Abstract
Torsin ATPases (Torsins) belong to the widespread AAA+ (ATPases associated with a variety of cellular activities) family of ATPases, which share structural similarity but have diverse cellular functions. Torsins are outliers in this family because they lack many characteristics of typical AAA+ proteins, and they are the only members of the AAA+ family located in the endoplasmic reticulum and contiguous perinuclear space. While it is clear that Torsins have essential roles in many, if not all metazoans, their precise cellular functions remain elusive. Studying Torsins has significant medical relevance since mutations in Torsins or Torsin-associated proteins result in a variety of congenital human disorders, the most frequent of which is Early Onset Torsion (DYT1) Dystonia, a severe movement disorder. A better understanding of the Torsin system is needed to define the molecular etiology of these diseases, potentially enabling corrective therapy. Here, we provide a comprehensive overview of the Torsin system in metazoans, discuss functional clues obtained from various model systems and organisms, and provide a phylogenetic and structural analysis of Torsins and their regulatory cofactors in relation to disease-causative mutations. Moreover, we review recent data that has led to a dramatically improved understanding of these machines at a molecular level, providing a foundation for investigating the molecular defects underlying the associated movement disorders. Lastly, we discuss our ideas on how recent progress may be utilized to inform future studies aimed at determining the cellular role(s) of these atypical molecular machines and their implications for dystonia treatment options.
Keywords: TorsinA (TorA), DYT1 dystonia, LAP1, LULL1, Endoplasmic Reticulum (ER), Nuclear Envelope (NE)
Structural features of AAA+ ATPases and Torsins: a brief introduction
Torsin ATPases are a family of proteins that belong to the AAA+ (ATPases associated with a variety of cellular activities) superfamily of ATPases (Neuwald et al., 1999). AAA+ ATPases utilize the energy of ATP hydrolysis to unfold or cause a conformational change in a substrate protein, energize motility in concert with cytoskeletal components, or remodel protein-protein or protein-nucleic acid complexes (Hanson and Whiteheart, 2005). Though their cellular functions and mechanism vary widely, AAA+ proteins share significant sequence similarity, and feature at least one AAA+ domain which is used as a common module that is repurposed in many different ways–often by the addition of accessory domains–to enable distinct functions. These are reviewed in detail elsewhere (Hanson and Whiteheart, 2005, Erzberger and Berger, 2006, Sauer and Baker, 2011); however a brief introduction is necessary to put Torsins in the structural context of related ATPases.
The AAA+ module consists of two different subdomains: a wedge-shaped α/β RecA fold at the N-terminus and an α-helical domain at the C-terminus (Lenzen et al., 1998, Zhang et al., 2000, Lee et al., 2003, Karlberg et al., 2009, Yu et al., 1998, Guenther et al., 1997). This tertiary structure is more conserved among AAA+ superfamily members than their primary amino acid sequences (Smith et al., 2004), although a number of motifs are highly conserved (cf. inset in Figure 1). The Walker A motif has a consensus sequence of GXXXXGK[T/S] (where X represents any amino acid) in which the lysine positioned on the P-loop directly contacts the β and γ phosphates of ATP, making it critical for ATP binding (Saraste et al., 1990). The Walker B motif consensus sequence is hhhhDE (where h represents a hydrophobic amino acid). The glutamate in this motif is required for ATP hydrolysis and mutating this residue to a glutamine or alanine abolishes this activity (Babst et al., 1998, Weibezahn et al., 2003, Dalal et al., 2004). A Walker B mutation functions as a “substrate trap” in Torsin-related Clp/HSP100 proteins because these mutant derivatives can bind but no longer hydrolyze ATP, effectively trapping the protein in the ATP-bound conformation which is the nucleotide state with highest affinity for substrate (Weibezahn et al., 2003, Schlieker et al., 2004). These substrate-trap mutants have proven useful for the functional study of AAA+ ATPases because they often exert a dominant-negative effect on the endogenous wild-type protein when overexpressed in the cell and in vitro (Whiteheart et al., 1994, Hanson and Whiteheart, 2005, Weibezahn et al., 2003, Dalal et al., 2004).
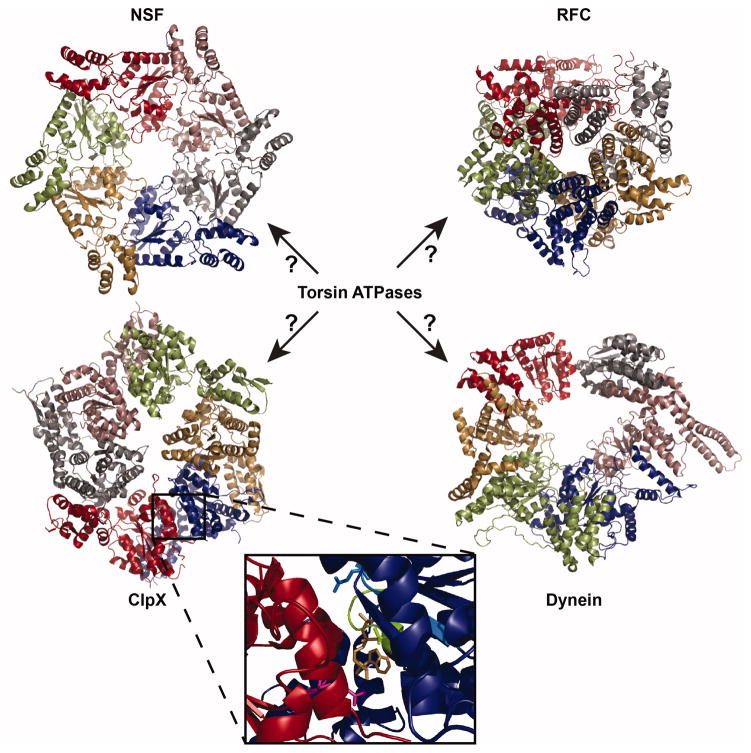
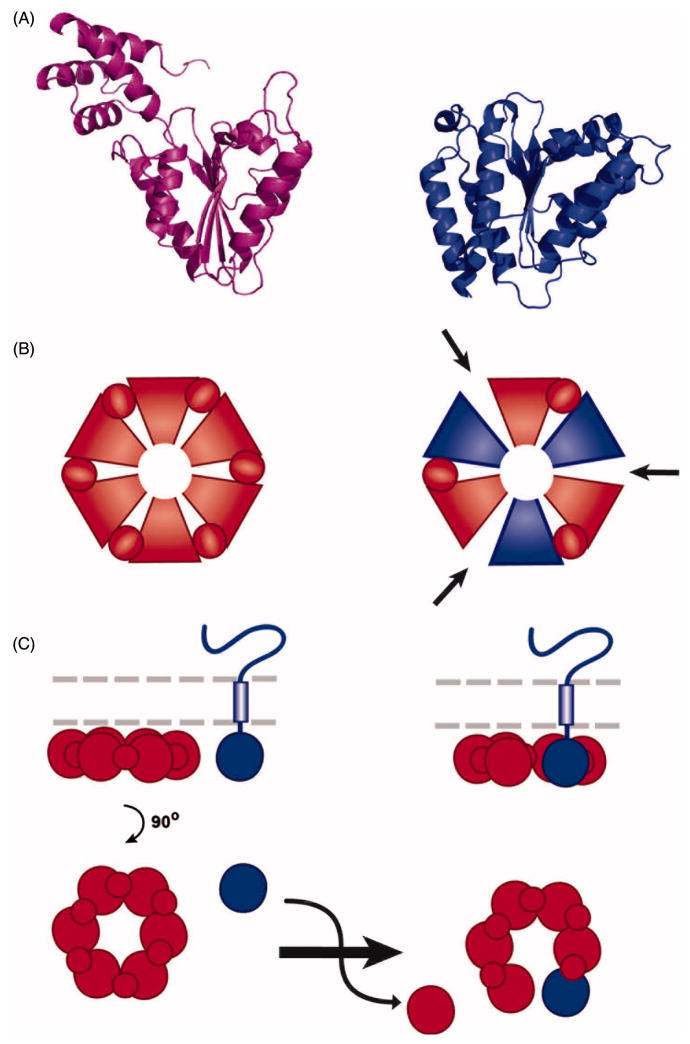
FIGURE 1.
Structural comparison of AAA+ ATPase assemblies. ClpX (PDB ID: 3HWS (Glynn et al. 2009)) and N-ethylmaleimide-sensitive factor (NSF) (PDB ID: 1D2N (Lenzen et al. 1998) are AAA+ ATPases that form homohexameric assemblies. Replication factor C (RFC) (PDB ID: 1SXJ (Bowman et al. 2004)) and dynein (4W8F (Bhabha et al. 2014)) are heteromeric AAA+ ATPases. RFC forms a heteropentameric assembly comprised of 5 different AAA subunits and dynein forms a heterohexameric assembly where all AAA domains are encoded in one continuous peptide. Individual AAA domains are depicted in different colors. Inset: Zoomed in image of the intersubunit interface of a ClpX dimer highlighting the Walker A site (green), Walker B site (light blue), and arginine finger (magenta) surrounding the nucleotide. Key residues K127, E185 (mutated to Q in the structure), and R307 are represented as sticks. ADP is shown in gold.
In addition to their conserved tertiary structure, most AAA+ ATPases share the common quaternary structural feature of assembling into higher-order oligomers that form either rings or spirals. The majority of AAA+ ATPases assemble into homotypic hexamers, but a small minority form heteromeric assemblies (Figure 1). These oligomers are key for function because ATP binding and hydrolysis occurs at the interfaces between subunits, driving large conformational changes that are utilized in different ways to apply force on their substrates. Within this oligomeric context, a highly conserved arginine residue (sometimes called an “arginine finger” (Ogura et al., 2004)) extends into the active site of a neighboring subunit in an AAA+ oligomer to facilitate ATP hydrolysis (inset in Figure 1). This structural motif acts similarly to the arginine residues provided by GTPase-activating proteins (GAPs) to activate their cognate GTPases via charge neutralization of the transition state (Scheffzek et al., 1998). Additionally, since these arginine residues occur at subunit-subunit interfaces, they aid in transmission of conformational changes between subunits of the AAA+ oligomer (Ogura et al., 2004, Augustin et al., 2009).
Another important consequence of the oligomerization is the formation of a central pore, which enables a substrate threading mechanism in a number of AAA+ ATPases, e.g. the Torsin-related Clp/Hsp100 proteins. These use the energy of ATP hydrolysis to unfold the substrate protein, enabling either refolding or degradation by associated proteases (Yamada-Inagawa et al., 2003, Schlieker et al., 2004, Weibezahn et al., 2004, Siddiqui et al., 2004, Weber-Ban et al., 1999).
The five members of the Torsin family found in human cells (TorsinA, TorsinB, Torsin2A, Torsin3A, and Torsin4A) (Figure 2A) are atypical members of the AAA+ superfamily for the following reasons: (i) they contain a non-canonical Walker A motif (GXXXXGKN) (Nagy et al., 2009) (ii) Torsins are the only AAA+ ATPases known to reside within the endoplasmic reticulum (ER) and contiguous nuclear envelope (NE) of mammalian cells (Figure 2C) (Jungwirth et al., 2010), and (iii) they lack the arginine finger that is otherwise conserved in closely related members of the AAA+ family. Therefore, intense debate had focused on whether Torsins are active ATPases or merely degenerated AAA+ modules, and conflicting results were obtained in measuring ATPase activity (Kustedjo et al., 2003, Pham et al., 2006, Konakova and Pulst, 2005, Zhao et al., 2013) since their discovery in 1997 by Breakefield and colleagues (Ozelius et al., 1997). This uncertainty had precluded a mechanistic understanding of Torsins and their malfunction in the disease state.
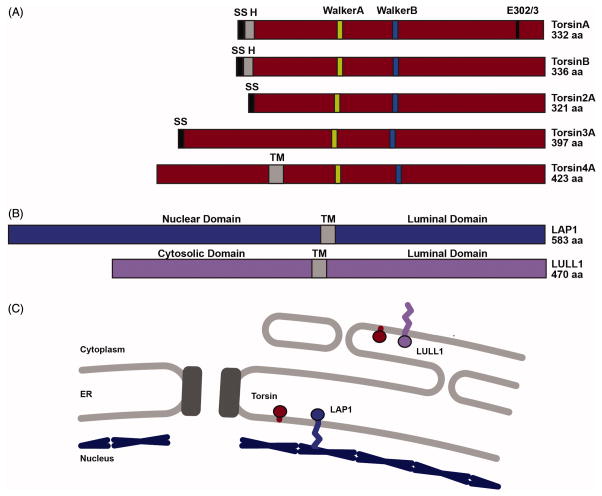
FIGURE 2.
Domain structure of Torsin family ATPases. (A) Graphical comparison of the five members of the Torsin family of AAA+ ATPases depicting locations of various features in the primary sequence. SS: Signal Sequence; H: Hydrophobic Patch; Walker A: ATP binding motif; Walker B: ATP hydrolysis motif; E302: Glutamate deleted in the TorsinA disease mutant; TM: Predicted transmembrane segment. (B) Graphical comparison of cofactors LAP1 and LULL1 depicting locations of transmembrane segments and activator arginines. TM: transmembrane segment; R563/R449: activator arginines. (C) Cartoon depicting the subcellular localization of Torsins, LAP1, and LULL1. Torsin is anchored to the luminal face of the ER and NE membranes. LAP1 resides in the inner nuclear membrane and LULL1 in the ER membrane. Black rectangles: nuclear pore complex. Blue lines: nuclear lamina.
Herein, we will review the literature on Torsin ATPases with a focus on TorsinA in the context of animal models and disease associations. We will then summarize our increasing knowledge pertaining to the molecular and biochemical properties of Torsins, and discuss how disease-causing mutations lead to Torsin dysfunction.
TorsinA: disease association and mutant phenotypes in animal models
TorsinA is the best-studied member of the Torsin family due to its association with the disease Early Onset Torsion Dystonia (EOTD) (also called DYT1 dystonia). This disorder is most strongly correlated with the deletion of one of two consecutive glutamate residues (E302/303; hereafter referred to as TorsinA ΔE). This glutamate is not the aforementioned glutamate residue in the Walker B motif, but instead is in the C-terminal α-helical domain of the AAA+ structure (Ozelius et al., 1997, Ozelius et al., 1998). A few dystonia cases have been reported to result from different TorsinA mutations (Leung et al., 2001, Zirn et al., 2008, Calakos et al., 2010, Cheng et al., 2014, Vulinovic et al., 2014). In the case of TorsinA ΔE, the disease is inherited in an autosomal dominant manner, but it is only approximately 30% penetrant (meaning not all carriers of the mutation show symptoms of the disease) (Bressman et al., 1989).
At a clinical level, EOTD manifests as involuntary twisting, sustained postures, and repetitive movements usually starting in the limbs with an average age of onset of approximately 12 years (Bressman et al., 1994), with little to no detectable neurodegeneration (Tanabe et al., 2009). The dystonic phenotype can be recapitulated in mice by conditional deletion of TorsinA or the isolated expression of TorsinA ΔE in the central nervous system in a TorA knockout background (Liang et al., 2014). Dystonic symptoms are also observed upon selective deletion of TorsinA in embryonic progenitor cells of cholinergic and GABAergic neurons in mouse forebrains (Pappas et al., 2015). In cultured cells, overexpression of the TorsinA ΔE mutant leads to whorled membrane inclusions in the cytoplasm derived from the NE (Hewett et al., 2000, Kustedjo et al., 2000, Bragg et al., 2004a, Gonzalez-Alegre and Paulson, 2004, Bragg et al., 2004b) and the mutant protein is enriched in the NE compared to wild-type TorsinA (Goodchild and Dauer, 2004). A homozygous knock-in of TorsinA ΔE in mice results in the accumulation of perinuclear herniations or blebs, which are similar to those observed in TorsinA null mice (Goodchild and Dauer, 2005, Liang et al., 2014).
TorsinA is ubiquitously expressed in all cell types but enriched in parts of the nervous system (Jungwirth et al., 2010). A homozygous knockout of TorsinA in mice causes early postnatal lethality, correlated with blebbing of the inner nuclear membrane (INM) into the perinuclear space in neurons only (Goodchild et al., 2005). A similar NE defect was observed in TorsinA-deficient Drosophila melanogaster larvae (Jokhi et al., 2013). These same perinuclear blebs also appear in TorsinA ΔE homozygous knock-in mice as well as in mice in which the ΔE knock-in allele is expressed on a TorsinA knockout background (Goodchild et al., 2005). The observation that the TorsinA ΔE knock-in fails to rescue the knockout phenotype strongly suggested that the ΔE mutation impairs TorsinA function (Goodchild et al., 2005), though the underlying molecular mechanism was unclear at the time. Early embryonic defects correlating with NE perturbations were also observed upon a Torsin loss-of-function mutation in Caenorhabditis elegans (VanGompel et al., 2015), again suggesting an essential and evolutionarily conserved role of Torsin ATPases for NE integrity or dynamics.
While the precise molecular function(s) of Torsins remain poorly understood, diverse functional clues were obtained in animal models or cell-based systems, suggesting four broader categories of Torsin function: molecular chaperone/protein quality control, vesicle/protein trafficking, cellular architecture, and NE vesiculation/trafficking.
Molecular Chaperone/Protein Quality Control
The earliest functional studies of Torsins implicated TorsinA as a molecular chaperone. This would be analogous to the function of the bacterial Clp family of proteins (Sauer and Baker, 2011) to which the Torsins are most similar. Several lines of evidence point towards the role of Torsins as molecular chaperones. First, α-synuclein aggregation in Lewy bodies associated with Parkinson’s Disease was shown to be suppressed by overexpression of TorsinA but not TorsinA ΔE (McLean et al., 2002). Furthermore, it was shown that overexpression of the C. elegans Torsin homolog Tor-2 can decrease the size of polyglutamine-GFP aggregates (Caldwell et al., 2003). However, given that human Torsins reside within the ER and polyQ toxicity arises in the cytosol or nucleus, it is difficult to translate these overexpression findings to mammalian pathophysiology. Additionally, TorsinA was shown to have chaperone activity in vitro in a citrate synthase aggregation assay, though this activity was independent of the presence of ATP or the TorsinA ΔE mutation (Burdette et al., 2010).
Similarly, it has also been suggested that TorsinA functions as a part of the ER-associated protein degradation (ERAD) pathway. Overexpression of wild-type TorsinA increases retrotranslocation and degradation of a mutant cystic fibrosis transmembrane conductance regulator (CFTR F508), while down-regulation of TorsinA or overexpression of TorsinA ΔE had the opposite effect (Nery et al., 2011). It was also shown that expression of wild-type human TorsinA suppressed the induction of the ER stress response, as measured by an increase in expression of GFP under the control of the BiP gene promoter, upon treatment with the classical ER stressor tunicamycin (Chen et al., 2010a). BiP is an hsc70 protein of the ER lumen and is known to be transcriptionally up-regulated by ER stress (Walter and Ron, 2011). Expression of human TorsinA ΔE increased BiP promoter-driven GFP expression in the absence of any other stressor, and knockout of endogenous TorsinA in mouse embryonic fibroblasts (MEFs) resulted in abnormally high steady-state levels of BiP (Chen et al., 2010a). In a follow-up study, knockdown of wild-type TorsinA was also shown to increase the sensitivity of MEFs and cortical neurons to oxidative stress (Chen et al., 2010b). Of note, TorsinA ΔE has an increased dependence on the ER chaperone BiP for proper folding (Zacchi et al., 2014), suggesting that the disease-causing mutation affects Torsin folding and stability.
Vesicle/Protein Trafficking
In addition to these potential roles in protein surveillance and quality control, TorsinA has also been implicated in vesicle/protein trafficking. Overexpression of TorsinA results in ER retention of polytopic membrane proteins normally trafficked through the Golgi to the plasma membrane (Torres et al., 2004). Likewise, secretion of a luciferase-YFP fusion protein is decreased in cells from dystonia patients (Hewett et al., 2007). A number of studies have also investigated the function of TorsinA in processes specific to neurons, since dystonia is primarily a neurological disease. For example, overexpression of wild-type TorsinA inhibits synaptic vesicle recycling via the interaction of TorsinA with SNARE-interacting protein snapin (Granata et al., 2008). TorsinA is also required for the metabolic stability of snapin and the related protein stonin-2 through interaction with the COP9 signalosome (CSN) complex (Granata et al., 2011), a multi-functional protein complex that regulates protein stability through modulation of components of the ubiquitin-proteasome system (Kato and Yoneda-Kato, 2009). Further, a homozygous knock-in of TorsinA ΔE results in increased exocytosis of synaptic vesicles as well as a decrease in size of the total recycling pool of synaptic vesicles (Kakazu et al., 2012a) and changes in glutamate release (Kakazu et al., 2012b).
Cellular Architecture
TorsinA has also been suggested to be important for the maintenance of cellular architecture. This proposition is based on several different lines of evidence. First, TorsinA has been found to interact with the light chain of cytoskeletal motor kinesin (KLC1) (Kamm et al., 2004) as well as nesprin-3 which is part of the linker of nucleoskeleton and cytoskeleton (LINC) complex (Crisp et al., 2006) that bridges the NE and connects the nuclear lamins with cytoskeletal filaments (Nery et al., 2008). Expression of a dominant-negative TorsinB variant induces the appearance of distinct Emerin- and Sun2-positive foci in the cytoplasm (Rose et al., 2014), and manipulation of TorsinA levels and mutation of C. elegans TorsinA homolog OOC-5 alter the localization of LINC components in tissue culture cells and C. elegans, respectively (Nery et al., 2008, Vander Heyden et al., 2009, VanGompel et al., 2015).
Additionally, TorsinA has been found to interact with the intermediate filament vimentin, and expression of TorsinA ΔE interferes with vimentin-dependent processes such as cellular adhesion and neurite extension (Hewett et al., 2006). Whether these interactions are direct or indirect is presently unclear; however, at least some of them must be indirect inasmuch as Torsins are restricted to the ER lumen and cannot access all of those clients directly. Lastly, knockdown of OOC-5 results in failure of nuclear rotation and spindle positioning at the embryonic two-cell stage (Basham and Rose, 1999, Basham and Rose, 2001).
Nuclear Envelope Vesiculation/Trafficking
The observation that knockout of TorsinA in mice results in vesicles in the perinuclear space (Goodchild et al., 2005, Liang et al., 2014) suggests that it may be important for regulating NE membrane dynamics. Recent data suggests that this phenotype is likely related to TorsinA participation in a nuclear transport pathway involving vesiculation of the nuclear envelope (Jokhi et al., 2013, Speese et al., 2012). Herpes Simplex Virus 1 (HSV-1) uses a morphologically similar pathway to exit the nucleus after capsid formation (Mettenleiter et al., 2009) and therefore can be used as a model system for studying transport via NE vesiculation. Overexpression of wild-type TorsinA inhibits replication of HSV-1, and the virus becomes trapped within the ER (Maric et al., 2011). This may occur due to the interruption of the established HSV-1 nuclear egress pathway (Mettenleiter et al., 2009). Furthermore, the D. melanogaster Torsin homolog was implicated in the newly discovered process of nuclear ribonucleoprotein (RNP) export via an NE budding mechanism (Jokhi et al., 2013, Speese et al., 2012). It has also been suggested that this Torsin-dependent membrane budding process could be involved in protein quality control by exporting misfolded protein species (Rose and Schlieker, 2012). Recently, a mutation in the gene for the C. elegans TorsinA homolog OOC-5 has been shown to result in mislocalization of nuclear pore complex components and impaired nuclear import kinetics (VanGompel et al., 2015). Given that perinuclear ubiquitin accumulates in TorA-deficient neurons within INM blebs, based on immunogold labeling (Liang et al., 2014), and the recent findings pertaining to the involvement of ESCRT (endosomal sorting complexes required for transport) machinery known to employ ubiquitin machinery (McCullough et al., 2013) in NE fusion (Olmos et al., 2015, Vietri et al., 2015) and NPC biogenesis (Webster et al., 2014), it is tempting to speculate that these phenomena may be functionally connected.
To conclude, while there is a significant amount of genetic data available, we still lack a definitive understanding of Torsin function(s). It does however provide some insight into the question of why TorsinA hypofunction results much more strongly affects neuronal cells, given that TorsinA expression is highest in those cells and cannot be readily compensated for by other Torsins (Kim et al., 2010). There have been a myriad of possible functions suggested for TorsinA, and it is possible that TorsinA is involved in several of these pathways. However, the fact that TorsinA resides entirely within the ER and contiguous perinuclear space would seem to require the use of membrane-spanning adaptor proteins in order to be involved in processes outside of the ER.
Other members of the Torsin family
Considerably less is known about TorsinB than TorsinA, despite the fact that TorsinB and TorsinA are 68% identical and 85% similar at the primary sequence level, and the fact that the TOR1B gene is immediately adjacent to TOR1A on chromosome 9q34 (Ozelius et al., 1999). Like TorsinA, TorsinB is a glycoprotein with two glycosylation sites (Hewett et al., 2004, O’Farrell et al., 2004) and is ubiquitously expressed in all cell types. However, TorsinB has not been found to be associated with any disease and it is much less abundant than TorsinA in neuronal tissues (Jungwirth et al., 2010, Kim et al., 2010). Interestingly, depletion of TorsinB in TorsinA-deficient MEFs results in the expansion of the otherwise neuronally restricted NE blebbing phenotype to non-neuronal cells (Kim et al., 2010). Additionally, overexpression of TorsinB or TorsinA in wild-type or TorsinA-deficient cells results in an increase in NE blebbing (Kim et al., 2010, Rose et al., 2014). This suggests that TorsinA and TorsinB may be functionally redundant, with TorsinA being more important in neuronal cells and TorsinB being more important in other tissues (Kim et al., 2010).
Despite these similarities, differences have been noted that suggest potentially unique roles for these proteins. First, TorsinB is significantly more basic than TorsinA, with a pI of 8.6 compared to TorsinA’s pI of 6.8 (O'Farrell et al., 2004), which could affect their cellular interaction partners. Also, in mouse neuronal tissue, TorsinB mRNA expression levels remain constant after P14, while TorsinA mRNA expression levels decrease, suggesting that TorsinA may play a more important role in early developmental stages (Vasudevan et al., 2006). Additionally, it has been noted that TorsinB has a tendency to form inclusions when overexpressed, while these inclusions are not seen with a similar level of TorsinA overexpression (Hewett et al., 2004, O'Farrell et al., 2004, Rose et al., 2014). Recently, it has been shown that overexpression of the Walker B mutant of TorsinB (TorsinB E178Q) results in sinusoidal organized smooth ER (OSER) structures (Rose et al., 2014). This suggests that TorsinB may be involved in regulating ER membrane morphology, but more conclusive experiments are needed. Additionally, overexpression of TorsinB in dystonia patient (DYT1) cells cannot rescue secretion of a Gaussia luciferase reporter, whereas TorsinA can rescue this phenotype. This indicates that TorsinB expression cannot complement the non-functional mutant TorsinA protein while wild-type TorsinA can (Hewett et al., 2008). Together, these data suggest that TorsinA and TorsinB have overlapping yet non-identical functions.
In Blue Native Polyacrylamide Gel Electrophoresis (BN-PAGE) analysis of cell extracts, TorsinB forms a higher-order assembly consistent with a stereotypical AAA+ hexamer (Jungwirth et al., 2010). However, it is unclear whether this higher-order species is a homo-oligomer of TorsinB or a hetero-oligomer of different Torsin proteins given that TorsinB is able to associate with both wild-type TorsinA and TorsinA ΔE (Naismith et al., 2009, Hewett et al., 2004, Goodchild and Dauer, 2004) and TorsinB and TorsinA have a similar molecular mass.
With only 321 amino acids, Torsin2A is the smallest of the Torsin proteins, and while it contains an N-terminal signal sequence, it lacks the hydrophobic domain common to both TorsinA and TorsinB (Figure 2A). Like the other Torsins, Torsin2A localizes to the ER, but whether the Walker B mutation E162Q causes Torsin2A enrichment in the NE is unclear, with groups reporting conflicting results (Jungwirth et al., 2010, Kim et al., 2010).
Torsin3A is an outlier among the other Torsin ATPases for several reasons. First, Torsin3A was originally discovered in a screen for genes upregulated by interferon treatment, where it was named ADIR (ATP-dependent interferon responsive) (Dron et al., 2002). No other Torsin proteins have been reported to respond to interferon treatment. Additionally, Torsin3A has almost no nucleotide sequence homology to other Torsins, though it does have strong protein sequence homology (Dron et al., 2002). It is also the only Torsin not located at the chromosome 9q34 locus, instead being located on chromosome 1q35. Lastly, Torsin3A runs as a monomer in BN-PAGE, indicating that it does not form the higher-order structures characteristic of the AAA+ superfamily including TorsinA, TorsinB, and Torsin2A (Jungwirth et al., 2010). Despite not appearing to form higher-order structures, Torsin3A has been found to co-immunoprecipitate with TorsinA (Naismith et al., 2009).
Torsin4A is a newly annotated member of the Torsin family (Figure 2A) that is predicted to have a full transmembrane domain in the middle of the protein while TorsinA and TorsinB only have an N-terminal hydrophobic domain. Also, unlike the other Torsins, Torsin4A has no signal sequence or predicted glycosylation sites (UniProt). While there is no published work specifically studying this protein, it has been identified in various screens under the generic name C9orf167. In a screen for mitotic defects via live-cell imaging, a small interfering RNA (siRNA) knockdown of Torsin4A resulted in poly-lobed nuclei with chromosomal segregation defects and a mitotic delay, which sometimes resulted in cell death (Neumann et al., 2010). Additionally, Torsin4A mRNA was found to be upregulated by overexpression of Krüppel-like Factor 9 (KLF9) (Simmen et al., 2008). Currently, the importance of these findings remains unclear and considerable work still remains to be done to characterize this newest Torsin family member.
Molecular properties of the Torsins and their membrane-spanning cofactors
TorsinA is a 332 amino acid protein that resides within the lumen of the ER and possesses an N-terminal hydrophobic domain. This hydrophobic domain anchors TorsinA to the luminal leaflet of the ER membrane via monotopic insertion and is required for TorsinA retention within the ER (Vander Heyden et al., 2011). Static ER retention, as opposed to cycling through the ER and Golgi, is also indicated by TorsinA glycans’ persistent sensitivity to Endoglycosidase H (Hewett et al., 2003). Thus Torsins are part of a very small group of ATPases residing in the ER aside from the Hsp70 chaperone BiP and Lhs1 (Osborne et al., 2005, Buck et al., 2013). Another distinctive feature of Torsins are the six conserved cysteine residues that are common to all members of the Torsin family but not found in closely related Clp/Hsp100 proteins. Of note, a subset of these residues have been implicated in redox sensing and regulation of nucleotide affinity (Zhu et al., 2010, Zhu et al., 2008).
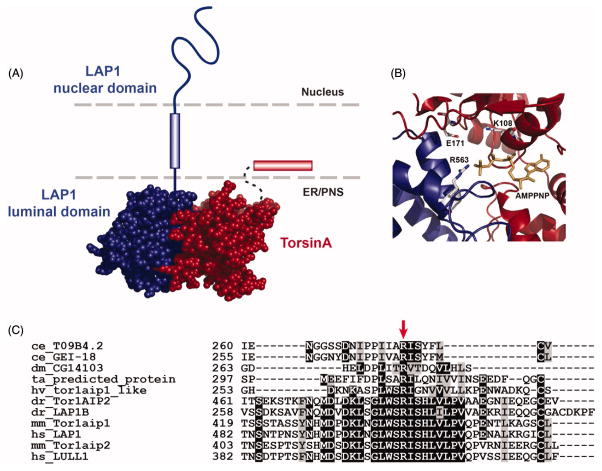
Given that Torsins are most closely related to the Clp/HSP100 family of AAA+ ATPases (Figure 3), it was widely assumed that Torsins form obligatory homohexameric assemblies. Indeed, oligomeric species consistent with the electrophoretic mobility of a homohexamer were observed via BN-PAGE (Jungwirth et al., 2010, Vander Heyden et al., 2009, Li et al., 2014). More recently, it is becoming clear that this view of Torsins is incomplete since the molecular assembly is not that of a stable, obligatory homohexamer, but instead includes LAP1 (Lamina-associated polypeptide 1) and LULL1 (Luminal domain like LAP1), two membrane-spanning cofactors (see below). These interaction partners were first discovered by the Dauer laboratory in a candidate approach to identify factors regulating Torsin localization: overexpression of LAP1 leads to an increase in NE staining of TorsinA (Goodchild and Dauer, 2005). LAP1, also known as TOR1AIP1 (Torsin-1A interacting protein 1), is a widely expressed (Goodchild and Dauer, 2005) type-II transmembrane protein that localizes to the INM due to its association with the nuclear lamina (Senior and Gerace, 1988, Foisner and Gerace, 1993). It contains two distinct domains, an N-terminal lamin-binding domain and a C-terminal domain located within the perinuclear space/ER lumen. LAP1 is present in three splicing variants in rats (LAP1A, B, and C) (Foisner and Gerace, 1993, Senior and Gerace, 1988), and two confirmed isoforms in humans (Santos et al., 2014) that differ only in their N-terminal nuclear domain. LAP1 and TorsinA interact directly through LAP1's luminal domain in vitro (Zhao et al., 2013), which is shared by all isoforms, although most work with LAP1 in human cells has focused on LAP1B.
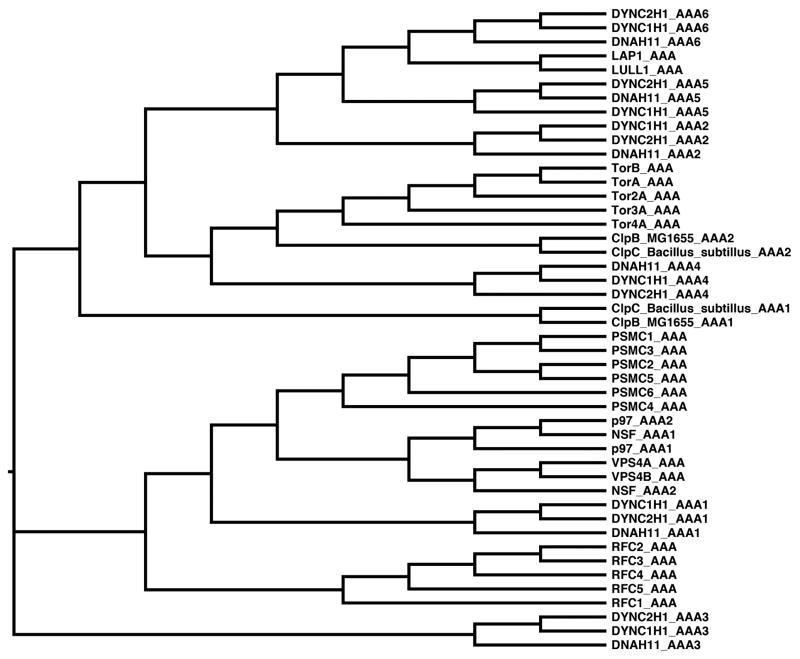
FIGURE 3.
Phylogenetic comparison of AAA domains from various AAA ATPases. Phylogenetic tree built by MetaPIGA (Helaers & Milinkovitch 2010). Input sequences were individual AAA domains from each protein as determined by NCBI Conserved Domain Database (Marchler-Bauer et al. 2011), UniProt (Dyneins), or HHpred (LAP1 and LULL1) (Soding et al. 2005). All sequences used represent the human protein unless otherwise stated.
A LAP1 knockout leads to perinatal lethality in mice (Kim et al., 2010), and cells from multiple tissues in these knockout mice display a NE blebbing phenotype. However, unlike TorsinA knockouts, the LAP1 knockout blebbing phenotype is not restricted to neuronal tissues (Kim et al., 2010). In an independent study, LAP1 has been found to be important for mitotic spindle assembly because a knockdown of LAP1 results in failure of spindle formation in prometaphase that eventually leads to aberrant mitotic exit and cell death (Neumann et al., 2010).
LAP1 has also been linked to disease. LAP1B has recently been shown to associate (via its nuclear domain) with Emerin, the protein mutated in Emery-Dreifuss Muscular Dystrophy (EDMD) (Shin et al., 2013). Also, in approximately 30% of LAP1 knockout mouse embryonic fibroblasts, both Emerin and LaminA are mislocalized. Conversely, Emerin deletion does not appear to affect LAP1 localization. Interestingly, Emerin deletion alone does not cause myopathy in mice, but a conditional knockout of LAP1 in an Emerin knockout background recapitulates EDMD symptoms, indicating that LAP1 is able to compensate for an Emerin knockout in muscle tissue (Shin et al., 2013). A different form of muscular dystrophy has also been associated with a deletion of guanine 186 of the LAP1 coding sequence, which is a mutation that causes a frame-shift resulting in a premature stop codon (Kayman-Kurekci et al., 2014). Lastly, a LAP1 E482A mutation has been connected to a phenotype of severe dystonia, cardiomyopathy, and cerebellar atrophy in a patient (Dorboz et al., 2014). However, as with TorsinA, the mechanistic connection between LAP1 and these pathologies awaits clarification.
LULL1 (also called TOR1AIP2: Torsin-1A Interacting Protein 2) was first identified by a bioinformatic search for proteins with a luminal domain similar to that of LAP1 (Goodchild and Dauer, 2005). Like LAP1, LULL1 is a widely expressed (Goodchild and Dauer, 2005), type-II integral membrane protein with a luminal domain that engages Torsins (Zhao et al., 2013). However, unlike LAP1, LULL1 resides in the ER with its N-terminal domain protruding into the cytoplasm, and there is considerably less LULL1 present in mouse embryonic fibroblasts than LAP1 (Goodchild and Dauer, 2005), though studies in human cell lines show approximately equal levels (Brown et al., 2014).
Overexpression of LULL1 leads to a redistribution of TorsinA WT from the ER to the NE, but Walker A (K108A) and Walker B (E171Q) mutants as well as TorsinA lacking its hydrophobic domain are not affected (Vander Heyden et al., 2009, Goodchild et al., 2015). This suggests that LULL1 may play a role in regulating the subcellular distribution of TorsinA. Consistent with its ER localization, depletion of LULL1 via siRNA, unlike depletion of LAP1, does not cause an NE blebbing phenotype (Vander Heyden et al., 2009). However, LULL1 depletion does decrease the formation of ER-derived sinusoidal membrane structures upon TorsinB overexpression (Rose et al., 2014), indicating a potential role for LULL1 in ER morphology. Recently, it has been found that LULL1 knockout in HeLa cells decreases the formation of infectious HSV-1 particles by an order of magnitude, while a LAP1B knockout has no effect (Turner et al., 2015). This again suggests that these proteins have different functions, which is consistent with their distinct subcellular localization.
Evolutionary conservation of Torsins and associated proteins
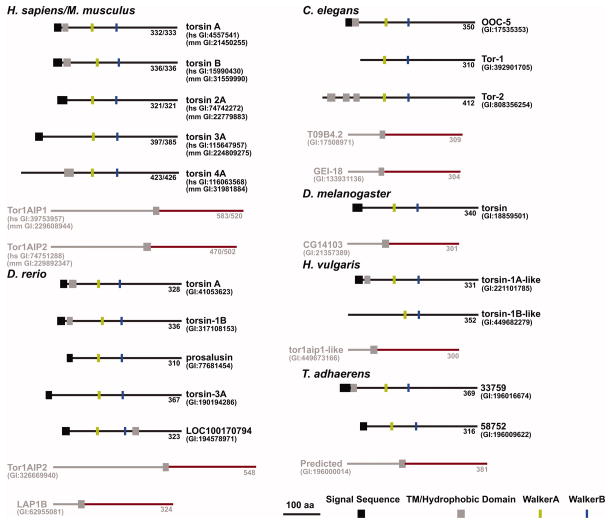
Torsins and LAP1/LULL1 homologs are well conserved in metazoans, with most organisms having multiple paralogs (Figure 4). However, no apparent homologs are present in yeast or any other single-celled organism according to HHpred or PSI-BLAST. Notably, Trichoplax adhaerens, among the simplest of extant metazoans, features two Torsin genes in its genome, while D. melanogaster appears to possess the simplest system with only one predicted paralog (Figure 4), a possible advantage for genetic approaches towards dissecting Torsin function relative to other organisms.
FIGURE 4.
Evolutionary conservation of Torsin and LAP1/LULL1. Graphical representation of the primary sequence characteristics of Torsin (black) and LAP1/LULL1 (gray/red) homologs from various species. Red portions of cofactor homologs denotes domains predicted to have a AAA-fold.
Many of the Torsin homologs include the N-terminal hydrophobic domain found in human TorsinA and TorsinB, and all are of a relatively similar size. The fact that these features are so well conserved suggests that they may be important to Torsin’s biolological function. Also, all organisms that have at least one Torsin protein also have at least one homolog of LAP1/LULL1 (Figure 4), suggesting the general importance of the regulation of Torsin ATPase activity by these cofactors (see below). This makes it important to fully understand the functional interplay between Torsins and LAP1/LULL1.
In the following section, we will outline how in vitro reconstitution and biochemical analysis have revised our understanding of the how Torsins and LAP1/LULL1 function together.
Regulation of Torsins by LAP1 and LULL1
LAP1 and LULL1 preferentially bind to TorsinA in its ATP-bound state (Goodchild and Dauer, 2005, Naismith et al., 2009, Zhao et al., 2013, Zhu et al., 2010), raising the question of whether these proteins are substrates or cofactors of TorsinA. Given that the TorsinA disease mutant TorsinA ΔE results in a loss of binding to LAP1 and LULL1 (Naismith et al., 2009, Zhao et al., 2013, Zhu et al., 2010), it is evident that this defect may contribute to disease pathology.
Unlike many related AAA+ ATPases, purified TorsinA by itself is unable to efficiently hydrolyze ATP provided that adequate negative controls are included to eliminate the contribution of contaminating ATPases (Zhao et al., 2013). Induction of a latent TorsinA ATPase activity strictly requires binding of either LAP1 or LULL1, with LULL1 being the more potent activator (Zhao et al., 2013). Specifically, LAP1 and LULL1 activate TorsinA’s ATPase activity by accelerating the hydrolysis step by up to two orders of magnitude (Zhao et al., 2013), arguing against a role as canonical substrates for global unfolding, which stimulate ATPase activity of related ATPases only moderately (Schlieker et al., 2004, Cashikar et al., 2002) (Burton et al., 2001). Consistent with this, TorsinA does not appear to cause unfolding of either LAP1 or LULL1 (Zhao et al., 2013). Interestingly, the LAP1/LULL1-mediated activation mechanism is conserved for TorsinB, while Torsin3A is only activated by LULL1, and no stimulation was observed for Torsin2A by either cofactor (Zhao et al., 2013).
Since activator binding is required to stimulate the ATPase activity of TorsinA, and the ΔE variant is unresponsive to either cofactor and is consequently catalytically inactive (Zhao et al., 2013), it seems reasonable to propose that this activation defect provides the molecular basis of DYT1 pathology (Brown et al., 2014, Zhao et al., 2013). These biochemical findings are in excellent agreement with a number of observations in animal models: First, they can explain the inability of a TorA ΔE knock-in to rescue the phenotype of a TorA −/− mouse model (Goodchild et al., 2005). Second, since two Torsins are activated by LAP1, these findings additionally explain why a LAP1 knockout provokes a more penetrant phenotype than a TorsinA knockout in isolation (Kim et al., 2010).
Molecular mechanism of Torsin activation
Two recent studies have provided convincing evidence for the role of LAP1 and LULL1 as cofactors by showing that the luminal domains of these proteins adopt AAA-like structures (Sosa et al., 2014, Brown et al., 2014). Structural homology searches suggested that the LAP1 and LULL1 luminal domains adopt AAA-like folds (Brown et al., 2014), a conclusion verified by concurrent crystallographic studies of LAP1’s luminal domain (Sosa et al., 2014). LAP1 displays a typical α/β domain (RecA fold) (Story et al., 1992) core with a five-stranded parallel β-sheet surrounded by ten α-helices (Sosa et al., 2014), but lacks the C-terminal α-subdomain common to AAA+ proteins (Figure 5A). LAP1’s C-terminus instead folds back towards the core of the protein to enable the formation of a disulfide bond between conserved cysteine residues C582 and C424 (Sosa et al., 2014). This disulfide could potentially serve as a redox switch for the Torsin/cofactor complex. In fact, it has been previously shown that mutation of cysteines in TorsinA affects both nucleotide and cofactor binding (Zhu et al., 2008, Zhu et al., 2010). LAP1 forms a complex with TorsinA in vitro (Brown et al., 2014, Zhao et al., 2013) (Figure 5A), but in vivo conditions (or crosslinking in vitro) are required to observe higher-order oligomers (Jungwirth et al., 2010, Vander Heyden et al., 2009, Li et al., 2014, Brown et al., 2014, Goodchild et al., 2015). The precise composition of these oligomers remains poorly understood. LAP1 and LULL1 lack residues required for nucleotide binding and hydrolysis. The structural similarity of these cofactors to AAA+ domains immediately suggested that Torsins and these proteins could potentially assemble into a mixed ring structure, in which LAP1 or LULL1 assume a position equivalent to a regular AAA+ subunit (Figure 5B) (Sosa et al., 2014, Brown et al., 2014).
FIGURE 5.
Crystal structure of LAP1’s luminal domain and its potential impact on the Torsin ATPase oligomeric assembly. (A) Left Cartoon representation of a single NSF AAA domain (D2) from Cricetulus griseus (PDB ID: 1D2N (Lenzen et al. 1998)) showing the alpha/beta Rossman/RecA fold with the C-terminal four helix bundle. Right Cartoon representation of LAP1’s luminal domain from Homo sapiens (PDB ID:4TVS (Sosa et al. 2014)). LAP1’s luminal domain adopts a AAA-like fold without the C-terminal helical bundle. (B) Left TorsinA’s proposed homohexameric ring assembly. Each Torsin monomer’s nucleotide binding domain is depicted as a wedge where each monomer’s C-terminal helical bundle is depicted as a ball (red). Right Proposed heterohexameric ring assembly of TorsinA (red) with cofactor LAP1/LULL1 (blue). LAP1/LULL1 lack C-terminal helical bundles, which may prevent contact between adjacent subunits at the non-activating interfaces (arrows). (C) TorsinA (red ball) may form a homohexameric ring that, upon binding LAP1/LULL1 (blue ball), could cause the ATPase to adopt a split lock-washer assembly since LAP1/LULL1 lack the C-terminal helical bundle. Different nucleotide states could impact opening/closing of the split ring, possibly enabling a gating-mechanism to access the central pore of the ring. Membrane, gray dashed lines; LAP1/LULL1’s transmembrane domain, blue rectangle; LAP1/LULL1 nuclear/cytoplasmic domain, blue curved line.
Importantly, both LAP1 and LULL1 contain a conserved arginine residue (R563 and R449, respectively) that is found at a position equivalent to the canonical arginine finger in many AAA+ ATPases (Sosa et al., 2014, Brown et al., 2014). As mentioned above, Torsins notably lack conservation of the equivalent arginine residue, explaining why Torsin ATPases are catalytically defective in isolation (Zhao et al., 2013, Brown et al., 2014, Sosa et al., 2014). However, this “deficient” active site can be “rescued” by the addition of LAP1 or LULL1, which can contribute their key arginine residue into Torsin’s ATP binding pocket to facilitate nucleotide hydrolysis via an active-site complementation mechanism (Figure 6A, B). This arginine in both cofactors is indeed required for induction of ATPase activity (Sosa et al., 2014, Brown et al., 2014) and is strictly conserved among all of the LAP1/LULL1 homologs identified in other organisms (Figure 6C). High-resolution structures of LULL1 and LAP1 in complex with Torsin bound to an ATP transition-state analog will be needed to prove that these cofactors do indeed contribute bona fide “arginine fingers.”
FIGURE 6.
Torsin activation by a strictly conserved arginine. (A) Surface representation of the heterodimeric model of LAP1 (PDB ID: 4TVS (Sosa et al. 2014)) and TorsinA (Phyre2 model (Brown et al. 2014)). The membrane is represented as dashed lines, membrane helices are represented as rectangular cylinders, and LAP1’s nuclear domain is shown as a curved line. (B) Zoomed in view of the active site showing the Walker A (K108) and Walker B (E171) residues of Torsin and the arginine (R563) provided by LAP1. AMPNP: gold (C) Sequence alignment (MUSCLE (Edgar 2004); colored by boxshade) of cofactor homologs diagrammed in Figure 4 showing the location of the strictly conserved arginine residue (arrow).
A similar active-site complementation mechanism applies to a small subset of other AAA+ proteins as well. The bacterial clamp loader contains three γ subunits that bind and hydrolyze ATP, while the δ and δ’ subunits are degenerate AAA domains that do not engage ATP (Jeruzalmi et al., 2001, Hedglin et al., 2013). However, the δ’ subunit uses an arginine finger to simulate ATP hydrolysis by its neighboring γ subunit (Johnson and O'Donnell, 2003). The presence of degenerate AAA+ domains extends to the Dynein ATPase family as well. Dynein is composed of six covalently linked AAA+ domains of which only four AAA+ domains can bind and/or hydrolyze ATP (AAA1–4) while the remaining two domains (AAA5–6) provide mainly structural roles for the ATPase machine (Cho and Vale, 2012). Interestingly, the AAA+ domains of LAP1 and LULL1 cluster near these degenerated AAA scaffolds in a phylogenetic tree (Figure 3). Since neither the clamp loader nor dynein operate via a threading mechanism, there is precedent for believing that Torsin action does not necessarily involve a threading activity, although this remains a formal possibility. Given the membrane-spanning nature of the Torsin-cofactor assembly, this activity would likely require a pore-forming component in the membrane. It is challenging to imagine such a proteinaceous pore that would not sterically clash with the membrane domains of the Torsins or their activators, unless the transmembrane domains themselves engage in pore formation.
Functional implications of the active site complementation mechanism
These recent structural and biochemical insights profoundly change our molecular view of the Torsin machinery (McCullough and Sundquist, 2014), and have important ramifications for Torsin regulation and function. As transmembrane proteins that are part of the core Torsin machinery, LAP1 and LULL1 could act as mechanical elements to transduce forces across the membrane (Brown et al., 2014). Additionally, LAP1 and LULL1 may themselves be subject to regulatory events (e.g. posttranslational modifications) or be controlled by association of cytosolic/nuclear interaction partners (Brown et al., 2014). LAP1 and LULL1 are known to be phosphorylated and can be modified by ubiquitin-like modifiers SUMO-1 and FAT10, respectively (Dephoure et al., 2008, Santos et al., 2014, Wang et al., 2008, Buchsbaum et al., 2012). Binding of interaction partners to LAP1 and LULL1 or their modification could induce conformational changes that propagate across the membrane to regulate Torsin activity on the luminal side (Brown et al., 2014). Indeed, recent evidence suggests that the cytosolic domain of LULL1 oligomerizes and that its oligomerization state affects the stoichiometry of the overall assembly as well as its subcellular localization (Goodchild et al., 2015). Therefore, only a thorough characterization of full-length components in the natural context of the membrane (e.g. by cryo-electron microscopy) will reveal the true range of structural states that the active complex may occupy, especially since interactions within the plane of the membrane may well be important to stabilize the assembly or constrain its stoichiometry. This structural information, in turn, will be crucial for models of substrate processing by the active assembly. Finally, the revelation that LAP1 and LULL1 form integral components of active Torsin complexes suggests that one must be cautious in interpreting studies relying on overexpression of Torsins in the absence of equal overexpression of these cofactors.
Although it is now clear that the LAP1 and LULL1 luminal domains adopt AAA-like folds, much remains to be learned about their nuclear and cytosolic domains, respectively. These domains are predicted to be mainly unstructured, although LULL1’s N-terminal domain has detectable structural homology to SNARE (soluble NSF attachment protein receptor) proteins Syntaxin1a and VAMP2 (vesicle-associated membrane protein 2) (Brown et al., 2014). Vesicle and target SNARE proteins, which also contain unstructured regions, “zipper” into a heteroligomeric assembly, bringing their respective biological membranes into close juxtaposition for fusion (Sudhof and Rothman, 2009). Therefore, this homology in LULL1 could be indicative of a function in membrane fusion or tethering (see below). Further investigations into the biochemical and structural properties of the nuclear/cytoplasmic domains will clearly be needed for a mechanistic understanding of how the Torsin machinery works.
Another question that still remains unanswered concerns the precise biological assembly of the Torsin ATPase machinery. Torsin:cofactor heterodimers (1:1) are stable in vitro only in the context of a Walker B mutation; they can form higher-order oligomeric ring assemblies that could potentially represent mixed rings consisting of a trimer of heterodimers (cf. Figure 5B, right panel) (Sosa et al., 2014, Brown et al., 2014). One could envision that a maximally efficient Torsin ATPase machine would consist of an alternating Torsin:cofactor assembly, such that every Torsin active site is matched with a cofactor arginine finger. However, this may not be the most abundant assembly in the cell given that other factors, such as local concentrations of proteins and the presence of competing interacting partners, could affect the assembly. Since Torsins can interact with one another in addition to the cofactor proteins, one can imagine a more dynamic assembly of mixed Torsin:cofactor rings in which an ensemble of different stoichiometric states can be populated at varying frequency. In this situation, multiple cellular functions of the ATPase machine could be achieved depending on which Torsin(s) and which cofactor(s) are present and at what relative levels. Indeed, Torsin-Torsin contacts incompatible with a strictly alternating, symmetric assembly of cofactor and Torsin subunits were observed in site-specific crosslinking experiments even when Torsins and cofactors were present in equimolar ratios, proving that assemblies of varying composition were forming (Brown et al., 2014). Further complicating the picture, several distinct Torsins may integrate into a higher-order assembly.
One appealing possibility is that LAP1 and LULL1 could serve to break the symmetry of a homomeric Torsin ring (Figure 5C). LAP1 and LULL1 lack the helical subdomain common to the AAA+ fold (Figure 5A), which is essential for inter-protomer contacts in related AAA+ ATPases (Mogk et al., 2003). Therefore, insertion of a cofactor into a Torsin assembly would likely result in oligomer destabilization due to loss of at least one such C-terminal four-helix bundle in the assembly (cf. Figure 1). The absence of this element may explain why a Torsin-LAP1 dimer is the only stable species observed by size-exclusion chromatography (Brown et al., 2014, Sosa et al., 2014). This destabilization of the ring structure could result in the opening of a lateral gate in the assembly similar to the “open washer” state observed for NSF (N-ethylmaleimide sensitive factor) (Zhao et al., 2015) or the clamp loader (O'Donnell and Kuriyan, 2006). Given that the Torsin/cofactor complex is membrane associated, opening of a lateral gate in the ring could allow for the diffusion of a transmembrane protein into the central pore of the ring where Torsin ATPase activity could act on it, or trigger the release of a substrate from the ring.
Functional Perspectives
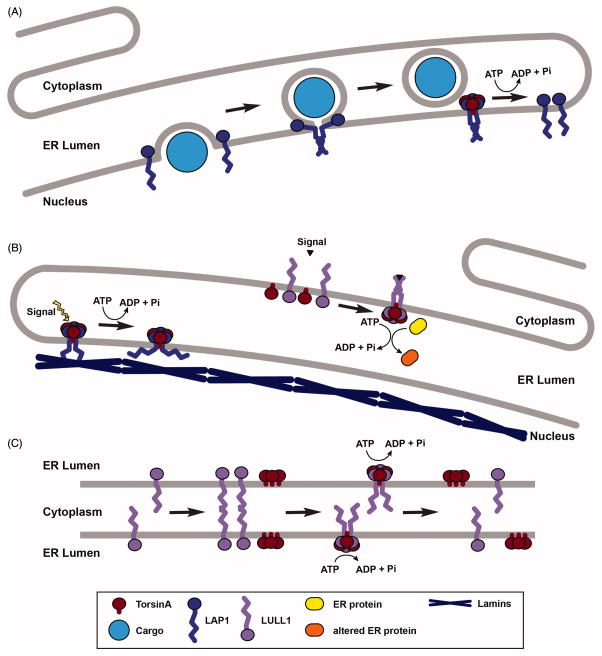
Despite the wealth of genetic and biochemical data on Torsins, their molecular function(s) remain unclear. However, many recent reports seem to support a role for Torsins in INM/ER dynamics. Jokhi and colleagues found that the D. melanogaster Torsin homolog is involved in the nuclear egress pathway of megaribonucleoprotein (megaRNP) granules (Jokhi et al., 2013). This pathway involves the vesicular budding of granules from the INM into the perinuclear space where the vesicles fuse with the outer nuclear membrane (ONM) and release the granule into the cytoplasm (Speese et al., 2012). When Torsin is depleted in D. melanogaster muscle cells, rows of single granules remain tethered to the INM within the perinuclear space. A Walker B mutant of Torsin tagged with mini-SOG (singlet oxygen generator) can be visualized as an electron-dense precipitate that localizes to the necks of the granule buds (Jokhi et al., 2013). These data suggest that Torsin may be involved in the membrane scission reaction that releases the buds from their attachment to the INM (Jokhi et al., 2013). These data would seem to be in excellent agreement with numerous studies implicating Torsins in processes that affect NE or ER morphology as described above.
With the new knowledge that Torsin and its cofactors LAP1 and LULL1 form heteromeric assemblies, it is possible to propose models for how the Torsin machinery could be involved in the process of NE budding. One possibility is that Torsin is involved in the membrane fission process, as is implied by the finding that D. melanogaster Torsin localizes to the bud necks of perinuclear blebs (Jokhi et al., 2013). The Torsin/LAP1 or Torsin/LULL1 machine could achieve membrane fission by the zippering of the cofactor’s nuclear or cytoplasmic domain, which is predicted to form a coiled coil (Brown et al., 2014). As with SNARE proteins, parallel multimerization of the LAP1 nuclear domain could bring two segments of the INM close together, which in this case would result in fission of the INM and the release of a vesicle into the perinuclear space. Torsin could then hydrolyze ATP to disassemble the LAP1 oligomer for reuse, similar to NSF-mediated recycling of SNAREs (Figure 7A). Alternatively, an as yet unknown protein may be responsible for the actual membrane fusion step of the reaction, but Torsin may still be required for their disassembly.
FIGURE 7.
Potential biological functions of the Torsin/cofactor assembly. (A) Model of nuclear membrane fission mediated by LAP1, with subsequent disassembly and recycling of LAP1 oligomers by Torsin ATPase activity. (B) Model of Torsin and cofactor-mediated transmembrane signaling between the ER and cytoplasm (LULL1) or nucleoplasm (LAP1). (C) Model of cytoplasmic membrane tethering by LULL1’s nuclear domain that is disassembled by Torsin ATPase activity.
An alternative role for the Torsin/cofactor complex may be as a mediator of transmembrane signaling between the ER lumen and the cytoplasm and/or nucleoplasm. Torsins and the cofactor luminal domains could respond to a signal within the ER lumen resulting in their association with each other and initiation of ATP hydrolysis (Figure 7B). The potential conformational changes associated with ATP hydrolysis could generate a mechanical force that is transduced through LAP1 and LULL1’s transmembrane domains to their nuclear/cytoplasmic domains, acting as mechanical elements to propagate force from one domain to another. This force could serve to disrupt binding between the nuclear/cytoplasmic domains and other proteins, such as LAP1 and the nuclear lamina. This transmembrane signaling could also potentially work in the opposite direction. Nuclear or cytoplasmic domains of LAP1 and LULL1 can be phosphorylated and modified by ubiquitin-like modifiers SUMO-1 and FAT10, respectively (Dephoure et al., 2008, Santos et al., 2014, Wang et al., 2008, Buchsbaum et al., 2012), either of which could serve as an initial cytoplasmic signal to be transduced to the luminal domains and initiate ATP hydrolysis (Figure 7B). This potential transmembrane signaling function could explain why TorsinA has been unexpectedly linked to a number of cytoplasmic proteins (see above).
Another putative function for Torsin ATPases could be to disassemble oligomers of LULL1 that serve to tether membranes via their cytoplasmic faces. LULL1’s cytoplasmic domain has been shown to oligomerize (Goodchild et al., 2015). Additionally, overexpression of the Walker B mutant of TorsinB results in sinusoidal membrane structures in which ER membranes are held tightly apposed via their cytoplasmic faces, and these structures are dependent on the presence of LULL1 (Rose et al., 2014). These pieces of data are in agreement with the idea that LULL1’s cytoplasmic domain can interact with other LULL1 molecules on opposing membranes to tether the two membranes together. Torsin could then bind these LULL1 oligomers from the luminal side and use its ATPase activity to disassemble the tethered state (Figure 7C). This membrane tethering activity would be loosely reminiscent of SNAREs to which LULL1 has been predicted to have structural homology (Brown et al., 2014), and also of Atlastin GTPases (Hu et al., 2011).
It must be noted, however, that while the potential mechanistic functions described above are consistent with recent data, they remain purely speculative and are mentioned solely to define working models for experimental attack. In this context, it is noteworthy that some AAA+ ATPases have multiple functions and this may be true of Torsins as well. p97, for example, has a vast number of cellular functions ranging from protein quality control to membrane fusion and genome stability, and these functions are determined by specific adaptors (Meyer et al., 2012). Torsins could have several cellular functions that are similarly dictated by its interaction partners and environment. LAP1 and LULL1 most likely specify different functions of the ATPase machine due to their different subcellular localizations and dissimilar N-terminal domains. To help define Torsin function, it will be critical to identify any potential interaction partners of the cofactors’ N-terminal domains, as well as additional interaction partners for Torsin itself.
Rationalizing disease-causing mutations
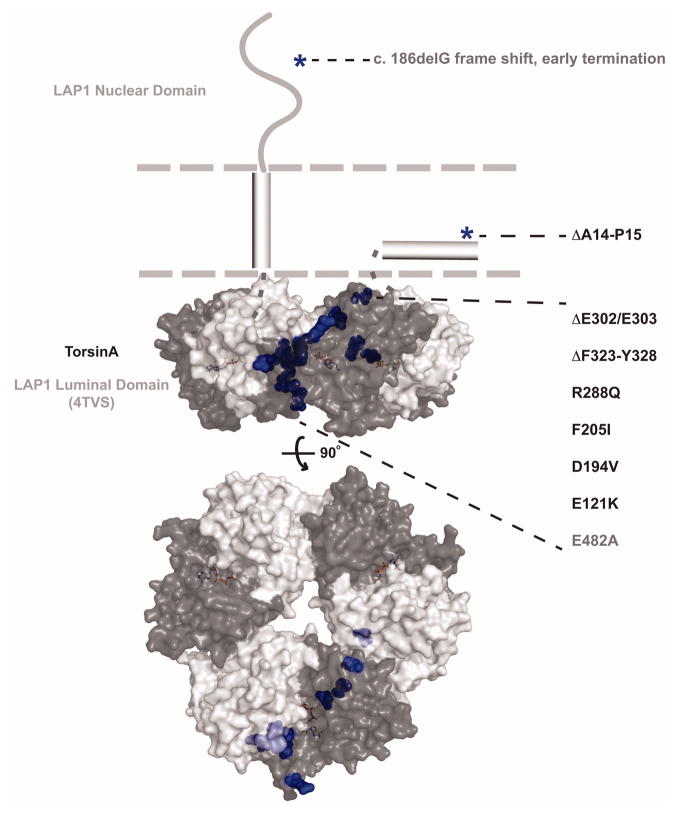
Despite our lack of functional understanding, the recent advances in our knowledge of LAP1/LULL1 structure and Torsin/cofactor assembly enhance our understanding of how mutations in the Torsin machinery could perturb its function (Figure 8). The TorsinA ΔE302/303 mutation is the most common Torsin mutation associated with disease phenotypes. However, other mutations in TorsinA as well as in the cofactor LAP1 have also been associated with disease in the absence of the TorsinA ΔE302/303 mutation (Table 1). TorsinA mutations ΔA14-P15, E121K, D194V, F205I, R288Q, and ΔF323-Y328 have all been linked to specific cases of dystonia (Vulinovic et al., 2014, Cheng et al., 2014, Calakos et al., 2010, Zirn et al., 2008, Leung et al., 2001), while a LAP1 E482A mutation has been associated with severe dystonia, cerebellar atrophy, and cardiomyopathy (Dorboz et al., 2014), and a LAP1 truncation results in muscular dystrophy (Kayman-Kurekci et al., 2014).
FIGURE 8.
Physical locations of disease mutations in TorsinA and LAP1. Semi-transparent surface representation of the heterohexameric model of LAP1 (PDB ID: 4TVS (Sosa et al. 2014)) and TorsinA (Phyre2 model (Brown et al. 2014)) bound to AMPPNP using a least-squares superposition of alpha-carbons (Coot) onto hexameric p97 (PDB ID: 3CF2 (Davies et al. 2008)). LAP1 is depicted in light gray, TorsinA in dark gray, and disease causing mutations in blue. Mutations that do not map to the luminal domains of these proteins are highlighted with blue asterisks. The membrane is represented as gray dashed lines, membrane helices are represented as rectangular cylinders, and LAP1’s nuclear domain is shown as a curved line.
Table 1.
Rose, Brown, Schlieker
| Protein | Mutation | Disease Phenotype | Reference |
|---|---|---|---|
| TorsinA | dE302/303 | early onset dystonia | (Ozelius et al. 1997) |
| TorsinA | dA14-P15 | early onset dystonia | (Vulinovic et al. 2014) |
| TorsinA | E121K | early onset dystonia | (Vulinovic et al. 2014) |
| TorsinA | D194V | early onset segmental dystonia | (Cheng et al. 2014) |
| TorsinA | F205I | idiopathic, late-onset focal dystonia | (Calakos et al. 2010) |
| TorsinA | R288Q | early onset dystonia | (Zirn et al. 2008) |
| TorsinA | dF323-Y328 | atypical early onset dystonia | (Leung et al. 2001) |
| LAP1 | c.186delG (p.E62fsTer25) | muscular dystrophy | (Kayman-Kurekci et al. 2014) |
| LAP1 | E482A | severe dystonia, cerebellar atrophy, and cardiomyopathy | (Dorboz et al. 2014) |
It is now possible to map the specific locations of these mutations on a potential TorsinA/LAP1 heterohexameric model to suggest mechanistic reasons for their pathological nature (Figure 8). TorsinA mutations R288Q, ΔE302/303, and ΔF323-Y328 are all predicted to lie at the TorsinA/LAP1 activating interface, and therefore it is reasonable to hypothesize that these mutations interfere with cofactor binding and ATPase activation. It has already been shown that ΔE302/303 fails to bind LAP1 and LULL1 (Naismith et al., 2009, Zhao et al., 2013, Zhu et al., 2010). Given that the ΔE302/303 mutation maps to the four-helix bundle in TorsinA that contributes to the Torsin-cofactor “activator” interface, it seems reasonable to assume that the ΔE302/303 deletion leads to a misalignment of this interface, consistent with the strong reduction with this disease variant of site-specific crosslinked products with its cofactors (Brown et al., 2014). Nevertheless, the ΔE302/303 retains a mild protective effect relative to a full TorA knockout in vivo, suggesting that it can act as a hypomorphic allele (Liang et al., 2014). This may also mean that secondary mutations or environmental factors contribute to the noted low penetrance of the mutation. Similarly to the ΔE302/303 mutation, since ΔF323-Y328 is located at TorsinA’s C-terminus (specifically the most C-terminal helix of the four-helix bundle), which is known to directly contact LAP1/LULL1 and is critical for LULL1 to bind TorsinB (Zhu et al., 2010, Rose et al., 2014, Brown et al., 2014), we expect that this mutation prevents LAP1/LULL1 binding and ATPase activation as well. TorsinA E121K is also predicted to be located at the activating interface, but at a location closer to the membrane. Similarly, TorsinA D194V and LAP1 E482A mutations are predicted to be at the back (non-activating) interface and therefore could interfere with oligomeric assembly of the ATPase machine. Interestingly, the LAP1 E482A mutations cause a more severe phenotype compared to typical DYT1 dystonia patients (Dorboz et al., 2014), which could be attributable to the loss of LAP1’s ability to activate multiple Torsin ATPases (Zhao et al., 2013). Since all mutations described lie at Torsin/cofactor interfaces, they most likely result in pathogenicity by failure to produce the active ATPase machine.
The TorsinA ΔA14-P15 and F205I mutations as well as a frameshift resulting in LAP1 truncation are likely harmful for different reasons. The TorsinA ΔA14-P15 mutation is within the signal sequence and most likely results in improper targeting of TorsinA to the ER. TorsinA F205I is buried deep within the folded core of the protein and may result in improper hydrophobic packing leading to misfolding. Lastly, the LAP1 truncation mutation would produce a highly-truncated protein that could no longer interact with Torsins and may therefore be functionally equivalent to a LAP1 deletion, which has been shown to cause muscular dystrophy in mice (Shin et al., 2013).
Since so many disease-associated mutations are predicted to lie at the Torsin/cofactor interface (Figure 8), a majority of these mutations likely result in a loss of Torsin activity. This presents a unique opportunity for potential therapeutic strategies to combat DYT1 dystonia as a whole, rather than on a case-by-case basis for each individual mutation. One strategy for therapeutic intervention would involve strengthening the association of TorsinA with its cofactors, potentially through the use of small molecules, thus ameliorating the disease phenotype. Additionally, if merely the loss of TorsinA activity due to impaired cofactor binding causes disease, then it may be possible to find a small molecule activator that would increase the ATPase activity of the non-affected TorA WT allele, thus compensating for the non-functional allele in vivo.
Conclusions and Future Perspectives
Our collective knowledge of Torsin ATPases has significantly advanced since its initial identification as the cause of DYT1 dystonia, but a number of questions still remain. Arguably the most important unresolved question for Torsin ATPases is the definition of their cellular function(s) and how this relates to disease presentation. It is clear from studies in several metazoans that Torsins are essential for life but why they are so important is unclear. It will be critical to identify cellular substrates of these ATPases in order to unambiguously pinpoint a biological function for them given that all known AAA+ proteins achieve their biological function by hydrolyzing ATP to perform work on specific cellular substrates.
Once the biological function of Torsin is known, an equally important goal will still be to determine how the Torsin ATPase machine works at a molecular level on its specific targets. In order to design therapeutic strategies to treat DYT1 dystonia and other diseases caused by mutations in Torsins and their cofactors, we need to understand how it achieves the most basic biochemical function of ATP hydrolysis. To do this, we will likely require high-resolution structures of the fully assembled ATPase machine in different nucleotide states. Ideally, these structures would include full-length proteins in the context of a membrane as well as the elusive cellular substrate(s). From these studies, we will gain a clearer understanding of Torsin function in the cell and will be in a much stronger position to treat diseases associated with its dysfunction.
Acknowledgments
We thank the members of the C.S. laboratory for critical reading of the manuscript and apologize to our colleagues whose work was not cited.
Footnotes
Declaration of Interest
The authors are funded by NIH (DP2 OD008624-01).
The authors report no declarations of interest.
References
- AUGUSTIN S, GERDES F, LEE S, TSAI FT, LANGER T, TATSUTA T. An intersubunit signaling network coordinates ATP hydrolysis by m-AAA proteases. Mol Cell. 2009;35:574–85. doi: 10.1016/j.molcel.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BABST M, WENDLAND B, ESTEPA EJ, EMR SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–93. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BASHAM SE, ROSE LS. Mutations in ooc-5 and ooc-3 disrupt oocyte formation and the reestablishment of asymmetric PAR protein localization in two-cell Caenorhabditis elegans embryos. Dev Biol. 1999;215:253–63. doi: 10.1006/dbio.1999.9447. [DOI] [PubMed] [Google Scholar]
- BASHAM SE, ROSE LS. The Caenorhabditis elegans polarity gene ooc-5 encodes a Torsin-related protein of the AAA ATPase superfamily. Development. 2001;128:4645–56. doi: 10.1242/dev.128.22.4645. [DOI] [PubMed] [Google Scholar]
- BHABHA G, CHENG HC, ZHANG N, MOELLER A, LIAO M, SPEIR JA, CHENG Y, VALE RD. Allosteric communication in the dynein motor domain. Cell. 2014;159:857–68. doi: 10.1016/j.cell.2014.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOWMAN GD, O’DONNELL M, KURIYAN J. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature. 2004;429:724–30. doi: 10.1038/nature02585. [DOI] [PubMed] [Google Scholar]
- BRAGG DC, CAMP SM, KAUFMAN CA, WILBUR JD, BOSTON H, SCHUBACK DE, HANSON PI, SENA-ESTEVES M, BREAKEFIELD XO. Perinuclear biogenesis of mutant torsin-A inclusions in cultured cells infected with tetracycline-regulated herpes simplex virus type 1 amplicon vectors. Neuroscience. 2004a;125:651–61. doi: 10.1016/j.neuroscience.2004.01.053. [DOI] [PubMed] [Google Scholar]
- BRAGG DC, KAUFMAN CA, KOCK N, BREAKEFIELD XO. Inhibition of N-linked glycosylation prevents inclusion formation by the dystonia-related mutant form of torsinA. Mol Cell Neurosci. 2004b;27:417–26. doi: 10.1016/j.mcn.2004.07.009. [DOI] [PubMed] [Google Scholar]
- BRESSMAN SB, DE LEON D, BRIN MF, RISCH N, BURKE RE, GREENE PE, SHALE H, FAHN S. Idiopathic dystonia among Ashkenazi Jews: evidence for autosomal dominant inheritance. Ann Neurol. 1989;26:612–20. doi: 10.1002/ana.410260505. [DOI] [PubMed] [Google Scholar]
- BRESSMAN SB, DE LEON D, KRAMER PL, OZELIUS LJ, BRIN MF, GREENE PE, FAHN S, BREAKEFIELD XO, RISCH NJ. Dystonia in Ashkenazi Jews: clinical characterization of a founder mutation. Ann Neurol. 1994;36:771–7. doi: 10.1002/ana.410360514. [DOI] [PubMed] [Google Scholar]
- BROWN RS, ZHAO C, CHASE AR, WANG J, SCHLIEKER C. The mechanism of Torsin ATPase activation. Proc Natl Acad Sci U S A. 2014;111:E4822–31. doi: 10.1073/pnas.1415271111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUCHSBAUM S, BERCOVICH B, ZIV T, CIECHANOVER A. Modification of the inflammatory mediator LRRFIP2 by the ubiquitin-like protein FAT10 inhibits its activity during cellular response to LPS. Biochem Biophys Res Commun. 2012;428:11–6. doi: 10.1016/j.bbrc.2012.09.110. [DOI] [PubMed] [Google Scholar]
- BUCK TM, PLAVCHAK L, ROY A, DONNELLY BF, KASHLAN OB, KLEYMAN TR, SUBRAMANYA AR, BRODSKY JL. The Lhs1/GRP170 chaperones facilitate the endoplasmic reticulum-associated degradation of the epithelial sodium channel. J Biol Chem. 2013;288:18366–80. doi: 10.1074/jbc.M113.469882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BURDETTE AJ, CHURCHILL PF, CALDWELL GA, CALDWELL KA. The early-onset torsion dystonia-associated protein, torsinA, displays molecular chaperone activity in vitro. Cell Stress Chaperones. 2010;15:605–17. doi: 10.1007/s12192-010-0173-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BURTON RE, SIDDIQUI SM, KIM YI, BAKER TA, SAUER RT. Effects of protein stability and structure on substrate processing by the ClpXP unfolding and degradation machine. EMBO J. 2001;20:3092–100. doi: 10.1093/emboj/20.12.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CALAKOS N, PATEL VD, GOTTRON M, WANG G, TRAN-VIET KN, BREWINGTON D, BEYER JL, STEFFENS DC, KRISHNAN RR, ZUCHNER S. Functional evidence implicating a novel TOR1A mutation in idiopathic, late-onset focal dystonia. J Med Genet. 2010;47:646–50. doi: 10.1136/jmg.2009.072082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CALDWELL GA, CAO S, SEXTON EG, GELWIX CC, BEVEL JP, CALDWELL KA. Suppression of polyglutamine-induced protein aggregation in Caenorhabditis elegans by torsin proteins. Hum Mol Genet. 2003;12:307–19. doi: 10.1093/hmg/ddg027. [DOI] [PubMed] [Google Scholar]
- CASHIKAR AG, SCHIRMER EC, HATTENDORF DA, GLOVER JR, RAMAKRISHNAN MS, WARE DM, LINDQUIST SL. Defining a pathway of communication from the C-terminal peptide binding domain to the N-terminal ATPase domain in a AAA protein. Mol Cell. 2002;9:751–60. doi: 10.1016/s1097-2765(02)00499-9. [DOI] [PubMed] [Google Scholar]
- CHEN P, BURDETTE AJ, PORTER JC, RICKETTS JC, FOX SA, NERY FC, HEWETT JW, BERKOWITZ LA, BREAKEFIELD XO, CALDWELL KA, CALDWELL GA. The early-onset torsion dystonia-associated protein, torsinA, is a homeostatic regulator of endoplasmic reticulum stress response. Hum Mol Genet. 2010a;19:3502–15. doi: 10.1093/hmg/ddq266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN XP, HU XH, WU SH, ZHANG YW, XIAO B, SHANG HF. RNA interference-mediated inhibition of wild-type Torsin A expression increases apoptosis caused by oxidative stress in cultured cells. Neurochem Res. 2010b;35:1214–23. doi: 10.1007/s11064-010-0177-4. [DOI] [PubMed] [Google Scholar]
- CHENG FB, FENG JC, MA LY, MIAO J, OTT T, WAN XH, GRUNDMANN K. Combined occurrence of a novel TOR1A and a THAP1 mutation in primary dystonia. Mov Disord. 2014;29:1079–83. doi: 10.1002/mds.25921. [DOI] [PubMed] [Google Scholar]
- CHO C, VALE RD. The mechanism of dynein motility: insight from crystal structures of the motor domain. Biochim Biophys Acta. 2012;1823:182–91. doi: 10.1016/j.bbamcr.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CRISP M, LIU Q, ROUX K, RATTNER JB, SHANAHAN C, BURKE B, STAHL PD, HODZIC D. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DALAL S, ROSSER MF, CYR DM, HANSON PI. Distinct roles for the AAA ATPases NSF and p97 in the secretory pathway. Mol Biol Cell. 2004;15:637–48. doi: 10.1091/mbc.E03-02-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVIES JM, BRUNGER AT, WEIS WI. Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change. Structure. 2008;16:715–26. doi: 10.1016/j.str.2008.02.010. [DOI] [PubMed] [Google Scholar]
- DEPHOURE N, ZHOU C, VILLEN J, BEAUSOLEIL SA, BAKALARSKI CE, ELLEDGE SJ, GYGI SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A. 2008;105:10762–7. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DORBOZ I, COUTELIER M, BERTRAND AT, CABERG JH, ELMALEH-BERGES M, LAINE J, STEVANIN G, BONNE G, BOESPFLUG-TANGUY O, SERVAIS L. Severe dystonia, cerebellar atrophy, and cardiomyopathy likely caused by a missense mutation in TOR1AIP1. Orphanet J Rare Dis. 2014;9:174. doi: 10.1186/s13023-014-0174-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DRON M, MERITET JF, DANDOY-DRON F, MEYNIEL JP, MAURY C, TOVEY MG. Molecular cloning of ADIR, a novel interferon responsive gene encoding a protein related to the torsins. Genomics. 2002;79:315–25. doi: 10.1006/geno.2002.6709. [DOI] [PubMed] [Google Scholar]
- EDGAR RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ERZBERGER JP, BERGER JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- FOISNER R, GERACE L. Integral membrane proteins of the nuclear envelope interact with lamins and chromosomes, and binding is modulated by mitotic phosphorylation. Cell. 1993;73:1267–79. doi: 10.1016/0092-8674(93)90355-t. [DOI] [PubMed] [Google Scholar]
- GLYNN SE, MARTIN A, NAGER AR, BAKER TA, SAUER RT. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell. 2009;139:744–56. doi: 10.1016/j.cell.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GONZALEZ-ALEGRE P, PAULSON HL. Aberrant cellular behavior of mutant torsinA implicates nuclear envelope dysfunction in DYT1 dystonia. J Neurosci. 2004;24:2593–601. doi: 10.1523/JNEUROSCI.4461-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOODCHILD RE, BUCHWALTER AL, NAISMITH TV, HOLBROOK K, BILLION K, DAUER WT, LIANG CC, DEAR ML, HANSON PI. Access of torsinA to the inner nuclear membrane is activity dependent and regulated in the endoplasmic reticulum. J Cell Sci. 2015;128:2854–65. doi: 10.1242/jcs.167452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOODCHILD RE, DAUER WT. Mislocalization to the nuclear envelope: an effect of the dystonia-causing torsinA mutation. Proc Natl Acad Sci U S A. 2004;101:847–52. doi: 10.1073/pnas.0304375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOODCHILD RE, DAUER WT. The AAA+ protein torsinA interacts with a conserved domain present in LAP1 and a novel ER protein. J Cell Biol. 2005;168:855–62. doi: 10.1083/jcb.200411026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOODCHILD RE, KIM CE, DAUER WT. Loss of the dystonia-associated protein torsinA selectively disrupts the neuronal nuclear envelope. Neuron. 2005;48:923–32. doi: 10.1016/j.neuron.2005.11.010. [DOI] [PubMed] [Google Scholar]
- GRANATA A, KOO SJ, HAUCKE V, SCHIAVO G, WARNER TT. CSN complex controls the stability of selected synaptic proteins via a torsinA-dependent process. EMBO J. 2011;30:181–93. doi: 10.1038/emboj.2010.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GRANATA A, WATSON R, COLLINSON LM, SCHIAVO G, WARNER TT. The dystonia-associated protein torsinA modulates synaptic vesicle recycling. J Biol Chem. 2008;283:7568–79. doi: 10.1074/jbc.M704097200. [DOI] [PubMed] [Google Scholar]
- GUENTHER B, ONRUST R, SALI A, O’DONNELL M, KURIYAN J. Crystal structure of the delta’ subunit of the clamp-loader complex of E. coli DNA polymerase III. Cell. 1997;91:335–45. doi: 10.1016/s0092-8674(00)80417-1. [DOI] [PubMed] [Google Scholar]
- HANSON PI, WHITEHEART SW. AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–29. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- HEDGLIN M, KUMAR R, BENKOVIC SJ. Replication clamps and clamp loaders. Cold Spring Harb Perspect Biol. 2013;5:a010165. doi: 10.1101/cshperspect.a010165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HELAERS R, MILINKOVITCH MC. MetaPIGA v2.0: maximum likelihood large phylogeny estimation using the metapopulation genetic algorithm and other stochastic heuristics. BMC Bioinformatics. 2010;11:379. doi: 10.1186/1471-2105-11-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HEWETT J, GONZALEZ-AGOSTI C, SLATER D, ZIEFER P, LI S, BERGERON D, JACOBY DJ, OZELIUS LJ, RAMESH V, BREAKEFIELD XO. Mutant torsinA, responsible for early-onset torsion dystonia, forms membrane inclusions in cultured neural cells. Hum Mol Genet. 2000;9:1403–13. doi: 10.1093/hmg/9.9.1403. [DOI] [PubMed] [Google Scholar]
- HEWETT J, ZIEFER P, BERGERON D, NAISMITH T, BOSTON H, SLATER D, WILBUR J, SCHUBACK D, KAMM C, SMITH N, CAMP S, OZELIUS LJ, RAMESH V, HANSON PI, BREAKEFIELD XO. TorsinA in PC12 cells: localization in the endoplasmic reticulum and response to stress. J Neurosci Res. 2003;72:158–68. doi: 10.1002/jnr.10567. [DOI] [PubMed] [Google Scholar]
- HEWETT JW, KAMM C, BOSTON H, BEAUCHAMP R, NAISMITH T, OZELIUS L, HANSON PI, BREAKEFIELD XO, RAMESH V. TorsinB--perinuclear location and association with torsinA. J Neurochem. 2004;89:1186–94. doi: 10.1111/j.1471-4159.2004.02404.x. [DOI] [PubMed] [Google Scholar]
- HEWETT JW, NERY FC, NILAND B, GE P, TAN P, HADWIGER P, TANNOUS BA, SAH DW, BREAKEFIELD XO. siRNA knock-down of mutant torsinA restores processing through secretory pathway in DYT1 dystonia cells. Hum Mol Genet. 2008;17:1436–45. doi: 10.1093/hmg/ddn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HEWETT JW, TANNOUS B, NILAND BP, NERY FC, ZENG J, LI Y, BREAKEFIELD XO. Mutant torsinA interferes with protein processing through the secretory pathway in DYT1 dystonia cells. Proc Natl Acad Sci U S A. 2007;104:7271–6. doi: 10.1073/pnas.0701185104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HEWETT JW, ZENG J, NILAND BP, BRAGG DC, BREAKEFIELD XO. Dystonia-causing mutant torsinA inhibits cell adhesion and neurite extension through interference with cytoskeletal dynamics. Neurobiol Dis. 2006;22:98–111. doi: 10.1016/j.nbd.2005.10.012. [DOI] [PubMed] [Google Scholar]
- HU J, PRINZ WA, RAPOPORT TA. Weaving the web of ER tubules. Cell. 2011;147:1226–31. doi: 10.1016/j.cell.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JERUZALMI D, O’DONNELL M, KURIYAN J. Crystal structure of the processivity clamp loader gamma (gamma) complex of E. coli DNA polymerase III. Cell. 2001;106:429–41. doi: 10.1016/s0092-8674(01)00463-9. [DOI] [PubMed] [Google Scholar]
- JOHNSON A, O’DONNELL M. Ordered ATP hydrolysis in the gamma complex clamp loader AAA+ machine. J Biol Chem. 2003;278:14406–13. doi: 10.1074/jbc.M212708200. [DOI] [PubMed] [Google Scholar]
- JOKHI V, ASHLEY J, NUNNARI J, NOMA A, ITO N, WAKABAYASHI-ITO N, MOORE MJ, BUDNIK V. Torsin mediates primary envelopment of large ribonucleoprotein granules at the nuclear envelope. Cell Rep. 2013;3:988–95. doi: 10.1016/j.celrep.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JUNGWIRTH M, DEAR ML, BROWN P, HOLBROOK K, GOODCHILD R. Relative tissue expression of homologous torsinB correlates with the neuronal specific importance of DYT1 dystonia-associated torsinA. Hum Mol Genet. 2010;19:888–900. doi: 10.1093/hmg/ddp557. [DOI] [PubMed] [Google Scholar]
- KAKAZU Y, KOH JY, HO KW, GONZALEZ-ALEGRE P, HARATA NC. Synaptic vesicle recycling is enhanced by torsinA that harbors the DYT1 dystonia mutation. Synapse. 2012a;66:453–64. doi: 10.1002/syn.21534. [DOI] [PubMed] [Google Scholar]
- KAKAZU Y, KOH JY, IWABUCHI S, GONZALEZ-ALEGRE P, HARATA NC. Miniature release events of glutamate from hippocampal neurons are influenced by the dystonia-associated protein torsinA. Synapse. 2012b;66:807–22. doi: 10.1002/syn.21571. [DOI] [PubMed] [Google Scholar]
- KAMM C, BOSTON H, HEWETT J, WILBUR J, COREY DP, HANSON PI, RAMESH V, BREAKEFIELD XO. The early onset dystonia protein torsinA interacts with kinesin light chain 1. J Biol Chem. 2004;279:19882–92. doi: 10.1074/jbc.M401332200. [DOI] [PubMed] [Google Scholar]
- KARLBERG T, VAN DEN BERG S, HAMMARSTROM M, SAGEMARK J, JOHANSSON I, HOLMBERG-SCHIAVONE L, SCHULER H. Crystal structure of the ATPase domain of the human AAA+ protein paraplegin/SPG7. PLoS One. 2009;4:e6975. doi: 10.1371/journal.pone.0006975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KATO JY, YONEDA-KATO N. Mammalian COP9 signalosome. Genes Cells. 2009;14:1209–25. doi: 10.1111/j.1365-2443.2009.01349.x. [DOI] [PubMed] [Google Scholar]
- KAYMAN-KUREKCI G, TALIM B, KORKUSUZ P, SAYAR N, SARIOGLU T, ONCEL I, SHARAFI P, GUNDESLI H, BALCI-HAYTA B, PURALI N, SERDAROGLU-OFLAZER P, TOPALOGLU H, DINCER P. Mutation in TOR1AIP1 encoding LAP1B in a form of muscular dystrophy: A novel gene related to nuclear envelopathies. Neuromuscul Disord. 2014 doi: 10.1016/j.nmd.2014.04.007. [DOI] [PubMed] [Google Scholar]
- KIM CE, PEREZ A, PERKINS G, ELLISMAN MH, DAUER WT. A molecular mechanism underlying the neural-specific defect in torsinA mutant mice. Proc Natl Acad Sci U S A. 2010;107:9861–6. doi: 10.1073/pnas.0912877107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KONAKOVA M, PULST SM. Dystonia-associated forms of torsinA are deficient in ATPase activity. J Mol Neurosci. 2005;25:105–17. doi: 10.1385/JMN:25:1:105. [DOI] [PubMed] [Google Scholar]
- KUSTEDJO K, BRACEY MH, CRAVATT BF. Torsin A and its torsion dystonia-associated mutant forms are lumenal glycoproteins that exhibit distinct subcellular localizations. J Biol Chem. 2000;275:27933–9. doi: 10.1074/jbc.M910025199. [DOI] [PubMed] [Google Scholar]
- KUSTEDJO K, DEECHONGKIT S, KELLY JW, CRAVATT BF. Recombinant expression, purification, and comparative characterization of torsinA and its torsion dystonia-associated variant Delta E-torsinA. Biochemistry. 2003;42:15333–41. doi: 10.1021/bi0349569. [DOI] [PubMed] [Google Scholar]
- LEE S, SOWA ME, WATANABE YH, SIGLER PB, CHIU W, YOSHIDA M, TSAI FT. The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell. 2003;115:229–40. doi: 10.1016/s0092-8674(03)00807-9. [DOI] [PubMed] [Google Scholar]
- LENZEN CU, STEINMANN D, WHITEHEART SW, WEIS WI. Crystal structure of the hexamerization domain of N-ethylmaleimide-sensitive fusion protein. Cell. 1998;94:525–36. doi: 10.1016/s0092-8674(00)81593-7. [DOI] [PubMed] [Google Scholar]
- LEUNG JC, KLEIN C, FRIEDMAN J, VIEREGGE P, JACOBS H, DOHENY D, KAMM C, DELEON D, PRAMSTALLER PP, PENNEY JB, EISENGART M, JANKOVIC J, GASSER T, BRESSMAN SB, COREY DP, KRAMER P, BRIN MF, OZELIUS LJ, BREAKEFIELD XO. Novel mutation in the TOR1A (DYT1) gene in atypical early onset dystonia and polymorphisms in dystonia and early onset parkinsonism. Neurogenetics. 2001;3:133–43. doi: 10.1007/s100480100111. [DOI] [PubMed] [Google Scholar]
- LI H, WU HC, LIU Z, ZACCHI LF, BRODSKY JL, ZOLKIEWSKI M. Intracellular complexes of the early-onset torsion dystonia-associated AAA+ ATPase TorsinA. Springerplus. 2014;3:743. doi: 10.1186/2193-1801-3-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIANG CC, TANABE LM, JOU S, CHI F, DAUER WT. TorsinA hypofunction causes abnormal twisting movements and sensorimotor circuit neurodegeneration. J Clin Invest. 2014;124:3080–92. doi: 10.1172/JCI72830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARCHLER-BAUER A, LU S, ANDERSON JB, CHITSAZ F, DERBYSHIRE MK, DEWEESE-SCOTT C, FONG JH, GEER LY, GEER RC, GONZALES NR, GWADZ M, HURWITZ DI, JACKSON JD, KE Z, LANCZYCKI CJ, LU F, MARCHLER GH, MULLOKANDOV M, OMELCHENKO MV, ROBERTSON CL, SONG JS, THANKI N, YAMASHITA RA, ZHANG D, ZHANG N, ZHENG C, BRYANT SH. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–9. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARIC M, SHAO J, RYAN RJ, WONG CS, GONZALEZ-ALEGRE P, ROLLER RJ. A functional role for TorsinA in herpes simplex virus 1 nuclear egress. J Virol. 2011;85:9667–79. doi: 10.1128/JVI.05314-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCCULLOUGH J, COLF LA, SUNDQUIST WI. Membrane fission reactions of the mammalian ESCRT pathway. Annu Rev Biochem. 2013;82:663–92. doi: 10.1146/annurev-biochem-072909-101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCCULLOUGH J, SUNDQUIST WI. Putting a finger in the ring. Nat Struct Mol Biol. 2014;21:1025–7. doi: 10.1038/nsmb.2928. [DOI] [PubMed] [Google Scholar]
- MCLEAN PJ, KAWAMATA H, SHARIFF S, HEWETT J, SHARMA N, UEDA K, BREAKEFIELD XO, HYMAN BT. TorsinA and heat shock proteins act as molecular chaperones: suppression of alpha-synuclein aggregation. J Neurochem. 2002;83:846–54. doi: 10.1046/j.1471-4159.2002.01190.x. [DOI] [PubMed] [Google Scholar]
- METTENLEITER TC, KLUPP BG, GRANZOW H. Herpesvirus assembly: an update. Virus Res. 2009;143:222–34. doi: 10.1016/j.virusres.2009.03.018. [DOI] [PubMed] [Google Scholar]
- MEYER H, BUG M, BREMER S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol. 2012;14:117–23. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- MOGK A, SCHLIEKER C, STRUB C, RIST W, WEIBEZAHN J, BUKAU B. Roles of individual domains and conserved motifs of the AAA+ chaperone ClpB in oligomerization, ATP hydrolysis, and chaperone activity. J Biol Chem. 2003;278:17615–24. doi: 10.1074/jbc.M209686200. [DOI] [PubMed] [Google Scholar]
- NAGY M, WU HC, LIU Z, KEDZIERSKA-MIESZKOWSKA S, ZOLKIEWSKI M. Walker-A threonine couples nucleotide occupancy with the chaperone activity of the AAA+ ATPase ClpB. Protein Sci. 2009;18:287–93. doi: 10.1002/pro.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NAISMITH TV, DALAL S, HANSON PI. Interaction of torsinA with its major binding partners is impaired by the dystonia-associated DeltaGAG deletion. J Biol Chem. 2009;284:27866–74. doi: 10.1074/jbc.M109.020164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NERY FC, ARMATA IA, FARLEY JE, CHO JA, YAQUB U, CHEN P, DA HORA CC, WANG Q, TAGAYA M, KLEIN C, TANNOUS B, CALDWELL KA, CALDWELL GA, LENCER WI, YE Y, BREAKEFIELD XO. TorsinA participates in endoplasmic reticulum-associated degradation. Nat Commun. 2011;2:393. doi: 10.1038/ncomms1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NERY FC, ZENG J, NILAND BP, HEWETT J, FARLEY J, IRIMIA D, LI Y, WICHE G, SONNENBERG A, BREAKEFIELD XO. TorsinA binds the KASH domain of nesprins and participates in linkage between nuclear envelope and cytoskeleton. J Cell Sci. 2008;121:3476–86. doi: 10.1242/jcs.029454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NEUMANN B, WALTER T, HERICHE JK, BULKESCHER J, ERFLE H, CONRAD C, ROGERS P, POSER I, HELD M, LIEBEL U, CETIN C, SIECKMANN F, PAU G, KABBE R, WUNSCHE A, SATAGOPAM V, SCHMITZ MH, CHAPUIS C, GERLICH DW, SCHNEIDER R, EILS R, HUBER W, PETERS JM, HYMAN AA, DURBIN R, PEPPERKOK R, ELLENBERG J. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–7. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NEUWALD AF, ARAVIND L, SPOUGE JL, KOONIN EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- O’DONNELL M, KURIYAN J. Clamp loaders and replication initiation. Curr Opin Struct Biol. 2006;16:35–41. doi: 10.1016/j.sbi.2005.12.004. [DOI] [PubMed] [Google Scholar]
- O’FARRELL C, LOCKHART PJ, LINCOLN S, DE LUCIA M, SINGLETON AB, DICKSON DW, COOKSON MR. Biochemical characterization of torsinB. Brain Res Mol Brain Res. 2004;127:1–9. doi: 10.1016/j.molbrainres.2004.05.005. [DOI] [PubMed] [Google Scholar]
- OGURA T, WHITEHEART SW, WILKINSON AJ. Conserved arginine residues implicated in ATP hydrolysis, nucleotide-sensing, and inter-subunit interactions in AAA and AAA+ ATPases. J Struct Biol. 2004;146:106–12. doi: 10.1016/j.jsb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- OLMOS Y, HODGSON L, MANTELL J, VERKADE P, CARLTON JG. ESCRT-III controls nuclear envelope reformation. Nature. 2015;522:236–9. doi: 10.1038/nature14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OSBORNE AR, RAPOPORT TA, VAN DEN BERG B. Protein translocation by the Sec61/SecY channel. Annu Rev Cell Dev Biol. 2005;21:529–50. doi: 10.1146/annurev.cellbio.21.012704.133214. [DOI] [PubMed] [Google Scholar]
- OZELIUS LJ, HEWETT JW, PAGE CE, BRESSMAN SB, KRAMER PL, SHALISH C, DE LEON D, BRIN MF, RAYMOND D, COREY DP, FAHN S, RISCH NJ, BUCKLER AJ, GUSELLA JF, BREAKEFIELD XO. The early-onset torsion dystonia gene (DYT1) encodes an ATP-binding protein. Nat Genet. 1997;17:40–8. doi: 10.1038/ng0997-40. [DOI] [PubMed] [Google Scholar]
- OZELIUS LJ, HEWETT JW, PAGE CE, BRESSMAN SB, KRAMER PL, SHALISH C, DE LEON D, BRIN MF, RAYMOND D, JACOBY D, PENNEY J, RISCH NJ, FAHN S, GUSELLA JF, BREAKEFIELD XO. The gene (DYT1) for early-onset torsion dystonia encodes a novel protein related to the Clp protease/heat shock family. Adv Neurol. 1998;78:93–105. [PubMed] [Google Scholar]
- OZELIUS LJ, PAGE CE, KLEIN C, HEWETT JW, MINETA M, LEUNG J, SHALISH C, BRESSMAN SB, DE LEON D, BRIN MF, FAHN S, COREY DP, BREAKEFIELD XO. The TOR1A (DYT1) gene family and its role in early onset torsion dystonia. Genomics. 1999;62:377–84. doi: 10.1006/geno.1999.6039. [DOI] [PubMed] [Google Scholar]
- PAPPAS SS, DARR K, HOLLEY SM, CEPEDA C, MABROUK OS, WONG JM, LEWITT TM, PAUDEL R, HOULDEN H, KENNEDY RT, LEVINE MS, DAUER WT. Forebrain deletion of the dystonia protein torsinA causes dystonic-like movements and loss of striatal cholinergic neurons. Elife. 2015;4 doi: 10.7554/eLife.08352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PHAM P, FREI KP, WOO W, TRUONG DD. Molecular defects of the dystonia-causing torsinA mutation. Neuroreport. 2006;17:1725–8. doi: 10.1097/WNR.0b013e3280101220. [DOI] [PubMed] [Google Scholar]
- ROSE A, SCHLIEKER C. Alternative nuclear transport for cellular protein quality control. Trends Cell Biol. 2012;22:509–14. doi: 10.1016/j.tcb.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROSE AE, ZHAO C, TURNER EM, STEYER AM, SCHLIEKER C. Arresting a Torsin ATPase reshapes the endoplasmic reticulum. J Biol Chem. 2014;289:552–64. doi: 10.1074/jbc.M113.515791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SANTOS M, DOMINGUES SC, COSTA P, MULLER T, GALOZZI S, MARCUS K, DA CRUZ E SILVA EF, DA CRUZ E SILVA OA, REBELO S. Identification of a novel human LAP1 isoform that is regulated by protein phosphorylation. PLoS One. 2014;9:e113732. doi: 10.1371/journal.pone.0113732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SARASTE M, SIBBALD PR, WITTINGHOFER A. The P-loop--a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990;15:430–4. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- SAUER RT, BAKER TA. AAA+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- SCHEFFZEK K, AHMADIAN MR, WITTINGHOFER A. GTPase-activating proteins: helping hands to complement an active site. Trends Biochem Sci. 1998;23:257–62. doi: 10.1016/s0968-0004(98)01224-9. [DOI] [PubMed] [Google Scholar]
- SCHLIEKER C, WEIBEZAHN J, PATZELT H, TESSARZ P, STRUB C, ZETH K, ERBSE A, SCHNEIDER-MERGENER J, CHIN JW, SCHULTZ PG, BUKAU B, MOGK A. Substrate recognition by the AAA+ chaperone ClpB. Nat Struct Mol Biol. 2004;11:607–15. doi: 10.1038/nsmb787. [DOI] [PubMed] [Google Scholar]
- SENIOR A, GERACE L. Integral membrane proteins specific to the inner nuclear membrane and associated with the nuclear lamina. J Cell Biol. 1988;107:2029–36. doi: 10.1083/jcb.107.6.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHIN JY, MENDEZ-LOPEZ I, WANG Y, HAYS AP, TANJI K, LEFKOWITCH JH, SCHULZE PC, WORMAN HJ, DAUER WT. Lamina-associated polypeptide-1 interacts with the muscular dystrophy protein emerin and is essential for skeletal muscle maintenance. Dev Cell. 2013;26:591–603. doi: 10.1016/j.devcel.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIDDIQUI SM, SAUER RT, BAKER TA. Role of the processing pore of the ClpX AAA+ ATPase in the recognition and engagement of specific protein substrates. Genes Dev. 2004;18:369–74. doi: 10.1101/gad.1170304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIMMEN FA, SU Y, XIAO R, ZENG Z, SIMMEN RC. The Kruppel-like factor 9 (KLF9) network in HEC-1-A endometrial carcinoma cells suggests the carcinogenic potential of dys-regulated KLF9 expression. Reprod Biol Endocrinol. 2008;6:41. doi: 10.1186/1477-7827-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SMITH GR, CONTRERAS-MOREIRA B, ZHANG X, BATES PA. A link between sequence conservation and domain motion within the AAA+ family. J Struct Biol. 2004;146:189–204. doi: 10.1016/j.jsb.2003.11.022. [DOI] [PubMed] [Google Scholar]
- SODING J, BIEGERT A, LUPAS AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–8. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SOSA BA, DEMIRCIOGLU FE, CHEN JZ, INGRAM J, PLOEGH H, SCHWARTZ TU. How lamina-associated polypeptide 1 (LAP1) activates Torsin. Elife. 2014:e03239. doi: 10.7554/eLife.03239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SPEESE SD, ASHLEY J, JOKHI V, NUNNARI J, BARRIA R, LI Y, ATAMAN B, KOON A, CHANG YT, LI Q, MOORE MJ, BUDNIK V. Nuclear Envelope Budding Enables Large Ribonucleoprotein Particle Export during Synaptic Wnt Signaling. Cell. 2012;149:832–46. doi: 10.1016/j.cell.2012.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STORY RM, WEBER IT, STEITZ TA. The structure of the E. coli recA protein monomer and polymer. Nature. 1992;355:318–25. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- SUDHOF TC, ROTHMAN JE. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323:474–7. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TANABE LM, KIM CE, ALAGEM N, DAUER WT. Primary dystonia: molecules and mechanisms. Nat Rev Neurol. 2009;5:598–609. doi: 10.1038/nrneurol.2009.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TORRES GE, SWEENEY AL, BEAULIEU JM, SHASHIDHARAN P, CARON MG. Effect of torsinA on membrane proteins reveals a loss of function and a dominant-negative phenotype of the dystonia-associated DeltaE-torsinA mutant. Proc Natl Acad Sci U S A. 2004;101:15650–5. doi: 10.1073/pnas.0308088101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TURNER EM, BROWN RS, LAUDERMILCH E, TSAI PL, SCHLIEKER C. The Torsin activator LULL1 is required for efficient growth of HSV-1. J Virol. 2015 doi: 10.1128/JVI.01143-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VANDER HEYDEN AB, NAISMITH TV, SNAPP EL, HANSON PI. Static retention of the lumenal monotopic membrane protein torsinA in the endoplasmic reticulum. EMBO J. 2011;30:3217–31. doi: 10.1038/emboj.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VANDER HEYDEN AB, NAISMITH TV, SNAPP EL, HODZIC D, HANSON PI. LULL1 retargets TorsinA to the nuclear envelope revealing an activity that is impaired by the DYT1 dystonia mutation. Mol Biol Cell. 2009;20:2661–72. doi: 10.1091/mbc.E09-01-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VANGOMPEL MJ, NGUYEN KC, HALL DH, DAUER WT, ROSE LS. A novel function for the Caenorhabditis elegans torsin OOC-5 in nucleoporin localization and nuclear import. Mol Biol Cell. 2015;26:1752–63. doi: 10.1091/mbc.E14-07-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VASUDEVAN A, BREAKEFIELD XO, BHIDE PG. Developmental patterns of torsinA and torsinB expression. Brain Res. 2006;1073–1074:139–45. doi: 10.1016/j.brainres.2005.12.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VIETRI M, SCHINK KO, CAMPSTEIJN C, WEGNER CS, SCHULTZ SW, CHRIST L, THORESEN SB, BRECH A, RAIBORG C, STENMARK H. Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature. 2015;522:231–5. doi: 10.1038/nature14408. [DOI] [PubMed] [Google Scholar]
- VULINOVIC F, LOHMANN K, RAKOVIC A, CAPETIAN P, ALVAREZ-FISCHER D, SCHMIDT A, WEISSBACH A, EROGULLARI A, KAISER FJ, WIEGERS K, FERBERT A, ROLFS A, KLEIN C, SEIBLER P. Unraveling cellular phenotypes of novel TorsinA/TOR1A mutations. Hum Mutat. 2014;35:1114–22. doi: 10.1002/humu.22604. [DOI] [PubMed] [Google Scholar]
- WALTER P, RON D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–6. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- WANG WL, LEE YC, YANG WM, CHANG WC, WANG JM. Sumoylation of LAP1 is involved in the HDAC4-mediated repression of COX-2 transcription. Nucleic Acids Res. 2008;36:6066–79. doi: 10.1093/nar/gkn607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WEBER-BAN EU, REID BG, MIRANKER AD, HORWICH AL. Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature. 1999;401:90–3. doi: 10.1038/43481. [DOI] [PubMed] [Google Scholar]
- WEBSTER BM, COLOMBI P, JAGER J, LUSK CP. Surveillance of nuclear pore complex assembly by ESCRT-III/Vps4. Cell. 2014;159:388–401. doi: 10.1016/j.cell.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WEIBEZAHN J, SCHLIEKER C, BUKAU B, MOGK A. Characterization of a trap mutant of the AAA+ chaperone ClpB. J Biol Chem. 2003;278:32608–17. doi: 10.1074/jbc.M303653200. [DOI] [PubMed] [Google Scholar]
- WEIBEZAHN J, TESSARZ P, SCHLIEKER C, ZAHN R, MAGLICA Z, LEE S, ZENTGRAF H, WEBER-BAN EU, DOUGAN DA, TSAI FT, MOGK A, BUKAU B. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119:653–65. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- WHITEHEART SW, ROSSNAGEL K, BUHROW SA, BRUNNER M, JAENICKE R, ROTHMAN JE. N-ethylmaleimide-sensitive fusion protein: a trimeric ATPase whose hydrolysis of ATP is required for membrane fusion. J Cell Biol. 1994;126:945–54. doi: 10.1083/jcb.126.4.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAMADA-INAGAWA T, OKUNO T, KARATA K, YAMANAKA K, OGURA T. Conserved pore residues in the AAA protease FtsH are important for proteolysis and its coupling to ATP hydrolysis. J Biol Chem. 2003;278:50182–7. doi: 10.1074/jbc.M308327200. [DOI] [PubMed] [Google Scholar]
- YU RC, HANSON PI, JAHN R, BRUNGER AT. Structure of the ATP-dependent oligomerization domain of N-ethylmaleimide sensitive factor complexed with ATP. Nat Struct Biol. 1998;5:803–11. doi: 10.1038/1843. [DOI] [PubMed] [Google Scholar]
- ZACCHI LF, WU HC, BELL SL, MILLEN L, PATON AW, PATON JC, THOMAS PJ, ZOLKIEWSKI M, BRODSKY JL. The BiP molecular chaperone plays multiple roles during the biogenesis of torsinA, an AAA+ ATPase associated with the neurological disease early-onset torsion dystonia. J Biol Chem. 2014;289:12727–47. doi: 10.1074/jbc.M113.529123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG X, SHAW A, BATES PA, NEWMAN RH, GOWEN B, ORLOVA E, GORMAN MA, KONDO H, DOKURNO P, LALLY J, LEONARD G, MEYER H, VAN HEEL M, FREEMONT PS. Structure of the AAA ATPase p97. Mol Cell. 2000;6:1473–84. doi: 10.1016/s1097-2765(00)00143-x. [DOI] [PubMed] [Google Scholar]
- ZHAO C, BROWN RS, CHASE AR, EISELE MR, SCHLIEKER C. Regulation of Torsin ATPases by LAP1 and LULL1. Proc Natl Acad Sci U S A. 2013;110:E1545–54. doi: 10.1073/pnas.1300676110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHAO M, WU S, ZHOU Q, VIVONA S, CIPRIANO DJ, CHENG Y, BRUNGER AT. Mechanistic insights into the recycling machine of the SNARE complex. Nature. 2015;518:61–7. doi: 10.1038/nature14148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHU L, MILLEN L, MENDOZA JL, THOMAS PJ. A unique redox-sensing sensor II motif in TorsinA plays a critical role in nucleotide and partner binding. J Biol Chem. 2010;285:37271–80. doi: 10.1074/jbc.M110.123471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHU L, WRABL JO, HAYASHI AP, ROSE LS, THOMAS PJ. The torsin-family AAA+ protein OOC-5 contains a critical disulfide adjacent to Sensor-II that couples redox state to nucleotide binding. Mol Biol Cell. 2008;19:3599–612. doi: 10.1091/mbc.E08-01-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZIRN B, GRUNDMANN K, HUPPKE P, PUTHENPARAMPIL J, WOLBURG H, RIESS O, MULLER U. Novel TOR1A mutation p.Arg288Gln in early-onset dystonia (DYT1) J Neurol Neurosurg Psychiatry. 2008;79:1327–30. doi: 10.1136/jnnp.2008.148270. [DOI] [PubMed] [Google Scholar]