Abstract
Angelica gigas Nakai (AGN) root ethanol extract exerts anti-cancer activity in several allograft and xenograft models. Here we examined its chemopreventive efficacy through gavage administration against primary carcinogenesis in the transgenic adenocarcinoma of mouse prostate (TRAMP) model. Male C57BL/6 TRAMP mice and wild type littermates were given a daily gavage (5 mg/mouse, Monday-Friday) of AGN or vehicle, beginning at 8 weeks of age (WOA). All mice were terminated at 24 WOA, unless earlier euthanasia was necessitated by large tumors. Whereas AGN-treated TRAMP mice decreased dorsolateral prostate lesion growth by 30% (P = 0.009), they developed fewer and smaller neuroendocrine-carcinomas (NE-Ca) (0.12 g/mouse) than vehicle-treated counterparts (0.81g/mouse, P = 0.037). We analyzed the proteome and transcriptome of banked NE-Ca to gain molecular insights. Angiogenesis-antibody array detected a substantial reduction in AGN-treated NE-Ca of basic fibroblast growth factor (FGF2), an angiogenesis stimulator. iTRAQ proteomics plus data mining suggested changes of genes upstream and downstream of FGF2 functionally consistent with AGN inhibiting FGF2/FGFR1 signaling at different levels of the transduction cascade. Moreover, AGN upregulated mRNA of genes related to immune responses, restored expression of many tumor suppressor genes, and prostate function and muscle differentiation genes. On the other hand, AGN down-regulated mRNA of genes related to neuron signaling, oncofetal antigens, inflammation and mast cells, Wnt signaling, embryonic morphogenesis, biosynthesis, cell adhesion, motility, invasion and angiogenesis. These changes suggest not only multiple cancer cell targeting actions of AGN but also impact on the tumor microenvironments such as angiogenesis, inflammation and immune surveillance.
Keywords: Angelica gigas Nakai, TRAMP model, prostate cancer, proteomics, microarray, transcriptomics
Introduction
Prostate cancer (PCA) is the second leading cause of cancer death in American men. It has been estimated that there will be some 28,000 deaths per year due to PCA in the United States [1]. Chemoprevention using naturally-occurring or synthetic chemicals is considered as a plausible approach to delay, block, or even reverse carcinogenesis and progression of PCA owing to its long latency and slow growth. Angelica gigas Nakai (AGN) is a traditional medicinal herb used in Korea [2]. Its dried root extract is marketed as a dietary supplement for pain relief and memory health in the United States and globally. Pyranocoumarin compound decursin (D) and its isomer decursinol angelate (DA) are the major non-polar chemical components of the alcoholic extract of the root of AGN [3, 4]. AGN extract as well as D and DA have been reported to exert neuro-protective and pain-killing activities in animal models as well as anti-cancer activities in several allograft and xenograft models (see our comprehensive review [2]).
We have earlier identified D and DA as novel anti-androgen signaling compounds [5, 6] and documented an in vivo inhibitory effect of AGN ethanol extract on the growth of androgen-independent DU145 and PC3 PCA xenografts [7]. However, AGN efficacy against primary carcinogenesis has yet to be established in any pre-clinical model. Therefore, in this study, we evaluated the effect of gavage administration of the extract of AGN root to inhibit the two lineages of carcinogenesis in the prostate of TRAMP (Transgenic Adenocarcinoma Mouse Prostate) mice [8]: i.e., the androgen receptor (AR)/probasin promoter/T-antigen (TAg)-mediated prostate epithelial atypical hyperplasia formation especially in the dorsal-lateral prostate (DLP), and the TAg-driven AR-independent neuroendocrine carcinomas (NE-Ca) predominantly originating in the ventral prostate (VP) [9–11]. We used a combination of proteomic and transcriptomic approaches to profile molecular changes that might inform potential pharmacodynamic targets in the NE-Ca lineage, owing to ample tumor tissue availability.
Material and Methods
AGN extract
Alcoholic extract of dried AGN root was prepared as described previously by the Kim group [5]. Chemical fingerprinting of AGN by HPLC-UV showed that content of D plus DA in this particular batch of AGN extract was approximately 27%.
Animal experiment
The animal study was approved by the IACUC of University of Minnesota and carried out at the Hormel Institute, Austin, MN. In-house bred (per genotyping protocol as reported before [11, 12]) male TRAMP mice (C57BL/6 background) (n=20 mice per group) and their wild type littermates (n=6 mice per group) were treated with ethanol extract of AGN (5 mg/mouse in 0.5 mL 1% Tween-80) or vehicle (0.5 mL 1% Tween-80) by gavage, 5 days per week, from 8 to 24 weeks of age (WOA). Animals were weighed weekly. Starting 16 weeks of age, TRAMP mice were palpated for abdominal mass indicative of prostate/genito-urinary (GU) tumors. All mice were terminated at 24 weeks of age, unless earlier euthanasia was necessitated by large tumor size.
At necropsy, the GU tract was removed en bloc and weighed. Tumors were dissected and weighed. The prostate lobes from mice without visible tumors were dissected and weighed to 0.1 mg precision. The organs were inspected for metastasis and, if visible, the lesions were dissected, weighed, and fixed in formalin for H&E confirmation of metastasis. Liver, kidney, and other major organs were inspected for health problems. Those mice with confirmed liver diseases were excluded from the final data set.
Misclassified “TRAMP” mice that failed to show TAg staining by immunohistochemistry IHC (see below) in the prostate were re-classified into the appropriate wild-type groups. After these adjustments, the final effective number of mice for the different groups was as follows: TRAMP-vehicle, n = 14; TRAMP-AGN, n=16; wild type mice-vehicle, n=8; and wild-type mice-AGN, n=7.
Histology and Immunohistochemical (IHC) analyses
Sections (5 μm) were cut from paraffin-embedded tumors, dried, and deparaffinized. Sections were stained with hematoxylin and eosin [12]. Immunostaining was done with antibody for TAg (Becton-Dickinson) at 1:100 dilution. Synaptophysin antibody was purchased from BD Biosciences. Normal horse serum was used as negative control. The biotinylated secondary antibody used was rabbit anti-mouse antibody IgG (1:200 in 10% normal rabbit serum; DakoCytomation). The slides were developed in diaminobenzidine and counterstained with a weak solution of hematoxylin. The stained slides were dehydrated and mounted in Permount. Images were captured and analyzed by ImagePro-plus software. Synaptophysin, AR and E-cadherin (epithelial) and vimentin (stromal) expression in all tumors were detected to confirm NE-carcinoma diagnoses as reported before [11, 12].
Reagents and Chemicals
Reagent kits for 4-plex iTRAQ™ were purchased from ABSCIEX (Foster City, CA, USA). Antibody for basic fibroblast growth factor (FGF2) was purchased from Santa Cruz (Santa Cruz, CA, USA). BCA protein quantitation kit was from Pierce (Rockford, IL, USA). Chrysin (internal standard), Tween-80 and ethyl acetate were purchased from Sigma-Aldrich Co. (St. Louis, MO, USA). HPLC grade methanol, ethanol and acetonitrile were from Fisher Scientific (Pittsburgh, PA, USA).
Tumor tissue sampling and protein/RNA extraction
IHC-confirmed NE-Cas from TRAMP mice were subjected to proteomic and transcripteomic analyses by antibody array and LC-MS/MS, and microarray. Four large tumors from control group (out of total 6 tumors), and all three tumors from AGN group were used. Tumors were symmetrically cut into pieces on a bed of dry ice. Within each group, pieces of similar weight from each tumor were combined to a pooled sample. For targeted analysis by antibody array, proteins were extracted using the 1X lysis buffer (RayBiotech Inc., Norcross, GA) according to manufacturer’s instruction. For proteomic analysis using LC-MS/MS, proteins were extracted as reported before [13, 14]. Protein concentration in lysates was determined by BCA method. For microarray analysis and RT-PCR, total RNAs were extracted using RNeasy Mini kit (QIAGEN) [14, 15].
Angiogenesis-targeted Antibody Array
Mouse Angiogenesis Antibody Arrays (glass slide) were purchased from RayBiotech Inc. The analysis of the levels of angiogenesis-related proteins was done according to the manufacture’s instruction. Briefly, 100μg protein from each group was diluted to 100μL with blocking buffer and applied to individual subarray on the glass slide. After final washing, the slide was scanned with Axon GenePix scanner using Cy3 channel (Excitation frequency = 532 nm) at the BioMedical Genomics Center of the University of Minnesota. The median signal intensity for all spots was normalized to the positive control spots on each sub-array and imported into an Excel based RayBio Analysis Tool supplied by the manufacture for further calculations. Each sample was analyzed at least twice using distinct antibody array slides on different days. Western-blot using the same protein extracts for the antibody array was carried out as previously described [10, 13] to validate the key data. Dilution of the primary antibody for FGF2 was 1:100.
iTRAQ Proteomic Analysis
Proteins (84 μg) from each group (in 15 μL lysis buffer) were denatured, reduced, alkylated and digested to peptides as described [13, 14]. The peptides were then labeled with iTRAQ™ Reagents: Tag 115 was used for the control sample whereas Tag 117 for AGN treatment group, respectively. Labeled peptides were mixed, cleaned up by solid phase extraction with MCX cartridge and analyzed by offline LC-MALDI-TOF/TOF at the Mass Spectrometry and Proteomics Facility of the University of Minnesota as reported by us [10, 15]. Briefly, peptide mixture was first fractioned by strong-cation-exchange chromatography and all 14 fractions collected were spotted onto MALDI targets in a 1232-spot format. MS data on all spots were acquired on a 4800 MALDI-TOF/TOF™ analyzer (ABSCIEX). Full scan MS spectra were acquired from 800 – 4000 m/z. Data dependent tandem MS settings included acquisition of the top 30 most intense ion signals per spot. ProteinPilot™ 3.0 software was used for data processing. The search was performed against NCBI database (version 2009–07). The quality of protein identification was further assessed by false-discovery-rate-analysis (FDR) and a 1% global FDR was set as a cut-off value for proteins subjected to quantization. Proteins identified as differentially expressed with p < 0.05 and EF<2, and expression ratio <0.8 or >1.2 against that of the corresponding control [13, 15] were reported here. The final results were subjected to analysis with Ingenuity Pathway Analysis (IPA, version 8.5), a literature based software (Ingenuity Systems, Mountain View, CA).
Microarray Analysis of mRNA transcriptome
The concentration, quality and integrity of the total RNA samples were determined using NanoDrop 8000 Spectrophotometer and Agilent 2100 Bioanalyzer. The mRNA expression profiles of NE-Ca from control and AGN treatment groups were analyzed using both Illumina Mouse WG-6 BeadChip whole genome expression array and Affymetrix GeneChip Mouse Genome 430 2.0 Array. All RNA labeling and hybridization was performed at the BioMedical Genomics Center of the University of Minnesota according to protocols specified by the manufacturers. Each Illumina Mouse WG-6 BeadChip array had six identical subarrays on the same slide, so it allowed the interrogation of six RNA samples in parallel and generates data that could be considered as from six independent microarrays. In our experiment, each of the two pooled samples (control, AGN) was hybridized to two Illumina subarrays as technical replicates. Affymetrix GeneChip Mouse Genome 430 2.0 Array only allowed the hybridization with one sample, so each of the samples was analyzed on one Affymetrix slide separately but the labeling and hybridization for all samples was performed at the same time under fully controlled conditions. Differentially expressed gene sets were subjected to bioinformatic analysis using DAVID (Database for Annotation, Visualization and Integrated Discovery [16]), version 6.7.
Real-time RT-PCR
Total RNA (1 μg) was used for cDNA synthesis in a 20μL reaction system, and 5μl of 1:20 diluted cDNA was used in each 25μl real-time PCR reaction using the Fast Start Universal SYBR Master with ROX (Roche) with an ABI 7500 or 7300 Real Time PCR System (Applied Biosystems) as described before [14, 15]. β-Actin was selected as an internal standard for normalization. Primers specific for each genes are listed in Supplemental Table S1.
Results
Effect of AGN on carcinogenesis outcomes
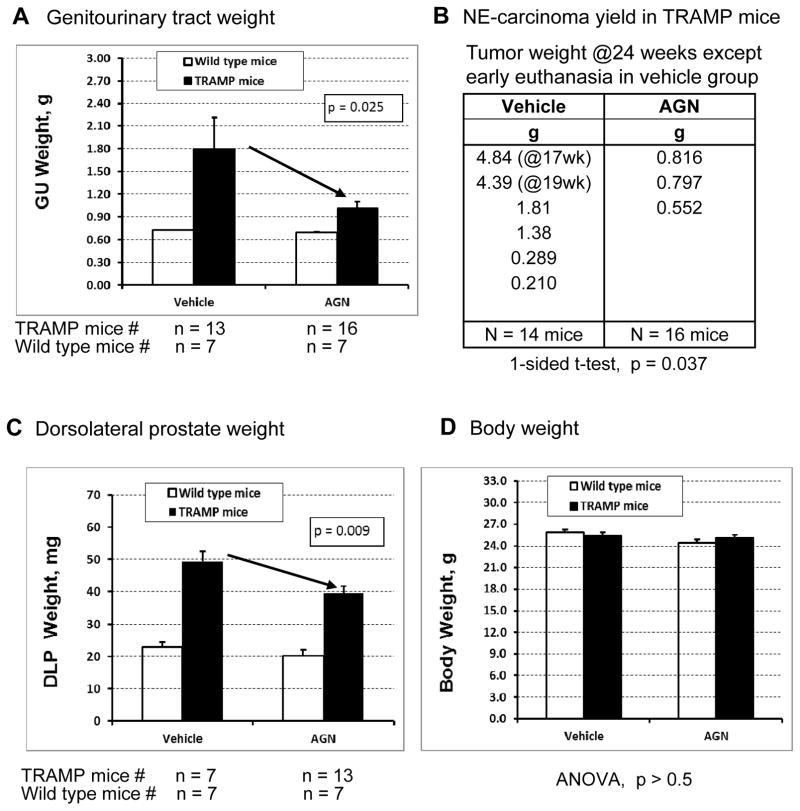
Compared to the vehicle group, AGN treatment reduced the genitourinary (GU) tract weight (P = 0.025, 1-sided t-test) (Fig. 1A) without significantly affecting body weight (Fig. 1D, P > 0.5) and major organs (liver, kidney, testes) in TRAMP mice (P > 0.5) (not shown). AGN-treated TRAMP mice had lower incidence (3 out of 16 vs. 6 out of 14 control mice) and smaller neuroendocrine-like prostate carcinomas (Fig. 1B) (NE-Ca, IHC verified as TAg+, synaptophysin+, AR−, E-cadherin−, see supplemental Figure S1) than vehicle-treated TRAMP mice in that the average tumor burden was 0.81g/mouse in control group (n=14 mice at risk) vs. 0.12 g/mouse in AGN group (n=16 mice at risk) (p = 0.037, 1-sided t-test). Considering the two largest tumors in the vehicle group were collected at 17 and 19 weeks of age (WOA) of the afflicted TRAMP mice when they had to be sacrificed (Fig. 1B), the tumor burden reduction could be under-estimated.
Figure 1.
Efficacy of AGN treatment on two lineages of carcinogenesis in TRAMP model. A) AGN gavage treatment reduced the genitourinary (GU) tract weight (p = 0.025, 1-sided t-test); B) AGN-treated TRAMP mice had lower incidence (3 out of 16 vs. 6 out of 14 control mice) and smaller NE-Ca; C) AGN-treated TRAMP mice underwent less epithelial growth in the dorsolateral prostate (DLP) (p = 0.009, 1-sided t-test); D) AGN treatment did not significantly affect body weight (p> 0.5).
Compared to vehicle-treated TRAMP mice, AGN-treated TRAMP mice underwent less growth in the dorsolateral prostate (DLP) weight (Fig. 1C, p = 0.009, 1-sided t-test), which reflected decreased epithelial lineage lesion expansion as reported before [11, 12]. In another recently completed experiment, we confirmed the substantial suppressing effect of AGN extract prepared by a different procedure on NE-Ca in TRAMP mice bred on-site in Texas Tech University Health Sciences Center, Amarillo, TX [17].
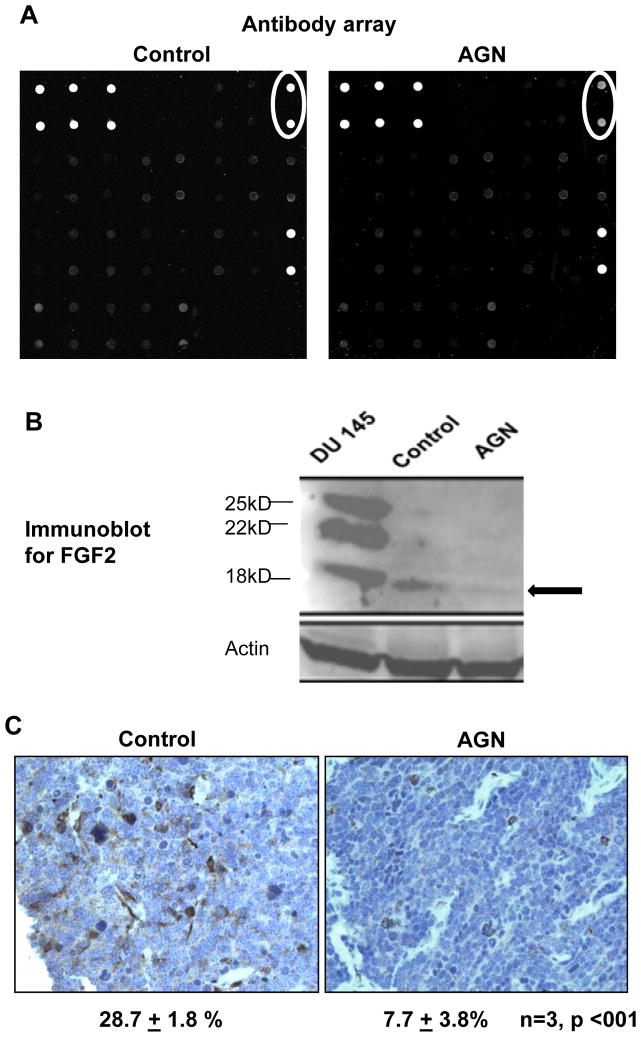
Antibody array revealed decreased FGF2 expression in AGN-treated NE-Ca
Given the extensive vascularity of NE-Ca from TRAMP mice and the reported anti-angiogenic properties of pyranocoumarins [2, 7, 9], we first profiled angiogenesis-related biomarker proteins with a targeted antibody array. Preliminary experiments were performed to optimize the experimental conditions and establish general quality control of the whole platform. We found that a 5 to 10 fold dilution of initial protein lysate with blocking buffer and 100μg protein per sample led to the best signal to background ratio. In order to estimate intra- and inter-slide variations, one sample was applied to two sub-arrays on the same or different slides. The intensity detected from different sub-arrays for all spots showed an excellent correlation and slope of close to unity (for intra-slide, r = 0.996, slope = 0.975; for inter-slides, r = 0.988, slope = 1.048), indicating that the variation could be kept at a minimal level. In addition, the average intensity of all spots was positively correlated with the amount of proteins applied in a linear manner (r = 0.998), further indicating this platform as an acceptable approach for semi-quantitative analysis.
As shown in Fig. 2A, the intensity of the duplicated spots (circled on up-right corner) representing FGF2 in NE-Ca of AGN-treated TRAMP mice was much lower (decreased by 57%) than those for the vehicle-treated NE-Ca. This finding was further confirmed by immunoblot detecting the 18-kD isoform (Fig. 2B, marked by black arrow) and immunohistochemical staining (Fig. 2C). The cytosol/membrane brown staining patterns suggested a possible vascular and stromal cell origin of FGF2 in NE-Ca. Quantitating % of positive-stained cells in 3 NE-Ca’s from each group showed a reduction to 7.7% from 28.7%.
Figure 2.
A) Angiogenesis antibody array detection of FGF2 (circles) in NE-Ca lysate of vehicle-treated control TRAMP mice and AGN-treated mice; B) Detection by Western blot of FGF2 isoforms in NE-Ca lysate of control TRAMP mice and AGN-treated mice. DU145 cell extract was used as a positive control for FGF2 isoforms. Black arrow marks the detected FGF2 isoform. C) Representative immunohistochemical detection of FGF2 in NE-Ca from control and AGN-treated mice. The cytosol/membrane staining patterns suggest a possible vascular, stromal cell and non NE-Ca origin of FGF2. The % of IHC-positive cells are shown as mean ± sem, One-sided t-test.
The FGF gene family consists of more than 20 different genes encoding related polypeptide mitogens, some of which play an important role in the growth and maintenance of the normal prostate [18]. It was reported that FGF2 was significantly increased in PCA when compared with uninvolved prostate. Its primary receptor, FGFR1, was overexpressed in cancerousepithelial cells in a subset of PCA and such overexpression correlated with poor differentiation [19]. N. Greenberg’s group reported differential expression patterns of FGF2 isoforms in the prostates of TRAMP mice [20] such that the expression of the 25-kDa isoform was 2-fold higher in the prostatic intraepithelial neoplasia (PIN), and well- and moderate-differentiated (WD and MD) tumors than in normal prostates. The expression of the 22-kDa isoform was not elevated in PIN lesions, but was observed to be increased in all the tumors. Interestingly, the low molecular weight FGF2 (18 kDa) was only expressed in poorly-differentiated (PD) and castration-resistant tumors (i.e., NE-Ca lineage). In our hands, only the 18kDa-isoform of FGF2 could be detected in the NE-Ca by immunoblot (Fig. 2B). The important role of FGF2 in PCA was further investigated genetically in FGF2 knockout mice [21] by Polnaszek et al. They found that even inactivation of one FGF2 allele resulted in increased mouse survival, less PD phenotype in primary tumors and a decrease in metastasis. R. Agarwal’s group reported that the chemopreventive effect of dietary silibinin against TRAMP carcinogenesis was associated with decreased plasma FGF2 level [22].
Profiling the TRAMP NE-Ca proteome and the effect of AGN treatment
We subsequently profiled the effect of AGN treatment on the proteome of the TRAMP NE-Ca using an unbiased approach by iTRAQ labeling [10, 13, 14]. In total, we identified 1379 proteins expressed in the NE-Ca of TRAMP mice. Among them, 318 and 585 proteins were identified at global FDRs of 1% and 5% respectively. Based on the criteria described in Methods, 5 proteins were significantly down-regulated and 5 proteins were significantly up-regulated by AGN treatment (Table 1). AGN treatment reversed the direction of changes of a number of proteins associated with TRAMP carcinogenesis reported by us previously [10]. For example, cysteine and glycine-rich protein 1 (Csrp1) was decreased in the DLP of TRAMP mice at 18 WOA [10], but it was up-regulated by AGN treatment in the NE-Ca by 49%. Myosin-11 (Myh11, SMMHC), a “good” stromal marker [23], was significantly decreased in both DLP and ventral prostate (VP) of TRAMP mice at 18 WOA [10] and was increased in AGN-treated NE-Ca by 65%. Actin alpha cardiac muscle 1 (Actc1) is a protein expressed in stromal cells. It was reported that mRNA level of Actc1 was four times lower in human prostate cancer tissues than normal prostate [24]. Integrative genomic profiling of PCA further supported the clinical significance of Actc1 [25] such that the mRNA level of Actc1 was significantly decreased in localized PCA compared to matched normal tissue, whereas it was further decreased in metastases compared to localized tumors (25). We found that Actc1 protein level was increased more than 130% and myosin-4 nearly 3 folds, respectively, in AGN-treated NE-Ca (Table 1). Their mRNA levels were up-regulated by AGN as indicated by microarray and real-time RT-PCR (described below in Fig. 3).
Table 1.
Proteins modulated by AGN treatment in TRAMP NE-Ca detected by iTRAQ-LC/MS.
| Protein | Expression Ratio
|
|---|---|
| AGN/Control | |
| Serotransferrin (Tf) | 0.62 |
| Serum albumin (Alb) | 0.71 |
| Apolipoprotein A-I (Apoa1) | 0.71 |
| Vimentin (Vim) * | 0.72 |
| Hemopexin (Hpx) | 0.77 |
| Phosphoserine aminotransferase (Psat1) | 1.28 |
| Cysteine and glycine-rich protein 1 (Csrp1)* | 1.49 |
| Myosin-11 (Myh11)* | 1.65 |
| Actin, alpha cardiac muscle 1 (Actc1)* | 2.31 |
| Myosin-4 (Myh4)* | 3.97 |
Concordant changes at mRNA level have been demonstrated by real-time PCR and/or microarray
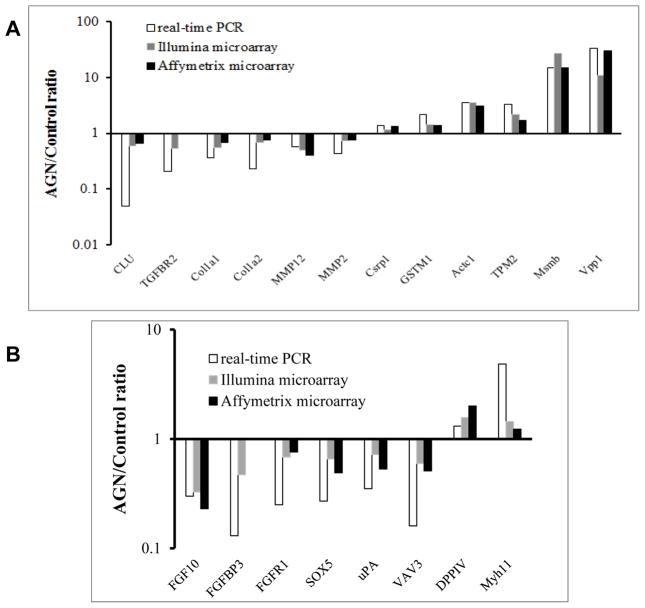
Figure 3.
Comparison of microarray platforms for mRNA expression detection with real-time RT-PCR. (A) Genes chosen based on their literature-documented functional significance in invasion, epithelial-mesenchymal transition and cancer progression as well as iTRAQ-detected proteomic changes; (B) Genes involved in FGF signaling pathway.
Vimentin is a major constituent of the intermediate filament family of proteins. As a marker for epithelial-mesenchymal transition (EMT), the overexpression of vimentin in cancer correlates well with accelerated tumor growth, invasion, and poor prognosis. It is being recognized as a potential molecular target for cancer therapy and prevention [26]. We found that the protein level of vimentin was decreased (28%) in AGN-treated NE-Ca, implying that anti-EMT might contribute to its efficacy. In addition, several major blood proteins (e.g. albumin and serotransferrin) were all decreased in AGN-treated group. Since the possible bias in protein loading across the samples has been minimized by dividing all the “raw” protein ratios to the average iTRAQ ratio throughout the whole experiment [13], the lower abundance of blood proteins in treated group could be a reflection of the reported anti-angiogenesis effect of AGN and decursin and its in vivo metabolite decursinol [7, 27].
Profiling the effect of AGN treatment on NE-Ca transcriptome
The integrity of total RNA extracted from TRAMP NE-Ca was adequate according to the data generated on Agilent 2100 Bioanalyzer (Supplemental Figure S2A). Two microarray platforms from Illumina and Affymetrix were used to profile the in vivo effects of AGN on the global mRNA expression levels. Each of the two pooled samples was hybridized to Illumina microarray in duplicate and the signal intensities for each probe acquired from two hybridization correlated highly (r = 0.9979, 0.9981 and slope = 0.9933, 1.043 for control, and AGN groups, respectively. See Supplemental Figure S2B for data from control group). This indicated very reproducible RNA amplification and hybridization conditions. The results from two microarray platforms (Illumina and Affymetrix) were highly correlated (data not shown).
We tested the correlation of expression changes detected by these microarray platforms with real time RT-PCR as a validation exercise. We chose a number of genes of varying extent of modulation by AGN detected by the Illumina and Affymetrix platforms. The choice of genes was based on our earlier reported proteomic and literature-reported TRAMP-associated gene alterations (Fig. 3A) and FGF-signaling axis suggested by the antibody array profiling above (Fig. 3B) (numerical data are presented in Supplemental Table 2). The results indicated that the microarray platforms often underestimated mRNA abundance detected by RT-PCR. Therefore, to focus on significant mRNA changes, we chose 2-fold change on microarray platforms as threshold for quantifiable gene expression landscape pattern changes. Using an up-regulation ratio of AGN/vehicle >2 or a reduction ratio of AGN/vehicle <0.5, we identified 106 up-regulated genes/ESTs (Table 2) and 124 down-regulated genes in AGN-treated NE-Ca (Table 3).
Table 2.
mRNAs up-regulated by AGN for more than two folds in TRAMP NE-Ca detected by Illumina microarray.
| GENE SYMBOL | DEFINITION | Ratio
|
Functional category | |
|---|---|---|---|---|
| AGN/Control | ||||
| Rank# | AGN-Up-regulated genes | |||
| Up29 | Abo | ABO blood group (transferase A, transferase B) | 3.31 | Cell adhesion |
| Up23 | Abpb | androgen binding protein beta | 3.56 | Prostate protein |
| Up63 | Acss1 | acyl-CoA synthetase short-chain family member 1, nuclear gene encoding mitochondrial protein,. | 2.30 | Mitochondrial energetics |
| Up5 | Acta1 | actin, alpha 1, skeletal muscle | 9.24 | Muscle |
| Up26 | Actc1 | actin, alpha, cardiac muscle 1 | 3.38 | Muscle |
| Up14 | Adh1 | alcohol dehydrogenase 1 (class I) | 5.08 | Behavior |
| Up35 | Amn | amnionless | 3.03 | Lipid digestion_ mobilization_ and transport |
| Up102 | Ankrd1 | ankyrin repeat domain 1 (cardiac muscle) | 2.04 | Muscle |
| Up8 | Apof | apolipoprotein F | 7.70 | Lipid transport |
| Up66 | Atad4 | ATPase family, AAA domain containing 4 | 2.26 | |
| Up50 | Atp2a1 | ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 | 2.47 | Ion transport |
| Up7 | Azgp1 | alpha-2-glycoprotein 1, zinc | 7.98 | Immunity |
| Up40 | B4galnt2 | beta-1,4-N-acetyl-galactosaminyl transferase 2 | 2.88 | Cell adhesion |
| Up51 | Bhlhb8 | basic helix-loop-helix domain containing, class B, 8 | 2.46 | Muscle, Calcium signaling |
| Up68 | Car12 | carbonic anyhydrase 12 | 2.23 | |
| Up70 | Casz1 | castor homolog 1, zinc finger (Drosophila) | 2.22 | |
| Up37 | Cbr2 | carbonyl reductase 2 | 3.00 | |
| Up94 | Cer1 | cerberus 1 homolog (Xenopus laevis) | 2.10 | Proliferation Suppressor |
| Up44 | Chrna4 | cholinergic receptor, nicotinic, alpha polypeptide 4 | 2.76 | Synaptic transmission |
| Up49 | Cntnap4 | contactin associated protein-like 4 | 2.62 | |
| Up87 | Cplx1 | complexin 1 | 2.14 | Synaptic transmission |
| Up57 | Csmd3 | CUB and Sushi multiple domains 3 | 2.37 | |
| Up11 | Cyp2f2 | cytochrome P450, family 2, subfamily f, polypeptide 2 | 6.13 | Drug Metabolism |
| Up83 | Dab1 | disabled homolog 1 (Drosophila), transcript variant 2 | 2.16 | Cell adhenison |
| Up17 | Dbx1 | developing brain homeobox 1 | 4.07 | Behavior |
| Up2 | Defb50 | defensin beta 50 | 22.39 | Immunity |
| Up98 | Egf | epidermal growth factor | 2.06 | Epithelial differentiation |
| Up92 | Erdr1 | erythroid differentiation regulator 1 | 2.11 | Epithelial differentiation |
| Up103 | Foxp2 | forkhead box P2, transcript variant 2,. | 2.04 | Muscle |
| Up28 | Fxyd3 | FXYD domain-containing ion transport regulator 3 | 3.31 | Ion transport |
| Up43 | Gata3 | GATA binding protein 3 | 2.77 | Proliferation Suppressor |
| Up32 | Gjd2 | gap junction protein, delta 2 | 3.07 | Tumor suppressor |
| Up95 | Golm1 | golgi membrane protein 1, transcript variant 2 | 2.10 | |
| Up72 | Gpm6a | glycoprotein m6a | 2.21 | |
| Up93 | Gpr22 | G protein-coupled receptor 22 | 2.11 | GPCR signaling pathway |
| Up42 | Grb7 | growth factor receptor bound protein 7 | 2.80 | Tumor suppressor |
| Up82 | Grm7 | glutamate receptor, metabotropic 7 | 2.16 | Synaptic transmission |
| Up31 | Gstt3 | glutathione S-transferase, theta 3 | 3.24 | Tumor suppressor |
| Up16 | Hamp2 | hepcidin antimicrobial peptide 2 | 4.38 | Immunity |
| Up38 | Hopx | HOP homeobox | 2.97 | hop pathway in cardiac development |
| Up97 | Igh-6 | immunoglobulin heavy constant mu | 2.06 | Immunity |
| Up10 | Igh-VJ558 | immunoglobulin heavy chain (J558 family) | 6.50 | Immunity |
| Up13 | Igk-C | immunoglobulin kappa constant | 5.38 | Immunity |
| Up24 | Igk-V5 | immunoglobulin kappa chain variable 5 (V5 family) | 3.47 | Immunity |
| Up15 | Igl-V1 | immunoglobulin lambda variable 1 | 4.67 | Immunity |
| Up106 | Irf6 | interferon regulatory factor 6 | 2.01 | Epithelial differentiation |
| Up78 | Jph4 | junctophilin 4, transcript variant a | 2.18 | |
| Up69 | Krt23 | keratin 23 | 2.22 | intermediate filament cytokeratin |
| Up67 | Lgals7 | lectin, galactose binding, soluble 7 | 2.25 | Cell adhenison |
| Up101 | Lingo2 | leucine rich repeat and Ig domain containing 2 | 2.04 | Immunity |
| Up74 | Liph | lipase, member H, transcript variant 2 | 2.20 | Lipid metabolism |
| Up6 | LOC546006 | similar to deleted in malignant brain tumors 1 | 9.15 | Tumor suppressor |
| Up33 | Lrrc26 | leucine rich repeat containing 26 | 3.06 | Ion transport |
| Up34 | Mb | myoglobin | 3.05 | Muscle |
| Up81 | Mist1 | muscle, intestine and stomach expression 1 | 2.16 | Muscle |
| Up1 | Msmb | beta-microseminoprotein | 27.64 | Prostate protein |
| Up30 | Myh8 | myosin, heavy polypeptide 8, skeletal muscle, perinatal | 3.27 | Muscle |
| Up3 | Myl1 | myosin, light polypeptide 1 | 12.99 | Muscle |
| Up62 | Myl3 | myosin, light polypeptide 3 | 2.32 | Muscle |
| Up79 | Mylpf | myosin light chain, phosphorylatable, fast skeletal muscle | 2.18 | Muscle |
| Up84 | Myrip | myosin VIIA and Rab interacting protein | 2.15 | Muscle |
| Up100 | Nmu | neuromedin U | 2.05 | GPCR signaling pathway |
| Up46 | Odz4 | odd Oz/ten-m homolog 4 (Drosophila) | 2.65 | |
| Up86 | Oxr1 | oxidation resistance 1, transcript variant 1 | 2.14 | |
| Up12 | Papss2 | 3′-phosphoadenosine 5′-phosphosulfate synthase 2 | 5.66 | Sulfur metabolism |
| Up19 | Pdzk1 | PDZ domain containing 1 | 3.80 | Ion transport |
| Up27 | Pdzrn4 | PDZ domain containing RING finger 4 | 3.35 | Ion transport |
| Up48 | Pou4f1 | POU domain, class 4, transcription factor 1 | 2.62 | Cell migration |
| Up58 | Ppargc1a | peroxisome proliferative activated receptor, gamma, coactivator 1 alpha | 2.37 | Drug Metabolism |
| Up64 | Ppp1r1b | protein phosphatase 1, regulatory (inhibitor) subunit 1B | 2.29 | GPCR signaling pathway |
| Up85 | Prima1 | proline rich membrane anchor 1 | 2.15 | Synaptic transmission |
| Up45 | Prlr | prolactin receptor | 2.74 | Epithelial differentiation |
| Up54 | Prom2 | prominin 2, transcript variant 2 | 2.41 | |
| Up80 | Ptprz1 | protein tyrosine phosphatase, receptor type Z, polypeptide 1 | 2.16 | Tumor suppressor |
| Up56 | Rab25 | RAB25, member RAS oncogene family | 2.39 | Tumor suppressor |
| Up89 | Rasd2 | RASD family, member 2 | 2.12 | Behavior |
| Up36 | Rcan2 | regulator of calcineurin 2, transcript variant 1 | 3.03 | Immunity |
| Up90 | Rnf182 | ring finger protein 182 | 2.12 | Immunity |
| Up96 | Runx1t1 | runt-related transcription factor 1; translocated to, 1 (cyclin D-related) | 2.07 | |
| Up39 | Scn3a | sodium channel, voltage-gated, type III, alpha | 2.90 | Ion transport |
| Up91 | Sct | secretin | 2.11 | GPCR signaling pathway |
| Up60 | Slc12a2 | solute carrier family 12, member 2 | 2.35 | Ion transport |
| Up22 | Slc17a8 | solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 8 | 3.57 | Ion transport |
| Up25 | Slc22a1 | solute carrier family 22 (organic cation transporter), member 1 | 3.38 | Ion transport |
| Up71 | Slc26a4 | solute carrier family 26, member 4 | 2.21 | Ion transport |
| Up47 | Slc38a11 | solute carrier family 38, member 11 | 2.65 | Ion transport |
| Up53 | Slc45a3 | solute carrier family 45, member 3 | 2.41 | Ion transport |
| Up41 | Slit2 | slit homolog 2 (Drosophila) | 2.82 | |
| Up61 | Smoc1 | SPARC related modular calcium binding 1 | 2.35 | Cell adhenison |
| Up104 | Spink2 | serine peptidase inhibitor, Kazal type 2 | 2.04 | Prostate protein |
| Up9 | Spink3 | serine peptidase inhibitor, Kazal type 3 | 7.37 | Prostate protein |
| Up77 | Spock2 | sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2 | 2.19 | Cell adhenison |
| Up76 | Tacstd2 | tumor-associated calcium signal transducer 2 | 2.20 | Motility Suppresor |
| Up105 | Tbx3 | T-box 3, transcript variant 1 | 2.01 | Epithelial differentiation |
| Up88 | Tcfcp2l1 | transcription factor CP2-like 1 | 2.13 | Epithelial differentiation |
| Up18 | Tecta | tectorin alpha | 3.95 | Mechanical Sensory perception |
| Up99 | Tmprss13 | transmembrane protease, serine 13 | 2.05 | |
| Up20 | Tnnc2 | troponin C2, fast | 3.62 | Muscle |
| Up21 | Tnni2 | troponin I, skeletal, fast 2 | 3.58 | Muscle |
| Up73 | Tox | thymocyte selection-associated high mobility group box | 2.21 | Immunity |
| Up75 | Tpm2 | tropomyosin 2, beta | 2.20 | Muscle |
| Up55 | Treh | trehalase (brush-border membrane glycoprotein) | 2.41 | Metabolism of carbohydrates |
| Up52 | Trhr | thyrotropin releasing hormone receptor | 2.45 | Calcium signaling |
| Up65 | Tspan8 | tetraspanin 8 | 2.27 | |
| Up59 | Upk1b | uroplakin 1B | 2.36 | Epithelial differentiation |
Table 3.
mRNAs down-regulated by AGN for more than two folds in TRAMP NE-Ca detected by Illumina microarray.
| DEFINITION | Ratio
|
Functional category |
|---|---|---|
| AGN/Control | ||
| AGN-down regulated genes | ||
| V-set and transmembrane domain containing 2A | 0.098 | Onco-fetal antigen |
| carboxypeptidase A3, mast cell | 0.137 | Inflammation |
| zinc finger protein 312 | 0.141 | Neuron signaling |
| synaptotagmin IV | 0.143 | Neuron signaling |
| homeo box D10 | 0.169 | Neuron differentiation |
| chymase 2, mast cell | 0.180 | Inflammation |
| corin | 0.184 | Neuron differentiation |
| small proline-rich protein 2A2 | 0.201 | Epithelium development |
| sclerostin domain containing 1 | 0.212 | Wnt signaling |
| carbonic anhydrase 4 | 0.222 | Oncogene, metabolic |
| distal-less homeobox 6 | 0.240 | Embryonic morphogenesis |
| otopetrin 1, transcript variant b | 0.241 | Embryonic morphogenesis |
| somatostatin receptor 2, transcript variant 2 | 0.243 | Neuron differentiation |
| TGFB-induced factor homeobox 1 | 0.247 | Proliferation |
| mast cell protease 6 | 0.247 | Inflammation |
| mast cell protease 4 | 0.261 | Inflammation |
| UDP galactosyltransferase 8A | 0.266 | Biosynthesis |
| transmembrane protein 100 | 0.273 | Neuron differentiation |
| chymase 1, mast cell | 0.282 | Inflammation |
| 1-acylglycerol-3-phosphate O-acyltransferase 9, transcript variant 1 | 0.288 | Biosynthesis |
| regulatory factor X, 4 (influences HLA class II expression), transcript variant 1 | 0.297 | Biosynthesis |
| cytochrome P450, family 19, subfamily a, polypeptide 1 | 0.301 | Drug metabolism |
| a disintegrin and metallopeptidase domain 23 | 0.302 | Cell adhesion |
| Fez family zinc finger 2 | 0.304 | Neuron differentiation |
| AHNAK nucleoprotein (desmoyokin), transcript variant 1 | 0.307 | Invasion |
| R-spondin 3 homolog (Xenopus laevis) | 0.308 | Wnt signaling |
| heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 | 0.309 | Biosynthesis |
| OCIA domain containing 2 | 0.320 | Oncogene |
| ankyrin 2, brain, transcript variant 3 | 0.324 | Neuron signaling |
| fibroblast growth factor 10 | 0.326 | Angiogenesis |
| G protein-coupled receptor 149 | 0.340 | Cell signaling |
| lumican | 0.347 | Cell signaling |
| adenylate cyclase 2 | 0.350 | Cell signaling |
| transmembrane protein 132E | 0.351 | Cell signaling |
| glutathione peroxidase 3, transcript variant 2 | 0.359 | Oncogene |
| basic helix-loop-helix domain containing, class B5 | 0.359 | Neuron differentiation |
| annexin A1 | 0.361 | Lipid transport |
| heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 | 0.362 | Biosynthesis |
| RAD52 motif 1 | 0.363 | DNA repair |
| mab-21-like 2 (C. elegans) | 0.371 | Embryonic morphogenesis |
| LIM and calponin homology domains 1 | 0.372 | Cell Adhesion |
| secreted frizzled-related protein 1 | 0.375 | Wnt signaling |
| thrombospondin 2 | 0.375 | Angiogenesis |
| crystallin, mu | 0.384 | hormone metabolic process |
| tachykinin receptor 1 | 0.388 | Neuron signaling |
| major facilitator superfamily domain containing 2 | 0.399 | |
| G protein-coupled receptor 120 | 0.400 | Cell signaling |
| short stature homeobox 2 | 0.406 | Embryonic morphogenesis |
| SRY-box containing gene 21 | 0.408 | Embryonic morphogenesis |
| eosinophil-associated, ribonuclease A family, member 2 | 0.410 | Inflammation |
| purinergic receptor P2Y, G-protein coupled 1 | 0.410 | Neuron signaling |
| trophoblast glycoprotein | 0.411 | Onco-fetal antigen |
| regulator of G-protein signalling 7 binding protein | 0.412 | Cell signaling |
| a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 9 | 0.414 | Invasion |
| GPI-anchored HDL-binding protein 1 | 0.415 | Lipid transport |
| progressive ankylosis | 0.417 | |
| quiescin Q6 sulfhydryl oxidase 1, transcript variant 2 | 0.417 | hormone metabolic process |
| gene trap locus 2 | 0.419 | |
| retinol binding protein 4, plasma | 0.422 | hormone metabolic process |
| dickkopf homolog 3 (Xenopus laevis) | 0.422 | Wnt signaling |
| adhesion molecule with Ig like domain 2 | 0.429 | Cell adhesion |
| solute carrier family 38, member 5 | 0.431 | Transport of amino acids/oligopeptides |
| claudin 5 | 0.432 | Cell adhension |
| growth factor receptor bound protein 10 | 0.434 | Cell signaling |
| calcitonin receptor, transcript variant a | 0.437 | Neuron signaling |
| Fc receptor, IgE, high affinity I, alpha polypeptide | 0.437 | Immunity |
| nucleoredoxin | 0.438 | Wnt signaling |
| decorin | 0.441 | sulfur metabolic process |
| endothelin receptor type B | 0.442 | Neuron differentiation |
| engrailed 1 | 0.444 | Neuron differentiation |
| heparan sulfate (glucosamine) 3-O-sulfotransferase 1 | 0.445 | heparan sulfate biosynthesis |
| insulin-like growth factor binding protein 3 | 0.446 | Anti-proliferation |
| aminolevulinic acid synthase 2, erythroid | 0.447 | Biosynthesis |
| transthyretin | 0.448 | Extracellular matrix organization |
| keratin 17 | 0.451 | regulation of developmental process |
| procollagen, type VI, alpha 1 | 0.451 | Cell adhension |
| gap junction membrane channel protein alpha 7 | 0.453 | Synaptic transmission |
| lymphoid enhancer binding factor 1 | 0.454 | Wnt signaling |
| sulfatase 1 | 0.454 | sulfur metabolic process |
| peptidase inhibitor 16 | 0.457 | |
| ets variant gene 1 | 0.459 | Neuron differentiation |
| insulin-like growth factor 2 | 0.459 | Proliferation |
| proprotein convertase subtilisin/kexin type 2 | 0.459 | hormone metabolic process |
| regulated endocrine-specific protein 18 | 0.463 | |
| CXXC finger 4 | 0.463 | Wnt signaling |
| diacylglycerol kinase, beta | 0.463 | Cell signaling |
| PDZ domain containing RING finger 3 | 0.464 | Neuron differentiation |
| kininogen 1 | 0.464 | GPCR ligand binding |
| claudin 3 | 0.465 | Cell adhension |
| eyes absent 1 homolog (Drosophila) | 0.465 | Embryonic morphogenesis |
| hemoglobin alpha, adult chain 1 | 0.465 | Angiogenesis |
| F-box protein 2 | 0.469 | Ubiquitin mediated proteolysis |
| G protein-coupled receptor 158 | 0.469 | Cell signaling |
| tachykinin 1 | 0.469 | GPCR ligand binding |
| ISL1 transcription factor, LIM/homeodomain | 0.469 | Neuron differentiation |
| fibroblast growth factor binding protein 3 | 0.471 | Angiogenesis |
| inhibitor of DNA binding 3 | 0.471 | Neuron differentiation |
| solute carrier family 15, member 2 | 0.472 | Transport of inorganic cations/anions and amino acids/oligopeptides |
| Paraneoplastic antigen MA3 | 0.473 | |
| UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 2 | 0.474 | Biosynthesis |
| hyaluronan synthase 2 | 0.475 | Biosynthesis |
| aquaporin 4 | 0.478 | Water transport |
| protein tyrosine phosphatase, receptor type, O | 0.479 | Cell signaling |
| regulator of G-protein signaling 17 | 0.483 | Cell signaling |
| tescalcin | 0.484 | Biosynthesis |
| nuclear factor I/B | 0.487 | Proliferation |
| stanniocalcin 1 | 0.488 | Cellular homeostasis |
| lipopolysaccharide binding protein | 0.489 | Inflammation |
| procollagen, type VI, alpha 2 | 0.490 | Cell adhension |
| transcription factor-like 5 (basic helix-loop-helix) | 0.490 | Proliferation |
| splA/ryanodine receptor domain and SOCS box containing 1 | 0.491 | Ubiquitination & Proteasome degradation |
| gap junction membrane channel protein alpha 1 | 0.491 | embryonic development |
| hemoglobin, beta adult major chain | 0.492 | Angiogenesis |
| microfibrillar-associated protein 4 | 0.493 | Extracellular matrix organization |
| Y box protein 3 | 0.495 | RalA downstream regulated genes |
| microfibrillar-associated protein 2 | 0.495 | Extracellular matrix organization |
| netrin G1 | 0.497 | Neuron differentiation |
| engulfment and cell motility 1, ced-12 homolog (C. elegans), transcript variant 2 | 0.497 | Motility |
| dual specificity phosphatase 23 | 0.497 | Oncogene |
| angiopoietin-like 4 | 0.498 | Angiogenesis |
| transmembrane protein 132C, transcript variant 4 | 0.499 | |
| zinc finger protein 533 | 0.499 | |
| matrix metallopeptidase 12 | 0.499 | Invasion |
| homeo box D9 | 0.500 | Neuron differentiation |
AGN up-regulated genes
Prominent among up-regulated gene categories were those involved in immune responses (Table 2, Up2, Up7, Up10, Up13, Up15, Up16, Up24, Up36, Up73, Up90, Up97, Up101); prostate function/differentiation (Table 2, Up1, Up4, Up9, Up23, Up104); muscle-related change (Table 2, Up3, Up5, Up20, Up21, Up30, Up62, Up79, Up102, Up103); ion transport (Table 2, Up19, Up22, Up27, Up28, Up33, Up39, Up47, Up50, Up53, Up60, Up71), tumor and proliferation suppressors (Table 2, Up6, Up31, Up32, Up42, Up43, Up56, Up76, Up80).
The immunity category of genes was particularly noteworthy. For example, Up2 entry Defb50 (defensin beta 50) and related members were substantially induced by AGN treatment. Defb50 was induced by AGN for more than 20 folds, whereas Defb1 (defensin beta 1, BD-1) was up-regulated for 5.7 folds by AGN (Affymetrix microarray data). Defensin genes code for a family of 3–4 kDa polycationic peptides, clustered on chromosome 8, and these peptides have been shown to act as antimicrobial agents through disrupting membrane integrity. Clinical specimens had high frequencies of loss of Defb1 in malignant prostatic tissue, while high levels of expression were maintained in adjacent benign regions [28]. The finding was in agreement with the mouse model in which the expression of Defb1 and other defensin members Defb2 and Defb4 were decreased in TRAMP tumors [29, 30]. In addition, ectopical expression of Defb1 in DU145 and PC3 PCA cells resulted in a decrease in cellular growth accomplished by cytolysis and caspase-mediated apoptosis [28]. Azbp1 (zinc alpha-2-glycoprotein 1, Up7) is a 41 kDa soluble protein synthesized by epithelial cells of many tissues including the prostate gland and is present in most body fluids. Its expression was reported to be decreased in TRAMP prostate and tumor at mRNA/protein levels by us [10] and Kela et al [29]. Recent findings indicated that absent/low Azgp1 expression was an independent predictor of recurrence in localized PCA after radical prostatectomy [31, 32]. Since Azgp1 has a major histocompatibility complex-1 (MHC-1)-like fold in its structure, it is reasonable to hypothesize that Azgp1 might be involved in host immune response to tumor by antigen processing and presentation to exert its tumor suppressing effect. These and many other up-regulated changes associated with immune responses suggest a potential promotion of immune surveillance by AGN treatment to inhibit NE-Ca growth.
Other notable changes were suggestive of restoration by AGN of genes inactivated during TRAMP lesion progression (some of which bona fide tumor suppressors). The prostate-specific proteins β-microseminoprotein (Msmb) (Table 2, Up1; also Fig. 3a) and serine peptidase inhibitor Kazal type 3 (Spink3) (Table 2, Up9) were 27.6 fold and 7.4 fold higher in AGN-treated NE-Ca, respectively, and we reported earlier their protein levels to be lower in TRAMP prostate than wild type prostate [10]. GSTM1 (Fig. 3a) and tropomyosin (Tpm2) (Table 2, Up75; also Fig. 3a), which we reported to be down-regulated in TRAMP prostate at protein level [10], and glutathione S-transferase, theta 3 (Table 2, Up31) were among tumor suppressor category of genes increased by AGN. Several other groups have profiled the dynamic changes of mRNA expression during TRAMP carcinogenesis in C57BL/6 TRAMP x FVB F1 mice [29, 33] or C57BL/6 TRAMP mice [30]. In addition to Msmb and Tpm2, they reported that Vpp1 (ventral prostate predominant 1) and Myh11 were down-regulated in TRAMP tumors. We found Vpp1 (Fig. 3a; Table 2 Up4) and Myh11 (Fig. 3b) were significantly induced by AGN treatment in the NE-Ca.
AGN down-regulated genes
In terms of down-regulated genes, notable categories include neuron signaling and differentiation (Table 3, Dn3, Dn4, Dn5, Dn7, Dn13, Dn18, Dn24, Dn29, Dn36, Dn45, Dn51, Dn65, Dn69, Dn70 etc.); oncogenes/oncofetal antigens and proliferation (Table 3, Dn1, Dn10, Dn14, Dn28, Dn35, Dn52, Dn82, Dn106, Dn110, Dn119, etc.); mast cells and inflammation (Table 3, Dn2, Dn6, Dn15, Dn16, Dn19, etc.); Wnt signaling (Table 3, Dn9, Dn26, Dn42, Dn60, Dn67, Dn78, Dn85, etc.); embryonic morphogenesis (Table 3, Dn11, Dn12, Dn40, Dn48, Dn49, Dn90, etc.); biosynthesis (Table 3, Dn17, Dn20, Dn21, Dn27, Dn38, Dn73, Dn100, Dn101, Dn105 etc.); cell adhesion, motility and invasion (Table 3, Dn23, Dn31, Dn32, Dn33, Dn34, Dn41, Dn47, Dn53, Dn54, Dn61, Dn63, Dn64, Dn76, etc.); and angiogenesis and hematopoiesis (Table 3, Dn30, Dn43, Dn96).
The suppressed expression of genes related to neuron signaling and differentiation was not un-expected because of the NE-Ca nature of the analyzed tissues and the significantly reduced NE-Ca burden (Fig. 1). In particular, bioinformatic analyses using different tools identified “neuroactive ligand-receptor interaction” pathway members SSTR2 (somatostatin receptor 2, Dn13), TACR1 (tachykinin receptor 1, Dn45), P2RY1 (purinergic receptor P2Y G-protein coupled 1; Dn51), Calcr (calcitonin receptor, Dn65) and EDNRB (endothelin receptor type B, Dn69). SYT4 (synaptotagmin IV, Dn4), known to play an important role in neurotransmitter secretion, was suppressed by AGN treatment as much as 7 folds (Table 3). Recently, increasing attention has been given to the diagnostic, prognostic and therapeutic utility of NE differentiation of prostate cancer. Lacking androgen receptor, the NE cells are considered one mechanism for hormone-refractory prostate cancer [34]. Clinically, C. Sawyer’s group reported that the expression of SYT4, SSTR2, CALCR and P2RY1 was increased in metastases compared to localized tumors [25]. Cytotoxic somatostatin conjugates that selectively bind to SSTR2 are being developed as novel chemotherapeutic agents [35].
The AGN-rendered suppression of genes in other categories, such as oncogenes/oncofetal antigens and proliferation, Wnt signaling, biosynthesis, cell adhesion, motility and invasion, and angiogenesis and hemopoiesis was consistent with them as potential contributors to the overall efficacy through not only direct action on cancer cells but also impact on their microenvironments. Along this rationale, several genes related to inflammatory mast cells, including mast cell carboxypeptidase A3 (Table 3, Dn2), proteases (Dn16, Mcpt4; Dn15, Mcpt6), mast cell chymases (Dn19, Cma1; Dn6, Cma2) and mast cell surface marker Fcer1a (immunoglobulin epsilon receptor subunit alpha) were substantially suppressed by AGN treatment. Mast cells (MC) are granulocytic immune cells best known for their role in allergy and anaphylaxis. They are implicated in pro-inflammatory responses to allergens but can also contribute to protection against pathogens. Pittoni et al reported that in tumors from TRAMP mice on C57BL/6 background and human patients, MCs were specifically enriched and degranulated in areas of “well-differentiated” adenocarcinoma but not around “poorly differentiated” foci that coexist in the same tumors [36]. Their additional experiments showed that MCs promoted “well-differentiated” adenocarcinoma growth by providing MMP9 but were dispensable for growth of “poorly differentiated” tumors [36]. Consistent with these findings, Morgenbesser et al [33] and Kela et al [29] showed that the expression of another MC surface marker c-kit was increased in TRAMP tumors. Though the histological classification of TRAMP lesions used in their paper was not identical as ours, these findings suggested the role of MC and inflammation in prostatic carcinogenesis and might be suppressed by AGN to contribute to inhibition of NE-Ca growth.
As part of the validation process, we examined by RT-PCR the expression changes of a number of genes known to be involved in prostate cancer aggressiveness, invasion and metastasis, even though their change magnitude did not meet the 2-fold threshold on microarray (Fig. 3A). They included CLU (Clusterin, TRPM2), TGFBR2 (transforming growth factor, β receptor II), Col1a1 and Col1a2 (as well as Col6a1), MMP2 and MMP12 (matrix metallopeptidase) (Fig. 3a). Since these genes play important roles in invasion and metastasis [37–39], the down-regulation of their expression by AGN suggested potential anti-invasion and anti-metastasis actions. Our follow-up experiment showed that AGN treatment decreased TRAMP NE-Ca metastasis to abdominal lymph nodes [17].
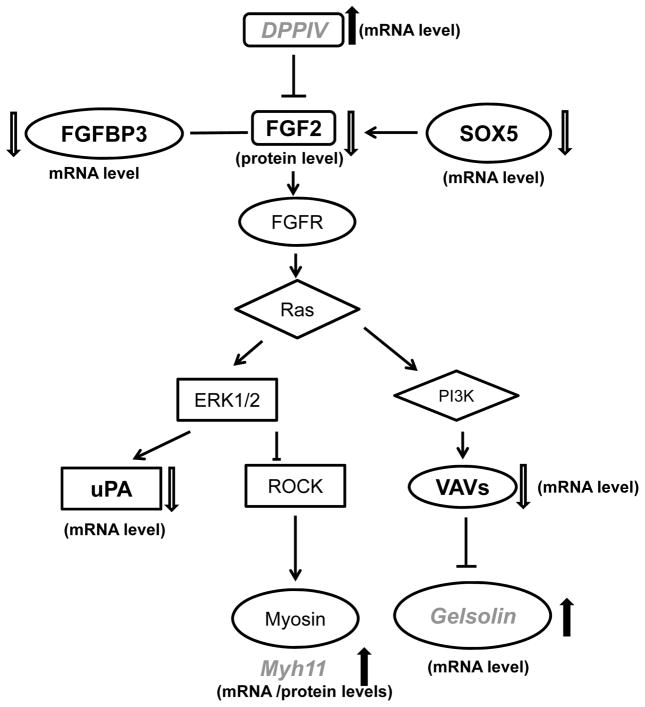
Integration of “omics” data centering on DPP4-FGF2-FGFR1 axis
Since the anti-angiogenesis effect of AGN and its pyranocoumarins has been reported [7, 27], and antibody array detected decreased FGF2 in AGN-treated NE-Ca (Fig. 2), we used FGF2 as the “seed” to integrate the pathway network of differentially expressed proteins and mRNAs associated with AGN treatment by IPA for pathway connections based on gene ontogeny and functionality. Distinct inferred network nodes included Akt, PTEN, PI3K, ERK1/2, VEGF and NF-κB. The identified customized pathways were then overlapped with FGF2 canonical pathways. By manually looking at the effect of AGN on the expression levels of the components in the overlapped pathway, we highlight DPP4-FGF2-FGFR1 axis as one possible mechanistic link to the chemopreventive efficacy of AGN. As illustrated in Fig. 4, the mRNA level of DDP4, which was reported to suppress the expression of FGF2 in DU145 cells [40], was up-regulated by AGN. Activation of ERK1/2 by FGF2 could increase the production of uPA proteinase [40, 41]. Overexpression of transcriptional factor sex determining region Y-box 5 (SOX5) has been reported to be associated with PCA progression and early development of distant metastasis [42]. Vav3, a Rho GTPase guanine nucleotide exchange factor (GEF), was overexpressed in human PCA, particularly in the castration-resistant stage [43]. FGFBP3, a FGF binding protein, could protect FGFs from degradation and present FGFs to high affinity cell surface receptors in an active form [44]. The expression of each of the above genes was down-regulated by AGN treatment (Fig. 3b).
Figure 4.
Integration of signal changes (dark bold, suppressed; soft shaded, increased) induced by AGN extract in TRAMP NE-Ca detected by mRNA microarray, antibody array (AB) and iTRAQ proteomics.
On the other hand, gelsolin and myh11, the “good” stromal markers reportedly down-regulated in both the prostate of 18-week old TRAMP mice and human PCA bio-specimens [10, 23], were up-regulated by AGN (Fig. 3b and microarray data). Functionally the observed changes were consistent with AGN extract inhibiting FGF2-FGFR1 signaling at different levels of the transduction cascade. We confirmed the effect of AGN on the expression of key components in this pathway (FGF2, FGFR1, SOX5, etc.), as well as other biomarkers (TGFBR2, etc.), in the NE-Ca of TRAMP mice at 28 WOA in our recently completed experiment ([17] to be published separately).
In addition, we interrogate the mRNA expression patterns of these genes in the clinical dataset published by C. Sawyers’ group [25] and others archived in Oncomine (www.oncomine.org [45]). DPP4 was dramatically decreased in metastases compared to localized PCA; Myh11 was significantly decreased in localized PCA compared to matched normal tissue and was further decreased in metastases compared to localized tumors by almost 20 folds; so was gelsolin (25). FGFBP3 was marginally increased in metastases [25]. The agreement between animal model and clinical specimens further indicated the translational potential of proposed signaling pathway.
Discussion
Our chemoprevention efficacy study was, to our knowledge, the first to test AGN extract in the TRAMP primary carcinogenesis model. We observed a statistically significant suppression of tumor burden as reflected by genitourinary tract weight (Fig. 1A) and NE-Ca yield and weight (Fig. 1B) and epithelial lesion expansion as represented by the reduction of dorsolateral prostate lobe weight (Fig. 1C). Enabled by the ample quantity of NE-Ca tissues, we subjected the banked tumor tissues to integrative “omics” analyses to seek insights into potential target cellular processes and molecular pathways associated with the in vivo efficacy against NE-Ca growth. Overall, the angiogenesis targeted antibody array (Fig. 2), the iTRAQ proteomic (Table 1) and transcriptomic analyses by microarrays (Fig. 3, Tables 2 and 3) were complementary and non-redundant in their abilities to detect protein/mRNA changes and provided a picture of the “systems biology” molecular changes in the AGN-treated NE-Ca.
Salient findings were the multitudes of changes observed at mRNA and protein levels implicating AGN affecting not only NE-Ca cells with respect to oncogenic signaling and restoration of tumor suppressors, but also tumor microenvironments as reflected by angiogenesis, inflammation, and immune responses. We used FGF2 signaling pathway as an illustrative example to integrate “omics” findings (Fig. 4), identifying inhibition of DPP4-FGF2-FGFR1 axis as one possible pathway associated with the chemopreventive efficacy. This was consistent with previous reports of the anti-angiogenesis activities of decursin and decursinol [7, 27, 46].
However, there are many unanswered questions. First, what are the active chemicals in AGN that afforded the in vivo efficacy against both epithelial and NE-lineages of carcinogenesis. Decursin and DA accounted for approximately 27% of the AGN extract tested in the current work. The “omics” profiling in the current work with NE-Ca provided us “signature” profiles to use for evaluating these compounds in comparison to the AGN extract and its D/DA-depleted counterpart in future studies. Second, what are the relative contributions of affected cellular processes and molecular pathways to overall efficacy? We anticipate time course design can help to address the temporal relationship issues, whereas dose response patterns will discriminate readily modulatable molecules at a low intake dose from those requiring high doses. Such knowledge plus information about active compounds can help design future studies with genetic models to address “cause-effect” and “mediator” relationships of these changes with efficacy.
Supplementary Material
Supplement Table 1. Sequences of Primers for real-time PCR.
Supplement Table 2. Relative gene expression abundance ratio numerical data (AGN/control) detected by real-time RT-PCR vs. the 2 microarray platforms.
Supplemental Figure S1. Representative images of NE-Ca biomarkers. A: T-Antigen; B: Synaptophysin; C: Androgen receptor; D: E-Cadherin. Insert shows enlarged image of NE-Ca staining. Trapped epithelial gland is marked by Epi.
Supplemental Figure S2: A) The integrity of total RNAs extracted from TRAMP NE-Ca analyzed by Agilent 2100 Bioanalyzer. Left to right: size marker; sample lane 1, RNA from vehicle-control; sample lane 3, RNA from AGN treated NE-Ca. B) Correlation of the signal intensities for each probe acquired from two hybridization of RNA from control group (Illumina microarray).
Acknowledgments
Grant supports This work was supported by grants R21AT005383 (Lu) and R01AT007395 (Lu, Xing) from US National Center for Complementary and Alternative Medicine and Korea Government (MEST) grant 2012-0005755 (Kim).
The authors thank Emily Quealy, Katai Nkata and Hormel Institute Animal Facility staff for assistance with mouse breeding and/or genotyping and animal care; Professor Joshua Liao of the Hormel Institute for assistance with histopathology. They thank Mr. Aaron D. Becker and Dr. LeeAnn Higgins of University of Minnesota for “omics” technical support. They thank Dr. Degeng Wang of University of Texas Health Sciences Center, San Antonio, TX for consultation on bioinformatic analyses of microarray data.
Abbreviations
- AGN
Angelica gigas Nakai
- FGF2
basic fibroblast growth factor
- D
decursin
- DA
Decursinol angelate
- NE-Ca
Neuroendocrine carcinoma
- PCA
Prostate cancer
- TRAMP
Transgenic adenocarcinoma mouse prostate
- WOA
Weeks of Age
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA: a cancer journal for clinicians. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Zhang J, Li L, Jiang C, et al. Anti-cancer and Other Bioactivities of Korean Angelica gigas Nakai (AGN) and Its Major Pyranocoumarin Compounds. Anti-cancer agents in medicinal chemistry. 2012;12:1239–1254. doi: 10.2174/187152012803833071. [DOI] [PubMed] [Google Scholar]
- 3.Ahn MJ, Lee MK, Kim YC, Sung SH. The simultaneous determination of coumarins in Angelica gigas root by high performance liquid chromatography-diode array detector coupled with electrospray ionization/mass spectrometry. J Pharm Biomed Anal. 2008;46:258–66. doi: 10.1016/j.jpba.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 4.Lu J, Kim SH, Jiang C, Lee H, Guo J. Oriental herbs as a source of novel anti-androgen and prostate cancer chemopreventive agents. Acta Pharmacol Sin. 2007;28:1365–72. doi: 10.1111/j.1745-7254.2007.00683.x. [DOI] [PubMed] [Google Scholar]
- 5.Jiang C, Lee HJ, Li GX, et al. Potent antiandrogen and androgen receptor activities of an Angelica gigas-containing herbal formulation: Identification of decursin as a novel and active compound with implications for prevention and treatment of prostate cancer. Cancer Research. 2006;66:453–463. doi: 10.1158/0008-5472.CAN-05-1865. [DOI] [PubMed] [Google Scholar]
- 6.Guo J, Jiang C, Wang Z, et al. A novel class of pyranocoumarin anti-androgen receptor signaling compounds. Mol Cancer Ther. 2007;6:907–17. doi: 10.1158/1535-7163.MCT-06-0231. [DOI] [PubMed] [Google Scholar]
- 7.Lee HJ, Lee EO, Lee JH, et al. In vivo anti-cancer activity of Korean Angelica gigas and its major pyranocoumarin decursin. Am J Chin Med. 2009;37:127–42. doi: 10.1142/S0192415X09006722. [DOI] [PubMed] [Google Scholar]
- 8.Greenberg NM, DeMayo F, Finegold MJ, et al. Prostate cancer in a transgenic mouse. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:3439–43. doi: 10.1073/pnas.92.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J, Wang L, Zhang Y, Lü J. Lobe-Specific Carcinogenesis in the Transgenic Adenocarcinoma of Mouse Prostate (TRAMP) Mouse Model. 2013 [Google Scholar]
- 10.Zhang J, Wang L, Zhang Y, et al. Lobe-specific proteome changes in the dorsal-lateral and ventral prostate of TRAMP mice versus wild-type mice. Proteomics. 2011;11:2542–9. doi: 10.1002/pmic.201100008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, Zhang J, Zhang Y, et al. Lobe-specific lineages of carcinogenesis in the transgenic adenocarcinoma of mouse prostate and their responses to chemopreventive selenium. Prostate. 2011;71:1429–40. doi: 10.1002/pros.21360. [DOI] [PubMed] [Google Scholar]
- 12.Wang L, Bonorden MJ, Li GX, et al. Methyl-selenium compounds inhibit prostate carcinogenesis in the transgenic adenocarcinoma of mouse prostate model with survival benefit. Cancer Prevention Research. 2009;2:484–95. doi: 10.1158/1940-6207.CAPR-08-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J, Wang L, Anderson LB, et al. Proteomic profiling of potential molecular targets of methyl-selenium compounds in the transgenic adenocarcinoma of mouse prostate model. Cancer prevention research. 2010;3:994–1006. doi: 10.1158/1940-6207.CAPR-09-0261. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J, Wang L, Li G, et al. Mouse prostate proteomes are differentially altered by supranutritional intake of four selenium compounds. Nutrition and cancer. 2011;63:778–89. doi: 10.1080/01635581.2011.563029. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J, Nkhata K, Shaik AA, et al. Mouse prostate proteome changes induced by oral pentagalloylglucose treatment suggest targets for cancer chemoprevention. Current cancer drug targets. 2011;11:787–98. doi: 10.2174/156800911796798959. [DOI] [PubMed] [Google Scholar]
- 16.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 17.Tang S, Zhang Y, Jiang P, et al. A paradigm of Carcinogenesis Lineage specificities of cancer chemoprevention: Korean Angelica and its pyranocoumarins in the transgenic adenocarcinoma of mouse prostate model. Proc AACR. 2013 Abstract #LB–184. [Google Scholar]
- 18.Dow JK, deVere White RW. Fibroblast growth factor 2: its structure and property, paracrine function, tumor angiogenesis, and prostate-related mitogenic and oncogenic functions. Urology. 2000;55:800–6. doi: 10.1016/s0090-4295(00)00457-x. [DOI] [PubMed] [Google Scholar]
- 19.Giri D, Ropiquet F, Ittmann M. Alterations in expression of basic fibroblast growth factor (FGF) 2 and its receptor FGFR-1 in human prostate cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 1999;5:1063–71. [PubMed] [Google Scholar]
- 20.Huss WJ, Barrios RJ, Foster BA, Greenberg NM. Differential expression of specific FGF ligand and receptor isoforms during angiogenesis associated with prostate cancer progression. Prostate. 2003;54:8–16. doi: 10.1002/pros.10163. [DOI] [PubMed] [Google Scholar]
- 21.Polnaszek N, Kwabi-Addo B, Peterson LE, et al. Fibroblast growth factor 2 promotes tumor progression in an autochthonous mouse model of prostate cancer. Cancer Res. 2003;63:5754–60. [PubMed] [Google Scholar]
- 22.Singh RP, Raina K, Sharma G, Agarwal R. Silibinin inhibits established prostate tumor growth, progression, invasion, and metastasis and suppresses tumor angiogenesis and epithelial-mesenchymal transition in transgenic adenocarcinoma of the mouse prostate model mice. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:7773–80. doi: 10.1158/1078-0432.CCR-08-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin JF, Xu J, Tian HY, et al. Identification of candidate prostate cancer biomarkers in prostate needle biopsy specimens using proteomic analysis. Int J Cancer. 2007;121:2596–605. doi: 10.1002/ijc.23016. [DOI] [PubMed] [Google Scholar]
- 24.Huang HC, Zheng S, VanBuren V, Zhao Z. Discovering disease-specific biomarker genes for cancer diagnosis and prognosis. Technology in cancer research & treatment. 2010;9:219–30. doi: 10.1177/153303461000900301. [DOI] [PubMed] [Google Scholar]
- 25.Taylor BS, Schultz N, Hieronymus H, et al. Integrative genomic profiling of human prostate cancer. Cancer cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Satelli A, Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cellular and molecular life sciences : CMLS. 2011;68:3033–46. doi: 10.1007/s00018-011-0735-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jung MH, Lee SH, Ahn EM, Lee YM. Decursin and decursinol angelate inhibit VEGF-induced angiogenesis via suppression of the VEGFR-2-signaling pathway. Carcinogenesis. 2009;30:655–661. doi: 10.1093/carcin/bgp039. [DOI] [PubMed] [Google Scholar]
- 28.Bullard RS, Gibson W, Bose SK, et al. Functional analysis of the host defense peptide Human Beta Defensin-1: new insight into its potential role in cancer. Molecular immunology. 2008;45:839–48. doi: 10.1016/j.molimm.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kela I, Harmelin A, Waks T, et al. Interspecies comparison of prostate cancer gene-expression profiles reveals genes associated with aggressive tumors. The Prostate. 2009;69:1034–44. doi: 10.1002/pros.20950. [DOI] [PubMed] [Google Scholar]
- 30.Haram KM, Peltier HJ, Lu B, et al. Gene expression profile of mouse prostate tumors reveals dysregulations in major biological processes and identifies potential murine targets for preclinical development of human prostate cancer therapy. The Prostate. 2008;68:1517–30. doi: 10.1002/pros.20803. [DOI] [PubMed] [Google Scholar]
- 31.Yip PY, Kench JG, Rasiah KK, et al. Low AZGP1 expression predicts for recurrence in margin-positive, localized prostate cancer. The Prostate. 2011;71:1638–45. doi: 10.1002/pros.21381. [DOI] [PubMed] [Google Scholar]
- 32.Henshall SM, Horvath LG, Quinn DI, et al. Zinc-alpha2-glycoprotein expression as a predictor of metastatic prostate cancer following radical prostatectomy. Journal of the National Cancer Institute. 2006;98:1420–4. doi: 10.1093/jnci/djj378. [DOI] [PubMed] [Google Scholar]
- 33.Morgenbesser SD, McLaren RP, Richards B, et al. Identification of genes potentially involved in the acquisition of androgen-independent and metastatic tumor growth in an autochthonous genetically engineered mouse prostate cancer model. Prostate. 2007;67:83–106. doi: 10.1002/pros.20505. [DOI] [PubMed] [Google Scholar]
- 34.Komiya A, Suzuki H, Imamoto T, et al. Neuroendocrine differentiation in the progression of prostate cancer. International journal of urology : official journal of the Japanese Urological Association. 2009;16:37–44. doi: 10.1111/j.1442-2042.2008.02175.x. [DOI] [PubMed] [Google Scholar]
- 35.Sun LC, Mackey LV, Luo J, Fuselier JA, Coy DH. Targeted chemotherapy using a cytotoxic somatostatin conjugate to inhibit tumor growth and metastasis in nude mice. Clinical medicine Oncology. 2008;2:491–9. doi: 10.4137/cmo.s970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pittoni P, Tripodo C, Piconese S, et al. Mast cell targeting hampers prostate adenocarcinoma development but promotes the occurrence of highly malignant neuroendocrine cancers. Cancer Research. 2011;71:5987–97. doi: 10.1158/0008-5472.CAN-11-1637. [DOI] [PubMed] [Google Scholar]
- 37.Lenferink AE, Cantin C, Nantel A, et al. Transcriptome profiling of a TGF-beta-induced epithelial-to-mesenchymal transition reveals extracellular clusterin as a target for therapeutic antibodies. Oncogene. 2010;29:831–44. doi: 10.1038/onc.2009.399. [DOI] [PubMed] [Google Scholar]
- 38.Son SH, Park KK, Park SK, et al. Decursin and Decursinol from Angelica gigas Inhibit the Lung Metastasis of Murine Colon Carcinoma. Phytother Res. 2010 doi: 10.1002/ptr.3372. [DOI] [PubMed] [Google Scholar]
- 39.Wang S, Gao J, Lei Q, et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer cell. 2003;4:209–21. doi: 10.1016/s1535-6108(03)00215-0. [DOI] [PubMed] [Google Scholar]
- 40.Wesley UV, McGroarty M, Homoyouni A. Dipeptidyl peptidase inhibits malignant phenotype of prostate cancer cells by blocking basic fibroblast growth factor signaling pathway. Cancer Research. 2005;65:1325–34. doi: 10.1158/0008-5472.CAN-04-1852. [DOI] [PubMed] [Google Scholar]
- 41.Giuliani R, Bastaki M, Coltrini D, Presta M. Role of endothelial cell extracellular signal-regulated kinase1/2 in urokinase-type plasminogen activator upregulation and in vitro angiogenesis by fibroblast growth factor-2. Journal of cell science. 1999;112(Pt 15):2597–606. doi: 10.1242/jcs.112.15.2597. [DOI] [PubMed] [Google Scholar]
- 42.Ma S, Chan YP, Woolcock B, et al. DNA fingerprinting tags novel altered chromosomal regions and identifies the involvement of SOX5 in the progression of prostate cancer. International journal of cancer Journal international du cancer. 2009;124:2323–32. doi: 10.1002/ijc.24243. [DOI] [PubMed] [Google Scholar]
- 43.Rao S, Lyons LS, Fahrenholtz CD, et al. A novel nuclear role for the Vav3 nucleotide exchange factor in androgen receptor coactivation in prostate cancer. Oncogene. 2011 doi: 10.1038/onc.2011.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang W, Chen Y, Swift MR, et al. Effect of FGF-binding protein 3 on vascular permeability. The Journal of biological chemistry. 2008;283:28329–37. doi: 10.1074/jbc.M802144200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Son SH, Kim MJ, Chung WY, et al. Decursin and decursinol inhibit VEGF-induced angiogenesis by blocking the activation of extracellular signal-regulated kinase and c-Jun N-terminal kinase. Cancer Letters. 2009;280:86–92. doi: 10.1016/j.canlet.2009.02.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement Table 1. Sequences of Primers for real-time PCR.
Supplement Table 2. Relative gene expression abundance ratio numerical data (AGN/control) detected by real-time RT-PCR vs. the 2 microarray platforms.
Supplemental Figure S1. Representative images of NE-Ca biomarkers. A: T-Antigen; B: Synaptophysin; C: Androgen receptor; D: E-Cadherin. Insert shows enlarged image of NE-Ca staining. Trapped epithelial gland is marked by Epi.
Supplemental Figure S2: A) The integrity of total RNAs extracted from TRAMP NE-Ca analyzed by Agilent 2100 Bioanalyzer. Left to right: size marker; sample lane 1, RNA from vehicle-control; sample lane 3, RNA from AGN treated NE-Ca. B) Correlation of the signal intensities for each probe acquired from two hybridization of RNA from control group (Illumina microarray).