Abstract
Alterations in the folate pathway have been related to both major depression and cognitive inflexibility; however, they have not been investigated in the genetic background of ruminative response style, which is a form of perseverative cognition and a risk factor for depression. In the present study, we explored the association of rumination (measured by the Ruminative Responses Scale) with polymorphisms of two distinct folate pathway genes, MTHFR rs1801133 (C677T) and MTHFD1L rs11754661, in a combined European white sample from Budapest, Hungary (n=895) and Manchester, United Kingdom (n=1309). Post hoc analysis investigated whether the association could be replicated in each of the two samples, and the relationship between folate pathway genes, rumination, lifetime depression and Brief Symptom Inventory depression score. Despite its functional effect on folate metabolism, the MTHFR rs1801133 showed no effect on rumination. However, the A allele of MTHFD1L rs11754661 was significantly associated with greater rumination, and this effect was replicated in both the Budapest and Manchester samples. In addition, rumination completely mediated the effects of MTHFD1L rs11754661 on depression phenotypes. These findings suggest that the MTHFD1L gene, and thus the C1-THF synthase enzyme of the folate pathway localized in mitochondria, has an important effect on the pathophysiology of depression through rumination, and maybe via this cognitive intermediate phenotype on other mental and physical disorders. Further research should unravel whether the reversible metabolic effect of MTHFD1L is responsible for increased rumination or other long-term effects on brain development.
Introduction
Major depressive disorder is an etiologically heterogeneous condition,1 in which a core and specific feature is depressive rumination.2
Ruminative response style, which is sometimes referred as depressive rumination, can be defined in several ways.3 In a broader sense, it is a form of cognitive inflexibility or perseverative cognition that prolongs the negative effect of everyday life stressors.4, 5 In addition, it may involve an impairment of the top-down cortical control on mnemonic processes, resulting in unwanted and uncontrollable dwelling on intrusive memories.6 Ruminative response style in association with depression is perceived as thinking repeatedly and passively about one's feelings and problems related to distress and depressed mood, thus exacerbating and prolonging depression.7 Indeed, it has been demonstrated that ruminative response style predicts the onset and level of future depression.7, 8 These facts suggest that ruminative response style or shortly rumination (as it is generally addressed) is a potential intermediate phenotype for depression.
Rumination is a moderately heritable trait with a 20–40% heritability rate based on twin studies.9, 10 Most importantly, phenotypic correlation between depressed mood and rumination appears to be explained mainly by shared genetic factors.9, 10 Thus, genetic risk factors for rumination are likely to share a pathophysiological role in the development of depression; however, hypothesis-free genome-wide association studies with rumination have not yet been reported. Using a candidate gene approach, both dopaminergic and serotonergic genes have been implicated in rumination (DRD2 (ref. 11) COMT (ref. 12) and serotonin transporter SLC6A4 (ref. 13)), and also extensively investigated in relation to cognitive flexibility and response inhibition (for review see Logue and Gould14). In addition, genes related to neuronal and synaptic plasticity (KCNJ6 (ref. 15), CREB1 (refs. 15, 16) and BDNF (refs. 13, 16) and stress response (NR3C2 (ref. 17)) showed significant associations with rumination.
Altered folate function and the linked one-carbon cycle have long been implicated in the pathogenesis of depression and also in cognitive inflexibility or perseverative cognition, yet their possible role in rumination has not been investigated. More specifically, folate is necessary to the catabolism of homocysteine, and folate deficiency and elevated homocysteine are related to both depression and inflexible cognition.18, 19, 20 These metabolic changes are also associated with altered brain monoamine metabolism and impaired neuronal plasticity.19, 20
The most investigated genetic variant of the folate pathway is the MTHFR C677T (rs1801133) polymorphism, which leads to an alanine (C allele) to valine (T allele) substitution in the 5,10-methylenetetrahydrofolate reductase (MTHFR) protein. The MTHFR 677 T allele codes a thermolabile and less active enzyme, which is associated with decreased folate and increased homocysteine levels.21 Despite its strong metabolic impact, this polymorphism has shown conflicting results in genetic association studies of inflexible cognition,19, 22 major depression1, 23 and other neuropsychiatric disorders, such as Alzheimer's disease (AD),24 bipolar disorder and schizophrenia.23
Three studies reported that the A allele of a polymorphism (rs11754661) in another gene involved in folate metabolism, MTHFD1L, showed a genome-wide significant association with late-onset AD,25, 26, 27 although one study was negative.28 MTHFD1L encodes the human mitochondrial monofunctional 10-formyl-tetrahydrofolate synthetase (C1-THF synthase) enzyme.29 The A allele, similar to the MTHFR 677 T allele, is associated with increased homocysteine concentrations.30 In addition, this enzyme is obligatory for the production of mitochondrial formate, the essential substrate for cytoplasmic purine and thymidylate biosynthesis, methionine biosynthesis and amino-acid metabolism.29, 31 Although the direct link between rumination and AD is as yet only hypothetical,32 the association of the MTHFD1L gene with age-related cognitive decline, together with its pivotal role in normal neuronal development,31, 33, 34 suggests that it could be relevant in cognitive processes throughout the life.
In the present study, we investigated the association of ruminative response style with MTHFR rs1801133 and MTHFD1L rs11754661. We hypothesized that genetic variants in the folate pathway are associated with rumination, which is a cognitive risk factor for depression. In addition, we examined the relationship between folate pathway genes, rumination and depression phenotypes.
Materials and methods
This study was part of the European Union-funded NewMood study (New Molecules in Mood Disorders, Sixth Framework Program of the EU, LSHM-CT-2004-503474), which was carried out in accordance with the Declaration of Helsinki and approved by local Ethics Committees (North Manchester Local Research Ethics Committee, Manchester, UK; Scientific and Research Ethics Committee of the Medical Research Council, Budapest, Hungary).
Participants
Participants aged 18–60 years were recruited through general practices and advertisements from Budapest, Hungary, and through general practices, advertisements and a website from Greater Manchester, UK. All participants provided written informed consent. N=2204 subjects (n=895 from Budapest and n=1309 from Manchester) provided information about gender, age and rumination by filling out the NewMood questionnaire pack (in English or Hungarian, as appropriate)16 and were successfully genotyped for MTHFR rs1801133 by providing DNA with a genetic saliva sampling kit. MTHFD1L rs11754661 was successfully genotyped in 2120 subjects among those who provided information about gender, age and rumination (n=862 from Budapest and n=1258 from Manchester). All subjects were of European white ethnic origin, and had no relatives participating in the study.
Phenotypic assessment
We used the 10-item Ruminative Responses Scale to measure rumination,8 and calculated rumination score as a continuous weighted score: the sum of item scores divided by the number of items completed. The NewMood questionnaire pack also included measures of two distinct depression phenotypes. Current depressive symptoms were measured by the depression items plus the additional items of the Brief Symptom Inventory (BSI),35 using a weighted score (see above, at rumination). Reported lifetime depression was derived from the background questionnaire and had been validated in a subpopulation with face-to-face diagnostic interviews.16
Genotyping
For genotyping we collected buccal mucosa cells and extracted genomic DNA according to a validated method.36 The two single-nucleotide polymorphisms (SNPs), MTHFR rs1801133 and MTHFD1L rs11754661, were genotyped with the Sequenom MassARRAY technology (Sequenom, San Diego, CA, USA, www.sequenom.com). All laboratory work was blinded with regard to phenotype and performed under the ISO 9001:2000 quality-management requirements.
Statistical analyses
PLINK v1.07 (http://pngu.mgh.harvard.edu/purcell/plink/) was used to calculate Hardy–Weinberg equilibrium for MTHFR rs1801133 and MTHFD1L rs11754661, and to build linear regression models for rumination score as an outcome variable. MTHFR rs1801133 or MTHFD1L rs11754661, respectively, and age, gender and population (Budapest or Manchester) were the predictor variables in all regression equations. With rs1801133, additive, dominant and recessive models were run in the combined sample. However, with rs11754661, we did not run the recessive model because of the low number of those homozygous for the minor allele. Bonferroni-corrected two-tailed P⩽0.010 was used as a significance threshold, and P⩽0.020 as a trend threshold, for the main analysis. As post hoc analysis, we investigated the significant effects separately in the Budapest and Manchester samples to test possible replications. In addition, we ran post hoc regression analyses similarly to the ones described above, for lifetime depression and current depression score. Furthermore, we tested the mediating role of depression phenotypes on rumination or the mediating role of rumination on depression phenotypes by including the mediating phenotype(s) as covariate(s) to test shared explained variance by the genes and these phenotypes. For post hoc statistical testing two-tailed P⩽0.05 threshold was used. We applied the parametric statistical methods based on the central limit theorem, as we have large samples (n>200).37 Descriptive statistics for the combined and separate samples were calculated with IBM SPSS 20.0 (IBM, Armonk, NY, USA) for Windows. We used Quanto for power calculations (http://biostats.usc.edu/Quanto.html), and OpenMeta[Analyst] for meta-analyses of genetic effects in the separate Budapest and Manchester samples (http://www.cebm.brown.edu/open_meta/download.html). To enhance the speed of the PLINK analysis, individually written R-scripts were used.38
Results
The minor allele is T for MTHFR rs1801133 and A for MTHFD1L rs11754661. Both SNPs were in Hardy–Weinberg equilibrium in Budapest, Manchester and in the combined sample. For rs1801133, P-values are as follows: P=0.384 in Budapest, P=0.670 in Manchester and P=0.852 in the combined sample. For rs11754661, P=1 in Budapest, P=0.064 in Manchester and P=0.112 in the combined sample. Description of total sample and for Budapest and Manchester separately is given in Table 1. As we can see in Table 1, the Budapest and Manchester samples differ significantly in age, rs11754661 genotype frequencies, rumination and both depression phenotypes, which makes it reasonable to include population as a predictor variable in the regression equations.
Table 1. Description of the population samples.
| Budapest | Manchester | Budapest+Manchester | Difference between Budapest and Manchester | ||
|---|---|---|---|---|---|
| Gender | |||||
| Female (%) | 624 (69.7%) | 916 (70%) | 1540 (69.9%) | X2=0.017; P=0.897 | |
| Male (%) | 271 (30.3%) | 393 (30%) | 664 (30.1%) | ||
| Age (mean±s.e.m.) | 31.26 (0.355) | 34.04 (0.284) | 32.91 (0.224) | t=−6.153; P<0.001 | |
| MTHFR rs1801133 | |||||
| TT (%) | 122 (13.6%) | 154 (11.8%) | 276 (12.5%) | X2=1.717; P=0.424 | |
| TC (%) | 400 (44.7%) | 602 (46%) | 1002 (45.5%) | ||
| CC (%) | 373 (41.7%) | 553 (42.2%) | 926 (42%) | ||
| MTHFD1L rs11754661 | |||||
| AA (%) | 1 (0.1%) | 10 (0.8%) | 11 (0.5%) | X2=9.914; P=0.007 | |
| GA (%) | 75 (8.7%) | 148 (11.8%) | 223 (10.5%) | ||
| GG (%) | 786 (91.2%) | 1100 (87.4%) | 1886 (89%) | ||
| Rumination score (mean±s.e.m.) | 1.94 (0.016) | 2.25 (0.017) | 2.13 (0.012) | t=−13.104; P<0.001 | |
| BSI depression score (mean±s.e.m.) | 0.56 (0.023) | 1.07 (0.028) | 0.86 (0.020) | t=−13.954; P<0.001 | |
| Lifetime depression | |||||
| Reported (%) | 192 (21.5%) | 734 (56.1%) | 926 (42%) | X2=261.521; P<0.001 | |
| Not reported (%) | 703 (78.5%) | 575 (43.9%) | 1278 (58%) | ||
Abbreviations: BSI, Brief Symptom Inventory; MTHFR, 5,10-methylenetetrahydrofolate reductase.
The Manchester sample shows significantly higher mean age, rumination score and BSI depression score, and higher frequencies of reported lifetime depression and of the MTHFD1L rs11754661 A allele than the Budapest sample.
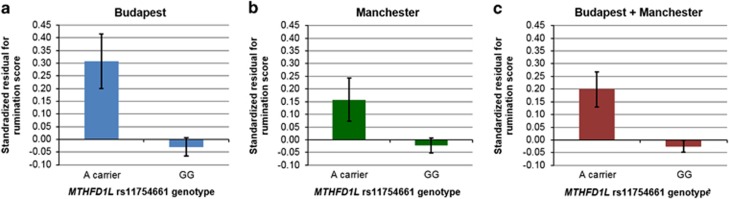
In our combined sample MTHFR rs1801133 did not show any significant effect on rumination (Table 2). Considering the effect of MTHFD1L rs11754661 on rumination, with age, gender and population as covariates in the regression equations, the A allele showed a significant positive association with rumination score in the combined sample, both in additive and dominant models (Table 2). These findings remained significant after Bonferroni correction for multiple testing. Post hoc analysis showed that the effect of the A allele remained statistically significant at a nominal (uncorrected) level in the Budapest and Manchester samples separately (Table 3). To visualize and meta-analyze these associations, standardized residuals were calculated for rumination score (separately in Budapest and Manchester and in the combined sample), by partialling out variance accounted for by age, gender and population (this latter only in the combined sample). The means (with s.e.'s) of these residuals are represented according to the rs11754661 genotype in Figure 1. A marked difference in rumination can be seen between A carriers and those with the GG genotype in Budapest (Figure 1a), Manchester (Figure 1b) and also in the combined sample (Figure 1c). We entered the means and s.d.'s of these standardized residuals of Budapest (0.31±0.930 in A carriers and −0.03±1.001 in the GG group) and Manchester (0.16±1.066 in A carriers and −0.02±0.988 in the GG group) into OpenMeta[Analyst] to calculate a combined mean difference between genotypes, in a continuous random-effects model. The combined mean difference and its s.e. is significant: 0.246±0.079 (P=0.002), underpinning our significant linear regression results in Budapest, Manchester and the combined sample (Tables 2 and 3). No significant between-study heterogeneity exists (tau2=0.002; Q=1.235; P=0.267; I2=19%), pointing out the validity of conducting a mega-analysis for the rs11754661 effect in the combined sample (Table 2).
Table 2. Linear regression models for rumination score as an outcome variable, separately with the two SNPs.
|
MTHFR rs1801133 |
MTHFD1L rs11754661 |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Predictor variable | N | Beta | s.e. | t | P | Predictor variable | N | Beta | s.e. | t | P | Predictor variable | N | Beta | s.e. | t | P | |
| Additive | MTHFR rs1801133 | 2204 | −0.023 | 0.017 | −1.358 | 0.175 | MTHFD1L rs11754661 | 2120 | 0.112 | 0.035 | 3.182 | 0.001 | MTHFD1L rs11754661 | 2117 | 0.070 | 0.029 | 2.388 | 0.017 |
| Age | 2204 | −0.006 | 0.001 | −5.296 | <0.001 | Age | 2120 | −0.006 | 0.001 | −5.214 | <0.001 | Age | 2117 | −0.006 | 0.001 | −6.347 | <0.001 | |
| Gender | 2204 | 0.273 | 0.025 | 10.710 | <0.001 | Gender | 2120 | 0.274 | 0.026 | 10.590 | <0.001 | Gender | 2117 | 0.177 | 0.022 | 8.161 | <0.001 | |
| Population | 2204 | 0.321 | 0.024 | 13.440 | <0.001 | Population | 2120 | 0.321 | 0.024 | 13.210 | <0.001 | Population | 2117 | 0.103 | 0.022 | 4.795 | <0.001 | |
| BSI depression score | 2117 | 0.284 | 0.012 | 23.570 | <0.001 | |||||||||||||
| Lifetime depression | 2117 | 0.217 | 0.024 | 9.199 | <0.001 | |||||||||||||
| Dominant | MTHFR rs1801133 | 2204 | −0.043 | 0.024 | −1.825 | 0.068 | MTHFD1L rs11754661 | 2120 | 0.122 | 0.038 | 3.228 | 0.001 | MTHFD1L rs11754661 | 2117 | 0.072 | 0.031 | 2.309 | 0.021 |
| Age | 2204 | −0.006 | 0.001 | −5.317 | <0.001 | Age | 2120 | −0.006 | 0.001 | −5.213 | <0.001 | Age | 2117 | −0.006 | 0.001 | -6.346 | <0.001 | |
| Gender | 2204 | 0.273 | 0.025 | 10.710 | <0.001 | Gender | 2120 | 0.274 | 0.026 | 10.570 | <0.001 | Gender | 2117 | 0.177 | 0.022 | 8.146 | <0.001 | |
| Population | 2204 | 0.321 | 0.024 | 13.460 | <0.001 | Population | 2120 | 0.322 | 0.024 | 13.230 | <0.001 | Population | 2117 | 0.104 | 0.022 | 4.815 | <0.001 | |
| BSI depression score | 2117 | 0.284 | 0.012 | 23.560 | <0.001 | |||||||||||||
| Lifetime depression | 2117 | 0.217 | 0.024 | 9.201 | <0.001 | |||||||||||||
| Recessive | MTHFR rs1801133 | 2204 | −0.002 | 0.035 | −0.058 | 0.954 | ||||||||||||
| Age | 2204 | −0.006 | 0.001 | −5.277 | <0.001 | |||||||||||||
| Gender | 2204 | 0.273 | 0.025 | 10.700 | <0.001 | |||||||||||||
| Population | 2204 | 0.321 | 0.024 | 13.450 | <0.001 | |||||||||||||
Abbreviations: BSI, Brief Symptom Inventory; MTHFR, 5,10-methylenetetrahydrofolate reductase; SNP, single-nucleotide polymorphism.
PLINK linear regression equations were constructed with the predictor variables displayed in the rows. Additive, dominant and recessive models were run in the combined sample, separately with both SNPs as predictors. T is the minor allele in case of MTHFR rs1801133; A in case of MTHFD1L rs11754661. Recessive models have not been run for rs11754661 because of low number in the AA group. Significance threshold is P⩽0.010 (Bonferroni-corrected) for the main analyses, and P⩽0.050 for post hoc analyses (those including also the two depression phenotypes as covariates). Significant findings for the SNPs as a predictor variable are marked with bold.
Table 3. Linear regression models of MTHFD1L rs11754661 for rumination score as an outcome variable, separately in Budapest and Manchester.
|
Budapest |
Manchester |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Predictor variable | N | Beta | s.e. | t | P | Predictor variable | N | Beta | s.e. | t | P | |
| Additive | MTHFD1L rs11754661 | 862 | 0.158 | 0.054 | 2.915 | 0.004 | MTHFD1L rs11754661 | 1258 | 0.095 | 0.046 | 2.049 | 0.041 |
| Age | 862 | −0.004 | 0.002 | −2.417 | 0.016 | Age | 1258 | −0.008 | 0.002 | −4.693 | <0.001 | |
| Gender | 862 | 0.220 | 0.034 | 6.409 | <0.001 | Gender | 1258 | 0.311 | 0.037 | 8.456 | <0.001 | |
| Dominant | MTHFD1L rs11754661 | 862 | 0.157 | 0.055 | 2.828 | 0.005 | MTHFD1L rs11754661 | 1258 | 0.107 | 0.050 | 2.120 | 0.034 |
| Age | 862 | −0.004 | 0.002 | −2.395 | 0.017 | Age | 1258 | −0.008 | 0.002 | −4.700 | <0.001 | |
| Gender | 862 | 0.220 | 0.034 | 6.419 | <0.001 | Gender | 1258 | 0.310 | 0.037 | 8.437 | <0.001 | |
PLINK linear regression equations were constructed with the predictor variables displayed in the rows. Additive and dominant models were run, separately in Budapest and Manchester; all with A as the minor allele. (Recessive models have not been run because of low number in AA groups.) Significant findings for MTHFD1L rs11754661 as a predictor variable are marked with bold.
Figure 1.
Means (and its s.e.'s) of standardized residuals for rumination score, according to the rs11754661 genotype. General linear models were created for rumination score as an outcome variable, separately in Budapest, Manchester (with age and gender as covariates) and in the combined sample (with age, gender and population as covariates). Standardized residuals of these models were then displayed according to the MTHFD1L rs11754661 genotype, thus representing the variance of rumination not accounted for by age, gender and population. A carriers show higher rumination than those with GG genotype in Budapest (a), Manchester (b) and also in the combined sample (c).
Similarly, we ran a meta-analysis for the combined mean difference between MTHFR rs1801133 T carriers and those with CC genotype. The means (and their s.d.'s) of the standardized residual for rumination score are as follows: −0.04±0.993 in T carriers in Budapest, 0.05±1.006 in the CC group in Budapest; and −0.03±1.002 in T carriers in Manchester, 0.05±0.995 in the CC group in Manchester. A continuous random-effects model yielded a combined mean difference (and its s.e.) of −0.084±0.043 (P=0.051), underpinning the result of the dominant model among linear regressions in that T carriers nominally tend to ruminate less than the CC group (Table 2). As in the case of rs11754661, the unsignificant heterogeneity test results between Budapest and Manchester (tau2<0.001; Q=0.013; P=0.909; I2=0%) also validate mega-analysis of the rs1801133 effect on rumination in the combined sample (Table 2).
We ran post hoc analyses in the combined sample to unravel whether the association between rs11754661 and rumination is mediated by depression phenotypes. Rumination shows a significant positive association with both of our depression phenotypes: Pearson correlation coefficient r=0.581 (N=2117; P<0.001) for BSI depression score and t=−22.022 (N=2120; P<0.001) for lifetime depression (mean rumination score is 1.909±0.014 in those who did not and 2.429±0.019 in those who did report lifetime depression). The MTHFD1L rs11754661 A allele also associates positively (either significantly or as a trend) to both depression phenotypes. Namely, for BSI depression score, its β=0.098; t=1.695; P=0.090 in an additive, and β=0.118; t=1.897; P=0.058 in a dominant PLINK linear regression model. For lifetime depression, its odds ratio (OR)=1.354; t=2.173; P=0.030 in an additive, and OR=1.405; t=2.271; P=0.023 in a dominant PLINK logistic regression model. N=2117 in BSI depression and N=2120 in lifetime depression models; and age, gender and population were covariates in all PLINK analyses. Because of their positive associations (either significantly or as a trend) with both the predictor rs11754661 A allele and the outcome rumination, we could include these two depression phenotypes as covariates (besides age, gender and population) in the linear regression equations described above. For the results see Table 2. Including the two depression phenotypes does not abolish the significant effect of MTHFD1L rs11754661 on rumination, but diminishes its effect size (beta) in either an additive or a dominant model (for comparisons also see Table 2). This suggests that depression is only partly responsible for the risk the A allele conveys for rumination.
On the other hand, rumination entirely explains the variance rs11754661 shares with each of the depression phenotypes. Including rumination as an additional predictor in the PLINK regression analyses discussed above, the effect of rs11754661 on depression is no longer statistically significant or a trend in the combined sample: β=−0.001; t=−0.018; P=0.986 in the additive, β=0.010; t=0.193; P=0.847 in the dominant model for BSI depression score, and OR=1.198; t=1.189; P=0.234 in the additive, OR=1.235; t=1.301; P=0.193 in the dominant model for lifetime depression.
With regard to the post hoc mediation analyses in the Budapest and Manchester samples separately (with age and gender as covariates in all models), in spite of the apparently replicable association with rumination, rs11754661 does not show a significant association with any of the depression phenotypes in Manchester (for BSI depression score, β=0.085; t=1.088; P=0.277 in an additive, and β=0.107; t=1.252; P=0.211 in a dominant model, and for lifetime depression, OR=1.230; t=1.276; P=0.202 in an additive, and OR=1.289; t=1.426; P=0.154 in a dominant model). However, we could implement the mediation analyses in the Budapest sample, as rs11754661 associates either significantly or as a trend to both depression phenotypes there (for BSI depression score, β=0.141; t=1.734; P=0.083 in an additive, and β=0.151; t=1.813; P=0.070 in a dominant model, and for lifetime depression, OR=1.775; t=2.226; P=0.026 in an additive, and OR=1.737; t=2.088; P=0.037 in a dominant model), and because rumination shows a significant positive association with both depression phenotypes (Pearson r=0.536; P<0.001 for BSI depression score; and t=−9.603; P<0.001 for lifetime depression, with a mean rumination score of 1.866±0.017 in those who did not, and of 2.226±0.035 in those who did report lifetime depression). In the mediation analyses in Budapest, we could replicate our findings seen in the combined sample. The two depression phenotypes as predictors diminish but do not abolish the effect of rs11754661 on rumination (β=0.094; t=2.051; P=0.041 in the additive, and β=0.090; t=1.921; P=0.055 in the dominant model; for comparisons see Table 3), whereas rumination as a predictor entirely abolishes the effect of rs11754661 on both depression phenotypes (for BSI depression score, β=0.014; t=0.210; P=0.834 in the additive, and β=0.025; t=0.361; P=0.719 in the dominant model, and for lifetime depression, OR=1.471; t=1.425; P=0.154 in the additive, and OR=1.443; t=1.322; P=0.186 in the dominant model).
The discrepancy in the detected effect of rs11754661 on rumination and the non-detected one of rs1801133 cannot be attributed to decreased power. Assuming an R2=1% and under a dominant model (mean rumination score is 2.13 and its s.d. is 0.58 in case of both SNPs), the power to detect an rs11754661 main effect on rumination (n=2120) is 99.6%, whereas the power of rs1801133 (n=2204) is 99.7%. In addition, rs1801133 has no effect on either depression phenotypes in the combined sample (BSI depression: β=0.004; t=0.135; P=0.892 in an additive, β=0.012; t=0.305; P=0.761 in a dominant, β=−0.010; t=−0.177; P=0.859 in a recessive model; lifetime depression: OR=1.029; t=0.407; P=0.684 in an additive, OR=1.047; t=0.482; P=0.630 in a dominant, OR=1.016; t=0.113; P=0.910 in a recessive model), suggesting that its lack of effect on ruminative response style is not spurious.
Discussion
Among polymorphisms of folate pathway genes, the widely investigated MTHFR rs1801133 is not associated with ruminative response style in our large combined European white sample, whereas the AD genome-wide marker MTHFD1L rs11754661 A allele represents a risk for higher ruminative response style. This association is replicated separately in the Budapest and Manchester cohorts. Moreover, this association is only partly mediated by current depression score and lifetime depression, but ruminative response style fully explains the variance that MTHFD1L rs11754661 shares with these depression phenotypes.
That there is an effect of the MTHFD1L variant but not of the MTHFR is in line with findings of the genome-wide mega-analysis on major depressive disorder by Ripke and colleagues,39 where the index SNP of the MTHFD1L gene (SNP with the highest significance) showed a more significant association (rs563440; P=0.004) with major depression than the index SNP from MTHFR (rs17037425; P=0.079).
Discrepancy in the effects of MTHFR and MTHFD1L
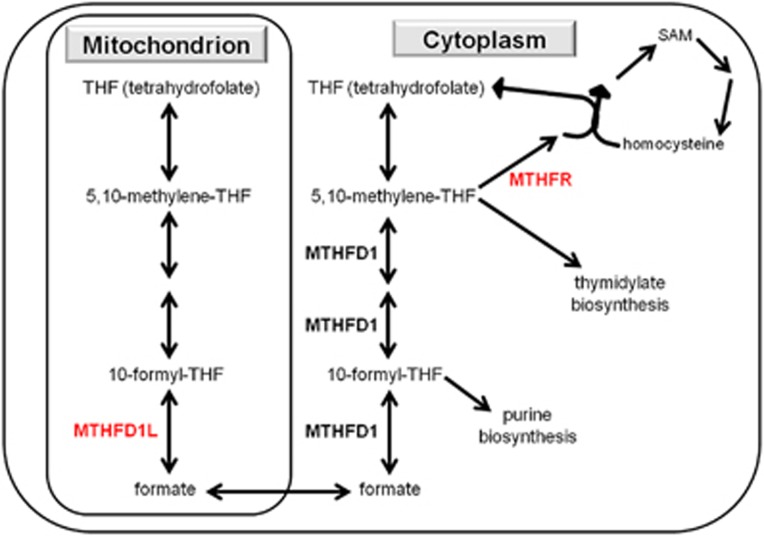
There may be several interrelated reasons for the discrepancy in the effects of MTHFD1L and MTHFR. First, the two enzymes have distinct biochemical roles (see Figure 2 and refs. 29, 40, 41). Specifically, MTHFD1L could enhance both the 10-formyl-THF generation and, by producing formate for the cytoplasm, the synthesis of S-adenosylmethionine (SAM). 10-formyl-THF generation is protective for mitochondria,22 whereas SAM is an important methyl donor in epigenetic regulation processes related to memory, learning, cognition and behavior,18 and is also crucial in the synthesis of dopamine, serotonin and noradrenaline in the brain.41 In contrast, MTHFR can support only one of these two directions, namely 10-formyl-THF generation or SAM synthesis, at the expense of the other one.22, 42
Figure 2.
Distinct roles of enzymes MTHFD1L and MTHFR in the folate-related one-carbon cycle. MTHFD1L: mitochondrial monofunctional C1-tetrahydrofolate synthase enzyme; MTHFD1: cytoplasmic trifunctional C1-tetrahydrofolate synthase enzyme; MTHFR: 5,10-methylenetetrahydrofolate reductase enzyme; SAM: S-adenosylmethionine (a methyl donor in numerous reactions). The arrows represent different reactions or a flow between the mitochondrion and cytoplasm (as appropriate), and the most important enzymes (with bold) and substrates are represented.
A second source of the discrepancy could be the distinct subcellular localization of the two enzymes.29, 40 As the protein C1-THF synthase coded by MTHFD1L is localized in mitochondria, whereas the MTHFR protein coded by MTHFR is present in the cytoplasm, we can conclude that the folate pathway in the mitochondria is essential in rumination and other cognitive processes. Furthermore, mitochondrial dysfunction has been associated with depression earlier.43, 44
A third reason for the discrepancy could be the distinct sensitivity of these enzymes to other factors, such as environmental effects. For example, it has been demonstrated that the effects of MTHFR rs1801133 genotype on the plasma homocysteine level,30, 45 DNA methylation level42 and cognitive performance22 are modulated by the folate status, namely this polymorphism has stronger effect in case of low level of folate compared with high level of folate. In contrast, the MTHFD1L gene has pleiotropic effects on the plasma homocysteine level and markers of genome-wide DNA methylation after controlling for nutrient status.30 Future research should reveal whether the effect of MTHFD1L rs11754661 on ruminative response style depends on the folate status.
Pathophysiological specificity of ruminative response style?
We found MTHFD1L rs11754661 more consistently associated with ruminative response style than with our two depression phenotypes, as rs11754661 does not predict depression in the Manchester sample. Moreover, in Budapest, and in the combined sample, the association of the MTHFD1L variant and ruminative response style is only partly mediated by depression, but completely accounts for the effect that the MTHFD1L variant exerts on depression. These findings correspond well with two reviews stating that rumination confers a risk not specifically for depression, but, for several psychopathologies, alterations in mental and physical health.3, 7 Taken together these observations, the MTHFD1L gene and thus the folate pathway may be important in the pathophysiology of other health conditions related to ruminative response style.
Regarding psychiatric disorders, the latest pathway-based genome-wide association studies by the Psychiatric Genomic Consortium46 found that methylation pathways, inclusive of the SAM-dependent methyltransferase activity, are among the most important in the common background of major depression, bipolar depression and schizophrenia. Interestingly, the 'one-carbon pool by folate' pathway, which contains the MTHFR and MTHFD1L genes, was nominally significant for bipolar disorder, showed a trend for major depressive disorder and was not significant for schizophrenia, suggesting that this pathway has distinct effect on mood disorders, probably through a common intermediate phenotype, such as ruminative response style. However, our positive findings with distinctive role of MTHFR and MTHFD1L polymorphisms still underline the importance of not only the pathway-, but also the gene- or polymorphism-based approach.
Taking into account non-psychiatric disorders, cardiovascular diseases could also be potential targets of future investigations because rumination, denoting a cognitive perseveration on distress, yields a prolonged stress response and slower cardiovascular recovery, and thus a risk for cardiovascular disease.4, 5 In addition, cardiovascular diseases share a pattern of alterations in the key one-carbon cycle components (levels of, for example, folate, homocysteine and the universal methyl donor SAM) with psychiatric disorders.18
Therapeutic implications
There is some evidence that methylfolate47 and SAM41 supplementations are effective in the treatment of major depression; however, the evidence that folate augments the efficacy of conventional antidepressant medication is mixed and includes a recent large negative study.48, 49 Our present results and previous genetic association studies may shed light on these contradictory findings. First, not the entire folate pathway is associated with depression39, 46 and treatment response,49 but elements with stronger influence on methylation processes30 have more consistent effects. Alterations in the DNA and histone methylations, which translate environmental exposures to specific gene expression patterns and are major factors in the regulation of brain development and synaptic plasticity, may cause long-term increased risk for depression,50 which is difficult to reverse by supplementation therapy. Second, ruminative response style, a trait-like risk factor for several psychiatric and physical disorders, represents an intermediate phenotype between MTHFD1L polymorphism and depression. Thus, methylfolate and SAM supplementations may be more effective in those with high ruminative response style, as an augmentation of targeted psychotherapies,51 although this hypothesis has not been tested yet.
Limitations
Our study is cross-sectional and cannot address the time course of an association between MTHFD1L rs11754661 and either ruminative response style or depression. In addition, it cannot account directly for reporting bias for past depressive episodes; however, we measured depression in two ways, one of which is current depression, allowing some confidence that reporting bias does not explain the association. Moreover, determining the precedence of rumination or lifetime depression episodes would be of crucial importance in our study, as we operationalized rumination specifically as ruminative response style, anchoring it to an answer to sadness or depressed mood. However, narrowing the concept of rumination like this makes it easier to interpret our findings. Our lifetime depression measure was not based on face-to-face diagnostic interviews, but had been validated in a subsample. In addition, further research covering the whole genes with haplotype tags or sequencing these regions is required to confirm the findings about the effects of single SNPs.
Conclusions and implications for future research
In conclusion, we have identified the MTHFD1L rs11754661 A allele as a genetic risk factor for ruminative response style, and this association may convey pathophysiological implications for not only depression but also other mental and physical disorders. This association, which replicated in two independent European white samples, enriches our knowledge about the genetic architecture of ruminative response style. In addition, the folate pathway can be linked to most of the previously described genetic risk factors for rumination. Therefore, future research is needed to shed light on the particular ways in which MTHFD1L rs11754661 might affect rumination, for example, via homocysteine levels, synaptic plasticity, methylation patterns of relevant genes and methylation-related dynamics of monoamine metabolism. It will also be crucial to determine whether MTHFD1L acts at specific points in neural development when the tendency to ruminate is established.
Acknowledgments
The study was supported by the Sixth Framework Program of the European Union, NewMood, LSHM-CT-2004-503474; by the National Institute for Health Research Manchester Biomedical Research Centre; by the TAMOP-4.2.1.B-09/1/KMR-2010-0001; by the Hungarian Brain Research Program (Grant KTIA_13_NAP-A-II/14) and National Development Agency (Grant KTIA_NAP_13-1-2013-0001); by the Hungarian Academy of Sciences (MTA-SE Neuropsychopharmacology and Neurochemistry Research Group); and by the Hungarian Academy of Sciences and the Hungarian Brain Research Program – Grant No. KTIA_NAP_13-2-2015-0001 (MTA-SE-NAP B Genetic Brain Imaging Migraine Research Group). XG is recipient of the Janos Bolyai Research Fellowship of the Hungarian Academy of Sciences. We thank Diana Chase, Emma J Thomas, Darragh Downey, Kathryn Lloyd-Williams and Zoltan G Toth for their assistance in the recruitment and data acquisition; Hazel Platt for her assistance in genotyping; Heaton Mersey Medical Practice and Cheadle Medical Practice for their assistance in the recruitment.
Disclaimer
The sponsors funded the work, but had no further role in the design of the study, in data collection or analysis, in the decision to publish, or in the preparation, review, or approval of the manuscript.
JFWD has variously performed consultancy, speaking engagements and research for Bristol-Myers Squibb, AstraZeneca, Eli Lilly, Schering Plough, Janssen-Cilag and Servier (all fees are paid to the University of Manchester to reimburse them for the time taken); he also has share options in P1vital. IMA has received consultancy fees from Servier, Alkermes, Lundbeck/Otsuka and Janssen, an honorarium for speaking from Lundbeck and grant support from Servier and AstraZeneca. RE has received consultancy fees from Cambridge Cognition and P1vital. The remaining authors declare no conflict of interest.
References
- Flint J, Kendler KS. The genetics of major depression. Neuron 2014; 81: 484–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott R, Zahn R, Deakin JF, Anderson IM. Affective cognition and its disruption in mood disorders. Neuropsychopharmacology 2011; 36: 153–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM, Alloy LB. A roadmap to rumination: a review of the definition, assessment, and conceptualization of this multifaceted construct. Clin Psychol Rev 2009; 29: 116–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosschot JF, Gerin W, Thayer JF. The perseverative cognition hypothesis: a review of worry, prolonged stress-related physiological activation, and health. J Psychosom Res 2006; 60: 113–124. [DOI] [PubMed] [Google Scholar]
- Larsen BA, Christenfeld NJ. Cardiovascular disease and psychiatric comorbidity: the potential role of perseverative cognition. Cardiovasc Psychiatry Neurol 2009; 2009: 791017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett JM, Benoit RG, Gagnepain P, Salman A, Bartholdy S, Bradley C et al. The origins of repetitive thought in rumination: separating cognitive style from deficits in inhibitory control over memory. J Behav Ther Exp Psychiatry 2015; 47: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen-Hoeksema S, Wisco BE, Lyubomirsky S. Rethinking rumination. Perspect Psychol Sci 2008; 3: 400–424. [DOI] [PubMed] [Google Scholar]
- Treynor W, Gonzalez R, Nolen-Hoeksema S. Rumination reconsidered: a psychometric analysis. Cognitive Ther Res 2003; 27: 247–259. [Google Scholar]
- Chen J, Li XY. Genetic and environmental influences on adolescent rumination and its association with depressive symptoms. J Abnorm Child Psychol 2013; 41: 1289–1298. [DOI] [PubMed] [Google Scholar]
- Moore MN, Salk RH, Van Hulle CA, Abramson LY, Hyde JS, Lemery-Chalfant K et al. Genetic and environmental influences on rumination, distraction, and depressed mood in adolescence. Clin Psychol Sci 2013; 1: 316–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmer AJ, Gotlib IH. Depressive rumination and the C957T polymorphism of the DRD2 gene. Cogn Affect Behav Neurosci 2012; 12: 741–747. [DOI] [PubMed] [Google Scholar]
- Pap D, Juhasz G, Bagdy G. Association between the COMT gene and rumination in a Hungarian sample. Neuropsychopharmacol Hung 2012; 14: 285–292. [PubMed] [Google Scholar]
- Clasen PC, Wells TT, Knopik VS, McGeary JE, Beevers CG. 5-HTTLPR and BDNF Val66Met polymorphisms moderate effects of stress on rumination. Genes Brain Behav 2011; 10: 740–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue SF, Gould TJ. The neural and genetic basis of executive function: attention, cognitive flexibility, and response inhibition. Pharmacol Biochem Behav 2014; 123: 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazary J, Juhasz G, Anderson IM, Jacob CP, Nguyen TT, Lesch KP et al. Epistatic interaction of CREB1 and KCNJ6 on rumination and negative emotionality. Eur Neuropsychopharmacol 2011; 21: 63–70. [DOI] [PubMed] [Google Scholar]
- Juhasz G, Dunham JS, McKie S, Thomas E, Downey D, Chase D et al. The CREB1-BDNF-NTRK2 pathway in depression: multiple gene-cognition-environment interactions. BiolPsychiatry 2011; 69: 762–771. [DOI] [PubMed] [Google Scholar]
- Klok MD, Giltay EJ, Van der Does AJ, Geleijnse JM, Antypa N, Penninx BW et al. A common and functional mineralocorticoid receptor haplotype enhances optimism and protects against depression in females. Transl Psychiatry 2011; 1: e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assies J, Mocking RJ, Lok A, Ruhe HG, Pouwer F, Schene AH. Effects of oxidative stress on fatty acid- and one-carbon-metabolism in psychiatric and cardiovascular disease comorbidity. Acta Psychiatr Scand 2014; 130: 163–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustafa AA, Hewedi DH, Eissa AM, Frydecka D, Misiak B. Homocysteine levels in schizophrenia and affective disorders-focus on cognition. Front Behav Neurosci 2014; 8: 343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds EH. Folic acid, ageing, depression, and dementia. Br Med J 2002; 324: 1512–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazki FH, Sameer AS, Ganaie BA. Folate: metabolism, genes, polymorphisms and the associated diseases. Gene 2014; 533: 11–20. [DOI] [PubMed] [Google Scholar]
- Durga J, van Boxtel MP, Schouten EG, Bots ML, Kok FJ, Verhoef P. Folate and the methylenetetrahydrofolate reductase 677C—>T mutation correlate with cognitive performance. Neurobiol Aging 2006; 27: 334–343. [DOI] [PubMed] [Google Scholar]
- Mitchell ES, Conus N, Kaput J. B vitamin polymorphisms and behavior: evidence of associations with neurodevelopment, depression, schizophrenia, bipolar disorder and cognitive decline. Neurosci Biobehav Rev 2014; 47: 307–320. [DOI] [PubMed] [Google Scholar]
- Zhang MY, Miao L, Li YS, Hu GY. Meta-analysis of the methylenetetrahydrofolate reductase C677T polymorphism and susceptibility to Alzheimer's disease. Neurosci Res 2010; 68: 142–150. [DOI] [PubMed] [Google Scholar]
- Ma XY, Yu JT, Wu ZC, Zhang Q, Liu QY, Wang HF et al. Replication of the MTHFD1L gene association with late-onset Alzheimer's disease in a Northern Han Chinese population. J Alzheimers Dis 2012; 29: 521–525. [DOI] [PubMed] [Google Scholar]
- Naj AC, Beecham GW, Martin ER, Gallins PJ, Powell EH, Konidari I et al. Dementia revealed: novel chromosome 6 locus for late-onset Alzheimer disease provides genetic evidence for folate-pathway abnormalities. PLoS Genet 2010; 6: e1001130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren RJ, Wang LL, Fang R, Liu LH, Wang Y, Tang HD et al. The MTHFD1L gene rs11754661 marker is associated with susceptibility to Alzheimer's disease in the Chinese Han population. J Neurol Sci 2011; 308: 32–34. [DOI] [PubMed] [Google Scholar]
- Ramirez-Lorca R, Boada M, Antunez C, Lopez-Arrieta J, Moreno-Rey C, Hernandez I et al. The MTHFD1L gene rs11754661 marker is not associated with Alzheimer's disease in a sample of the Spanish population. J Alzheimers Dis 2011; 25: 47–50. [DOI] [PubMed] [Google Scholar]
- Prasannan P, Appling DR. Human mitochondrial C1-tetrahydrofolate synthase: submitochondrial localization of the full-length enzyme and characterization of a short isoform. Arch Biochem Biophys 2009; 481: 86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernimont SM, Clark AG, Stover PJ, Wells MT, Litonjua AA, Weiss ST et al. Folate network genetic variation, plasma homocysteine, and global genomic methylation content: a genetic association study. BMC Med Genet 2011; 12: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minguzzi S, Selcuklu SD, Spillane C, Parle-McDermott A. An NTD-associated polymorphism in the 3' UTR of MTHFD1L can affect disease risk by altering miRNA binding. Hum Mutat 2014; 35: 96–104. [DOI] [PubMed] [Google Scholar]
- Marchant NL, Howard RJ. Cognitive debt and Alzheimer's disease. J Alzheimers Dis 2015; 44: 755–770. [DOI] [PubMed] [Google Scholar]
- Momb J, Lewandowski JP, Bryant JD, Fitch R, Surman DR, Vokes SA et al. Deletion of Mthfd1l causes embryonic lethality and neural tube and craniofacial defects in mice. Proc Natl Acad Sci USA 2013; 110: 549–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parle-McDermott A, Pangilinan F, O'Brien KK, Mills JL, Magee AM, Troendle J et al. A common variant in MTHFD1L is associated with neural tube defects and mRNA splicing efficiency. Hum Mutat 2009; 30: 1650–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derogatis LR. BSI: Brief Symptom Inventory: Administration, Scoring, and Procedures Manual. National Computer Systems Pearson Inc.: : Minneapolis, 1993. [Google Scholar]
- Freeman B, Smith N, Curtis C, Huckett L, Mill J, Craig IW. DNA from buccal swabs recruited by mail: evaluation of storage effects on long-term stability and suitability for multiplex polymerase chain reaction genotyping. Behav Genet 2003; 33: 67–72. [DOI] [PubMed] [Google Scholar]
- Field A. Discovering Statistics Using SPSS. Sage Publications: London, UK, 2005. [Google Scholar]
- Team RC. R: A language and environment for statistical computing. R Foundation for Statistical Computing: : Vienna, Austria, 2013. [Google Scholar]
- Ripke S, Wray NR, Lewis CM, Hamilton SP, Weissman MM, Breen G et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry 2013; 18: 497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike ST, Rajendra R, Artzt K, Appling DR. Mitochondrial C1-tetrahydrofolate synthase (MTHFD1L) supports the flow of mitochondrial one-carbon units into the methyl cycle in embryos. J Biol Chem 2010; 285: 4612–4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanger O, Fowler B, Piertzik K, Huemer M, Haschke-Becher E, Semmler A et al. Homocysteine, folate and vitamin B12 in neuropsychiatric diseases: review and treatment recommendations. Expert Rev Neurother 2009; 9: 1393–1412. [DOI] [PubMed] [Google Scholar]
- Friso S, Choi SW, Girelli D, Mason JB, Dolnikowski GG, Bagley PJ et al. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc Natl Acad Sci USA 2002; 99: 5606–5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anglin RE, Garside SL, Tarnopolsky MA, Mazurek MF, Rosebush PI. The psychiatric manifestations of mitochondrial disorders: a case and review of the literature. J Clin Psychiatry 2012; 73: 506–512. [DOI] [PubMed] [Google Scholar]
- McFarquhar M, Elliott R, McKie S, Thomas E, Downey D, Mekli K et al. TOMM40 rs2075650 may represent a new candidate gene for vulnerability to major depressive disorder. Neuropsychopharmacology 2014; 39: 1743–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bree A, Verschuren WM, Bjorke-Monsen AL, van der Put NM, Heil SG, Trijbels FJ et al. Effect of the methylenetetrahydrofolate reductase 677C—>T mutation on the relations among folate intake and plasma folate and homocysteine concentrations in a general population sample. Am J Clin Nutr 2003; 77: 687–693. [DOI] [PubMed] [Google Scholar]
- The Network and Pathway Analysis Subgroup of the Psychiatric Genomics Consortium. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci 2015; 18: 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papakostas GI, Shelton RC, Zajecka JM, Etemad B, Rickels K, Clain A et al. L-methylfolate as adjunctive therapy for SSRI-resistant major depression: results of two randomized, double-blind, parallel-sequential trials. Am J Psychiatry 2012; 169: 1267–1274. [DOI] [PubMed] [Google Scholar]
- Taylor MJ, Carney SM, Goodwin GM, Geddes JR. Folate for depressive disorders: systematic review and meta-analysis of randomized controlled trials. J Psychopharmacol 2004; 18: 251–256. [DOI] [PubMed] [Google Scholar]
- Bedson E, Bell D, Carr D, Carter B, Hughes D, Jorgensen A et al. Folate augmentation of treatment—evaluation for depression (FolATED): randomised trial and economic evaluation. Health Technol Assess 2014; 18, vii-viii 1–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC. Integrating early life experience, gene expression, brain development, and emergent phenotypes: unraveling the thread of nature via nurture. Adv Genet 2014; 86: 277–307. [DOI] [PubMed] [Google Scholar]
- Watkins E. Psychological treatment of depressive rumination. Curr Opin Psychol 2015; 4: 32–36. [Google Scholar]