Abstract
Background
In contrast to the complexity found in mammals, only two Bcl-2 family genes have been found in Drosophila melanogaster including the pro-cell survival, human Bok-related orthologue, Buffy. The directed expression of α-synuclein, the first gene identified to contribute to inherited forms of Parkinson disease (PD), in the dopaminergic neurons (DA) of flies has provided a robust and well-studied Drosophila model of PD complete with the loss of neurons and accompanying motor defects. To more fully understand the biological basis of Bcl-2 genes in PD, we altered the expression of Buffy in the dopamine producing neurons with and without the expression of α-synuclein, and in the developing neuron-rich eye.
Results
To alter the expression of Buffy in the dopaminergic neurons of Drosophila, the Ddc-Gal4 transgene was used. The directed expression of Buffy in the dopamine producing neurons resulted in flies with increased climbing ability and enhanced survival, while the inhibition of Buffy in the dopaminergic neurons reduced climbing ability over time prematurely, similar to the phenotype observed in the α-synuclein-induced Drosophila model of PD. Subsequently, the expression of Buffy was altered in the α-synuclein-induced Drosophila model of PD. Analysis revealed that Buffy acted to rescue the associated loss of locomotor ability observed in the α-synuclein-induced model of PD, while Buffy RNA interference resulted in an enhanced α-synuclein-induced loss of climbing ability. In complementary experiments the overexpression of Buffy in the developing eye suppressed the mild rough eye phenotype that results from Gal4 expression and from α-synuclein expression. When Buffy is inhibited the roughened eye phenotype is enhanced.
Conclusions
The inhibition of Buffy in DA neurons produces a novel model of PD in Drosophila. The directed expression of Buffy in DA neurons provide protection and counteracts the α-synuclein-induced Parkinson disease-like phenotypes. Taken all together this demonstrates a role for Buffy, a Bcl-2 pro-cell survival gene, in neuroprotection.
Background
Parkinson disease (PD) is the most common human movement disorder and the second most common neurodegenerative disease; afflicting about 1–2 % of the population over 50 years of age [1, 2]. PD is strongly associated with the selective and profound loss of dopaminergic (DA) neurons to result in marked clinical features which include muscle rigidity, resting tremors, postural instability, bradykinesia as well as non-motoric symptoms like autonomic, cognitive and psychiatric problems [2]. The neuropathological hallmarks exhibited by PD patients include the presence of Lewy Bodies (LB) and Lewy Neurites (LN) in surviving neurons. This is due to the loss of DA neurons in the substantia nigra pars compacta (SNpc) region of the brain, coupled with the presence of eosinophilic, intracytoplasmic proteinaceous inclusions comprised of the α-synuclein and ubiquitin proteins, among others [2–4]. This unusual protein accumulation is believed to lead to cellular toxicity and, eventually, the PD pathogenesis. Other associated pathological mechanisms include aberrant protein aggregation and mitochondrial damage [5–7]. Although the majority of PD cases are considered to be sporadic, familial forms have been documented and much has been discovered through study of the associated gene loci in model organisms [8–10]. PARK1/4 was the first gene associated with PD to be identified [3], and it encodes for a small soluble protein of unknown function predominantly found in neural tissues [3, 8, 11]. The first Drosophila model of PD utilized a human α-synuclein transgene to induce the PD-like symptoms [12]. The success of this model is its ability to recapitulate features of human PD such as (1) age-dependent loss in locomotor function (2) LB-like inclusions and (3) age-dependent loss of DA neurons; and therefore has found wide use for studying the molecular basis of α-synuclein-induced neurodegeneration [12–19]. The utilization of the UAS/GAL4 spatio-temporal expression system [20], and the availability of a plethora of promoters or enhancers of which TH-Gal4, elav-Gal4 and Ddc-Gal4 are employed in modelling PD in flies, makes Drosophila a powerful model organism [12–19]. Mitochondrial dysfunction due to the accumulation of α-synuclein has been implicated as one of the mechanisms leading to PD [21–24]. The association of α-synuclein with components of the mitochondria is thought to lead to oxidative stress, apoptosis, autophagy and the eventual neurodegeneration.
The Bcl-2 family of genes are key regulators of cell death and survival in animals and are functionally composed of proapoptotic and pro-cell survival (antiapoptotic) members [25–28]. The pro-survival proteins protect the mitochondria in part, from disruption by the proapoptotic proteins [26, 29–32]. In mammals, the antiapoptotic members possess four Bcl-2 homology (BH) domains—BH1, BH2, BH3 and BH4—and include Bcl-2, BclXL, Mcl-1 among others. The proapoptotic members, Bax, Bak and Bok, have three BH domains: BH1, BH2 and BH3. A BH3-only domain class of proapoptotic proteins is present and includes Bid, Bim, Bad, Bik, Hrk, Noxa and Puma [33–35]. The multi-domain proapoptotic proteins are required for mitochondrial outer membrane (MOM) permeabilization and the subsequent release of apoptogenic factors into the cytosol [36–39]. As thus, the antiapoptotic members guard the mitochondria and stop the release of a plethora of death causing molecules that initiate apoptosis.
The Bcl-2 family of proteins are thought to be the “guardians” of the mitochondria, involved in the life and death decisions at the cellular level by initiating mitochondrial remodelling, mitochondrial outer membrane permeabilization and the release of apoptotic factors from the mitochondria [40]. This delicate balance is maintained by the activity of the pro-survival and anti-survival members of the protein family. Many of the apoptotic pathway proteins that participate in the intrinsic and extrinsic cell death pathways have been identified in Drosophila [41–43]. The Bcl-2 family member homologues in Drosophila are limited to the single pro-cell survival Buffy and the sole pro-cell death Debcl [44–48]. These two proteins share a strong similarity within their domains and with the mammalian pore-forming proapoptotic member Bok/Mtd.
In previous studies, the overexpression of Buffy has been shown to suppress Pink1 mutant phenotypes [49] and suggest a role for this protein (1) in interacting with the Pink1 protein and other mitochondrial proteins or (2) in a pathway that regulates mitochondrial function and integrity. Studies show that Buffy has little involvement in cell death during development [50], though it has a role in regulating cell death that occurs in response to external stimuli and a role in the mitochondrial pathway for the activation of cell death during Drosophila oogenesis [51], all which point to an important role for this protein in aspects of cell death. Indeed, early studies have demonstrated that Buffy plays roles in both anti- and pro-survival [52, 53] depending upon the stimuli.
A direct role for the Bcl-2 proteins in mitochondrial dynamics has been shown in the activation of cell death in Drosophila melanogaster during mid-oogenesis [51] and in the Pink1 loss-of-function Parkinson disease model [49]. The predicted role of the mitochondria in PD pathogenesis makes the α-synuclein-induced model of PD [12] a very attractive model for the investigation of the role of Bcl-2 proteins. First we examine the effects of increasing and decreasing Buffy activity in DA neurons and, secondly, we investigate the potential suppression of the α-synuclein-induced PD phenotypes by the overexpression of the pro-survival Bcl-2 homologue Buffy.
Results
Buffy is similar to the human proapoptotic Bok
Bioinformatic analysis of the protein sequences encoded by the Buffy and Bok genes reveal 33 % identity. The Buffy protein consists of 299 amino acids and reveals the presence of the BH1, BH2, BH3, BH4 and TM domains (Fig. 1). The Eukaryotic Linear Motif (ELM) resource search for functional sites indicates the presence of a monopartite variant of a basically charged NLS between amino acids 101 and 106. There is a number of BH3-homology region binding sites in the central region of the protein. Bok has 212 amino acids and similarly shows the presence of the BH1, BH2, BH3, and BH4 domains. Although, the two proteins are determined to be antiapoptotic and proapoptotic respectively, both show the presence of a BH4 domain, the homology domain that is associated with pro-survival proteins.
Fig. 1.

Buffy is closely related to human Bcl-2 ovarian killer (Bok). When Buffy protein is aligned with human Bok, the Bcl-2 homology (BH) domains show strong conservation. Clustal Omega multiple sequence alignment [64, 65] of Drosophila melanogaster Buffy protein (Drosophila melanogaster NP_523702.1) with the human Bok (Homo sapiens NP_115904.1) showing the highlighted conserved BH domains, the BH3-homology region binding site, and the TM (trans-membrane) helices. Buffy possesses a monopartite basically charged nuclear localisation signal (NLS) region. The domains were identified using NCBI Conserved Domain Database Search (CDD) [66]. “Asterisks” indicate the residues that are identical, “colon” indicate the conserved substitutions, “dot” indicate the semi-conserved substitutions. Colours show the chemical nature of amino acids: red is small hydrophobic (including aromatic), blue is acidic, magenta is basic, and green is basic with hydroxyl or amine groups
Loss of Buffy decreases lifespan and climbing ability
When Buffy is inhibited in the DA neurons by RNA interference, the resulting flies have a shortened lifespan and impaired climbing ability. The median lifespan for these flies is 58 days compared to 64 days for the controls (Fig. 2a). The nonlinear fitting of the climbing curves resulted in a slope gradient that was significantly different at 95 % confidence interval (Fig. 2b). This suggests that Buffy is required for the normal functioning of DA neurons and inhibition in DA neurons confers a disadvantage by reducing lifespan and impairing the locomotor ability of these flies.
Fig. 2.
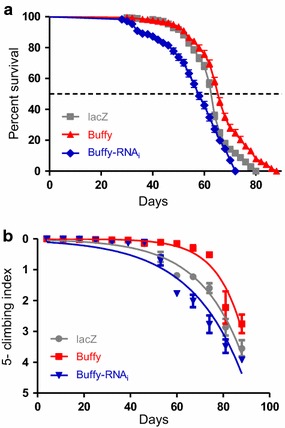
Buffy alters lifespan and climbing ability when mis-expressed in DA neurons. a Directed expression of UAS-Buffy in the DA neurons driven by Ddc-Gal4 results in increased survival compared to the controls overexpressing UAS-lacZ, while inhibition via Buffy-RNAi results in reduced survival. The genotypes are UAS-lacZ/Ddc-Gal4; UAS-Buffy/Ddc-Gal4; and UAS-Buffy-RNAi/Ddc-Gal4. Longevity is shown as percent survival (P < 0.01, determined by log-rank and n ≥ 200). b Directed expression of UAS-Buffy results in increased climbing ability as determined by non-linear fitting of the climbing curves and comparing at 95 % confidence intervals. Inhibition by Buffy-RNAi decreased the locomotor ability when expressed in dopaminergic neurons. The genotypes are UAS-lacZ/Ddc-Gal4; UAS-Buffy/Ddc-Gal4; and UAS-Buffy-RNAi/Ddc-Gal4. Error bars indicate the standard error of the mean (SEM), asterisks represents statistically significant result and n = 50
Buffy increases lifespan and climbing ability when overexpressed in DA neurons
When Buffy is overexpressed in DA neurons, the survival parameters of these flies differ slightly (Fig. 2a), with Buffy-overexpressing flies having a median lifespan of 68 days compared to 64 days for the controls. The overexpression of Buffy in DA neurons led to an increased climbing ability as indicated by the nonlinear fitting of the curve with 95 % CI (Fig. 2b). This suggests that Buffy improves longevity and markedly improves climbing ability in Drosophila when expressed in DA neurons to improve the general healthspan of these flies.
Inhibition of Buffy enhances the α-synuclein-induced phenotypes
The inhibition of Buffy by RNA interference when co-expressed with α-synuclein, under the directions of Ddc-Gal4, results in short-lived flies with strongly impaired locomotor function. The median lifespan of the α-synuclein-expressing control flies was 60 days, while that of those co-expressing the Buffy-RNAi inhibitory transgene along with α-synuclein was 50 days (Fig. 3a). The climbing ability of these flies was more impaired than those expressing α-synuclein alone, as indicated by the nonlinear fitting of the climbing curves (Fig. 3b).
Fig. 3.
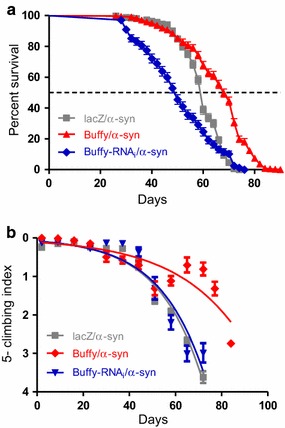
Buffy rescues the α-synuclein-induced phenotypes of decreased lifespan and climbing ability. a Directed overexpression of Buffy in the DA neurons increase longevity whereas flies with Buffy loss-of-function show a decline in lifespan. Genotypes are UAS-α-synuclein, Ddc-Gal4/UAS-lacZ; UAS-α-synuclein, Ddc-Gal4/UAS-Buffy; and UAS-α-synuclein, Ddc-Gal4/UAS-Buffy-RNAi. Longevity is shown as percent survival (P < 0.01, determined by log-rank and n ≥ 200). b The co-expression of Buffy in the α-synuclein model of PD rescued the age-dependent loss in climbing ability. The directed overexpression of Buffy in the DA neurons remarkably increased the climbing ability over time compared to the control, while the suppression of Buffy resulted in flies that climbed similar to the control. The genotypes are UAS-α-synuclein; Ddc-Gal4/UAS-lacZ, UAS-α-synuclein; Ddc-Gal4/UAS-Buffy, and UAS-α-synuclein; Ddc-Gal4/UAS-Buffy-RNAi. Analysis of the climbing curves and significance was determined by comparing the 95 % confidence intervals. Error bars indicate the SEM, asterisks represents statistically significant result and n = 50
Overexpression of Buffy in DA neurons rescues the α-synuclein-induced loss of climbing ability
The overexpression of Buffy in DA neurons expressing α-synuclein results in an increased median lifespan of 68 days, compared to 60 days for the control flies (Fig. 3a). The climbing curves indicate that there was a significant improvement in the climbing ability of the flies when Buffy was overexpressed (Fig. 3b) and thus, suppressing the phenotypes observed when α-synuclein is expressed in DA neurons. This suggests that the overexpression of Buffy confers protection to DA neurons, as a result of the expression of α-synuclein.
Overexpression of Buffy suppresses the α-synuclein-induced developmental defects in the eye
The expression of α-synuclein in the developing eye results in developmental defects. When Buffy is overexpressed in the developing eye, developmental defects resulting from GMR-Gal4 (Fig. 4b, I) and from GMR-Gal4 and α-synuclein expression (Fig. 4b, IV) are suppressed. The disruption of the ommatidial array is restored to control levels in both cases (Fig. 4b). These results suggest that overexpression of Buffy is able to counteract the toxic effects of α-synuclein in the developing eye in addition to the effects of GMR-Gal4.
Fig. 4.
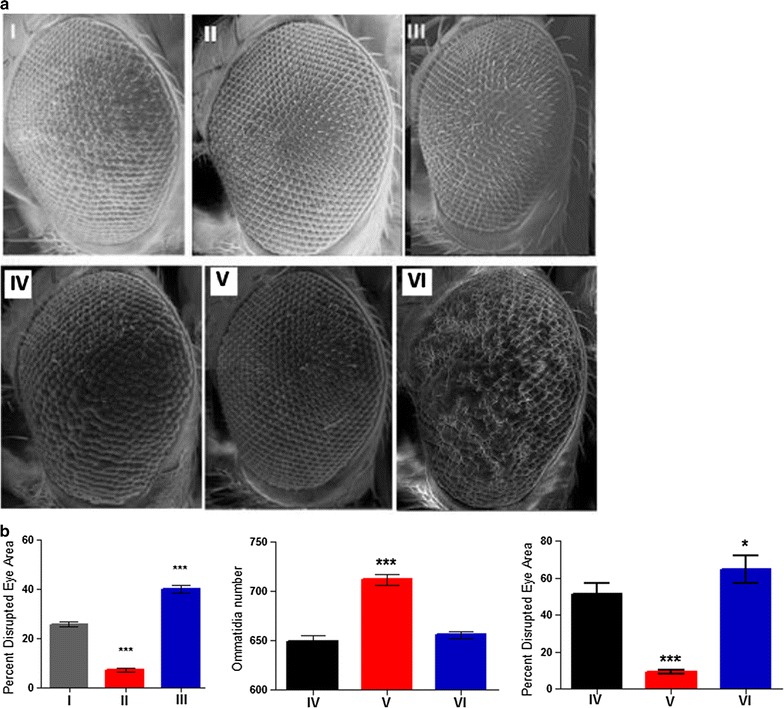
Buffy suppresses the α-synuclein-induced developmental defects in the eye. a Scanning electron micrographs when Buffy is overexpressed or inhibited in the eye with the eye-specific GMR-Gal4 transgene; (I) GMR-Gal4/UAS-lacZ; (II) GMR-Gal4/UAS-Buffy; (III) GMR-Gal4/UAS-Buffy-RNAi and when co-expressed with α-synuclein; (IV) UAS-α-synuclein; GMR-Gal4/UAS-lacZ; (V) UAS-α-synuclein; GMR-Gal4/UAS-Buffy; and (VI) UAS-α-synuclein; GMR-Gal4/UAS-Buffy-RNAi. b Biometric analysis showed a significant difference in the disrupted area of the eye when Buffy was inhibited in the developing eye (I–III). Biometric analysis shows a marked difference when Buffy is inhibited in an α-synuclein expressing background (IV–VI) with decreased ommatidia number and highly degenerated ommatidial array whereas when Buffy is overexpressed in the α-synuclein background, there is a rescue of the α-synuclein-induced phenotypes as determined by one-way ANOVA and Dunnett’s multiple comparison test (P < 0.05 and 95 % CI), error bars indicate the SEM, asterisks represents statistically significant result and n = 10
Discussion
The recapitulation of PD-like symptoms in Drosophila melanogaster and especially the age-dependent loss of climbing ability led to the investigation of gene products that could suppress this phenotype [12, 13, 54]. Mitochondrial dysfunction as a result of α-synuclein accumulation has been implicated in PD pathogenesis and, thus, we have investigated the consequences of the overexpression of the Drosophila Bcl-2 homologue Buffy. The analysis of climbing over the lifespan of the flies has been applied to determine the role of the various gene products in rescuing the α-synuclein-induced phenotypes [12, 54–56]. This assay allows for scoring of flies based on their loss of climbing ability and is a key indicator of the effect the overexpressed gene has on the phenotype.
The α-synuclein-expressing models of PD in Drosophila show little difference in lifespan between the control and wild type, A53T and A30P α-synuclein flies [12]. In our study, when Buffy is overexpressed in the DA neurons under the control of the Ddc-Gal4 driver, there is a significant increase in their longevity. This may be partly due to defects in mitochondrial complex I function, the pro-survival Buffy likely plays a mitochondrial protective role to increase longevity. The inhibition of Buffy in the DA neurons resulted in a marked decrease in survival. This inhibition of Buffy is sufficient to negate its protective role and thus promote cell death as has recently been shown by Clavier and colleagues [57]. Thus, the pro-survival properties of Buffy are evident.
Locomotor dysfunction is one of the associated symptoms of PD, the α-synuclein-expressing model demonstrated a clear age-dependent loss in climbing ability [12]. When we overexpressed Buffy in the DA neurons under the control of Ddc-Gal4, the flies produced a climbing index significantly different from that of control flies. The Buffy flies lost the climbing ability later than the control flies and is likely due to the protective role that Buffy confers to the mitochondria. In contrast, the inhibition of Buffy results in a highly compromised climbing ability when compared to the controls. The degree of locomotor dysfunction seemed to be similar to that observed when α-synuclein is overexpressed in DA neurons. Taken together, these results would indicate an early protective role for Buffy in the DA neurons even in the absence of induced cellular stress.
The inhibition of Buffy in the DA neurons of the α-synuclein-induced PD model significantly decreased lifespan, indicating that low levels of Buffy compromise the health of DA neurons. When Buffy was overexpressed along with α-synuclein, there was a marked improvement in the climbing ability of these flies. These results suggest that overexpressing Buffy in the DA neurons counteracts the α-synuclein-induced phenotype of locomotor dysfunction over their lifespan. The Buffy loss-of-function flies displayed a reduced climbing ability compared to the control flies. Therefore, expression of the pro-survival Buffy can rescue the α-synuclein-dependent model of PD from climbing dysfunction.
Directed overexpression of Buffy in the developing eye rescues the roughened eye phenotypes caused by Gal4 and α-synuclein expression, whereas the inhibition of Buffy results in a more disrupted ommatidial array. This indicates that elevated levels of Buffy in the developing eye offers protection from toxic protein insults to normalize cellular differentiation, neurogenesis and cell survival.
Conclusions
Buffy inhibition results in shortened lifespan and impaired locomotor function and represents a novel model of PD in Drosophila melanogaster. The overexpression of Buffy improves healthspan and counteracts the effects of α-synuclein expression to demonstrate its protective function. Further studies are required to fully elucidate how Buffy may interact with the other PD genes, and how these interactions fit into the regulation of mitochondrial integrity by the Bcl-2 proteins.
Methods
Drosophila media and culture
Stocks and crosses were maintained on a standard medium containing cornmeal, molasses, yeast, agar, water and treated with propionic acid and methylparaben. Seven millilitre aliquots of media was poured into vials, allowed to solidify, and refrigerated at 4–6 °C. Stocks were maintained on solid media for two to 3 weeks before transfer onto new media to re-culture. Stocks were kept at room temperature (22 ± 2 °C) while crosses and experiments were carried out at 25 and 29 °C.
Drosophila stocks and derivative lines
UAS-Buffy [52] was generously provided by Dr. Leonie Quinn (University of Melbourne), UAS-α-synuclein [12] was provided by Dr. M. Feany of Harvard Medical School and Dr. J. Hirsch (University of Virginia) provided Ddc-Gal4 flies [58]. UAS-Buffy-RNAi (w[*]; P{w[+mC] = UAS-Buffy.RNAi}3), GMR-Gal4 [59] and UAS-lacZ flies were obtained from the Bloomington Drosophila Stock Center at Indiana University. The UAS-α-synuclein/CyO; Ddc-Gal4/TM3 was generated using standard homologous recombination methods and was used to overexpress α-synuclein in the dopaminergic neurons using the dopa decarboxylase (Ddc) transgene. The UAS-α-synuclein/CyO; GMR-Gal4 line was used to overexpress α-synuclein in the developing eye using the Glass Multiple Reporter (GMR) elements. PCR reactions and gel electrophoresis were used for analysis of recombination events.
Ageing assay
Several single vial matings of five females and three males of each genotype were made and a cohort of critical class male flies were collected upon eclosion. At least two hundred flies were aged per genotype at a density of ≤20 flies per vial on fresh media which was replenished every other day to avoid crowding. Flies were observed and scored every 2 days for the presence of deceased adults. Flies were considered dead when they did not display any movement upon agitation [60]. Longevity data was analysed using the GraphPad Prism version 5.04 and survival curves were compared using the log-rank (Mantel-Cox) test. Significance was determined at 95 %, at a P value less than or equal to 0.05 with Bonferroni correction.
Climbing assay
A cohort of critical class male flies was collected upon eclosion and scored for their ability to climb over their lifetime [56, 61]. Every 7 days, 50 males from every genotype were assayed for their ability to climb 10 cm in 10 s in a clean climbing apparatus in ten repetitions. Analysis was performed using the GraphPad Prism version 5.04 and climbing curves were fitted using non-linear regression and compared using 95 % confidence interval with a 0.05 P value.
Scanning electron microscopy of the Drosophila eye
Several single vial matings were made at 29 °C and a cohort of adult male flies collected upon eclosion and aged for 3 days before being frozen at −80 °C. Whole flies were mounted on scanning electron microscope stubs, desiccated overnight and photographed with a FEI Mineral Liberation Analyzer 650F scanning electron microscope. For each cross at least 10 eye images were analysed using the National Institutes of Health (NIH) ImageJ software [62] and biometric analysis performed using GraphPad Prism version 5.04. The percent area of eye disruption was calculated as previously described [63].
Authors’ contributions
PGM performed the bioinformatic, survival, climbing, biometric and statistical analyses. BES conceived and participated in the design, supervision of the study and revisions to the final draft of the manuscript. Both authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Availability of data and material
The datasets supporting the conclusions of this article are included within the article.
Ethics approval
This study has been conducted under the approval of the Animal Care Committee of Memorial University of Newfoundland as a Category of Invasiveness Level A protocol under the project title of “Genetic, biochemical and molecular analysis of cell survival and cell death in Drosophila melanogaster” (protocol number: 15-09-BS). Consent was not applicable for this study.
Funding
PGM has been partially funded by Department of Biology Teaching Assistantships and a School of Graduate Studies Fellowship from Memorial University of Newfoundland. BES has been funded by a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant. Funding organizations were not involved in the design of the study, nor in the collection, analysis, interpretation of data or in the writing of the manuscript.
Abbreviations
- BH
Bcl-2 homology
- Bcl-2
B cell lymphoma 2
- DA
dopaminergic
- Ddc
dopa decarboxylase
- PD
Parkinson disease
- RNAi
ribonucleic acid interference
- SEM
standard error of the mean
Contributor Information
P. Githure M’Angale, Email: mgithure@mun.ca.
Brian E. Staveley, Email: bestave@mun.ca
References
- 1.Forno LS. Neuropathologic features of Parkinson’s, Huntington’s, and Alzheimer’s diseases. Ann N Y Acad Sci. 1992;648:6–16. doi: 10.1111/j.1749-6632.1992.tb24519.x. [DOI] [PubMed] [Google Scholar]
- 2.Forno LS. Neuropathology of Parkinson’s disease. J Neuropathol Exp Neurol. 1996;55:259–272. doi: 10.1097/00005072-199603000-00001. [DOI] [PubMed] [Google Scholar]
- 3.Polymeropoulos MH. Mutation in the α-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 4.Leroy E, Boyer R, Auburger G, Leube B, Ulm G, Mezey E, et al. The ubiquitin pathway in Parkinson’s disease. Nature. 1998;395:451–452. doi: 10.1038/26652. [DOI] [PubMed] [Google Scholar]
- 5.Gupta A, Dawson VL, Dawson TM. What causes cell death in Parkinson’s disease? Ann Neurol. 2008 doi: 10.1002/ana.21573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schulz JB. Mechanisms of neurodegeneration in idiopathic Parkinson's disease. Parkinsonism Relat Disord. 2007;13(Suppl 3):S306–8. doi: 10.1016/S1353-8020(08)70021-X. [DOI] [PubMed] [Google Scholar]
- 7.Whitworth AJ. Drosophila models of Parkinson’s disease. Adv Genet. 2011;73:1–50. doi: 10.1016/B978-0-12-380860-8.00001-X. [DOI] [PubMed] [Google Scholar]
- 8.Ambegaokar SS, Roy B, Jackson GR. Neurodegenerative models in Drosophila: polyglutamine disorders, Parkinson disease, and amyotrophic lateral sclerosis. Neurobiol Dis. 2010;40:29–39. doi: 10.1016/j.nbd.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gasser T. Molecular pathogenesis of Parkinson disease: insights from genetic studies. Expert Rev Mol Med. 2009;11:e22. doi: 10.1017/S1462399409001148. [DOI] [PubMed] [Google Scholar]
- 10.Guo M. Drosophila as a model to study mitochondrial dysfunction in Parkinson's disease. Cold Spring Harb Perspect Med. 2012;2(11). doi:10.1101/cshperspect.a009944. [DOI] [PMC free article] [PubMed]
- 11.Dehay B, Vila M, Bezard E, Brundin P, Kordower JH. Alpha-synuclein propagation: new insights from animal models. Mov Disord. 2015 doi: 10.1002/mds.26370. [DOI] [PubMed] [Google Scholar]
- 12.Feany MB, Bender WW. A Drosophila model of Parkinson’s disease. Nature. 2000;404:394–398. doi: 10.1038/35006074. [DOI] [PubMed] [Google Scholar]
- 13.Auluck PK, Chan HY, Trojanowski JQ, Lee VM, Bonini NM. Chaperone suppression of alpha-synuclein toxicity in a Drosophila model for Parkinson’s disease. Science. 2002;295:865–868. doi: 10.1126/science.1067389. [DOI] [PubMed] [Google Scholar]
- 14.Buttner S, Broeskamp F, Sommer C, Markaki M, Habernig L, Alavian-Ghavanini A, et al. Spermidine protects against alpha-synuclein neurotoxicity. Cell Cycle (Georgetown, Tex) 2014;13:3903–3908. doi: 10.4161/15384101.2014.973309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong Y, Liang X, Liu L, Zhang D, Wan C, Gan Z, et al. High throughput sequencing identifies microRNAs mediating α-synuclein toxicity by targeting neuroactive-ligand receptor interaction pathway in early stage of Drosophila Parkinson’s disease model. PLoS One. 2015;10:e0137432. doi: 10.1371/journal.pone.0137432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang B, Liu Q, Shan H, Xia C, Liu Z. Nrf2 inducer and cncC overexpression attenuates neurodegeneration due to alpha-synuclein in Drosophila. Biochem Cell Biol. 2015;93:351–358. doi: 10.1139/bcb-2015-0015. [DOI] [PubMed] [Google Scholar]
- 17.Zhu ZJ, Wu KC, Yung WH, Qian ZM, Ke Y. Differential interaction between iron and mutant alpha-synuclein causes distinctive Parkinsonian phenotypes in Drosophila. Biochim Biophys Acta. 2016 doi: 10.1016/j.bbadis.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 18.Staveley BE. Drosophila models of Parkinson disease. In: LeDoux MS, editor. Movement disorders: genetics and models. 2. Amsterdam: Elsevier; 2014. pp. 345–354. [Google Scholar]
- 19.Botella JAA, Bayersdorfer F, Gmeiner F, Schneuwly S. Modelling Parkinson’s disease in Drosophila. Neuromolecular Med. 2009;11:268–280. doi: 10.1007/s12017-009-8098-6. [DOI] [PubMed] [Google Scholar]
- 20.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 21.Chinta SJ, Mallajosyula JK, Rane A, Andersen JK. Mitochondrial alpha-synuclein accumulation impairs complex I function in dopaminergic neurons and results in increased mitophagy in vivo. Neurosci Lett. 2010;486:235–239. doi: 10.1016/j.neulet.2010.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choubey V, Safiulina D, Vaarmann A, Cagalinec M, Wareski P, Kuum M, et al. Mutant A53T alpha-synuclein induces neuronal death by increasing mitochondrial autophagy. J Biol Chem. 2011;286:10814–10824. doi: 10.1074/jbc.M110.132514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Esteves AR, Arduino DM, Silva DF, Oliveira CR, Cardoso SM. Mitochondrial dysfunction: the road to alpha-synuclein oligomerization in PD. Parkinson’s Dis. 2011;2011:693761. doi: 10.4061/2011/693761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu Y, Duan C, Lu L, Gao H, Zhao C, Yu S, et al. α-Synuclein overexpression impairs mitochondrial function by associating with adenylate translocator. Int J Biochem Cell Biol. 2011;43:732–741. doi: 10.1016/j.biocel.2011.01.014. [DOI] [PubMed] [Google Scholar]
- 25.Adams JM, Cory S. The Bcl-2 protein family: arbiters of cell survival. Science. 1998;281:1322–1326. doi: 10.1126/science.281.5381.1322. [DOI] [PubMed] [Google Scholar]
- 26.Cory S, Adams JM. The Bcl2 family: regulators of the cellular life-or-death switch. Nat Rev Cancer. 2002;2:647–656. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- 27.Fu YF, Fan TJ. Bcl-2 family proteins and apoptosis. Sheng wu hua xue yu sheng wu wu li xue bao Acta biochimica et biophysica Sinica. 2002;34:389–394. [PubMed] [Google Scholar]
- 28.Siddiqui WA, Ahad A, Ahsan H. The mystery of BCL2 family: Bcl-2 proteins and apoptosis: an update. Arch Toxicol. 2015;89:289–317. doi: 10.1007/s00204-014-1448-7. [DOI] [PubMed] [Google Scholar]
- 29.Tsujimoto Y. Bcl-2 family of proteins: life-or-death switch in mitochondria. Biosci Rep. 2002;22:47–58. doi: 10.1023/A:1016061006256. [DOI] [PubMed] [Google Scholar]
- 30.Suen DF, Norris KL, Youle RJ. Mitochondrial dynamics and apoptosis. Genes Dev. 2008;22:1577–1590. doi: 10.1101/gad.1658508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Colin J, Gaumer S, Guenal I, Mignotte B. Mitochondria, Bcl-2 family proteins and apoptosomes: of worms, flies and men. Front Biosci (Landmark edition) 2009;14:4127–4137. doi: 10.2741/3517. [DOI] [PubMed] [Google Scholar]
- 32.Martinou JC, Youle RJ. Mitochondria in apoptosis: Bcl-2 family members and mitochondrial dynamics. Dev Cell. 2011;21:92–101. doi: 10.1016/j.devcel.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siddiqui WA, Ahad A, Ahsan H. The mystery of BCL2 family: Bcl-2 proteins and apoptosis: an update. Arch Toxicol. 2015;89:289–317. doi: 10.1007/s00204-014-1448-7. [DOI] [PubMed] [Google Scholar]
- 34.Vela L, Marzo I. Bcl-2 family of proteins as drug targets for cancer chemotherapy: the long way of BH3 mimetics from bench to bedside. Curr Opin Pharmacol. 2015;23:74–81. doi: 10.1016/j.coph.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 35.Zheng J, Viacava Follis A, Kriwacki RW, Moldoveanu T. Discoveries and controversies in BCL-2 proteins-mediated apoptosis. FEBS J. 2015 doi: 10.1111/febs.13527. [DOI] [PubMed] [Google Scholar]
- 36.Delbridge AR, Strasser A. The BCL-2 protein family, BH3-mimetics and cancer therapy. Cell Death Differ. 2015;22:1071–1080. doi: 10.1038/cdd.2015.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doerflinger M, Glab JA, Puthalakath H. BH3-only proteins: a 20-year stock-take. FEBS J. 2015;282:1006–1016. doi: 10.1111/febs.13190. [DOI] [PubMed] [Google Scholar]
- 38.Li MX, Dewson G. Mitochondria and apoptosis: emerging concepts. F1000prime reports. 2015;7:42. [DOI] [PMC free article] [PubMed]
- 39.Lopez J, Tait SW. Mitochondrial apoptosis: killing cancer using the enemy within. Br J Cancer. 2015;112:957–962. doi: 10.1038/bjc.2015.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang C, Youle RJ. The role of mitochondria in apoptosis*. Annu Rev Genet. 2009;43:95–118. doi: 10.1146/annurev-genet-102108-134850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCall K, Steller H. Facing death in the fly: genetic analysis of apoptosis in Drosophila. Trends Genet TIG. 1997;13:222–226. doi: 10.1016/S0168-9525(97)01126-8. [DOI] [PubMed] [Google Scholar]
- 42.Richardson H, Kumar S. Death to flies: Drosophila as a model system to study programmed cell death. J Immunol Methods. 2002;265:21–38. doi: 10.1016/S0022-1759(02)00068-6. [DOI] [PubMed] [Google Scholar]
- 43.Kornbluth S, White K. Apoptosis in Drosophila: neither fish nor fowl (nor man, nor worm) J Cell Sci. 2005;118:1779–1787. doi: 10.1242/jcs.02377. [DOI] [PubMed] [Google Scholar]
- 44.Brachmann CB, Jassim OW, Wachsmuth BD, Cagan RL. The Drosophila bcl-2 family member dBorg-1 functions in the apoptotic response to UV-irradiation. Curr Biol. 2000;10:547–550. doi: 10.1016/S0960-9822(00)00474-7. [DOI] [PubMed] [Google Scholar]
- 45.Colussi PA, Quinn LM, Huang DC, Coombe M, Read SH, Richardson H, et al. Debcl, a proapoptotic Bcl-2 homologue, is a component of the Drosophila melanogaster cell death machinery. J Cell Biol. 2000;148:703–714. doi: 10.1083/jcb.148.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Igaki T, Kanuka H, Inohara N, Sawamoto K, Nunez G, Okano H, et al. Drob-1, a Drosophila member of the Bcl-2/CED-9 family that promotes cell death. Proc Natl Acad Sci USA. 2000;97:662–667. doi: 10.1073/pnas.97.2.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang H, Huang Q, Ke N, Matsuyama S, Hammock B, Godzik A, et al. Drosophila pro-apoptotic Bcl-2/Bax homologue reveals evolutionary conservation of cell death mechanisms. J Biol Chem. 2000;275:27303–27306. doi: 10.1074/jbc.M002846200. [DOI] [PubMed] [Google Scholar]
- 48.Quinn L, Coombe M, Mills K, Daish T, Colussi P, Kumar S, et al. Buffy, a Drosophila Bcl-2 protein, has anti-apoptotic and cell cycle inhibitory functions. EMBO J. 2003;22:3568–3579. doi: 10.1093/emboj/cdg355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 50.Sevrioukov EA, Burr J, Huang EW, Assi HH, Monserrate JP, Purves DC, et al. Drosophila Bcl-2 proteins participate in stress-induced apoptosis, but are not required for normal development. Genesis. 2007;45:184–193. doi: 10.1002/dvg.20279. [DOI] [PubMed] [Google Scholar]
- 51.Tanner EA, Blute TA, Brachmann CB, McCall K. Bcl-2 proteins and autophagy regulate mitochondrial dynamics during programmed cell death in the Drosophila ovary. Development. 2011;138:327–338. doi: 10.1242/dev.057943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quinn L, Coombe M, Mills K, Daish T, Colussi P, Kumar S, et al. Buffy, a Drosophila Bcl-2 protein, has anti-apoptotic and cell cycle inhibitory functions. EMBO J. 2003;22:3568–3579. doi: 10.1093/emboj/cdg355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu JN, Nguyen N, Aghazarian M, Tan Y, Sevrioukov EA, Mabuchi M, et al. Grim promotes programmed cell death of Drosophila microchaete glial cells. Mech Dev. 2010;127:407–417. doi: 10.1016/j.mod.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haywood AF, Staveley BE. Parkin counteracts symptoms in a Drosophila model of Parkinson’s disease. BMC Neurosci. 2004;5:14. doi: 10.1186/1471-2202-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haywood AF, Staveley BE. Mutant alpha-synuclein-induced degeneration is reduced by parkin in a fly model of Parkinson’s disease. Genome. 2006;49:505–510. doi: 10.1139/G06-011. [DOI] [PubMed] [Google Scholar]
- 56.Todd AM, Staveley BE. Pink1 suppresses alpha-synuclein-induced phenotypes in a Drosophila model of Parkinson’s disease. Genome. 2008;51:1040–1046. doi: 10.1139/G08-085. [DOI] [PubMed] [Google Scholar]
- 57.Clavier A, Baillet A, Rincheval-Arnold A, Coleno-Costes A, Lasbleiz C, Mignotte B, et al. The pro-apoptotic activity of Drosophila Rbf1 involves dE2F2-dependent downregulation of diap1 and buffy mRNA. Cell Death Dis. 2014;5:e1405. doi: 10.1038/cddis.2014.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H, Chaney S, Roberts IJ, Forte M, Hirsh J. Ectopic G-protein expression in dopamine and serotonin neurons blocks cocaine sensitization in Drosophila melanogaster. Curr Biol. 2000;10:211–214. doi: 10.1016/S0960-9822(00)00340-7. [DOI] [PubMed] [Google Scholar]
- 59.Freeman M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell. 1996;87:651–660. doi: 10.1016/S0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- 60.Staveley BE, Phillips JP, Hilliker AJ. Phenotypic consequences of copper-zinc superoxide dismutase overexpression in Drosophila melanogaster. Genome. 1990;33:867–872. doi: 10.1139/g90-130. [DOI] [PubMed] [Google Scholar]
- 61.Todd AM, Staveley BE. Novel assay and analysis for measuring climbing ability in Drosophila. Drosoph Inf Serv. 2004;87:101–107. [Google Scholar]
- 62.Schneider CA, Rasband WS, Eliceiri KW. NIH image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.M’Angale PG, Staveley BE. Effects of α-synuclein expression in the developing Drosophila eye. Drosoph Inf Serv. 2012;95:85–89. [Google Scholar]
- 64.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, et al. A new bioinformatics analysis tools framework at EMBL–EBI. Nucleic Acids Res. 2010;38:W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]


