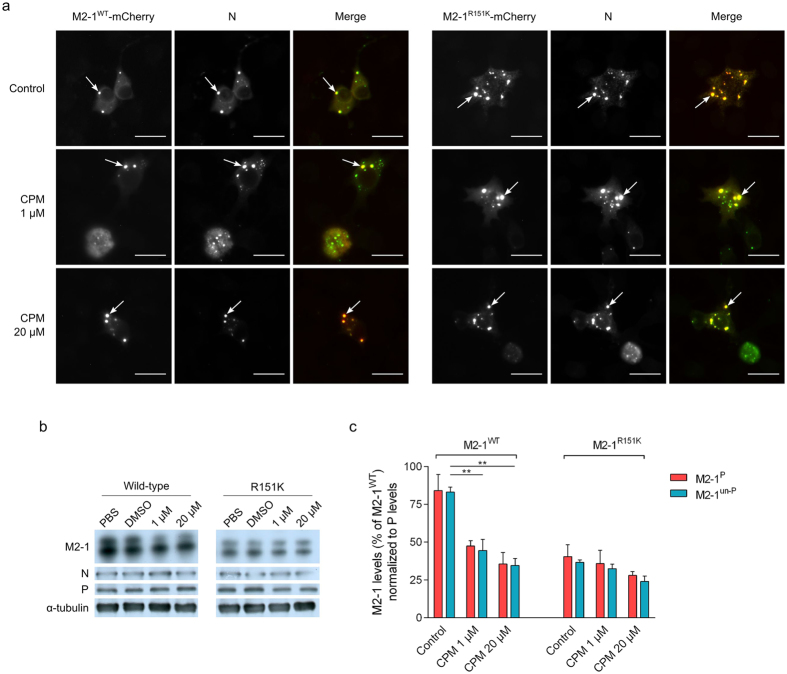
We performed immunofluorescence microscopy on minigenome-transfected cells to determine if CPM had an effect on M2-1 protein expression and subcellular localization. As shown in Fig. 5a, M2-1WT and M2-1R151K were readily detected in inclusion bodies together with N in the presence of increasing concentrations of compound. Overall expression levels of M2-1WT, however, were reduced by CPM while levels of N and P remained unaffected (Fig. 5b). Figure 5c shows that both the phosphorylated (M2-1P) and unphosphorylated (M2-1un-P) forms of M2-1 were reduced by CPM in a dose-dependent manner. Expression levels of M2-1R151K were lower compared to M2-1WT, but were not significantly affected by CPM treatment (Fig. 5b,c). These results show that treatment of cells with CPM is correlated with a selective reduction of M2-1 protein levels, and that M2-1R151K is resistant to CPM-mediated down-expression.
Figure 5. Effect of CPM on M2-1 expression.
(a) Immuno-fluorescence labelling of N and direct fluorescence of M2-1WT-mCherry or M2-1R151K-mCherry in minigenome-transfected BSR-T7/5 cells. Transfected cells were treated for 24 h with either DMSO or the indicated concentrations of CPM. N (green) was visualized using a mouse monoclonal anti-N primary antibody. Scale bars: 20 μm. The white arrows point to the location of cytoplasmic inclusion bodies. Merged images were created using the software Fiji. (b) Expression of M2-1, P, N and α-tubulin in minigenome-transfected BSR-T7/5 cells with the indicated concentrations of CPM, using M2-1WT or M2-1R151K, resolved by SDS-PAGE 24 h post-transfection and revealed by Western-blot using rabbit polyclonal anti-M2-1, P and N and mouse monoclonal anti-α-tubulin antibodies. Levels of α-tubulin are shown as a control of protein expression and DMSO served as control. The results are representative of 3 independent experiments (c) Associated levels of phosphorylated (M2-1P) and unphosphorylated (M2-1un-P) M2-1WT or M2-1R151K, normalized to levels of P in the absence of CPM or DMSO, and expressed as a percentage of M2-1WT in the absence of CPM. Proteins levels were measured from western blots using a GeneGnome Digital Imager (Syngene, Frederick, MD, USA). Bars are the mean of 3 independent experiments ± SEM. **p < 0.01 (Two-tailed t-test).
Antiviral efficacy of CPM in a mouse model of hRSV infection
Bioavailability and tissue distribution of CPM, CPM-analogues and Smo antagonists have been widely documented35,36. We therefore tested if the in vitro anti-hRSV activity of CPM could be observed in an experimental mouse model of hRSV infection. CPM was able to reduce lung hRSV titers by 1.5 logs when administered at 100 mg/kg for four days post infection (Fig. 6). The lung titer reduction was statistically significant (p > 0.001) and comparable to that observed with the hRSV fusion inhibitor BMS-47733115,37 at 50 mg/kg. Importantly, the magnitude of infection inhibition in mice was dose dependent. The animal data extend our in vitro observations and suggest that CPM and CPM analogues targeting M2-1 may be a promising avenue for the development of targeted hRSV-specific therapy.
Figure 6. Efficacy of CPM against hRSV in the mouse BALB/c host model of infection.
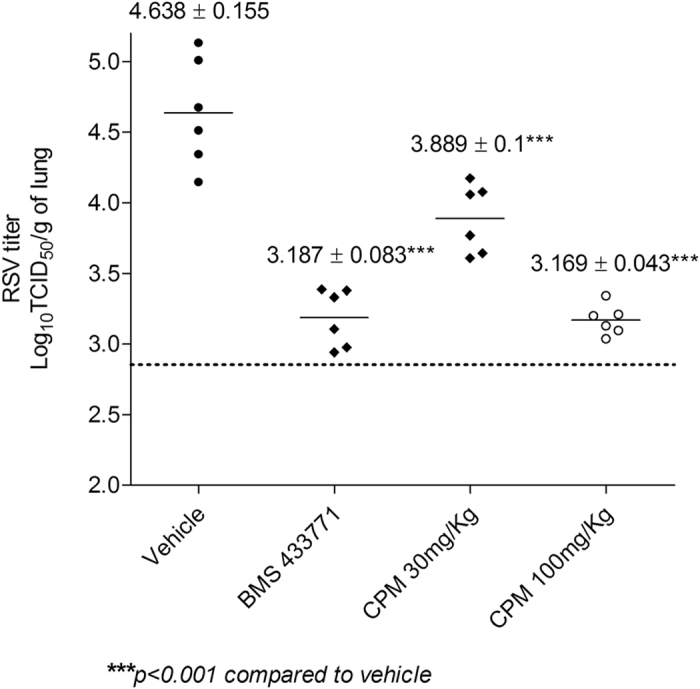
Animals were inoculated intranasally with hRSV Long strain. CPM and BMS-4337714 were administered intraperitoneally and by oral gavage, respectively, as a 4-day b.i.d. regimen in which the first dose was given 1 h before virus inoculation. Treatment cohorts are shown on the abscissa; animals of the infection control group were inoculated with virus and treated with vehicle only. The infectious hRSV lung titers are shown on the ordinate as log10 TCID50 per gram of lung. Each data point represents the hRSV titer for each individual animal of the respective treatment cohort. The horizontal line, drawn in each cohort, marks the geometric mean hRSV titer of the group, with the corresponding viral titer ± SD stated above. The horizontal hatched line, across the graph, represents the hRSV titer at the limit of assay detection. N = 6 mice per group.
Discussion
We identified the known Smo antagonist CPM as a potent post-entry inhibitor of hRSV replication by phenotypic screening of compound libraries. Several Veratrum steroidal alkaloids were evaluated in vitro to assess the structure-activity relationship of CPM. Only the Smo antagonist jervine38, a close structural analogue of CPM that possesses a keto-group in C-11 position on the D-ring, was able to inhibit viral replication. This keto-group leads to a 30-fold reduction of potency, providing important clues for future structure-activity relationship (SAR) using CPM analogues. In view of these results, it is also expected that the potent cyclopamine analogue IPI-926 (saridegib)39,40 would have a similar level of inhibition towards hRSV infection, which could add value to further SAR studies.
We hypothesized that the anti-hRSV activity of CPM may be mediated by Shhp and the Smo receptor. CPM binds to the same domain of Smo as antagonists GDC-0449 (vismodegib, Erivedge®) and LY2940680, currently in clinical trials, as well as the Smo-agonist purmorphamine. The 3 compounds, known competitors of CPM for binding to Smo27,32,34,41,42, had no effect on infection in vitro neither individually nor in combination with CPM, strongly suggesting that Smo is not involved in CPM-mediated inhibition of hRSV replication. Interestingly, an anti-Sonic hedgehog antibody was previously shown to reduce hepatitis C virus replication43. We did not observe any effect of this antibody on hRSV replication (data not shown), corroborating our conclusion that CPM inhibition of hRSV was independent of Smo and the Shhp.
To characterize the mechanism of action of CPM against hRSV, we selected CPM-resistant viruses and identified that a single R151K mutation on the hRSV M2-1 protein was able to confer resistance to the compound. Our data are consistent with previous studies which demonstrated that this residue is critical for RdRp activity. Indeed, an R151D substitution in M2-1 resulted in 70% reduction of M2-1-dependent transcription21,26. In an hRSV RdRp-specific minigenome functional assay CPM potently inhibited RdRp activity. The R151K variant was significantly less sensitive to CPM, demonstrating that the R151K mutation is responsible for CPM resistance. An analysis of 486 wild-type sequences reported in GenBank showed that R151 is fully conserved in all human M2-1 sequences (data not shown). Furthermore, virus supernatants were unable to recover from a sharp increase of cyclopamine concentration. This strongly suggests that pre-existing resistant R151K mutant quasispecies are unlikely to circulate in significant proportion in the population, and that they are unlikely to recover from a CPM treatment. The role of M2-1 in CPM-mediated hRSV inhibition is also consistent with our observation that CPM had no effect on hPIV-3 replication, which does not encode an M2-1-like protein (data not shown).
CPM significantly reduced M2-1 expression levels in minigenome-transfected cells expressing M2-1WT, while M2-1R151K was unaffected. Both phosphorylated and non-phosphorylated forms of M2-1WT were significantly reduced by CPM, in a dose-dependent manner compared to M2-1R151K. The overall levels of M2-1R151K, however, were lower than the ones of M2-1WT. This could be due to a poorer stability of the mutant protein, or a lower expression resulting from the mutation. Nevertheless, since the R151K variant had a phenotype similar to that of the WTp virus, it is possible that CPM uses an additional mechanism other than a reduction of M2-1 levels alone to inhibit viral replication. Although we could not demonstrate a direct effect of CPM on M2-1:P or M2-1:RNA interactions in vitro using recombinant proteins (data not shown), we cannot exclude that the reduction of M2-1 levels could be due to a specific effect of CPM on M2-1 binding to specific RNA and/or P, resulting in exclusion of M2-1 from inclusion bodies and protein degradation. By analogy with rhabdoviruses, inclusion bodies are considered as sanctuaries where viral RNA synthesis occurs44,45. Thus, although M2-1WT and M2-1R151K could still be detected in inclusion bodies in the presence of CPM (Fig. 5a), the levels of M2-1WT (but not M2-1R151K) were strongly reduced as compared to untreated cells (Fig. 5c). The overall reduction in M2-1 expression levels induced by CPM is expected to result in reduced viral RNA transcription and consequently virus production. Determining the exact molecular mechanism by which CPM interferes with the M2-1 function in hRSV transcription will require further investigation.
We showed that CPM is able to reduce lung hRSV titers by 1.5 logs in the BALB/c model of infection. Several small compounds inhibiting hRSV infection are currently under development, some of them targeting proteins of the hRSV RdRp complex such as L or N18,19,46,47,48. Other compounds inhibiting the critical F protein-mediated viral fusion have reached clinical development: the pyrazolopyrimidine GS-5806 has shown efficacy in phase 2a clinical trials13 while the azabenzimidazole BMS-433771, when administered orally in the BALB/c mouse model, was able to reduce lung titers of hRSV by 1.5 logs37. BMS-433771 efficacy in mice was predictive of efficacy in cotton rats despite different pharmacokinetic and pharmacodynamics properties between the models15.
Mousseau et al.49 have previously reported the discovery of a chemical analogue of the naturally occurring steroidal alkaloid cortistatin A as an inhibitor of human immunodeficiency virus Tat-dependent viral genome replication. Other studies have reported the discovery of inhibitors of the hRSV RdRp complex using small molecule libraries and hRSV minireplicon systems46,47,48,50. Compounds have also been used to target the hRSV M2-1 zinc-binding domain, but show limited in vivo efficacy and their mode of action remains unclear51,52.
To our knowledge, our study is the first to report a natural steroidal alkaloid as a specifically targeted antiviral compound showing in vivo efficacy. Our results also demonstrate that M2-1 constitutes a promising target for inhibitors of hRSV replication, and pave the way for new approaches in the design of antiviral compounds against hRSV.
Materials and Methods
Cells and viruses
Human respiratory syncytial virus Long strain (ATCC reference VR-26) and HEp-2 cells (ATCC reference CCL-23) were used for infection assays. Cells were maintained in DMEM, supplemented with penicillin/streptomycin and 10% FBS. Virus was passaged in HEp-2 cells in similar conditions, with 2% FBS. Virus stocks were prepared using HEp-2 cells, at low multiplicity of infection (MOI) for two to three days. Cells and virus-containing supernatant were then subjected to a single freeze/thaw cycle to release cell-bound virus, and samples were clarified by centrifugation at 2000 × g for 10 min at 4 °C. The supernatants were homogenized, aliquoted and stored at −80 °C.
Phenotypic screening and compound libraries
The screenings were performed by CPE-based assays in hRSV-infected Hep-2 cells. The 3244 compound library (CombinatoRx Inc.53) was screened in a 384-well format at a final concentration of 10 μM and was added to cells 1 h prior to inoculation with hRSV. CPE was evaluated 96 h post-infection using an ATPlite Luminescence Assay System (PerkinElmer Inc., Waltham, MA) following the manufacturer’s instructions. The cytotoxicity of compounds was assessed following the same method in absence of virus. Compounds were considered as hits if they inhibited more than 25% CPE at 10 μM, while reducing less than 30% of cell proliferation at active concentrations. The 502 compound library (Screen-Well® Natural Products Library, Enzo Life Sciences Inc., Farmingdale, NY) was screened in a 96-well format at a final concentration of 0.25 μg/mL and compounds were added to cells 1.5 h post-inoculation with hRSV. Virus-induced CPE was evaluated 96 h post-infection using a CellTiter-Glo Luminescent Cell Viability Assay (Promega, Madison, WI) following the manufacturer’s instructions. The cytotoxicity of compounds was assessed following the same method in absence of virus. The percentage of CPE inhibition was calculated using the formula (signal background – signal virus-induced CPE)/(signal cytotoxicity –signal virus-induced CPE), and hits were selected if the resulting value was superior to 30%.
Cyclopamine was purchased from Logan Natural Products (Plano, TX), GDC-0449 from Selleck Chemicals (Shanghai, P.R. China), LY2940680 from Biochempartner (Shanghai, P.R. China), tomatidine from Yingxuan Pharmaceutical (Shanghai, P.R. China) and BMS-433771 from ChemPartner (Shanghai, P.R. China). The Veratrum steroidal alkaloids were part of the Screen-Well® Natural Product Library from Enzo Life Sciences (Farmingdale, NY).
Virus titration by focus forming assay
Dilutions of virus were adsorbed onto confluent HEp-2 cells in 24-well plates for 1.5 h at 37 °C followed by two washing steps with PBS. Cells were subsequently overlaid with 0.75% carboxymethyl cellulose in DMEM supplemented with antibiotics and 2% FBS and incubated at 37 °C for 72 h. The culture supernatant was discarded and the cells were washed 3 × 5 min with PBS before being fixed for 30 min with 4% PFA in PBS at room temperature. Cells were washed and incubated for 1 h at room temperature with a primary anti-hRSV F antibody (mouse – Fitzgerald, Acton, MA) in PBS-5% skim milk, followed by a washing step with PBS-0.02% Tween 20. Antigen-antibody complexes were revealed by incubating cells for 1 h at room temperature with a secondary goat anti-mouse HRP-conjugated antibody (Bethyl, Montgomery, TX) in PBS-5% skim milk. Cells were then washed with PBS-0.02% Tween 20 and overlaid with TrueBlue Peroxidase Substrate (KPL, Gaithersburg, MD) until blue foci appeared. The plates were then rinsed with water, dried and scanned using a Canon CanoScan 5600 F table-top scanner for focus counting and size measurement.
Immuno-stained focus reduction assays
The anti-hRSV potency of compounds was evaluated by focus reduction assay, as previously described54, following the focus forming assay method and using 50 to 100 focus forming units per well. Compounds were tested during virus adsorption for 1 h at 4 °C or 37 °C to investigate inhibition of viral entry or propagation and replication, respectively, or during all stages of infection. IC50-90-95-99 values were determined by focus counting (viral entry) or focus size measurement (viral propagation, treatment post-adsorption) and were obtained using non-linear regression analysis using GraphPad Prism (GraphPad Software, La Jolla California USA). Compound cytotoxicity was assessed using a CellTiter-Glo Luminescent Cell Viability Assay (Promega, Madison, WI), following the manufacturer’s instruction in the conditions of the focus reduction assay.
Selection of hRSV resistance to CPM
CPM-resistant viruses were generated by blind-passages with increasing concentration of compound. Infections were started at an MOI of 0.1 for the first round and infections were stopped after 72 h or when about 50% CPE was observed. The infection media were subsequently clarified and 100 μL of supernatant were used to inoculate cells during the next passage. A total of 8 blind passages were sequentially performed in the presence of 100 nM, 200 nM, 400 nM, 600 nM, 800 nM and 1 μM CPM. A wild-type virus passaged with DMSO only was used as control.
Viral genome extraction, amplification and sequencing
A detailed procedure for the amplification and sequencing can be found in Supplementary Methods. Briefly, the total vRNA of each resistant supernatant was extracted using a TIANamp Virus RNA extraction Kit (Tiangen Biotech, Beijing) following the manufacturer’s instructions. The genomes were amplified using a SuperScript III One-Step RT-PCR System with Platinum Taq High Fidelity DNA Polymerase (Invitrogen, Carlsbad, CA). The fragments were checked by agarose gel electrophoresis and retrieved by gel extraction using a TIANgel Midi Purification Kit (Tiangen Biotech, Beijing), following the manufacturer’s instructions. An aliquot of each fragment was sequenced (Sangon Biotech, Shanghai), and the sequences were analyzed using the software Lasergene SeqMan Pro v.7.1 (DNASTAR, Madison, WI).
Minigenome assay
Minigenome assays in BSR-T7/5 cells were performed as described previously21,22, using M2-1WT or M2-1R151K sequences. Compounds were added during transfection, and left to incubate with cells for 24 h. Briefly, BSR-T7/5 cells stably expressing the T7 RNA polymerase55 were transfected in triplicates in 48-well plates for 24 h in the presence of Lipofectamine 2000 (Invitrogen, Carlsbad, CA) and 125 ng of pN, 125 ng of pP, 62.5 ng of pL and 37.5 ng of pM2-1WT or pM2-1R151K plasmids encoding hRSV N, P, L and M2-1WT or M2-1R151K proteins, respectively, under control of the T7 promoter. They were co-transfected with 125 ng of a pM/Luc subgenomic minireplicon expressing the firefly luciferase gene downstream of a hRSV M/SH gene junction, as well as with 37.5 ng of a pβ-Gal plasmid coding for the β-galactosidase as a reference gene. After 24 h of transfection, cells were lysed in luciferase lysis buffer (30 mM Tris pH 7.9, 10 mM MgCl2, 1 mM DTT, 1% Triton X-1000, 15% glycerol). Luciferase and β-Gal activities were measured for each well as previously described (26) using a Tecan Infinite 200 PRO (Tecan Group LTD, Männedorf). Compound cytotoxicity towards BSR-T7/5 cells was assessed using a CellTiter-Glo Luminescent Cell Viability Assay (Promega, Madison, WI), following the manufacturer’s instruction in the conditions of the minigenome assay.
Immunofluorescence microscopy
BSR-T7/5 cells were transfected as described above, using a plasmid encoding for wild-type or R151K-mutant M2-1-mCherry fusion proteins56 in place of the pM2-1 plasmid. The transfected cells were fixed, permeabilized and incubated with a primary mouse monoclonal anti-N antibody (AbD Serotec, clone B023) for 1 h, followed by incubation with a secondary goat anti-mouse Alexa Fluor 488 antibody for 1 h. Coverslips were mounted in Prolong Gold Antifade reagent (Invitrogen, Carlsbad, CA), observed with a Nikon TE200 microscope equipped with a CoolSNAP ES2 (Photometrics, Tucson, AZ) camera, and images were processed using the Meta-Vue (Molecular Devices, Sunnyvale, CA) and Fiji57 softwares.
Immunoassays
Proteins from SDS-PAGE were transferred onto nitrocellulose membranes which were incubated with primary rabbit polyclonal anti-P58, -N58; -M2-126, and mouse monoclonal anti-tubulin-α antibodies (Sigma-Aldrich, Saint-Quentin Fallavier, France) for 1 h, followed by appropriate HRP-conjugated secondary antibodies for 1 h. Immunodetections were performed using an enhanced chemiluminescence (ECL) substrate (GE Healthcare GMB, Vélizy, France). Protein levels were determined from Western blots using the GeneTools software after acquisition of the signals with a GeneGnome Digital Imager (Syngene, Frederick, MD, USA).
Ethics Statement
The protocol for use of animals was approved by the Biological Resource Center, Singapore Animal Care and Use Committee (Protocol 070236). It was established following the Guidelines on the Care and Use of Animals for Scientific Purposes as setup by the National Advisory Committee for Laboratory Animal Research, the Animal and Birds Act (Chapter 7) and the Guide for the Care and Use of Laboratory Animals 8th Edition.
Animal model
The antiviral efficacy of CPM was examined in the BALB/c mouse model of infection. Female BALB/c mice were obtained from the Biological Resource Center, Singapore, and used at a body weight of 16–20 g. All animals were housed in a certified, specific pathogen-free facility and were fed and watered ad libitum. CPM was formulated as a 0.5% hydroxypropyl methyl cellulose suspension and was administered at a dose of 30 and 100 mg/kg intraperitoneally b.i.d. for four days. BMS-433771 was dissolved in sterile water, adjusted to pH 2 to 3.5 and administered at a dose of 50 mg/kg by oral gavage b.i.d. for four days15. Treatments were initiated 1 h prior to hRSV inoculation. Mice were anesthetized by intraperitoneal injection of ketamine (70 mg/kg) and xylazine (20 mg/kg) and inoculated via intranasal route with 8 × 105 TCID50 of hRSV Long Strain in 100 μL of EMEM. The control group received vehicle only. On day 4 after inoculation and 1 h after last compound administration, mice (N = 6 per group) were euthanized by CO2 gas asphyxiation and lungs were excised and weighed. They were homogenized (10% wt/vol) in HBSS containing 0.21 M sucrose, 25 mM HEPES, 5 mM sodium l-glutamate, 20 U/mL penicillin G, 20 μg/mL streptomycin and 0.05 μg/mL amphotericin B (GIBCO/BRL, Carlsbad, Calif.). Homogenates were frozen and thawed to release cell-associated virus and held on ice until clarification by 10 min centrifugation at 300 × g at 4 °C. The supernatants were immediately titrated for hRSV load in HEp-2 cells as described previously15. Final hRSV lung titers were expressed as log10 TCID50 per gram of lung. Infectious hRSV lung titers for the treatment groups, or cohorts, were calculated as mean titers and expressed as log10 TCID50 per gram of lung.
Additional Information
How to cite this article: Bailly, B. et al. Targeting human respiratory syncytial virus transcription anti-termination factor M2-1 to inhibit in vivo viral replication. Sci. Rep. 6, 25806; doi: 10.1038/srep25806 (2016).
Supplementary Material
Acknowledgments
We thank Christina Sizun (Gif-sur-Yvette) for NMR assays with M2-1 and CPM, Jill Grenier for assistance with high throughput screenings, Rong Chen for providing HEp-2 cells and hRSV Long strain and José Melero (Madrid) for providing M2-1 22K4 antibodies. This work was supported by the Institut Pasteur Shanghai – Chinese Academy of Sciences and the Li Ka Shing Foundation [R.A.], the French Agence Nationale de la Recherche, specific program ANR Blanc 2011 “Bronchioliteaserl” [ANR-11-BSV8-0024, J.F.E.], and in part by funding from National Natural Science Foundation of China [31400148 to G.Z.] and the Science and Technology Commission of Shanghai Municipality [14YF1407600 to G.Z.] A Doctoral Grant from the Institut Pasteur International Network and a Griffith University Post-Graduate Scholarship are also acknowledged [B.B].
Footnotes
Author Contributions R.A. conceived and supervised the project. B.B. conducted mechanism of action studies. L.W., Y.W. and G.Z. generated resistant mutants and L.J., R.C. and B.B. performed screening assays. C.-A.R., B.B., M.G. and J.C. performed M2-1-related biochemical and minigenome assays. G.S., D.C.S. and V.P. performed in vivo studies. The manuscript was written by B.B. and R.A. and was reviewed by J.-F.E., P.G., M.v.I., M.G., G.Z., L.J., G.S., V.P. and D.C.S.
References
- Karron R. & Collins P. In Fields Virology (eds. Knipe D. & Howley P.) 1497–1526 (Lippincott Williams & Wilkins, 2007). [Google Scholar]
- Nair H. et al. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. The Lancet 375, 1545–1555 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C. B. Respiratory syncytial virus and parainfluenza virus. N. Engl. J. Med. 344, 1917–1928 (2001). [DOI] [PubMed] [Google Scholar]
- Johnstone J., Majumdar S. R., Fox J. D. & Marrie T. J. Viral infection in adults hospitalized with community-acquired pneumonia: prevalence, pathogens, and presentation. Chest 134, 1141–1148 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsey A. R., Hennessey P. A., Formica M. A., Cox C. & Walsh E. E. Respiratory syncytial virus infection in elderly and high-risk adults. N. Engl. J. Med. 352, 1749–1759 (2005). [DOI] [PubMed] [Google Scholar]
- Falsey A. R. & Walsh E. E. Respiratory syncytial virus infection in adults. Clin. Microbiol. Rev. 13, 371–384 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resch B., Kurath S. & Manzoni P. Epidemiology of Respiratory Syncytial Virus Infection in Preterm Infants. Open Microbiol. J. 5, 135–143 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H. Y. & Englund J. A. Respiratory syncytial virus disease: prevention and treatment. Curr. Top. Microbiol. Immunol. 372, 235–258 (2013). [DOI] [PubMed] [Google Scholar]
- Hussman J. M., Li A., Paes B. & Lanctôt K. L. A review of cost–effectiveness of palivizumab for respiratory syncytial virus. Expert Rev. Pharmacoecon. Outcomes Res. 12, 553–567 (2012). [DOI] [PubMed] [Google Scholar]
- Smith D. W. et al. A controlled trial of aerosolized ribavirin in infants receiving mechanical ventilation for severe respiratory syncytial virus infection. N. Engl. J. Med. 325, 24–29 (1991). [DOI] [PubMed] [Google Scholar]
- Wyde P. R. Respiratory syncytial virus (RSV) disease and prospects for its control. Antiviral Res. 39, 63–79 (1998). [DOI] [PubMed] [Google Scholar]
- Wyde P. R. et al. Antiviral efficacy of VP14637 against respiratory syncytial virus in vitro and in cotton rats following delivery by small droplet aerosol. Antiviral Res. 68, 18–26 (2005). [DOI] [PubMed] [Google Scholar]
- DeVincenzo J. P. et al. Oral GS-5806 Activity in a Respiratory Syncytial Virus Challenge Study. N. Engl. J. Med. 371, 711–722 (2014). [DOI] [PubMed] [Google Scholar]
- Mackman R. L. et al. Discovery of an oral respiratory syncytial virus (RSV) fusion inhibitor (GS-5806) and clinical proof of concept in a human RSV challenge study. J. Med. Chem. 58, 1630–1643 (2015). [DOI] [PubMed] [Google Scholar]
- Cianci C. et al. Oral Efficacy of a Respiratory Syncytial Virus Inhibitor in Rodent Models of Infection. Antimicrob. Agents Chemother. 48, 2448–2454 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewska W. et al. Antiviral and lung protective activity of a novel respiratory syncytial virus fusion inhibitor in a mouse model. Eur. Respir. J. 38, 401–408 (2011). [DOI] [PubMed] [Google Scholar]
- Chapman J. et al. RSV604, a novel inhibitor of respiratory syncytial virus replication. Antimicrob. Agents Chemother. 51, 3346–3353 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challa S. et al. Mechanism of action for respiratory syncytial virus inhibitor RSV604. Antimicrob. Agents Chemother. 59, 1080–1087 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. et al. Discovery of 4′-chloromethyl-2′-deoxy-3′,5′-di-O-isobutyryl-2′-fluorocytidine (ALS-8176), a first-in-class RSV polymerase inhibitor for treatment of human respiratory syncytial virus infection. J. Med. Chem. 58, 1862–1878 (2015). [DOI] [PubMed] [Google Scholar]
- Collins P. L., Hill M. G., Cristina J. & Grosfeld H. Transcription elongation factor of respiratory syncytial virus, a nonsegmented negative-strand RNA virus. Proc. Natl. Acad. Sci. USA 93, 81–85 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondot M.-L. et al. Structure and Functional Analysis of the RNA- and Viral Phosphoprotein-Binding Domain of Respiratory Syncytial Virus M2-1 Protein. PLoS Pathog. 8, e1002734 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran T.-L. et al. The Respiratory Syncytial Virus M2-1 Protein Forms Tetramers and Interacts with RNA and P in a Competitive Manner. J. Virol. 83, 6363–6374 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawar R. G. et al. Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science 326, 1279–1283 (2009). [DOI] [PubMed] [Google Scholar]
- Fearns R. & Collins P. L. Role of the M2-1 Transcription Antitermination Protein of Respiratory Syncytial Virus in Sequential Transcription. J. Virol. 73, 5852–5864 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy R. W. & Wertz G. W. The product of the respiratory syncytial virus M2 gene ORF1 enhances readthrough of intergenic junctions during viral transcription. J. Virol. 72, 520–526 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner S. J. et al. Crystal structure of the essential transcription antiterminator M2-1 protein of human respiratory syncytial virus and implications of its phosphorylation. Proc. Natl. Acad. Sci. USA 111, 1580–1585 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale J. et al. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature 406, 1005–1009 (2000). [DOI] [PubMed] [Google Scholar]
- Chen J. K., Taipale J., Cooper M. K. & Beachy P. A. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 16, 2743–2748 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham P. W. & McMahon A. P. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 15, 3059–3087 (2001). [DOI] [PubMed] [Google Scholar]
- Dessinioti C., Plaka M. & Stratigos A. J. Vismodegib for the treatment of basal cell carcinoma: results and implications of the ERIVANCE BCC trial. Future Oncol. Lond. Engl. 10, 927–936 (2014). [DOI] [PubMed] [Google Scholar]
- Bender M. H. et al. Abstract 2819: Identification and characterization of a novel smoothened antagonist for the treatment of cancer with deregulated hedgehog signaling. Cancer Res. 71, 2819–2819 (2011). [Google Scholar]
- Wang C. et al. Structure of the human smoothened receptor bound to an antitumour agent. Nature 497, 338–343 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rominger C. M. et al. Evidence for Allosteric Interactions of Antagonist Binding to the Smoothened Receptor. J. Pharmacol. Exp. Ther. 329, 995–1005 (2009). [DOI] [PubMed] [Google Scholar]
- Sinha S. & Chen J. K. Purmorphamine activates the Hedgehog pathway by targeting Smoothened. Nat. Chem. Biol. 2, 29–30 (2006). [DOI] [PubMed] [Google Scholar]
- Lin T. L. & Matsui W. Hedgehog pathway as a drug target: Smoothened inhibitors in development. OncoTargets Ther. 5, 47–58 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heretsch P., Tzagkaroulaki L. & Giannis A. Cyclopamine and hedgehog signaling: chemistry, biology, medical perspectives. Angew. Chem. Int. Ed Engl. 49, 3418–3427 (2010). [DOI] [PubMed] [Google Scholar]
- Cianci C. et al. Orally Active Fusion Inhibitor of Respiratory Syncytial Virus. Antimicrob. Agents Chemother. 48, 413–422 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper M. K., Porter J. A., Young K. E. & Beachy P. A. Teratogen-mediated inhibition of target tissue response to Shh signaling. Science 280, 1603–1607 (1998). [DOI] [PubMed] [Google Scholar]
- Jimeno A. et al. Phase I Study of the Hedgehog Pathway Inhibitor IPI-926 in Adult Patients with Solid Tumors. Clin. Cancer Res. 19, 2766–2774 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay M. R. et al. Discovery of a potent and orally active hedgehog pathway antagonist (IPI-926). J. Med. Chem. 52, 4400–4418 (2009). [DOI] [PubMed] [Google Scholar]
- Robarge K. D. et al. GDC-0449—A potent inhibitor of the hedgehog pathway. Bioorg. Med. Chem. Lett. 19, 5576–5581 (2009). [DOI] [PubMed] [Google Scholar]
- Stanton B. Z. et al. A small molecule that binds Hedgehog and blocks its signaling in human cells. Nat. Chem. Biol. 5, 154–156 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S. S. et al. Upregulation of Hedgehog Pathway is Associated with Cellular Permissiveness for Hepatitis C Virus Replication. Hepatology 54, 1580–1590 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahaye X. et al. Functional Characterization of Negri Bodies (NBs) in Rabies Virus-Infected Cells: Evidence that NBs Are Sites of Viral Transcription and Replication. J. Virol. 83, 7948–7958 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich B. S., Cureton D. K., Rahmeh A. A. & Whelan S. P. J. Protein Expression Redirects Vesicular Stomatitis Virus RNA Synthesis to Cytoplasmic Inclusions. PLoS Pathog 6, e1000958 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiong-Yip C.-L. et al. Characterization of a respiratory syncytial virus L protein inhibitor. Antimicrob. Agents Chemother. 58, 3867–3873 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laganas V. A. et al. Characterization of novel respiratory syncytial virus inhibitors identified by high throughput screen. Antiviral Res. 115, 71–74 (2015). [DOI] [PubMed] [Google Scholar]
- Xiong H. et al. Discovery of a potent respiratory syncytial virus RNA polymerase inhibitor. Bioorg. Med. Chem. Lett. 23, 6789–6793 (2013). [DOI] [PubMed] [Google Scholar]
- Mousseau G. et al. An Analog of the Natural Steroidal Alkaloid Cortistatin A Potently Suppresses Tat-Dependent HIV Transcription. Cell Host Microbe 12, 97–108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant H. et al. High-Throughput Hit Screening Cascade to Identify Respiratory Syncytial Virus (RSV) Inhibitors. J. Biomol. Screen. 20, 597–605 (2015). [DOI] [PubMed] [Google Scholar]
- Boukhvalova M. S., Prince G. A. & Blanco J. C. Inactivation of respiratory syncytial virus by zinc finger reactive compounds. Virol. J. 7, 20 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancellieri M. et al. In silico structure-based design and synthesis of novel anti-RSV compounds. Antiviral Res. 122, 46–50 (2015). [DOI] [PubMed] [Google Scholar]
- Owens C. M. et al. Chemical combinations elucidate pathway interactions and regulation relevant to Hepatitis C replication. Mol. Syst. Biol. 6, 375 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly B. et al. A dual drug regimen synergistically blocks human parainfluenza virus infection. Sci. Rep. 6, 24138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz U. J., Finke S. & Conzelmann K. K. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73, 251–259 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloux M. et al. Identification and characterization of the binding site of the respiratory syncytial virus phosphoprotein to RNA-free nucleoprotein. J. Virol. 89, 3484–3496 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castagné N. et al. Biochemical characterization of the respiratory syncytial virus P-P and P-N protein complexes and localization of the P protein oligomerization domain. J. Gen. Virol. 85, 1643–1653 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.